An upstream open reading frame in the 5ʹ untranslated region of a phosphate transporter gene inhibits its translation in a phosphate-independent manner, and this regulation can be harnessed to increase shoot phosphate accumulation and improve growth under phosphate deficiency.
Abstract
Inorganic orthophosphate (Pi) is an essential nutrient for plant growth, and its availability strongly impacts crop yield. PHOSPHATE1 (PHO1) transfers Pi from root to shoot via Pi export into root xylem vessels. In this work, we demonstrate that an upstream open reading frame (uORF) present in the 5′ untranslated region of the Arabidopsis (Arabidopsis thaliana) PHO1 inhibits its translation and influences Pi homeostasis. The presence of the uORF strongly inhibited the translation of a PHO1 5′UTR-luciferase construct in protoplasts. A point mutation removing the PHO1 uORF (ΔuORF) in transgenic Arabidopsis resulted in increased association of its mRNA with polysomes and led to higher PHO1 protein levels, independent of Pi availability. Interestingly, deletion of the uORF led to higher shoot Pi content and was associated with improved shoot growth under low external Pi supply and no deleterious effects under Pi-sufficient conditions. We further show that natural accessions lacking the PHO1 uORF exhibit higher PHO1 protein levels and shoot Pi content. Increased shoot Pi content was linked to the absence of the PHO1 uORF in a population of F2 segregants. We identified the PHO1 uORF in genomes of crops such as rice (Oryza sativa), maize (Zea mays), barley (Hordeum vulgare), and wheat (Triticum aesativum), and we verified the inhibitory effect of the rice PHO1 uORF on translation in protoplasts. Our work suggests that regulation of PHO1 expression via its uORF might be a genetic resource useful—both in natural populations and in the context of genome editing—toward improving plant growth under Pi-deficient conditions.
Phosphorus (P) is one of the most important elements limiting plant growth, and success in obtaining high crop yield is often dependent on the use of fertilizers containing inorganic orthophosphate (Pi; Roy et al., 2016). However, relying on Pi fertilizers to maintain or increase crop yield to meet the needs of a growing population will be challenging, since P-rich rocks are a finite nonrenewable resource and Pi overuse leads to environmental problems such as eutrophication (Sattari et al., 2016). There is thus a need to increase the ability of plants to better acquire Pi from the soil and optimize its internal use to achieve high yield with lower fertilizer input.
Pi deficiency induces broad changes in gene and protein expression, leading to a series of developmental and metabolic adaptations aimed at improving Pi acquisition and use. For example, Pi deficiency leads to changes in root architecture, with a reduction in primary root growth, enhancement of secondary roots, and elongation of root hairs, features that enhance the ability of the root system to explore the soil for nutrients (Gutiérrez-Alanís et al., 2018). Examples of Pi deficiency-induced metabolic changes include a shift from the synthesis of phospholipids to galactolipids in membranes and the secretion of organic acids and enzymes in the root apoplast to help acquire Pi from both organic and inorganic sources (Zhang et al., 2014). Pi-deficient plants typically enhance the expression of high-affinity Pi transporters of the PHOSPHATE TRANSPORTER1 (PHT1) family. The Arabidopsis (Arabidopsis thaliana) genome contains nine PHT1 genes encoding H+/Pi cotransporters (Mudge et al., 2002). Most PHT1 genes are primarily expressed in root epidermal and cortical cells, and several are transcriptionally induced by Pi starvation (Morcuende et al., 2007). PHT1 expression is influenced by several posttranscriptional mechanisms that impact their localization and activity. For example, localization of at least a subset of PHT1 transporters to the plasma membrane is regulated by phosphorylation and interaction with the PHOSHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1; Bayle et al., 2011). PHT1 protein turnover is also controlled by ubiquitination via the E2 conjugase PHOSPHATE2 (PHO2) and the E3 ligase NITROGEN LIMITATION ADAPTATION1 (NLA1) protein (Liu et al., 2012; Huang et al., 2013; Pan et al., 2019). Considering their critical role as the primary transporters responsible for the initial uptake of Pi from the soil into plants, modulation of PHT1 expression has been a focus of several studies aimed at improving Pi uptake and use in transgenic plants (Gu et al., 2016).
Following its acquisition by roots, Pi needs to be transported to the shoot. PHO1 is the main root-to-shoot Pi exporter involved in loading Pi into the xylem cells (Hamburger et al., 2002). pho1 mutants thus have low shoot Pi but high amounts of Pi in roots. PHO1 expression is under the partial control of the WRKY6 and WRKY42 transcription factors, which act as repressors (Chen et al., 2009; Su et al., 2015). PHO1 is only weakly regulated at the transcriptional level by Pi deficiency, while its protein stability is controlled by ubiquitination via PHO2 (Liu et al., 2012). pho2 mutants thus show enhanced expression of both PHT1 and PHO1 proteins, and exhibit a constitutively active phosphate-deficiency starvation response, leading to reduced growth associated with excessive shoot Pi content (Aung et al., 2006; Bari et al., 2006). PHO1 protein has a N-terminal hydrophilic region containing a SPX domain, a middle region with four transmembrane α-helices, followed by another hydrophobic region harboring an EXS domain (Wege et al., 2016). The SPX domain of PHO1 is involved in binding inositol polyphosphates, the concentrations of which change in response to Pi availability (Wild et al., 2016; Jung et al., 2018). pho1 null mutants exhibit the typical features associated with strong Pi deficiency, including poor growth (Poirier et al., 1991). While low shoot Pi in pho1 was initially thought to be the main cause of its stunted growth, transgenic plants with low expression of PHO1 showed normal growth despite having low shoot Pi content similar to that of pho1 null mutants (Rouached et al., 2011). Interestingly, root expression of the PHO1 EXS domain alone, which lacks Pi transport activity, was enough to rescue the stunted shoot growth of pho1 mutants, despite shoot Pi levels remaining low (Wege et al., 2016). Both these studies showed that low Pi content can be dissociated from its main effect on growth and that PHO1 could be involved in modulating the Pi-deficiency signaling cascade and its effect on growth. While ectopic overexpression of PHO1 in leaves has been shown to lead to stunted growth associated with aberrant Pi export in leaf apoplasts (Stefanovic et al., 2011), the effects of enhanced PHO1 expression in its endogenous root tissues have not been studied with respect to Pi homeostasis and growth.
In this work, we demonstrated that the Arabidopsis PHO1 mRNA harbors a small upstream open reading frame (uORF) in the 5′ untranslated region (UTR) that inhibits its translation. A point mutation that removes the uORF (∆uORF) led to increased PHO1 mRNA translation and protein accumulation. Expression of a PHO1 ∆uORF in the pho1-2 mutant resulted in increased shoot Pi content compared to wild-type plants, without signs of Pi toxicity symptoms. Interestingly, plants with PHO1 ΔuORF grown in Pi-deficient conditions showed improved growth. Increased PHO1 protein levels and higher shoot Pi accumulation were also observed in natural accessions lacking a PHO1 uORF. Furthermore, analysis of a segregating population from a cross between ecotype Columbia of Arabidopsis (Col-0), which contains a uORF, and an accession devoid of uORF (Ice50) supported the conclusion that absence of the uORF leads to enhanced shoot Pi content. Altogether, our results show that the PHO1 uORF can be used as a genetic resource in both natural populations and genetic engineering to modulate PHO1 protein expression and improve Pi acquisition and adaptation to Pi deficiency.
RESULTS
Presence of uORFs in PHO1 Genes in Dicots and Monocots
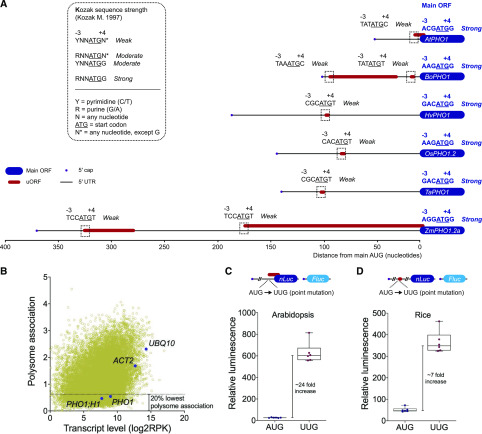
Analysis of PHO1 5′ UTR sequences revealed that uORFs are readily found in PHO1 transcripts of several dicots and monocots, including Arabidopsis (AtPHO1) and its close relative Brassica oleracea (BoPHO1), as well as in barley (Hordeum vulgare; HvPHO1), rice (Oryza sativa; OsPHO1.2), wheat (Triticum aestivum; TaPHO1), and maize (Zea mayz; ZmPHO1.2a; Fig. 1A; Supplemental Dataset S1). These uORFs vary in both size and position relative to the main ORF (mORF) start codon. The PHO1 5′ UTRs in rice, barley, and wheat all have a diminutive AUG-STOP uORF ∼100 nucleotides upstream of the mORF. In contrast, maize and B. oleracea both have a long and a short uORF, while Arabidopsis has a single short uORF. In both maize and Arabidopsis, a uORF overlaps with the mORF, with the stop codon being four nucleotides downstream of the mORF AUG (Fig. 1A).
Figure 1.
PHO1 transcripts harbor an inhibitory uORF. A, Schematic of the PHO1 5′ UTR in various plant species. 5′ UTRs (black line) and associated uORFs (red bars) are shown to scale. The beginnings of mORFs are shown in blue. Kozak sequence strength associated with the AUG start codon (uORF and mORF) is indicated (Kozak, 1986; Orr et al., 2019). uORF Kozak regions are in dotted boxes. Sequences (GenBank no., species) were obtained for AtPHO1 (NM_113246, Arabidopsis), BoPHO1 (XM_013762946, B. oleracea), HvPHO1 (AK364904, H. vulgare), OsPHO1.2 (XM_015770667, O. sativa), TaPHO1 (AK331635, T. aestivum), and ZmPHO1.2 (XM_008680959, Z. mays). B, Relationship between polysome association and transcript level for the Arabidopsis transcriptome. The data were obtained from supplemental table S1 of Deforges et al. (2019a) and represent mean values across several experimental conditions, i.e. Pi deprivation; treatment with auxin (indole acetic acid), abscisic acid, methyl-jasmonate, and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid; and comparison between root and shoot. Indicated genes are PHO1, PHO1;H1, ACT2, and UBQ10. C and D, Relative luminescence in transfected protoplasts (n = 6 biological replicates); the associated uORF is depicted with a red bar. Luminescence values produced in Arabidopsis (C) and rice (D) protoplasts, from the expression of nLuc fused to either Arabidopsis PHO1 5′ UTR (C) or rice PHO1.2 5′ UTR (D), were normalized against luminescence from Fluc. Both nLuc and Fluc luciferase genes were expressed from the same plasmid.
Ribosomes encountering a uORF can (1) translate the uORF and stall, triggering mRNA decay; (2) translate the uORF and then reinitiate to translate the downstream ORF; or (3) simply skip the uORF (Orr et al., 2019). The effect of a uORF on translation of the mORF depends on factors such as uORF context (including the strength of the Kozak sequence), conservation, distance from the cap and the mORF, and number of uORFs in the 5′ UTR (Calvo et al., 2009). Interestingly, for the PHO1 gene set analyzed, the Kozak strength was weak for all uORFs but strong for the mORF (Fig. 1A), suggesting that the mORF may be more translationally active and the uORF may have a more limited effect on translation efficiency.
PHO1 uORF Inhibits Translation
To gain insight into the translational efficiency of AtPHO1, we analyzed its polysome association and transcript levels across several experimental conditions (Fig. 1B) using a previously published dataset (from supplemental table S1 of Deforges et al., 2019a). Highly abundant transcripts that are also found at high protein abundancy, such as actin (ACT2) and ubiquitin (UBQ10), showed good association between transcript and translation. On the other hand, both AtPHO1 and its closest homolog, AtPHO1;H1, showed transcript levels in a moderate range relative to other mRNAs, while their polysome associations were among the 20% most poorly associated transcripts, suggesting a low translation efficiency. Similar poor translation efficiency has been reported for the rice OsPHO1.2 (Jabnoune et al., 2013). To examine whether the uORFs found in AtPHO1 and OsPHO1.2 could influence translational levels of a heterologous gene in Arabidopsis and rice, respectively, we fused the 5′ UTRs of each gene, including the entire uORF sequences, to a nano-luciferase gene (nLuc) and compared the effect of point mutations removing the uORF in transfected protoplasts (Fig. 1, C and D). The AUG to UUG mutation in the AtPHO1 uORF start codon resulted in a >20-fold increase in relative luminescence in Arabidopsis protoplasts, while a similar mutation in the OsPHO1.2 uORF start codon resulted in a 7-fold increase in rice protoplasts, without significant changes in nLuc mRNA levels (Supplemental Fig. S1). Altogether, these results indicate that the uORFs present in the rice and Arabidopsis PHO1 5′ UTRs act as translation inhibitory elements.
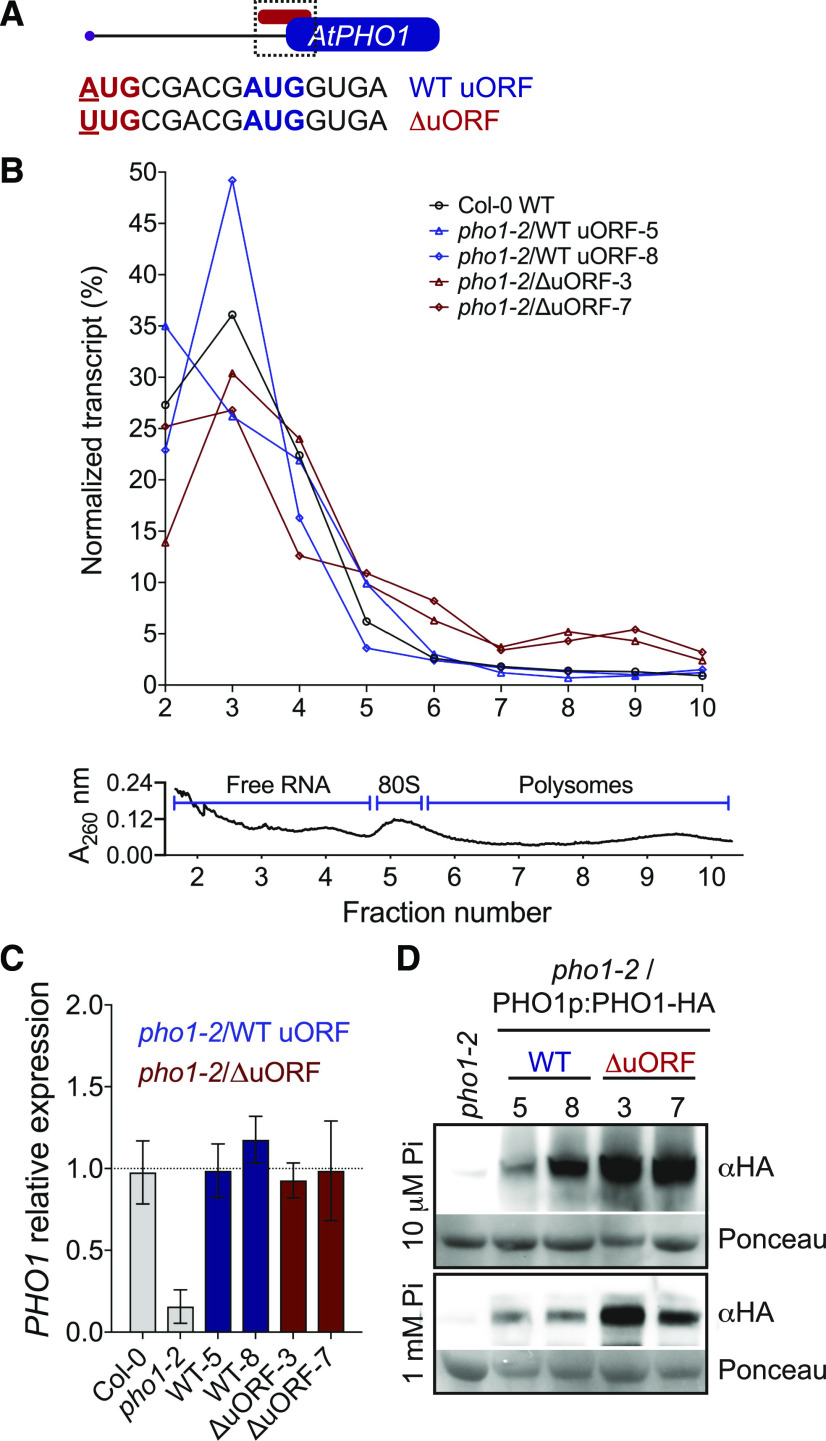
To study the putative inhibitory effect of PHO1 uORF on Pi homeostasis, we focused on the well characterized Arabidopsis PHO1. Its uORF codes for a four-amino acid peptide (MRRW) overlapping with the mORF’s AUG (Fig. 2A). We transformed the pho1-2 null mutant with a construct containing ∼6.5 kb of genomic PHO1 (1 kb of promoter region and the entire coding sequence) fused at the C terminus to a human influenza hemagglutinin (HA) tag (referred to as wild-type uORF), as well as a similar construct harboring an AUG to UUG point mutation in the uORF start codon (ΔuORF). Both wild-type and ΔuORF constructs complemented the pho1-2 growth phenotype associated with Pi deficiency (Supplemental Fig. S2). We then analyzed the effect of the uORF mutation on translation efficiency of PHO1 mRNA (Fig. 2B). We observed that the wild-type PHO1 transcript is poorly associated with ribosomes (monosomes and polysomes) and at least two-thirds of its transcript pool appear not engaged in translation. While the ΔuORF transcript showed a substantial increase in polysomal association compared to the wild type, the overall PHO1 transcript pool remained poorly associated with ribosomes. Importantly, PHO1 transcripts were at wild-type levels in all transgenic lines analyzed (Fig. 2C), indicating that ΔuORF point mutants retain wild-type mRNA stability. The observed increases in translation efficiency in plants lacking PHO1 uORF, without changes in transcript levels, were further confirmed by increased protein levels in plants expressing ΔuORF, as compared with the wild type (Fig. 2D). PHO1 protein was higher in ΔuORF plants growing under standard and low Pi concentration, indicating that the uORF inhibits PHO1 translation in a Pi-independent manner.
Figure 2.
Arabidopsis PHO1 uORF represses translation of the mORF. A, Schematic representation of the wild-type (WT) Arabidopsis PHO1 uORF (AUG start codon shown in red) overlapping with the PHO1 mORF AUG start codon (blue). Removal of the uORF in the construct ∆uORF was obtained by changing the uORF AUG to UUG. B to D, Analysis of PHO1 RNA and protein from root tissue of wild-type Col-0 (B and C only), two independent transformants of pho1-2 expressing PHO1 with a wild-type (lines 5 and 8) and mutated (∆uORF; lines 3 and 7) uORF, and pho1-2 (A and C only). Distribution of PHO1 transcript across fractions from a Suc density gradient (B) yielded a total RNA profile (A260) across fractions (bottom) showing the location of free RNA, 80S ribosomes, and polysomal fraction. PHO1 transcript expression levels (C) were determined relative to that in Col-0 wild type plants (normalized against ACT2 expression). For both B and C, plants were grown on one-half strength Murashige and Skoog medium with 1 mm Pi. D, PHO1 protein accumulation in roots of plants grown in one-half strength Murashige and Skoog medium with either high (1 mm) or low (10 μm) Pi was determined using antibody against HA tag.
PHO1 uORF Deletion Enhances Shoot Pi Accumulation and Improves Growth under Pi Deficiency
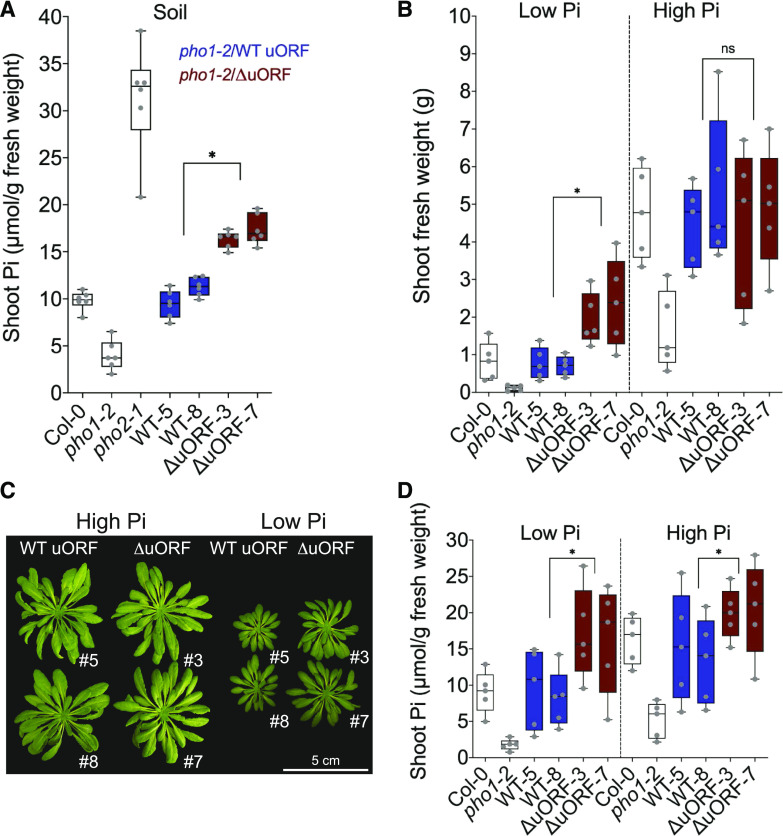
To test whether the increased level of PHO1 in plants expressing the gene with a mutated uORF affects Pi homeostasis, we analyzed Pi content in shoots of pho1-2 lines transformed with the ΔuORF and wild-type uORF constructs, grown in fertilized soil (Fig. 3A). As expected, the pho1-2 mutant showed low levels of Pi in the shoot, while pho2 showed about three times more shoot Pi than Col-0 wild type plants. Transgenic pho1-2 lines expressing the wild-type uORF transcript showed Pi levels similar to that of Col-0. In contrast, lines expressing the ΔuORF transcript showed an ∼1.7-fold increase in shoot Pi content relative to Col-0. As previously reported for both mutant lines (Poirier et al., 1991; Bari et al., 2006), pho1-2 and pho2-1 mutants showed strong and mild reduction, respectively, in shoot growth compared to Col-0, while transgenic pho1-2 lines expressing either the wild-type uORF or ΔuORF PHO1 transcript showed shoot growth similar to Col-0 control plants (Supplemental Fig. S2).
Figure 3.
Deletion of PHO1 uORF improves response to Pi deprivation. A, Shoot Pi content of plants cultivated in soil (n = 6 biological replicates; *P < 0.05, Student’s t test). B, Shoot fresh weight (n = 5 biological replicates; *P < 0.05, Student’s t test; ns, not significant) of plants grown in clay with low or high Pi. C, Representative pictures of plants from the indicated lines grown as described in B. D, Shoot Pi content (n = 5 biological replicates; *P < 0.05, Student’s t test) of plants grown as described in B. For A, B, and D, each gray dot corresponds to a biological replicate for either Col-0, pho1-2, two independent transformants of pho1-2 expressing PHO1 with a wild-type (WT; lines 5 and 8) and mutated (∆uORF; lines 3 and 7) uORF.
We next tested the effect of higher PHO1 protein levels on growth under Pi-deficient conditions. Plants were grown in fertilized clay until the four-rosette-leaf stage, after which they were transferred to either high- or low-Pi conditions and further cultivated for 5 weeks, until inflorescence emergence. Under low Pi conditions, PHO1 uORF deletion resulted in plants with approximately twice the shoot mass of those carrying the wild-type PHO1 uORF, either in the transgenic pho1-2 background or in Col-0 (Fig. 3, B and C). In contrast, plants growing on high external Pi showed no significant differences in shoot mass, except for reduced growth of pho1-2 mutant. Interestingly, shoot Pi content was significantly higher in transgenic plants expressing ΔuORF PHO1 transcript in both the low- and high-Pi treatments than in plants expressing a wild-type PHO1 transcript (Fig. 3D). However, root weight and root Pi accumulation were similar in plants expressing PHO1 with or without the uORF (Supplemental Fig. S3). The improved shoot growth of plants expressing the ΔuORF PHO1 transcript was not accompanied by changes in gene expression of phosphate starvation markers (Supplemental Fig. S4). These results show that a deletion of the uORF in PHO1 improves growth of plants undergoing Pi deprivation mainly by enhancing the transfer of Pi from soil to shoot.
Lack of PHO1 uORF in Natural Accessions Is Associated with Higher Shoot Pi Levels
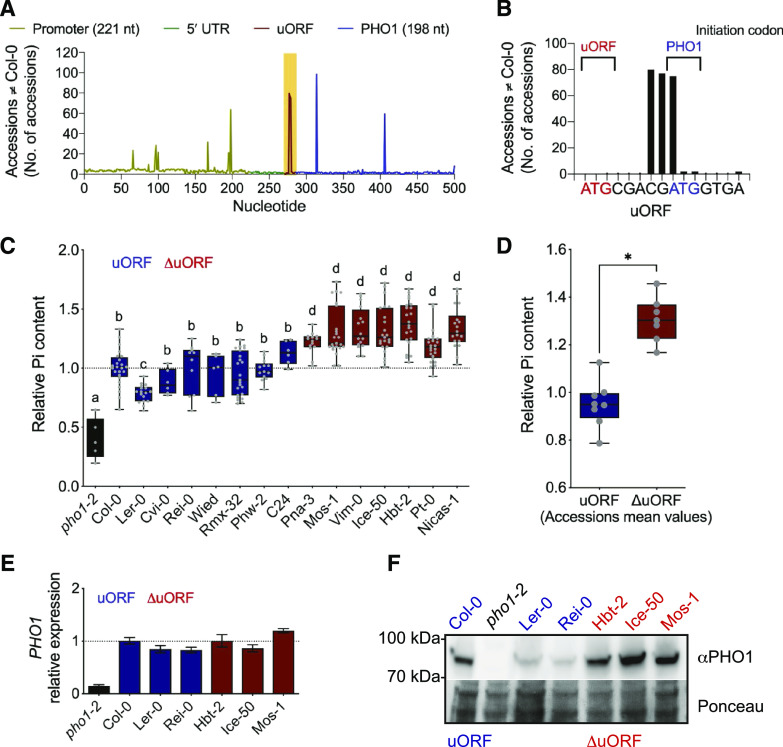
To look at the extent of the presence of PHO1 uORF in natural populations of Arabidopsis, we retrieved the 500-nucleotide sequences centered around the PHO1 5′ UTR from all available accessions (854 in total, as per December 2015) and aligned them against Col-0 as reference (Fig. 4A; Cao et al., 2011). Single-nucleotide polymorphisms (SNPs) could be found throughout the 500-nucleotide region, but mostly at very low frequency. However, a few nucleotides and/or regions exhibited relatively higher SNP frequency, including nucleotides within the uORF. Surprisingly, the uORF appears to be a region, rather than isolated nucleotides, of high variability across natural accessions. Indeed, three sequential uORF nucleotides (of its 15 nucleotides) showed high SNP frequency, and unexpectedly, the adenine of the mORF’s AUG start codon was among those nucleotides (Fig. 4B), indicating that in many accessions PHO1 mRNA is translated from an alternative start codon. We also observed that all accessions harboring a uORF SNP have deletions, and often a given accession has more than one SNP: one accession had four deletions, 67 had three deletions, 10 had two deletions, and eight had one deletion (Supplemental Dataset S2). Thus, a total of 86 accessions have at least one SNP within the PHO1 uORF, corresponding to ∼10% of all analyzed accessions. Most of these, however, retained the uORF either because they lack a nucleotide triplet (67 accessions had a three-base deletion), and hence the uORF maintains its frame, or because the uORF is extended to a downstream stop codon. Only eight of these 86 accessions lost the uORF, and that occurred by the same process: deletion of two nucleotides resulting in translational “fusion” of the uORF AUG with the mORF, which coincided with the loss of the mORF’s AUG in three accessions (Table 1). Therefore, the PHO1 uORF is absent in 0.9% of all accessions analyzed.
Figure 4.
Shoot Pi accumulation in natural accessions lacking the PHO1 uORF. A, Accessions differing from the Col-0 reference genome in a 500-nucleotide window around the PHO1 5′ UTR. Sequences for 854 accessions were aligned, and peaks refer to the number of accessions found with SNPs relative to the Col-0 reference sequence. SNPs within the uORF are highlighted in yellow. B, SNPs in accessions differing from the Col-0 reference within the PHO1 uORF sequence. The AUG start codon of PHO1 mORF is shown in blue, while the AUG start codon of the uORF is shown in red. C, Shoot Pi content of various accessions cultivated in soil for 3 weeks. Each gray dot corresponds to a biological replicate. Statistically significant differences (P < 0.05, ANOVA; 5–15 biological replicates) between accessions are indicated by lowercase letters above the boxes. D, Comparison of mean shoot Pi content in accessions with (blue) and without (red) the uORF (*P < 0.001, Student’s t test,). E and F, Analysis of root tissue of plants cultivated in one-half strength Murashige and Skoog medium for 10 d. Accessions with and without a PHO1 uORF are shown in blue and red, respectively. E, PHO1 transcript levels relative to expression in Col-0 wild-type plants (normalized against ACT2 expression). F, PHO1 protein accumulation determined using a polyclonal antibody against the endogenous PHO1.
Table 1. PHO1 uORF sequence in selected natural accessions.
The ATG start codon of the PHO1 mORF in Col-0 is shown in uppercase letters.
| Accession | PHO1 uORF |
|---|---|
| Col-0 | atgcgacgATG |
| Ice-50 | atgcgac–TG |
| Vim-0 | atgcgac–TG |
| Hbt-2 | atgcga–ATG |
| Cad-0 | atgcga–ATG |
| Mos-1 | atgcga–ATG |
| Nicas-1 | atgcga–ATG |
| Pna-3 | atgcga–ATG |
| Pt-0 | atgcga-g-TG |
Given the availability of natural accessions lacking the PHO1 uORF, we compared them with randomly selected accessions that have a uORF. Accessions with the PHO1 uORF showed shoot Pi content similar to that of Col-0, with the exception of lower content in Ler-0 (Fig. 4C). Strikingly, all accessions lacking the PHO1 uORF showed higher Pi content in the shoot compared with Col-0. On average, absence of the uORF resulted in an ∼30% increase in shoot Pi relative to the average content in accessions with the PHO1 uORF (Fig. 4D), suggesting a positive correlation between lack of uORF and Pi accumulation in shoot tissue. PHO1 mRNA levels were found to be similar in selected accessions (Fig. 4E), although protein levels were higher in accessions lacking the uORF as compared with those that have the PHO1 uORF (Fig. 4F). These results indicate that the higher levels of PHO1 associated with the absence of uORF in certain Arabidopsis accessions result in increased amounts of Pi in shoots.
Lack of PHO1 uORF and Its Associated Higher Shoot Pi Is Inheritable
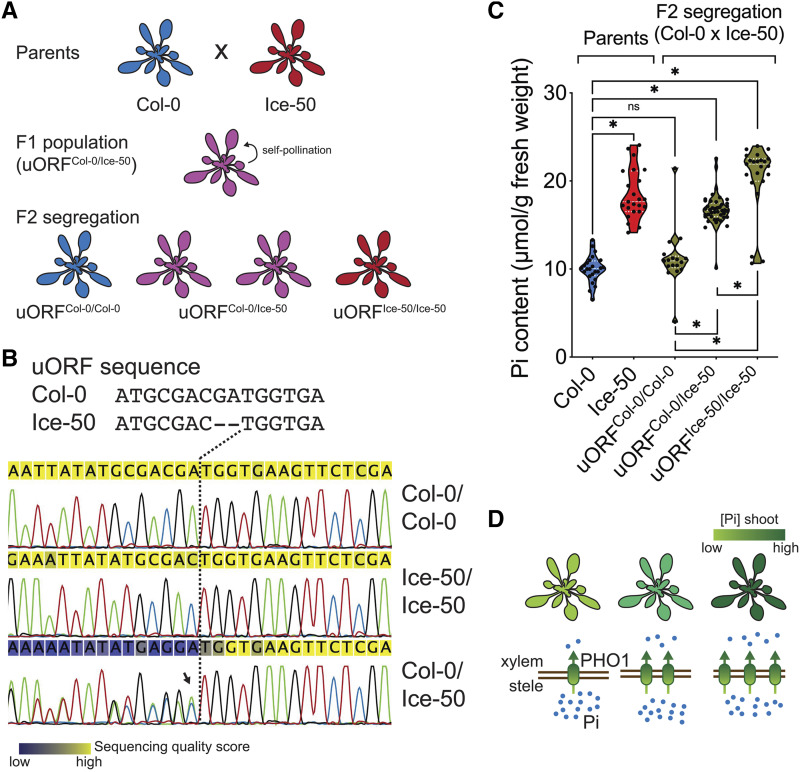
Arabidopsis accessions exhibit a large number of SNPs that can be translated into phenotypical and environmental adaptation heterogeneity (Alonso-Blanco et al., 2016). To untangle the contribution of SNPs within the PHO1 uORF from other SNPs across the genome that might also play a role in Pi homeostasis, we crossed Col-0 (uORF present) with the Ice-50 (also referred to as Toufl-1) accession (uORF absent) and analyzed the F2 segregating population (Fig. 5A). Col-0 and Ice-50 uORFs differ by two nucleotides, placing the uORF’s AUG in frame with the PHO1 coding sequence in Ice-50, thus eliminating the uORF in this ecotype (Fig. 5B, top). In order to genotype the F2 population, we sequenced the PHO1 uORF of each individual plant of a total of 82 segregants, using conventional Sanger sequencing of PCR products. Unambiguous results matching the Col-0 or Ice-50 uORF sequence were assigned as homozygous (uORFCol-0/Col-0 and uORFIce-50/Ice-50), while heterozygous uORFs (uORFCol-0/Ice-50) were assigned to sequencing spectra showing overlapping peaks at the precise position where Col-0 and Ice-50 differ (Fig. 5B, bottom). We identified 17 uORFCol-0/Col-0, 23 uORFIce-50/Ice-50, and 42 uORFCol-0/Ice-50 plants in the F2 population, indicating Mendelian segregation (χ2 = 0.93, degree of freedom = 1). Shoot Pi content was measured for each genotyped F2 segregant, as well as for the Col-0 and Ice-50 parental accessions (Fig. 5C). uORFCol-0/Col-0 and uORFIce-50/Ice-50 homozygous plants showed shoot Pi content similar to that of the corresponding parental lines, while uORFCol-0/Ice-50 heterozygous showed an intermediate accumulation of Pi. Although Col-0 and Ice-50 may exhibit a large number of SNPs, these results demonstrated that higher shoot Pi levels are linked to SNPs within the PHO1 uORF. In agreement with increased PHO1 protein in the absence of the uORF and its effect on shoot Pi accumulation, we observed a dose-dependent increase in shoot Pi, with the lowest levels seen in uORFCol-0/Col-0, intermediate levels in uORFCol-0/Ice-50, and highest levels in uORFIce-50/Ice-50. Altogether, our results support a model in which shoot Pi content correlates with PHO1 levels (Fig. 5D).
Figure 5.
Shoot Pi accumulation in F2 segregation from a cross of natural accessions with or without the PHO1 uORF. A, Schematic of crossing between Col-0 and Ice-50 and subsequent F1 and F2 populations. B, Genotyping strategy. Sanger sequence profiles are aligned against the Col-0 and Ice-50 uORF region. Parental genotypes are readily assigned and heterozygous loci are identified by overlapping peaks immediately upstream of the first polymorphism between Col-0 and Ice-50 uORF sequence (highlighted with an arrow and showing a distinct sequencing-quality score shift in the Col-0/Ice-50 profile). C, Shoot Pi content of plants cultivated in soil for 3 weeks. Genotypes of plants at the PHO1 uORF are shown for the F2 plants. Each dot corresponds to a biological replicate; n > 20 of each parental accession (Col-0 and Ice-50); n = 82 F2 segregation, i.e. 17 uORFCol-0/Col-0, 23 uORFIce-50/Ice-50, and 42 uORFCol-0/Ice-50; *P < 0.001, Student’s t test; ns, not significant). D, Model proposing an association between PHO1 protein accumulation and shoot Pi levels.
DISCUSSION
Translation of uORFs is most commonly associated with a reduction of translation of the mORF, as a result of reduced ribosome (re)initiation at the downstream mORF start codon. At least one-third of the Arabidopsis transcriptome (>9,000 mRNAs) have one or more uORFs, and their impact on translation regulation can be substantial (Srivastava et al., 2018). Analysis of changes in mRNA translation by ribosome footprinting revealed that ∼30% of mRNAs translationally regulated by phosphate deficiency in roots have one or more uORFs, and changes in mORF-to-uORF ribosome occupancy were associated with lower translation of the mORF (Bazin et al., 2017). While most uORFs appear to code for small, nonconserved peptides, some uORFs were found to be evolutionarily conserved and could code for functional peptides (Andrews and Rothnagel, 2014). However, only a small fraction of uORF-derived peptides identified by ribosome profiling have been validated by mass spectrometry (Menschaert et al., 2013), suggesting that in many cases ribosome footprinting may indicate ribosome stalling instead of synthesis of functional peptides. In some apparently rare cases, more than one functional protein is coded from a single bicistronic or polycistronic transcript (Mouilleron et al., 2016; Lorenzo-Orts et al., 2019), defying the concept of the uORF as upstream to a main ORF. In addition to playing a role in translation, uORFs are also involved in nonsense-mediated mRNA decay, a surveillance pathway involved in the degradation of aberrant transcripts with premature stop codons.
The Arabidopsis PHO1 contains a uORF that overlaps, in a different reading frame, with the mORF. Overlapping uORFs are often more repressive than those with an upstream stop codon, because they hinder the possibility of a ribosome translating the uORF to reinitiate translation at the start codon of the mORF (Yun et al., 2012; Torrance and Lydall, 2018). The PHO1 uORF has a weak Kozak context, and thus, leaky scanning is likely to be the main mechanism by which PHO1 mORF is translated. Protoplast transformation using the PHO1 5′ UTR fused to a dual luciferase system showed that mutation of the PHO1 uORF resulted in a pronounced translation enhancement, revealing the potential of the uORF to inhibit translation. In transgenic plants, mutation of the uORF enhanced the association of PHO1 mRNA with polysomes, leading to enhanced protein synthesis. However, a large fraction of PHO1 mRNA remained poorly associated with ribosomes, indicating that there might be an element(s) within PHO1 mRNA other than the uORF that limits its translation. The PHO1 uORF codes for a four-amino acid peptide (MRRW), which is too short for a meaningful conventional search for conservation and putative function and is below the minimum size of most known functional peptides. Thus, it is unlikely that the uORF-coded peptide is required for translation inhibition of PHO1. In addition, it is also unlikely that nonsense-mediated mRNA decay is involved in this process, because we found no evidence of mRNA decay in transcripts with or without the PHO1 uORF, including in protoplasts, transgenic lines, and natural accessions.
Translation efficiency of uORFs largely defines their translation inhibition on the mORF, and this process is regulated by their sequences, but can also be modulated by stress conditions and, in some cases, specific metabolites. Examples of the latter modulation include regulation of the uORF that impacts translation of mRNA encoding phosphoethanolamine-aminomethyltransferase by phosphocholine (Alatorre-Cobos et al., 2012), S-adenosyl-Met decarboxylase by polyamine (Hanfrey et al., 2005), the ZIP11 transcription factor by Suc (Wiese et al., 2004), and GDP-l-Gal phosphorylase by ascorbate (Laing et al., 2015). Metabolite-dependent uORF inhibition of mORF also plays an important role in boron homeostasis, as NIP5;1 and BOR1 boron transporter mRNAs are regulated by uORF in a boron-dependent manner. Boron is an essential element for plants, but toxic at high concentrations (Reid et al., 2004). Both NIP5;1, a boric acid channel essential for its uptake from soil, and BOR1, a borate exporter that facilitates its root-to-shoot translocation, are translationally modulated by a uORF in a boron-dependent manner (Tanaka et al., 2016; Aibara et al., 2018). To our knowledge, Pi-dependent regulation of mORF translation via a regulatory uORF has not yet been described. Although mutation of an uORF present in the rice NLA1 5′ UTR was recently shown to increase expression of a reporter gene in a Pi-dependent manner, it is unclear whether the effect was transcriptional or translational (Yang et al., 2019). Transgenic plants expressing the PHO1 ΔuORF transcript showed increased PHO1 protein without changes in the steady-state mRNA level in both standard and Pi-depriving conditions, suggesting that PHO1 uORF is not regulated by low Pi in Arabidopsis. Whether toxic levels of Pi could affect uORFs is elusive, as observed in boron homeostasis, particularly because plants tolerate many orders of magnitude more Pi than boron.
The PHO1 uORF(s) is readily found in monocots and dicots, although its overall structure, position, and potential peptide encoded are not conserved. However, it is striking that an AUG-STOP uORF is found at approximately the same location, relative to the AUG in the mORF, in the 5′ UTR of PHO1 in more phylogenetically related rice (OsPHO1.2), barley (HvPHO1), and wheat (TaPHO1), in contrast to the divergent uORF found in the more distantly related maize (ZmPHO1.2a). This suggests that there might be an evolutionary constraint on the PHO1 uORF in grasses, further indicating a conserved putative function.
This study shows that removal of the Arabidopsis PHO1 uORF, either via mutagenesis in transgenic plants or naturally in various accessions, increases PHO1 protein expression and results in higher Pi accumulation in shoots. Importantly, enhanced PHO1 expression and higher shoot Pi observed in transgenic plants led to improved growth of plants under Pi-deficient conditions, without deleterious effects under Pi-sufficient conditions. This contrasts with the pho2 mutant, which has a constitutively active Pi-deficiency signal transduction pathway, overexpresses both PHT1 and PHO1 proteins, and has a Pi-toxicity phenotype (Bari et al., 2006; Liu et al., 2012). uORF disruption appears to have an overall very specific impact on plants, because despite the abundance of uORFs in plant genomes, very few examples of the deleterious impact of uORF disruption on plant development and physiology have been shown. These include the negative effect on growth of uORF disruption in the adenosyl-Met decarboxylase gene (Hanfrey et al., 2005) and the pleotropic effects of overexpression of the transcription factor HB1, caused by uORF deletion, on hypocotyl, leaf, and flower development (Ribone et al., 2017).
Improvement of plant tolerance of Pi deficiency is an important goal in the quest to maintain high crop yields under reduced fertilizer input. Numerous studies have focused on overexpression of the primary Pi-H+ cotransporters encoded by the PHT1 family, with limited success (Gu et al., 2016). Indeed, overexpression of rice PHT1 has often been associated with Pi-toxicity symptoms, which negatively impact growth (Jia et al., 2011; Wang et al., 2014). Such undesirable phenotypes may be associated with the use of strong constitutive promoters, such as CaMV35S and Ubiquitin, which enhance PHT1 expression ectopically. Overexpression of genes encoding central transcription factors involved in Pi-deficiency signal transduction, such as PHOSPHATE STARVATION RESPONSE1 (PHR1) in Arabidopsis and PHOSPHATE STARVATION RESPONSE2 (PHR2) in rice, also leads to Pi-toxicity symptoms and decreased plant growth (Nilsson et al., 2007; Zhou et al., 2008).
Lack of the PHO1 uORF in some natural Arabidopsis accessions, associated with increased shoot Pi, suggests that regulation of PHO1 via its uORF might be a genetic resource for plants to cope with an unreliable availability of Pi in the soil. We envisage that a beneficial impact of the increased expression of PHO1 via mutation of its uORF might be obtainable with other plant species, in particular crops, either through conventional breeding using natural accessions lacking an uORF or with the aid of genome editing tools (Zhang et al., 2018).
MATERIAL AND METHODS
Plant Material, Growth Conditions, and Constructs
Null mutants pho1-2 and pho2-1 have been previously described (Poirier et al., 1991; Delhaize and Randall, 1995). Natural accessions were obtained from the Nottingham Arabidopsis Stock Centre: Hbt-2 (N76899), Ice-50 (N76348, also referred to as Toufl-1), Cad-0 (N76739), Mos-1 (N77108), Vim-0 (N78844), Nicas-1 (N77127), Pna-3 (N77184), Pt-0 (N78915), Ler-0 (N77020), Phw-2 (N77173), Vig-1 (N78843), Rei-0 (N77208) and Rmx-3 (N77219). An ∼6.5-kb genomic sequence of PHO1 locus (∼1 kb upstream of the transcription start site and ∼5.5 kb of coding sequence until the last nucleotide before the translation stop codon) was PCR-amplified using a reverse oligo containing a sequence for the HA epitope, thus introducing a C-terminal HA tag to PHO1. An ATG-to-TTG point mutation was introduced via overlapping PCR to modify the PHO1 uORF start codon, using the cloned wild-type PHO1 pEntry vector as template. Both unmodified (wild-type uORF) and mutated (ΔuORF) pEntry-cloned sequences were LR-recombined into the pFASTR01 destination vector (Shimada et al., 2010). The final constructs were introduced into Agrobacterium tumefaciens pGV3101 and used to transform the pho1-2 mutant. The list of primers can be found in Supplemental Table S1.
Protoplast Transformation and Luminescence Analysis
Plasmids used for protoplast transfection have been previously described and validated for translation assay (Deforges et al., 2019a, 2019b). The dual luciferase system was modified to express either the Arabidopsis (Arabidopsis thaliana) or rice (Oryza sativa) PHO1 5′ UTR fused to nLuc, while firefly luciferase (Fluc) was expressed from an independent promoter and used as the loading control.
Arabidopsis protoplasts were produced and transformed as previously described (Yoo et al., 2007). In brief, wild type Col-0 plants were grown on soil in a short photoperiod (8 h light and 16 h dark at 21°C) for 4 to 5 weeks and leaves were cut with razor blades to produce 0.5- to 1-mm leaf strips. These were submerged in enzyme solution (1% [w/v] cellulase, 0.25% [w/v] macerozyme, 0.4 m mannitol, 20 mm KCl, 20 mm MES, and 10 mm CaCl2), vacuum infiltrated, and incubated at room temperature for 2 h. Protoplasts were harvested by centrifugation at 100g for 3 min, washed with W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES) and resuspended in MMG solution (4 mm MES [pH 5.7], 0.4 m mannitol, and 15 mm MgCl2) at 1 × 106 protoplasts/mL. Protoplast transformation was performed by combining ∼1.5 × 105 protoplasts, 5 μg of plasmid, and polyethylene glycol (PEG) solution (40% [w/v] PEG4000, 0.2 m mannitol, and 100 mm CaCl2). After replacing PEG solution with W5 solution by consecutive washings, protoplasts were kept in the dark for ∼16 h at 21°C.
Rice protoplasts were isolated and transformed as previously described (Zhang et al., 2011). Briefly, rice seeds were surface sterilized with 0.05% (w/v) Tween-80 and 7% (v/v) hypochlorite solution for 3 min, washed with sterile water, and germinated on water-soaked paper in a magenta box at 28°C for 7 d in the dark, then 3 d under a 12-h light/12-h dark cycle. The stems and sheathes of seedlings were cut into small strips (∼0.5 mm) and incubated in solution containing cell wall-digesting enzymes (1.5% [w/v] cellulase RS, 0.75% [w/v] macerozyme R-10, 0.6 m mannitol, 20 mm MES [pH 5.7], 20 mm KCl, and 10 mm CaCl2) for 3 to 5 h in the dark with gentle shaking. The released protoplasts were collected and washed with W5 solution, pelleted again and resuspended in MMG solution before PEG-mediated transfection. Protoplast transfection was performed by combining ∼105 protoplasts, 5 μg of dual luciferase plasmid, and PEG solution. After replacing PEG solution with W5 solution by consecutive washings, protoplasts were kept in the dark for ∼16 h at 21°C.
Transformed protoplasts were harvested by centrifugation at 6,000g for 1 min, and resuspended in 1X Passive Lysis buffer (E1941, Promega). The lysate was cleared by centrifugation and used for luminescence quantification using the Nano-Glo Dual-Luciferase Reporter Assay System (N1610, Promega). Luminescence values for nLuc were normalized against Fluc. Statistically significant differences (P < 0.05, Student’s t test) in relative luminescence (nLuc:Fluc) were used to assess cotransfection effects.
Polysome Fractionation
Arabidopsis roots were frozen, then ground, and the polysomes were extracted essentially as previously described (Mustroph et al., 2009), with minor modifications. Briefly, the powder was resuspended in two volumes of polysome extraction buffer (200 mm Tris [pH 9.0], 200 mm KCl, 1% [w/v] deoxycholate, 25 mm EGTA, 1% [v/v] detergent mix containing equal proportions of Brij-35, Triton X-100, octylphenyl-polyethylene glycol, and Tween 20, 1% [w/v] polyoxyethylene 10 tridecyl ether, 35 mm MgCl2, 1 mm dithiothreitol [DTT] and 100 μg mL−1 cyclohexomide), and incubated on ice for 15 min. The mixture was then centrifuged at 4°C for 15 min at 20,000g, and the supernatant was layered on top of a 10-mL Suc cushion (60% [w/v] Suc, 400 mm Tris [pH 9.0], 200 mm KCl, 25 mm EGTA, 35 mm MgCl2, 1 mm DTT, and 100 μg mL−1 cyclohexomide) and centrifuged for 3 h at 170,000g. The pellet was resuspended in 10 μL of the same Suc cushion buffer without Suc, and loaded on top of 5-mL 15% to 60% (w/v) continuous Suc gradients. The gradients were centrifuged for 75 min at 237,000g in swinging buckets and fractionated using a gradient-holder apparatus (Brandel) into 12 fractions. During gradient fractionation, the UV absorbance was continuously monitored in order to detect the position of the different complexes within the gradient. RNA was extracted from each collected fraction using the RNA Clean and Concentrator-25 kit according to the manufacturer’s instruction (R1017, Zymo Research). Reverse transcription quantitative PCR (RT-qPCR) was performed as described above. Polysome association was calculated as the relative proportion of mRNA in each fraction of the gradient, as previously described (Faye et al., 2014).
RNA Isolation and RT-qPCR
Total RNA was extracted from roots using an RNA purification kit according to the manufacturer’s instructions (PP-210, Jena Bioscience), followed by DNase I treatment. Complementary DNA was synthesized from 0.5 μg RNA using M-MLV Reverse Transcriptase (M3681, Promega) and oligo d(T)15 according to the manufacturer's instructions. RT-qPCR analysis was performed using SYBR Select Master Mix (4472908, Applied Biosystems) with primer pairs specific to genes of interest and ACT2 used for data normalization. Primer sequences are listed in Supplemental Table S1.
Immunoblot Analysis
For immunoblot using anti-PHO1, we performed the extraction and blotting as previously described (Liu et al., 2012). Briefly, ∼0.5 to 1 g of root material was ground in liquid nitrogen and dissolved in protein lysis buffer containing 2% (w/w) SDS, 60 mm Tris-HCl (pH 8.5), 2.5% (v/v) glycerol, 0.13 mm EDTA, and 1× complete protease inhibitor (Roche). Around 50 to 80 μg of total protein was separated in a 4% to 12%-gradient SDS-PAGE gel, transferred to a polyvinylidine fluoride membrane, incubated with anti-PHO1 (Liu et al., 2012), and incubated with antirabbit secondary antibody before chemiluminescence image capture. For immunoblot using anti-HA, total protein was extracted from nitrogen-ground root material using 10 mm phosphate buffer (pH 7.4), 300 mm Suc, 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 1 mm DTT, 20 mm NaF, and 1× protease inhibitor without EDTA (Roche). Homogenized samples were sonicated for 10 min in an ice-cold batch and lysate was centrifuged at 6,000g for 5 min to remove debris. Supernatant was centrifuged at 21,000g for 1 h and the membrane-enriched pellet was resuspended in 2% (w/v) SDS, 60 mm Tris-HCl (pH 8.5), 2.5% (v/v) glycerol, 0.13 mm EDTA, and 1× complete protease inhibitor (Roche). Around 50 μg of membrane-enriched protein was separated in a 4% to 12%-gradient SDS-PAGE gel, transferred to a polyvinylidine fluoride membrane, incubated with anti-HA (3F10 clone, Roche), and incubated with antirat secondary antibody before chemiluminescence image capture.
Phosphate Quantification
Quantification of Pi was performed as previously described (Ames, 1966). Shoot or root material was placed in water and at least three freeze-thaw cycles were applied to release the Pi content before quantification by molybdate assay using a standard curve.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_113246 (AtPHO1), AK331635 (TaPHO1), XM_013762946 (BoPHO1), AK364904 (HvPHO1), XM_015770667 (OsPHO1.2), and XM_008680959 (ZmPHO1.2) and are shown in Supplemental Dataset S1. Sequences (∼500 nucleotides) around the PHO1 ATG start codon in various Arabidopsis accessions are shown in Supplemental Dataset S2.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article.
Supplemental Figure S1. Levels of luciferase transcripts in transfected protoplast.
Supplemental Figure S2. Phenotype of Col-0, pho1-2, pho2, and transformants of pho1-2 expressing PHO1 with a wild type and mutated uORF.
Supplemental Figure S3. Effect of PHO1 uORF deletion on root and shoot growth and Pi content.
Supplemental Figure S4. Effect of PHO1 uORF deletion on the expression of phosphate starvation markers.
Supplemental Table S1. Primer list.
Supplemental Dataset S1. PHO1 complementary DNA sequence in various plant species.
Supplemental Dataset S2. Aligned sequences around the main PHO1 ATG start codon in natural accessions.
Acknowledgments
The authors thank Tzyy-Jen Chiou (Academia Sinica) for the PHO1 antibody and Syndie Delessert and Dominique Jacques Vuarambon (University of Lausanne) for technical assistance.
Footnotes
This work was supported by the Swiss National Science Foundation (Schweizerische Nationalfonds; grant nos. CRSII3_154471 and 31003A–182462 to Y.P.).
Articles can be viewed without a subscription.
References
- Aibara I, Hirai T, Kasai K, Takano J, Onouchi H, Naito S, Fujiwara T, Miwa K(2018) Boron-dependent translational suppression of the borate exporter BOR1 contributes to the avoidance of boron toxicity. Plant Physiol 177: 759–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatorre-Cobos F, Cruz-Ramírez A, Hayden CA, Pérez-Torres C-A, Chauvin A-L, Ibarra-Laclette E, Alva-Cortés E, Jorgensen RA, Herrera-Estrella L(2012) Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J Exp Bot 63: 5203–5221 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KMM, Cao J, Chae E, Dezwaan TMM, Ding W, et al. ; 1001 Genomes Consortium (2016) 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN.(1966) Assay of inorganic phosphate, total phosphate and phosphatases In Neufeld EF, and Ginsburg V, eds, Complex Carbohydrates, Methods in Enzymology, volume 80. Academic Press, Amsterdam, pp 115–118 [Google Scholar]
- Andrews SJ, Rothnagel JA(2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 15: 193–204 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ(2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R(2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V, Arrighi J-F, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L(2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J(2017) Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA 114: E10018–E10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK(2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH(2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforges J, Reis RS, Jacquet P, Sheppard S, Gadekar VP, Hart-Smith G, Tanzer A, Hofacker IL, Iseli C, Xenarios I, et al. (2019a) Control of cognate sense mRNA translation by cis-natural antisense RNAs. Plant Physiol 180: 305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforges J, Reis RS, Jacquet P, Vuarambon DJ, Poirier Y(2019b) Prediction of regulatory long intergenic non-coding RNAs acting in trans through base-pairing interactions. BMC Genomics 20: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ(1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye MD, Graber TE, Holcik M(2014) Assessment of selective mRNA translation in mammalian cells by polysome profiling. J Vis Exp 92: 52295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Chen A, Sun S, Xu G(2016) Complex regulation of plant phosphate rransporters and the gap between molecular mechanisms and practical application: What is missing? Mol Plant 9: 396–416 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Ojeda-Rivera JO, Yong-Villalobos L, Cárdenas-Torres L, Herrera-Estrella L(2018) Adaptation to phosphate scarcity: Tips from Arabidopsis roots. Trends Plant Sci 23: 721–730 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y(2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ(2005) A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J Biol Chem 280: 39229–39237 [DOI] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY, et al. (2013) Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y(2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G(2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Ried MK, Hothorn M, Poirier Y(2018) Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr Opin Biotechnol 49: 156–162 [DOI] [PubMed] [Google Scholar]
- Kozak M.(1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 [DOI] [PubMed] [Google Scholar]
- Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. (2015) An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27: 772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Huang T-K, Tseng C-Y, Lai Y-S, Lin S-I, Lin W-Y, Chen J-W, Chiou T-J(2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Orts L, Witthoeft J, Deforges J, Martinez J, Loubéry S, Placzek A, Poirier Y, Hothorn LA, Jaillais Y, Hothorn M(2019) Concerted expression of a cell cycle regulator and a metabolic enzyme from a bicistronic transcript in plants. Nat Plants 5: 184–193 [DOI] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappé J, Gevaert K, Van Damme P(2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol Cell Proteomics 12: 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Mouilleron H, Delcourt V, Roucou X(2016) Death of a dogma: Eukaryotic mRNAs can code for more than one protein. Nucleic Acids Res 44: 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW(2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJHH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J(2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH(2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Orr MW, Mao Y, Storz G, Qian S-B(2019) Alternative ORFs and small ORFs: Shedding light on the dark proteome. Nucleic Acids Res 48: 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Wu Y, Xie Q(2019) Regulation of ubiquitination is central to the phosphate starvation response. Trends Plant Sci 24: 755–769 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J(1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJ, Hayes JE, Post A, Stangoulis JCR, Graham RD(2004) A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ 27: 1405–1414 [Google Scholar]
- Ribone PA, Capella M, Arce AL, Chan RL(2017) A uORF represses the transcription factor AtHB1 in aerial tissues to avoid a deleterious phenotype. Plant Physiol 175: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, Bligny R, Poirier Y(2011) Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J 65: 557–570 [DOI] [PubMed] [Google Scholar]
- Roy ED, Richards PD, Martinelli LA, Coletta LD, Lins SRM, Vazquez FF, Willig E, Spera SA, VanWey LK, Porder S(2016) The phosphorus cost of agricultural intensification in the tropics. Nat Plants 2: 16043. [DOI] [PubMed] [Google Scholar]
- Sattari SZ, Bouwman AF, Martinez Rodríguez R, Beusen AHW, van Ittersum MK(2016) Negative global phosphorus budgets challenge sustainable intensification of grasslands. Nat Commun 7: 10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I(2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu JK(2018) UTR-dependent control of gene expression in plants. Trends Plant Sci 23: 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y(2011) Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF(2015) WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol 167: 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, et al. (2016) The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28: 2830–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance V, Lydall D(2018) Overlapping open reading frames strongly reduce human and yeast STN1 gene expression and affect telomere function. PLoS Genet 14: e1007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Piñeros MA, Wang Z, Wang W, Li C, Wu Z, Kochian LV, Wu P(2014) Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ 37: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Wege S, Khan GA, Jung J-Y, Vogiatzaki E, Pradervand S, Aller I, Meyer AJ, Poirier Y(2016) The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol 170: 385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S(2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung J-Y, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, et al. (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352: 986–990 [DOI] [PubMed] [Google Scholar]
- Yang S, Lu W, Ko S-S, Sun C, Hung J, Chiou T-J(2019) uORF and phosphate-regulated expression of rice OsNLA1 controls phosphate transport and reproduction. Plant Physiol 182: 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yun Y, Adesanya TMA, Mitra RD(2012) A systematic study of gene expression variation at single-nucleotide resolution reveals widespread regulatory roles for uAUGs. Genome Res 22: 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Si X, Ji X, Fan R, Liu J, Chen K, Wang D, Gao C(2018) Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol 36: 894–898 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ(2014) Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol 56: 192–220 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P(2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]