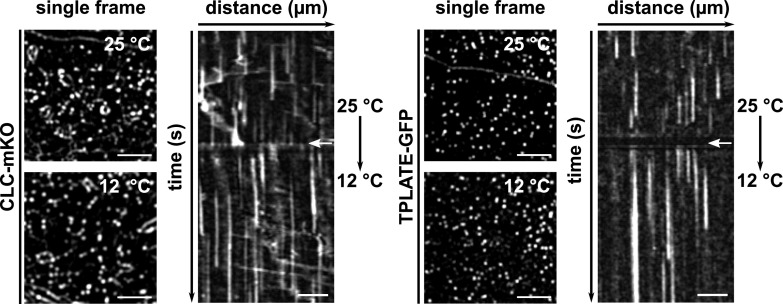
Lowering the temperature increases the temporal resolution of visualizing protein recruitment at the plasma membrane, demonstrated by dual-color live-cell imaging of endocytic complex subunits.
Abstract
The TPLATE complex (TPC) is a key endocytic adaptor protein complex in plants. TPC in Arabidopsis (Arabidopsis thaliana) contains six evolutionarily conserved subunits and two plant-specific subunits (AtEH1/Pan1 and AtEH2/Pan1) which, although cytoplasmic proteins, are not associated with the hexameric subcomplex in the cytoplasm. To investigate the dynamic assembly of the octameric TPC at the plasma membrane (PM), we performed state-of-the-art dual-color live-cell imaging at physiological and lowered temperatures. Lowering the temperature slowed down endocytosis, thereby enhancing the temporal resolution of the differential recruitment of endocytic components. Under both normal and lowered temperature conditions, the core TPC subunit TPLATE and the AtEH/Pan1 proteins exhibited simultaneous recruitment at the PM. These results, together with colocalization analysis of different TPC subunits, allow us to conclude that the TPC in plant cells is not recruited to the PM sequentially but as an octameric complex.
Clathrin-mediated endocytosis (CME) is the best-studied and predominant endocytic pathway in eukaryotes to internalize plasma membrane (PM) proteins and extracellular materials, commonly termed cargo, into cells (Dhonukshe et al., 2007; Robert et al., 2010; Kitakura et al., 2011; Bitsikas et al., 2014). The formation of clathrin-coated vesicles requires several highly coordinated stages: initiation, cargo selection, coat assembly, scission, and vesicle uncoating (McMahon and Boucrot, 2011). Although the initiation of CME at the PM remains poorly understood in plants, similar to other systems, adaptor proteins are presumed to recognize cargo proteins via cargo-recognition motifs and subsequently recruit clathrin triskelia through clathrin-binding motifs (Zhang et al., 2015). As clathrin does not bind directly to the PM or to the cargo proteins, adaptor proteins thus play an essential role to link the PM and the clathrin cage (McMahon and Boucrot, 2011).
Two early-arriving adaptor protein complexes function at the PM in plants: the heterotetrameric Adaptor Protein-2 complex (AP-2) and the octameric TPLATE complex (TPC). AP-2 comprises two large (AP2A and AP2B or α and β), one medium (AP2M or μ), and one small (AP2S or σ) subunit (Di Rubbo et al., 2013). TPC contains eight components: six that can be traced back to the last eukaryotic common ancestor (TPLATE, TML, TASH3, LOLITA, TWD40-1, and TWD40-2) and two components, AtEH1/Pan1 and AtEH2/Pan1, which are plant-specific TPC subunits (Gadeyne et al., 2014; Hirst et al., 2014). AP-2 function is important for somatic plant development, as single Arabidopsis (Arabidopsis thaliana) ap-2 mutants display developmental defects yet still result in viable plants (Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013). TPC is essential for both pollen and somatic plant development, as knockout or knockdown of single subunits of TPC in Arabidopsis leads to pollen or seedling lethality, respectively (Van Damme et al., 2006; Gadeyne et al., 2014).
TPC is an evolutionarily ancient protein complex that has been so far experimentally characterized only in plants and in an amoeba, Dictyostelium discoideum (Gadeyne et al., 2014; Hirst et al., 2014). TPC in plants was identified as an octameric complex by tandem affinity purification experiments (Gadeyne et al., 2014). Recently, we also identified an important role for the AtEH/Pan1 proteins in actin-regulated autophagy and in recruiting several components of the endocytic machinery to the AtEH/Pan1-positive autophagosomes (Gadeyne et al., 2014; Wang et al., 2019). TPC in D. discoideum (described as TSET), however, was identified as a hexameric complex lacking the AtEH/Pan1 homologs (Hirst et al., 2014). Although Amoebozoa contain homologous proteins to AtEH/Pan1, these resemble more closely Ede1 than Pan1 (Gadeyne et al., 2014; Wang et al., 2019).
Truncation of the TML subunit of TPC forces the complex into the cytoplasm, and this correlates with the dissociation of AtEH/Pan1 proteins from the complex (Gadeyne et al., 2014). It therefore appears that TPC composition depends on its localization and that the two AtEH/Pan1 subunits might be peripherally associated with a hexameric subcomplex of TPC, which would resemble TSET in D. discoideum. To investigate whether there is a differential order of recruitment between both AtEH/Pan1 proteins and the remaining hexameric subcomplex, we performed dual-color time-lapse microscopy of CME in etiolated Arabidopsis epidermal hypocotyl cells. We also took advantage of lowering the temperature of our samples to slow down CME and thereby increase the temporal resolution. Altogether, our data strongly suggest that TPC is recruited as the octameric complex to the PM, where it functions as the early adaptor complex for plant CME.
RESULTS
Lowering the Experimental Temperature Reduces CME Kinetics
TPC is proposed to serve as an early adaptor complex (Gadeyne et al., 2014); however, temporal resolution remains the biggest challenge to monitor the dynamic recruitment of different endocytic protein players (Fujimoto et al., 2010; Fan et al., 2013; Gadeyne et al., 2014; Bashline et al., 2015). As lowering the temperature generally slows down dynamics of cellular processes (Das et al., 1966), we hypothesized that by lowering the temperature we could also slow down dynamics of endocytic events and therefore enhance the temporal visualization of the differential recruitment of the endocytic players at the PM.
To time how intracellular dynamics respond to decreasing the temperature in plant cells, we first imaged Arabidopsis plants expressing the microtubule-binding protein EB1a-GFP (Van Damme et al., 2004). Hypocotyl cells of Arabidopsis seedlings expressing EB1a-GFP were imaged at 20°C for 5 min, then the temperature was lowered to 10°C with the aid of the CherryTemp Heater Cooler system and the effect on microtubule polymerization rate was assessed. Lowering the temperature had an immediate effect on the microtubule polymerization speed, measured as EB1a-labeled tracks in time-projection images (Supplemental Fig. S1A). Visualization and quantification of the microtubule growth dynamics further confirmed that the temperature shift was transmitted to the seedling almost instantaneously (Supplemental Fig. S1).
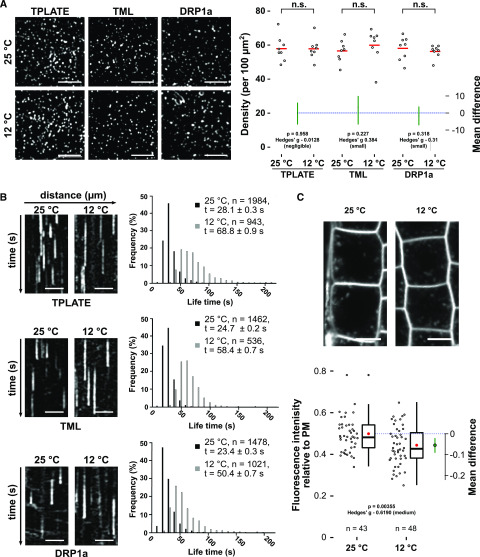
To examine the capacity of lowering the temperature to slow down endocytosis, we used etiolated hypocotyl cells expressing the endocytic markers CLC2-mKO and TPLATE-GFP and shifted the temperature during image acquisition; no obvious defect in the recruitment was observed. Density analysis of early-arriving TPLATE and TML as well as late-arriving DRP1A furthermore showed that lowering the temperature did not visually affect the PM recruitment of these endocytic markers, while it prolonged their lifetime at the PM instantly (Figs. 1 and 2A).
Figure 1.
Endocytic dynamics alter immediately upon lowering the temperature. Spinning-disk images and representative kymographs show endocytic foci and lifetimes of CLC2 (left) or TPLATE (right) in Arabidopsis etiolated hypocotyl epidermal cells at different temperatures. Cells were imaged at 25°C for 2 min and then imaged at 12°C for an additional 3 min. Images were acquired with a 1-s interval. White arrows on the kymographs indicate the position of the temperature shift. Total time = 300 s. Bars = 5 µm for spinning-disk images and 30 µm for kymographs.
Figure 2.
Lowering the temperature slows down CME. A, Spinning-disk images and density quantification of TPLATE-, TML-, and DRP1a-marked foci in epidermal hypocotyl cells at the permissive (25°C) and restrictive (12°C) temperatures. The density of the foci is independent of the tested temperatures (n.s., not significant, Mann-Whitney U test). The mean difference with the bootstrap 95% confidence interval (green circles and green lines) is shown as a part of the plot. Hedges’ g value is a standardized effect size indicating how much two groups differ from each other. For each transgenic line, eight cells from eight seedlings were imaged at both temperatures. Bars = 5 µm. B, Representative kymographs and histograms showing the lifetime distribution of TPLATE-, TML-, and DRP1a-positive foci at both temperatures. Number of events (n) and mean lifetime ± se (t) are indicated. Total time = 120 s. Bars = 25 µm. C, Representative confocal images and Jitter and box-plot quantification showing internalization of the FM4-64 dye (4 μm, 30 min) in Columbia-0 (Col-0) root epidermal cells at both temperatures. Red circles represent the mean. The P value was calculated by the Welch two-sample t test. The mean difference with the bootstrap 95% confidence interval (green circle and green line) is shown on the right side of the plot. Hedges’ g value is a standardized effect size. n represents the number of measurements (n = 43 cells for 25°C and n = 48 cells for 12°C) from 11 individual roots. Bars = 10 µm.
To further evaluate how lowering the temperature affects the dynamic behavior of endocytic proteins, we carefully measured the dynamics of TPLATE, TML, and DRP1a at different temperatures. Kymograph and histogram analyses of measured lifetimes confirmed that lowering the temperature from 25°C to 10°C correlated with a gradual increase in the lifetime of endocytic proteins at the PM (Supplemental Fig. S2). For TPLATE and TML, we observed a strong increase in average lifetime between 15°C and 12°C. Therefore, we selected 25°C and 12°C as the two temperatures for our future experiments; the fold change in the mean lifetime for each studied protein between these two temperatures was around 2.2.
Visualizing the individual lifetimes of the three markers at both temperatures in a histogram showed a clear shift of the distribution to longer lifetimes (Fig. 2B), and this correlated with a significant reduction in the internalization of the styryl dye FM4-64, which is used as a proxy for endocytic flux (Fig. 2C; Van Damme et al., 2011; Bashline et al., 2013; Fan et al., 2013; Gadeyne et al., 2014; Dejonghe et al., 2016, 2019). Furthermore, photobleaching experiments showed a dramatic reduction in the recovery of endocytic foci at the PM at the low temperature, in agreement with reduced dynamics of the process (Supplemental Fig. S3). These results together show that lowering the temperature in etiolated seedlings using our experimental setup slows down CME efficiently and rapidly.
Lowering the Temperature Enhances the Temporal Resolution of PM Recruitment
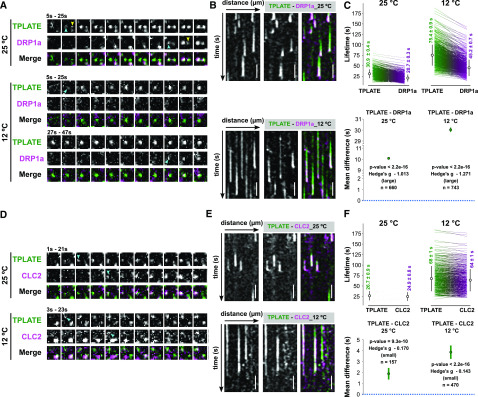
Having established the effect of lowering the temperature on endocytic dynamics in Arabidopsis cells, we then examined whether we would be able to enhance the temporal difference between an early player (TPLATE) and a late one (DRP1A). We therefore generated a dual marker line of TPLATE-GFP (Van Damme et al., 2006) and DRP1a-mRFP (Mravec et al., 2011) and compared the temporal behavior of each protein at both temperatures. When imaged at 25°C, the time-lapse images and kymographs showed multiple independent events where TPLATE-GFP foci clearly appeared earlier than DRP1a-mRFP at the PM, while they disappeared together (Fig. 3, A and B; Supplemental Fig. S4). Lowering the temperature prolonged the lifetime of both TPLATE-GFP and DRP1a-mRFP (the fold change in the mean lifetime was around 2.2; Fig. 3, A and B), whereas their departure remained synchronized. Overall, the temporal difference between their PM recruitment was dramatically enhanced (Fig. 3, A and B; Supplemental Fig. S4), which resulted in a very significant mean difference of the paired lifetimes (Hedges’ g value of −1.271; Fig. 3C).
Figure 3.
Lowering the temperature enhances the temporal resolution of plasma membrane recruitment. A and D, Representative time series of dual-color spinning-disk movies (2 s per frame) showing the sequential recruitment between TPLATE and DRP1a or CLC2 at the different temperatures. Arrowheads mark the appearance of TPLATE or DRP1a and CLC2 at the PM. B and E, Representative kymographs displaying the differential recruitment between TPLATE and DRP1a or CLC2 at different temperatures. Total time = 120s. Bars = 25 µm. C and F, Paired comparison of the lifetimes of particular protein pairs at the different temperatures. On the sides of each plot for different combinations of constructs and temperatures, each line represents an individual pair with the mean (white circle) ± sd (black line), and the number represents the mean lifetime ± se. The P value was calculated using the Wilcoxon signed rank test. The green circles in the bottom graphs represent the paired mean difference with the bootstrap 95% confidence interval (green line). Hedges’ g value is a standardized effect size indicating how much two groups differ from each other. n represents the number of events.
We performed a similar experiment comparing TPLATE and CLC2, which were previously shown only to have minor differences in their PM recruitment (Gadeyne et al., 2014; Narasimhan et al., 2020). Here, we also observed a temperature-enhancing effect of their differential recruitment, with TPLATE being recruited before CLC2, yet less pronounced than the difference between TPLATE and DRP1a (Fig. 3, D–F; Supplemental Fig. S4).
Taken together, these results show that lowering the temperature slows down dynamics of endocytosis as well as enhances the temporal resolution of recruitment of the endocytic proteins at the PM.
TPLATE Is Closely Associated with the AtEH/Pan1 Proteins at the PM
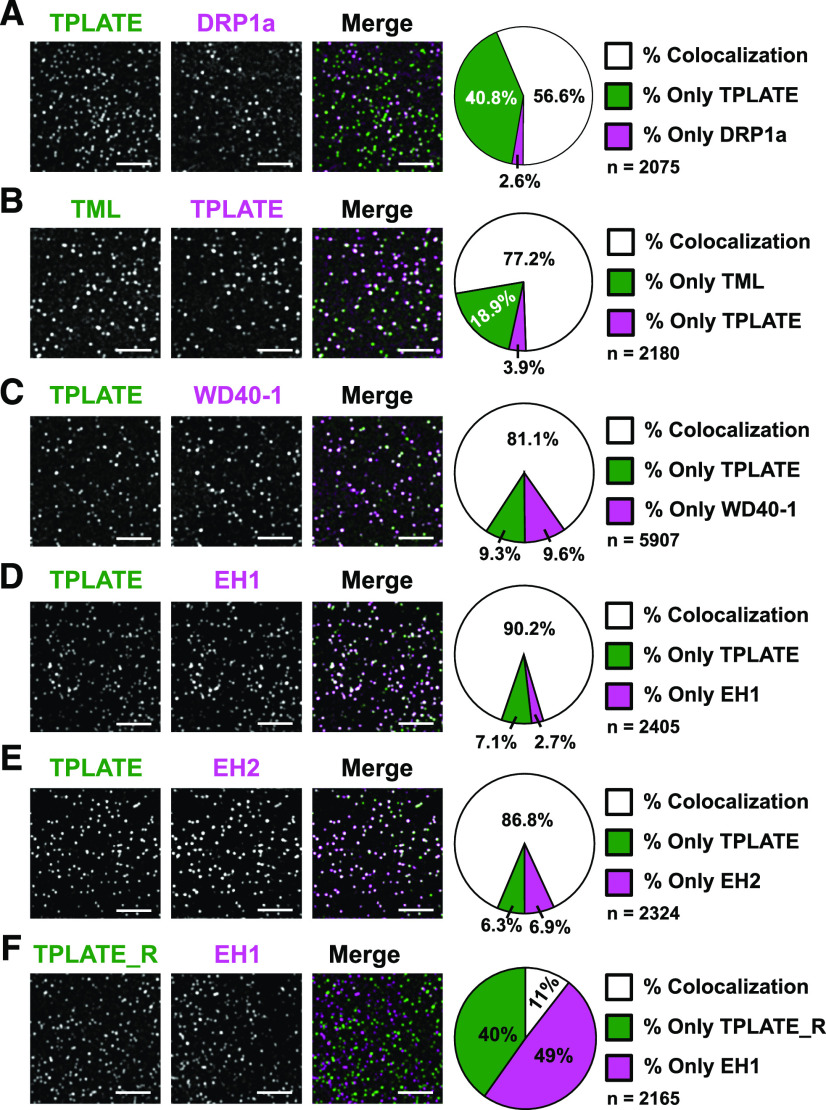
The absence of AtEH1/Pan1 and AtEH2/Pan1 as subunits of TSET (Hirst et al., 2014), together with the observation that inducing mislocalization of TPC to the cytoplasm leads to loss of those two subunits from the complex (Gadeyne et al., 2014) and the observation that AtEH/Pan1 proteins have a specific role in promoting autophagy (Wang et al., 2019), suggest that TPC could be in essence a hexameric complex, which temporarily gains two additional subunits. To reveal the temporal relationship among the TPC subunits, we visualized their dynamic behavior at the PM using multiple dual-color-labeled lines. To evaluate the functional association of TPC subunits at the PM, we crossed complemented mutant lines of core subunits (TPLATE, TML, and TWD40-1) and generated double complemented homozygous mutant lines. We also combined complemented mutants of the core subunit TPLATE with AtEH1/Pan1 or AtEH2/Pan1 and generated the respective double complemented, double homozygous mutant lines. As a first indication for differential recruitment, we monitored the steady-state percentage of colocalization between the markers using an average projection of five consecutive frames. Comparing an early endocytosis marker such as TPLATE with a late marker such as DRP1A revealed that the differential arrival between those markers leads to a substantial noncolocalizing fraction (roughly 40%; Fig. 4A).
Figure 4.
Colocalization analysis hints at a tight association between core and peripheral TPC subunits at the PM. Representative spinning-disk dual-color images and corresponding pie charts displaying the percentage of colocalization of dual-labeled endocytic foci are shown. TPLATE was compared with a late endocytic marker, DRP1a (A), with both TML (B) and TWD40-1 (C), and with both AtEHs/Pan1 (D and E). A quality check for the used algorithm was performed by comparing rotated TPLATE (90° rotation; TPLATE_R) with AtEH1/Pan1 to control for random association of foci (F). Z-projections of five consecutive frames with average intensity were used for quantification. Quantification (percent) of colocalization was calculated using the DiAna algorithm. The high percentage of overlap between TPLATE and the AtEH/Pan1 proteins suggests a tight connection between those proteins at the plasma membrane. Bars = 5 µm.
Combining TPLATE-TagRFP with TML-GFP resulted in around 77% of total foci where TML and TPLATE overlapped (Fig. 4B). We also tested functional association when combining TPLATE-GFP with a TWD40-1-mRuby3 complemented line (Table 1; Supplemental Fig. S5). Similarly as for TML and TPLATE, combining TPLATE-GFP and TWD40-1-mRuby3 resulted in a very high percentage of colocalizing foci (Fig. 4C), confirming their intrinsic behavior as part of the same complex.
Table 1. twd40-1-1 plants expressing C-terminal fusions of TWD40-1 with mRuby3 allow transfer of the T-DNA via the male.
The results of the analysis of backcross experiments between Col-0 as female (♀) and the twd40-1-1 heterozygous plants with or without expression of TWD40-1-mRuby3 as male (♂) are shown. The analysis clearly shows that the mutation blocks transfer via the male and that this block is lifted by the presence of the fluorescent fusion construct, indicating that it is functional.
| Backcross to Col-0 (♀) | No. of Plants | T-DNA Transfer via ♂ |
|---|---|---|
| twd40-1-1 (+/−) | 12 | 0 |
| twd40-1-1 (−/−) + TWD40-1-mRuby3 ♂ | 12 | 12 |
In Arabidopsis seedlings, AtEH1/Pan1 and AtEH2/Pan1 have so far been shown to be functionally associated with TPLATE at autophagosomes and to be delivered to the vacuole after carbon starvation and Concanamycin A treatment (Wang et al., 2019). To address the recruitment of AtEH/Pan1 and TPLATE in endocytic foci at the PM, we combined AtEH/Pan1 with TPLATE in their respective complemented mutant backgrounds. Similar to the combination of the core subunits, both AtEH1/Pan1-mRuby3 and AtEH2/Pan1-mRuby3 showed very high degrees of colocalization with TPLATE-GFP at the PM (Fig. 4, D and E). To exclude the possibility that TPLATE and AtEH/Pan1 foci overlapped due to the high density of endocytic foci, we performed a quality check of the algorithm used by comparing the colocalization percentage between TPLATE and AtEH1/Pan1 on images where the TPLATE channel was rotated 90°. This rotation resulted in only around 10% of foci colocalizing (Fig. 4F), confirming that the observed high degree of colocalization is not random. These results also suggest a tight association between TPLATE and the AtEH/Pan1 proteins at the PM.
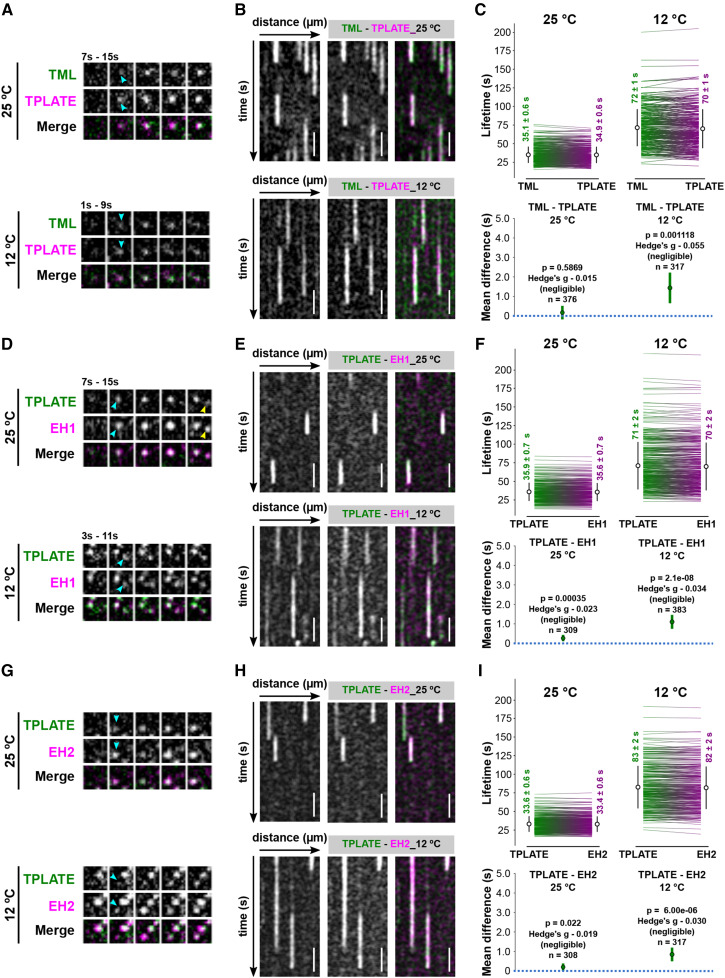
AtEHs/Pan1 and the Core TPC Subunits Are Simultaneously Recruited to the PM
To further investigate TPC assembly at the PM at the level of the different subunits, we compared the recruitment and disassociation behavior among pairs of TPC subunits at 25°C and 12°C. As a benchmark, we compared the behavior of two closely related TPC subunits, TPLATE and TML, which by homology to other adaptor complexes are presumed to be part of the core of TPC.
Time-lapse imaging and kymograph analyses showed that dual-labeled TML-GFP and TPLATE-TagRFP foci appear and disappear simultaneously at the PM when imaged at 25°C (Fig. 5, A and B; Supplemental Fig. S6). Lowering the temperature maintained this simultaneous appearing and disappearing behavior. However, there were very small changes in average lifetime, which were found to be negligible by their effect size (expressed as the Hedges’ g value). These small changes could be explained by there being more signal noise and differential bleaching effects of the fluorophores during the image acquisition at the lower temperature conditions (Fig. 5, A–C; Supplemental Fig. S6).
Figure 5.
The AtEHs/Pan1 and the core TPC subunits are simultaneously recruited to the PM. A, D, and G, Representative time series of dual-color spinning-disk movies (2 s per frame) showing the recruitment between TPLATE and TML, EH1/Pan1, and EH2/Pan1 at different temperatures. Arrowheads mark the appearance of TPLATE or TML, EH1/Pan1, and EH2/Pan1 on the PM. B, E, and H, Representative kymographs displaying the recruitment of TPLATE versus TML, EH1/Pan1, and EH2/Pan1 at different temperatures. Total time = 120s. Bars = 25 µm. C, F, and I, Paired comparison of the lifetimes of particular protein pairs at different temperatures. Each line on the sides of each plot represents an individual pair with the mean (white circle) ± sd (black line) for different combinations of constructs and temperatures. Each number on the sides of each plot represents the mean lifetime ± se. The P value was calculated by the Wilcoxon signed rank test. The green circles in the bottom graphs represent the paired mean difference with the bootstrap 95% confidence interval (green line). Hedges’ g value is a standardized effect size indicating how much two groups differ from each other. n represents the number of events.
We next compared the PM appearance and disappearance behavior of AtEH/Pan1 and TPLATE. Similar to the recruitment behavior of TPLATE and TML foci, when imaged at 25°C, both AtEH1/Pan1 and AtEH2/Pan1 foci simultaneously appeared and disappeared with TPLATE at the PM (Fig. 5, D, E, G, and H; Supplemental Fig. S7). Lowering the temperature did not alter the simultaneous recruitment of TPLATE and AtEH/Pan1, and the respective Hedges’ value remained very low (Fig. 5, D–I–I; Supplemental Fig. S7). These results, together with our colocalization analysis, strongly suggest that AtEH/Pan1 subunits are simultaneously recruited to, and maintained, at the PM with the other TPC subunits.
DISCUSSION
Clathrin-mediated endocytosis is a highly dynamic process. It is best understood in mammalian and yeast model systems, while little is known in plants. Although very fast endocytic events were observed in neurons (Balaji and Ryan, 2007), in yeast or cultured animal cells the whole endocytic process takes up to minutes, and the physiological role and precise temporal dynamics of endocytic proteins are well defined (Taylor et al., 2011; Lu et al., 2016; Ma et al., 2016; Picco and Kaksonen, 2018; Pascolutti et al., 2019; Wrobel et al., 2019). In plant cells, the endocytic process is much more dynamic, with larger clathrin cages being formed than those in yeast in shorter time periods (Narasimhan et al., 2020), which brings about more technical difficulties in monitoring the temporal recruitment of the diverse endocytic players (Zhang et al., 2015). To better understand the dynamics of endocytosis in plant cells, new approaches are required, which should provide us with enhanced temporal resolution.
Here, we report that lowering the temperature using a microfluidics-based on-slide approach can be used as a rapid and efficient tool to slow down intracellular dynamic processes in plant cells similar to earlier reports demonstrating temperature effects on endocytosis and synaptic vesicle recycling in animal cells (Pyott and Rosenmund, 2002; Bui and Glavinović, 2014). The effect on microtubule growth rate was immediate, and the dampening effect on the dynamics of endocytosis, as measured by the dwell time at the PM of several markers, FM uptake, as well as measuring recruitment by fluorescence recovery after photobleaching, correlated with the reduction in temperature. Lowering the temperature, however, had no significant effect on the density of endocytic foci, suggesting that cargo proteins could still be adequately recognized.
Our data also show that the temporal resolution of the differential recruitment of endocytic players could be enhanced at lower temperatures, which was most apparent between early- and late-arriving endocytic proteins. Lowering the temperature also enabled us to slightly enhance the temporal difference between two early-arriving proteins, TPLATE and CLC2, indicating that our approach worked. Given the temperature effect on the dynamics of plant processes, awareness should be increased to perform live-cell imaging in plants under accurate temperature control. No doubt this will substantially increase the robustness and reproducibility of future image data collection.
TPC is an evolutionarily ancient adaptor complex that was not retained in yeast or animal cells (Hirst et al., 2014). It is proposed to function as an adaptor complex during CME and has been shown to be recruited to the PM earlier than dynamin-related proteins while only slightly earlier than clathrin (Gadeyne et al., 2014; Narasimhan et al., 2020). D. discoideum contains a similar complex, which however lacks the AtEH/Pan1 proteins, and in contrast to TPC, TSET is not essential (Gadeyne et al., 2014; Hirst et al., 2014). These AtEH/Pan1 proteins both share their male sterility phenotype with the other TPC subunits (Van Damme et al., 2006; Gadeyne et al., 2014; Wang et al., 2019), they are dynamically recruited to the PM (Gadeyne et al., 2014; Wang et al., 2019), associated with the other TPC subunits as well as AP-2, but not when TPC is forced into the cytoplasm using proteomic manipulations (Gadeyne et al., 2014), and are functionally associated with several endocytic players during autophagy (Wang et al., 2019). Here, we investigated how these AtEH/Pan1 proteins are recruited to the PM in relation to the other TPC subunits. Our live-cell analysis could not identify any deviation in recruitment dynamics between AtEH/Pan1 and TPLATE, mirroring the results observed between the other core subunits TPLATE and TML. The small differences reported could be attributed to technical issues, including the differential bleaching effects of fluorophores. The close association, as well as the simultaneous recruitment between TPLATE- and AtEH/Pan1-labeled endocytic foci at the PM, therefore allow us to hypothesize that TPC is recruited to the PM as an octameric complex in plants. When combining TPLATE-TagRFP with TML-GFP, we observed an unexpectedly high percentage of TML recruitment to the PM without TPLATE (Fig. 4B). As this occurs in a double mutant, double complemented background, we interpret this recruitment as independent of TPLATE and, by extension, independent of the complex. Under this hypothesis, and in agreement with the cytoplasmic localization of the complex containing a truncated TML (Gadeyne et al., 2014), this subunit likely plays a role in PM recruitment of TPC.
Altering the temperature will likely affect many plant processes, such as cytoskeleton dynamics, phosphoinositide production/conversion, or membrane fluidity. However, our conclusions on the simultaneous recruitment of the various TPC subunits are independent of these temperature effects. Under normal conditions, there is a difference in recruitment between TPLATE and DRP1a or clathrin (Gadeyne et al., 2014; Narasimhan et al., 2020). Here, we describe that this difference increases upon lowering the temperature. We did not observe any significant differences between TPLATE and AtEH/Pan1 proteins at either temperature, although this all happens in the same context of altered cytoskeleton, lipid conversion, or membrane fluidity. The lack of statistical difference in the density of endocytic foci at 12°C or 25°C indicates that effects on membrane fluidity, phosphoinositide levels, and saturation degree of lipid acyl chains in our setup and are not majorly affected within the time window that we use.
In plants, lifetimes of endocytic proteins are measured using time-lapse imaging of cells expressing fluorescence-labeled proteins imaged via spinning-disk microscopy (Bashline et al., 2013, 2015; Gadeyne et al., 2014; Dejonghe et al., 2016, 2019; Zhou et al., 2018) or total internal reflection fluorescence microscopy or variable angle epifluoresence microscopy (Konopka et al., 2008; Konopka and Bednarek, 2008a, 2008b; Ito et al., 2012; Johnson and Vert, 2017; Narasimhan et al., 2020). Based on earlier work in the animal field (Jaqaman et al., 2008; Aguet et al., 2013; Compeer et al., 2018; Pascolutti et al., 2019; Wrobel et al., 2019), automated image-analysis tools have recently been employed to analyze CME images unbiasedly and rapidly (Johnson and Vert, 2017; Narasimhan et al., 2020). Accurate detection of endocytic events requires high signal-to-noise ratio (SNR) images, a quality that is determined by multiple factors such as the susceptibility of the fluorophore to bleaching and the expression level of the fluorescence reporter, and close-to-endogenous levels of fluorescence-labeled endocytic proteins often failed to provide sufficient SNR for accurate lifetime measurements (Aguet et al., 2013). Here, we aimed at visualizing the recruitment dynamics of various TPC subunits at the PM. In order to avoid competition with endogenous subunits and to avoid overexpression effects such as induction of autophagy (Wang et al., 2019), we opted to work with double-labeled endocytic proteins in either the single or double complemented tpc mutant background rather than use overexpression lines. Although our approach represents the optimal strategy from a biological perspective, our transgenic lines, especially those fused with red fluorescent proteins, combined with extended imaging times under low-temperature conditions failed to generate sufficient SNR images for automatic quantification. Actually, the only combination that yielded acceptable data when automatically quantified was our TPLATE-DRP1A combination, as the DRP1A is actually 35S driven.
We therefore generated kymographs of our dual-color endocytic spinning-disk movies and measured the dwell times of the proteins manually. We aimed at measuring several hundred events per experiment and combined measurements of independent persons to achieve an unbiased assessment of the data. We were unable to identify any recruitment of AtEH/Pan1 proteins at the PM that was independent of another TPC subunit. Our findings therefore show that the TPLATE adaptor protein complex is likely recruited to the PM as an octameric unit, both at normal and lowered temperature conditions. We cannot, however, exclude the possibility that the differential recruitment between the individual TPC subunits is too dynamic to monitor under our limiting, 1-s temporal resolution, conditions. However, due to the fact that we were able to detect differences in the CLC2 and TPLATE dynamics, which are of the order of a few seconds, we hypothesize that TPC is recruited as an octameric unit at the PM during CME. The continuous development of novel microscopy as well as labeling techniques will help to overcome our current limitations of working with low-expressing functional fusions and will allow us to understand how endocytosis in plants is executed at high spatiotemporal resolution.
MATERIALS AND METHODS
Molecular Cloning
To yield the expression construct for TWD40-1, entry clones of TWD40-1 without a stop codon in pDONR221 (Gadeyne et al., 2014) were combined with pB7m34GW (Karimi et al., 2007), pDONRP4-P1R-Histone3p (Ingouff et al., 2017), and pDONRP2-P3R-mRuby3 (Wang et al., 2019) in a triple gateway LR reaction (Invitrogen) to generate Histone3p::TWD40-1-mRuby3. To yield a red fluorophore-tagged TPLATE, TPLATE without stop in pDONR207 (Van Damme et al., 2006) was combined with the Lat52 promotor in pDONRP4P1R (Van Damme et al., 2006), TagRFP in pDONRP2P3R (Mylle et al., 2013), and pH7m34GW (Karimi et al., 2007) in a triple gateway LR reaction (Invitrogen) to generate Lat52p::TPLATE-TagRFP.
Plant Material
The Arabidopsis (Arabidopsis thaliana) lines expressing EB1a-GFP (Van Damme et al., 2004), Lat52p::TPLATE-GFP (Van Damme et al., 2006; Gadeyne et al., 2014), TMLp::TML-GFP (Gadeyne et al., 2014), 35Sp::DRP1a-mRFP (Mravec et al., 2011), and 35Sp::CLC2(At2g40060)-mKO (Ito et al., 2012) were previously described. The dual-color line expressing TPLATE-GFP/CLC2-TagRFP in the tplate homozygous background was reported previously (Gadeyne et al., 2014). The dual-marker lines expressing TPLATE-GFP/AtEH1/Pan1-mRuby3 or TPLATE-GFP/AtEH2/Pan1-mRuby3 in a double homozygous mutant (tplate ateh1/pan1 or tplate ateh2/pan1) background were reported before (Wang et al., 2019).
To generate TWD40-1-mRuby3 complemented lines, heterozygous mutants of twd40-1-1 (Gadeyne et al., 2014) were transformed with Histone3p::TWD40-1-mRuby3 by floral dip. The T1 plants were selected for the complementation constructs on one-half-strength Murashige and Skoog (MS) plates (Duchefa) supplemented with 10 mg L−1 Basta (Labconsult). Resistant plants were genotyped to identify those with a twd40-1-1 heterozygous mutant background. T2 plants expressing TWD40-1-mRuby3 were tested by genotyping PCR to identify homozygous lines for the twd40-1-1 insertion mutations. Genotyping PCR was performed on genomic DNA isolated from rosette leaves. Genotyping primers for twd40-1-1 were described before (Gadeyne et al., 2014).
For backcross experiments, the complemented lines of TWD40-1-mRuby3 as well as the heterozygous mutant plants of twd40-1-1 were used as male to cross with Col-0 as female. The transfer of the T-DNA, causal to the functionality of the complementing fusion, was analyzed by genotyping PCR on F1 plants.
To generate dual-marker lines of TPLATE core subunits, a complemented tplate line expressing Lat52p::TPLATE-tagRFP was generated by dipping tplate(+/−) heterozygous plants with Lat52p::TPLATE-TagRFP and selecting for the transformants. Homozygous tplate(−/−) plants were identified in the next generations by genotyping PCR.
Homozygous tplate mutant plants carrying Lat52p::TPLATE-TagRFP were crossed with the complemented tml-1 line expressing TMLp::TML-GFP (Gadeyne et al., 2014). The complemented twd40-1-1 lines expressing TWD40-1-mRuby3 were crossed with the complemented tplate line expressing Lat52p::TPLATE-GFP. F2 plants in a double homozygous mutant background (tml-1/tplate or tplate/twd40-1-1) were identified by genotyping PCR. For the dual-color TPLATE-GFP/DRP1a-mRFP lines, the complemented tplate line expressing Lat52p::TPLATE-GFP was crossed with a 35Sp::DRP1a-mRFP-expressing line, and F2 plants in a tplate homozygous background were identified by genotyping PCR. Genotyping primers for tplate, tml-1, and twd40-1-1 were described before (Gadeyne et al., 2014).
Temperature Modulation Using the CherryTemp System
The CherryTemp system (Cherrybiotech), which enables ultra-fast temperature shifts between 5°C and 45°C, was used to modulate and maintain the temperature during the spinning-disk imaging (https://www.cherrybiotech.com/heater-cooler-for-microscopy). Prior to imaging, a single etiolated seedling was positioned between two cover slips with one-half-strength MS liquid medium and incubated with the CherryTemp Heater Cooler device for 5 min prior to imaging to stabilize the temperature of the seedling (https://www.cherrybiotech.com/heater-cooler-for-microscopy/temperature-control-for-plant-microscopy).
FM4-64 Uptake
Prior to imaging, whole 6-d-old Col-0 seedlings were incubated with 4 μm FM4-64 (Invitrogen) solution in one-half-strength MS liquid medium between two cover slips at 25°C or 12°C for 30 min.
Live-Cell Imaging
A Nikon Ti microscope with the Perfect Focus System (PFSIII) for Z-drift compensation, equipped with an Ultraview spinning-disk system (Perkin-Elmer) and two 512 × 512 Hamamatsu ImagEM C9100-13 EMccd cameras, was used to image endocytic dynamics. Images of hypocotyl epidermal cells of 4-d-old etiolated seedlings expressing single- or dual-color fluorophore-labeled proteins were acquired with a 100× oil-immersion objective (Plan Apo; numerical aperture = 1.45). During imaging, the CherryTemp system was used to maintain the temperature of the samples constant.
Seedlings expressing GFP-fused proteins were imaged with 488-nm excitation light and an emission window between 500 and 530 nm in single-camera mode or 500 and 550 nm in dual-camera mode. Seedlings expressing mKO-, mRFP-, and tagRFP-labeled proteins were imaged with 561-nm excitation light and an emission window between 570 and 625 nm in single-camera mode or 580 and 630 nm in dual-camera mode. Single-marker line movies were acquired with an exposure time of 500 ms per frame. Movies of at least 2 min in total were made. Dual-color lines were acquired either sequentially (one-camera mode) or simultaneously (two-camera mode) with an exposure time of 500 ms per frame. Single-camera mode was used for density, colocalization (TML-TPLATE and TPLATE-DRP1A), and lifetime (TML-DRP1A and TML-TPLATE 25°C) measurements. Dual-camera mode was used for colocalization (TPLATE-AtEH1/Pan1, TPLATE-AtEH2/Pan1, TPLATE-TWD40-1, and TPLATE-TWD40-2) and lifetime (TPLATE-AtEH1/Pan1, TPLATE-AtEH2/Pan1, and TML-TPLATE 12°C) measurements. For photobleaching experiments, seedlings were exposed to 100% laser power for around 2 s during the imaging, using the Photokinesis unit of the Perkin-Elmer spinning-disk system.
Root meristematic epidermal cells of 6-d-old seedlings were acquired with a Zeiss 710 inverted confocal microscope with the ZEN 2009 software package and equipped with a C-Apochromat 40× water Korr M27 objective (numerical aperture = 1.2). FM4-64 was visualized using 561-nm laser excitation and a 650- to 750-nm detection window.
Quantification of Measurements
Dynamics of EB1a-GFP were analyzed using ImageJ equipped with the Trackmate (v4.0.1) plugin (Tinevez et al., 2017). We used the LoG detector with an estimated blob diameter of 10 pixels and a threshold value of 1, using the median filter and subpixel localization. An auto initial thresholding, followed by a linear motion LAP tracker with as parameters a five-pixel initial search radius, a five-pixel search radius, and a two-frame maximum frame gap correction, were applied. Tracks with a duration of less than 10 frames were excluded, and the obtained median speed per track was converted to µm min−1 using the pixel size values. Outliers for the median speed were defined by the iterative median absolute deviation method (Leys et al., 2013), and their values were excluded. Three movies coming from two different seedlings were analyzed.
The density of labeled endocytic markers was measured in Matlab 2017b using the detection and tracking parts of the cmeAnalysis package and further processed as described (Johnson and Vert, 2017; Narasimhan et al., 2020). Density calculations were obtained from all the tracks within the region of interest over certain frames of the movies, which were used to produce an average density. The region of interest was selected on the middle of the image. The middle frame was used as a reference, and the temporal range is based on the middle frame. We used the pixel size and the area to convert this to spots per µm2. For each analyzed sample set, eight cells from eight different seedlings were analyzed.
Object-based colocalization was performed using the ImageJ plugin DiAna (Gilles et al., 2017). Prior to analyzing with DiAna, images were processed with ImageJ. Each channel was processed with a walking average of four and then merged (also rotated if required). Regions of interest were selected based on whether they excluded the border of the cells and still represented a good number of objects. Z-projection images were generated from five slices with average intensity. Each channel of Z-projected images was processed using Morphological filters from the MorphoLibJ plugin (Legland et al., 2016), using the parameters white top-hat, disk element, and a two-pixel radius. A first step in the DiAna plugin is to segment the objects for each channel, which is done by selecting the 3D Spot segmentation tool of the DiAna-Segment plugin. If requested, adapt the calibration by changing the pixel size to 1.00001 for all dimensions. Both the noise and seed threshold value were obtained by averaging the maximum intensity of three regions covering only background signal. The spot was defined with a minimum value of four and a maximum value of 36 pixels. The option to exclude objects on xy edges was activated. Default values were used for the other parameters. Results for number of total objects (Tot) or touching objects (Tou) in image A/B obtained from DiAna were recorded. The colocalization ratio of objects was calculated as follows:
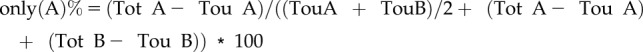 |
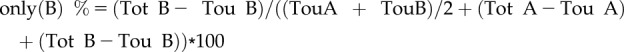 |
As a control, one of the channels was rotated 90° (no interpolation) and analyzed similarly as described above. For each analyzed sample set, a minimum of six cells coming from three different seedlings were analyzed.
Lifetimes of individual endocytic events were measured from kymographs generated by the Volocity software package (Perkin-Elmer). To measure paired lifetimes of dual-color kymographs, individual events showing good SNR in both channels were marked. Following the measurement of the lifetimes of the marker in the red channel, the lifetime of the marker in the green channel was analyzed. Data were further analyzed in Excel (Microsoft Office) by checking the start position of X from each line of the kymograph to avoid mistakes in pairing the red and green channel values. Calculation to time (ns) of each line was done. Outliers for the lifetime differences were defined by the iterative median absolute deviation method (Leys et al., 2013), and their values were excluded. For each of the analyzed sample sets, a minimum of nine movies spread among a minimum of three seedlings were analyzed, except for TPLATE-GFP/CLC2-tagRFP, for which seven movies over three seedlings (12°C) and five movies over two seedlings (25°C) were analyzed. To generate unbiased data, paired lifetimes of endocytic events labeled by dual-color fluorophores were measured by independent persons.
Statistical Analysis
The results were analyzed with the estimation method to calculate mean, mean differences, confidence intervals, and Hedges’ g (Claridge-Chang and Assam, 2016; Ho et al., 2019). The 95% confidence intervals for the mean differences were calculated using bootstrap methods (resampled 5,000 times, bias corrected, and accelerated). Effect size was measured using Hedges’ g and according to the standard practice is referred to as negligible (g < 0.2), small (0.2 < g < 0.5), medium (0.5 < g < 0.8), or large (g > 0.8; Cumming, 2012). Hedges’ g is a quantitative measurement for the difference between means indicating how much two groups differ from each other: if Hedges’ g = 1, the two groups differ by 1 sd. R version 3.6.2 and Rstudio 1.2.5001 were used to calculate the Wilcoxon signed rank test (paired samples), the Mann-Whitney U test, and the Welch two-sample t test (R Core Team, 2019). All plots and figures were generated with Rstudio 1.2.5001 and Inkscape (version 0.92; https://inkscape.org/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The CherryTemp system can be effectively used to quickly alter intracellular dynamics in plant cells.
Supplemental Figure S2. Decreasing temperature increases the lifetime of endocytic proteins at the PM.
Supplemental Figure S3. Lowering the temperature decreases the recruitment dynamics of the endocytic proteins.
Supplemental Figure S4. Lowering the temperature enhances the temporal resolution of recruitment.
Supplemental Figure S5. TWD40-1 localizes to the PM as endocytic foci.
Supplemental Figure S6. TPLATE and TML are recruited to the PM simultaneously.
Supplemental Figure S7. The AtEHs/Pan1 and the core TPC subunits are simultaneously recruited to the PM.
Footnotes
This work was supported by the European Research Council (grant no. 682436), the China Scholarship Council (grant no. 201508440249 to J.W.), and by Ghent University BOF CSC co-funding (grant no. ST01511051 to J.W.). The acquisition of the CherryTemp Heater Cooler system (Cherrybiotech) was supported by the VIB Tech Watch Initiative and by the Research Foundation Flanders (grant no. 1511817N to D.V.D.). A.J. was supported by the Austrian Science Fund (grant no. I3630B25 to J.F.).
Articles can be viewed without a subscription.
References
- Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G(2013) Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell 26: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji J, Ryan TA(2007) Single-vesicle imaging reveals that synaptic vesicle exocytosis and endocytosis are coupled by a single stochastic mode. Proc Natl Acad Sci USA 104: 20576–20581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Li S, Anderson CT, Lei L, Gu Y(2013) The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol 163: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Li S, Zhu X, Gu Y(2015) The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc Natl Acad Sci USA 112: 12870–12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitsikas V, Corrêa IR Jr., Nichols BJ(2014) Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife 3: e03970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui L, Glavinović MI(2014) Temperature dependence of vesicular dynamics at excitatory synapses of rat hippocampus. Cogn Neurodyn 8: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Assam PN(2016) Estimation statistics should replace significance testing. Nat Methods 13: 108–109 [DOI] [PubMed] [Google Scholar]
- Compeer EB, Kraus F, Ecker M, Redpath G, Amiezer M, Rother N, Nicovich PR, Kapoor-Kaushik N, Deng Q, Samson GPB, et al. (2018) A mobile endocytic network connects clathrin-independent receptor endocytosis to recycling and promotes T cell activation. Nat Commun 9: 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming G.(2012) Understanding the New Statistics: Effect Sizes, Confidence Intervals, and Meta-Analysis. Routledge/Taylor & Francis, New York [Google Scholar]
- Das TM, Hildebrandt AC, Riker AJ(1966) Cine-photomicrography of low temperature effects on cytoplasmic streaming, nucleolar activity and mitosis in single tobacco cells in microculture. Am J Bot 53: 253–259 [PubMed] [Google Scholar]
- Dejonghe W, Kuenen S, Mylle E, Vasileva M, Keech O, Viotti C, Swerts J, Fendrych M, Ortiz-Morea FA, Mishev K, et al. (2016) Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat Commun 7: 11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejonghe W, Sharma I, Denoo B, De Munck S, Lu Q, Mishev K, Bulut H, Mylle E, De Rycke R, Vasileva M, et al. (2019) Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat Chem Biol 15: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J(2007) Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Kim SY, Xu ZY, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, et al. (2013) The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, Botella MA, Wang H, Lin J(2013) Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140: 3826–3837 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N(2010) Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc Natl Acad Sci USA 107: 6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadeyne A, Sánchez-Rodríguez C, Vanneste S, Di Rubbo S, Zauber H, Vanneste K, Van Leene J, De Winne N, Eeckhout D, Persiau G, et al. (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704 [DOI] [PubMed] [Google Scholar]
- Gilles JF, Dos Santos M, Boudier T, Bolte S, Heck N(2017) DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115: 55–64 [DOI] [PubMed] [Google Scholar]
- Hirst J, Schlacht A, Norcott JP, Traynor D, Bloomfield G, Antrobus R, Kay RR, Dacks JB, Robinson MS(2014) Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife 3: e02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A(2019) Moving beyond P values: Data analysis with estimation graphics. Nat Methods 16: 565–566 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Selles B, Michaud C, Vu TM, Berger F, Schorn AJ, Autran D, Van Durme M, Nowack MK, Martienssen RA, et al. (2017) Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev 31: 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E, Fujimoto M, Ebine K, Uemura T, Ueda T, Nakano A(2012) Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J 69: 204–216 [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G(2008) Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods 5: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Vert G(2017) Single event resolution of plant plasma membrane protein endocytosis by TIRF microscopy. Front Plant Sci 8: 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P(2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Xu ZY, Song K, Kim DH, Kang H, Reichardt I, Sohn EJ, Friml J, Juergens G, Hwang I(2013) Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25: 2970–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Löfke C, Teichmann T, Tanaka H, Friml J(2011) Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Backues SK, Bednarek SY(2008) Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY(2008a) Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol 147: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY(2008b) Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J 53: 186–196 [DOI] [PubMed] [Google Scholar]
- Legland D, Arganda-Carreras I, Andrey P(2016) MorphoLibJ: Integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32: 3532–3534 [DOI] [PubMed] [Google Scholar]
- Leys C, Ley C, Klein O, Bernard P, Licata L(2013) Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. J Exp Soc Psychol 49: 764–766 [Google Scholar]
- Lu R, Drubin DG, Sun Y(2016) Clathrin-mediated endocytosis in budding yeast at a glance. J Cell Sci 129: 1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Umasankar PK, Wrobel AG, Lymar A, McCoy AJ, Holkar SS, Jha A, Pradhan-Sundd T, Watkins SC, Owen DJ, et al. (2016) Transient Fcho1/2⋅Eps15/R⋅AP-2 nanoclusters prime the AP-2 clathrin adaptor for cargo binding. Dev Cell 37: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E(2011) Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 12: 517–533 [DOI] [PubMed] [Google Scholar]
- Mravec J, Petrášek J, Li N, Boeren S, Karlova R, Kitakura S, Pařezová M, Naramoto S, Nodzyński T, Dhonukshe P, et al. (2011) Cell plate restricted association of DRP1A and PIN proteins is required for cell polarity establishment in Arabidopsis. Curr Biol 21: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mylle E, Codreanu MC, Boruc J, Russinova E(2013) Emission spectra profiling of fluorescent proteins in living plant cells. Plant Methods 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan M, Johnson A, Prizak R, Kaufmann WA, Tan S, Casillas-Pérez B, Friml J(2020) Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife 9: e52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolutti R, Algisi V, Conte A, Raimondi A, Pasham M, Upadhyayula S, Gaudin R, Maritzen T, Barbieri E, Caldieri G, et al. (2019) Molecularly distinct clathrin-coated pits differentially impact EGFR fate and signaling. Cell Rep 27: 3049–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco A, Kaksonen M(2018) Quantitative imaging of clathrin-mediated endocytosis. Curr Opin Cell Biol 53: 105–110 [DOI] [PubMed] [Google Scholar]
- Pyott SJ, Rosenmund C(2002) The effects of temperature on vesicular supply and release in autaptic cultures of rat and mouse hippocampal neurons. J Physiol 539: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019) R: A language and environment for statistical computing. http://www.r-project.org/index.html
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. (2010) ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ(2011) A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol 9: e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW(2017) TrackMate: An open and extensible platform for single-particle tracking. Methods 115: 80–90 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Bouget FY, Van Poucke K, Inzé D, Geelen D(2004) Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J 40: 386–398 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Coutuer S, De Rycke R, Bouget FY, Inzé D, Geelen D(2006) Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell 18: 3502–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Gadeyne A, Vanstraelen M, Inzé D, Van Montagu MC, De Jaeger G, Russinova E, Geelen D(2011) Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc Natl Acad Sci USA 108: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K, Gong J, Guan Y, et al. (2019) Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun 10: 5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel AG, Kadlecova Z, Kamenicky J, Yang JC, Herrmann T, Kelly BT, McCoy AJ, Evans PR, Martin S, Muller S, et al. (2019) Temporal ordering in endocytic clathrin-coated vesicle formation via AP2 phosphorylation. Dev Cell 50: 494–508.e411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Shimono Y, Shirakawa M, Fukao Y, Kawase T, Hatsugai N, Tamura K, Shimada T, Hara-Nishimura I(2013) Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25: 2958–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Persson S, Hirst J, Robinson MS, van Damme D, Sánchez-Rodríguez C(2015) Change your TPLATE, change your fate: Plant CME and beyond. Trends Plant Sci 20: 41–48 [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu D, Wang P, Ma X, Lin W, Chen S, Mishev K, Lu D, Kumar R, Vanhoutte I, et al. (2018) Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc Natl Acad Sci USA 115: E1906–E1915 [DOI] [PMC free article] [PubMed] [Google Scholar]