Structure–function modeling explores the basis of interallelic complementation in multiple combinations of rooty alleles.
Abstract
Auxin is a crucial plant growth regulator. Forward genetic screens for auxin-related mutants have led to the identification of key genes involved in auxin biosynthesis, transport, and signaling. Loss-of-function mutations in genes involved in glucosinolate biosynthesis, a metabolically related route that produces defense compounds, result in auxin overproduction. We identified an allelic series of fertile, hypomorphic Arabidopsis (Arabidopsis thaliana) mutants for the essential glucosinolate biosynthetic gene ROOTY (RTY) that exhibit a range of phenotypic defects characteristic of enhanced auxin production. Genetic characterization of these lines uncovered phenotypic suppression by cyp79b2 cyp79b3, wei2, and wei7 mutations and revealed the phenomenon of interallelic complementation in several RTY transheterozygotes. Structural modeling of RTY elucidated the relationships between structure and function in the RTY homo- and heterodimers, and unveiled the likely structural basis of interallelic complementation. This work underscores the importance of employing true null mutants in genetic complementation studies.
Forward genetic screens in the model plant Arabidopsis (Arabidopsis thaliana) proved to be a powerful tool for identifying key players of the auxin biosynthesis, transport, and signaling machineries (Estelle et al., 2011; Band et al., 2014; Brumos et al., 2014; Zhao, 2014; Adamowski and Friml, 2015; Kasahara, 2016; Leyser 2018). In the past 30 years, the main components of these pathways have been uncovered and functionally characterized using a wide array of genetic, biochemical, and molecular approaches (Estelle et al., 2011; Brumos et al., 2014; Zhao, 2014; Adamowski and Friml, 2015; Kasahara, 2016; Leyser, 2018).
In Arabidopsis, the predominant biologically active form of auxin, indole-3-acetic acid (IAA), is synthesized from the amino acid Trp via indole-3-pyruvic acid (IPyA) by the sequential action of two enzyme families, Trp aminotransferases, represented by TRP AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), TAA1-RELATED1 (TAR1), and TAR2, and flavin-containing monooxygenases, represented by YUCCA1 (YUC1) through YUC11 (Supplemental Fig. S1A; Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). Loss-of-function mutations that inactivate multiple TAA1/TAR or YUC genes lead to an array of auxin-deficient phenotypes, such as dwarfism, loss of apical dominance, reduced leaf venation, abnormal flower development and, in more severe cases, defective embryogenesis resulting in monopteros-like seedlings that lack hypocotyls and roots and develop a single cotyledon (Cheng et al., 2007; Stepanova et al., 2008, 2011; Chen et al., 2014).
Ethyl methanesulfonate (EMS) alleles of TAA1 that show mild auxin deficiency were originally isolated in genetic screens for reduced ethylene sensitivity (weak ethylene insensitive8 [wei8]; Stepanova et al., 2008), loss of shade avoidance (shade avoidance3 [sav3]; Tao et al., 2008), and resistance to auxin transport inhibitors (transport inhibitor response2 [tir2]; Yamada et al., 2009), whereas higher-order mutant combinations of the TAA1 and/or YUC gene families were generated using T-DNA mutant alleles identified via reverse genetics (Mashiguchi et al., 2011; Stepanova et al., 2011). Furthermore, gain-of-function mutants of the YUC1 gene have been found by screening a collection of activation-tagged lines and then recapitulated using transgenic approaches by expressing YUC1 or other YUC genes under strong promoters (Zhao et al., 2001). YUC overexpression results in extreme overproduction of auxin, and leads to plants with epinastic (i.e. downward curling) cotyledons and leaves, a greater number of lateral and adventitious roots, and enhanced apical dominance in the inflorescences (Zhao et al., 2001).
Although the IPyA pathway is thought to be the main route of auxin synthesis in plants (Mashiguchi et al., 2011; Stepanova et al., 2011; Brumos et al., 2018), alternative branches may contribute to total IAA pools, at least at some developmental stages, or in some tissues, or under specific environmental conditions.
Loss-of-function mutations in several genes involved in glucosinolate production, specifically SUPERROOT2 (SUR2; also known as CYTOCHROME P450 83B1 [CYP83B1], RED ELONGATED1 [RED1], RUNT1 [RNT1], and ALTERED TRP REGULATION4 [ATR4]; Barlier et al., 2000; Bak et al., 2001), ROOTY (RTY; also known as SUR1, ABERRANT LATERAL ROOT FORMATION1 [ALF1], INVASIVE ROOT1 [IVR1], and HOOKLESS3 [HLS3]; King et al., 1995; Mikkelsen et al., 2004), and UDP-GLUCOSYL TRANSFERASE74B1 (UGT74B1; also known as WEI9;Grubb et al., 2004), result in reduced glucosinolate biosynthesis and enhanced auxin production.
Glucosinolates are sulfur-containing aliphatic and aromatic secondary metabolites that are produced by some plants, mostly from the order Brassicales, to defend themselves against herbivorous insects, pathogens, and other pests (Kliebenstein, 2004; Halkier and Gershenzon, 2006; Katz et al., 2015; Barco and Clay, 2019). The morphological defects of these glucosinolate loss-of-function mutants are phenotypically similar to that of YUC gain-of-function lines, with mutant plants displaying epinastic cotyledons and leaves and large numbers of adventitious and lateral roots (Bak et al., 2001; Grubb et al., 2004; Mikkelsen et al., 2004). However, unlike YUC gain-of-function mutants, the SUR2, RTY, and UGT74B1 knockouts not only overproduce auxin, but also hamper the global production of glucosinolates, including Trp-derived indole glucosinolates (IGs), and hinder the homeostasis of their biosynthetic precursors. These mutations are thought to hyperactivate one or more of the putative IPyA-independent auxin biosynthetic branches by rerouting the flux of the pathway intermediate, indole-3-acetaldoxime (IAOx), which is normally channeled into IG and camalexin production, toward making extra IAA (Halkier and Gershenzon, 2006; Sugawara et al., 2009; Brumos et al., 2014). However, the molecular mechanisms linking these defective glucosinolate pathway enzymes to the synthesis of excess auxin remain uncharacterized, and the metabolic intermediates involved in these processes are not firmly established (Supplemental Fig. S1A). Likewise, the physiological significance of the auxin production route utilized in these glucosinolate mutants remains controversial (Bak et al., 2001; Mikkelsen et al., 2004; Halkier and Gershenzon, 2006; Nafisi et al., 2007; Müller et al., 2015; Mucha et al., 2019). Furthermore, it remains to be determined if this metabolic route has any role outside of the order Brassicales, as glucosinolates are believed to be produced nearly exclusively in this order (Kliebenstein, 2004; Halkier and Gershenzon, 2006; Barco and Clay, 2019).
In this study, we describe an allelic series of hypomorphic mutants for one of the genes involved in glucosinolate production, RTY. The RTY protein is a C-S lyase that catalyzes the conversion of S-(alkylacetohydroximoyl)-l-Cys to thiohydroximate in the synthesis of both aliphatic and aromatic glucosinolates (Mikkelsen et al., 2004). Partial inactivation of this enzymatic step due to missense mutations in RTY leads to milder defects than previously reported for the seedling-lethal null mutants of this gene (King 1994; King et al., 1995), with the developmentally stunted but fertile new alleles of RTY in our collection ranging in their severity from moderate to mild. Unexpectedly, genetic characterization of these lines uncovered several instances of interallelic complementation between missense mutants of RTY. Computational modeling of the RTY protein structure and mapping of RTY mutations onto the structural model of the RTY protein not only highlighted key residues essential for the formation of potential dimers and binding of the substrate, but also shed light on structure-to-function relationships, revealing the likely structural basis of interallelic complementation. The combination of genetics and modeling employed in this work provided an effective, synergistic strategy to investigating the molecular basis of the complementation phenomenon in F1 transheterozygotes. Given the scarcity of reports describing interallelic complementation despite the ubiquitous use of allelic testing in the genetic characterization of mutants, this study serves as a cautionary tale that underscores the need of utilizing true null alleles of genes of interest in all complementation crosses.
RESULTS
Characterization of a New Allele of the RTY Locus
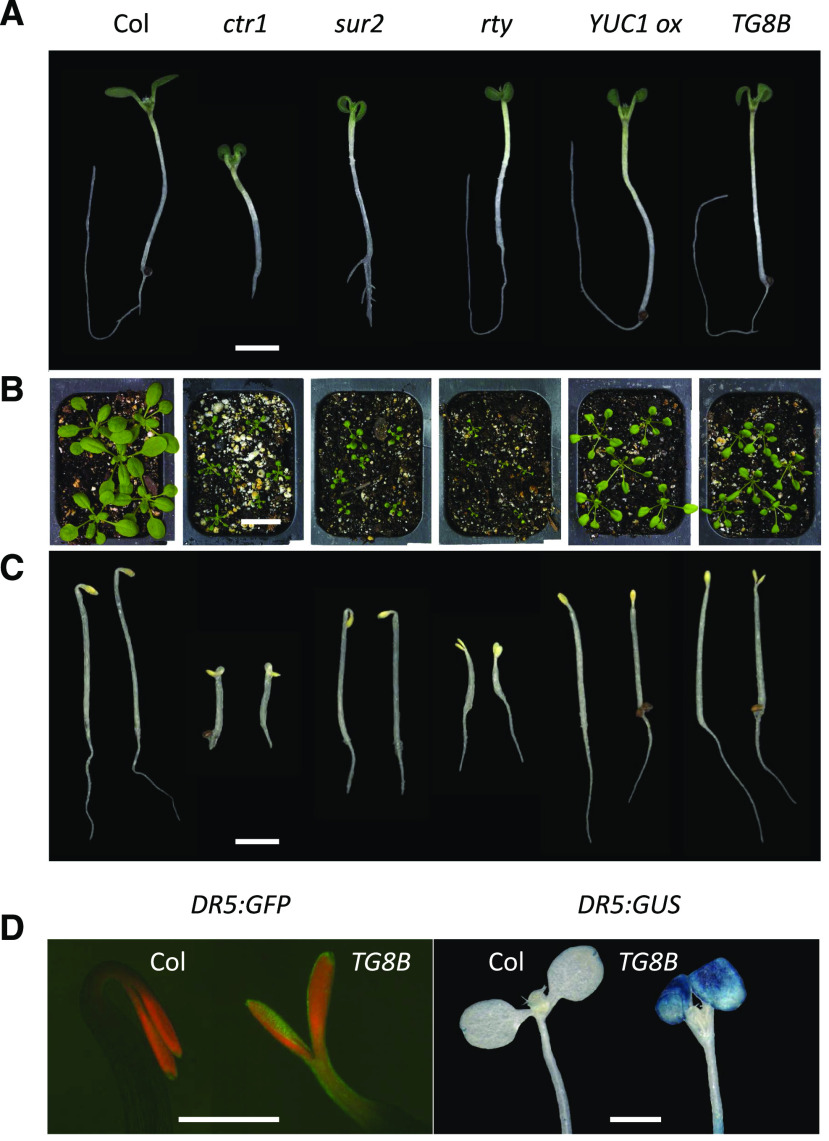
Downward curling (i.e. epinasty) of cotyledons (Fig. 1A) and true leaves (Fig. 1B) is a characteristic feature of auxin biosynthesis mutants that produce excess IAA, such as gain-of-function mutants in the key IPyA pathway genes of the YUC family (Zhao et al., 2001) and loss-of-function alleles of the SUR2 and RTY genes (Fig. 1A; Bak et al., 2001; Mikkelsen et al., 2004) disrupted in glucosinolate production and, hence, in the IG branch of auxin biosynthesis (Supplemental Fig. S1A).
Figure 1.
Phenotypes of mutants that share an epinastic cotyledon defect. Auxin overproducing and constitutive ethylene signaling mutants display profound cotyledon (A) and leaf (B) epinasty. A, Eight-day-old seedlings grown in plates for 3 d in the dark followed by 5 d in the light. B, Four-week-old adults grown in soil. C, Three-day-old plate-grown seedlings germinated in the dark. D, Elevated DR5 reporter activity in the new epinastic mutant TG8B relative to wild-type Col. Three-day-old dark-grown plate-germinated seedlings harboring DR5:GFP (left) and 8-d-old plants grown in plates for 3 d in the dark followed by 5 d in the light harboring DR5:GUS (right) are shown. In A, C, and D, multiple images were digitally abstracted and combined into a single composite image for comparison. Scale bars = 2 mm (A and C), 2 cm (B), and 1 mm (D).
Epinasty is, however, also observed in other classes of hormonal mutants, e.g. in the constitutive ethylene signaling mutant ctr1 (Fig. 1A; Kieber et al., 1993) or brassinosteroid-insensitive mutant bri1 (Clouse et al., 1996). In the search for new ethylene and auxin mutants, we performed forward genetic screens that led to the isolation of dozens of lines with highly epinastic cotyledons, including multiple new alleles of known genes, such as CTR1 and SUR2. One distinct subclass of mutants phenotypically resembled rty loss-of-function mutants (Mikkelsen et al., 2004) and YUC gene overexpression lines (Zhao et al., 2001), but showed varying degrees of epinasty and dwarfism in adults (see below). To gain insights into the nature of the affected gene(s), we performed phenotypic and genetic characterization of these lines.
The first mutant of that subclass characterized was TG8B, which showed characteristic cotyledon and leaf epinasty in light-exposed seedlings and soil-grown adults (Fig. 1, A and B) and lacked apical hooks in etiolated seedlings (Fig. 1C), displaying phenotypes similar to that of YUC1 overexpression lines (Zhao et al., 2001) and rty loss-of-function mutants (Mikkelsen et al., 2004). The expression of the auxin-response reporters DR5:GFP and DR5:GUS in this line was dramatically enhanced, consistent with the increased auxin activity in TG8B (Fig. 1D), which was comparable to that of other auxin overproducers (Supplemental Fig. S1, B and C). Genetic analysis of TG8B in the F1 generation of crosses with Columbia (Col) DR:GFP and Col DR5:GUS (reporter introgression backcrosses) and Landsberg erecta (Ler; mapping) revealed the recessive nature of the mutation (Table 1), suggesting that the TG8B phenotype is unlikely to be caused by a direct activation of one of the YUC genes.
Table 1. Crosses of new epinastic mutants.
| Cross (Female × Male) | Phenotype in F1 |
|---|---|
| TG8B selfed | Weak rty-like (see Figs. 3 and 4) |
| Col DR5:GFP x TG8B | Wild type-like |
| Col DR5:GUS x TG8B | Wild type-like |
| Ler x TG8B | Wild type-like |
| sur2 DR5:GUS x TG8B | Wild type-like |
| wei2-1 rty x TG8B | Wild type-like |
| TG8B DR5:GFP x wei2-1 rty | Wild type-like (see Fig. 4) |
| TH5H selfed | Moderate rty-like (see Figs. 3 and 4) |
| Col x TH5H | Wild type-like |
| sur2 DR5:GUS x TH5H | Wild type-like |
| Ler x TH5H | Wild type-like |
| wei2-1 rty x TH5H | Wild type-like (see Fig. 4) |
| TF5D selfed | Moderate rty-like (see Figs. 3 and 4) |
| Col DR5:GFP x TF5D | Wild type-like |
| Ler x TF5D | Wild type-like |
| wei2-1 rty x TF5D DR5:GFP | Weak rty-like (see Fig. 4) |
| TD11B selfed | Very weak rty-like (see Figs. 3 and 4) |
| Col DR5:GFP x TD11B | Wild type-like |
| Ler x TD11B | Wild type-like |
| TD11B x sur2 DR5:GUS | Wild type-like |
| TD11B DR5:GFP x wei2-1 rty | Very weak rty-like (see Fig. 4) |
| TJ12H selfed | Very weak rty-like |
| Col DR5:GFP x TJ12H | Wild type-like |
| sur2 DR5:GUS x TJ12H | Wild type-like |
| IK6G12 selfed | Moderate rty-like (see Figs. 3 and 4) |
| Col x IK6G12 | Wild type-like |
| Wei2-1 rty x IK6G12 | Wild type-like (see Fig. 4) |
| CS878294 selfed | Strong rty-like (see Figs. 3 and 4) |
| CS878294 x wei2-1 rty | Strong rty-like (see Fig. 4) |
Accordingly, we next tested TG8B for potential allelism with known recessive auxin overproducing mutants, sur2 and rty. The F1 progeny of the cross between TG8B and sur2 DR5:GUS, a T-DNA null mutant with the introgressed auxin reporter (Ulmasov et al., 1997; Stepanova et al., 2005), was phenotypically wild type (Table 1), suggesting that the two mutants are not allelic. In the complementation test with rty, we utilized the fertile rty wei2-1 (Stepanova et al., 2005) double mutant combination as a parent, because the rty single mutant is lethal. As in the case with sur2, the F1 generation of reciprocal crosses between TG8B and rty wei2-1 was phenotypically wild type (Table 1). This suggested that TG8B may not be allelic to rty. We therefore performed marker-based mapping on the phenotype-selected F2 population derived from a cross between TG8B (Col) and Ler. To our surprise, despite the complementation test results, TG8B was found to be tightly linked to the RTY locus (At2g20610), with 0 recombinants observed in 58 chromosomes for the F26H11-1 indel PCR marker located at 9.065 Mb on chromosome 2 next to the RTY gene (8.878–8.880 Mb; Supplemental Table S1).
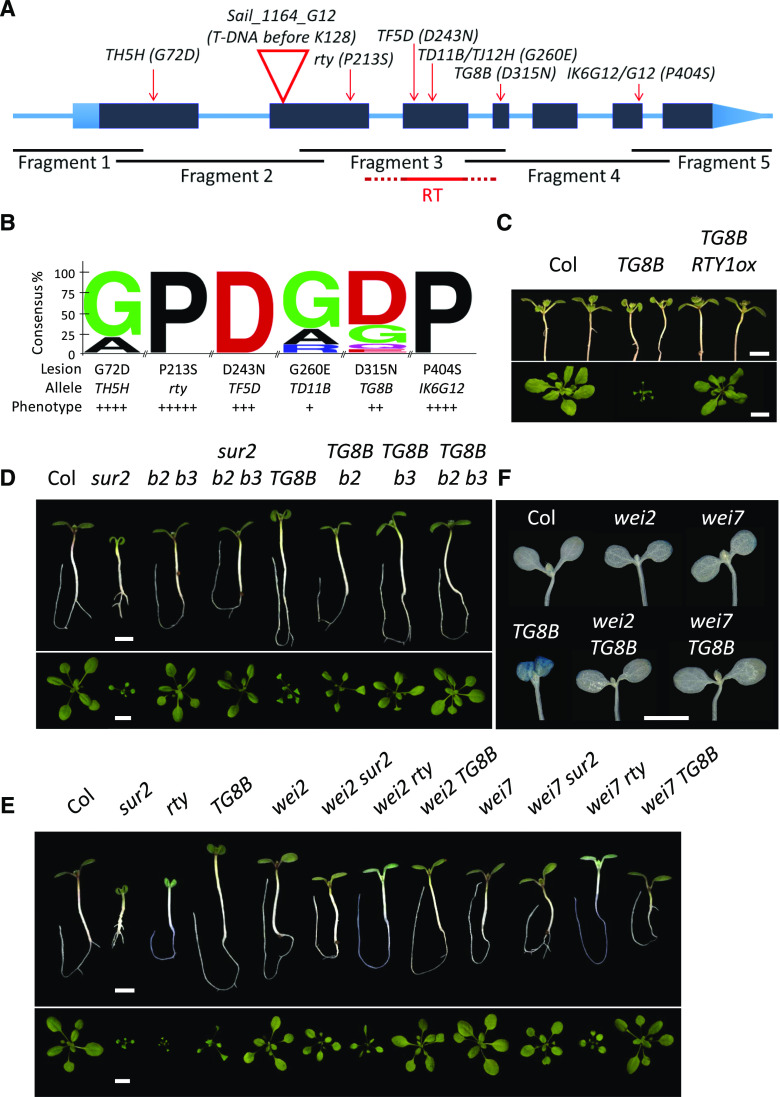
To determine whether TG8B is, indeed, an allele of RTY, we developed a set of five primer pairs that amplify five overlapping gene fragments and cover the entire RTY gene (Fig. 2A; Supplemental Table S1). We PCR-amplified RTY from TG8B, Col, and rty genomic DNA and performed a CEL1 assay on each mutant against the Col RTY sample. CEL1 is a crude mixture of DNases from celery that preferentially cut mismatched bases in a DNA heteroduplex formed by annealing of denatured PCR-amplified wild-type and mutant samples (Till et al., 2004). The CEL1 assay enables identification of polymorphisms in an amplified mutant gene fragment with respect to the corresponding wild-type fragment upon separating the products of the CEL1 digestion reaction by regular agarose gel electrophoresis (Till et al., 2004). Herein, polymorphisms were detected by CEL1 assay for both mutants: in fragment 3 for rty, and in fragment 4 for TG8B (Fig. 2A).
Figure 2.
Identification of TG8B as a hypomorphic allele of RTY. A, Schematic of the RTY gene displaying its exon (boxes) and intron (inner lines) structure. Dark-gray boxes correspond to coding regions, whereas light-blue boxes represent the 5′ and 3′ untranslated regions. Locations of mutations are marked with red arrows (substitutions) and a triangle (T-DNA insertion) and the nature of mutations is indicated next to the mutant line name. Overlapping PCR fragments amplified for CEL1 assay and sequencing are shown as black lines below the gene model. The fragment shown in red corresponds to the reverse transcription product used in the quantification of RTY gene expression in Supplemental Figure S5, with the dashed flanks corresponding to the introns (which lack in the cDNA fragment amplified with primers spanning the exon–exon junctions). B, MSA among RTY-like proteins was performed to analyze the conservation of the mutated amino acids of RTY. Each amino acid is written in a different color for clarity. The sequence logo reveals a functional correlation between amino acid conservation and the severity of missense mutant phenotypes, with the weaker alleles of RTY, TD11B, and TG8B, showing the least amino acid conservation, and the strongest allele, rty, showing full conservation. C, Complementation of TG8B with a cDNA construct of RTY (RTY1ox). Ten-day-old T2-generation seedlings grown in plates for 3 d in the dark followed by 7 d in the light and 4-week-old soil-grown adults are shown next to age-matched control plants. D to F, The cyp79b2 cyp79b3 double mutant (b2 b3; D) and the single mutants wei2 (E) and wei7 (F) can suppress the high-auxin phenotypes of TG8B. Shown here are 8-d-old seedlings, grown in plates for 3 d in the dark, followed by 5 d in the light (upper), and soil-grown, 4-week-old adults (lower). F, GUS staining in the TG8B wei2 and TG8B wei7 double mutants is reduced relative to TG8B itself. In C to F, multiple images were digitally abstracted and combined into a single composite image for comparison. Scale bars = 2 mm (C–F, upper) and 2 cm (C–F, lower).
The presence of mutations was then confirmed by Sanger sequencing. rty was found to harbor a C893T transition in the second exon of RTY genomic DNA, which results in a P213S amino acid substitution; whereas TG8B had a polymorphism later in the gene, in the fourth exon, mutating G1412A in the genomic DNA, and thus replacing an Asp with an Asn (D315N) in the RTY protein. To examine the degree of conservation of these amino acids and thus, the potential impact of the mutated residues on the enzyme structure or activity, the frequency of respective amino acids in proteins with high sequence homology to RTY was examined (Fig. 2B). The two mutations affect strongly and moderately conserved amino acids (Fig. 2B) and (based on the strength of mutant phenotypes) result in the complete or partial loss of RTY function in rty and TG8B, respectively. To further confirm the molecular identity of TG8B as an allele of RTY, we transformed the mutant with a complementary DNA (cDNA) construct of RTY driven by the 35S promoter and observed complementation of the cotyledon and leaf epinasty and dwarfism rescue (Fig. 2C).
Consistent with TG8B being an allele of rty, double mutant cyp79b2 cyp79b3, which is disrupted in the conversion of Trp to IAOx (Supplemental Fig. S1A) and thus devoid of the indole-glucosinolate pathway precursor IAOx (Zhao et al., 2002), could fully suppress the high-auxin phenotypes of TG8B (Fig. 2D). Single cyp79b2 or cyp79b3 mutants could only partially rescue TG8B (with the cyp79b3 being the more effective suppressor of the two), a finding that is in agreement with the partial functional redundancy of CYP79B2 and CYP79B3 (Zhao et al., 2002). The phenotypic complementation of TG8B by mutations in the first committed step of the IAOx route catalyzed by the CYP79B family is in agreement with a previous report that showed the ability of cyp79b2 cyp79b3 to also revert sur2 (Fig. 2D; Stepanova et al., 2011), another auxin-overproducing, glucosinolate mutant that in the IG branch of the auxin pathway maps downstream of the CYP79B step but upstream of RTY (Supplemental Fig. S1A; Bender and Celenza, 2009), back to wild type. Furthermore, wei2 and wei7, the anthranilate synthase alpha and beta subunit mutants defective in the Trp biosynthesis pathway (Supplemental Fig. S1A) that were previously shown to suppress the high-auxin phenotypes of sur2 and rty by limiting the availability of the auxin precursor Trp (Stepanova et al., 2005), could also suppress TG8B, resulting in improved plant growth (Fig. 2E) and normalized auxin response (Fig. 2F).
Possible Genetic Mechanisms of RTY Complementation in the F1
We next decided to investigate why the F1 generation of the cross between the classical rty mutant (King et al., 1995) and the new allele, TG8B, showed full complementation. We could envision two possibilities. One possible scenario explaining the wild-type morphology of the rty/TG8B transheterozygote is that the rty phenotype might have been suppressed by one copy of the wei2-1 mutation, because a rty wei2 double mutant was utilized for this cross and, therefore, the F1 was genotypically rty/TG8B wei2/+. This is, however, an unlikely possibility, as two copies of the wei2-1 mutant allele are required for the suppression of the high-auxin defects of sur2 and rty (Stepanova et al., 2005) or of TG8B (Fig. 2, E and F), and the sesquimutants TG8B/TG8B wei2/+, rty/rty wei2/+ (Supplemental Fig. S2, A and B) and sur2/sur2 wei2/+ (Supplemental Fig. S2C) are phenotypically similar to TG8B, rty, and sur2, respectively.
The second possible scenario that may explain interallelic complementation is that the RTY protein is expected to function as a dimer (or a higher-order multimeric complex), as all RTY-related enzymes with a defined crystal structure are known to form dimers or tetramers (Supplemental Tables S2 and S3). Thus, in that scenario, the F1 transheterozygote that harbors an interallelic combination of two different RTY alleles, rty and TG8B, forms a fully functional heteromer. In other words, if the two RTY monomers in the F1 are defective in different domains of the protein, they may be able to complement one another by supplying the missing activity to the potential dimer/oligomer. In fact, RTY belongs to a large family of C-S lyases/transaminases/alliinases, several of which have been shown to function as dimers or tetramers (Nock and Mazelis, 1987; John, 1995; Breitinger et al., 2001), with the aforementioned crystal structure analyses of extended family members across multiple domains of life, from bacteria to humans, supporting this notion (Supplemental Tables S2 and S3). For example, the closest related Arabidopsis protein in the database for which the crystal structure is available, At2g22250, an Asp/prephenate aminotransferase MATERNAL EFFECT EMBRYO ARREST17 (MEE17), is inferred to function as a dimer (Holland et al., 2018). Consistent with the possibility of transheterozygote complementation, the amino acid substitutions in rty and TG8B map to different domains of the protein (see below), suggesting that they may compromise different activities. Thus, transheterozygote complementation in RTY is a plausible explanation.
Interallelic Complementation in the RTY Locus Is Not Uncommon
To further explore the basis of mutant phenotype complementation in the RTY transheterozygote, we analyzed several additional putative rty alleles identified in our mutant screen. TD11B, TF5D, and TH5H show varying degree of cotyledon and leaf epinasty in light-grown plants (Fig. 3, A and B) and lack apical hooks in dark-grown seedlings (Fig. 3C), as do rty and TG8B. Adult phenotypes range in severity in terms of rosette size and fertility, with TD11B being the weakest of all mutants, and TH5H being the strongest in this subset, but not as severe as rty itself (Fig. 3, B and D). It is not surprising that neither of the novel mutants is as strong as the classical rty allele (King et al., 1995), as only fertile mutants have been recovered from our epinastic cotyledon genetic screens. In fact, several additional rty-like lines were initially selected in seedlings but lost during propagation due to their lethality in young adults.
Figure 3.
An allelic series of hypomorphic to null alleles of RTY. A, Eight-day-old seedlings grown in plates for 3 d in the dark followed by 5 d in the light. B, Five-week-old adults grown in soil. C, Three-day-old plate-grown seedlings germinated in the dark. D, Twelve-week-old adult plants grown in soil. Note that the control wild-type Col plant was staked and its inflorescences tied and folded to prevent seed loss during harvesting. In A to C, multiple images were digitally abstracted and combined into a single composite image for comparison. Scale bars = 1 mm (A and C) and 2 cm (B and D).
Backcrosses of TD11B, TF5D, and TH5H to wild-type Col and Ler revealed that all three mutants are recessive (Table 1). CEL1 assay, sequencing, and/or mappings of these mutants were performed, and they demonstrated that all of these lines are, indeed, new alleles of rty (Fig. 2A). TD11B harbors a G1161A mutation in the third exon of RTY genomic DNA that alters Gly 260 to Glu in the RTY protein. Rough mapping of TD11B (Col) in the F2 generation of a cross to Ler supports the notion that TD11B is an allele of RTY, as 0 out of 40 chromosomes were recombinant with the F26H11-1 indel PCR marker (Supplemental Table S1) that is <200 Kb away from the RTY locus. TF5D shows a CEL1-assay-detectable polymorphism in the fragment 3 of RTY (Fig. 2A). Sequencing of this fragment in the TF5D mutant background has uncovered a G1109A nucleotide substitution in RTY genomic DNA that corresponds to a D243N mutation in the third exon. Furthermore, another recessive rty-like mutant, TJ12H, that harbors an identical mutation to that of TD11B (Fig. 2A) but has been isolated from a different EMS family and thus has arisen independently from TD11B, is phenotypically indistinguishable from TD11B (Table 1), further suggesting that the high-auxin phenotypes of both of these mutants are a consequence of the G260E amino acid substitution in the RTY protein and not some other mutation. Finally, the TH5H mutant has been found to harbor a G215A missense mutation in the first exon of RTY genomic DNA, resulting in a G72D amino acid substitution (Fig. 2A). Taken together, these results demonstrate that an allelic series of the RTY locus has been identified. Not surprisingly, the severity of the high-auxin phenotypes in RTY mutants (Fig. 3) is in agreement with the degree of conservation of the specific amino acid affected by the mutation (Fig. 2B).
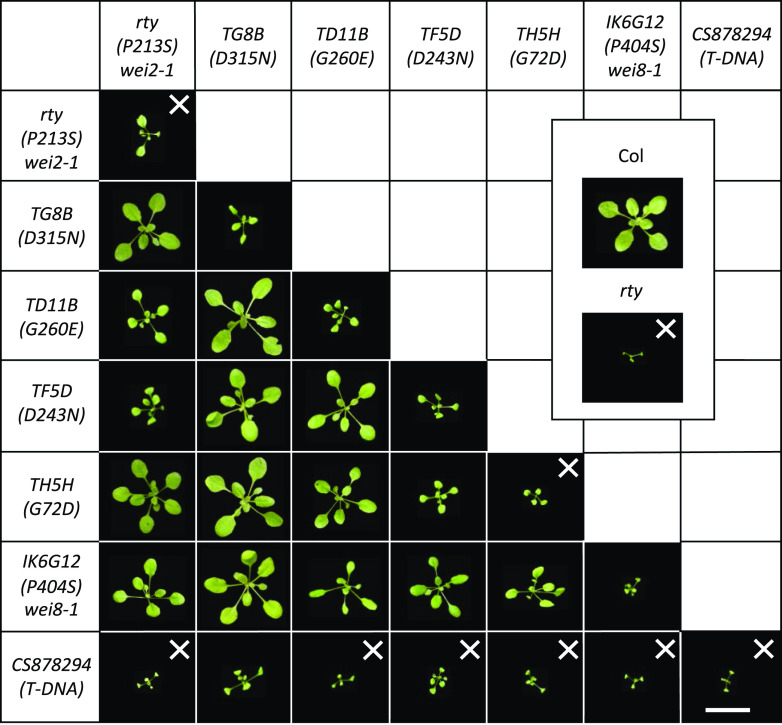
To test the ability of the new rty alleles to form functional heteromers in F1 transheterozygotes, we intercrossed all of the available RTY locus mutants, i.e. rty (in the wei2-1 background for fertility reasons), TG8B, TD11B, TF5D, and TH5H, and examined the morphology of the F1 generation (Fig. 4). Remarkably, some but not all interallelelic combinations were able to complement each other, potentially offering insights into the structure–function relationships in RTY heteromers. For example, the TG8B amino acid substitution mutant was able to complement all other missense alleles of RTY to wild-type level (Fig. 4), demonstrating full interallelic complementation in F1 transheterozygotes. Complementation was also observed between two stronger RTY alleles, the classical rty and TH5H, whereas no or only very weak complementation was seen between rty or TH5H and the moderate allele TF5D (Fig. 4). Several RTY allele intercrosses (e.g. rty/TD11B, TH5H/TD11B, and TF5D/TD11B F1 transheterozygotes) displayed phenotypes less severe than that seen in the parents (in this case, TD11B; Fig. 4; Supplemental Fig. S3), yet the morphology of these F1s was also distinct from that of wild-type plants, suggesting partial complementation.
Figure 4.
Interallelic combinations of RTY mutants. Phenotypes of 4-week-old soil-grown F1 and parental plants are shown. “X” marks the lines that failed to set seeds in two independent F1 propagation experiments. Note that the rty wei2-1 and TH5H parental lines are able to set a limited amount of seeds in some trials. Multiple rosette images were digitally abstracted and combined into a single composite image for comparison. Scale bar = 2 cm.
In summary, we found that the phenomenon of interallelic complementation is not uncommon in the RTY gene, with all missense mutations examined complementable by at least one other missense allele. More importantly, although we found that the strength of the homozygous mutants is consistent with the degree of conservation of the affected amino acid, there was no clear relation between the strength of the individual alleles in the homozygous plants and the severity of the defects in the heteroallelic combinations in the F1s of the intercrosses. This suggests that in some heteroallelic combinations, specific structural changes induced by the monomer of one allele can compensate for the changes imposed by a different allele in the second monomer. To examine this possibility, we determined the predicted structural changes of several homo- and heteroallelic combinations of RTY.
Structural Details Inferred from Molecular Models of RTY
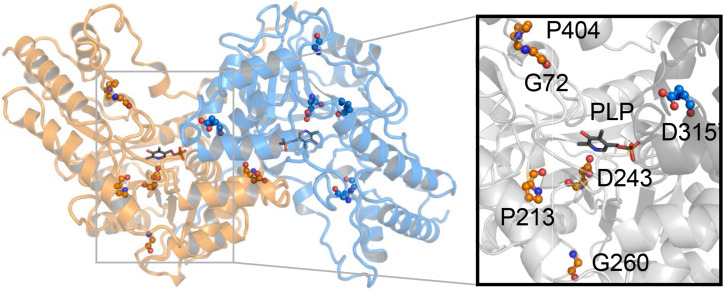
We next employed molecular modeling to provide structural reasoning for the ability of transheterozygous mutations in RTY to restore the RTY activity and thus lead to a phenotype that is milder than either of the homozygous parents. A three-dimensional (3D) structural model of dimeric RTY was generated using the crystal structure of the closest RTY homolog from Arabidopsis, the aforementioned aminotransferase MEE17 (PDB: 5WMH; Supplemental Fig. S4; Supplemental Table S2; Holland et al., 2018). The mutations described above were then mapped onto the wild-type dimeric RTY model (Fig. 5; Table 2), landing in varied locations throughout the dimeric structures. In particular, the D315 residue (mutated in TG8B) is located directly in the predicted dimerization interface, suggesting that an amino acid substitution at this site may affect the efficiency of RTY protein dimerization. This site also falls within 8 Å from the pyridoxal phosphate (PLP) cofactor and substrate binding pocket of the other monomer (across the predicted dimerization interface) and lines the binding pocket entrance.
Figure 5.
Dimeric model of RTY. Locations of mutated residues and a detailed view of the PLP binding pocket in the RTY protein model are displayed. The mutations are labeled and shown in sphere-and-stick format on a cartoon-format background of the protein structure. One monomer is colored orange while the other is colored blue. PLP ligand, shown in stick-and-ball format, is included for clarity. The inset provides a detailed view of the substrate binding pocket and the location of the mutations that line the entrance and bottom of the PLP binding pocket. The surface is colored gray, while the mutations are colored orange and blue, and shown in sphere-and-stick format. The dimeric RTY model was generated by employing the MODELER software.
Table 2. RTY mutations and the distances to the binding pocket and dimerization interface in the RTY protein model.
| Residue | Distance to Binding Pocket within Monomer | Distance to Binding Pocket to Monomer Partner | Distance to Dimerization Interface |
|---|---|---|---|
| Å | |||
| 72 | ∼15 | ∼36 | ∼10 Å–Y104’ |
| 213 | ∼12 | ∼37 | ∼12.5 Å–Y104’ |
| 243 | ∼7.5 | ∼31 | ∼9.3 Å– Y104’ |
| 260 | ∼20 | ∼38 | ∼16 Å–D69’ |
| 315 | ∼18 | ∼12 | ∼3 Å–K284’ |
| 404 | ∼19 | ∼40 | ∼16.5 Å–N308’ |
To experimentally test the possible involvement of the D315N in the dimer formation, wild-type RTY and the D315N (TG8B) mutant were expressed in a heterologous system as part of bait and prey hybrid proteins in yeast, one fused with a transcriptional activation domain (AD) and another with a DNA binding domain (BD) of GAL4 (Supplemental Fig. S5). We observed that the activity of the LacZ reporter in plates was dramatically reduced when both the bait and the prey carry the D315N mutation (TG8B/TG8B), whereas a nearly wild-type level of the reporter activity was seen for the interallelic combination with wild-type RTY (wild type/TG8B; Supplemental Fig. S5A). Quantification of the beta-galactosidase activity in yeast using liquid assays revealed that the interallelic wild-type/TG8B combination yielded ∼80% of the wild-type/wild-type activity (Supplemental Fig. S5B), consistent with the recessive nature of TG8B in plants.
These results provide a plausible explanation for the observed interallelic complementation by D315N in pairwise combinations with other missense mutations of RTY in Arabidopsis. Specifically, the phenotype of the homozygous D315N mutant is most likely the result of its inability to form homodimers, but dimer formation can be largely restored if only one of the monomers has the D315N mutation. The fact that all interallelic combinations of TG8B (D315N) with other missense alleles in our collection display wild-type morphology suggests that those other alleles of RTY do not interfere with the dimerization process. These results also imply that the D315N mutation does not directly affect the catalytic activity of the protein besides preventing the formation of functionally active RTY homodimers. Therefore, if a heterodimer forms between D315N and a catalytically impaired monomer such as D243N (see next paragraph), the resulting dimer should still be able to carry out the C-S lyase reaction (due to the catalytic functionality of D315N).
The second RTY mutation for which the structural model provides a likely explanation is D243N (defective in TF5D), for which the side chain of D243 falls in the binding pocket of the PLP cofactor (between 1 and 2 Å; Fig. 5) and thus most likely affects the ability of RTY subunits to recruit PLP and the substrate, potentially resulting in reduced activity of mutant RTY. Consistent with this explanation, the heteroallelic combination D315N/D243N shows a wild-type plant phenotype, as we anticipate that this allelic combination would be able to form dimers allowing for the catalytic domain of D315N to function even if the catalytic domain of D243N is largely inactive.
Other RTY mutations, G72D (TH5H), P213S (rty), and G260E (TD11B/TJ12H), map away from the catalytic pocket or the dimerization interface and, therefore, are harder to interpret with respect to the mutant defects that they trigger (Fig. 5; Table 2). In these cases, we wondered whether the phenotypes of different interallelic combinations could provide some clues on the structure–function relationship of these mutations. The wild-type phenotype of the interallelic combination between G72D and the dimerization mutant D315N suggests that the G72D is unlikely to directly affect dimerization. On the other hand, the strong transheterozygous mutant defects of the G72D and the PLP-binding–impaired D243N combination would suggest that these two mutants may affect the same function of the protein (i.e. the PLP binding and/or substrate recruitment). In fact, this is not so far-fetched based on the structure analysis, as the residue G72 is solvent-exposed and lines the entrance to the PLP binding channel in our structural model (Fig. 5).
The positions in the 3D model of the two other missense mutations, P213S (which is found in our strongest allele, rty) and G260E (seen in the weakest allele in the series, TD11B), do not provide strong clues on the possible structure–function alterations in these mutants, as they are located distal to the PLP binding pocket. P213 and G260 occupy a center of mass (a point representing the mean position of the matter in a protein) that is spaced ∼7 and 13 Å away from D243 (the residue in the substrate binding pocket likely implicated in PLP binding), respectively (Fig. 5).
Experimental Testing of the 3D Structural Model of RTY
To test the potential capacity of the computational model of RTY to explain the phenotypes (complemented or not complemented) of F1 transheterozygotes, we have mapped the amino acid substitution of another novel RTY allele, IK6G12, which has been recently identified in our laboratory from a genetic screen in the wei8-1 mutant background. From prior studies, we knew that wei8-1 is deficient in the Trp aminotransferase activity in the parallel IPyA branch of auxin biosynthesis (Supplemental Fig. S1), and that this mutation does not suppress the epinastic cotyledon and leaf phenotypes of rty and sur2 (Stepanova et al., 2008). IK6G12 is a moderately strong recessive allele of RTY (Fig. 3) that harbors a C1890T missense mutation in the sixth exon, which alters Pro404 to Ser (Fig. 2, A and B). Based on our 3D model, P404 is located ∼19 Å from the PLP binding pocket, ∼40 Å from the PLP binding pocket within the other monomer, and ∼16 Å from the nearest residue in the dimerization interface (Y104; Fig. 5; Table 2). Thus, the structural effects of P404S are likely to be indirect through creating conformational changes to the entrance of the PLP binding pocket or alteration to the dimerization interface and, therefore, should be functionally different than the structural consequences of all other missense mutants characterized in our study. Accordingly, we predicted that the IK6G12 allele should be able to, at least in part, complement all of the available missense mutants. In fact, when we performed crosses between IK6G12 and other alleles of RTY to experimentally test our predictions, we found that indeed IK6G12 was able to fully complement TG8B and partially all other missense mutants in the F1 transheterozygotes (Fig. 4). These observations support the idea that interallelic complementation is possible for mutant combinations that disable functionally nonoverlapping protein domains. Our findings are also consistent with the notion that lack of complementation may be indicative of the two allelic mutations affecting domains of similar functions.
As the genetic analyses and computational modeling of RTY described above strongly indicate that the use of missense mutations in complementation tests can be misleading, we decided to also utilize a true null allele of RTY. Our expectation was that a mutant that makes no full-length RTY protein should be unable to complement any of the amino acid substitution alleles of rty. We identified a previously uncharacterized insertional allele of RTY, Sail_1164_G12 (CS878294), which harbors a T-DNA at the beginning of the second exon of RTY immediately upstream of the nucleotide 638 in the RTY genomic DNA (Fig. 2A). This T-DNA line can still make partial 3′ RTY mRNA, as detected by RT-qPCR with primers that anneal downstream of the insertion site (Supplemental Fig. S6), but the levels of the 3′ product are reduced more than 3-fold. The corresponding truncated protein, if produced, should be nonfunctional as it is expected to lack at least the first 207 amino acids, because the first available Met downstream of the T-DNA insertion site in frame with the RTY protein is M208. Based on our mutant analysis, even a single amino acid substitution in the N terminus, G72D (TH5H), leads to severe functional consequences, and the deletion of the entire N-terminal part of the protein would remove substantial portions of the dimerization interface (86%) as well as nearly half of the PLP binding pocket. It is highly unlikely that this protein can fold correctly, let alone be able to dimerize and function.
Consistent with the notion that the Sail_1164_G12 allele is null, the mutant was unable to rescue any of the missense RTY alleles in interallelic combinations (Fig. 4). This lack of complementation is not simply due to a more severe phenotype of Sail_1164_G12 compared to the other alleles of RTY employed in this study, as the strength of this T-DNA mutant is similar to that of the classical missense allele, rty, utilized herein (Fig. 3). Because Sail_1164_G12 is an exonic insertional mutant that cannot produce full-length RTY RNA, in F1 transheterozygotes this mutant should not be able to form RTY protein heteromers and should only produce homomeric mutant RTY protein complexes. Consistent with that expectation, for the majority of RTY mutants, the severity of the phenotypes in the F1 generation of interallelic crosses with the T-DNA allele of RTY was found to be enhanced with respect to that of the homozygous missense mutant parents (Fig. 4), suggesting that these interallelic RTY mutants may be haploinsufficient. Surprisingly, the TG8B/Sail_1164_G12 F1 transheterozygotes were phenotypically indistinguishable from TG8B itself (Fig. 4), suggesting that in this mutant the number of the oligomers formed may not be rate-limiting despite the reduced ability of this mutant to dimerize relative to wild-type RTY.
DISCUSSION
Genetic Analysis
In this study, we identified and characterized an allelic series of missense mutants of RTY derived from EMS screens for epinastic ethylene- and auxin-related mutants. While complementation analyses combined with phenotyping proved to be a very useful and efficient technique for subgrouping the CTR1 and SUR2 mutant alleles identified in these screens, complementation tests largely failed when trying to categorize new alleles of RTY using a previously characterized presumed null allele, rty (King et al., 1995). In contrast, standard molecular biology approaches, including classical PCR-based mapping, CEL1 assay, and Sanger sequencing, were successful at identifying RTY as the causal gene for the six novel amino acid substitution mutants. Mutant complementation by a cDNA construct confirmed the identity of RTY as the gene responsible for TG8B epinasty. Genetic suppression of the high-auxin phenotypes of TG8B by the double cyp79b2 cyp79b3 mutant provided additional support for attributing the auxin phenotype of this novel mutant to a defect in the production of IG and thus overaccumulation of IAA. This result is consistent with the previously described ability of cyp79b2 cyp79b3 to suppress sur2 (Stepanova et al., 2011), i.e. another mutant with impaired IG biosynthesis, and further confirms the role of RTY in the IAOx-mediated auxin production. Likewise, the elevated levels of DR5 reporter activity and the suppression of auxin-related phenotypic defects of TG8B by wei2 and wei7, mutants deficient in the rate-limiting biosynthetic step of the auxin precursor Trp (Stepanova et al., 2005), are also in agreement with the notion that the phenotypic defects of this RTY mutant arise from auxin overproduction.
In contrast with the problems encountered when employing missense RTY mutants in complementation testing, utilization of the true null T-DNA allele of RTY, Sail_1164_G12, was very helpful at establishing (or, in this case, confirming) the allelic relationships between all RTY gene mutants. This discovery emphasizes the critical importance of using true (i.e. full-length RNA) null alleles for all complementation tests, at least for the genes whose products may form dimers or higher-order complexes and, therefore, in which interallelic complementation is possible.
Interallelic (also known as heteroallelic) complementation in transheterozygotes has been observed in many species, from yeast to humans, but is not a very commonly reported, nor widely known, phenomenon. In flowering plants, there is only a handful of cases attributed to interallelic complementation. Some of the first reports are in the shrunken Suc synthetase (Chourey, 1971; Chourey and Nelson, 1979) and aldehyde dehydrogenase (Schwartz, 1975) loci of maize (Zea mays). The best-known examples of intragenic complementation in plants, however, come from the analysis of numerous GNOM alleles in Arabidopsis (Mayer et al., 1993; Busch et al., 1996; Grebe et al., 2000). GNOM encodes a guanine nucleotide exchange factor that regenerates ARF-GTPase involved in vesicle trafficking (Shevell et al., 1994, Busch et al., 1996). Mutations that abolish or severely reduce GNOM activity lead to dramatic defects in the establishment of apical–basal polarity during embryogenesis, at least in part due to the mislocalization of the auxin transporter PIN1 (Steinmann et al., 1999). Mayer et al. (1993) classified 24 different alleles of GNOM into three classes—A, B, and C. The A- and B-class mutants display at least partial interallelic complementation in transheterozygotes, whereas all other allelic combinations (i.e. within the A and B classes, between A and C alleles, or between B and C alleles) result in plants phenotypically similar to their parents. The ability of different A- and B-class gnom alleles to complement each other in transheterozygotes has been attributed to homomeric interactions between the GNOM subunits and/or partial stop-codon read-through in gnom nonsense alleles (Busch et al., 1996).
More recently, Goldraij et al. (2003) reported interallelic complementation of the ubiquitous urease gene, known as AJ6/EU4, in soybean (Glycine max). de León et al. (2004) described a similar phenomenon for the cytokinin receptor CRE1/WOL1 locus in Arabidopsis. Likewise, Shang et al. (2011) observed interallelic complementation of the BRI1 brassinosteroid receptor alleles in Arabidopsis that harbor mutations in extracellular and cytoplasmic (kinase-inactive) domains of the protein. Finally, Pysh (2015) reported intragenic complementation in transheterozygotes between two Arabidopsis alleles of the cellulose synthase gene, AtCESA3. Again, the common theme in the interpretation of all of these results is that the affected proteins function as dimers or higher-order multimeric complexes: Their ability to di- or multimerize leads to the complementation and, hence, restoration of missing activities in the complex. In fact, the Theologis group (Tarun et al., 1998; Tsuchisaka et al., 2009) took advantage of this trans-subunit complementation phenomenon to explore functional homo- and heterodimerization between two tomato (Solanum lycopersicum; Tarun et al., 1998) and nine Arabidopsis (Tsuchisaka et al., 2009) ACC synthase (ACS) family members by expressing pairs of recombinant ACS proteins with complementary mutations in vitro or in Escherichia coli, and accessing restoration of the ACS enzymatic activity.
Another example of interallelic complementation that does not require dimerization is found in genes coding for proteins that possess more than one enzymatic activity. For example, in Nicotiana plumbaginifolia, mutations in the nitrate reductase gene that target different functional domains of the protein complement each other, but not mutations that inactivate the same domain (Caboche and Rouzé, 1990). We have used this concept to functionally group the different alleles based on 3D structural information and the phenotypes of the transheterozygous mutant combinations. Theoretically, another way two mutant alleles of the same gene can generate a fully functional product in an interallelic combination is through mRNA trans-splicing (Mongelard et al., 2002; Lasda and Blumenthal, 2011). This mechanism splices together two or more mRNA molecules in trans, and therefore theoretically can correct mutant phenotypes of the missense alleles that are defective in different exons. Trans-splicing events for nuclear-encoded genes have been reported to occur in rice (Oryza sativa; Zhang et al., 2010; Mathew et al., 2016), maize (Lu et al., 2013), and tomato (Chen et al., 2019), but the frequency of these RNA processing events appears to be much lower in plants than that in, for example, Drosophila (McManus et al., 2010). Thus, the physical separation of the different mutations in the cross-complementing RTY gene is consistent with three possibilities: complementation via protein dimerization, functional specialization of different protein domains, and mRNA trans-splicing. We do, however, favor a combination of the first two possibilities, as these are supported by our computational modeling.
It is important to disclose here that multiple strong alleles of RTY have been previously isolated by different groups, yet no concerns about the interallelic complementation have been raised. The classical rty allele utilized as a reference in our study was first reported in a PhD thesis by King (1994) and published a year later by King et al. (1995). Shortly after, Celenza et al. (1995) reported the identification of a phenotypically similar alf1-1 mutant, Boerjan et al. (1995) described seven additional alleles of this gene under the name of sur1-1 through sur1-7, and Lehman et al. (1996) published an hls3 allele of RTY. King et al. (1995) reported the results of complementation crosses between rty and hls3, as well as rty and two unpublished alleles of this gene, ivr-1 and ivr-2, with all three crosses revealing lack of complementation and, hence, suggesting allelism. Likewise, Celenza et al. (1995) stated lack of complementation in three crosses between alf1-1, rty, and hls3 mutant, again suggesting that these mutations affect the same gene. Finally, the seven sur1 alleles have each been crossed to one other sur1 allele, and also found to be noncomplementing (Boerjan et al., 1995). One obvious difference between these previously reported alleles of RTY and our new allelic series is the strength of the mutations. The EMS mutants described herein are phenotypically milder relative to the rty allele and are partially fertile (Fig. 3D), as these were derived from large M2 families from which all strong mutants (that were initially selected but failed to set seeds) have been simply lost. Unlike our missense alleles, many (but not all) of the previously published RTY alleles may be null. Nonetheless, given that the phenotypically null rty mutant could complement several of our weaker alleles of RTY, the cautionary take-home message of this study stays the same: Allelism tests require the use of a full-length RNA-null mutant as a reference in complementation crosses, or can otherwise provide misleading results. In our case, in light of the complementation observed in rty crosses, we chose to invest time into the characterization of these mutants because, initially, we erroneously concluded that these mutants were novel.
It is also important to note that this work relied exclusively on auxin-related phenotypes as a developmental readout of the severity of the mutant defects and no direct measurements of RTY’s C-S lyase activity or glucosinolate accumulation was performed. It remains to be determined how well the strength of the RTY mutants’ developmental abnormalities correlates with the reduction in the activity of mutant enzymes, what metabolic intermediates accumulate in various RTY alleles, and whether any of the rty phenotypes are caused by the abnormal homeostasis of specific metabolic intermediates of the glucosinolate or auxin biosynthetic pathways (other than auxin itself). The latter possibility is, perhaps, unlikely, given that excessive endogenous (Zhao et al., 2001; Mashiguchi et al., 2011) or exogenous (Boerjan et al., 1995; King et al., 1995) auxin can phenocopy rty mutant defects. Nonetheless, in vitro enzymatic assays on recombinant RTY proteins and in vivo measurements of glucosinolates, IAA, and their biosynthetic precursors, should furnish a more comprehensive picture of the metabolic routes that link glucosinolate biosynthetic defects with auxin overproduction. Having an allelic series of viable RTY alleles and their heteroallelic combinations will provide an important genetic resource to start tackling the long-standing questions in the underexplored IG–auxin crosstalk.
Molecular Modeling of RTY
The computational modeling of the 3D RTY structure provided important clues to explain the F1 generation transheterozygote complementation in the RTY heterodimers and also demonstrated potential predictive power of the model. To our knowledge, this is the first time that these types of approaches have been used to structurally characterize transheterozygous mutations in plants. In other model organisms, the structural basis of interallelic complementation has been explored only in a handful of cases, such as for myosin light chain protein Cdc4p in Schizosaccharomyces pombe and acetylcholine receptor AChR in humans (Slupsky et al., 2001; Shen et al., 2016). We argue that combining genetics with structural modeling yields unique insights into the function of proteins harboring amino acid substitutions, as demonstrated by the characterization of mutant proteins encoded by the missense alleles of RTY and their interallelic combinations (see below).
Discerning structure-to-function relationships requires quality structural data (e.g. from nuclear magnetic resonance, X-ray crystallography, or cryo-electron microscopy) for the protein of interest or a related protein. To our knowledge, no protein structure is currently available for RTY from Arabidopsis, but several related proteins have been structurally characterized through crystallization. Although the RTY protein sequence aligns with Tyr and other aromatic amino acid aminotransferases from mouse, human, and bacterial proteomes, we chose the sequence of MEE17 (PDB: 5WMH) from Arabidopsis, a bifunctional Asp aminotransferase and Glu/Asp-prephenate aminotransferase (Holland et al., 2018), as the template for our analysis. MEE17 is a far better template for this molecular modeling study, as it is derived from the same species as RTY, harbors PLP in the binding pocket; includes an extra alpha-helical structure that is lacking in the related human Tyr aminotransferase (PDB: 3DYD; Supplemental Fig. S4); contains coordinates for 399 out of 475 residues; and aligns well with the RTY sequence, with an E-value of 5.7E-17.
The structural interrogation of the RTY molecular model presented here allowed us to infer possible structure-to-function relationships for the mutations characterized in this study. We worked under the premise that mutations that affect the same functional domain should not complement each other in the transheterozygous plants. This basic assumption, together with the structural models, provides possible mechanism(s) for the disruption of the dimerization interface, cofactor binding pocket, or alteration of the catalytic domain. For example, D243 is likely directly involved in PLP/substrate binding and any disruption in this region is likely to interfere with RTY’s ability to recruit substrates. When examining the phenotypes of the transheterozygous F1 plants, we observed that G72D has the strongest phenotype when combined with the D243N mutation, suggesting that both affect the same functional domain, which in this case is the PLP binding pocket. This is further supported by the fact that although G72 is not directly located in the cofactor binding pocket, it is a solvent-exposed residue that lines the channel entrance for this cofactor. Similarly, while the very different phenotypes of the P213 (strong) and G260 (weak) mutants do not suggest that these two mutations affect the same functional domain, their location immediately adjacent to the substrate-binding pocket and the fact that the strongest phenotype of any of the G260E-containing interallelic combinations is the one with P213S, insinuates that these two mutations may disable the same protein functionality.
Residue D315 is positioned directly in the dimerization interface in our 3D model and any disruption here could lead to alteration of the interface and a decrease in the dimer’s ability to form. In addition, this mutation is also in close proximity to the substrate binding pocket of the adjacent monomer (∼8 Å away). Of these two nonmutually exclusive possibilities, our analysis suggests that the D315N mutation predominantly affects dimerization and is less likely to affect substrate binding. This possibility is supported by the yeast two-hybrid data that revealed reduced binding of the mutant monomers to one another, as well as by the fact that D315N complemented all other missense mutations examined, including those predicted to affect the cofactor and substrate binding, thus disfavoring the possibility of D315N interfering with substrate recruitment. Conversely, the position of P404 in the 3D model failed to provide convincing support for a specific functional alteration in this mutant. Again, the combination of structural-data analysis and transheterozygous phenotypes provided a reasonable hypothesis for the structure–function relationship of these mutations. In fact, distinct location of the P404S in the 3D model far away from the dimerization interface and the substrate binding pocket suggested that this mutation may disable a functional domain distinct from those affected by the other missense mutations analyzed. Consistent with this line of thought, we discovered partial complementation in all transheterozygotes containing the P404S mutations, suggesting that it impairs an activity not critically disturbed in any of the other alleles.
The 3D molecular model of RTY described above has provided structure-to-function reasons for the observed phenotypes for a majority of the RTY mutant alleles characterized in this study. We cannot rule out more nuanced structural reasons for the phenotypes observed, such as complementary effects of the mutations stabilizing either the substrate-binding pocket, the dimerization interface, and/or interactions with potential partner proteins in some of the transheterozygous mutant combinations. Future experimental high-resolution structural data or molecular dynamic simulations could provide greater insight into the structural effects of these mutations, such as altered conformations within the dimerization interface leading to weaker dimers or within the substrate binding pocket, reducing or abolishing substrate recruitment. In a meantime, a combination of genetics and 3D computational models enabled us to find a plausible explanation for the molecular consequences of several RTY missense mutations and their interallelic combinations. Future experimental validation of these findings could provide more definitive support for the structure–function relationships of RTY heteromers.
MATERIALS AND METHODS
Mutant and Complementation Lines, Growth Conditions, and EMS Mutagenesis
All of the new Arabidopsis (Arabidopsis thaliana) mutants described in this work are in the Col background. rty (CS8156), Sail_1164_G12 (CS878294), sur2 (Salk_028573), and Col DR5:GFP (CS9361) were obtained from the Arabidopsis Biological Resource Center (ABRC). The Col DR5:GUS reporter (Ulmasov et al., 1997), YUC1 overexpression line (Zhao et al., 2001), and cyp79b2 cyp79b3 T-DNA knockout (Zhao et al., 2002) were provided by Drs. Thomas Guilfoyle, Yunde Zhao, and John Celenza, respectively. The mutants ctr1-1 (Kieber et al., 1993), wei2-1, wei2-1 DR5:GUS, wei7-2, wei7-4, wei7-4 DR5:GUS, wei2-1 sur2, wei7-4 rty, wei2-1 rty (Stepanova et al., 2005), rty DR5:GUS (Stepanova et al., 2007), and sur2 cyp79b2 cyp79b3 (Stepanova et al., 2011) were previously reported. New mutant combinations were generated by crossing various mutants to each other and phenotyping/genotyping the lines in the F2 and F3 generation (see below). Reporter introgression was also done by crosses and phenotyping of progeny in F2 and F3. Seeds were germinated in plates with Murashige and Skoog media (1× Murashige and Skoog salts, 10 g L−1 Suc, pH 6.0, with 1 m of KOH, and 7 g L−1 Bacto-Agar) in the dark for 3 d and then moved to constant light, as indicated. For propagation, 10- to 14-d-old seedlings were transplanted to soil and grown under 16-h light/8-h dark cycle in 1:1 mix of Fafard superfine germinating mix and Fafard 4P mix.
A RTY cDNA complementation construct driven by the 35S promoter and fused to a C-terminal TAP-tag in LIC6 vector was ordered from the ABRC (DKLAT2g20610) and transformed into TG8B via the floral dip method (Clough and Bent, 1998). T1s were selected in gentamycin and propagated, with images of complemented lines taken in the T2 generation.
EMS mutagenesis was performed on imbibed Col seeds as described in Guzmán and Ecker (1990). The M1 generation was propagated in families of ∼1,000 plants each. A total of 27 M1 families was generated and screened in the M2 generation. M2 seeds were plated at 1,000 to 2,000 plants per 15-cm petri plate filled with 50 mL of Murashige and Skoog media (two plates per family), stratified in the cold for 2 to 4 d, germinated in the dark for 3 d, and then transferred to light for additional 5 to 10 d. The plates were periodically screened visually for cotyledon epinasty. Putative mutants were picked, propagated in soil, and then retested in the M3 generation. One additional RTY allele, IK6G12, was derived from an equivalent EMS mutagenesis carried out in the wei8-1 mutant background.
PCR-Based Mutant Mapping
Physical mapping of new mutants was performed in the F2 generation of crosses to Ler. Epinastic F2 seedlings were picked from plates and transplanted to soil. A single leaf per plant was harvested, its genomic DNA extracted, and short genomic fragments that vary in length between Col and Ler were PCR-amplified in 10-μL reactions (1 μL of genomic DNA, 1 μL of 10× PCR buffer, 0.25 μL of 2 mm dNTPs, 0.25 μL of homemade Taq polymerase, 0.25 μL of 20-μm forward primer, 0.25 μL of 20-μm reverse primer, and 7 μL of water) using 40 cycles of the following PCR program: 30 s at 94°C, 30 s at 56°C, 1 min at 72°C. PCR products were separated on 3% to 4% (w/v) agarose 1× Tris/acetic acid/ EDTA buffer (TAE) gels, photographed, and scored. The indel PCR marker primers F26H11-1-F1 and F26H11-1-R1 used in mapping of RTY mutants are listed in Supplemental Table S1.
Mutant Genotyping
Plant genotyping was performed by CEL1 assay and/or Sanger sequencing (for the EMS mutants) and by PCR (for T-DNA mutants). PCR primers are listed in Supplemental Table S1. PCR conditions were the same as described for mapping, except longer amplification times were allowed (1 min per kb).
For the CEL1 assay, gene-specific primers were used to amplify overlapping gene fragments from the mutant and wild-type Col samples in 20-μL PCR reactions (2 μL of genomic DNA, 2 μL 10× PCR buffer, 0.5 μL of 2-mm dNTPs, 0.5 μL of homemade Taq polymerase, 0.5 μL of 20-μm forward primer, 0.5 μL of 20-μm reverse primer, and 14 μL of water) using 40 cycles of the following PCR program: 30 s at 94°C, 30 s at 56°C, 1 to 2 min (1 min per kb) at 72°C. Five microliters of each mutant and 5 μL of the wild-type sample for each fragment were combined, and then denatured and annealed to each other in a thermocycler by heating the mixture for 10 min at 99°C, reducing the temperature to 70°C, and running 70 steps of the program reducing the temperature 0.3°C per step, keeping the samples for 20 s at each temperature, going from 70°C to 49°C. Annealed 10-μL samples were combined with 10 μL of CEL1 mix (2 μL of 10× CEL1 buffer, 1 μL of CEL1, and 7 μL of distilled water), incubated at 45°C for 15 to 20 min, and then immediately moved to ice. Digested chilled samples were resolved on a 1% (w/v) agarose 1× TAE gel by gel-electrophoresis, photographed, and scored.
GUS Staining and Basic GFP Microscopy
Samples harboring the DR5:GUS transgene were harvested in ice-cold 90% (v/v) acetone and stored at −20°C overnight or longer. To stain for GUS, samples were washed once with Rinse buffer [50 mm Sodium Phosphate buffer at pH 7, 0.5 mm of K3Fe(CN)6, and 0.5 mm of K4Fe(CN)6] and stained overnight in Rinse buffer supplemented with 1 μg mL−1 of X-gluc (with X-gluc dissolved in dimethyl sulfoxide at 5 mg per 100 μL and diluted with Rinse buffer to 1 μg mL−1 of final concentration). Staining reactions were stopped with 15% (v/v) ethanol. To remove chlorophyll, GUS-stained seedlings were incubated for 2 to 3 d in 70% (v/v) ethanol at 37°C to 50°C. Samples were photographed using the software QCapture with a QImaging digital camera (Teledyne Imaging) attached to a dissection scope (Leica). To visualize expression of DR5:GFP, an AxioPlan epifluorescence microscope (Zeiss) was utilized. Images were captured with a Diagnostic Instruments Color Mosaic camera (www.spotimaging.com) using the software Spot Insight (www.spotimaging.com).
Yeast Two-Hybrid Assay
To test the level of interaction between RTY monomers, we employed the yeast two‐hybrid assay. Binding among wild-type monomers, TG8B mutant monomers, and wild type with TG8B monomers, was evaluated. The cDNA clone G10872 (ABRC) was employed as RTY-wild type and used as template for the PCR-based approach to generate the TG8B mutant open reading frame. To obtain TG8B, three PCR reactions (iProof; Bio-Rad) were performed: PCR 1 using primers SUR1_For and sur1_RevTG8B_Internal; PCR 2 using sur1_ForTG8B_Internal and SUR1_Rev (No STOP); and fusion PCR 3 (to join both fragments harboring the mutation) using the external primers SUR1_For and SUR1_Rev (No STOP; see Supplemental Table S1 for primer information). The final PCR product was subcloned into pENTR/d-Topo. Both wild type and TG8B RTY were transferred to pACT-GW (GAL4-AD) and pAS-GW (GAL4-BD; Nakagawa et al., 2007; Shimoda et al., 2008) by Gateway LR Clonase recombination. The Y190 yeast strain was transformed with the resulting bait constructs, harboring the GAL4-BD. Strains were tested for lack of transactivation and then transformed with the prey constructs containing the GAL4-AD. Interaction between bait and prey was inferred from the activity of the LacZ reporter (Deplancke et al., 2006). Qualitative colony-lift assays were performed to assess the activity of β-galactosidase by visual scoring of the levels of the resulting blue staining in the presence of X-gal (GoldBio) substrate (Deplancke et al., 2006). To quantify the β-galactosidase activity, lysed yeast cells were resuspended in Z-buffer (Deplancke et al., 2006), supplemented with o-nitrophenyl-beta-d-galactopyranoside (Sigma-Aldrich), and the amount of yellow product obtained from the o-nitrophenyl-beta-d-galactopyranoside catalysis was measured by determining the absorbance at 420 nm (Smale, 2010) using a SpectraMax M2 microplate reader (Molecular Devices).
Analysis of Conserved Residues in the RTY Protein
Conservation of the amino acid residues mutated in our allelic series of RTY was analyzed using the tool Multiple Sequence Alignment (MSA; Phytozome; Goodstein et al., 2012; https://phytozome.jgi.doe.gov) by comparing RTY-like proteins of the Brassicales and Malvales orders of flowering plants from the following species: Arabidopsis halleri, Arabidopsis lyrata, Arabidopsis, Boechera stricta, Brassica oleracea, Brassica rapa, Capsella grandiflora, Capsella rubella, Carica papaya, Eutrema salsugineum, Gossypium raimondii, and Theobroma cacao. Utilizing the MSA results, a sequence logo was generated to represent the level of conservation of each mutated residue by employing the program WebLogo (Crooks et al., 2004; http://weblogo.berkeley.edu/logo.cgi).
RNA Extraction and RT-qPCR
Eight-day-old seedlings of the indicated genotypes grown for 3 d in the dark followed by 5 d in constant light were collected into 1.5-mL Sarstedt screw-cap tubes (Thermo Fisher Scientific) prefilled with four 2.4-mm Zirconia/Silica beads each, frozen in liquid nitrogen, and then ground in a Vivadent shaker (Ivoclar Vivident). For rty and Sail_1164_G12 mutants that are sterile, homozygous seedlings were phenotypically selected from the progeny of heterozygous parents. Total RNAs were extracted (Reuber and Ausubel, 1996) and reverse-transcribed with random hexamer primers using a TaqMan Reverse Transcription Kit (Applied Biosystems). SYBR Green qPCRs were performed in an Applied Biosystems StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) with Power SYBR Green Master Mix (Applied Biosystems) using primers For1RTY-qRT and Rev1RTY-qRT for RTY, and At5g44200F and At5g44200R for the control gene, At5g44200 (see Supplemental Table S1 for primer information). Normalized mean values for RTY levels ±sd were plotted on a bar graph.
Molecular Modeling
The software tool MODELER v9.18 (https://salilab.org/modeller/) was used to construct a model of the dimeric RTY wild-type and transheterozygotes using Arabidopsis prephenate aminotransferase (PDB: 5WMH; Holland et al., 2018). During the model building process, we employed an optimization method involving conjugate gradients and molecular dynamics to minimize violations of the spatial restraints. Five-hundred models were generated from an alignment of the Arabidopsis sequence and scored by the internal MODELER scoring method Discrete Optimized Protein Energy (Shen and Sali, 2006). The structure with the lowest Discrete Optimized Protein Energy score was subsequently run through the programs PROCHECK (https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/) and WHATCHECK (which checks the stereochemical quality of a protein structure; https://swift.cmbi.umcn.nl/gv/whatcheck/) for quality (Laskowski et al., 1993; Hooft et al., 1996). All images were produced with the software PyMOL (Baker et al., 2001; The PyMOL Molecular Graphics System, v2.0; Schrödinger).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank/European Molecular Biology Laboratory databases under accession numbers: RTY (At2g20610), CYP79B2 (At4g39950), CYP79B3 (At2g22330), WEI2 (At5g05730), WEI7 (At1g25220), WEI8 (At1g07560), CTR1 (At5g03730), SUR2 (At4g31500), YUC1 (At4g32540), CBP80 (At5g44200), and MEE17 (At2g22250).
Supplemental Data
The following materials are available.
Supplemental Figure S1. Increased levels of auxin responses in auxin-overproducing mutants.
Supplemental Figure S2. Lack of suppression of the TG8B, rty, and sur2 defects by one copy of wei2-1.
Supplemental Figure S3. Interallelic combinations of RTY mutants.
Supplemental Figure S4. RTY protein model based on the crystal structure of MEE17.
Supplemental Figure S5. Reduced dimerization capacity of the TG8B (D315N) version of RTY in yeast.
Supplemental Figure S6. Quantification of RTY expression in various alleles of RTY by RT-qPCR.
Supplemental Table S1. Primers employed in this study.
Supplemental Table S2. RTY-related enzymes function as dimers or tetramers.
Supplemental Table S3. RTY-related enzymes using PLP as cofactor.
Acknowledgments
We thank Jeonga Yun for technical assistance with sequencing RTY alleles, and C. Douglas Grubb for helpful discussions on interallelic complementation. We are grateful to the National Science Foundation for the continuous support of our research and outreach.
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB0923727, IOS1444561, and IOS1650139 to A.N.S. and J.M.A., grant no. MCB1158181 to J.M.A., and grant no.IOS1750006 to A.N.S.).
Articles can be viewed without a subscription.
References
- Adamowski M, Friml J(2015) PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R(2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA(2001) Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP, Wilson MH, Yu L, Li W, Hijazi HI, et al. (2014) Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco B, Clay NK(2019) Evolution of glucosinolate diversity via whole-genome duplications, gene rearrangements, and substrate promiscuity. Annu Rev Plant Biol 70: 585–604 [DOI] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C(2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Celenza JL(2009) Indolic glucosinolates at the crossroads of tryptophan metabolism. Phytochem Rev 8: 25–37 [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, van Onckelen H, van Montagu M, Inzé D(1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger U, Clausen T, Ehlert S, Huber R, Laber B, Schmidt F, Pohl E, Messerschmidt A(2001) The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity. Plant Physiol 126: 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Alonso JM, Stepanova AN(2014) Genetic aspects of auxin biosynthesis and its regulation. Physiol Plant 151: 3–12 [DOI] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN(2018) Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47: 306–318.e5 [DOI] [PubMed] [Google Scholar]
- Busch M, Mayer U, Jürgens G(1996) Molecular analysis of the Arabidopsis pattern formation of gene GNOM: Gene structure and intragenic complementation. Mol Gen Genet 250: 681–691 [DOI] [PubMed] [Google Scholar]
- Caboche M, Rouzé P(1990) Nitrate reductase: A target for molecular and cellular studies in higher plants. Trends Genet 6: 187–192 [DOI] [PubMed] [Google Scholar]
- Celenza JL Jr., Grisafi PL, Fink GR(1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Chen L, Li W, Li Y, Feng X, Du K, Wang G, Zhao L(2019) Identified trans-splicing of YELLOW-FRUITED TOMATO 2 encoding the PHYTOENE SYNTHASE 1 protein alters fruit color by map-based cloning, functional complementation and RACE. Plant Mol Biol 100: 647–658 [DOI] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y(2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y(2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS.(1971) Interallelic complementation at the sh locus in maize. Genetics 68: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Nelson OE(1979) Interallelic complementation at the sh locus in maize at the enzyme level. Genetics 91: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC(1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE(2004) WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de León BG, Zorrilla JMF, Rubio V, Dahiya P, Paz-Ares J, Leyva A(2004) Interallelic complementation at the Arabidopsis CRE1 locus uncovers independent pathways for the proliferation of vascular initials and canonical cytokinin signalling. Plant J 38: 70–79 [DOI] [PubMed] [Google Scholar]
- Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJM(2006) Gateway-compatible yeast one-hybrid screens. CSH Protocols 2006: pdb.prot4590. [DOI] [PubMed] [Google Scholar]
- Estelle M, Weijers D, Ljung K, Leyser O(2011) Auxin Signaling: From Synthesis to Systems Biology. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- Goldraij A, Beamer LJ, Polacco JC(2003) Interallelic complementation at the ubiquitous urease coding locus of soybean. Plant Physiol 132: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jürgens G(2000) A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Müller J, Masuno MN, Molinski TF, Abel S(2004) Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40: 893–908 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR(1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J(2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Holland CK, Berkovich DA, Kohn ML, Maeda H, Jez JM(2018) Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis. Plant J 94: 304–314 [DOI] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE(1996) Errors in protein structures. Nature 381: 272. [DOI] [PubMed] [Google Scholar]
- John RA.(1995) Pyridoxal phosphate-dependent enzymes. Biochim Biophys Acta 1248: 81–96 [DOI] [PubMed] [Google Scholar]
- Kasahara H.(2016) Current aspects of auxin biosynthesis in plants. Biosci Biotechnol Biochem 80: 34–42 [DOI] [PubMed] [Google Scholar]
- Katz E, Nisani S, Yadav BS, Woldemariam MG, Shai B, Obolski U, Ehrlich M, Shani E, Jander G, Chamovitz DA(2015) The glucosinolate breakdown product indole-3-carbinol acts as an auxin antagonist in roots of Arabidopsis thaliana. Plant J 82: 547–555 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR(1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- King J. (1994) Adventitious root formation and auxin homeostasis in Arabidopsis thaliana L. Heynh.: Genetic and physiological analyses. PhD thesis. University of Wisconsin-Madison, Madison, WI [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB(1995) a mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7: 2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ.(2004) Secondary metabolites and plant/environment interactions: A view through Arabidopsis thaliana tinged glasses. Plant Cell Environ 27: 675–684 [Google Scholar]
- Lasda EL, Blumenthal T(2011) Trans-splicing. Wiley Interdiscip Rev RNA 2: 417–434 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM(1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR(1996) HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194 [DOI] [PubMed] [Google Scholar]
- Leyser O.(2018) Auxin signaling. Plant Physiol 176: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Chen D, Shu D, Zhang Z, Wang W, Klukas C, Chen LL, Fan Y, Chen M, Zhang C(2013) The differential transcription network between embryo and endosperm in the early developing maize seed. Plant Physiol 162: 440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew IE, Das S, Mahto A, Agarwal P(2016) Three rice NAC transcription factors heteromerize and are associated with seed size. Front Plant Sci 7: 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Buttner G, Jurgens G(1993) Apical-basal pattern-formation in the Arabidopsis embryo—studies on the role of the GNOM gene. Development 117: 149–162 [Google Scholar]
- McManus CJ, Duff MO, Eipper-Mains J, Graveley BR(2010) Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci USA 107: 12975–12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, Halkier BA(2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37: 770–777 [DOI] [PubMed] [Google Scholar]
- Mongelard F, Labrador M, Baxter EM, Gerasimova TI, Corces VG(2002) Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160: 1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucha S, Heinzlmeir S, Kriechbaumer V, Strickland B, Kirchhelle C, Choudhary M, Kowalski N, Eichmann R, Hückelhoven R, Grill E, et al. (2019) The formation of a camalexin biosynthetic metabolon. Plant Cell 31: 2697–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller TM, Böttcher C, Morbitzer R, Götz CC, Lehmann J, Lahaye T, Glawischnig E(2015) TRANSCRIPTION ACTIVATOR-LIKE EFFECTOR NUCLEASE-mediated generation and metabolic analysis of camalexin-deficient cyp71a12 cyp71a13 double knockout lines. Plant Physiol 168: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J(2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nock LP, Mazelis M(1987) The C-S lyases of higher plants: Direct comparison of the physical properties of homogeneous alliin lyase of garlic (Allium sativum) and onion (Allium cepa). Plant Physiol 85: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD.(2015) Two alleles of the AtCesA3 gene in Arabidopsis thaliana display intragenic complementation. Am J Bot 102: 1434–1441 [DOI] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM(1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.(1975) The molecular basis for allelic complementation of alcohol dehydrogenase mutants of maize. Genetics 79: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Lee MM, Li J, Nam KH(2011) Characterization of cp3 reveals a new bri1 allele, bri1-120, and the importance of the LRR domain of BRI1 mediating BR signaling. BMC Plant Biol 11: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MY, Sali A(2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15: 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XM, Brengman J, Neubauer D, Sine SM, Engel AG(2016) Investigation of congenital myasthenia reveals functional asymmetry of invariant acetylcholine receptor (AChR) Cys-loop aspartates. J Biol Chem 291: 3291–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell DE, Leu WM, Gillmor CS, Xia G, Feldmann KA, Chua NH(1994) EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Shinpo S, Kohara M, Nakamura Y, Tabata S, Sato S(2008) A large scale analysis of protein-protein interactions in the nitrogen-fixing bacterium Mesorhizobium loti. DNA Res 15: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupsky CM, Desautels M, Huebert T, Zhao R, Hemmingsen SM, McIntosh LP(2001) Structure of Cdc4p, a contractile ring protein essential for cytokinesis in Schizosaccharomyces pombe. J Biol Chem 276: 5943–5951 [DOI] [PubMed] [Google Scholar]
- Smale ST.(2010) Beta-galactosidase assay. Cold Spring Harb Protoc 2010: pdb.prot5423. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G(1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM(2005) A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM(2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM(2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM(2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H(2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun AS, Lee JS, Theologis A(1998) Random mutagenesis of 1-aminocyclopropane-1-carboxylate synthase: a key enzyme in ethylene biosynthesis. Proc Natl Acad Sci USA 95: 9796–9801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Burtner C, Comai L, Henikoff S(2004) Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res 32: 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A(2009) A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183: 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ(1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y(2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M(2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20: 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.(2014) Auxin biosynthesis. The Arabidopsis Book 12: e0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J(2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL(2002) Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]