The mutation of FBL17 leads to a strong upregulation of genes involved in DNA damage and repair processes, and FBL17 protein is recruited at nuclear DNA lesion foci upon double-strand break induction.
Abstract
In Arabidopsis (Arabidopsis thaliana), the F-box protein F-BOX-LIKE17 (FBL17) was previously identified as an important cell-cycle regulatory protein. FBL17 is required for cell division during pollen development and for normal cell-cycle progression and endoreplication during the diploid sporophyte phase. FBL17 was reported to control the stability of the CYCLIN-DEPENDENT KINASE inhibitor KIP-RELATED PROTEIN (KRP), which may underlie the drastic reduction in cell division activity in both shoot and root apical meristems observed in fbl17 loss-of-function mutants. However, whether FBL17 has other substrates and functions besides degrading KRPs remains poorly understood. Here we show that mutation of FBL17 leads not only to misregulation of cell cycle genes, but also to a strong upregulation of genes involved in DNA damage and repair processes. This phenotype is associated with a higher frequency of DNA lesions in fbl17 and increased cell death in the root meristem, even in the absence of genotoxic stress. Notably, the constitutive activation of DNA damage response genes is largely SOG1-independent in fbl17. In addition, through analyses of root elongation, accumulation of cell death, and occurrence of γH2AX foci, we found that fbl17 mutants are hypersensitive to DNA double-strand break-induced genotoxic stress. Notably, we observed that the FBL17 protein is recruited at nuclear foci upon double-strand break induction and colocalizes with γH2AX, but only in the presence of RETINOBLASTOMA RELATED1. Altogether, our results highlight a role for FBL17 in DNA damage response, likely by ubiquitylating proteins involved in DNA-damage signaling or repair.
The eukaryotic cell cycle is composed of four phases. In DNA synthesis (S) phase, DNA replication occurs, and in mitosis (M) phase, chromosomes segregate into two nuclei, followed by cytokinesis, allowing cells to be divided into two daughter cells (Nurse, 2000). These two phases are separated by two gap phases (G1 and G2) during which cells increase in size and in number of organelles, and are subjected to cell-cycle checkpoints. The proper orchestration of the cell cycle requires numerous levels of control. In particular, cyclin-dependent kinases (CDKs), activated by cyclins, are crucial players in this process and their activity is strictly regulated (Malumbres and Barbacid, 2005; De Veylder et al., 2007). For instance, several CDKs are inactivated by CDK inhibitors (CKIs; Denicourt and Dowdy, 2004), and in both fungi and metazoans, it has been established that CKI degradation at the G1-to-S transition releases CDK activity, which in turn is required to enter S phase. In budding yeast, this is achieved by the ubiquitin E3 ligase complex SCFCdc4 (Skp1, Cdc53/CULLIN, and Cdc4, a WD40-type F-box protein), which ubiquitylates the CKI Sic1 protein, leading to its proteolysis shortly before S phase (Schwob et al., 1994; Feldman et al., 1997). Similarly, in mammalian cells, the CKI protein p27Kip1 becomes unstable when cells enter S phase, as targeted by the SCFSkp2 (Skp2 being a Leu rich repeat-containing F-box protein) ubiquitin ligase (for a review, see Starostina and Kipreos, 2012). Notably, the human SCFSkp2 E3 also targets several other essential regulators of S-phase progression as well as other regulatory proteins.
Whether a similar regulation also occurs in plants is still not fully understood, but the Arabidopsis (Arabidopsis thaliana) F-box protein FBL17 has been proposed to mediate such a process. FBL17 loss-of-function mutants fail to undergo pollen mitosis II, which normally generates the two sperm cells in a mature pollen grain (Kim et al., 2008; Gusti et al., 2009). This major cell-cycle defect could be at least partially suppressed by the mutation of some CKI genes, called KIP-RELATED PROTEINs (KRPs; Gusti et al., 2009; Zhao et al., 2012). As some viable, though sterile, fbl17 loss-of-function plants could be recovered, it was possible to show that these mutants accumulate a higher level of the KRP2 CKI protein and share some phenotypic characteristics with plants overexpressing KRP proteins (Noir et al., 2015). However, it also appeared that fbl17 mutant plants exhibited some characteristics not observed in KRP overexpressors, suggesting that this F-box protein might have other protein targets and functions. In particular, we observed the occurrence of cell death and abnormal chromosome segregation in fbl17 mutant root tips, suggesting defects in genome stability (Noir et al., 2015).
The maintenance of genome integrity requires efficient DNA damage sensing and repair mechanisms (Cools and De Veylder, 2009; Nisa et al., 2019). Cells are constantly subjected to DNA damage arising from multiple origins, such as replication errors, mutations induced by the production of reactive oxygen species, or exposure to UV light, among others. However, most DNA damage will be detected and efficiently repaired by several DNA repair pathways (for review, see Spampinato, 2017). For cells, the most deleterious type of DNA damage is a double-strand break (DSB), which can lead to chromosomal rearrangements, loss of genetic information, and eventually cell death (Amiard et al., 2013). DSBs induce a DNA-damage response (DDR) that activates both cell-cycle checkpoints and DNA repair pathways (Hu et al., 2016). At the molecular level, when DSBs occur on chromatids, they are recognized by the MRE11-RAD50-NBS1 (MRN) complex (Syed and Tainer, 2018), which recruits ataxia telangiectasia mutated (ATM) kinase. Note that another kinase, ATM- and RAD3-related (ATR), is not activated by DSBs but rather by single-stranded DNA damage and replication fork stalling. Upon ATM activation, the kinase phosphorylates a multitude of downstream proteins involved in DDR. Among these, ATM phosphorylates the plant-specific transcription factor SUPPRESSOR OF GAMMA RESPONSE1 (SOG1; Yoshiyama et al., 2013), which plays a central role in DDR by activating the expression of genes that participate in DNA repair, cell cycle arrest, and cell death (Yoshiyama et al., 2009; Bourbousse et al., 2018). For instance, SOG1 binds to the promoters and induces the expression of B1-type cyclin CYCB1;1 (Weimer et al., 2016), CDK inhibitors SIAMESE-RELATED5 (SMR5) and SMR7 (Yi et al., 2014), and the DNA repair protein BRCA1 (Sjogren et al., 2015). Another important target of ATM is the histone variant H2AX, which upon phosphorylation becomes γH2AX (Friesner et al., 2005; Dickey et al., 2009). γH2AXs form foci at DSB sites that are important for recruitment of DNA-repair proteins such as RADIATION SENSITIVE51 (RAD51) and BRCA1 (Biedermann et al., 2017; Horvath et al., 2017). Strikingly, cell-cycle regulators not only are transcriptionally regulated during DDR, but may also directly participate in the repair mechanism. Indeed, it has recently been reported that upon DNA damage, the Arabidopsis RETINOBLASTOMA RELATED (RBR1) protein and its binding partner E2FA are recruited to γH2AX-labeled foci in an ATM- and ATR-dependent manner, and that RBR1 and BRCA1 even physically interact (Lang et al., 2012; Biedermann et al., 2017; Horvath et al., 2017). Moreover, RBR1 also partially colocalizes at DNA break sites with RAD51, a recombinase involved in homology-dependent DNA repair (Biedermann et al., 2017). However, the functional relevance for genome integrity of the specific association of RBR1 with DNA repair proteins remains to be elucidated. Notably, silencing of RBR1 leads to the upregulation of several genes involved in DDR, and for at least one of these, BRCA1, it represses expression through DNA-binding of the E2FA transcription factor (Horvath et al., 2017).
Here we show that the F-box protein FBL17, previously cited for its functions in cell-cycle regulation, is also involved in DDR processes. FBL17 loss of function is associated with a constitutive activation of DDR gene expression, a higher frequency of DNA lesions, and increased cell death in the root meristem, even in the absence of genotoxic stress. Moreover, the FBL17 mutation leads to hypersensitivity to DSB-induced genotoxic stress. Notably, the FBL17 protein is recruited at nuclear foci upon DSB induction and partially colocalizes with γH2AX. The possible roles of FBL17 in DNA damage-signaling or repair are discussed.
RESULTS
The fbl17 Mutant Transcriptome Exhibits a Strong Upregulation of Genes Related to DDR Processes
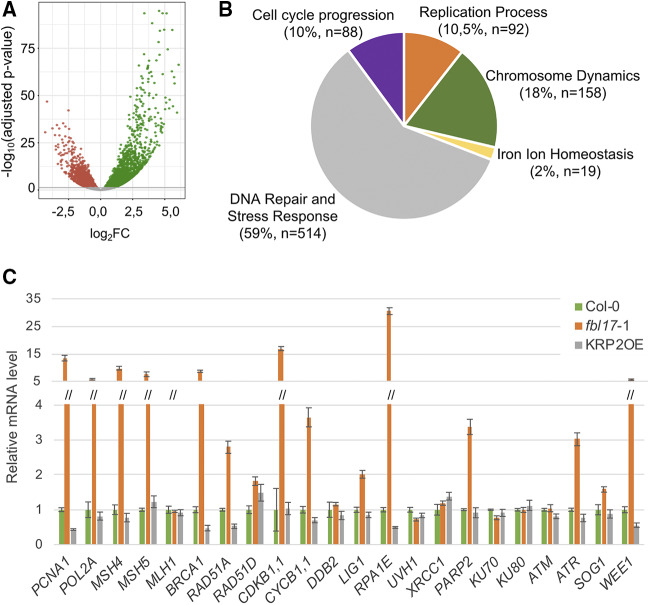
Previous analyses have shown that Arabidopsis fbl17 mutation leads to reduced leaf size, the appearance of serrated leaves, and sterility, likely caused by multiple cell-cycle defects (Noir et al., 2015). To further investigate the molecular basis of this phenotype, we performed a RNA sequencing (RNA-seq) analysis based on three biological replicates to identify differentially expressed genes (DEGs) between Col-0 and fbl17-1 homozygous seedlings (Fig. 1A; Supplemental Table S1). The comparative analysis of Col-0 and fbl17-1 transcriptome data revealed that there are 6,804 DEGs (i.e. ∼25% of the whole transcriptome) in the fbl17-1 mutant (adjusted P-value of < 0.05), with almost 54% of those upregulated (Fig. 1A). Considering all DEGs in fbl17, a Gene Ontology (GO) term enrichment analysis based on the Biological Process category showed an overrepresentation of genes involved in primary metabolic pathways, such as photosynthesis, and other cellular responses, most of them related to stress conditions (Supplemental Table S2), in line with the severe global phenotypic alterations of the mutant plants.
Figure 1.
The whole transcriptome of the fbl17 mutant reveals misexpression of numerous cell-cycle and DDR genes. A, All DEGs in fbl17 compared with Col-0 wild-type plants (log2FC [x axis]) were plotted against the −log10(adjusted P-value). The horizontal line indicates the significance threshold for DEGs (P < 0.05). Up- and downregulated genes are shown with green and red dots, respectively. Non-DEGs are shown with gray dots. B, GO functional analysis of DEGs in fbl17 exhibiting a log2FC absolute value >1.5 (i.e. 1,443 genes). The GO enrichment analysis is based on Biological Process functional categories of ShinyGo v0.61 software. The five major functional groups are based on the 50 most significant terms taken into account from the hierarchical clustering tree summarizing the correlation among pathways with many shared genes (Supplemental Fig. S1). The number of nonredundant genes (n) per functional group and the corresponding percentage are indicated in parentheses. C, Relative expression levels of gene transcripts from 8-d-old in vitro grown plants of the indicated genotype as determined by RT-qPCR. The bar graph depicts expression level mean values of the indicated transcripts of one independent replicate (± se of the technical triplicate). The experiment was repeated two times, giving the same tendency.
Remarkably, by filtering DEGs based on the log2 fold change (i.e. log2FC absolute value >1.5; Fig. 1B), the comparative RNA-seq analysis revealed that there are still >1,400 DEGs in the fbl17 mutant and that their GO term enrichment analysis highlighted their involvement in cell cycle progression, DNA replication mechanisms, chromosome dynamics, and, in an unexpectedly extended manner, DNA repair and stress response. The latter category represents 59% of the DEGs (Fig. 1B, Supplemental Fig. S1), with 405 genes (∼79%) exhibiting upregulation, suggesting a constitutive induction of genes linked to DNA damage and stress response. More precisely, using the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2017) enrichment analysis, six enriched pathways were identified (Supplemental Table S3). One corresponds to pyrimidine metabolism, involving modifications of both DNA and RNA nucleic acids. The five others are related to DNA metabolism, in particular to DNA replication and DNA repair mechanisms, including mismatch repair, homologous recombination, nucleotide excision repair, and base excision repair. Remarkably, in the six enriched pathways, the identified genes were all upregulated in fbl17-1 compared to Col-0.
Finally, to validate the RNA-seq approach, genes implicated in distinct DNA damage pathways, suggested by the KEGG analysis and some other genes, were selected and their expression was monitored for comparison by reverse transcription quantitative PCR (RT-qPCR) in wild-type Col-0 and the fbl17-1 homozygous mutant, as well as in the KRP2 overexpressor line (KRP2OE; Noir et al., 2015), under standard culture conditions (Fig. 1C). Each of the 22 tested genes revealed the same tendency in terms of expression level in both analyses, thus validating the data of the RNA-seq analysis. Furthermore, besides the expression of WEE1, ATR, CYCB1;1, CDKB1;1, and BRCA1 already reported (Noir et al., 2015), the selected genes PARP2, RPA1E, RAD51A, MSH5, MSH4, POL2A, and PCNA1 are also upregulated under standard growth conditions in fbl17 but not in the control, Col-0 (Fig. 1C). Given that fbl17 mutants present an accumulation of the CDK inhibitor KRP2, it is expected that the KRP2OE line might mimic some of the fbl17 mutant phenotypes (Noir et al., 2015). Interestingly, in this analysis, the KRP2OE line exhibited an expression pattern comparable to that of Col-0, indicating that the constitutive upregulation of DDR genes in fbl17 is not a direct consequence of KRP2 overaccumulation. Altogether, this analysis indicates that loss of FBL17 function causes a constitutive and rather global induction of the DDR.
fbl17 Mutants Reveal an Increased Frequency of DNA Lesions
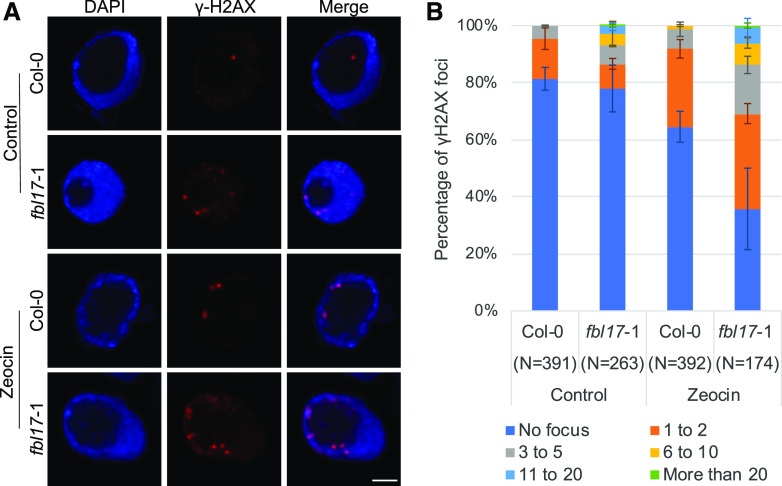
The constitutive transcriptional DDR suggests that fbl17 mutants are subjected to genome instability, which is further supported by the occurrence of micronuclei and chromosome bridges previously observed in dividing mutant cells (Noir et al., 2015). To further investigate this issue, we used the sensitive and highly specific γH2AX marker, which detected by immunolabeling can reveal DNA break sites (Fig. 2A). Interestingly, accumulation of γH2AX foci in the fbl17 mutant background was observed. More precisely, whereas the frequency of root nuclei exhibiting constitutive γH2AX foci in fbl17 was only slightly increased in comparison to Col-0 (i.e. ∼30% in fbl17 versus 20% in Col-0; Fig. 2A), the number of γH2AX-marked foci per nucleus was much higher in fbl17 than in Col-0 (Fig. 2B), reflecting an excessive frequency of DNA lesions even in the absence of genotoxic stress.
Figure 2.
Increased accumulation of γH2AX foci in fbl17. A, Representative images of Col-0 and fbl17-1 after immunostaining of γH2AX foci (red) in root-tip nuclei from seedlings under control conditions or treated for 16 h with 5 μm zeocin. Nuclei were counterstained with 4′,6-diamino-phenylindole (blue). Scale bar = 2 μm. B, Quantification of γH2AX foci in nuclei from Col-0 and fbl17-1 seedlings under control conditions or treated with zeocin. Between 79 and 233 nuclei per line per replicate were analyzed and categorized into six types: no focus, 1 to 2, 3 to 5, 6 to 10, 11 to 20, or >20 γH2AX foci/nucleus, respectively. Two independent replicates were performed. Error bars indicate the sd.
fbl17 Mutants Are Hypersensitive to Drug-Induced DSB DNA Lesions
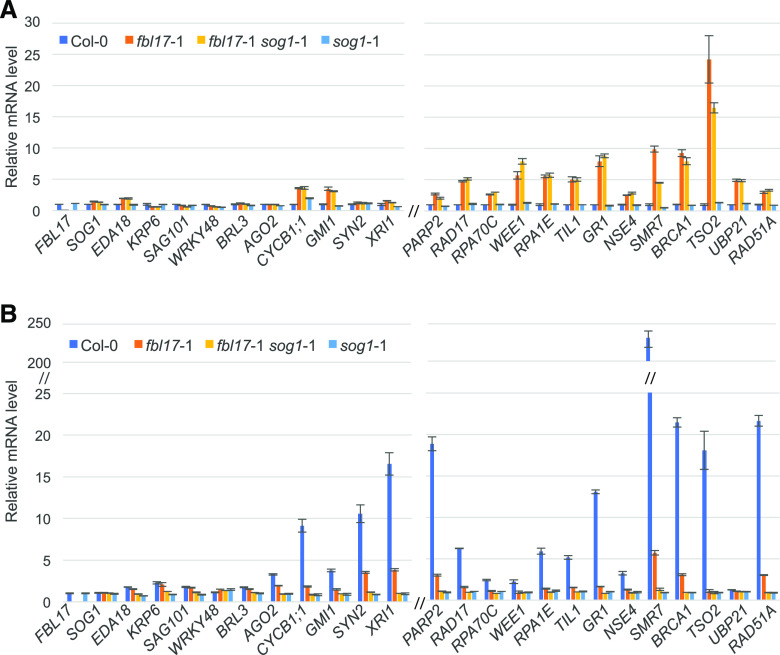
Given the specificity of γH2AX recruitment at DNA lesion sites and their accumulation in the fbl17 loss-of-function mutant, and also considering that this type of DNA lesion especially can result in loss of genetic information, we turned our attention to genotoxic conditions triggering DSB DNA lesions. To begin with, transcript levels of the previously tested DNA-damage genes were evaluated after treatment of seedlings with zeocin, an antibiotic of the bleomycin (BLM) family widely used as an inducer of DSBs. It should be mentioned that the FBL17 gene itself is not differentially regulated upon genotoxic stress or in the tested DDR mutant background sog1-1 (Fig. 3). Under zeocin treatment, a number of genes known to be involved in the DDR pathway, such as RAD51A, TSO2, BRCA1, SMR7, GR1, RPA1E, RAD17, PARP2, XRI1, SYN2, CYCB1;1, and SIP4, among others, appeared strongly induced in Col-0 (i.e. log2FC up to 200; Fig. 3B). According to the literature, many of these genes are known to be induced after DSB-induced stress, and are targeted by the transcription factor SOG1 (Culligan et al., 2006; Ogita et al., 2018). Notably, for several genes, such as RAD51A, BRCA1, SMR7, PARP2, and XRI1, whereas they were also induced in fbl17 (i.e. log2FC between 2 and 6), expression induction was lower compared to the Col-0 control, possibly due to the preexisting constitutive induction of these genes in the mutant (Fig. 3A). In addition, TSO2, NSE4, GR1, TIL1, RPA1E, WEE1, RPA70C, and RAD17 did not appear to be induced by zeocin in fbl17, likely because they were already at maximal gene expression levels in the mutant even in the absence of the drug. Lastly, some genes (among which some were constitutively upregulated in fbl17) were not upregulated after zeocin treatment in the fbl17 mutant or in the Col-0 control (i.e. LIG4, PCNA1, FAN1, ATM, and ATR; Supplemental Fig. S2). This is in accordance with previous analyses showing that these genes are not induced by DSB stress and are not targeted by SOG1 (Culligan et al., 2006).
Figure 3.
DDR gene expression levels in Col-0, fbl17, the sog1 single-mutant, and the fbl17 sog1 double-mutant backgrounds with and without zeocin treatment. A, Relative expression levels of genes in 8-d-old in vitro grown plants (standard conditions) of the indicated genotypes as determined by RT-qPCR. Data are compared to the Col-0 value normalized at 1. B, Relative expression levels of genes in 8-d-old in vitro grown plants of the indicated genotypes after 3 h of 20 μm zeocin treatment as determined by RT-qPCR. Data are compared to the value of the same genotype under standard conditions (A) normalized at 1. Bar graphs depict expression level mean values of the indicated transcripts of one independent replicate (± se of the technical triplicate). The experiment was repeated two times with similar results.
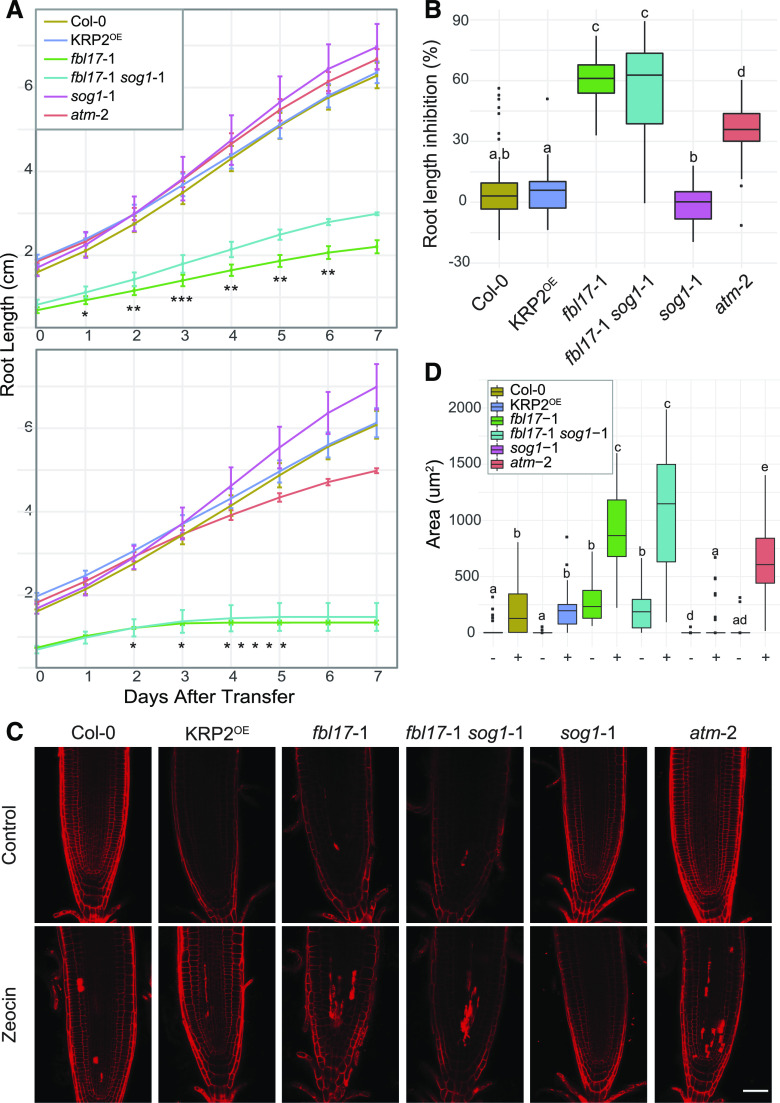
Next, we investigated the sensitivity of the fbl17 mutant to zeocin treatment using a root elongation assay. Severe delay of fbl17 primary root elongation, observed previously, was confirmed under standard conditions (Fig. 4A). The atm-2 mutant was used as a sensitive control (Garcia et al., 2003) and, as expected, the primary root elongation of this mutant started to be slightly delayed after 4 d of zeocin treatment (Fig. 4A), confirming the efficacy of the treatment. At that time point, Col-0 and KRP2OE were not yet affected by the chemical treatment, whereas fbl17 root elongation had completely stopped already by day 3. After 7 d of zeocin treatment, atm-2 exhibited an intermediate phenotype of root length inhibition (i.e. 40%, median value), which lay between the respective ratios of ∼5% to 6% for Col-0 and KRP2OE lines and 60% for the fbl17 mutant line (Fig. 4B).
Figure 4.
The fbl17 mutant exhibits hypersensitivity to zeocin treatment. A, Root growth elongation of the indicated genotypes in 5-d-old seedlings grown under standard conditions and then transferred onto control medium (top) or medium containing 5 μm zeocin (bottom) for a further 7 d of culture. Error bars indicate the mean ± sd of three independent experiments (4 < N [per genotype] < 37). Asterisks indicate significant difference between fbl17-1 and fbl17-1 sog1-1: *P < 0.05, **P < 0.01, and ***P < 0.001 by Wilcoxon-Mann-Whitney test. Complete statistical analyses are given in Supplemental Tables S4 and S5. B, Percentage of root length inhibition for the experiment described in A. Statistical significance was calculated by Wilcoxon-Mann-Whitney test. Box whiskers with different lowercase letters denote statistical differences determined by one-way ANOVA (P < 0.05 at least). Complete statistical analysis is given in Supplemental Table S6. C, Representative images of root tips of 5-d-old seedlings transferred onto control medium or medium containing 5 μm zeocin for a further 3 d of growth before propidium iodide staining. Scale bar = 50 μm. Three independent replicates were performed (4 < N [per genotype] < 11). D, Cell death quantification of the root samples illustrated in C on control medium (−) or medium containing 5 μm zeocin (+) for a further 3 d. Statistical significance analysis was calculated by Wilcoxon-Mann-Whitney test. Box whiskers with different lowercase letters denote statistical differences determined by one-way ANOVA (P < 0.05 at least). Complete statistical analysis is given in Supplemental Table S7.
A strategy undertaken by multicellular organisms to eliminate damaged cells is to actively trigger cell death (Hu et al., 2016). Under the same experimental culture conditions, occurrence of cell death was estimated after 3 d of zeocin exposure. As expected, the atm-2 sensitive mutant exhibited more cell death than the control, Col-0 (Fulcher and Sablowski, 2009), and the KRP2OE line. Remarkably, whereas fbl17 root tips already exhibited constitutive cell death, further accumulation was noticed upon zeocin treatment, which corresponded to an even wider area of dead cells than observed in the atm-2 mutant (Fig. 4, C and D). In addition, whereas cell death observed in Col-0 root tips was qualitatively mainly located at the level of the quiescent center (QC), cell death in fbl17 occurred both at the QC and in more distant tissues of the root. Finally, the frequency of γH2AX foci was monitored after zeocin treatment in Col-0 and fbl17 (Fig. 2A). Under this condition, the frequency of γH2AX-marked nuclei in fbl17 was significantly higher (∼70%) than in Col-0 (∼30%; Fig. 2B). Moreover, γH2AX-marked nuclei accumulated a larger number of foci per nucleus in fbl17 than in Col-0; in some cases, >20 γH2AX foci were observed in a single nucleus.
Altogether, the impaired root meristem activity, the accumulation of cell death, and the increased number of γH2AX foci upon zeocin treatment indicate that the fbl17 mutant is hypersensitive to DSB-induced genotoxic stress. This phenotype is not the consequence of KRP overaccumulation occurring in fbl17 mutants, but rather suggests that FBL17 might be involved in DDR beyond its cell-cycle regulatory functions.
fbl17 Constitutive Overexpression of DDR Genes and Hypersensitivity to Drug-Induced DSBs Are SOG1 Independent
Mechanisms for sensing DNA lesions and initiating DDR involve massive gene regulation, ultimately leading to DNA repair. At this level of control, the Arabidopsis SOG1 transcription factor of the NAC family has been shown to be a master regulator controlling multiple DDR pathways (Yoshiyama, 2016; Bourbousse et al., 2018; Ogita et al., 2018). As already mentioned above (see Fig. 3), SOG1 is not differentially regulated in fbl17 at the transcriptional level. In order to investigate the putative implication of SOG1 in the DDR observed in the fbl17 mutant, the double mutant fbl17-1 sog1-1 was generated. At a macroscopic level, the developmental phenotype of the double mutant was similar to that of the fbl17 single mutant (Supplemental Fig. S3), although, regarding its root growth, the double mutant exhibits a minor rescue of root length (Fig. 4A). Upon genotoxic stress, whereas sog1-1 exhibits a slight resistance to zeocin in our experimental conditions (Fig. 4B; see also Adachi et al., 2011), the fbl17-1 sog1-1 double mutant shares a similar level of sensitivity with the fbl17 single mutant in terms of root growth inhibition (Fig. 4B) and cell death occurrence (Fig. 4, C and D). Despite the absence of obvious rescue of the fbl17 phenotype by the sog1 mutation, we next asked whether SOG1 loss of function could at least partially attenuate the global upregulation of DNA-damage genes observed in the fbl17 mutant background.
At first, by testing some gene targets of SOG1 that are not implicated in DDR according to Ogita et al. (2018), e.g. EDA18, KRP6, SAG101, BRL3, AGO2, we verified that they are not constitutively upregulated in the fbl17 single mutant and, as expected, in fbl17-1 sog1-1 (Fig. 3A). Nevertheless, most of the DDR genes constitutively induced without genotoxic stress were similarly differentially expressed in fbl17-1 sog1-1 and fbl17 (Fig. 3A); the exceptions were TSO2 and SMR7, which showed only a slight decrease of expression. By contrast, and as expected, induction of these genes by zeocin was fully suppressed in the sog1-1 single mutant and also in the fbl17-1 sog1-1 double mutant (Fig. 3B). It is noteworthy, however, that upon zeocin treatment in fbl17, only the additive increase of expression of some DDR genes (e.g. RAD51A, BRCA1, SMR7, PARP2) was dependent on SOG1. Altogether, these results indicate that the constitutive DDR and hypersensitivity to DSB-induced genotoxic stress observed in fbl17 do not depend on SOG1 and likely involve other transcriptional regulatory mechanisms.
FBL17 Is Recruited at Nuclear Foci upon DSB Induction
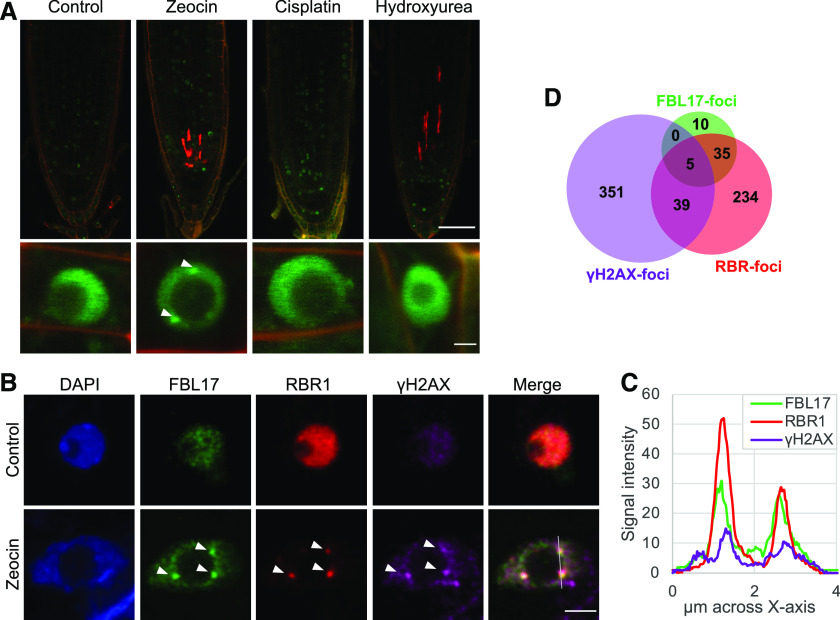
It was previously shown that FBL17 is a nuclear F-box protein restricted to a few cells in the root meristem that shows a cell cycle phase-dependent expression pattern (Noir et al., 2015; Desvoyes et al., 2019). We next investigated whether DNA damage affects subcellular distribution of FBL17. To answer this question, we took advantage of the previously established fbl17-1 pFBL17:FBL17-GFP line (Noir et al., 2015). At first, the sensitivity of this reporter line to zeocin was monitored using a root elongation assay (Supplemental Fig. S4, A and B) and by RT-qPCR analysis (Supplemental Fig. S4D). In the tested conditions, the reporter line exhibited behavior similar to that of the Col-0 wild-type control, establishing that the FBL17-GFP protein is functional and could confidently be used for our analyses. Consequently, the GFP-reporter line was exposed to distinct genotoxic stresses and the distribution of the fusion protein was imaged by confocal microscopy (Fig. 5A). For this assay, we used zeocin to induce DSB DNA lesions, and due to the implication of FBL17 in DNA replication, cisplatin and hydroxyurea (HU) treatments were also applied to trigger DNA crosslinking and stalled replication forks, respectively. Under these conditions, no obvious differential distribution of the FBL17-GFP protein at a tissue level was observed among the three treatments tested. However, focusing at a subcellular nuclear level, the formation of FBL17 nuclear foci could be observed only with the zeocin treatment, and not with cisplatin or HU, suggesting that the formation of FBL17 foci might be specific to DSB-type DNA lesions.
Figure 5.
FBL17 proteins are recruited at γH2AX foci and colocalize with RBR1 upon DSB-induced stress. A, Live microscopy of fbl17-1, pFBL17:FBL17-GFP after 16 h of genotoxic treatment (zeocin 20 μm, cisplatin 15 μm, or HU 5 μm). Scale bars = 50 μm (top) and 2 µm (bottom). Three independent experiments were analyzed (5 < N [per genotype] < 10). B, Representative images of fbl17-1, pFBL17:FBL17-GFP root nuclei with triple immunolocalization of γH2AX (purple), RBR1 (red), and FBL17-GFP (anti-GFP; green), showing colocalization of the three signals (arrowheads) after 16 h of zeocin treatment (20 μm). Nuclei were counterstained with 4′,6-diamino-phenylindole (blue). Scale bar = 2 µm. C, Signal intensity distribution of the total amount of pixels at the x axis shown in the zeocin-treated nucleus in B. Statistical significance analysis of the signal colocalization is given in Supplemental Table S8. D, Venn diagram showing the frequency of the different colocalization combinations of FBL17, RBR1, and γH2AX foci in the nuclei of one replicate (total number of foci = 758). Complete frequency analysis of the three independent replicates is given in Supplemental Table S9.
DSB-Type DNA Lesions Recruit FBL17 at γH2AX Foci
The observation of FBL17 foci upon genotoxic stress was reminiscent of the formation of γH2AX foci. Intriguingly, it was recently shown that in addition to the expected proteins from the DNA-damage machinery, some cell-cycle transcriptional regulators, both Arabidopsis E2FA and Nicotiana tabacum E2F transcription factors and RETINOBLASTOMA RELATED1 (RBR1), also localized at DNA damage sites (Lang et al., 2012; Biedermann et al., 2017; Horvath et al., 2017). To better define the FBL17 foci, we used an immunostaining approach that, despite revealing a low frequency of nuclei with FBL17 foci (Supplemental Table S9), allowed us to investigate whether they colocalize with γH2AX foci and/or RBR1. Indeed, we observed the colocalization of FBL17 with γH2AX foci in some nuclear foci, supporting the idea that the F-box protein is recruited at DNA lesion sites upon zeocin treatment (Fig. 5B). More interestingly, we also observed a clear colocalization of FBL17 and RBR1 (Fig. 5, B and C). In fact, quantification of these nuclear foci under zeocin treatment (Fig. 5D) revealed a mean value of 5% colocalization of RBR1 with γH2AX and 5% colocalization of FBL17 with RBR1 (Supplemental Table S9). Note that FBL17 and γH2AX never colocalize if RBR1 is not itself detected at these foci (Fig. 5D; Supplemental Table S9) and colocalization of all three proteins together represented only 1% of our observations. These results suggest that FBL17 and RBR1 follow a dynamic recruitment at the DNA damage sites, where they likely contribute to DNA repair and genome integrity.
DISCUSSION
We have previously shown that Arabidopsis FBL17 loss of function slows plant growth by decreasing cell proliferation and also suppressing endoreplication (Noir et al., 2015). At the molecular level, fbl17 mutant plants showed increased accumulation of the KRP2 protein, which is known to switch off CDKA;1 kinase activity (Verkest et al., 2005), and phenotypically resembled the cdka;1 null mutant (Nowack et al., 2012), indicating that a main function of FBL17 is to positively regulate CDKA;1 activity. In line with such a role, the loss of FBL17 delayed, or even blocked, S-phase in some cells and led to differential expression of cell-cycle genes, among them, consistently, genes involved in the process of DNA replication (Noir et al., 2015). However, our transcriptomic approach revealed that the fbl17 mutation also leads to a broader activation of numerous DDR genes beyond those solely linked to DNA replication stress. Note that genome-wide transcriptional studies of synchronized plant cells revealed that several DDR genes have their expression maximum in S-phase (Menges et al., 2005; Trolet et al., 2019).
In plants, a major regulator of DDR is the transcription factor SOG1, which has been functionally compared to the mammalian tumor suppressor p53 (Yoshiyama et al., 2009; Yoshiyama, 2016). SOG1 is directly phosphorylated by ATM, and its mutation impairs DNA repair, cell cycle arrest, and activation of cell death (Preuss and Britt, 2003; Yoshiyama et al., 2009; Furukawa et al., 2010). Whereas posttranslational regulation of SOG1 by FBL17 cannot be fully excluded, according to our analysis, FBL17 does not seem to act in SOG1 transcription regulation. First, the constitutive transcriptional activation of DDR genes in fbl17 is not suppressed by the sog1 mutation. Second, many of the SOG1 target genes can still be induced upon zeocin treatment in the fbl17 mutant background. Third, fbl17 mutant hypersensitivity to DSB-inducing agents and increased amount of cell death are not dependent on SOG1.
Interestingly, it has recently been shown that several genes involved in DDR are induced when RBR1 is downregulated by RNA interference or in the hypomorphic rbr1-2 mutant (Biedermann et al., 2017; Horvath et al., 2017). At least for Arabidopsis BRCA1, it was shown that RBR1 directly represses this gene through the E2FA transcription factor (Horvath et al., 2017). A genome-wide RBR1 chromatin immunoprecipitation analysis further indicated that RBR1 is recruited to E2F sites present in the promoters of many DDR genes (Bouyer et al., 2018). According to our analysis, combining the information of E2FA-binding sites (Verkest et al., 2014) with the RBR1 chromatin immunoprecipitation dataset (Bouyer et al., 2018), several of the DDR genes constitutively induced in fbl17 are likely targets of RBR1/E2FA.
At the functional level, it was reported that rbr1 mutants exhibit an elevated level of DNA damage in normal growth conditions, whereas after BLM treatment, these mutants show a significantly higher level of DNA fragmentation (Biedermann et al., 2017; Horvath et al., 2017). Moreover, both the lack of functional RBR1 and the loss of E2FA resulted in hypersensitivity against DNA DSB-inducing agents (Roa et al., 2009; Lang et al., 2012; Biedermann et al., 2017). Thus, the loss of FBL17 function shares many similarities with the phenotype of RBR1/E2FA-deficient plants, suggesting that the F-box protein could act at this level. Indeed, we observed the colocalization of FBL17 and RBR1 at DNA damage sites. The recruitment of RBR1 and E2FA to γH2AX foci has been previously reported (Lang et al., 2012; Biedermann et al., 2017; Horvath et al., 2017), suggesting that these proteins might play a more direct role in DNA repair besides their known transcriptional regulatory function. Interestingly, RBR1 colocalizes with the RADIATION SENSITIVE51 (RAD51) protein and is necessary for RAD51 localization to DNA after BLM treatment (Biedermann et al., 2017). RBR1 also colocalizes with BRCA1 foci upon DNA stress, although RBR1 recruitment to γH2AX foci was found independent of BRCA1 (Horvath et al., 2017). As shown in mammals, where retinoblastoma physically interacts with the BRCA1 (Aprelikova et al., 1999), RBR1 also directly interacts with BRCA1 in plants (Horvath et al., 2017), suggesting a structural role of retinoblastoma-related proteins in the DDR machinery. Our observation that FBL17 and RBR1 colocalize in nuclear foci after DNA damage generating DSBs suggests that the F-box protein directly participates in the process of DNA repair. Since FBL17 association with γH2AX seems to depend on RBR1, it is possible that the latter recruits FBL17 at the DNA damage sites in a dynamic manner. This raises the question of which proteins might be ubiquitylated by FBL17 at the sites of DNA lesions.
In mammalian cells, DSB repair implies a complex interplay between ubiquitylation and SUMOylation for faithful repair of such damage (Schwertman et al., 2016). In particular, it has been shown that ubiquitylation of proteins in the vicinity of DNA lesions functions as a recruitment signal for DSB repair factors. Ubiquitylation and deubiquitylation cycles also control the steady-state level of DSB repair factors and/or their interactions. Of particular interest for our study is the mammalian F-box protein Skp2. Whereas Skp2 is a main regulator of the cell cycle (Frescas and Pagano, 2008; Starostina and Kipreos, 2012), it has also been involved in DDR. Thus, it was shown that Skp2 is required for the activation and recruitment of the ATM kinase to DNA damage foci (Wu et al., 2012). At the molecular level, in response to DSBs, Skp2 triggers the K63-dependent ubiquitylation of NBS1, a component of the MRN complex, which in turn facilitates ATM recruitment to DNA foci for activation. Skp2 also ubiquitylates other proteins at DSBs, such as BRCA1, which is important for the timing of end resection (Parameswaran et al., 2015). Similar to Skp2, the Arabidopsis FBL17 F-box protein is able to degrade CKI proteins (Noir et al., 2015 and references therein), but whether it also ubiquitylates components of the DNA repair machinery is presently unknown. Note that at the DNA damage sites, RBR1 itself and/or E2FA are also possible candidate substrates of the SCFFBL17 ubiquitin E3 ligase. Therefore, further experiments will be necessary to elucidate the function of this plant F-box protein at DSB sites.
MATERIALS AND METHODS
Plant Material
The following Arabidopsis (Arabidopsis thaliana) lines were used in this study: the transfer DNA insertion lines fbl17-1 (Gabi-KAT_170-E02; Noir et al., 2015), atm-2 (SALK_006953; Garcia et al., 2003), the EMS mutant line sog1-1 (Yoshiyama et al., 2009), and the Arabidopsis reporter and/or overexpressor lines fbl17-1, pFBL17:FBL17-GFP, 35S:FBL17-GFP, and GFP-KRP2OE described in Noir et al. (2015). Transfer DNA insertions and mutations were confirmed by PCR-based genotyping and by further sequencing for the sog1-1 allele. The fbl17-1 sog1-1 double mutant was generated by performing crosses and genotyping/sequencing of the resulting F2 and/or F3 progenies by PCR-based approaches. Primers designed for this purpose are listed in Supplemental Table S10.
Plant Growth Conditions
For in vitro growth conditions, seed sterilization, stratification, and in vitro culture were performed as described previously (Noir et al., 2015) with or without a supplemented genotoxic agent. To obtain flowering plants and seeds, seedlings initially grown under in vitro culture were transferred onto soil at days 6 to 8 and kept in 16-h light/8-h dark growth chambers under fluorescent light (49W/965, Osram Biolux).
For monitoring of root growth, seedlings were germinated and grown in vitro on vertical plates using 1% (w/v) Murashige and Skoog agar medium and transferred at day 5 to 1% (w/v) Murashige and Skoog agar medium with or without 5 μm zeocin (Invitrogen). Root elongation was scored each day for 7 d and root length was measured using Fiji software (ImageJ 1.52p; http://imageJ.nih.gov/ij). The final values were calculated using R software (version 3.6.1) to determine the arithmetic mean of the root length values of three biological replicates, which were themselves the average of 4 to 37 plants.
RT-qPCR
Purification of total RNA from 8-d-old seedlings grown under in vitro conditions was performed using Tri-Reagent (Molecular Research Center) according to the manufacturer’s instructions. Complementary DNAs (cDNAs) were prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed using gene-specific primers and SYBR Green Master Mix (Roche) on a LightCycler LC480 apparatus (Roche) according to the manufacturer’s instructions. The mean value of three replicates was normalized using the TIP4.1 (AT4G34270) and SAND (AT2G28390) genes as internal controls. All primers used in RT-qPCR analyses are listed in Supplemental Table S10.
Nucleic Acid Isolation, cDNA Library Preparation, Sequencing, and Data Analysis
Total RNA was extracted using Trizol solution (Invitrogen) from 10-d-old fbl17-1 and Col-0 seedlings grown in vitro, with extraction performed as described above and completed by a second phenol/chloroform treatment. Three biological replicates were used as starting material. RNA concentrations were determined with a QuBit Fluorometer (Thermo Fisher Scientific). RNA integrities were checked using the 2100 Bioanalyzer (Agilent). mRNA was isolated from total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB) for mRNA library preparation. Sequencing libraries were prepared using the Collibri stranded RNA library kit for Illumina (Invitrogen). The libraries were sequenced using an Illumina Nextseq 500 system (single-end mode 1 × 75 bp). RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-9050.
For the bioinformatics analysis, the preprocessing of the sequencing data were performed using TrimGalore (v0.5.0; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore); reads were processed by removing the Illumina adaptor sequences using Cutadapt v1.18 and quality was assessed using FastQC v0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads with quality >30 and minimal read length of 50 pb were kept. The data were mapped to the Arabidopsis genome (TAIR10) using Hisat2 (v2.1.0) software (Kim et al., 2015) and sorted with Samtools v1.9 (Li et al., 2009). For each gene, read quantification was performed using HTSeq-count v0.11.0 software (with the parameter “intersection nonempty”; Anders et al., 2015). Differential expression analysis by pairwise comparison was performed using the R package DESeq2 (v1.24.0; Anders and Huber, 2010) and the betaprior parameter set to true. GO and KEGG enrichment analysis were performed using ShinyGo v0.61 software (Ge et al., 2019).
Immunolabeling
Fixation and immunostaining were performed as previously described (Batzenschlager et al., 2015) using 6-d-old in vitro grown seedlings. The primary antibodies used were the rabbit polyclonal anti-γH2AX (diluted at 1:500; Davids biotechnologie) against the synthetic phosphopeptide VGKNKGDIGSA(p)SQGEF as described in Friesner et al. (2005), mouse monoclonal anti-GFP (1:500; Life Technologies), and chicken polyclonal anti-RBR1 (1:7,000; Agrisera). Depending on the experiments, the conjugated secondary antibodies for γH2AX detection were either the goat anti-rabbit Alexa Fluor 568 (1:300; Life Technologies) for red signals or the goat anti-rabbit Cyn5 (1:500; Life Technologies) for purple signals. For GFP and RBR1 detection, the conjugated secondary antibodies used were, respectively, goat anti-mouse Alexa Fluor 488 (1:200; Interchim) for the green signal and goat anti-chicken Alexa Fluor 568 (1:300; Life Technologies) for the red signal.
Confocal Microscopy Analyses and Image Treatments
All confocal microscopy observations were performed using the Leica TCS SP8 microscope. Roots of seedlings expressing fluorescent reporter constructs were observed after treatment with 20 μm zeocin (Invitrogen), 15 μm cisplatin (Sigma), or 5 mm HU (Sigma), or after transfer to standard conditions for 16 h, and just before observation were counterstained in 75 mg mL−1 propidium iodide (Fluka). To score cell death, 8-d-old seedlings not treated or treated for 3 d with 5 μm zeocin were stained as described previously (Biedermann et al., 2017). Dead cell quantification was performed at the quiescent center (QC) plan considering a fixed area of 15,000 µm2 (200 µm length long, from the QC toward the elongation zone, and 75 µm wide) using Fiji software (ImageJ 1.52p; http://imageJ.nih.gov/ij). The final values were calculated by determining the arithmetic mean of three biological replicates (4 < N per genotype < 11) using R software (v3.6.1). For immunolabeling imaging, confocal images of fixed nuclei were taken as a consecutive series along the z axis. Microscope settings were kept the same for image acquisition of each genotype and/or condition, and signal colocalization was evaluated using Fiji software.
Accession Numbers
Gene data from this article can be found in The Arabidopsis Information Resource database (www.arabidopsis.org) under accession numbers: AT1G02970 (WEE1), AT1G07370 (PCNA1), AT1G07745 (RAD51D), AT1G08130 (LIG1), AT1G08260 (TIL1), AT1G08260.1 (POL2A), AT1G16970 (KU70), AT1G25580 (SOG1), AT1G31280 (AGO2), AT1G48050 (KU80), AT1G48360 (FAN1), AT1G51130 (NSE4A), AT1G80420 (XRCC1), AT2G28390 (SAND), AT2G34920 (EDA18), AT3G13380 (BRL3), AT3G19150 (KRP6), AT3G20475 (MSH5), AT3G27060 (TSO2), AT3G27630 (SMR7), AT3G48190 (ATM), AT3G52115 (GR1), AT3G54180 (CDKB1;1), AT3G54650 (FBL17), AT4G02390 (PARP2), AT4G09140 (MLH1), AT4G17380 (MSH4), AT4G19130 (RPA1E), AT4G21070 (BRCA1), AT4G34270 (TIP41), AT4G37490 (CYCB1;1), AT5G14930 (SAG101), AT5G20850 (RAD51A), AT5G24280 (GMI1), AT5G40820 (ATR), AT5G41150 (UVH1), AT5G45400 (RPA70C), AT5G46740 (UBP21), AT5G48720.02 (XRI1), AT5G49520 (WRKY48), AT5G57160 (LIG4), AT5G58760 (DDB2), AT5G66130 (RAD17), and AtT5G40840.2 (SYN2).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Details of the GO functional analysis of DEGs in fbl17 exhibiting a log2FC absolute value >1.5 (i.e. 1,443 genes; see Fig. 1B).
Supplemental Figure S2. Gene expression analysis under standard conditions and after zeocin treatment.
Supplemental Figure S3. The fbl17-1 sog1-1 double mutant exhibits the same phenotype as the fbl17 single mutant.
Supplemental Figure S4. FBL17-GFP reporter lines exhibit the same sensitivity as Col-0 under zeocin treatment.
Supplemental Table S1. Set of genes differentially regulated in fbl17-1 seedlings compared with Col-0 plants (i.e. 6,804 DEGs).
Supplemental Table S2. GO enrichment analysis of all DEGs in fbl17 compared to Col-0 (i.e. 6,804 DEGs).
Supplemental Table S3. KEGG pathway enrichment analysis based on DEGs in fbl17 compared to Col-0 (log2FC absolute value >1.5; i.e. 1,443 genes).
Supplemental Table S4. Statistical significance for Figure 4A, regarding the root length of fbl17-1 and fbl17-1 sog1-1 under standard conditions (Wilcoxon-Mann-Whitney test).
Supplemental Table S5. Statistical significance for Figure 4A, regarding the root length of fbl17-1 and fbl17-1 sog1-1 under zeocin conditions (Wilcoxon-Mann-Whitney test).
Supplemental Table S6. Statistical significance for Figure 4B, root length inhibition (Wilcoxon-Mann-Whitney test).
Supplemental Table S7. Statistical significance table for Figure 4D, cell death quantification (Wilcoxon-Mann-Whitney test).
Supplemental Table S8. Statistical significance analysis of the FBL17-, RBR1-, and γH2AX-signal colocalization (Pearson’s coefficient and Mander’s coefficient).
Supplemental Table S9. Frequency of nuclei showing γH2AX, RBR1, and/or FBL17 foci compared to the total number of nuclei with foci, and frequency of foci showing γH2AX, RBR1, and/or FBL17 foci compared to the total number of foci.
Supplemental Table S10. Primer combinations used for genotyping and RT-qPCR analyses.
Acknowledgments
We thank Jérôme Mutterer (Institut de Biologie Moléculaire des Plantes, Microscopy and Cellular Imaging platform) for confocal microscopy support; Sandrine Koechler and Abdelmalek Alioua (Institut de Biologie Moléculaire des Plantes, Gene Expression Analysis platform), and Anne Molitor, Antoine Hanauer, and Raphael Carapito (Institut National de la Santé et de la Recherche Médicale, GENOMAX platform, UMRS_1109) for Next-Generation Sequencing experiments.
Footnotes
This work was supported by the Grand Est Région (grant no. 168947) and the Agence Nationale de la Recherche (ANR) International Collaborative Research Project (PRCI; ANR-RHiD grant no. ANR–19–CE13–0032) and ANR LabEx program (grant no. ANR–10–LABX–0036_NETRNA).
Articles can be viewed without a subscription.
References
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. (2011) Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Gallego ME, White C(2013) Signaling of double strand breaks and deprotected telomeres in Arabidopsis. Front Plant Sci 4: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W(2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W(2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprelikova ON, Fang BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen RA, Liu ET(1999) BRCA1-associated growth arrest is RB-dependent. Proc Natl Acad Sci USA 96: 11866–11871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzenschlager M, Lermontova I, Schubert V, Fuchs J, Berr A, Koini MA, Houlné G, Herzog E, Rutten T, Alioua A, et al. (2015) Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc Natl Acad Sci USA 112: 8656–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann S, Harashima H, Chen P, Heese M, Bouyer D, Sofroni K, Schnittger A(2017) The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J 36: 1279–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbousse C, Vegesna N, Law JA(2018) SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc Natl Acad Sci USA 115: E12453–E12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Heese M, Chen P, Harashima H, Roudier F, Grüttner C, Schnittger A(2018) Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet 14: e1007797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, De Veylder L(2009) DNA stress checkpoint control and plant development. Curr Opin Plant Biol 12: 23–28 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB(2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Denicourt C, Dowdy SF(2004) Cip/Kip proteins: More than just CDK inhibitors. Genes Dev 18: 851–855 [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Noir S, Masoud K, Lopéz MI, Genschik P, Gutierrez C(2019) FBL17 targets CDT1a for degradation in early S-phase to prevent Arabidopsis genome instability. biorXiv 774109 [Google Scholar]
- Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM(2009) H2AX: Functional roles and potential applications. Chromosoma 118: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ(1997) A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M(2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat Rev Cancer 8: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB(2005) Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R(2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BWL, Aggoune D, Hays JB, Britt AB(2010) A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 9: 940–948 [DOI] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A(2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Jung D, Yao R(2019) ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36: 2628–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P(2009) The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4: e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath BM, Kourova H, Nagy S, Nemeth E, Magyar Z, Papdi C, Ahmad Z, Sanchez-Perez GF, Perilli S, Blilou I, et al. (2017) Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J 36: 1261–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Cools T, De Veylder L(2016) Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol 67: 439–462 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K(2017) KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(D1): D353–D361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL(2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG(2008) Control of plant germline proliferation by SCF FBL17 degradation of cell cycle inhibitors. Nature 455: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Lang J, Smetana O, Sanchez-Calderon L, Lincker F, Genestier J, Schmit AC, Houlné G, Chabouté ME(2012) Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol 194: 353–363 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M(2005) Mammalian cyclin-dependent kinases. Trends Biochem Sci 30: 630–641 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH(2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Nisa MU, Huang Y, Benhamed M, Raynaud C(2019) The plant DNA damage response: Signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front Plant Sci 10: 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P(2015) The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-box protein FBL17. Plant Cell 27: 1461–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A(2012) Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev Cell 22: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Nurse P.(2000) A long twentieth century of the cell cycle and beyond. Cell 100: 71–78 [DOI] [PubMed] [Google Scholar]
- Ogita N, Okushima Y, Tokizawa M, Yamamoto YY, Tanaka M, Seki M, Makita Y, Matsui M, Okamoto-Yoshiyama K, Sakamoto T, et al. (2018) Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J 94: 439–453 [DOI] [PubMed] [Google Scholar]
- Parameswaran B, Chiang HC, Lu Y, Coates J, Deng CX, Baer R, Lin HK, Li R, Paull TT, Hu Y(2015) Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle 14: 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Britt AB(2003) A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 164: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montané MH, Houlné G, Chabouté ME(2009) Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman P, Bekker-Jensen S, Mailand N(2016) Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol 17: 379–394 [DOI] [PubMed] [Google Scholar]
- Schwob E, Böhm T, Mendenhall MD, Nasmyth K(1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Sjogren CA, Bolaris SC, Larsen PB(2015) Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell 27: 2501–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampinato CP.(2017) Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell Mol Life Sci 74: 1693–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina NG, Kipreos ET(2012) Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends Cell Biol 22: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A, Tainer JA(2018) The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem 87: 263–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolet A, Baldrich P, Criqui M-C, Dubois M, Clavel M, Meyers BC, Genschik P(2019) Cell cycle-dependent regulation and function of ARGONAUTE1 in plants. Plant Cell 31: 1734–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Abeel T, Heyndrickx KS, Van Leene J, Lanz C, Van De Slijke E, De Winne N, Eeckhout D, Persiau G, Van Breusegem F, et al. (2014) A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol 164: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes C-L, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L(2005) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D(2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D, et al. (2016) The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J 35: 2068–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH, Li JM, Wang J, Gao Y, Han F, et al. (2012) Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol Cell 46: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama K, Larkin J, Van den Daele H, et al. (2014) The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K, Conklin P, Huefner ND, Britt AB(2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO.(2016) SOG1: A master regulator of the DNA damage response in plants. Genes Genet Syst 90: 209–216 [DOI] [PubMed] [Google Scholar]
- Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M(2013) ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 14: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Harashima H, Dissmeyer N, Pusch S, Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B, et al. (2012) A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet 8: e1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]