Alteration of the C terminus of 1-aminocyclopropane-1-carboxylic acid (ACC) Synthase 7 (ZmACS7), an ethylene biosynthetic protein, enhances the stability of this enzyme, thus increasing ACC and ethylene production, which in turn alters plant architecture in maize.
Abstract
Plant height and leaf angle are two crucial determinants of plant architecture in maize (Zea mays) and are closely related to lodging resistance and canopy photosynthesis at high planting density. These two traits are primarily regulated by several phytohormones. However, the mechanism of ethylene in regulating plant architecture in maize, especially plant height and leaf angle, is unclear. Here, we characterized a maize mutant, Semidwarf3 (Sdw3), which exhibits shorter stature and larger leaf angle than the wild type. Histological analysis showed that inhibition of longitudinal cell elongation in the internode and promotion in the auricle were mainly responsible for reduced plant height and enlarged leaf angle in the Sdw3 mutant. Through positional cloning, we identified a transposon insertion in the candidate gene ZmACS7, encoding 1-aminocyclopropane-1-carboxylic acid (ACC) Synthase 7 in ethylene biosynthesis of maize. The transposon alters the C terminus of ZmACS7. Transgenic analysis confirmed that the mutant ZmACS7 gene confers the phenotypes of the Sdw3 mutant. Enzyme activity and protein degradation assays indicated that the altered C terminus of ZmACS7 in the Sdw3 mutant increases this protein’s stability but does not affect its catalytic activity. The ACC and ethylene contents are dramatically elevated in the Sdw3 mutant, leading to reduced plant height and increased leaf angle. In addition, we demonstrated that ZmACS7 plays crucial roles in root development, flowering time, and leaf number, indicating that ZmACS7 is an important gene with pleiotropic effects during maize growth and development.
Maize (Zea mays) is one of the most widely cultivated cereal crops in the world. During the past six decades, the grain yield per unit area of maize worldwide exhibited a linear increase of 66.9 kg ha−1 yr−1 (FAOstat database, 1961−2018). This annual increase in maize yield per unit area is not attributed to increased yield potential per plant; instead, it is mainly due to increased planting density (Duvick, 2004). However, high planting density can heighten interplant competition for environmental resources, especially light, leading to increased shoot elongation at the expense of internode thickening and root development, thereby increasing plant susceptibility to lodging (Hébert et al., 2001; Xue et al., 2016). Moderately reducing plant height is an effective strategy for improving lodging resistance in maize grown at high density (Tang et al., 2007). The incredible increases in rice (Oryza sativa) and wheat (Triticum aestivum) production during the “Green Revolution” were largely due to the use of semidwarf genes (semidwarf1 in rice and Reduced height-B1b and Reduced height-D1b in wheat; Hedden, 2003). In addition, breeding strategies aimed at optimizing leaf angle could increase canopy photosynthesis and grain yield at high planting density (Pendleton et al., 1968; Tian et al., 2011).
Several plant hormones, including gibberellins (GAs), brassinosteroids (BRs), and auxin, play important roles in regulating plant height and/or leaf angle, as demonstrated in rice (Wang et al., 2018a). To date, many genes responsible for plant height in maize have been identified. Maize mutants defective in GA biosynthesis and signaling, including anther ear1, dwarf1 (d1), d3, Dwarf8 (D8), and D9, show various levels of plant height reduction (Bensen et al., 1995; Winkler and Helentjaris, 1995; Peng et al., 1999; Cassani et al., 2009; Teng et al., 2013). Several BR-deficient or insensitive maize mutants, such as nana plant1, nana plant2, and the BRASSINOSTEROID INSENSITIVE1 knockdown line, are characterized by severe dwarfism (Hartwig et al., 2011; Kir et al., 2015; Best et al., 2016), and nana plant1 and the BRASSINOSTEROID INSENSITIVE1 knockdown line also have erect leaves (Hartwig et al., 2011; Kir et al., 2015). Genes involved in polar auxin transport, such as Brachytic2 and ZmPIN1a, are also implicated in the genetic control of plant height in maize (Multani et al., 2003; Xing et al., 2015; Li et al., 2018; Wei et al., 2018). Moreover, some genes responsible for flowering time affect plant height in maize, largely due to variations in internode number, such as delayed flowering1, ZmRap2.7, ZmCCT, and indeterminate1 (Colasanti et al., 1998; Muszynski et al., 2006; Salvi et al., 2007; Yang et al., 2013). Furthermore, in maize production, ethephon (2-chloroethylphosphonic acid), an ethylene-releasing compound, is widely used to increase lodging resistance by reducing plant height and ear height (Langan and Oplinger, 1987).
In maize, the ligular region consists of a ligule and two wedge-like auricles. The ligule is an adaxial fringe that protects the axillary bud from water, debris, and predators, whereas the auricles serve as hinges between the leaf blade and leaf sheath, thereby controlling leaf angle (Moreno et al., 1997; Foster and Timmermans, 2009; Moon et al., 2013). liguleless1 (LG1) and LG2 are the most informative genes responsible for ligule and auricle development. The loss-of-function mutants lg1 and lg2 have upright leaves (Moreno et al., 1997; Walsh et al., 1998; Tian et al., 2011). Liguleless narrow, encoding a kinase, also regulates ligule and auricle development and leaf angle formation (Moon et al., 2013). Although many maize mutants related to plant architecture have been characterized and their underlying genes cloned, the role and regulatory mechanism of ethylene in plant architecture, especially plant height and leaf angle, are unclear.
Ethylene, a gaseous phytohormone, plays important roles in various physiological processes, including seed germination, cell expansion, adventitious root formation, sex determination, leaf abscission, flower fading, fruit ripening, and biotic/abiotic stress responses (Mattoo and Suttle, 1991). Ethylene is produced in essentially all parts of higher plants, and its biosynthesis begins with the conversion of Met to S-adenosyl-l-Met (SAM), a process catalyzed by SAM synthase. SAM is then converted into 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS). ACC is ultimately oxidized to ethylene in the catalysis of ACC oxidase (ACO; Yang and Hoffman, 1984). ACS, the key rate-limiting enzyme in this pathway (Yu et al., 1979), is encoded by a multigene family in every plant species investigated (Wang et al., 2002; Gallie and Young, 2004; Lin et al., 2009).
Arabidopsis (Arabidopsis thaliana) contains eight enzymatically active ACS isoforms (AtACS2, AtACS4 to AtACS9, and AtACS11) and an inactive isoform (AtACS1; Yamagami et al., 2003). These nine AtACS isoforms are divided into three types based on the hypervariable C-terminal sequence. Type I ACS isoforms (AtACS1, AtACS2, and AtACS6) have a long C-terminal domain (CTD), carrying three Ser residues that are phosphorylation sites for mitogen-activated protein kinase (MAPK) and a Ser residue that can be phosphorylated by calcium-dependent protein kinase. Type II isoforms (AtACS4, AtACS5, AtACS8, AtACS9, and AtACS11) have a relatively short CTD that includes only the calcium-dependent protein kinase phosphorylation site, and the Type III isoform (AtACS7) has the shortest CTD, which lacks both phosphorylation sites (Argueso et al., 2007; Luo et al., 2014). In general, ACS is present in low abundance in tissues (Sato and Theologis, 1989) and could be regulated transcriptionally and posttranslationally in response to various developmental, environmental, and hormonal factors (Yip et al., 1992; Spanu et al., 1994; Botella et al., 1995; Lelièvre et al., 1997; Chae et al., 2003; Luo et al., 2014; Lee et al., 2017). Increasing studies demonstrated that the CTD of ACS plays a pivotal role in posttranslationally regulating ACS stability (Joo et al., 2008; Choudhury et al., 2013; Xu and Zhang, 2015). In addition to phosphorylation sites, the CTD of ACS also carries the recognition sites for ACS degradation mediated by the ubiquitin-proteasome system, which could be impeded by phosphorylation of the CTD and promoted by dephosphorylation of the CTD (Yoshida et al., 2006; Joo et al., 2008; Han et al., 2010; Skottke et al., 2011; Ludwików et al., 2014; Xu and Zhang, 2015). Several studies on ethylene-overproducer (eto) mutants showed that CTD mutations of AtACS5 and AtACS9 in eto2 and eto3 mutants, respectively, resulted in increased ACS stability and ethylene production due to damage of protein interaction with ETO1, a component of E3 ligase, and its homologs ETO1-like 1 (EOL1) and EOL2 (Vogel et al., 1998; Chae et al., 2003; Wang et al., 2004; Christians et al., 2009; Xu and Zhang, 2015).
In maize, three ACS genes, ZmACS2, ZmACS6, and ZmACS7, were identified by PCR amplification from the genome of the commonly used maize inbred line B73 (Gallie and Young, 2004). It had been reported that ZmACS2 and ZmACS6 were involved in many physiological processes of maize, including leaf senescence, root growth in response to hypoxia and physical resistance presented by soils, and kernel development in response to pathogen infection (Young et al., 2004; Gallie et al., 2009; Geisler-Lee et al., 2010; Wang et al., 2017). However, little is known about the physiological roles of ZmACS7, especially in regulating plant architecture, due to the lack of available mutants (Young et al., 2004). In this study, we characterized a semidominant maize mutant, Semidwarf3 (Sdw3), which exhibits shorter stature and larger leaf angle than the wild type. Taking a map-based cloning strategy, we limited the target gene into an 8-kb interval on maize chromosome 10 and identified a transposon insertion into the only candidate gene, ZmACS7. The transposon insertion alters the C terminus of ZmACS7 and increases the stability of this protein but does not affect its catalytic activity. Thus, more ACC and ethylene are produced in the Sdw3 mutant than those in the wild type, leading to reduced plant height and enlarged leaf angle. In addition, we showed that ZmACS7 plays crucial roles in root development, flowering time, and leaf number in maize.
RESULTS
The Semidominant Sdw3 Mutant Displays Reduced Plant Height and Enlarged Leaf Angle
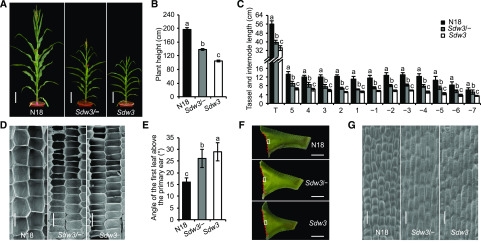
We identified a spontaneous maize mutant in the breeding field, Sdw3, which exhibits short stature and a large leaf inclination angle (Fig. 1A). Quantitative measurements at anthesis indicated that plant height and ear height were markedly reduced in the Sdw3 mutant compared with the wild-type near-isogenic line (N18; Fig. 1B; Supplemental Fig. S1A). Gratifyingly, these changes could increase stalk lodging resistance in the Sdw3 plants (Supplemental Fig. S1B). To investigate the dwarfism of the Sdw3 mutant, we measured the aboveground internode number and internode length of the plants. Unexpectedly, the aboveground internode number was significantly increased by 1.75 on average in the Sdw3 plants compared with N18 (Supplemental Fig. S1C). By contrast, each measured internode and tassel of the Sdw3 mutant were almost uniformly shortened (42% to 66%) than those of N18 (Fig. 1C). Histological observation by scanning electron microscopy (sem) indicated that the longitudinal parenchyma cells of the second internode above the primary ear were dramatically shorter (∼58%) in Sdw3 than in N18 (Fig. 1D; Supplemental Fig. S1D), and the internode cell number was slightly reduced in Sdw3 (Supplemental Fig. S1E). These results demonstrate that the semidwarf phenotype of the Sdw3 mutant is mainly attributed to reduced cell elongation in the internode.
Figure 1.
Phenotypic characterization of the Sdw3 mutant. A, Overall morphology of three NILs of Sdw3 at anthesis. The wild type (N18), heterozygous mutant (Sdw3/−), and homozygous mutant (Sdw3) are shown. Scale bars = 20 cm. B, Quantitative measurements of plant height of N18, Sdw3/−, and Sdw3 at anthesis. Error bars represent ± sd (n = 24). C, Quantitative measurements of the length of tassels and aboveground internodes at anthesis. Internodes above the primary ear are labeled 1, 2, 3, 4, and 5, and internodes below the ear are labeled −1, −2, −3, −4, −5, −6, and −7. All internodes were measured except the internodes below −7 due to obstruction from strong brace roots. Error bars represent ± sd (n = 20). T, Tassels. D, sem observation of longitudinal sections of parenchyma tissue in the middle region of the second internode above the primary ear of N18, Sdw3/−, and Sdw3 at anthesis. Scale bars = 100 μm. E, Quantitative measurements of the angle of the first leaf above the primary ear of N18, Sdw3/−, and Sdw3 at anthesis. Error bars represent ± sd (n = 24). F, Adaxial auricles of the first leaf above the primary ear of N18, Sdw3/−, and Sdw3 at anthesis. The red dotted lines indicate the outer edge of the auricles. Scale bars = 1 cm. G, sem observation of adaxial epidermal cells in auricles in the middle region near the outer edge (framed by a white rectangle in Fig. 1F) of the first leaf above the primary ear of N18, Sdw3/−, and Sdw3 at anthesis. Scale bars = 50 μm. Different letters above the error bars in B, C and E indicate significant differences at P < 0.05 using Tukey’s multiple comparison test. Images in A and F were digitally extracted from their original photos and placed on a black background.
To investigate leaf inclination, we measured the angles of the first leaf above the primary ear (Supplemental Fig. S1F), the third leaf below the primary ear, and the flag leaf. All three leaf angles were significantly increased in the Sdw3 mutant compared with N18 (Fig. 1E; Supplemental Fig. S1, G and H). We found that the outer edge of the auricle was markedly longer in the Sdw3 mutant than in N18 (Fig. 1F; Supplemental Fig. S1I), thereby leading to an increased leaf angle. Further investigation indicated that epidermal cells in the auricle were much longer in the Sdw3 mutant than those in N18 (Fig. 1G; Supplemental Fig. S1J), but there were fewer such cells in Sdw3 than in N18 (Supplemental Fig. S1K). These results indicate that the enlarged leaf angle in the Sdw3 mutant is mainly due to increased cell elongation in the auricles, in contrast with the inhibited cell elongation observed in the internode.
In addition, we analyzed the phenotypic variations in the heterozygous mutant Sdw3/−. Most of the phenotypes and quantitative data for the Sdw3/− mutant (Fig. 1; Supplemental Fig. S1) were intermediate to those of N18 and the Sdw3 mutant, but closer to Sdw3. Moreover, plant height in the F2 generation plants derived from self-pollination of the Sdw3/− mutant segregated at an approximate ratio of 1:2:1 (18 high:37 intermediate:20 dwarf, X2 = 0.12 < X2 (0.05, 2) = 5.99). Collectively, these findings demonstrate that the Sdw3 phenotype is controlled by a single semidominant nuclear gene.
ZmACS7, Encoding an ACC synthase, Is the Candidate Gene of the Sdw3 Phenotype
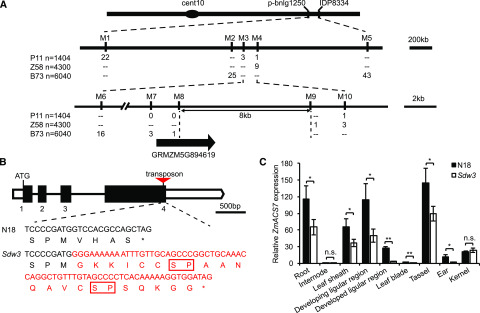
We applied a map-based cloning strategy to identify the gene responsible for the Sdw3 phenotype. With use of a small P11 (Productive11) population of 96 individuals, the target gene was initially mapped to the long arm of maize chromosome 10 between molecular markers p-bnlg1250 and IDP8334. We screened three large populations of P11, Z58 (Zheng58), and B73 plants containing over 10,000 individuals and localized the Sdw3 gene to an 8-kb interval between markers M8 and M9 (Fig. 2A). This region contains a partial sequence of only one candidate gene, ZmACS7 (GRMZM5G894619), based on a database search (www.gramene.org). ZmACS7 encodes an ACC synthase, a key enzyme catalyzing the biosynthesis of ACC, the precursor for ethylene biosynthesis. A possible 205-bp nonautonomous MuDR transposon was inserted in the fourth exon of ZmACS7 in the Sdw3 mutant locating 11 bp upstream of the stop codon (Fig. 2B; Supplemental Fig. S2; www.girinst.org/censor/). Phylogenetic analysis showed that ZmACS7 belongs to Type I ACSs and its closest ortholog is AtACS6 (Supplemental Fig. S3A). Like AtACS6, ZmACS7 and its closest paralog ZmACS2 harbor potential phosphorylation sites in their CTDs, that is, Ser residues in the RLSL (Arg [R] - Leu [L] - Ser [S] - Leu [L]) and three SP (Ser [S] - Pro [P]) motifs (Supplemental Fig. S3B; Choudhury et al., 2013). The transposon brought about a frameshift mutation that deleted the last four amino acids VHAS of ZmACS7, but extended its CTD, with two additional SP motifs (Fig. 2B; Supplemental Fig. S3B). Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that ZmACS7 was widely expressed in various tissues, including the root, internode, leaf sheath, ligular region, leaf blade, tassel, ear, and kernel (Fig. 2C). ZmACS7 was highly expressed in the developing ligular region, which is consistent with the changes in leaf angle observed in the Sdw3 mutant. In addition, ZmACS7 was expressed at markedly lower levels in most tissues of the Sdw3 mutant compared with N18 (Fig. 2C). However, ZmACS7 transcript levels were low in the internodes of both lines, with no significant difference between Sdw3 and N18 plants (Fig. 2C). These results suggest that the phenotypic variations in the Sdw3 mutant may be not directly caused by reduced ZmACS7 transcript levels.
Figure 2.
Map-based cloning of the Sdw3 mutant and mRNA expression pattern of ZmACS7. A, Fine mapping of the Sdw3 mutant. M1 to M10 indicate molecular markers. Numbers below the markers indicate the number of recombinants between the locus and the markers shown. Double hyphens below the markers indicate that the markers shown displayed no molecular polymorphism between the Sdw3 mutant and the corresponding inbred line. The final mapping interval (M8 to M9) overlaps with a partial sequence of only one candidate gene, GRMZM5G89619 (ZmACS7). B, Gene structure and mutated sequences of ZmACS7. ZmACS7 has four exons and three introns. The boxes indicate exons, and the lines between boxes indicate introns. The black boxes represent the coding sequence. The empty boxes represent the 5′ untranslated region (left) and 3′ untranslated region (right). The red triangle indicates the 205-bp transposon. The DNA sequence and the deduced amino acid sequence of mutated ZmACS7 in the Sdw3 mutant are in red. The transposon deleted the last four amino acids VHAS of ZmACS7, but extended the C terminus, containing two additional SP motifs (encircled by red rectangles). C, RT-qPCR analysis of ZmACS7 in multiple N18 and Sdw3 tissues. The tissues and stages used for RNA extraction are described in the legend of Supplemental Fig. S14B. ZmActin1 (GRMZM2G126010) served as the reference gene for normalization of RT-qPCR data. Error bars represent ± sd (n = 3). Asterisks indicate significant differences between N18 and Sdw3 using Student’s t test (*P < 0.05 and **P < 0.01; n.s. = no significant difference).
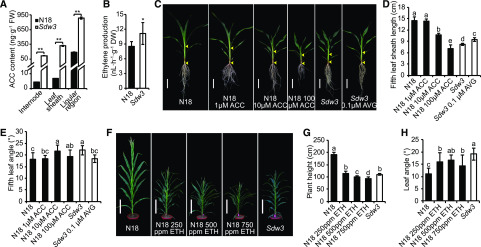
Elevated ACC and Ethylene Levels Are Responsible for the Phenotypes of the Sdw3 Mutant
To investigate whether the mutation of ZmACS7 in the Sdw3 plants influences its metabolite composition, we measured endogenous ACC levels. The Sdw3 mutant had dramatically higher endogenous ACC levels than N18 in all tissues examined, including the developing internode, leaf sheath, and developing ligular region at the V7 stage (Fig. 3A). We reasoned that this excessive ACC might be responsible for the phenotypic differences between Sdw3 and N18 plants. We tested the responses of N18 plants to exogenous ACC treatment in a hydroponic assay. After cultivation, we measured the length of the fifth leaf sheath and the angle of the fifth leaf of N18, and found that the length of the leaf sheath obviously decreased, whereas the leaf angle significantly increased, with increasing ACC concentration (Fig. 3, C–E), indicating that treatment with high concentrations of ACC mimicked the phenotypes of the Sdw3 mutant. We also examined the effects of aminoethoxyvinyl-Gly (AVG), a potent competitive inhibitor of ACS, on the Sdw3 mutant. The leaf sheaths of Sdw3 plants treated with 0.1 μm AVG were significantly elongated, whereas the leaf angle of the Sdw3 mutant was markedly reduced after this treatment (Fig. 3, C–E), suggesting that inhibiting ACS using 0.1 μm AVG at least partially restored the wild-type phenotypes in the Sdw3 plants.
Figure 3.
Excessive ACC and ethylene confer reduced plant height and enlarged leaf angle in the Sdw3 mutant. A, Quantitative measurements of endogenous ACC content in three tissues between N18 and Sdw3 seedlings at the V7 stage. Error bars represent ± sd (n = 3). FW, fresh weight. B, Quantitative measurements of endogenous ethylene production in the leaf blade between N18 and Sdw3 seedlings at the V7 stage. Error bars represent ± sd (n = 9). DW, dry weight. C, Overall morphology of N18 and Sdw3 seedlings after hydroponic cultivation. The distance between two yellow arrowheads refers to the length of the fifth leaf sheath. Scale bars = 10 cm. D and E, Quantitative measurements of the fifth leaf sheath length (D) and the fifth leaf angle (E) of N18 and Sdw3 plants after hydroponic cultivation. Error bars represent ± sd (n = 12). F, Overall morphology of various concentrations of ethephon (ETH)-treated N18 plants at anthesis. Scale bars = 30 cm. G and H, Quantitative measurements of plant height (G) and the first leaf angle above the primary ear (H) of ETH-treated N18 plants at anthesis. Error bars represent ± sd (n = 20). Asterisks in A and B indicate significant differences in the corresponding tissue between N18 and Sdw3 (*P < 0.05 and **P < 0.01; Student’s t test). Different letters above the error bars in D, E, G, and H indicate significant differences at P < 0.05 using Tukey’s multiple comparison test. Images in C and F were digitally extracted from their original photos and placed on a black background.
Because ACC is the precursor of ethylene biosynthesis, we measured endogenous ethylene levels in leaves at the V7 stage. Consistent with the changes in ACC content, endogenous ethylene production dramatically increased in the Sdw3 mutant (11.2 nL h−1 g−1 DW) compared with N18 (8.5 nL h−1 g−1 DW; Fig. 3B). To simulate the higher ethylene levels in the Sdw3 mutant, we sprayed N18 plants with different concentrations of ethephon weekly from the jointing stage to the tasseling stage. Indeed, N18 plants continuously treated with ethephon mimicked the phenotypes of the Sdw3 mutant (Fig. 3F). At anthesis, the plant height, ear height, and internode length of N18 were dramatically reduced with increasing ethephon concentration (Fig. 3, F and G; Supplemental Fig. S4, A, C, and D), whereas the leaf angle was markedly increased (Fig. 3, F and H; Supplemental Fig. S4B). Collectively, these results demonstrate that the mutation of ZmACS7 increases ACC and ethylene levels in the Sdw3 mutant, thus conferring reduced plant height and enlarged leaf angle in this mutant.
ZmACS7 Confers Reduced Plant Height and Enlarged Leaf Angle in the Sdw3 Mutant
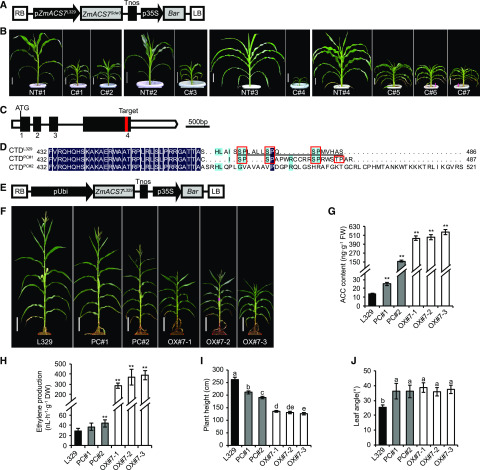
To determine whether the ZmACS7 mutation was responsible for the Sdw3 phenotype, a construct with a recombinant fragment including the deduced coding sequence of ZmACS7 in the Sdw3 mutant under the native promoter of ZmACS7 from the transgenic acceptor L329 (pZmACS7L329:ZmACS7Sdw3) was transformed (Fig. 4A). Three positive transgenic T0 plants (C#1 to C#3) were identified from two separate transformations to L329 (Fig. 4B; Supplemental Fig. S5, A and B). The phenotypes of all three transgenic T0 plants mimicked the Sdw3 phenotypes, with obviously reduced stature and flat leaves compared with the corresponding nontransgenic plants (NT#1 and NT#2). However, we were not able to obtain any higher generation of all three T0 plants due to excessively defective growth and development. We further introduced the pZmACS7L329:ZmACS7Sdw3 construct into another acceptor B104. The four positive T0 plants C#4 to C#7 from two transformations also mimicked the Sdw3 phenotypes to different degrees (Fig. 4B; Supplemental Fig. S5B). RT-qPCR analysis showed that C#4 to C#7 plants harbored much higher mRNA of ZmACS7Sdw3 compared with the parental mutant Sdw3 (Supplemental Fig. S5C). The increased times of ACC content in C plants were much greater than that of the Sdw3 mutant compared with their corresponding wild types (Supplemental Fig. S5, D–F), which probably led to excessively defective growth and development in these T0 plants than that in Sdw3. Taken together, these results at least indicated that the mutated ZmACS7 confers reduced plant height and enlarged leaf angle in the Sdw3 mutant.
Figure 4.
ZmACS7 is responsible for reduced plant height and enlarged leaf angle in the Sdw3 mutant. A, Schematic diagram of the pZmACS7L329:ZmACS7Sdw3 construct including the coding sequence of ZmACS7 from the Sdw3 mutant under the native promoter of ZmACS7 from L329. RB, right border; Tnos, terminator of nopaline synthase gene; p35S, cauliflower mosaic virus 35S promoter; Bar, (bialaphos resistance gene) serves as the selective marker; LB, left border. B, Overall morphology of seven positive pZmACS7L329:ZmACS7Sdw3 T0 plants (C#1 to C#7) grown in the greenhouse from four batches of transformations. NT#1 (nontransgenic plant 1) is the control for C#1 and C#2, whereas NT#2 is the control for C#3; all these lines share the same genetic background of germplasm L329. NT#3 is the control of C#4, and NT#4 is the control of C#5, C#6, and C#7 with genetic background of B104. Scale bars = 15 cm. C, Gene structure of ZmACS7 with the target (marked in red) for CRISPR/Cas9-specific binding to perturb the C terminus of ZmACS7. D, Alignment of C-terminal domains of ZmACS7 in L329, PC#1, and PC#2. Background colors indicate the similarity level of amino acid sequences (dark blue = 100%; baby blue <75% and ≥50%). Red rectangles were used to mark the SP and TP motifs in the CTDs of ZmACS7. The homologous sequence of the last 16 amino acids of AtACS6 in ZmACS7 was marked by a black line. E, Schematic diagram of the construct used for transgenic overexpression of ZmACS7. The maize ubiquitin promoter was applied to dominate the expression of the ZmACS7 coding sequence from L329. The Bar gene serves as the selective marker. pUbi, the maize ubiquitin-1 promoter. F, Overall morphology of PC and ZmACS7 OX lines at anthesis. Scale bars = 30 cm. The Cas9 constructs were screened out from PC lines. The wild-type parent L329 served as the control of both PC and ZmACS7 OX lines, because L329 displayed similar phenotypes with L329’, a transgenic line introduced by an empty vector, similar to pBCXUN (Supplemental Fig. S6). G, Quantitative measurements of endogenous ACC content of the shoot at V1 stage in PC and ZmACS7 OX seedlings. Error bars represent ± sd (n = 3). FW, fresh weight. H, Quantitative measurements of endogenous ethylene production in leaf 8 and leaf 9 at V7 stage in PC and ZmACS7 OX seedlings. Error bars represent ± sd (n = 5). DW, dry weight. I and J, Quantitative measurements of plant height (I) and angle of the first leaf above the primary ear (J) in PC and ZmACS7 OX lines at anthesis. Error bars represent ± sd (n = 20). Asterisks above the error bars in G and H indicate significant differences between the corresponding line and L329 (**P < 0.01; Student’s t test). Different letters above the error bars in I and J indicate significant differences at P < 0.05 using Tukey’s multiple comparison test. Images in B and F were digitally extracted from their original photos and placed on a black background.
To further validate the function of ZmACS7, we created lines with similar C-terminal mutations of ZmACS7 to that of the Sdw3 mutant using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome editing (Fig. 4C). We identified two lines (perturbation of the CTD of ZmACS7 [PC]#1 and PC#2) with perturbed ZmACS7 C termini due to frameshift mutations (Fig. 4D; Supplemental Fig. S7A). We also produced transgenic plants overexpressing ZmACS7 to investigate its functions at the transcriptional level (Fig. 4E). We identified three positive overexpression lines (OX#7-1, OX#7-2, and OX#7-3) with ZmACS7 mRNA levels 200- to 300-fold higher than L329 (Supplemental Fig. S7B). When compared with L329, all the transgenic PC and OX lines had markedly increased ACC contents and ethylene production (Fig. 4, G and H) and showed an obvious decrease in plant height, ear height, and internode length (Fig. 4, F and I; Supplemental Fig. S7, C–E) but a significant increase in stalk lodging resistance, aboveground internode number, and leaf angle (Fig. 4, F and J; Supplemental Fig. S7, F–H). All these transgene-generated lines recapitulated the phenotypes of the Sdw3 mutant to different degrees, suggesting that ZmACS7 indeed corresponds to the Sdw3 locus.
Altering the C-terminal Domain Enhances the Protein Stability of ZmACS7
In general, increased ACC levels due to the mutation of ACS are correlated with a rise in ACS transcript levels. However, we found that the steady-state level of ZmACS7 mRNA was reduced in most Sdw3 tissues examined (Fig. 2C), suggesting that the elevated ACC levels in the Sdw3 mutant are not due to an increase in ZmACS7 transcript levels. Indeed, in some cases, altering the CTD of ACS proteins in Arabidopsis increased their protein stability rather than affecting their specific activity (Vogel et al., 1998; Chae et al., 2003). These findings prompted us to explore the protein stability and catalytic activity of the mutated ZmACS7 in the Sdw3 mutant.
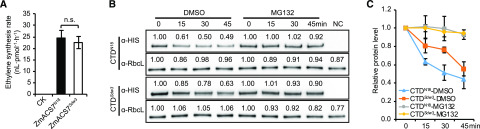
We expressed HIS-tagged intact ZmACS7N18 and ZmACS7Sdw3 proteins and their corresponding C-terminal domains, CTDN18 and CTDSdw3, in Escherichia coli (Supplemental Fig. S8, A–C) and evaluated the catalytic activities of ZmACS7N18 and ZmACS7Sdw3 based on the ethylene synthesis rate in vitro. Enzyme activity assay showed that the specific activities of ZmACS7Sdw3 and ZmACS7N18 were similar (Fig. 5A), suggesting that the altered CTD in the Sdw3 mutant does not affect the specific activity of ZmACS7. To further confirm the effect of the CTD mutation on ZmACS7 stability, we compared the turnover rates of HIS-tagged CTDN18 and CTDSdw3 in a cell-free protein degradation system. Immunoblot analysis revealed that CTDSdw3 was comparatively harder to be degraded than CTDN18 (Fig. 5, B and C). Moreover, the degradation rates of CTDN18 and CTDSdw3 were markedly inhibited by MG132, a specific inhibitor of the 26S proteasome (Fig. 5, B and C), suggesting that the degradation of ZmACS7 is mediated by the ubiquitin-26S proteasome pathway. These results demonstrate that the altered CTD of ZmACS7 (caused by a transposon insertion) in the Sdw3 mutant enhances ZmACS7’s function by increasing its stability but not affecting the specific activity.
Figure 5.
Specific activity and stability of ZmACS7 in N18 and Sdw3 plants. A, Specific activity of ZmACS7N18 and ZmACS7Sdw3 measured based on the ethylene synthesis rate in vitro. CK refers to HIS-tagged protein from the empty vector, which served as the negative control. Error bars represent ± sd (n = 9); n.s. indicates no significant difference between ZmACS7N18 and ZmACS7Sdw3 (Student’s t test). B, Immunoblot analysis of HIS-tagged CTDN18 and CTDSdw3 proteins after incubation in a cell-free degradation system at different time points. NC indicates the negative control (sample without adding the target protein to the cell-free extract). RbcL (Rubisco large subunit) was used as a nondegraded control to indicate the same loading quantity. All the samples were equally diluted to test protein quantity of RbcL by Western blotting. Numbers above the Western blotting bands refer to relative protein content measured by ImageJ software. C, Quantitative measurements of HIS-tagged CTDN18 and CTDSdw3 proteins obtained by immunoblot analysis using ImageJ software. The mean values from three biological replicates are shown. Error bars represent ± sd (n = 3). DMSO, dimethyl sulfoxide.
It is worth noting that the CTD variant in the Sdw3 mutant ablated the last four amino acids VHAS of the wild-type ZmACS7, but harbors a prolonged terminus. To determine whether the deletion of the VHAS motif confers ZmACS7 stabilization in the Sdw3 mutant, we compared the protein turnover rates of HIS-tagged CTDN18 and CTDΔVHAS that lacks the VHAS motif. Immunoblot analysis revealed comparable protein turnover rates of CTDΔVHAS and CTDN18 (Supplemental Fig. S8, D and E), suggesting that the deletion of the VHAS motif is not responsible for ZmACS7 stabilization in the Sdw3 mutant.
Transcriptome Changes in the Developing Internode and the Developing Ligular Region of the Sdw3 Mutant
The reduced plant height and enlarged leaf angle of the Sdw3 mutant are mainly attributed to a reduction in longitudinal cell elongation in the internode and an increase in the auricle (Fig. 1, D and G; Supplemental Fig. S1, D and J). To investigate the transcriptome changes during internode and auricle development in Sdw3 versus N18 plants, we performed RNA sequencing (RNA-seq) analysis. We identified 462 differentially expressed genes (DEGs) in the developing internode and 1588 DEGs in the developing ligular region of the Sdw3 mutant versus N18 using a cutoff of twofold change or greater in expression and a false discovery rate of q value <0.05. Verification of the expression patterns of various DEGs via RT-qPCR revealed highly positive correlations between the RNA-seq data and RT-qPCR results for both the internode and ligular region (Pearson correlation coefficients of 0.97 for the internode transcriptome and 0.95 for the ligular region), indicating that our RNA-seq data were quite reliable (Supplemental Fig. S9, A and B).
A global view of the pathways based on MapMan ontology (Thimm et al., 2004) revealed that DEGs in the internode and ligular region were mapped to a wide range of functional categories, including cell wall development, lipid metabolism, cell cycle, and hormone metabolism. Further analysis showed that an ACC oxidase gene (ACO35), an ETHYLENE INSENSITIVE3-like2 gene (EIL2), and 11 ethylene-responsive genes (Chao et al., 1997; Riechmann and Meyerowitz, 1998; Gallie and Young, 2004) were significantly up-regulated in both the internode and ligular region (Supplemental Fig. S10A), suggesting that the ethylene biosynthesis, signaling, and response pathway is highly activated in the Sdw3 mutant. Several negative regulators in this pathway, such as REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1)-like genes and ETHYLENE INSENSITIVE3-Binding F-box genes (EBF1; Potuschak et al., 2003; Resnick et al., 2008), were also up-regulated in the Sdw3 mutant, perhaps due to negative feedback regulation.
Plant kinesins, a family of microtubule-based motor proteins, are essential for cell division and cell elongation (Li et al., 2011; Deng et al., 2015). Our RNA-seq data showed that 12 kinesin-related genes were up-regulated in the ligular region but down-regulated in the internode of the Sdw3 mutant (Supplemental Fig. S10B), which perhaps lead to the promotion of cell elongation in the auricle and inhibition in the internode of the Sdw3 mutant, although they displayed an insignificant decrease in the developing Sdw3 internode. Notably, LG2 transcript levels increased by 2.18-fold in the ligular region of the Sdw3 mutant (Supplemental Fig. S10A), which may play an important role in the auricle extension and enlarged leaf angle observed in the Sdw3 mutant.
Further investigation showed that flowering time (days to first pollen shed) was delayed by 2 d in the Sdw3 mutant (Sdw3, 68.4 ± 1.0; N18, 66.2 ± 1.7), and days to first silk emergence was delayed by 6 d in the mutant (Sdw3, 70.4 ± 1.6; N18, 64.4 ± 2.1). In addition, Sdw3 plants produced an average of 1.1 more leaves than N18 (Sdw3, 21.9 ± 0.8; N18, 20.8 ± 0.5) due to an increase in leaf number below the primary ear (Sdw3, 16.7 ± 0.7; N18, 15.6 ± 0.5). These results are in accordance with the highly positive correlation found between days to anthesis and the number of leaves below the primary ear (Li et al., 2016). Intriguingly, some DEGs involved in flowering time in maize were enriched in the internode and ligular regions of the Sdw3 mutant. ZmRap2.7, encoding a negative regulator of flowering (Salvi et al., 2007), and its homolog (GRMZM2G416701) were up-regulated in the internode and ligular region of the Sdw3 mutant (Supplemental Fig. S10A). Overexpression of ZmRap2.7 causes late flowering and an increased number of leaves (Salvi et al., 2007). By contrast, the transcript levels of ZMM4 (Z. mays MADS-box 4), encoding a MADS box transcription factor that promotes flowering and functions downstream of ZmRap2.7 (Danilevskaya et al., 2008; Romero Navarro et al., 2017), as well as ZmMADS67, were reduced in the internode and ligular region of the Sdw3 mutant (Supplemental Fig. S10A). The transcriptional changes in these genes might explain the late flowering and increased leaf number in the Sdw3 mutant.
Increased ZmACS7 Function Alters Root Morphology and Causes Grain Yield Reduction in Maize
The maize root is a vital organ responsible for the uptake of water and nutrients from soil and providing anchorage (Aiken and Smucker, 1996). Interestingly, in addition to variation in plant height, ear height, leaf angle, flowering time, and leaf number, we found that Sdw3 seedlings harbored more embryonic seminal roots and postembryonic crown roots at the V1 stage than N18 (Supplemental Fig. S11, A–C). The Sdw3 mutant also displayed more layers of brace roots, but with smaller root angle at the mature stage compared with N18 (Supplemental Fig. S11, D–F). These phenotypic variations of the root system in the Sdw3 mutant are in accordance with the enrichment of ZmACS7 transcript in roots (Fig. 2C; Supplemental Fig. S12). The PC and ZmACS7 OX lines also mimicked the root architecture of the Sdw3 mutant compared with L329 (Supplemental Fig. S11, G–I). Grain yield is the highlight for cereal crops. In this study, all yield-related traits investigated, including ear length (EL), ear diameter (ED), ear row number (ERN), ear grain number (EGN), ear grain weight (EGW), and hundred kernel weight (HKW) were decreased at different levels in ZmACS7 overexpressors than those in their wild types (Supplemental Fig. S13). EGW, which best represents grain yield, was significantly reduced ranging from 26.7% to 77.2% in the ZmACS7 overexpressors compared with that in their controls (Supplemental Fig. S13). All together, these results indicated that increased ZmACS7 function could alter root morphology and could also cause grain yield reduction in maize.
ZmACS2 Has Similar Expression Patterns and Functions with ZmACS7
Phylogenetic analysis indicated that ZmACS2 is the closest paralog of ZmACS7 (Supplemental Fig. S3A). Sequence alignment revealed only a few single-amino acid substitutions and deletions between ZmACS7 and ZmACS2 (Supplemental Fig. S3B). Additionally, ZmACS2 and ZmACS7 share highly similar mRNA expression patterns based on analysis of publicly available transcriptome data of maize at 79 different tissues/stages (Supplemental Fig. S12, Pearson correlation coefficient of 0.72; MaizeGDB, www.maizegdb.org; Stelpflug et al., 2016) and RT-qPCR analysis of various tissues and developmental stages (Supplemental Fig. S14A, Pearson correlation coefficient, 0.92). Interestingly, expression levels of ZmACS7 and ZmACS2 were both reduced in most Sdw3 tissues than those in N18 (Fig. 2C; Supplemental Fig. S14B), probably due to negative feedback regulation in response to elevated ACC levels. The high identity of protein sequences and similar expression patterns of ZmACS2 and ZmACS7 imply that they may share conserved functions. Thus, we generated transgenic plants overexpressing ZmACS2 (Supplemental Fig. S14, C and D) and found that they were markedly shorter and had flatter leaves than L329 (Supplemental Fig. S14E), similar to the transgenic plants overexpressing ZmACS7 (Fig. 4F). These results suggest that ZmACS2 harbors similar expression patterns and functions with ZmACS7.
We further generated mutants (knockout mutants [KO]#1 and KO#2) in which ZmACS7 is knocked out using CRISPR/Cas9 (Supplemental Fig. S15, A and B). However, KO#2 displayed very similar plant architecture with the wild-type L329 (Supplemental Fig. S15, C–J). KO#1 showed a statistically significant but mild increase in plant height (4.0%), ear height (14.1%), above internode number (3.3%), and leaf angle (10.5%) compared with L329 (Supplemental Fig. S15, C–J). The loss-of-function mutation of ZmACS7 in the KO lines might trigger a genetic compensation response from other genes in the same pathway (El-Brolosy and Stainier, 2017), particularly its closest homolog, ZmACS2.
DISCUSSION
The Functional Divergence and the Regulatory Complexity of ACS Isoforms in Maize
Ethylene plays multiple roles in plant growth and development (Mattoo and Suttle, 1991). ACS is encoded by a multigene family in every plant species investigated (Wang et al., 2002; Gallie and Young, 2004; Lin et al., 2009). Arabidopsis contains eight enzymatically active ACS isoforms (AtACS2, AtACS4 to AtACS9, and AtACS11) and an inactive ACS isoform (AtACS1; Yamagami et al., 2003). These various ACS isoforms have different enzymatic properties and expression patterns and can be induced by diverse stimuli (Yamagami et al., 2003; Argueso et al., 2007). Five ACSs have been annotated in the maize genome: ZmACS1, ZmACS2, ZmACS3, ZmACS6, and ZmACS7 (www.gramene.org). Our phylogenetic analysis of ACS isoforms from maize and Arabidopsis showed that the five maize ACSs are grouped among the three defined ACS types and the putative aminotransferase group (Supplemental Fig. S3A). ZmACS7 belongs to the Type I clade along with its closest paralog, ZmACS2, which share 94.3% identity at the protein level. ZmACS6 and ZmACS1 are grouped into Type II and Type III ACSs, respectively (Supplemental Fig. S3A), whereas ZmACS3 belongs to the putative aminotransferase group (Supplemental Fig. S3A), suggesting that ZmACS3 may not be an ACC synthase but an aminotransferase. Noticeably, classification of maize ACS isoforms in our study was consistent with Gallie and Young (2004)'s research, although ZmACS1 and ZmACS3 were not mentioned in their research. However, Zhang et al. (2012) placed ZmACS6 in a monocot Type I ACS clade. This discrepancy was likely attributed to the difference of sequence types (protein sequences were used in Gallie and Young [2004] and our studies, while nucleotide sequences and species number in Zhang et al. [2012] used many more and divergent species) for phylogenic analyses. In our study, further alignment of protein sequences of ZmACS6 with ACS isoforms in Arabidopsis showed that ZmACS6 lacks the three SP motifs that often exist in Type I ACS, whereas it has the WAN and RLTP motifs that are similar to the WVF and RLSF motifs of Type II ACS (Supplemental Fig. S16), suggesting that ZmACS6 tends to be a type II ACS.
Publicly available transcriptome data of maize at 79 different tissues/stages showed that ZmACS1 and ZmACS6 are mainly expressed at various stages during root development (Supplemental Fig. S12), whereas ZmACS2 and ZmACS7 are widely expressed in various tissues and stages throughout the whole growth period (Supplemental Figs. S12 and 14A). Consistent with the expression patterns, previous studies revealed that ZmACS6 and ZmACS2 regulated root growth in response to hypoxia and physical resistance presented by soils (Gallie et al., 2009; Geisler-Lee et al., 2010). Additionally, ZmACS6 is sporadically expressed during leaf and seed development, which conforms with that ZmACS6 and ZmACS2 were involved in leaf senescence and response to pathogen infection in maize kernels (Young et al., 2004; Habben et al., 2014; Wang et al., 2017). ZmACS6 could also influence grain yield under drought and low nitrogen conditions (Habben et al., 2014). However, little was known about the physiological roles of ZmACS7 although it’s widely expressed due to the lack of available mutants (Young et al., 2004). In our study, through positional cloning of a semidominant plant architecture mutant, Sdw3, we demonstrated that ZmACS7 regulates many morphological and physiological traits of maize, including plant height, ear height, leaf angle, root development, flowering time, leaf number, and grain yield, which corresponds to its wide expression during the maize lifespan. High similarity of protein sequences, expression patterns, and transgenic overexpression plants between ZmACS2 and ZmACS7 suggested that ZmACS2 may play similar roles in plant architecture of maize, like ZmACS7. This may explain why ZmACS7 gene KO displayed mild changes compared with the wild type L329 (Supplemental Fig. S15), probably due to genetic compensation (El-Brolosy and Stainier, 2017) from other paralogues within the same network, particularly ZmACS2.
Regulation of the CTD of ZmACS7 in the Stability of this Protein
ACS isoforms have very low abundance in vivo and could be regulated transcriptionally and posttranslationally in response to various endogenous and environmental factors. Reversible protein phosphorylation is thought to play a role in regulating ACS stability in Arabidopsis (Liu and Zhang, 2004; Skottke et al., 2011; Ludwików et al., 2014). The CTD of AtACS6 contains the phosphorylation sites that mediate the stabilization and accumulation of AtACS6 as well as the cis-determinants that function in the rapid degradation of AtACS6 by the ubiquitin-proteasome pathway (Chae et al., 2003; Joo et al., 2008). The disruption of the CTDs of AtACS5 and AtACS9 hampers the proteasome-mediated degradation of these proteins in the dominant mutants eto2 and eto3, respectively, thus resulting in increased ACS stability and ethylene production (Vogel et al., 1998; Chae et al., 2003; Wang et al., 2004; Yoshida et al., 2005). Similarly, in our study, a transposon leads to a frameshift mutation of ZmACS7. The resulting CTD alteration increases ZmACS7 stability (Fig. 5, B and C) but does not affect the specific activity of this enzyme (Fig. 5A), thereby increasing ACC and ethylene production in the Sdw3 mutant (Fig. 3, A and B). However, unlike replacement of the last 12 C-terminal amino acids of AtACS5 in the eto2 mutant or the missense mutation of the conserved WVF motif of AtACS9 in the eto3 mutant that hampered the proteasome-mediated degradation (Chae et al., 2003; Wang et al., 2004; Yoshida et al., 2005), the CTD variant in the Sdw3 mutant only ablated the last four amino acids VHAS of the wild-type ZmACS7. The VHAS motif appears similar to the C-terminal motif VRAQt in AtACS6, which had not been specially linked to ACS stability (Joo et al., 2008). Indeed, we found that the deletion of the VHAS motif does not affect ZmACS7 stability (Supplemental Fig. S8, D and E). Although the prolonged CTD of ZmACS7 in the Sdw3 mutant contains two Ser residues in the additional SP motifs, which may serve as potential MAPK phosphorylation sites to increase ZmACS7 stability.
Two CRISPR/Cas9-induced CTD variants of ZmACS7 in the PC mutants partially recapitulated the phenotypes of the Sdw3 mutant; however, their mutation sites are different from that in Sdw3 plants due to lack of the Cas9 guide sequence to precisely change the VHAS motif. Owing to frameshift mutations, both CTD variants in the PC#1 and PC#2 lines ablated the last 22 amino acids of the wild-type ZmACS7 (Fig. 4D). In Arabidopsis, the C terminus of AtACS6 served as the targeting site of the degradation machinery and the deletion of the last 16 amino acids (Supplemental Fig. S3B) was sufficient to stabilize AtACS6 due to interference with its degradation (Joo et al., 2008). Similarly, CTD mutations ablating the last 22 amino acids of ZmACS7 in the PC#1 and PC#2 lines, which include the intact homologous sequence of the last 16 amino acids of AtACS6 (Fig. 4D), might increase ZmACS7 stability. The CTD variant of ZmACS7 in the PC#2 line owned a longer but very different tail lacking three SP motifs, whereas the unelongated CTD variant in PC#1 retained the three SP motifs although the flanking sequences were changed and harbored an additional TP motif, which should be subject to MAPK-mediated phosphorylation (Fig. 4D; Pitzschke, 2015). This distinction of CTD mutations may confer the differences of increased ACC and ethylene production and phenotypic severity in the PC#1 and PC#2 lines (Fig. 4, F–J; Supplemental Fig. S7, C–H). It would be interesting to further understand the fine mechanism underlying ZmACS7 stabilization and turnover.
Pleiotropy of ZmACS7 on Maize Growth and Development
Ethylene has been widely used in agriculture to control lodging in cereals mainly by shortening plant height and ear height (Langan and Oplinger, 1987; Simmons et al., 1988). Here, we present a key gene in ethylene biosynthesis, ZmACS7, with pleiotropic effects on leaf angle, root architecture, leaf number, and flowering time, besides plant height and ear height. ZmACS7 overexpressors, including the Sdw3, PC, and ZmACS7 OX lines, exhibited reduced plant height and ear height, larger leaf angle, and more layers of brace roots with smaller angles compared with their corresponding wild types. Indeed, stalk lodging resistance was enhanced in ZmACS7 overexpressors, probably due to the reduction of plant height and ear height. Besides, the Sdw3 mutant displayed increased leaf number and delayed flowering due to increased ZmACS7 stability. It is possible that the far-ranging expression of ZmACS7 in multiple tissues and stages during the lifespan resulted in pleiotropy of ZmACS7 on maize growth and development. Generally, erect upper leaves could improve maize tolerance to high plant density (Tian et al., 2011). Surprisingly, ZmACS7 KO mutants did not show reduced leaf angle but rather a mild increase in leaf angle, which may originate from a genetic compensation effect by other genes, particularly ZmACS2 (El-Brolosy and Stainier, 2017). However, enlarged maize leaf angle is not totally disadvantageous. Prior reports showed that maize plants with erect leaves above the ear and intermediate or horizontal leaf orientation below the ear could yield better than the plants with all leaves upright (Pendleton et al., 1968; Mock and Pearce, 1975). In this study, our results also indicated that increased ZmACS7 function could cause alteration of root morphology and reduction of grain yield in maize (Supplemental Figs. S11 and S13), which is in accordance with the reports that ACC and ethylene promoted the emergence of nodal roots, including aboveground brace roots and underground crown roots (Hochholdinger, 2009; Shi et al., 2019), and the report that ethephon caused grain yield reduction when lodging was not a severe problem (Cox and Andrade, 1988).
Ethylene is generally considered as a growth inhibitor; however, accumulating evidences revealed a biphasic response of ethylene (Smalle et al., 1997; Hattori et al., 2009). The inhibitory or stimulatory effect of ethylene on growth, especially cell elongation, had been reported to depend on species (wetland or terrestrial plants), growing environments (the presence or absence of light, light quality, and the condition of nutrients), and ethylene dose (Pierik et al., 2006; Dugardeyn and Van Der Straeten, 2008). In our study, inhibition of longitudinal cell elongation in the internode and promotion in the auricle are mainly responsible for the reduced plant height and the enlarged leaf angle in the ethylene-overproducing mutant Sdw3, suggesting a tissue-specific growth response of ethylene. The underlying mechanism of the opposite effects of ethylene on cell elongation in different tissues of the same plant needs further investigation. In addition, ethylene also displayed biphasic effects on flowering time in different species, for example, stimulating flowering in pineapple (Ananas comosus) and rice but inhibiting flowering in Arabidopsis (Burg and Burg, 1966; Achard et al., 2007; Wuriyanghan et al., 2009; Schaller, 2012). The observation of delayed flowering in maize due to increased ethylene level in this study enriched our knowledge of the regulation of ethylene in plant flowering time.
ZmACS7 overexpressors using its own promoter or the ubiquitin promoter exhibited reduced plant height and ear height, enhanced brace root development, and enlarged leaf angles below the ear, thereby facilitating lodging resistance and photosynthesis below the ear, but would also be associated with some adverse traits, such as increased leaf angles above the ear and reduction of grain yield. Some tissue- and stage-specific promoters should be applied to overexpress ZmACS7 individually or collectively, in order to decrease ear height, promote brace root development, and enlarge leaf angles below the ear, but not negatively affect grain yield of maize. Equally important, using a specific promoter to decrease leaf angles above the ear (e.g. by knocking down ZmACS7 and ZmACS2 expression in the auricles above the ear) is worth further investigation. Orchestrating ZmACS7 expression may help maize adaptation to higher plant density, thereby increasing maize grain yield per unit area.
MATERIALS AND METHODS
Plant Materials
The maize (Zea mays) Sdw3 mutant originated from a spontaneous mutation in the field. The heterozygous mutant Sdw3/− was selected to self for five generations to generate the near-isogenic lines (NILs) N18 (the wild type), Sdw3/− (the heterozygous mutant), and Sdw3 (the homozygous mutant). The NILs of Sdw3 and transgene-generated lines, including the PC lines, ZmACS7 OX lines, and ZmACS2 OX lines, were grown in Beijing (40.1°N, 116.2°E), China, in the summers of 2015 and 2017. The transgenic pZmACS7L329:ZmACS7Sdw3 T0 plants were grown in the greenhouse. ZmACS7 KO lines and the transgenic L329’ line (Supplemental Fig. S6) were grown in Beijing (40.1°N, 116.2°E), China, in the summer of 2019.
Positional Cloning
Sdw3 was crossed with inbred lines P11, B73, and Z58, and the F1 progenies were backcrossed with their corresponding wild-type parents to develop three BC1 populations. Initial mapping was performed using 86 molecular markers that uniformly covered the entire maize genome (www.maizegdb.org). With use of a small P11 population of 96 individuals, the target gene was initially mapped to the long arm of maize chromosome 10 between molecular markers p-bnlg1250 and IDP8334. Ten molecular markers, M1 to M10, were developed for fine mapping, as shown in Supplemental Table S1. With use of a large P11 population of 1404 individuals, the Sdw3 locus was further mapped between molecular markers M3 and M10. Subsequently, Z58 and B73 populations of over 10,000 individuals were screened, and the Sdw3 locus was ultimately narrowed down to an 8-kb interval between markers M8 and M9.
RNA Extraction and RT-qPCR Analysis
Various tissues from different maize lines at different developmental stages were collected separately, cut into small pieces, immediately frozen in liquid nitrogen, and stored at −80°C. Each tissue sample had three independent biological replicates with at least six individual plants per biological replicate. Total RNA was extracted from the samples using a Plant RNA Isolation Kit (www.huayueyang.com.cn/) according to the manufacturer’s instructions. Total RNA (1 μg) was reversely transcribed using a FastKing RT Kit (www.tiangen.com/). RT-qPCR was performed using the 7500 Fast and 7500 Real-Time PCR Systems (Applied Biosystems) with a SYBR Premix Ex Taq Kit (www.takara.com.cn/). ZmActin1 (GRMZM2G126010) was used for normalization among samples. The primers used for RT-qPCR are listed in Supplemental Table S1. The ΔΔCT method was used to measure the relative expression levels of the target gene (Livak and Schmittgen, 2001). All data are based on three independent biological replicates and four technical replicates.
Phylogenetic Analysis and Protein Sequences Alignment
The full-length protein sequences of the five annotated ACS isoforms from maize and the 11 ACS isoforms from Arabidopsis (Arabidopsis thaliana) were obtained from Gramene (www.gramene.org); AtACS3 was excluded as a pseudogene. The 16 sequences were aligned using ClustalW embedded in MEGA version 6.0 with default parameters. Phylogenetic trees were constructed using the neighbor-joining method with a Poisson model and 1000 bootstrap replications (Tamura et al., 2013). The program for multiple sequences alignment embedded in the DNAMAN v6 software (www.lynnon.com/) was used to perform protein sequences alignment. In some cases, fine manual adjustment after automatic alignment was conducted to generate an anchoring alignment to highlight important motifs.
Quantification of Endogenous ACC and Ethylene Levels
For endogenous ACC measurements, the developing internode between leaf 7 and leaf 8, the sheath of leaf 8, and the developing ligular regions of leaf 8 and leaf 9 of Sdw3 and N18 seedlings at the V7 stage were separately harvested. Each sample was collected from six individual plants and pulverized in liquid nitrogen. With use of the internal standard method, approximately 200 mg of fresh plant tissue per sample was used to extract endogenous ACC, which was measured using the QTRAP 5500 LC-MS/MS system (AB SCIEX; Xin et al., 2020).
Endogenous ethylene levels were assayed as described previously, with some modifications (Habben et al., 2014). The seventh and eighth leaf blades of N18 and Sdw3 seedlings were harvested at the V7 stage, and a hole punch was used to create small discs (6.33 mm in diameter). The discs were incubated for 2 h at 25°C to allow wounding-induced ethylene production to subside. One hundred discs per sample were sealed in a 20-mL headspace vial with a rubber cap. After incubation for 16 h at 25°C, ethylene levels in the vials were assayed by gas chromatography (Shimadzu GC-17A). Finally, the leaf discs were dried and weighed to evaluate normalized ethylene levels with nine biological replicates.
Exogenous ACC and Ethephon Treatment
N18 and Sdw3 seeds were surface-sterilized in 10% (v/v) H2O2 solution for 30 min, washed five times in sterile water, and germinated at 28°C for 36 h in the dark on moist filter paper. The germinated seeds were sown in aseptic vermiculite and cultured in an illumination incubator for 1 week (60% relative humidity; light, 26°C, 14 h; dark, 24°C, 10 h). The endosperm was removed from the seedlings by hand, and uniform seedlings with one developed leaf were transferred into an 18-l lightproof plastic case filled with full-strength Hoagland solution (pH 6.0) containing 0.5 mm (NH4)2SO4, 2 mm Ca(NO3)2, 0.25 mm KH2PO4, 0.75 mm K2SO4, 0.65 mm MgSO4, 0.1 mm KCl, 1 μm MnSO4, 1 μm ZnSO4, 0.1 μm CuSO4, 0.005 μm (NH4)6Mo7O24, 1 μm H3BO3, 0.1 mm FeSO4, and 0.1 mm EDTA-Na. Stock solutions of ACC or AVG were added to the plastic cases and diluted in Hoagland solution to the corresponding concentrations (0, 1, 10, and 100 μm ACC; 0 and 0.1 μm AVG). The seedlings were cultured in an artificial climate chamber for 3 weeks (60% relative humidity; light, 26°C, 14 h; dark, 24°C, 10 h); the culture solution was renewed every 3 d. All treated lines were harvested for morphology observation and quantitative measurements after the fifth leaves had fully developed. Because the height of maize seedlings is largely determined by the length of leaf sheaths, the length of the fifth leaf sheath and the angle of the fifth leaf were measured.
N18 and Sdw3 seeds were planted in Beijing (40.1°N, 116.2°E), China, in the summer of 2019. Beginning at the V7 stage, the leaves of N18 plants were sprayed weekly with various concentrations of ethephon (250, 500, and 750 ppm; 20 mL per plant) until the tassels emerged. The control plants were sprayed with the same amount of acidic double-distilled water (pH = 2.5), because the pH of working solutions containing 250, 500, and 750 ppm ethephon are 2.79, 2.56 and 2.42, respectively. At anthesis, plant height, ear height, the length of the second internode above the primary ear, and the angle of the first leaf above the primary ear were measured.
Plasmid Construction and Plant Transformation
To generate the pZmACS7L329:ZmACS7Sdw3 construct, a 4172-bp DNA fragment containing a 2660-bp putative native promoter of ZmACS7 amplified from the genome of the transgenic acceptor L329 and the 1512-bp coding sequence of ZmACS7 amplified from the complementary DNA library of the Sdw3 mutant was cloned into the pCAMBIA3300M vector. Notably, the putative promoter of ZmACS7 used in our study had been submitted to National Center for Biotechnology Information (NCBI)’s GenBank database. The CRISPR/Cas9 plasmids to alter the CTD and knockout ZmACS7 were constructed as previously described (Xing et al., 2014). Briefly, two specific 19-bp Cas9 guide sequences (Supplemental Figs. S7A and S15A) near the transposon insertion site and the translation start site of ZmACS7, respectively, were designed using CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) and introduced into the pBUE411 vector. The introduced sequences from the Cas9 constructs in the transgene-derived PC and KO mutants were screened and excluded. In addition, to construct the overexpression plasmids (Ubi:ZmACS7 and Ubi:ZmACS2), the full coding sequences of ZmACS7 and ZmACS2 from B73 were fused into the pBCXUN vector (from the Center for Crop Functional Genomics and Molecular Breeding of China Agricultural University [CAU]) driven by the maize ubiquitin promoter. The primers used for cloning are listed in Supplemental Table S1. The pZmACS7L329:ZmACS7Sdw3 construct, the CRISPR/Cas9 plasmids used for perturbation of the CTD of ZmACS7 (PC) and knockout of ZmACS7 (KO), and the Ubi:ZmACS7 and Ubi:ZmACS2 constructs overexpressing ZmACS7 and ZmACS2 (OX) were transformed into L329 by the Center for Crop Functional Genomics and Molecular Breeding at CAU. The pZmACS7L329:ZmACS7Sdw3 construct was also introduced into the transgenic acceptor B104.
Preparation of Recombinant Proteins
The overall coding sequences of ZmACS7 from N18 and Sdw3 plants and truncated DNA sequences expressing CTDs of ZmACS7 from N18 and Sdw3, and the CTD that lacks the VHAS motif, were independently fused in-frame with the HIS tag sequence in the pCold TF vector (www.takarabiomed.com.cn/). The primers used are listed in Supplemental Table S1. The empty vector and five recombinant plasmids were subsequently transformed into Escherichia coli strain BL21 for inducible expression. The expressed proteins were purified using ProteinIso Ni-IDA Resin (www.transgen.com.cn/) according to the product manual. The concentrations of purified proteins were measured using an Enhanced BCA Protein Assay Kit (www.beyotime.com/).
Assay of the Specific Enzyme Activity of ZmACS7
The specific enzyme activity assay was conducted using ∼8 pmol of newly purified HIS-ZmACS7N18 and HIS-ZmACS7Sdw3 fusion proteins in a 11.7-mL headspace bottle containing 0.5 mL of reaction buffer (50 mm Tris-HCl, 4 mm dithiothreitol, 20 μm pyridoxal 5′-phosphate, and 150 μm SAM [pH 8.0]) at 30°C for 30 min. After 100 μL of 20 mm HgCl2 was added to terminate the reaction, the sample was precooled on ice for 3 min. Following the addition of 100 μL of a 1:1 (v/v) cold mixture of commercial NaClO (10% [w/v] available chlorine) and saturated NaOH, the vial was immediately sealed with a rubber cap and incubated on ice for 3 min, shaking once during the incubation. The ethylene levels in the vial headspace were detected by gas chromatography (GC-17A, Shimadzu; Luo et al., 2014).
Cell-Free Protein Degradation Assay
A cell-free protein degradation assay was carried out as described (Wang et al., 2018b), with some modifications. Ten-day-old wild-type B73 seedlings were harvested and ground into fine powder in liquid nitrogen. Total proteins were extracted with degradation buffer (pH 8.0) containing 50 mm Tris-HCl, 0.5 m Suc, 1 mm MgCl2, 10 mm EDTA, 5 mm dithiothreitol, and 1 mm ATP. Cell debris was removed by two 10-min centrifugations at 12,000g and 4°C, and the supernatant was filtered through Miracloth (Calbiochem). The extract was collected and supplemented with 50 μm MG132 (APExBIO) or dimethyl sulfoxide as a control. The newly purified HIS-CTDN18 (300 ng), HIS-CTDSdw3 (308.5 ng), and HIS-CTDΔVHAS (298.1 ng, the same molar amount as HIS-CTDN18 and HIS-CTDSdw3) proteins were incubated with 100 μL of total protein extracts (∼500 μg total proteins). The protein degradation reactions were carried out in a 25°C water bath, and SDS-PAGE loading buffer was added to terminate the reaction at different time points. Anti-HIS antibody was used to detect HIS-CTD protein abundance by immunoblot analysis, and the band intensity was quantified using ImageJ software (imagej.nih.gov/ij/).
Transcriptome Analysis
The developing internode between leaf 7 and leaf 8 and the developing ligular regions of leaf 8 and leaf 9 of N18 and Sdw3 seedlings at the V7 stage were separately sampled for total RNA extraction, with three biological replicates. Library construction was performed using the Illumina HiSeq mRNA construction method and sequenced on the Illumina Hiseq 4000 platform. RNA-seq data analysis was conducted as previously described (Trapnell et al., 2012). Briefly, after low-quality bases and adapter sequences were removed, the clean data were mapped to the maize genome (AGPv3) with TopHat v2.1.0 (ccb.jhu.edu/software/tophat/). RNA-seq data were normalized as fragments per kilobase of exon per million fragments mapped. The Cufflinks package was used to assemble transcripts, compare and merge assembles, and identify DEGs. When Sdw3 is compared with N18, a cutoff of twofold change or greater in expression and a false discovery rate of q value <0.05 were used to screen the DEGs. Two subsets of 15 DEGs were selected at random from the transcriptomes of the internode and ligular region to confirm the reliability of the RNA-seq data via RT-qPCR. The primers used for verification of the relative mRNA levels of the DEGs via RT-qPCR are listed in Supplemental Table S1. MapMan software first was used to perform a global view of the pathways of DEGs in the internode and ligular region based on the functional annotation file “Zm_B73_5b_FGS_cds_2012” (https://mapman.gabipd.org), and detailed gene annotations were obtained from the Gramene database (www.gramene.org). We then artificially selected and grouped the genes related to ethylene biosynthesis, signaling and response, the genes determining cell elongation, the genes reported to regulate leaf angle, and the genes that have been reported to determine flowering time in maize to analyze the correlation of expression patterns of DEGs and phenotypic changes.
Statistical Analysis
Microsoft Excel 2016 was used for data processing. F-test and Student’s t test were performed to compare two samples using the data analysis module in Excel. Tukey’s multiple comparison method was used after one-way ANOVA to compare the means of three or more samples using IBM SPSS software.
Accession Numbers
Some DNA and protein sequences from this study can be found in the NCBI’s GenBank database under the following accession numbers: the transposon including target-site duplication sequence: MK450314; ZmACS7N18 and CTDN18, MK450317; CTDΔVHAS, MN815769; ZmACSSdw3 and CTDSdw3, MK450315; the putative promoter sequence of ZmACS7, MK450316. RNA-seq data from this article can be found in the NCBI's Short Read Archive sequence database under the following accession number: GSE127448.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypic comparison of N18, Sdw3/−, and Sdw3 plants.
Supplemental Figure S2. Verification of the transposon insertion by PCR and Sanger sequencing.
Supplemental Figure S3. Phylogenetic analysis of maize and Arabidopsis ACS isoforms and protein sequence alignment of ZmACS7 and ZmACS2 with AtACS6.
Supplemental Figure S4. Internode and leaf morphology in ethephon-treated N18 plants.
Supplemental Figure S5. Identification of positive T0 plants of the pZmACS7L329:ZmACS7Sdw3 construct.
Supplemental Figure S6. Phenotypic comparison of L329 and L329’.
Supplemental Figure S7. Phenotypic analyses of PC and ZmACS7 OX lines.
Supplemental Figure S8. Purified HIS-tagged proteins and protein turnover of CTDN18 and CTDΔVHAS.
Supplemental Figure S9. Verification of DEGs (based on RNA-seq data) by RT-qPCR analysis.
Supplemental Figure S10. Heat map representing expression patterns of DEGs obtained by RNA-seq.
Supplemental Figure S11. Increased ZmACS7 function alters root morphology in maize.
Supplemental Figure S12. Expression atlas of four ACS isoforms in maize.
Supplemental Figure S13. Analyses of ear traits in Sdw3 NILs, PC and ZmACS7 OX lines.
Supplemental Figure S14. Similarity of expression patterns and functions of ZmACS2 and ZmACS7.
Supplemental Figure S15. Analysis of transgene-derived lines by knocking out ZmACS7.
Supplemental Figure S16. Alignment of protein sequences of ZmACS6 and Arabidopsis ACS isoforms.
Supplemental Table S1. The primers used in this study.
Acknowledgments
We are grateful to Dr. Zhongwei Lin (China Agricultural University) for critically reading and commenting on our manuscript. We appreciate Dr. Hongxu Dong (University of Georgia) for polishing this article and Hongbing Liu and Yumin Huang (China Agricultural University) for help in the preliminary processing of the transcriptome data. We appreciate Dr. Junping Gao (China Agricultural University) for sharing equipment and expertise for ethylene measurement and Drs. Peiyong Xin and Jinfang Chu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for their expertise in ACC measurement.
Footnotes
This work was supported by National Natural Science Foundation of China (grant nos. 91735305 and 91535206 to W.J., and 31271734 to B.L.).
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP(2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken RM, Smucker AJ(1996) Root system regulation of whole plant growth. Annu Rev Phytopathol 34: 325–346 [DOI] [PubMed] [Google Scholar]
- Argueso CT, Hansen M, Kieber JJ(2007) Regulation of ethylene biosynthesis. J Plant Growth Regul 26: 92–105 [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP(1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best NB, Hartwig T, Budka J, Fujioka S, Johal G, Schulz B, Dilkes BP(2016) Nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol 171: 2633–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella JR, Arteca RN, Frangos JA(1995) A mechanical strain-induced 1-aminocyclopropane-1-carboxylic acid synthase gene. Proc Natl Acad Sci USA 92: 1595–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg SP, Burg EA(1966) Auxin-induced ethylene formation: Its relation to flowering in the pineapple. Science 152: 1269. [DOI] [PubMed] [Google Scholar]
- Cassani E, Bertolini E, Badone CF, Landoni M, Gavina D, Sirizzotti A, Pilu R(2009) Characterization of the first dominant dwarf maize mutant carrying a single amino acid insertion in the VHYNP domain of the dwarf8 gene. Mol Breed 24: 375–385 [Google Scholar]
- Chae HS, Faure F, Kieber JJ(2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR(1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Choudhury SR, Roy S, Sengupta DN(2013) C-terminal phosphorylation is essential for regulation of ethylene synthesizing ACC synthase enzyme. Plant Signal Behav 8: e23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD(2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V(1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Cox WJ, Andrade HF(1988) Growth, yield, and yield components of maize as influenced by ethephon. Crop Sci 2: 536–542 [Google Scholar]
- Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG(2008) Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol 147: 2054–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng ZY, Liu LT, Li T, Yan S, Kuang BJ, Huang SJ, Yan CJ, Wang T(2015) OsKinesin-13A is an active microtubule depolymerase involved in glume length regulation via affecting cell elongation. Sci Rep 5: 9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugardeyn J, Van Der Straeten D(2008) Ethylene: Inhibitor and stimulator of plant growth In Bögre L, and Beemster G, eds, Plant Growth and Signaling, Plant Cell Monographs, Vol 10 Springer, Berlin, Heidelberg, pp 199–221 [Google Scholar]
- Duvick DN.(2004) Long-term selection in a commercial hybrid maize breeding program. Plant Breed Rev 24: 109–151 [Google Scholar]
- El-Brolosy MA, Stainier DYR(2017) Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet 13: e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TM, Timmermans M(2009) Axial patterning of the maize leaf In Bennetzen JL, and Hake S, eds, Handbook of Maize: Its Biology. Springer, New York, pp 161–178 [Google Scholar]
- Gallie DR, Geisler-Lee J, Chen J, Jolley B(2009) Tissue-specific expression of the ethylene biosynthetic machinery regulates root growth in maize. Plant Mol Biol 69: 195–211 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Young TE(2004) The ethylene biosynthetic and perception machinery is differentially expressed during endosperm and embryo development in maize. Mol Genet Genomics 271: 267–281 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, Caldwell C, Gallie DR(2010) Expression of the ethylene biosynthetic machinery in maize roots is regulated in response to hypoxia. J Exp Bot 61: 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben JE, Bao X, Bate NJ, DeBruin JL, Dolan D, Hasegawa D, Helentjaris TG, Lafitte RH, Lovan N, Mo H, et al. (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought-stress conditions. Plant Biotechnol J 12: 685–693 [DOI] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S(2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B(2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108: 19814–19819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hedden P.(2003) The genes of the green revolution. Trends Genet 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Hébert Y, Guingo E, Loudet O(2001) The response of root/shoot partitioning and root morphology to light reduction in maize genotypes. Crop Sci 41: 363–371 [Google Scholar]
- Hochholdinger F.(2009) The maize root system: Morphology, anatomy, and genetics In Bennetzen JL, and Hake SC, eds, Handbook of Maize: Its Biology. Spring, New York, pp 145–160 [Google Scholar]
- Joo S, Liu Y, Lueth A, Zhang S(2008) MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Kir G, Ye H, Nelissen H, Neelakandan AK, Kusnandar AS, Luo A, Inzé D, Sylvester AW, Yin Y, Becraft PW(2015) RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol 169: 826–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan TD, Oplinger ES(1987) Growth and yield of ethephon treated maize. Agron J 79: 130–134 [Google Scholar]
- Lee HY, Chen YC, Kieber JJ, Yoon GM(2017) Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. Plant J 91: 491–504 [DOI] [PubMed] [Google Scholar]
- Lelièvre JM, Latché A, Jones B, Bouzayen M, Pech JC(1997) Ethylene and fruit ripening. Physiol Plant 101: 727–739 [Google Scholar]
- Li D, Wang X, Zhang X, Chen Q, Xu G, Xu D, Wang C, Liang Y, Wu L, Huang C, et al. (2016) The genetic architecture of leaf number and its genetic relationship to flowering time in maize. New Phytol 210: 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang J, Qian Q, Xu Y, Zhang C, Xiao J, Du C, Luo W, Zou G, Chen M, et al. (2011) Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang X, Zhao Y, Li Y, Zhang G, Peng Z, Zhang J(2018) Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol J 16: 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D(2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S(2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ C(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ludwików A, Cieśla A, Kasprowicz-Maluśki A, Mituła F, Tajdel M, Gałgański Ł, Ziółkowski PA, Kubiak P, Małecka A, Piechalak A, et al. (2014) Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol Plant 7: 960–976 [DOI] [PubMed] [Google Scholar]
- Luo X, Chen Z, Gao J, Gong Z(2014) Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J 79: 44–55 [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Suttle JC(1991) The plant hormone ethylene. CRC Press, Boca Raton, FL [Google Scholar]
- Mock JJ, Pearce RB(1975) An ideotype of maize. Euphytica 24: 613–623 [Google Scholar]
- Moon J, Candela H, Hake S(2013) The Liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development 140: 405–412 [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M(1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11: 616–628 [DOI] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS(2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84 [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON(2006) Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol 142: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ(1968) Field investigations of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agron J 60: 422–424 [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Pierik R, Tholen D, Poorter H, Visser EJ, Voesenek LA(2006) The Janus face of ethylene: Growth inhibition and stimulation. Trends Plant Sci 11: 176–183 [DOI] [PubMed] [Google Scholar]
- Pitzschke A.(2015) Modes of MAPK substrate recognition and control. Trends Plant Sci 20: 49–55 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P(2003) EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Resnick JS, Rivarola M, Chang C(2008) Involvement of RTE1 in conformational changes promoting ETR1 ethylene receptor signaling in Arabidopsis. Plant J 56: 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM(1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Romero Navarro JA, Willcox M, Burgueño J, Romay C, Swarts K, Trachsel S, Preciado E, Terron A, Delgado HV, Vidal V, et al. (2017) A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nat Genet 49: 476–480 [DOI] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann E, et al. (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104: 11376–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Theologis A(1989) Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc Natl Acad Sci USA 86: 6621–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE.(2012) Ethylene and the regulation of plant development. BMC Biol 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Drummond BJ, Habben JE, Brugire N, Weers BP, Hakimi SM, Lafitte HR, Schussler JR, Mo H, Beatty M, et al. (2019) Ectopic expression of ARGOS8 reveals a role for ethylene in root-lodging resistance in maize. Plant J 97: 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SR, Oelke EA, Wiersma JV, Lueschen WE, Warnes DD(1988) Spring wheat and barley responses to ethephon. Agron J 80: 829–834 [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A(2011) Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet 7: e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Straeten DV, Van Der Straeten D(1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu P, Grosskopf DG, Felix G, Boller T(1994) The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiol 106: 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM(2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S(2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JH, Teng WT, Yan JB, Ma XQ, Meng YJ, Dai JR, Li JS(2007) Genetic dissection of plant height by molecular markers using a population of recombinant inbred lines in maize. Euphytica 155: 117–124 [Google Scholar]
- Teng F, Zhai L, Liu R, Bai W, Wang L, Huo D, Tao Y, Zheng Y, Zhang Z(2013) ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J 73: 405–416 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M(2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES(2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L(2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Woeste KE, Theologis A, Kieber JJ(1998) Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci USA 95: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Waters CA, Freeling M(1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev 12: 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Smith SM, Li J(2018a) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69: 437–468 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang S, Hu K, Yang J, Xin X, Zhou W, Fan J, Cui F, Mou B, Zhang S, et al. (2018b) The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiol 176: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR(2002) Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR(2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wang S, Park YS, Yang Y, Borrego EJ, Isakeit T, Gao X, Kolomiets MV(2017) Seed-derived ethylene facilitates colonization but not aflatoxin production by Aspergillus flavus in maize. Front Plant Sci 8: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Zhang X, Zhang Z, Liu H, Lin Z(2018) A new allele of the Brachytic2 gene in maize can efficiently modify plant architecture. Heredity 121: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Helentjaris T(1995) The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell 7: 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuriyanghan H, Zhang B, Cao WH, Ma B, Lei G, Liu YF, Wei W, Wu HJ, Chen LJ, Chen HW, et al. (2009) The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21: 1473–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin P, Guo Q, Li B, Cheng S, Yan J, Chu J(2020) Tailored high-efficiency sample pretreatment method for simultaneous quantification of 10 classes of known endogenous phytohormones. Plant Comm 1: 100047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A, Gao Y, Ye L, Zhang W, Cai L, Ching A, Llaca V, Johnson B, Liu L, Yang X, et al. (2015) A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J Exp Bot 66: 3791–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ(2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S(2015) Ethylene biosynthesis and regulation in plants. Ethylene in Plants 1–25 [Google Scholar]
- Xue J, Zhao Y, Gou L, Shi Z, Yao M, Zhang W(2016) How high plant density of maize affects basal internode development and strength formation. Crop Sci 56: 3295–3306 [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A(2003) Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem 278: 49102–49112 [DOI] [PubMed] [Google Scholar]
- Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, Li K, Yang N, Li Y, Zhong T, et al. (2013) CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA 110: 16969–16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE(1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 115–189 [Google Scholar]
- Yip WK, Moore T, Yang SF(1992) Differential accumulation of transcripts for four tomato 1-aminocyclopropane-1-carboxylate synthase homologs under various conditions. Proc Natl Acad Sci USA 89: 2475–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR(2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Wang KL, Chang CM, Mori K, Uchida E, Ecker JR(2006) The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Mol Biol 62: 427–437 [DOI] [PubMed] [Google Scholar]
- Young TE, Meeley RB, Gallie DR(2004) ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J 40: 813–825 [DOI] [PubMed] [Google Scholar]
- Yu YB, Adams DO, Yang SF(1979) 1-Aminocyclopropanecarboxylate synthase, a key enzyme in ethylene biosynthesis. Arch Biochem Biophys 198: 280–286 [DOI] [PubMed] [Google Scholar]
- Zhang TC, Qiao Q, Zhong Y(2012) Detecting adaptive evolution and functional divergence in aminocyclopropane-1-carboxylate synthase (ACS) gene family. Comput Biol Chem 38: 10–16 [DOI] [PubMed] [Google Scholar]