Thousands of lncRNAs with ecotype-specific expression, including two that likely regulate primary root growth, are potentially linked to the evolution of regulatory mechanisms among ecotypes.
Abstract
Root architecture varies widely between species; it even varies between ecotypes of the same species, despite strong conservation of the coding portion of their genomes. By contrast, noncoding RNAs evolve rapidly between ecotypes and may control their differential responses to the environment, since several long noncoding RNAs (lncRNAs) are known to quantitatively regulate gene expression. Roots from ecotypes Columbia and Landsberg erecta of Arabidopsis (Arabidopsis thaliana) respond differently to phosphate starvation. Here, we compared transcriptomes (mRNAs, lncRNAs, and small RNAs) of root tips from these two ecotypes during early phosphate starvation. We identified thousands of lncRNAs that were largely conserved at the DNA level in these ecotypes. In contrast to coding genes, many lncRNAs were specifically transcribed in one ecotype and/or differentially expressed between ecotypes independent of phosphate availability. We further characterized these ecotype-related lncRNAs and studied their link with small interfering RNAs. Our analysis identified 675 lncRNAs differentially expressed between the two ecotypes, including antisense RNAs targeting key regulators of root-growth responses. Misregulation of several lincRNAs showed that at least two ecotype-related lncRNAs regulate primary root growth in ecotype Columbia. RNA-sequencing analysis following deregulation of lncRNA NPC48 revealed a potential link with root growth and transport functions. This exploration of the noncoding transcriptome identified ecotype-specific lncRNA-mediated regulation in root apexes. The noncoding genome may harbor further mechanisms involved in ecotype adaptation of roots to different soil environments.
Over the last decade, genome-wide transcriptomics has revealed that a large intergenic part of eukaryotic genomes is transcribed. These transcripts, globally known as noncoding RNAs (Ariel et al., 2015), can regulate genome expression at transcriptional, posttranscriptional, and epigenetic levels, and are generally classified as small (21–24 nucleotides [nt]), long (>200 nt, <100 kb), and circular noncoding RNAs. Plant small RNAs (sRNAs) are produced by processing longer noncoding transcripts that generally contain a hairpin structure or lead to double-strand RNA formation. Plant sRNAs include microRNAs (miRNAs), endogenous small interfering RNAs (siRNAs; generally 21–22 nt long), and, most abundantly, heterochromatin siRNA (24 nt long; Borges and Martienssen, 2015). On the other hand, long noncoding RNAs (lncRNAs) are a heterogeneous group of RNA molecules with a coding capacity <50 amino acids (Chekanova, 2015). lncRNA transcripts are generally polyadenylated and can be intergenic (lincRNAs), intronic, or natural antisense (NATs) with respect to protein-coding genes (Ariel et al., 2015). When compared to mRNAs, lncRNAs are expressed at low levels in a tissue-specific manner or in response to environmental stresses (Liu et al., 2012) and are more frequently accumulated in the nucleus (Derrien et al., 2012), where they can regulate nuclear organization or function (Ariel et al., 2015).
lncRNAs utilize both cis- and trans-modalities of action to regulate gene expression through interactions with ribonucleoproteins and can form scaffolds and/or sequester proteins or RNA molecules as decoys or sponges. However, molecular functions have only been identified for a few lncRNAs in plants. As lncRNA genes lack regions with high primary sequence constraints (Derrien et al., 2012), it is difficult to use sequence conservation to identify potential functions. Even though the sequences of lncRNAs are not particularly conserved between plant species, they may show relative positional conservation in genomes (Mohammadin et al., 2015).
Finally, lncRNAs could be simply transcriptional by-products; in this framework, the sole act of their transcription rather than their sequence per se would be the source of the regulatory activity (Kopp and Mendell, 2018).
Resequencing approaches in model species have allowed the determination of whole-genome variations and evolution, from which it has been possible to provide the characterization of pan-genomes composed of “core” genomes (present in all accessions) and “dispensable” genomes (those specific to two or more accessions or even unique sequences specific to only one accession). Core genes are frequently highly expressed whereas dispensable genes are variably expressed, and generally in a tissue-specific manner (Contreras-Moreira et al., 2017). The dispensable genomes may play important roles in the capacity of individual organisms to cope with environmental conditions (Vernikos et al., 2015). Indeed, identification of natural variations in large worldwide populations (accessions) of Arabidopsis (Arabidopsis thaliana) showed an average of one SNP per 10 bp more frequently located in intergenic regions than in coding mRNAs (The 1001 Genomes Consortium, 2016). This has also been observed recently in rice (Oryza sativa), for which only 3.5% of SNPs and 2.5% of small insertion-deletions (InDels) were located in coding regions (Zhao et al., 2018). This latter observation would explain why lncRNAs differ even between closely related plant species (Nelson et al., 2017). In plants, three mechanisms have been proposed for the origin of lncRNAs: evolution from transposable element (TE) sequences, pseudogenization of protein-coding gene sequences, or duplication of preexisting lncRNAs (Kapusta and Feschotte, 2014)
The inorganic phosphate (Pi) accumulated in the upper soil layer is perceived by plants at the root apex (Svistoonoff et al., 2007). Accessions of ecotypes Columbia (Col) and Landsberg erecta (Ler) of Arabidopsis display different primary root growth and architecture in response to Pi starvation (Reymond et al., 2006). The identification of LOW PHOSPHATE ROOT1 (LPR1), a major quantitative trait locus, has been done in recombinant inbred lines obtained by crosses of accessions presenting this opposite root response to low Pi and it has been linked to differential expression of LPR1 in root apexes (Reymond et al., 2006; Svistoonoff et al., 2007). When the primary root tip of a Col seedling encounters a low-Pi medium, cell elongation in the transition zone rapidly decreases and cell proliferation in the root apical meristem (RAM) progressively ceases as callose deposition occurs in RAM plasmodesmata (Müller et al., 2015; Abel, 2017; Gutiérrez-Alanís et al., 2018). Root growth inhibition in low Pi depends on iron (Fe) availability in soil or media (Svistoonoff et al., 2007; Ward et al., 2008), as Fe concentrations clearly increase in Col plants during Pi starvation (Misson et al., 2005; Hirsch et al., 2006; Baxter et al., 2008). Indeed, inhibition of cell elongation and the RAM arrest are Fe-dependent (Svistoonoff et al., 2007; Ward et al., 2008; Müller et al., 2015; Abel, 2017; Balzergue et al., 2017; Mora-Macías et al., 2017; Gutiérrez-Alanís et al., 2018). Interestingly, in low-Pi conditions, Fe accumulates in the elongation zone, but not in the RAM, and more generally, in Col plants, Fe is redistributed among tissues (Mora-Macías et al., 2017; Gutiérrez-Alanís et al., 2018). By contrast, in Ler seedlings that are subject to low Pi, elongation and proliferation of root cells in the root apex continue, thereby sustaining root growth (Reymond et al., 2006). The corresponding regulatory system controlling root inhibition involves LPR1, PHOSPHATE DEFICIENCY RESPONSE2 (PDR2), SENSITIVE TO PROTON RHIZOTOXICITY1(STOP1), and ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (ALMT1). The LPR1-PDR2 and STOP1-ALMT1 modules allow Fe accumulation in roots under low Pi (Ticconi et al., 2009; Abel, 2017; Balzergue et al., 2017; Gutiérrez-Alanís et al., 2018). From these results, one concludes that interactions between Pi and Fe determine the differential growth response of Col and Ler ecotypes. In this work, we identified and characterized the noncoding transcriptomes of Col and Ler root apexes during early Pi starvation responses. Thousands of Arabidopsis lncRNAs, notably in the Ler accession, were identified, with only a minor fraction linked to sRNA production. Several “ecotype-specific” or “ecotype-enriched” variants were highly conserved at the DNA level and showed expression variation correlated with changes in the expression of key regulators of the Pi-starvation response. Functional analysis of five lncRNAs in Col revealed two further regulators of primary root growth, allowing us to hypothesize that lncRNA expression patterns contribute to the modulation of environmental responses in different ecotypes.
RESULTS
Col and Ler Root Tip Transcriptome Assemblies
We characterized the root-tip transcriptome of Col and Ler ecotypes, which present contrasting root phenotypes in response to Pi deficiency (Reymond et al., 2006). We performed comparative whole-genome transcriptomic analyses using paired-end sequencing of three biological replicates of root tips during a short kinetics (0, 1, and 2 h) of low (10 μm) Pi treatment (Supplemental Table S1). To avoid possible differences related to the erecta mutation present in the Ler ecotype, we used the Coler105 mutant here. For each ecotype, the reads were independently mapped to their reference genome (Supplemental Fig. S1A): TAIR10 for Col (Lamesch et al., 2012) and Ler v7 for Ler (Gan et al., 2011) that shared the same TAIR10 annotation (unlike Ler v8; Zapata et al. [2016]). We predicted previously unannotated transcripts by comparing our data to TAIR10. The homology of these predicted transcripts in Col and Ler was determined by mapping them onto the other genome. We retained as transcripts only RNA molecules of at least 200 nt. When these previously uncharacterized transcripts overlapped with pre-existing annotations, fusions were generated. Transcripts identified by this pipeline (Supplemental Fig. S1A) were compared with those from different Arabidopsis databases: Araport 11 (Cheng et al., 2017), RepTas (Liu et al., 2012), CANTATAdb (Szcześniak et al., 2016), miRBase v21 (Kozomara and Griffiths-Jones, 2014), and with those from two previous studies concerning lncRNAs (Ben Amor et al., 2009; Li et al., 2016). Finally, we used COME software to determine the potential coding capacity of identified transcripts (Hu et al., 2017). On the basis of both database information and COME predictions we classified the corresponding genes as coding or noncoding.
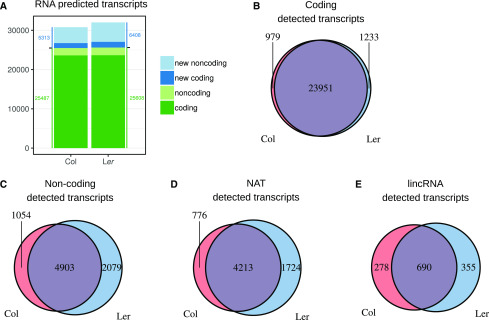
In total, we identified 5,313 and 6,408 previously uncharacterized putative genes in Col and Ler ecotypes, respectively (Fig. 1A; Supplemental Table S2; Supplemental Files S1 and S2). In root apexes, these identified genes were predominantly noncoding RNAs: 76% and 77% of the total previously uncharacterized genes in the case of Col and Ler, respectively (Fig. 1A; Supplemental Fig. S1, B and C). As expected, noncoding genes were globally less expressed than coding genes (Supplemental Fig. S2, A and B). Genes specifically detected in one ecotype belong much more often to the noncoding (>40% of the total noncoding genes) than to the coding class (<8% of the total coding genes; Fig. 1, B and C), notably true for lincRNA genes (52% of the total lncRNAs) as compared to NATs (34% of the total NATs; Fig. 1, D and E). Overall, expression of noncoding genes is more ecotype specific than that of coding genes.
Figure 1.
Identification of the transcripts and their occurrence across the two ecotypes. A, Number of predicted coding and noncoding transcripts in the two ecotypes, classified by type. New transcripts refers to genes not characterized in previously published studies. B to E, Predicted transcripts in each ecotype were classified as coding (B) or noncoding (C). For the latter case, two subclasses are defined: antisense of another annotation (NAT; D) and intergenic (lincRNA; E). In contrast to coding genes, many noncoding RNAs, notably lincRNAs, were detected only in one ecotype despite the high DNA sequence similarity in both ecotypes.
We detected a greater number of previously uncharacterized genes in the Ler ecotype (Fig. 1, A and C). Such differences do not result from library sequencing saturation. Indeed, in the last 2% of sequencing reads, <10 additional genes of this type were detected (Supplemental Fig. S2C). Thus, sequencing was deep enough to detect expressed genes, and the difference in gene detection between Col and Ler does not result from a sequencing bias.
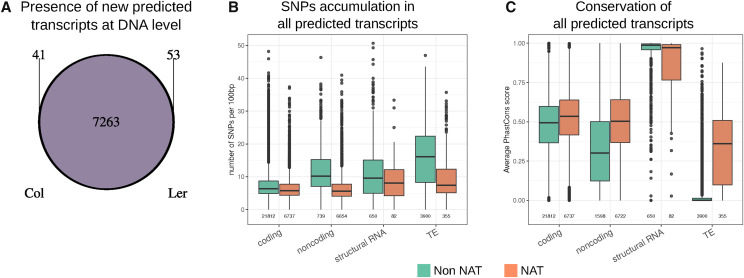
We next sought to determine whether these previously uncharacterized detected genes expressed in Ler (coding or noncoding) could correspond to specific parts of the Ler genome missed or rearranged in the Col genome. Out of these 7,357 genes, only 41 and 53 genes in Col and Ler, respectively, coincided with missing DNA sequences in the other ecotype (Fig. 2A), showing that the DNA sequence of the different previously uncharacterized genes is largely conserved apart from a few SNPs. Thus, the ecotype differences in transcript accumulation came from a shift in transcription that could be due to the deregulation of gene regulators, the accumulation of small sequence differences in promoters, or to specific differences in epigenetic status in the lncRNA-producing region due to TE insertions or other rearrangements possibly at large distance from the differentially expressed loci.
Figure 2.
Characterization of transcripts at the DNA level. A, Detection of the DNA sequences of previously uncharacterized predicted transcripts in the two ecotypes (minimum of 90% sequence identity along 90% of the RNA length). The large majority of RNAs come from common DNA regions from both ecotypes. B, SNP accumulation per 100 bp of transcript length for each type of transcript according to data from The 1001 Genomes Project (The 1001 Genomes Consortium, 2016). C, Conservation among plant species (average PhasCons score) of each type of transcript according to genomic position in relation to other annotations. In B and C, Non-NAT refers to transcripts which do not overlap with annotations on the other DNA strands, independent of annotation type (coding, noncoding, and structural RNA or TE).
Evolutionary Analysis of lncRNA Genes Expressed in Root Tips
Mechanisms modifying root architecture result from local signaling that occurs at the root tip (Svistoonoff et al., 2007; Thibaud et al., 2010; Müller et al., 2015; Balzergue et al., 2017). We therefore characterized the Arabidopsis genes expressed in the root apex, taking advantage of the extensive sequence information in Arabidopsis accessions (The 1001 Genomes Consortium, 2016). According to current annotations, genes were considered as non-NAT (no gene on the other strand) or NAT (presence of a gene on the other strand). For Arabidopsis species, we calculated the rate of SNPs accumulated in the different types of genes among all accessions (Fig. 2B). As expected, TEs accumulated many more SNPs than coding genes, whereas non-NAT lncRNAs and structural RNA genes showed an intermediate level of SNPs between TEs and coding genes. By contrast, the amount of SNPs was generally similar for NAT lncRNA and coding genes, as can be justified by the fact that the coding regions are under strong selection pressure.
To investigate sequence evolution at a larger scale, we used the PhastCons score that represents an interspecies level of nt conservation (normalized between 0 and 1) according to the alignment of 20 angiosperm genomes (Fig. 2C; Hupalo and Kern, 2013). As expected, structural RNA genes were strongly conserved (median score of 1), whereas TEs were not (median score of 0). Coding genes presented a score between these two extremes (∼0.5). Interestingly, non-NAT lncRNA genes showed an intermediate score between those of coding and transposable genes (median score of ∼0.3), whereas NAT lncRNA genes again showed the same degree of conservation as coding genes. These observations further suggest that NAT lncRNA genes are strongly constrained, whereas intergenic noncoding genes allow more variability even though they are more constrained than TEs.
Few lncRNA Transcripts Colocalize with Small RNA-Generating Loci
In animals and plants, some lncRNA loci colocalize with regions producing sRNA molecules (Matzke and Mosher, 2014). Therefore, we asked whether the lncRNA loci identified could generate sRNAs. Using similar samples previously prepared for the lncRNA studies, we prepared sRNA libraries for each ecotype and sequenced sRNAs to obtain a full description of the sRNAome mapped on each ecotype genome. Only a minority of the lncRNAs accumulate sRNAs, but of those, most contained sequences capable of generating nonphased RNA molecules of 21/22 or 24 nt (Supplemental Fig. S3, A and B) and only a small fraction of lncRNAs overlapped with phased siRNAs or were miRNA precursors.
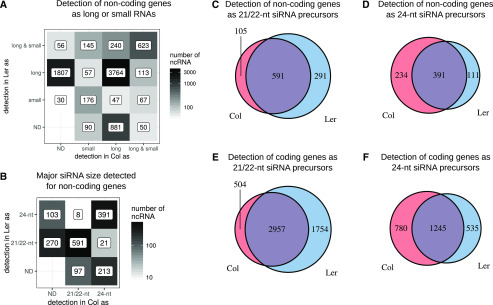
We then analyzed the potential link between siRNAs and lncRNAs in each ecotype. The majority of lncRNAs (6,452 genes of 7,850 detected) did not lead to accumulation of siRNAs. This is also true for the lncRNAs specifically detected only in one ecotype (2,688 genes out of 3,110 ecotype-specific genes), since many of them did not generate any siRNA in either ecotype (long in Col and not detected [ND] in Ler, or vice versa; Fig. 3A). Thus, the differential detection of lncRNA between ecotypes could not be linked to a change in the processing of siRNA by the encoding lncRNA loci.
Figure 3.
LncRNAs as sRNA precursors. The major specificity difference between Col and Ler is the lncRNA component of the transcriptome. A, Identification of noncoding transcripts as siRNAs or long RNAs. B, Distribution of the major siRNA sizes for noncoding transcripts detected as long in the two ecotypes. There is no major change of siRNA size between the two accessions. C, Detection of noncoding RNAs as 21-nt and 22-nt siRNA precursors. D, Detection of noncoding RNA as 24-nt siRNA precursors. E, Detection of coding RNA as 21-nt and 22-nt siRNAs precursors. F, Detection of coding RNA as 24-nt siRNAs precursors. Detection threshold for small RNA set at 1 read per million. ND, Not detected; ncRNA, noncoding RNA.
We then wondered whether different sRNA processing by lncRNAs could occur between the two ecotypes. We first looked at lncRNAs that accumulated sRNAs in only one accession: (1) lncRNAs that could generate siRNAs in only one ecotype while being detected as lncRNAs in both ecotypes (long in Ler and long and small in Col [113 genes], and vice versa [240 genes]); and (2) loci that produce siRNAs only in one ecotype yet are detected as lncRNAs only in the other (long in Ler and small in Col [57 genes], or vice versa [47 genes]). It is known that 21/22-nt siRNAs act on gene transcripts, whereas 24-nt siRNAs mediate chromatin modifications (Matzke and Mosher, 2014). Thus, a difference in the size of accumulated siRNAs for a given gene in one ecotype could indicate a modification of posttranscriptional (21/22 nt) or epigenetic (24 nt) regulation in the other ecotype. Among the lncRNA genes accumulating siRNA, a large portion accumulated the same size in both ecotypes (591 for 21/22-nt siRNA and 391 for 24-nt siRNA) or produced siRNA in just one ecotype (367 for 21/22-nt and 316 for 24-nt; Fig. 3B). Among the 1,694 lncRNA genes accumulating siRNAs, only 29 accumulated a different size of siRNAs between the two ecotypes. Therefore, no major change of reciprocal posttranscriptional or transcriptional regulation of lncRNA by sRNAs could be established between ecotypes.
Finally, we investigated the specificity of detection of sRNAs between the two ecotypes. First, we studied lncRNA genes predicted to produce phased 21/22-nt siRNAs. Among the seven predicted lncRNAs, only two were specific to Ler (Supplemental Fig. S3C). Second, searching for miRNA loci, we found that 23 and 12 of the 191 detected miRNAs were specifically detected in Col and Ler, respectively (Supplemental Fig. S3D), a proportion related to the variation detected for protein-coding genes. Third, we analyzed the proportion of specific expression for the vast majority of 21/22-nt and 24-nt siRNAs located in coding or noncoding genes. Altogether, the Ler ecotype produces a larger number of 21/22-nt siRNAs specifically linked to this ecotype (Fig. 3, C and E), whereas Col is more enriched in ecotype-specific 24-nt siRNAs (Fig. 3, D and F), suggesting that in these loci, links with differential posttranscriptional and epigenetic regulations among ecotypes occurred.
Overall, the major difference in the noncoding transcriptome of the two ecotypes was linked to lncRNAs and not associated with small RNAs, even though in certain cases sRNAs may be involved in ecotype-specific regulation.
Differential Accumulation of Transcripts between Ecotypes in Early Response to Pi Deficiency
Root growth arrest in the Col ecotype occurs in the first hours of low-Pi sensing by the root tip (Balzergue et al., 2017), whereas root growth continues in Ler. To determine the effects of short kinetics in Pi deficiency, we examined gene expression patterns in the two ecotypes in response to this stress. Principal component analysis (PCA) showed a data dispersion that allowed a clear distinction between effects of the ecotype (first axis; Supplemental Fig. S4A) and of the kinetics (second axis; Supplemental Fig. S4A). Thus, we used a multifactor analysis that takes into account the ecotype, the kinetics, and their interaction to investigate differential gene expression independent of coding classification, as coding and noncoding genes had comparable dispersion in our experiments. For each comparison, we confirmed the distribution of P-values as a criterion of statistical robustness (Rigaill et al., 2018). After processing the differential analyses, we interpreted the results by separating the genes as “coding” or “noncoding” as defined above.
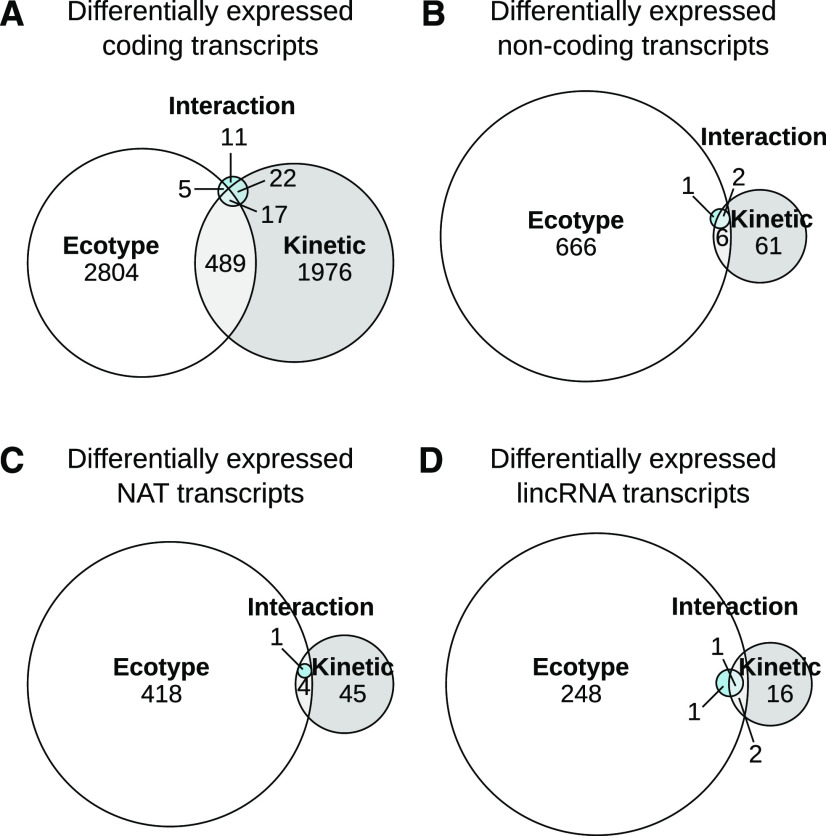
For coding genes, we observed 3,315 genes differentially expressed between the two ecotypes over the kinetics, with 2,504 differentially expressed between at least two kinetics points over the two ecotypes (Fig. 4A; Supplemental Fig. S4B; Supplemental Table S3). The number of differentially expressed coding genes between ecotypes or along the stress kinetics was similar. However, the response to phosphate starvation was significantly impacted by ecotype in only 55 genes (“interaction” of the two factors; Fig. 4A). Upregulation was observed in 1,566 and 1,749 coding genes along the kinetics in Col and Ler, respectively (Supplemental Fig. S4B). Interestingly, a clear bias of expression between ecotypes could be observed for noncoding genes (Fig. 4B). Indeed, 675 (666 + 6 + 2 +1) noncoding genes were differentially expressed between the two ecotypes, whereas only 70 (61 + 6 + 2) were differentially expressed along at least one point of the kinetics. Comparable biases were observed for both classes of noncoding genes, lincRNAs and NATs (Fig. 4, C and D). Globally, 146 lincRNAs and 236 NATs were significantly upregulated in Col compared to Ler and 106 lincRNAs and 187 NATs in Ler compared to Col (Supplemental Fig. S4C).
Figure 4.
Differentially expressed genes according to ecotype and kinetic effects. Statistical analysis revealed differentially expressed genes between ecotypes and kinetics during phosphate starvation treatments for coding and noncoding genes. The differentially expressed genes can be grouped according to their significant link with genotype effect (different level between the two ecotypes), kinetic effect (differential between any pair of time points in the phosphate starvation kinetics), and the interaction of the two effects (showing differential expression in response to phosphate stress according to genotype). After determining the global distribution, genes were partitioned between coding (A) and noncoding (B) transcripts. Among noncoding genes, we sorted transcripts according to their being antisense to another annotation (C) or intergenic (D).
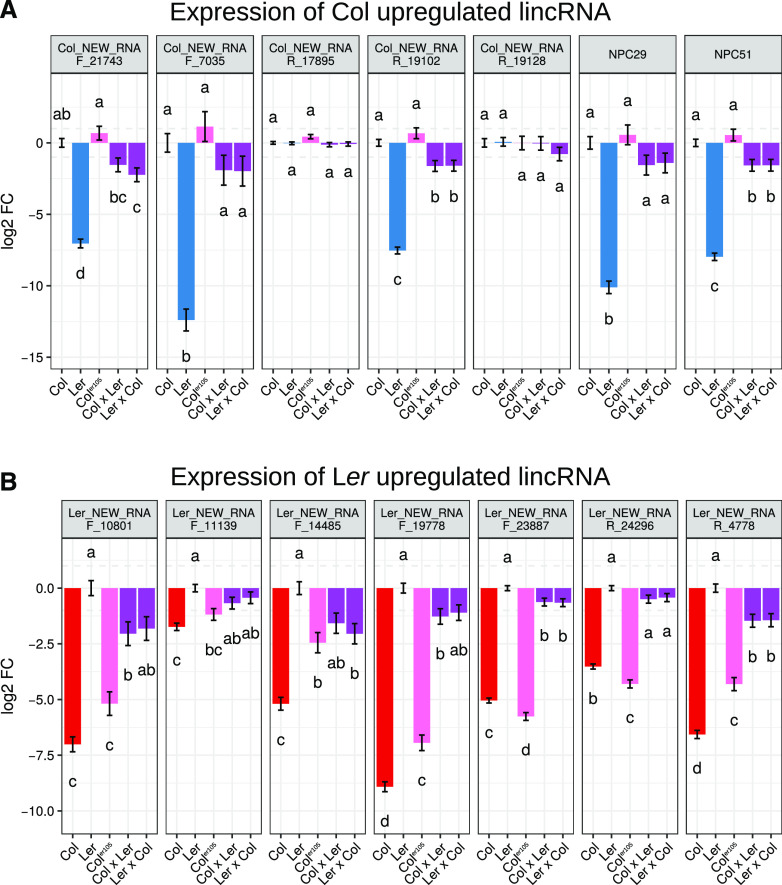
We used reverse transcription quantitative PCR (RT-qPCR) analysis of independent replicates of Col, Coler105, and Ler to confirm the differential expression of 14 lincRNA genes (seven in Col and seven in Ler) previously identified in the RNA-sequencing (RNA-seq) analysis. We were thereby able to confirm the differential expression of 12 lncRNAs (Fig. 5). Globally, Col and Coler105 showed similar expression levels despite minor differences. To investigate any dominant expression effect from one ecotype, we investigated the level of expression of these lncRNAs in the F1 offspring of Col and Ler crosses. Among the 12 differentially expressed genes, an intermediate level of expression was always detected (of which eight were statistically significant; Fig. 5). This suggests independent regulation of lncRNAs between the parental genomes and discards major dominant “trans” regulatory effects of lncRNA expression between genomes.
Figure 5.
Expression of strongly deregulated lncRNAs between the two ecotypes. The level of expression of strongly deregulated lncRNAs between ecotypes was investigated by RT-qPCR in roots of 11-d-old plants of Col, Ler, Coler105, and hybrids between Col and Ler grown under control conditions. A, LincRNAs upregulated in Col relative to Ler. B, LincRNAs upregulated in Ler relative to Col. Measurements represent the log 2-fold change (FC) compared to Col (A) or Ler (B) grown in the same high-phosphate conditions. Error bars represent the se (n = 8; for details, see Supplemental Table S5). Results were analyzed by one-way ANOVA followed by Tukey’s honestly significant difference (HSD) mean-separation test. Lowercase letters indicate statistical difference among groups (P ≤ 0.05).
One interesting possibility is that specific lncRNAs may be expressed in the Col and Ler genomes in relation to known regulators of the Pi-starvation response. As a first such case, we were able to identify two specific Ler antisense lncRNAs to the Pie transporter AT5G43370/PHT1.2 gene (Mudge et al., 2002), which is expressed at a higher level in Ler compared to Col (Supplemental Fig. S5A). The increase of Pi transporter expression in Ler might impart an increased Pi uptake. As a second case, we found that a Col-expressed NAT RNA is complementary to SPX4, a critical regulator of phosphate responses (Duan et al., 2008); in our analysis, it shows reduced expression in Col compared to Ler (Supplemental Fig. S5B). In other cases, we observed that two consecutive coding transcripts showing differential levels of expression among ecotypes flank a lincRNA with an ecotype-specific expression pattern (Supplemental Fig. S5C), suggesting that various cis effects may be involved in these differential ecotype-linked expression patterns.
Differential Accumulation of sRNAs between Ecotypes
The differential accumulation of sRNAs of 21/22and 24 nt was also examined in each ecotype and during the Pi starvation response. PCA of these sequencing data again clearly separated sRNA abundance between ecotypes but not at the level of the kinetics response (Supplemental Fig. S6, A and B; Supplemental Table S3). We identified 416 coding and 211 noncoding genes that accumulated 21/22-nt siRNAs differentially between ecotypes, with generally more siRNAs in Ler (298 coding genes, 83 lincRNAs, and 40 NATs) than in Col (118 coding genes, 49 lincRNAs, and 39 NATs; Supplemental Fig. S6D; Supplemental Table S3). A greater number of genes accumulated 24-nt siRNAs between ecotypes differentially, showing upregulated siRNAs in Ler (758 coding genes, 189 lincRNAs, and 67 NATs) compared to Col (391 coding genes, 104 lincRNAs, and 69 NATs; Supplemental Fig. S6E).
Concerning miRNA, as for other small RNAs, the PCA analysis showed only differences between ecotypes (Supplemental Fig. S6C). Indeed, 38 miRNAs were differentially expressed between the two ecotypes (15 and 23 for Col and Ler, respectively, Supplemental Fig. S6F; Supplemental Table S3). Interestingly, the families of miR399 and miR397 specifically accumulated in the Ler ecotype. These miRNAs target the PHOSPHATE2 (PHO2) and NITROGEN LIMITATION ADAPTATION (NLA) transcripts, the encoded proteins of which are known to act together to allow degradation of the Pi transporter PHT1;4 (Park et al., 2014). In Ler, the higher amount of miR399 and miR397 might be expected to lead to a lower level of PHO2 and NLA and therefore a higher level of PHT1;4 protein; upregulation could increase Pi uptake if there were no counteracting posttranslational regulations affecting Pi transporters (Bayle et al., 2011; Nussaume et al., 2011; Ayadi et al., 2015). However, we observed no difference between ecotypes in accumulation of transcripts of those targets, namely PHO2 and NLA, nor of PHT1;4 (as also previously reported; Shin et al., 2004; Ayadi et al., 2015). This result suggests that the promoter activity of PHO2 and NLA may compensate for the increased accumulation of these miRNAs in Ler.
Misregulation of lncRNA Expression Affects Primary Root Growth in Col
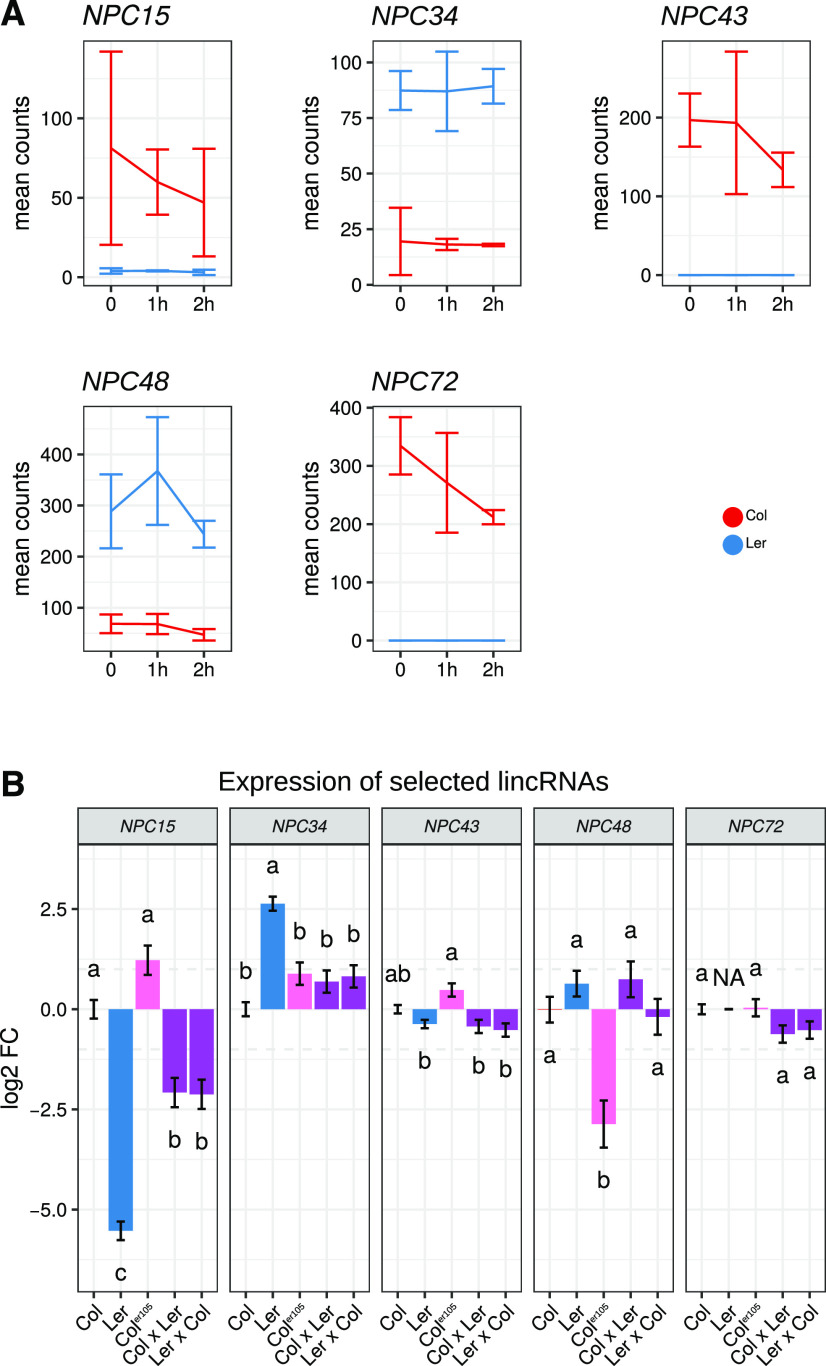
The different patterns of lncRNA expression between ecotypes may induce regulation of root-growth responses. We selected five lncRNA genes, NPC15, NPC34, NPC43, NPC48, and NPC72, that showed differential expression among ecotypes to study the impact of their expression on Col primary root growth. In RNA-seq data, three of these lncRNA genes were more highly expressed in Col (NPC15, NPC43, and NPC72) and two in Ler (NPC34 and NPC48; Fig. 6A). The potential dominant expression patterns of these lncRNA genes were evaluated in two F1s of Col × Ler reciprocal crosses. Expression analysis by RT-qPCR confirmed the RNA-seq results for NPC15, NPC34, and NPC72 genes (Fig. 6B), whereas for NPC48, the differential expression was only detected in the Coler105 mutant, suggesting that the erecta mutation affects the expression of this gene. No differential expression could be detected for the NPC43 gene. This latter result might be due to the concomitant accumulation of antisense transcripts (NPC504) in this locus. The differential expression of lncRNA genes between ecotypes could be linked to genetic changes at their loci. No significant modifications (except a few SNPs) were detected between Col and Ler for the NPC34, NPC43, and NPC48 loci (Supplemental Fig. S7). By contrast, the NPC15 locus contains an insertion of 2,417 nt in Ler v8 and NPC72 is completely missing in Ler v7 and v8 genomes (Supplemental Fig. S7, A–E). Therefore, genome modifications at the locus level could explain the specific expression pattern of NPC72 and NPC15 genes in Ler.
Figure 6.
Expression of selected lncRNAs in Col and Ler. A, Expression profiles of selected lincRNAs in Col and Ler for early Pi starvation kinetics (RNA-seq data, average expression ± sd, and three replicates). Five selected lincRNAs showed differential expression between Col and Ler at each time point. B, Level of expression of selected lincRNAs in roots of 11-d-old plants grown under high-Pi conditions in Col, Ler, Coler105, and hybrids between Col and Ler. Measurements represent corrected means of log 2-fold changes (FC) compared to Col measured by RT-qPCR. Error bars represent the se (n ≥ 7; for details, see Supplemental Table S5). Results were analyzed by one-way ANOVA followed by Tukey’s HSD mean-separation test. Lowercase letters indicate statistical difference among groups (P ≤ 0.05).
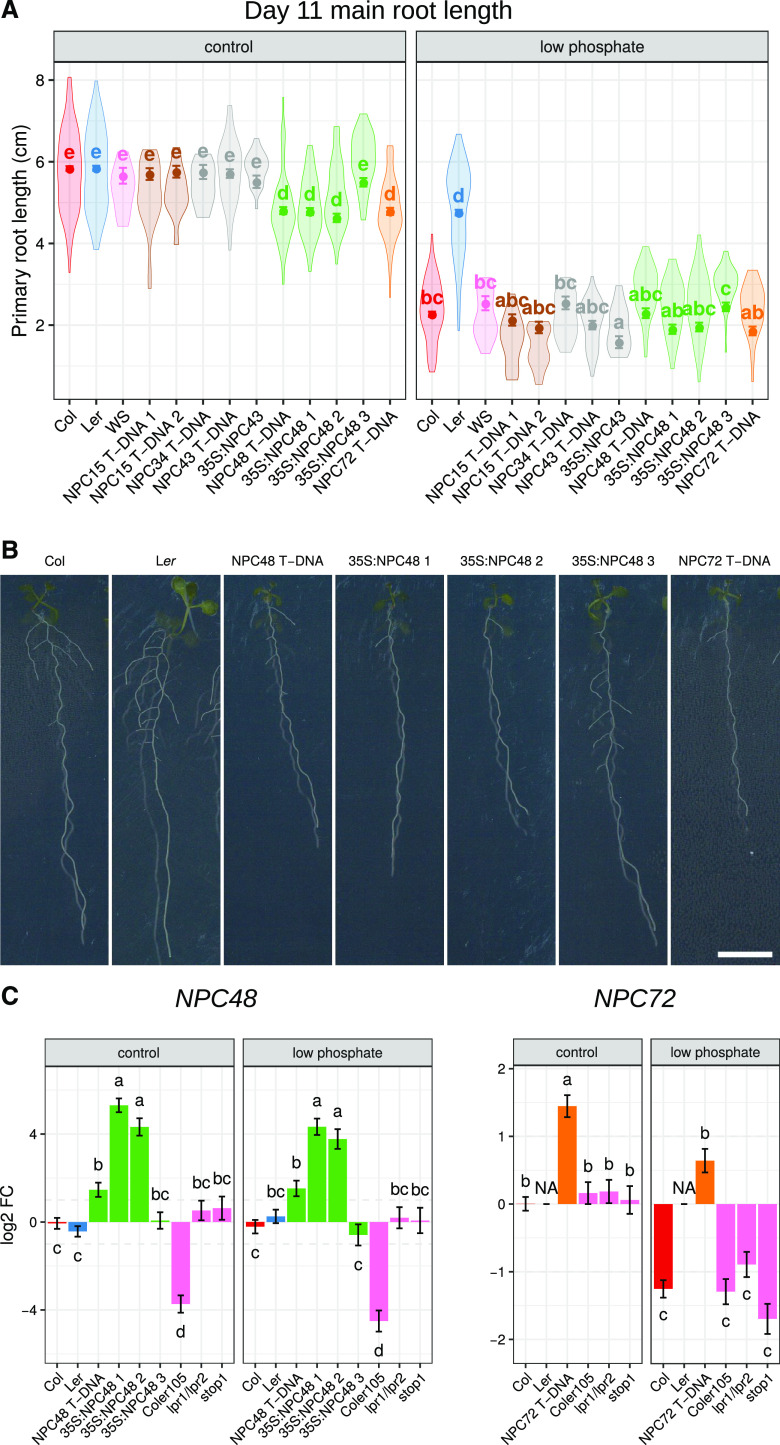
To support the potential actions of lncRNAs at a phenotypic level, we used overexpressing (35S promoter) and T-DNA insertion lines for the same five genes (NPC15, NPC34, NPC43, NPC48, and NPC72) in Col to monitor the effects on root growth in control and low-Pi conditions. In control conditions, only NPC48 and NPC72 overexpression lines led to significant root growth reduction compared to Col (Fig. 7, A and B; Supplemental Fig. S8, A–E). The T-DNA insertions have been mapped in the 5′ region of the NPC48 and NPC72 loci. In these lines, the lncRNAs were overexpressed (Fig. 7C). Furthermore the npc48 T-DNA line strongly supported the phenotype observed with the 35S:NPC48 lines. Repeating the analysis in low-Pi conditions known to inhibit root growth in Col but not in Ler, we observed minor differences in root length across the different lines. A priori, the ratio of root growth in control and low-Pi conditions should highlight potential differences in Pi sensitivity of transgenic lines. This ratio was significantly increased for NPC48 and NPC72 lines compared to Col (Supplemental Fig. S8F).
Figure 7.
Overexpression of the lincRNAs NPC48 and NPC72 affects primary root growth. A, Mean primary root length according to genotype and Pi condition at the age of 11 d after sowing (n ≥ 23; for details, see Supplemental Table S6). B, Representative pictures of roots of each genotype 11 d after sowing under high-Pi conditions. Scale bar = 1 cm. C, Expression levels of NPC48 and NPC72 in roots of 11-d-old plants grown under high-Pi conditions in lines deregulated in NPC48 or NPC72 and mutants affected in Pi-related root arrest. Measurements represent the log 2-fold changes (FC) compared to Col (n ≥ 4; for details, see Supplemental Table S5). Measurements represent corrected means of primary root growth (A) or of the fold change compared to Col (C). Error bars represent the se. Results were analyzed by two-way ANOVA (A) or one-way ANOVA (C) followed by Tukey’s HSD mean-separation test. Lowercase letters indicate statistical difference among groups (P ≤ 0.05).
NPC48 and NPC72 deregulated lines presented a significant decrease in root length in control conditions, but not in low-Pi conditions. Perhaps even in control conditions the mutants act as if they are partially limited in Pi. Hence, we asked whether these phenotypes could be linked to a root growth arrest due to oversensitive perception of Pi starvation under control conditions or an alteration of Pi systemic sensing (which would affect Pi uptake). This does not seem to be the case, since (1) known Pi-starvation markers were not deregulated in roots under control conditions; and (2) these markers were induced in these lines to the same extent as in Col (Supplemental Fig. S9, A and B). Then, we investigated the local Pi signaling response, exploiting the genes LOW PHOSPHATE ROOT1 (LPR1) and LPR2 and the transcription factor STOP1, which are known to be locally involved in primary root growth arrest under low Pi (Svistoonoff et al., 2007; Ticconi et al., 2009; Müller et al., 2015; Balzergue et al., 2017; Mora-Macías et al., 2017). Expression analysis of NPC48 and NPC72 genes in lpr1/lpr2 and stop1 mutant lines revealed no significant variation of these lncRNA expression patterns (Fig. 7C). Reciprocally, no significant expression variation was detected for the LPR1/LPR2 pathway in NPC48 or NPC72 lines (LPR1, LPR2, STOP1, ALMT1, and MATE genes; Supplemental Fig. S9, C to G).
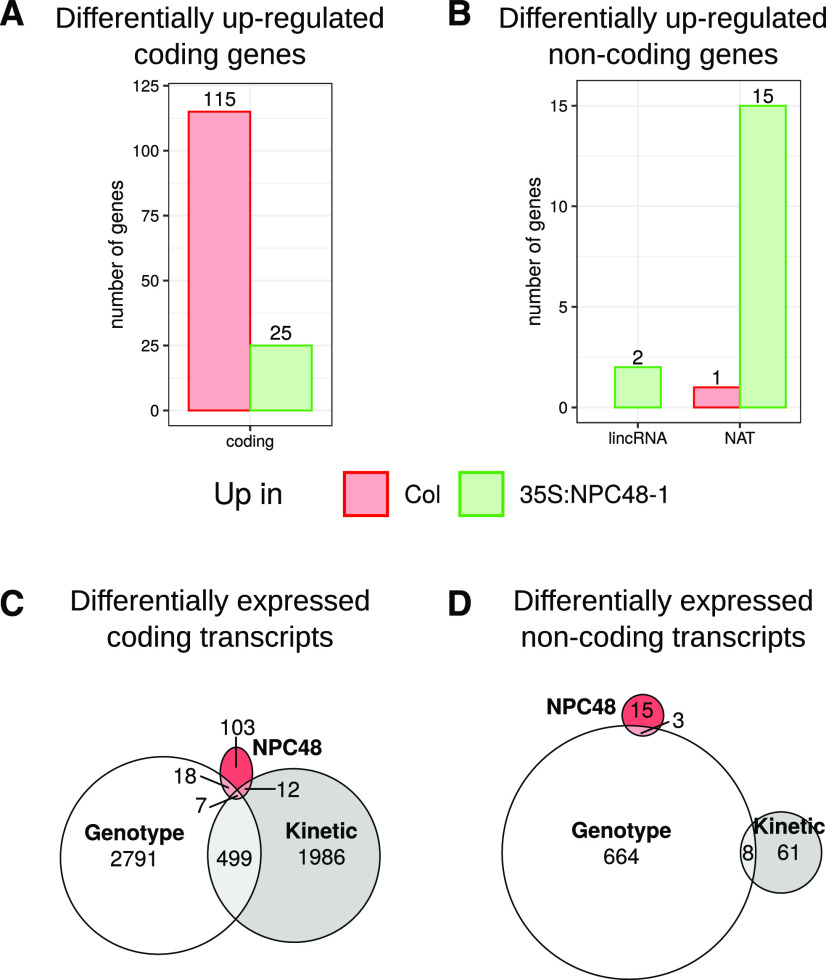
To gain further insight into the function of NPC48, we performed an RNA-seq analysis of Col ecotype overexpressing NPC48 (35S:NPC48-1) or not under control conditions (where the phenotype was observed) in order to assess the impact of NPC48 deregulation on the genome-wide transcriptome (Fig. 8). Among the 158 differentially expressed genes, 140 were coding genes, and the majority of these were upregulated in correlation with increased NPC48 expression (Fig. 8, A and B). In contrast, the great majority of noncoding genes were downregulated in 35S:NPC48-1, including two lincRNAs and 15 NATs, but none of their antisense coding genes. Since NPC48 was upregulated in the Ler ecotype, we asked whether, among the deregulated genes, several could be linked to ecotype- or phosphate kinetics-related variations (Fig. 8, C and D). However no such direct link could be made, as the large majority of deregulated genes are not linked to any of these categories. To further confirm the deregulation of specific targets, we also tested another 35S:NPC48 line showing an intermediate level of expression compared to 35S:NPC48-1 (Supplemental Fig. S10). Several genes could not be confirmed in this second line, notably the strongly downregulated gene in 35S:NPC48-1 encoding the iron-regulated gene AT3G01260 (Rodríguez-Celma et al., 2013). Among the genes differentially expressed, we could identify several linked to nutrient transport and root hair growth, such as ABCB3 (Shibata et al., 2018), CPL1 (Zhang et al., 2016), and JAL22 (Diet et al., 2006). ABCB3 and CPL1 were repressed by overexpression of NPC48, in contrast to JAL22 (Supplemental Fig. S10). There were also several genes encoding known growth regulators of primary root growth that might be linked to the NPC48 overexpression phenotype (Supplemental Table S8). These include RGF7, BIG, RPK2, and CASP5, which were upregulated. Differentially expressed genes are significantly enriched in genes regulated during iron starvation (25 of 138 genes; P = 1.62 × 10−3; Rodríguez-Celma et al., 2013). Hence, we tested, in the two different NPC48 overexpressing lines, whether overexpression of NPC48 might lead to iron-related phenotypes such as (1) modification of the expression of genes related to iron responses (Supplemental Fig. S11, A and B), (2) significant root growth in response to changes in the iron concentration of the growth medium (Supplemental Fig. S11, C and D), or (3) modification of iron accumulation in root tips (Supplemental Fig. S11, E and F). However, no clear link could be made with any alteration of Fe-related responses dependent on NPC48 misregulation.
Figure 8.
Differentially expressed genes in plants overexpressing NPC48. A and B, Numbers of coding (A) and noncoding (B) genes showing statistically different expression levels in RNA-seq data between Col and 35S:NPC48-1. C and D, Venn diagrams showing how differentially expressed coding (C) and noncoding (D) genes in 35S:NPC48-1 correlate with those between Col and Ler or those found during the phosphate starvation kinetics.
Altogether, the identification of ecotype-related lncRNAs allowed us to characterize further regulators of primary root growth. However, overexpression of NPC48 did not affect the Col transcriptome in a manner that could be linked to Ler expression patterns. Nevertheless, several genes related to transport, growth regulation, and root hair function are deregulated by misregulation of this ecotype-specific lncRNA.
DISCUSSION
Until recently, transcriptome studies were mainly focused on protein-coding gene transcripts and ignored lncRNAs. Variation in the nt sequences or expression patterns of the noncoding genome can have less pleiotropic effects than changes in the protein sequence of critical regulators. However, it is now commonly accepted that lncRNAs can play central roles in development and response to environmental conditions by their expression in a particular cell-type (Ariel et al., 2015). In the current study, using strand-specific RNA-seq analysis in root tips, we identified thousands of previously uncharacterized lncRNAs (lincRNAs and NATs) expressed at low levels from two Arabidopsis accessions. We focused on root growth, as it is a complex trait that is responsive to the soil environment (Petricka et al., 2012) and impacts a large number of loci spread across the genome. Interestingly, in our study, we observed many lncRNA genes that were differentially or specifically expressed in Col or Ler, in contrast to the number of protein-coding genes. It had already been shown in other systems that intraspecies variation is strongly linked to the noncoding part of the genome. For example, ∼45% of disease-associated human SNPs mapped to noncoding regions of the genome (Ning et al., 2017). In chicken (Gallus gallus), domestication traits governing body morphology or behavior are under selection and often associated with lncRNA genes (Wang et al., 2017). Similarly, in plants, the comparison of SNPs associated with fruit phenotypes in two tomato (Solanum lycopersicum) cultivars also corresponded to noncoding genomic regions (Scarano et al., 2017). The SNPs could act directly at the level of lncRNA expression or affect the expression of lncRNA-neighboring genes (Kopp and Mendell, 2018). lncRNAs are thus elements to be considered in genetic association studies.
Surprisingly, very few ecotype-specific lncRNAs coincided with the deletion of specific DNA sequences in one particular ecotype (Fig. 3). Hence, we propose that the lncRNA differential expression of a relatively similar DNA molecule results from shifts in transcription rate or stability of lncRNAs that could be connected to SNP or InDel polymorphisms in promoters and/or rearrangements distant from lncRNA loci in the two ecotypes (e.g. transposon insertions). As lncRNAs can repress or activate the transcription of other genes, the expression polymorphisms observed between the two ecotypes could also result in a cascade of cis-local or trans-distal action on target genes (Ariel et al., 2015; Marchese et al., 2017). It is noteworthy that the majority of ecotype-specific lncRNAs identified did not colocalize with siRNAs and thus could not reflect putative gene silencing differences between ecotypes (either transcriptional or posttranscriptional processes; Matzke and Mosher, 2014). This points to the lncRNA itself, or its transcription, being linked to the quantitative regulation of target gene expression (Marchese et al., 2017).
We were able to confirm, by RT-qPCR, the expression levels of 12 lincRNAs among the 14 chosen for validation, supporting the expression variation identified by RNA-seq (Fig. 5). Allele-specific expression is known to affect productivity in plants (Springer and Stupar, 2007). Moreover, in Arabidopsis, heterosis has been reported for different traits, such as flowering time (Seymour et al., 2016) or phosphate acquisition (Narang and Altmann, 2001). As lncRNAs are able to modify chromatin and thus alter gene expression, we added, in our expression analysis, the study of the F1 resulting from reciprocal crosses between Col and Ler. In the F1 hybrid, the 12 confirmed differentially expressed lncRNA genes chosen for validation globally exhibited an additive expression pattern compared to their parents. This is consistent with results obtained in maize (Zea mays) F1 hybrids, where additivity is frequently observed for lncRNAs (Li et al., 2014).
Analysis of the pan-genome (restricted to coding genes) of 19 Arabidopsis ecotypes showed that at least 70 accessory genes could be identified in each ecotype (Contreras-Moreira et al., 2017). In response to stress, accessory genes can explain at least part of the phenotypic difference of behavior observed among ecotypes (Gan et al., 2011). Since we find that lncRNAs have a lower selection pressure (Fig. 2), our results give further evidence that they might play a similar role as accessory coding genes in response to environmental changes.
Root apexes play an important role in sensing external stimuli. We examined the gene expression profile soon after stress application (at 1 and 2 h) in two ecotypes that present differences in response to Pi starvation. In the two ecotypes, the number of differentially expressed coding genes during stress kinetics was similar to that between ecotypes. By contrast, a clear bias of specific expression of lncRNAs and siRNAs was identified. However, among the coding genes that are differentially expressed along the Pi kinetics, one-quarter are also differentially expressed between the two ecotypes, whereas for the noncoding genes, this fraction is one-eighth (Fig. 4). This suggests that the main part of the ecotype-specific phosphate response comes from the coding portion of the genome. However, despite their lack of response to early stress application, lncRNAs may still influence the plant response to stress by priming the chromatin conformation for a fast response of the coding part of the genome.
The general adaptation of root architecture in response to low Pi consists of an arrest of primary root growth after the perception of Pi limitation. In Arabidopsis, studies concerning plant Pi homeostasis during Pi deficiency characterized the IPS1/AT4 lncRNA controlling the distribution of Pi from root to shoot. It acts as a target mimic for miR399, which regulates PHO2 mRNAs (Lin et al., 2008). Moreover, Yuan et al. (2016) identified lncRNAs differentially expressed in roots and shoots of plants grown in the presence or absence of Pi for 10 d. Those authors suggested that a coexpression between lncRNAs and adjacent coding genes may be linked to cis-regulation by lncRNAs of target genes involved in Pi-starvation processes. Interestingly, in fission yeast (Schizosaccharomyces pombe), two of the three genes of the Pi regulon are repressed in Pi-rich medium by the transcription of lncRNA genes (Shah et al., 2014) that are present in the 5′ region (cis-regulation). The molecular mechanisms that govern root growth modification by Pi have been mostly elucidated in Col plants. For the local impact of Pi (restricted to root architecture), Pi deficiency is sensed by the root tips and primary root growth inhibition is induced by both the reduction of cell elongation (STOP1 and LPR1/LPR2 pathways) and the progressive arrest of meristem division (LPR1/LPR2 pathway), notably linked to the presence of iron in the medium. Expression analysis in response to Pi deficiency in our mutant lines NPC48 and NPC72 did not link these two lncRNAs to Pi starvation root growth arrest mediated by LPR1/LPR2 and STOP1 pathways.
The overexpression of NPC48 leads to reduction of the main root growth. Transcriptome analysis of the strongest deregulated line (35S:NPC48-1) shows enrichment in genes that are deregulated during iron starvation (Rodríguez-Celma et al., 2013). However, no visible relation to iron homeostasis could be confirmed in the lines overexpressing NPC48. Apart from the link to iron starvation, no enrichment of any pathway could be demonstrated. Few identified deregulated genes could be confirmed in the second line, possibly because of the lower level of expression of NPC48 in this line or because of the specificity of the T-DNA insertions of both lines. Notably, the iron-regulated gene AT3G01260 was only deregulated in the 35S:NPC48-1 line. Few potential regulators of root growth or genes related to root hair growth and nutrient transport are modified. Nevertheless, we confirmed the presence of these latter genes in the second overexpressing line, supporting their link with NPC48 overexpression. Overexpression of NPC48 may decrease the absorption of essential nutrients, leading to a restriction of root growth by global nutrition deficiency. This lncRNA is a quantitative regulator of primary root growth, but its overexpression did not show any major alteration in the transcriptome. However, the overexpression did modify expression of several genes dealing with root growth or nutrient assimilation, and this is likely linked to its quantitative phenotype. Only a few core regulators of root growth rate have been identified up to now (Satbhai et al., 2015; Motte et al., 2019). Clearly, one can expect more subtle regulators to exist, and NPC48 might be one of them.
In Arabidopsis, using grafts between ecotypes presenting a high frequency of SNPs, Thieme et al. (2015) showed that about 2,000 mRNAs, among which 9,300 contain SNPs, could move in plants that were subjected to Pi deficiency for 2 weeks. These mRNAs were transported from root to shoot or shoot to root. The authors suggested that these mobile mRNAs might function widely as specific signaling molecules coordinating growth, cell differentiation, and stress adaptation of distant plant organs. As the lncRNAs described here have 3′ polyadenyylated tails and are probably 5′ capped, it is tempting to assume that at least some of them can be transported through the xylem and/or the phloem and may contribute to systemic signaling responses.
Globally, the in-depth exploration of the noncoding transcriptome of two ecotypes presented in this work identified thousands of previously uncharacterized lncRNAs with ecotype-specific expression. Statistical analysis among ecotypes identified several coregulations between coding and noncoding genes (including sRNAs). These coregulations are likely linked to the evolution of different regulatory mechanisms among ecotypes grown in diverse soil environments, and our detailed study of specific cases has provided two ecotype-related lncRNAs that are potentially involved in regulating primary root growth.
MATERIALS AND METHODS
Plant Growth
Seeds were surface-sterilized and sown on a horizontal line in plates vertically disposed in a growing chamber (16-h photoperiod; intensity 90 μE; 21°C). The growth medium was previously described in Balzergue et al. (2017). The −Pi and +Pi agar medium contained 10 and 500 μm Pi, respectively.
For the root apex isolation, seeds were sown on 1-cm bands of nylon membrane (Nitex 100 μm). After 1 week on +Pi agar medium, the membranes were transferred to −Pi agar medium. Plants were sampled at time points 0, 1, and 2 h after transfer. Each biological replicate is a pool of >100 root apexes cut at 0.5 cm from the root tip.
Arabidopsis Lines
The stop1 (SALK_114108, Nottingham Arabidopsis Stock Center [NASC] reference N666684), lpr1;lpr2 (Svistoonoff et al., 2007), npc15 T-DNA1 (SALK_027817; NASC reference N527817), npc15 T-DNA2 (SALK_090867; NASC reference N590867), npc43 T-DNA (SALK_007967; NASC reference N507967), npc48 T-DNA (SAIL_1165_H01; NASC reference N843057), and npc72 T-DNA (SAIL_571_C12; NASC reference N824316) lines are in the ecotype Columbia (Col) of Arabidopsis (Arabidopsis thaliana) background. Overexpressing lines 35S:NPC43, 35S:NPC48-1, 35S:NPC48-2, and 35S:NPC48-3 were retrieved from Ben Amor et al. (2009) and are in the Col-0 background. npc34 T-DNA (FLAG_223D08 or FLAG_228A07) is in the Wassilewskija background. Coler105 is in the Columbia background (Col-0) with the null allele erecta-105.
Library Construction and Sequencing
For each time point (0, 1, and 2 h), total RNA of three biological replicates of the Coler105 and Ler pool of root apexes were extracted following the RNeasy micro kit (Qiagen) protocol. One microgram of total RNA of each sample was used for mRNA library preparation using the Illumina TruSeq Stranded mRNA library preparation kit according to the manufacturer’s instructions. Libraries were sequenced on a HiSeq 2000 Sequencing System (Illumina) using 100-nt paired-end reads.
For 35S:NPC48-1 RNA-seq analysis, total RNA of three biological replicates of whole roots from Col and 35S:NPC48-1 were extracted using a Quick-RNA Miniprep kit (Zymo Research). One microgram of total RNA of each sample was used for mRNA library preparation using an Illumina Truseq Stranded mRNA library preparation kit according to the manufacturer’s instructions. Libraries were sequenced on a NextSeq 500 Sequencing System (Illumina) using 75-nt single-end reads.
sRNAs of root apexes were extracted using the mirVana miRNA Isolation Kit (Ambion). sRNA libraries were constructed using the Ion Total RNA-Seq Kit v2 (Ion Torrent, Life Technologies) according to the manufacturer’s instructions. Libraries were then sequenced using IonProton and the adapters.
Previously Uncharacterized Transcript Identification
According to their ecotype of origin, transcript reads were aligned to the TAIR10 (Lamesch et al., 2012) or Ler v7 (Gan et al., 2011) genome. The previously uncharacterized transcripts were predicted in each ecotype independently and then cross-positioned on the other genome to analyze homology. Using the information available on the Col genome from databases, the different transcripts were classified as coding and noncoding (lncRNAs), lncRNAs included lincRNAs and NATs. See the detailed protocol in Supplemental Materials and Methods.
sRNA Analysis
The cleaned sRNA reads were aligned on the TAIR10 or Ler v7 genome. For Araport11 annotations and previously uncharacterized genes predicted here, the accumulation of sRNA was analyzed to classify them as miRNA, phased siRNA cluster, or siRNA cluster and determine the main siRNA size. See detailed protocol in Supplemental Materials and Methods.
Expression Analysis
For each annotation, mRNA reads were counted to estimate the level of expression of each gene. These counts were used for differential gene expression analysis. Using siRNA accumulation on each annotation, differential accumulation of 21/22-nt and 24-nt sRNAs on coding and noncoding genes was computed independently. Bonferroni correction of the P-value was used for each analysis. Differentially expressed genes or siRNA accumulations were defined as having an adjusted P-value inferior to 0.01. See the detailed protocol in Supplemental Materials and Methods.
Measurement of the Primary Root Length
Images were taken with a flat scanner and root lengths were measured using RootNav software (Pound et al., 2013).
RT-qPCR
Total RNA was extracted from whole roots using the Quick-RNA MiniPrep kit (Zymo Research, USA) according to the manufacturer’s instructions. Reverse transcription was performed on 500 ng total RNA using the Maxima Reverse Transcriptase (Thermo Scientific). RT-qPCR was performed on a 480 LightCycler thermocycler (Roche) according to the manufacturer’s instructions with Light cycler 480 SYBR Green I Master (Roche) and the primers listed in Supplemental Table S4. We used PP2A subunit PD (AT1G13320) as a reference gene for normalization.
Statistics and Reproducibility of Experiments
For each measure (root length, qPCR expression level), the least-squares means were computed. This approach makes it possible to correct for interrepetition variation. Data are presented as least-squares means ± se. The statistical significance tests are included in the legend of each figure. See detailed protocol in Supplemental Materials and Methods.
Accession Numbers
Sequence files generated during this study have been deposited in the NCBI GEO database under the accessions GSE128250, GSE128256 and GSE151005. Names and accession numbers of genes mentioned are listed in Supplemental Table S9.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characteristics of identified transcripts.
Supplemental Figure S2. Expression level and detection of coding and noncoding genes.
Supplemental Figure S3. Ecotype-specific classification of lncRNAs as siRNA precursors.
Supplemental Figure S4. Ecotype effect on gene expression.
Supplemental Figure S5. Genome organization and correlation of expression at selected loci.
Supplemental Figure S6. Ecotype effect on siRNA accumulation.
Supplemental Figure S7. Genome homology at selected ncRNA loci
Supplemental Figure S8. Deregulation of selected noncoding RNAs.
Supplemental Figure S9. Deregulation of NPC48 and NPC72 does not change the expression of phosphate starvation-related genes.
Supplemental Figure S10. Expression analysis of genes regulated in the 35S:NPC48-1 line in a different independent transgenic line.
Supplemental Figure S11. Overexpression of NPC48 does not affect iron homeostasis and/or phenotypic responses.
Supplemental Table S1.Mapping efficiency for each sequence sample.
Supplemental Table S2. Genomic information of previously uncharacterized transcripts compared to TAIR10.
Supplemental Table S3. Differential gene expression analysis.
Supplemental Table S4. Sequence of primers used in this study.
Supplemental Table S5. Number of samples used for each genotype and condition in the RT-qPCR experiments.
Supplemental Table S6. Number of samples used for each genotype and condition in root-length measurements.
Supplemental Table S7. Software used for the bioinformatics analyses.
Supplemental Table S8. Differentially expressed genes in 35S:NPC48-1.
Supplemental Table S9. Gene name abbreviations.
Supplemental File S1. Identified Col transcripts.
Supplemental File S2. Identified Ler transcripts.
Supplemental Materials and Methods. Detailed bioinformatic and extra materials and methods.
Acknowledgments
We thank Ambre Miassod and Janina Lüders (Institute of Plant Sciences Paris-Saclay, Centre Nationale de la Recherche Scientifique, Institut National de la Recherche Agronomique, Université Evry, Université Paris-Saclay) for technical assistance with gene expression experiments. We thank Olivier Martin (Institute of Plant Sciences Paris-Saclay, Centre Nationale de la Recherche Scientifique, Institut National de la Recherche Agronomique, Université Evry, Université Paris-Saclay) for critical reading of the manuscript.
Footnotes
This work was supported by the Agence Nationale pour la Recherche (grant nos. ANR–12–ADAP–0019 [RNAdapt], ANR–16–CE12–0032 [SPLISIL], and ANR–17–EUR–0007 [Saclay Plant Sciences Graduate School of Research] managed under an Investments for the Future program [grant no. ANR–11–IDEX–0003–02]) and The King Abdulla University of Science and Technology (KAUST) International Program (grant no. OCRF–2014–CRG4).
References
- Abel S.(2017) Phosphate scouting by root tips. Curr Opin Plant Biol 39: 168–177 [DOI] [PubMed] [Google Scholar]
- Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M(2015) Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci 20: 362–371 [DOI] [PubMed] [Google Scholar]
- Ayadi A, David P, Arrighi J-F, Chiarenza S, Thibaud M-C, Nussaume L, Marin E(2015) Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol 167: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon J-M, Creff A, Bissler M, Brouchoud C, Hagège A, et al. (2017) Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun 8: 15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, Guerinot ML, Salt DE(2008) The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc Natl Acad Sci USA 105: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V, Arrighi J-FJ-F, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L(2011) Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B, Wirth S, Merchan F, Laporte P, d’Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, et al. (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA(2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA.(2015) Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD(2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Contreras-Moreira B, Cantalapiedra CP, García-Pereira MJ, Gordon SP, Vogel JP, Igartua E, Casas AM, Vinuesa P(2017) Analysis of plant pan-genomes and transcriptomes with GET_HOMOLOGUES-EST, a clustering solution for sequences of the same species. Front Plant Sci 8: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter WD, Ringli C(2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P(2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Ojeda-Rivera JO, Yong-Villalobos L, Cárdenas-Torres L, Herrera-Estrella L(2018) Adaptation to phosphate scarcity: Tips from Arabidopsis roots. Trends Plant Sci 23: 721–730 [DOI] [PubMed] [Google Scholar]
- Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC(2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Hu L, Xu Z, Hu B, Lu ZJ(2017) COME: A robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res 45: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalo D, Kern AD(2013) Conservation and functional element discovery in 20 angiosperm plant genomes. Mol Biol Evol 30: 1729–1744 [DOI] [PubMed] [Google Scholar]
- Kapusta A, Feschotte C(2014) Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet 30: 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT(2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172: 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S(2014) miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yamada M, Han X, Ohler U, Benfey PN(2016) High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell 39: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ(2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua N-H(2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Raimondi I, Huarte M(2017) The multidimensional mechanisms of long noncoding RNA function. Genome Biol 18: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA(2014) RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadin S, Edger PP, Pires JC, Schranz ME(2015) Positionally-conserved but sequence-diverged: Identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol 15: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Macías J, Ojeda-Rivera JO, Gutiérrez-Alanís D, Yong-Villalobos L, Oropeza-Aburto A, Raya-González J, Jiménez-Domínguez G, Chávez-Calvillo G, Rellán-Álvarez R, Herrera-Estrella L(2017) Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc Natl Acad Sci USA 114: E3563–E3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte H, Vanneste S, Beeckman T(2019) Molecular and environmental regulation of root development. Annu Rev Plant Biol 70: 465–488 [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW(2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S(2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33: 216–230 [DOI] [PubMed] [Google Scholar]
- Narang RA, Altmann T(2001) Phosphate acquisition heterosis in Arabidopsis thaliana: A morphological and physiological analysis. Plant Soil 234: 91–97 [Google Scholar]
- Nelson ADL, Devisetty UK, Palos K, Haug-Baltzell AK, Lyons E, Beilstein MA(2017) Evolinc: A tool for the identification and evolutionary comparison of long intergenic non-coding RNAs. Front Genet 8: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S, Yue M, Wang P, Liu Y, Zhi H, Zhang Y, Zhang J, Gao Y, Guo M, Zhou D, et al. (2017) LincSNP 2.0: An updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res 45(D1): D74–D78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC(2011) Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua N-H(2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN(2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound MP, French AP, Atkinson JA, Wells DM, Bennett MJ, Pridmore T(2013) RootNav: Navigating images of complex root architectures. Plant Physiol 162: 1802–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T(2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Rigaill G, Balzergue S, Brunaud V, Blondet E, Rau A, Rogier O, Caius J, Maugis-Rabusseau C, Soubigou-Taconnat L, Aubourg S, et al. (2018) Synthetic data sets for the identification of key ingredients for RNA-seq differential analysis. Brief Bioinform 19: 65–76 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, Schmidt W(2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satbhai SB, Ristova D, Busch W(2015) Underground tuning: Quantitative regulation of root growth. J Exp Bot 66: 1099–1112 [DOI] [PubMed] [Google Scholar]
- Scarano D, Rao R, Corrado G(2017) In silico identification and annotation of non-coding RNAs by RNA-seq and de novo assembly of the transcriptome of tomato fruits. PLoS One 12: e0171504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour DK, Chae E, Grimm DG, Martín Pizarro C, Habring-Müller A, Vasseur F, Rakitsch B, Borgwardt KM, Koenig D, Weigel D(2016) Genetic architecture of nonadditive inheritance in Arabidopsis thaliana hybrids. Proc Natl Acad Sci USA 113: E7317–E7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Wittmann S, Kilchert C, Vasiljeva L(2014) lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev 28: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Breuer C, Kawamura A, Clark NM, Rymen B, Braidwood L, Morohashi K, Busch W, Benfey PN, Sozzani R, et al. (2018) GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 145: dev159707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ(2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM(2007) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T(2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Szcześniak MW, Rosikiewicz W, Makałowska I(2016) CANTATAdb: A collection of plant long non-coding RNAs. Plant Cell Physiol 57: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1001 Genomes Consortium (2016) 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L(2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, et al. (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S(2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA 106: 14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G, Medini D, Riley DR, Tettelin H(2015) Ten years of pan-genome analyses. Curr Opin Microbiol 23: 148–154 [DOI] [PubMed] [Google Scholar]
- Wang YM, Xu HB, Wang MS, Otecko NO, Ye LQ, Wu DD, Zhang YP(2017) Annotating long intergenic non-coding RNAs under artificial selection during chicken domestication. BMC Evol Biol 17: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG(2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhang Y, Dong J, Sun Y, Lim BL, Liu D, Lu ZJ(2016) Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics 17: 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata L, Ding J, Willing E-M, Hartwig B, Bezdan D, Jiao W-B, Patel V, Velikkakam James G, Koornneef M, Ossowski S, et al. (2016) Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc Natl Acad Sci USA 113: E4052–E4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Yang G, Chen Y, Zhao Y, Gao P, Liu B, Wang H, Zheng ZL(2016) C-terminal domain (CTD) phosphatase links Rho GTPase signaling to Pol II CTD phosphorylation in Arabidopsis and yeast. Proc Natl Acad Sci USA 113: E8197–E8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, et al. (2018) Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50: 278–284 [DOI] [PubMed] [Google Scholar]