Astaxanthin, derived from the ketolation of zeaxanthin, is esterified mainly with oleic acid, and accumulates independently of de novo fatty acid synthesis in the green alga Chromochloris zofingiensis.
Abstract
The biosynthesis of astaxanthin, a high-value keto-carotenoid with broad industrial applications, remains unambiguous in algae. Here, we dissected the astaxanthin biosynthetic pathway and the coordination between astaxanthin and triacylglycerol (TAG) biosynthesis in the emerging model alga Chromochloris zofingiensis. In vivo and in vitro experiments demonstrated that astaxanthin, utilizing the methylerythritol phosphate pathway-derived isopentenyl diphosphate as the building block, was synthesized from β-carotenoid ketolase-mediated ketolation of zeaxanthin rather than β-carotenoid hydroxylase-mediated hydroxylation of canthaxanthin, thus leading to the buildup of astaxanthin and canthaxanthin as end products in C. zofingiensis. The synthesized astaxanthin, stored in TAG-filled lipid droplets, was esterified mainly with the fatty acid C18:1, which was not catalyzed by any acyltransferase previously proposed. Astaxanthin accumulated in a well-coordinated manner with TAG, supported by the coordinated up-regulation of both biosynthetic pathways at the transcriptional level. Nevertheless, astaxanthin and TAG showed no interdependence: inhibition of de novo fatty acid biosynthesis severely attenuated TAG biosynthesis but promoted the accumulation of astaxanthin, particularly in the diester form, leading to a fivefold increase in the astaxanthin/TAG ratio; however, inhibition of astaxanthin biosynthesis showed little effect on TAG accumulation. Our data suggest that an increase in astaxanthin accumulation following inhibition of de novo fatty acid biosynthesis, which is not regulated at the transcriptional level, is likely derived from the conversion of other carotenoids rather than from a shunt of carbon flux from lipid biosynthesis. Combined, these findings further our understanding of astaxanthin biosynthesis and provide a feasible strategy for promoting astaxanthin content and purity in algae.
Microalgae have the potential to produce a broad range of compounds, including oils, functional proteins, ω-3 polyunsaturated fatty acids, and carotenoids, depending on the algal species/strains (Hu et al., 2008; Tang et al., 2016). Astaxanthin is a high-value keto-carotenoid with the highest antioxidant activity found in nature and is believed to have broad applications in food, feed, nutraceutical, and pharmaceutical industries (Guerin et al., 2003; Ambati et al., 2014; Dose et al., 2016; Galasso et al., 2018; Fang et al., 2019). Although many algae can synthesize astaxanthin, astaxanthin biosynthesis and regulation are most comprehensively understood in Haematococcus pluvialis and Chromochloris zofingiensis (Boussiba, 2000; Kopecky et al., 2000; Lemoine and Schoefs, 2010; Liu et al., 2014; Shah et al., 2016; Solovchenko and Neverov, 2017). Note that the latter was previously named as Chlorella zofingiensis or Muriella zofingiensis and has now been assigned under the genus Chromochloris (Fucikova and Lewis, 2012).
H. pluvialis and C. zofingiensis are closely related green algae, yet the latter distinguishes itself from the former in that it is capable of utilizing sugars, in particular Glc, as the sole carbon and energy source for growth. C. zofingiensis can achieve ultra-high cell densities of more than 70 g L−1, which is around 10-fold higher than H. pluvialis (Ip and Chen, 2005; Liu et al., 2010a; Sun et al., 2019). Its growth robustness under multitrophic culture conditions has enabled C. zofingiensis to become an alternative to H. pluvialis as a promising natural astaxanthin producer (Rise et al., 1994; Bar et al., 1995; Del Campo et al., 2004; Ip et al., 2004; Liu et al., 2014; Mulders et al., 2014; Sun et al., 2019). However, C. zofingiensis has a lower astaxanthin content (0.7% versus 4% in H. pluvialis; Orosa et al., 2001; Ranjbar et al., 2008; Liu et al., 2014). Elucidating the carotenogenic pathways and regulatory mechanisms for astaxanthin biosynthesis may help uncover the bottlenecks of astaxanthin accumulation in C. zofingiensis and allow rational genetic engineering of this alga for improved astaxanthin production.
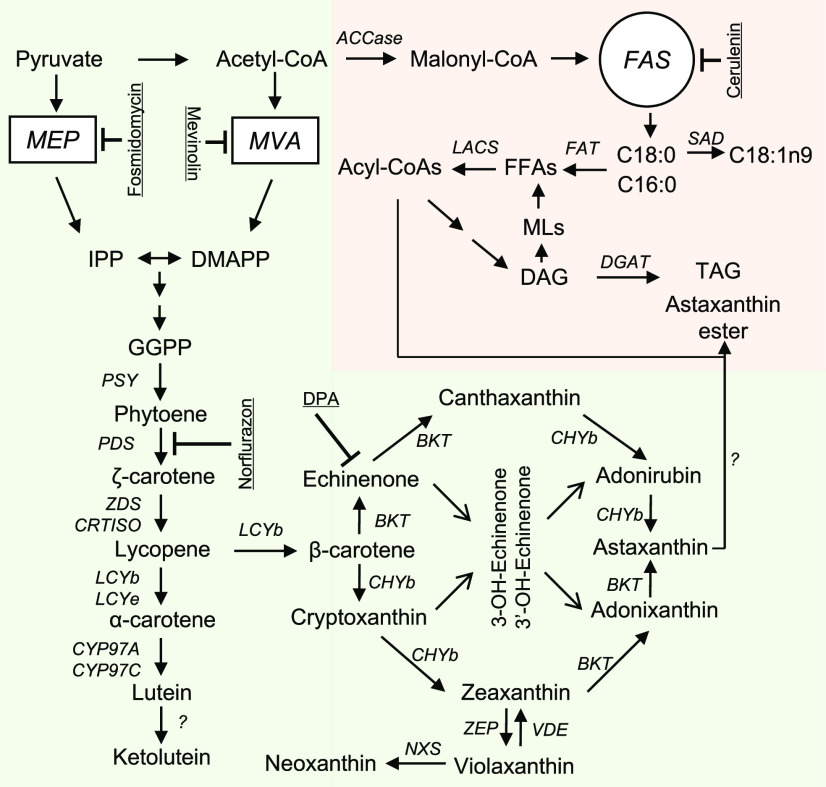
Generally, algae use similar pathways to land plants for primary carotenoid biosynthesis (Sandmann et al., 2006; Rodriguez-Concepcion et al., 2018), which include (1) synthesis of isopentenyl diphosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), the five-carbon precursors; (2) condensation of isoprene units to form the colorless C40 carotene phytoene; (3) step-wise desaturation of phytoene to lycopene; (4) cyclization of lycopene to α-carotene and β-carotene; and (5) formation of hydroxylated carotenoids such as lutein and zeaxanthin (Fig. 1). Aside from the primary carotenoids, H. pluvialis and C. zofingiensis are able to synthesize secondary carotenoids (keto derivatives of certain primary carotenoids) via additional ketolation and hydroxylation steps (Liu et al., 2014; Mulders et al., 2015; Shah et al., 2016), which are typically absent in land plants except for Adonis aestivalis (Cunningham and Gantt, 2011). These ketolation steps are mediated by β-carotenoid ketolase (BKT) and have been shown to occur outside of the chloroplast in H. pluvialis (Grünewald et al., 2001; Grunewald and Hagen, 2001; Schoefs et al., 2001).
Figure 1.
Schematic illustration of astaxanthin and TAG synthesis. Specific inhibitors are underlined. AAT, Long-chain-alcohol O-fatty-acyltransferase; ACCase, acetyl-CoA carboxylase; BKT, β-carotenoid ketolase; CHYb, β-carotenoid hydroxylase; CRTISO, carotenoid isomerase; CYP97A, cytochrome P450 β-hydroxylase; CYP97C, cytochrome P450 ε-hydroxylase; DAG, diacylglycerol; DGAT, diacylglycerol acyltransferase; DMAPP, dimethylallyl pyrophosphate; DPA, diphenylamine; FAS, fatty acid synthesis; GGPP, geranylgeranyl diphosphate; IPP, isopentenyl diphosphate; LACS, long-chain acyl-CoA synthetase; LCYb, lycopene β-cyclase; LCYe, lycopene ε-cyclase; MEP, methylerythritol phosphate pathway; MLs, membrane lipids; MVA, mevalonate pathway; NXS, neoxanthin synthase; PDS, phytoene desaturase; PSY, phytoene synthase; SAD, stearoyl-ACP desaturase; TAG, triacylglycerol; VDE, violaxanthin deepoxidase; ZDS, ζ-carotene desaturase; ZEP, zeaxanthin epoxidase.
There are multiple routes for the synthesis of astaxanthin from β-carotene, involving two hydroxylation and two ketolation steps in total (Fraser et al., 1997, 1998). The route with two step-wise ketolation reactions followed by two step-wise hydroxylation reactions is thought to contribute to the major astaxanthin biosynthesis and accumulation in H. pluvialis (Fraser et al., 1997; Schoefs et al., 2001). Unlike the free form in the red yeast Xanthophyllomyces dendrorhous (Schmidt et al., 2011), astaxanthin in H. pluvialis is present predominantly in the form esterified with fatty acids at either or both hydroxyl groups (Miao et al., 2006; Holtin et al., 2009; Gwak et al., 2014), catalyzed by an acyltransferase yet to be clarified (Chen et al., 2015). Inhibition of de novo fatty acid synthesis by specific inhibitors severely attenuates triacylglycerol (TAG) accumulation and meanwhile blocks astaxanthin biosynthesis in H. pluvialis (Schoefs et al., 2001; Zhekisheva et al., 2005; Chen et al., 2015), suggesting the presence of potential cross-talk between the two biosynthetic pathways.
Compared with H. pluvialis, only a limited number of carotenogenic genes from C. zofingiensis have been cloned and partially characterized, including those encoding phytoene synthase, phytoene desaturase, lycopene β-cyclase, lycopene ε-cyclase, zeaxanthin epoxidase, β-carotenoid hydroxylase (CHYb), and BKT (Huang et al., 2006, 2008; Li et al., 2009; Cordero et al., 2010, 2011, 2012; Liu et al., 2010b; Couso et al., 2012). Recently, the availability of whole-genome sequence and transcriptomic data has benefited the reconstruction of carotenogenic pathways in C. zofingiensis (Huang et al., 2016; Roth et al., 2017, 2019; Liu et al., 2019; Ye and Huang, 2019; Zhang et al., 2019; Mao et al., 2020). Previous work suggests that C. zofingiensis may differ from H. pluvialis with respect to the astaxanthin biosynthetic route (Wang and Chen, 2008; Liu et al., 2014) and cross-talk between astaxanthin and TAG biosynthesis (Liu et al., 2016b); however, the exact molecular mechanisms remain ambiguous.
To further decipher these questions, we performed an in-depth investigation by integrating lipidomics, carotenoid profiling, and other biochemical analyses in the presence of various chemical inhibitors; in vitro enzymatic assays; and a transcriptional analysis with selected inhibitor treatment. The in vitro enzymatic assay demonstrated that CHYb had no activity on the conversion of canthaxanthin to astaxanthin whereas BKT was able to convert zeaxanthin to astaxanthin via the intermediate adonixanthin, supporting that, in C. zofingiensis, astaxanthin is synthesized from the ketolation of zeaxanthin rather than the hydroxylation of canthaxanthin. Astaxanthin biosynthesis correlated well with TAG accumulation, as supported by the coordinated up-regulation of both biosynthetic pathways at the transcriptional level. Inhibition of de novo fatty acid biosynthesis that attenuated both total fatty acid (TFA) and TAG accumulation unexpectedly stimulated astaxanthin accumulation, particularly diester, leading to an approximately 5-fold increase in the astaxanthin/TFA ratio. We discuss the insights of our findings into astaxanthin biosynthesis and regulation in C. zofingiensis, an emerging model alga for studying carotenogenesis and lipogenesis.
RESULTS
C. zofingiensis Utilizes the Methylerythritol Phosphate Pathway for Carotenoid Biosynthesis
There are two pathways present in land plants for IPP/DMAPP biosynthesis, the cytosolic mevalonate (MVA) pathway starting from acetyl-CoA and the plastidial methylerythritol phosphate (MEP) pathway starting from pyruvate and glyceraldehyde-3-phosphate (Rodriguez-Concepcion et al., 2018). The MEP pathway rather than the MVA pathway has been suggested to contribute to IPP/DMAPP precursors for carotenoid biosynthesis in the green algae H. pluvialis and Chlamydomonas reinhardtii (Hagen and Grunewald, 2000; Lohr et al., 2005). Accordingly, genes involved in the MEP pathway rather than the MVA pathway are all present in the two algae (Lohr et al., 2005; Gwak et al., 2014).
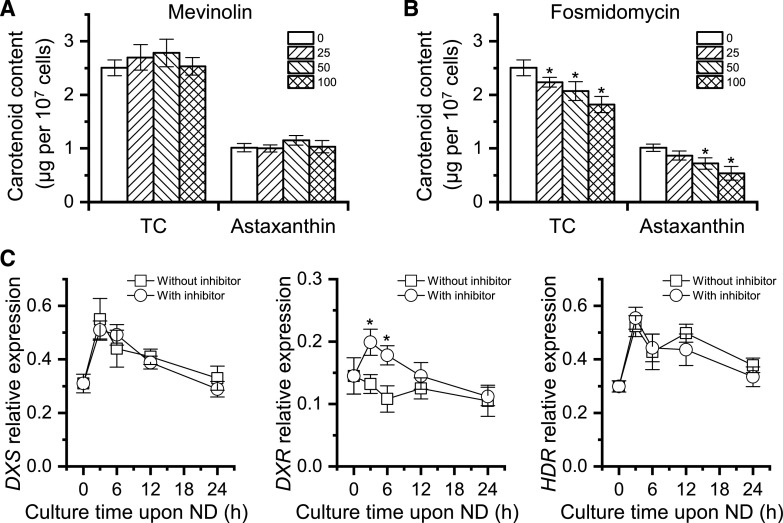
We searched the genome of C. zofingiensis and identified all genes involved in the MEP pathway, but it lacks some genes involved in the MVA pathway (Supplemental Fig. S1). Two inhibitors, mevinolin and fosmidomycin that target the MVA and MEP pathway, respectively, were applied to C. zofingiensis cultures under nitrogen deprivation (ND) conditions. As indicated by Figure 2A, mevinolin showed no impact on either total carotenoids (TC) or astaxanthin, even when applied at 100 µm. By contrast, fosmidomycin affected the accumulation of both TC and astaxanthin in a concentration-dependent manner, leading to decreases of 27% and 47% for TC and astaxanthin, respectively, at the concentration of 100 µm (Fig. 2B).
Figure 2.
Effects of mevinolin and fosmidomycin on carotenoid synthesis in C. zofingiensis. A and B, TC and astaxanthin as affected by different concentrations (micromolar) of mevinolin (A) and fosmidomycin (B). Cells were cultured under ND conditions for 48 h. Asterisks indicate significant differences (Student’s t test, *P < 0.05) compared with the control without inhibitors (0 μm). C, Transcriptional expression of DXS, DXR, and HDR genes with or without 100 μm fosmidomycin (inhibitor). The gene expression level was normalized to the internal control β-actin gene. Asterisks indicate significant differences (Student’s t test, *P < 0.05) in gene expression between samples treated with or without fosmidomycin. Data represent means ± sd (n = 3).
To understand the expression pattern of the MEP pathway, reverse transcription quantitative PCR (RT-qPCR) analysis was performed for the genes encoding 1-deoxy-d-xylulose 5-phosphate (DXP) synthase (DXS), DXP reductoisomerase (DXR), and 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (HDR), which are generally considered as key enzymes controlling carbon flux of the MEP pathway (Vranová et al., 2013). Upon ND induction, DXS and HDR showed mild increases, whereas DXR had little change at the transcriptional level (Fig. 2C), indicative of just a slight stimulation of the MEP pathway in C. zofingiensis. This differs from H. pluvialis, in which the MEP pathway is considerably up-regulated under carotenogenic conditions (Gwak et al., 2014; Huang et al., 2019). Although exhibiting little effect on the expression of DXS and HDR, fosmidomycin stimulated DXR expression (Fig. 2C), probably induced by the substrate DXP that may accumulate when the inhibitor is present.
These data suggest that, similar to H. pluvialis, C. zofingiensis is likely to employ the MEP pathway to supply IPP/DMAPP for the biosynthesis of carotenoids, including astaxanthin. It is possible that the MVA pathway was lost during the evolution of green algae (Vranová et al., 2013). However, we cannot completely rule out the presence of the MVA pathway or an MVA-like pathway in these algae.
Astaxanthin Is from Ketolation of Zeaxanthin Rather Than Hydroxylation of Canthaxanthin
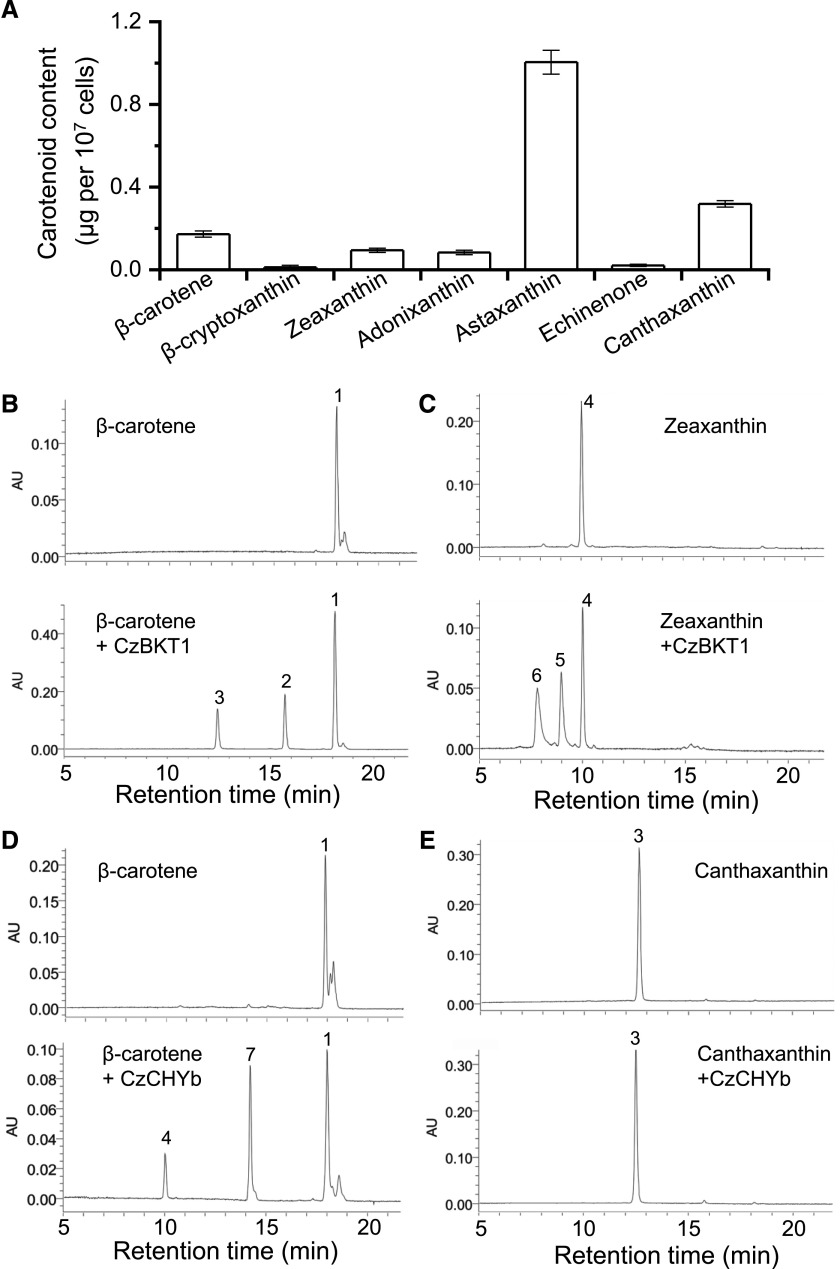
Carotenoid profiling of ND-induced C. zofingiensis cells confirmed the presence of intermediates in the astaxanthin biosynthetic pathway, including β-cryptoxanthin, zeaxanthin, adonixanthin, echinenone, and canthaxanthin, which are hydroxylated and/or ketolated derivatives of β-carotene (Fig. 3A). It is worth mentioning that adonirubin, the intermediate of canthaxanthin hydroxylation for astaxanthin formation, was not detected. Notably, astaxanthin represents the most abundant carotenoid, followed by canthaxanthin. In this context, we infer that in C. zofingiensis astaxanthin may be synthesized via zeaxanthin ketolation instead of canthaxanthin hydroxylation, leading to the accumulation of canthaxanthin as an end product as well. In corroboration, when C. zofingiensis was exposed to astaxanthin induction conditions, the addition of diphenylamine (DPA), a specific inhibitor of BKT, or the dysfunction of BKT caused by random mutagenesis both resulted in an increase in zeaxanthin and a decrease in astaxanthin (Wang and Chen, 2008; Roth et al., 2017; Huang et al., 2018; Ye and Huang, 2019).
Figure 3.
Clarification of astaxanthin biosynthetic route in C. zofingiensis. A, Levels of β-carotene and its derivatives in C. zofingiensis under ND conditions for 48 h. Data represent means ± sd (n = 3). B to E, HPLC analysis of in vitro enzymatic reaction products of CzBKT1 and CzCHYb: β-carotene + CzBKT1 (B), zeaxanthin + CzBKT1 (C), β-carotene + CzCHYb (D), and canthaxanthin + CzCHYb (E). Peaks are as follows: 1, β-carotene; 2, echinenone; 3, canthaxanthin; 4, zeaxanthin; 5, adonixanthin; 6, astaxanthin; 7, β-cryptoxanthin. AU, Arbitrary units.
To elucidate the substrate specificity of BKT and CHYb, we performed an in vitro enzymatic assay. Although C. zofingiensis harbors two BKT genes, BKT1 rather than BKT2 contributes to astaxanthin biosynthesis (Roth et al., 2017; Huang et al., 2018; Ye and Huang, 2019). Therefore, only BKT1 was investigated in our experiment. When incubating BKT1 with β-carotene, echinenone and canthaxanthin were produced (Fig. 3B). When incubating BKT1 with zeaxanthin, adonixanthin and astaxanthin were formed (Fig. 3C). On the other hand, CHYb was able to catalyze the production of β-cryptoxanthin and zeaxanthin from β-carotene but had no observed activity on canthaxanthin (Fig. 3, D and E). Nevertheless, we cannot exclude the possibility that the in vitro assay may lack certain cofactors causing the failure of CHYb to show activity on canthaxanthin. Overall, the in vitro data were consistent with the in vivo results shown in Figure 3A and strongly supported that astaxanthin in C. zofingiensis is derived from ketolation of zeaxanthin rather than hydroxylation of canthaxanthin. This differs from H. pluvialis, in which astaxanthin is mainly derived from the hydroxylation of canthaxanthin (Boussiba, 2000; Schoefs et al., 2001).
Induced Astaxanthin Is Esterified Mainly with Oleic Acid and Stored in Lipid Droplets
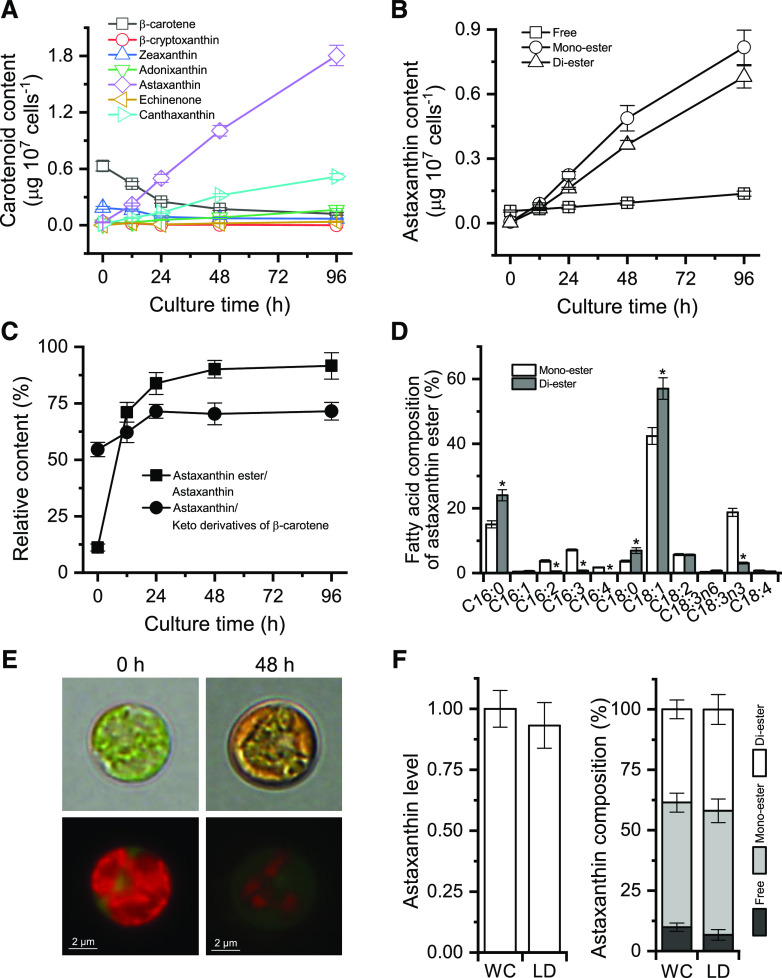
To investigate the pattern of accumulation and compartmentalization of astaxanthin, ND-induced C. zofingiensis cells over a 96-h period were examined. Quantification of carotenoids by HPLC analysis showed that under the favorable growth condition (0 h of ND), primary carotenoids such as β-carotene dominated, with the presence of trace amounts of secondary carotenoids (Fig. 4A). Upon ND induction, β-carotene exhibited a drastic decline within the first 24 h and thereafter maintained a relatively stable level. Similarly, zeaxanthin showed a decrease during the first 24 h of ND, yet to a lesser extent than β-carotene. By contrast, the secondary carotenoids astaxanthin and canthaxanthin increased considerably in response to ND and did not reach a maximum level even at the end of the culture period (96 h).
Figure 4.
Properties of astaxanthin synthesis and accumulation in C. zofingiensis. A, Time-course changes in the contents of β-carotene and its derivatives. B, Time courses of astaxanthin content in the form of free, monoester, and diester. C, Relative contents of astaxanthin ester to astaxanthin and of astaxanthin to keto derivatives of β-carotene. D, Fatty acid composition of astaxanthin monoester and diester at 48 h of ND. Asterisks indicate significant differences (Student’s t test, *P < 0.05) between monoester and diester. E, Microscopic views of C. zofingiensis cells at 0 and 48 h of ND. Top row, Bright field; bottom row, fluorescent field. Red indicates chlorophyll autofluorescence, whereas green indicates LDs stained with BODIPY. F, Comparison of astaxanthin levels and composition from whole-cell (WC) and LD fractions of C. zofingiensis cells after 48 h of ND. Equal amounts of cells were used for whole-cell and LD experiments. The LD astaxanthin level was normalized to whole-cell astaxanthin, which was set as 1. Data in A to D and F represent means ± sd (n = 3).
Clearly, during the late ND period (48–96 h), the decreased amount of β-carotene was much less than the increased amount of astaxanthin, supporting the presence of de novo β-carotene synthesis for sustaining continued astaxanthin accumulation (Schoefs et al., 2001). Free astaxanthin, present in a trace amount, showed only a slight increase during the whole culture period. By contrast, astaxanthin ester (monoester and diester), which was less than free astaxanthin at 0 h, exhibited a considerable increase in response to ND (Fig. 4B). Accordingly, the ratio of esterified astaxanthin to total astaxanthin increased over time and reached 92% after 96 h of ND (Fig. 4C). ND also stimulated the ratio of astaxanthin to keto derivatives of β-carotene (including astaxanthin, canthaxanthin, echinenone, and adonixanthin; Fig. 4C).
The fatty acid moiety of astaxanthin monoester consisted mainly of C18:1, C18:3n3, and C16:0. By contrast, C18:1 and C16:0 represented the predominant fatty acids of astaxanthin diester, accounting for 80% of the fatty acid moiety (Fig. 4D). In comparison with the vegetative cells under favorable growth conditions (Fig. 4E, 0 h of ND), stressed cells appeared orange peripherally due likely to the accumulation of astaxanthin in lipid droplets (LDs; Fig. 4E, 48 h of ND). Observations using a fluorescence microscope revealed that LDs (stained with the green fluorescence dye BODIPY) were situated peripherally and surrounded the shrunken chloroplast (Fig. 4E).
To confirm the compartmentalization of astaxanthin, LDs from algal cells after 48 h of ND were isolated and purified, which had minimized chlorophyll contamination (Supplemental Fig. S2). Carotenoid analysis showed no significant difference in either astaxanthin level or composition observed between the isolated LD fraction and whole cells (Fig. 4F), indicating that the bulk of astaxanthin is located in LDs. This is similar to H. pluvialis, in which astaxanthin is stored predominantly in LDs (Boussiba, 2000; Grunewald and Hagen, 2001; Peled et al., 2011).
Accumulation of Astaxanthin Is Accompanied by an Increase of TAG
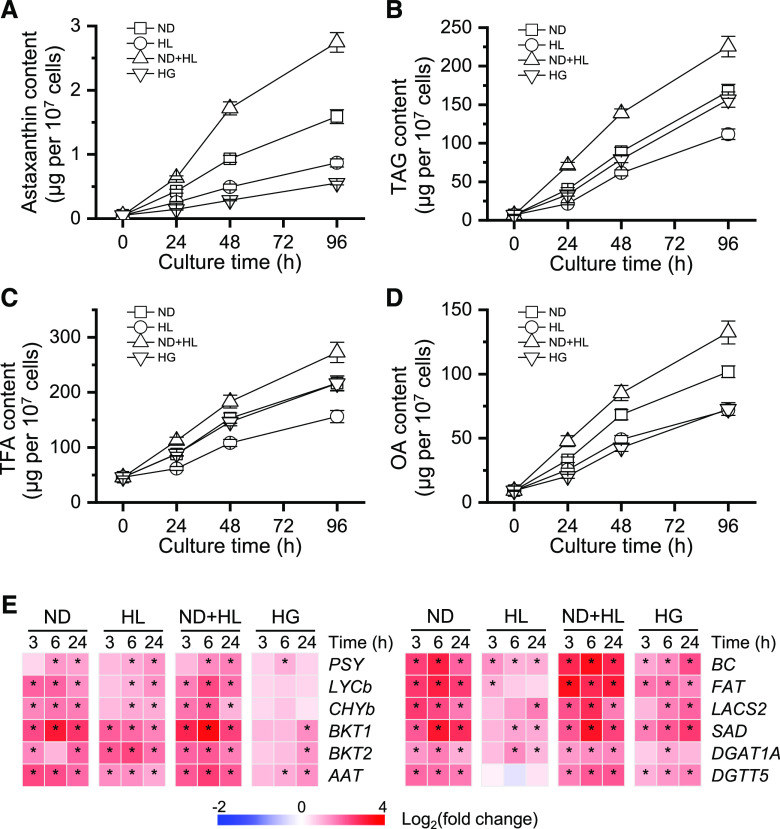
We investigated the relationship between the accumulation of astaxanthin and lipids under four culture conditions, ND, high light (HL), ND+HL, and heterotrophic growth with Glc (HG), which have been proven to be effective for the induction of astaxanthin in C. zofingiensis (Orosa et al., 2001; Li et al., 2009; Liu et al., 2010b, 2013; Mulders et al., 2014). As indicated by Figure 5, A and B, under nonstress conditions (0 h), trace amounts of astaxanthin (0.05 μg per 107 cells) and TAG (7 μg per 107 cells) were detected. Under ND stress conditions, C. zofingiensis showed a drastic increase in astaxanthin content, reaching 0.43 μg per 107 cells at 24 h and as high as 1.59 μg per 107 cells at 96 h. A sizable increase was also observed in TAG content, which was 167.4 μg per 107 cells at 96 h and approximately 24-fold higher than that at 0 h. Similarly, HL and HG enhanced the accumulation of both astaxanthin and TAG, but to a lesser extent as compared with ND. ND+HL, on the other hand, led to the highest levels of astaxanthin and TAG, reaching 2.75 and 225.5 μg per 107 cells, respectively. Clearly, all four examined conditions stimulated both astaxanthin and TAG accumulation, with a concomitant increase in the contents of TFA and oleic acid (OA; C18:1; Fig. 5, A–D). The plotting of astaxanthin and TAG contents revealed a good linear relationship, with R2 being over 0.987; the correlation between astaxanthin and OA contents was also obvious (R2 > 0.988; Supplemental Fig. S3). Furthermore, C. zofingiensis astaxanthin ester and TAG showed highly similar fatty acid profiles (Supplemental Fig. S4), consistent with the data in H. pluvialis (Zhekisheva et al., 2005; Lemoine et al., 2008). These results suggest that the biosynthesis and accumulation of astaxanthin and TAG are well coordinated in these algae.
Figure 5.
Accumulation of astaxanthin and lipids under various inducing conditions of ND, HL, ND+HL, and HG. A to D, Time courses of astaxanthin content (A), TAG content (B), TFA content (C), and OA content (D). E, Heat map showing the transcript dynamics of selected genes involved in astaxanthin and TAG synthesis under different inducing conditions. The data are expressed as log2 (fold change) values of transcripts relative to the control (0 h), determined by RT-qPCR. Significant differences [absolute log2 (fold change) value > 1 and P < 0.01] are indicated with asterisks. Time refers to the duration in the induction conditions. Data represent means ± sd (n = 3).
To correlate the synthesis of astaxanthin and lipid with certain genes, 12 genes were analyzed at the transcriptional level, including six carotenogenic genes encoding phytoene synthase, lycopene β-cyclase, CHYb, BKT1 and BKT2, and the putative AAT (Roth et al., 2017; Zhang et al., 2019) and six TAG synthesis-related genes encoding biotin carboxylase (BC; a subunit of ACCase), acyl-ACP thioesterase (FAT), long-chain acyl-CoA synthetase (LCAS2), stearoyl ACP desaturase, and diacylglycerol acyltransferase (DGAT1A and DGTT5; Liu et al., 2019; Mao et al., 2019). All 12 genes were up-regulated by stress conditions, with ND+HL causing the greatest expression induction (Fig. 5E). The expression of LYCb, CHYb, BKT1, and AAT showed high correlation (R2 > 0.9) with astaxanthin content (Supplemental Fig. S5), indicating that these genes play important roles in astaxanthin synthesis. Similarly, the transcript levels of BC, FAT, and LCAS correlated well with TAG content (Supplemental Fig. S5), suggesting their function in TAG accumulation.
Inhibition of Astaxanthin Synthesis Has Little Effect on Lipid Accumulation
Carotenoid biosynthesis is susceptible to certain carotenogenic inhibitors such as norflurazon and DPA that specifically target phytoene desaturase and BKT, respectively (Harker and Young, 1995; Grunewald and Hagen, 2001; Schoefs et al., 2001; Zhekisheva et al., 2005; Liu et al., 2010b). Three concentrations of each inhibitor were tested for C. zofingiensis, none of which showed any compromising effect on cell growth (Supplemental Fig. S6A). Both norflurazon and DPA resulted in a sizable decrease in astaxanthin content in a concentration-dependent manner (Supplemental Fig. S7). However, the inhibition of astaxanthin (up to 67% decrease by norflurazon and 89% decrease by DPA) had little effect on TAG synthesis (Supplemental Fig. S7). Moreover, the two carotenogenic inhibitors only impacted the fatty acid profiles of TFA and TAG slightly (Supplemental Table S1).
Inhibition of de Novo Fatty Acid Biosynthesis Enhances Astaxanthin Accumulation
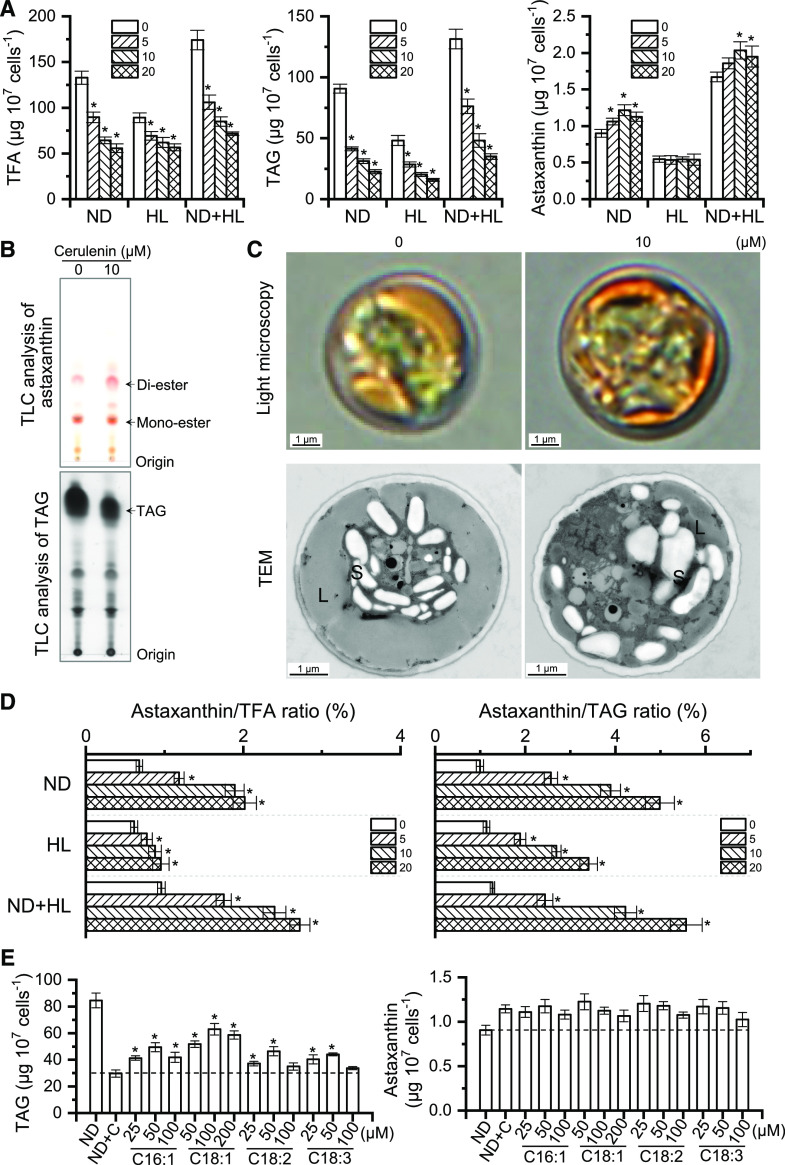
Specific inhibitors against fatty acid biosynthesis, such as cerulenin and sethoxydim, are reported to attenuate TAG accumulation and at the same time abolish astaxanthin synthesis in H. pluvialis under HL conditions (Schoefs et al., 2001; Zhekisheva et al., 2005; Chen et al., 2015). In this study, cerulenin with three concentrations (5, 10, and 20 μm) was employed to evaluate the responses of lipids and carotenoids in C. zofingiensis under various stress conditions, specifically ND, HL, and ND+HL (Fig. 6; Supplemental Fig. S8). Clearly, cerulenin inhibited fatty acid synthesis in a concentration-dependent manner under each stress conditions (Fig. 6A). The inhibition of TAG accumulation by cerulenin was also observed, as supported by gas chromatography-mass spectrometry (GC-MS) quantification (Fig. 6A), thin-layer chromatography (TLC) analysis (Fig. 6B), and transmission electron microscopy (less LDs; Fig. 6C). As for the relative abundance of fatty acids, cerulenin led to a great decrease in C18:1 with concomitant increases in C16:0, C18:2, and C18:3n3 under all stress conditions (Supplemental Table S2).
Figure 6.
Inhibition of de novo fatty acid synthesis enhanced astaxanthin accumulation in C. zofingiensis. A, Quantification of TFA, TAG, and astaxanthin contents in the presence of different cerulenin concentrations (0, 5, 10, and 20 µm) under ND, HL, or ND+HL conditions. Asterisks indicate significant differences (Student’s t test, *P < 0.05) compared with 0 µm cerulenin. B, TLC analysis of astaxanthin and TAG under ND with (10 µm) or without (0 µm) cerulenin. C, Representative images of algal cells under light microscopy and transmission electron microscopy (TEM) under ND with (10 µm) or without (0 µm) cerulenin. L, Lipid droplet; S, starch granule. D, Effects of cerulenin concentration on the astaxanthin/TFA (left) and astaxanthin/TAG (right) ratios. Asterisks indicate significant differences (Student’s t test, *P < 0.05) compared with 0 µm cerulenin. E, Effects of FFA feeding on TAG (left) and astaxanthin (right) synthesis under ND conditions. ND+C, Supplementation of 10 µm cerulenin under ND conditions. FFAs were fed into the cultures upon treatment with ND+C. Asterisks indicate significant differences (Student’s t test, *P < 0.05) compared with ND+C. Cultures after 48 h of ND induction were used for analysis. Data in A, D, and E represent means ± sd (n = 3).
Similar to TFA, TAG exhibited a considerable change in the fatty acid composition in response to cerulenin treatment, namely increased C16:0, C18:2, and C18:3n3 at the expense of C18:1 (Supplemental Table S2). Although phospholipids showed little change, the glycolipids monogalactosyl diacylglycerol (MGDG) and digalactosyl diacylglycerol (DGDG), particularly the polyunsaturated fatty acids (PUFAs) such as C16:2, C16:3, C18:2, and C18:3n3 in MGDG, were attenuated by cerulenin treatment (Supplemental Fig. S9).
Astaxanthin, on the other hand, did not decrease in response to cerulenin treatment (Fig. 6A; Supplemental Fig. S8). Specifically, under HL conditions, although TAG content was reduced by over 60%, cerulenin had little effect on astaxanthin accumulation (Fig. 6A). This is inconsistent with the phenomenon observed in H. pluvialis wherein the inhibition of TAG by approximately 40% caused a nearly complete block of astaxanthin biosynthesis (Schoefs et al., 2001; Zhekisheva et al., 2005). Interestingly, under ND conditions, astaxanthin accumulation was stimulated by cerulenin, as suggested by the HPLC quantification (approximately 35% more; Fig. 6A), TLC analysis (Fig. 6B), and microscopic observation (more intense orange color; Fig. 6C). The enhanced astaxanthin synthesis caused by cerulenin treatment was also observed under ND+HL conditions, although less prominently as compared with that under ND conditions (Fig. 6A; Supplemental Fig. S8).
It is worth noting that in C. zofingiensis, enhanced astaxanthin is attributed primarily to the increase of astaxanthin diester rather than the free astaxanthin or monoester, which showed only a slight change; canthaxanthin and β-carotene, on the other hand, exhibited considerable decreases (Table 1). The astaxanthin abundance relative to TFA or TAG, which is expressed as astaxanthin/TFA or astaxanthin/TAG ratio, exhibited a sizable increase upon cerulenin treatment regardless of the stress conditions (Fig. 6D). Specifically, the highest increase was achieved under ND conditions, 3- and 5-fold increases for astaxanthin/TFA and astaxanthin/TAG ratios, respectively. As TAG constitutes the hydrophobic core of LDs (Nguyen et al., 2011; Yoneda et al., 2016; Wang et al., 2019), the considerable increase in astaxanthin/TAG ratio in C. zofingiensis cells indicates that astaxanthin is highly concentrated in LDs, which was also reflected by the color intensity of LDs (Fig. 6C). By contrast, H. pluvialis showed a severe impairment of astaxanthin accumulation and astaxanthin/TFA and astaxanthin/TAG ratios when treated with the de novo fatty acid synthesis inhibitor under ND conditions (Supplemental Fig. S10). Therefore, C. zofingiensis, in contrast to H. pluvialis, synthesizes and accumulates astaxanthin independently of de novo fatty acid synthesis.
Table 1. Carotenoids as affected by cerulenin under ND conditions for 48 h.
Carotenoid contents are expressed as μg per 107 cells. Asterisks indicate the significance between control without cerulenin (0 µm) and the treatment (Student’s t test, *P < 0.05).
| Carotenoids | Cerulenin | |||
|---|---|---|---|---|
| 0 µm | 5 µm | 10 µm | 20 µm | |
| Astaxanthin free | 0.08 ± 0.01 | 0.09 ± 0.01 | 0.10 ± 0.01* | 0.10 ± 0.01 |
| Astaxanthin monoester | 0.46 ± 0.03 | 0.50 ± 0.03 | 0.50 ± 0.03 | 0.46 ± 0.02 |
| Astaxanthin diester | 0.36 ± 0.01 | 0.47 ± 0.02* | 0.62 ± 0.04* | 0.57 ± 0.03* |
| Adonixanthin | 0.10 ± 0.00 | 0.15 ± 0.01* | 0.21 ± 0.01* | 0.21 ± 0.02* |
| Canthaxanthin | 0.39 ± 0.02 | 0.38 ± 0.02 | 0.22 ± 0.02* | 0.25 ± 0.01* |
| β-Carotene | 0.13 ± 0.01 | 0.13 ± 0.02 | 0.09 ± 0.01* | 0.08 ± 0.01* |
| TC | 2.65 ± 0.14 | 2.56 ± 0.21 | 2.35 ± 0.18 | 2.29 ± 0.12* |
To investigate the effect of free fatty acid (FFA) feeding on astaxanthin synthesis, four fatty acids (i.e. C16:1, C18:1, C18:2, and C18:3n3) were applied to C. zofingiensis cells treated with cerulenin under ND conditions. Each fatty acid was tested with three concentrations that showed no significant impact on the algal growth (Supplemental Fig. S6B). FFA feeding led to the restoration of TAG to differential levels depending on FFAs and their concentrations (Fig. 6E). The enhanced astaxanthin biosynthesis caused by cerulenin treatment, however, was not affected by the feeding of FFAs (Fig. 6E).
Comparative Transcriptomic Analysis under Cerulenin Treatment
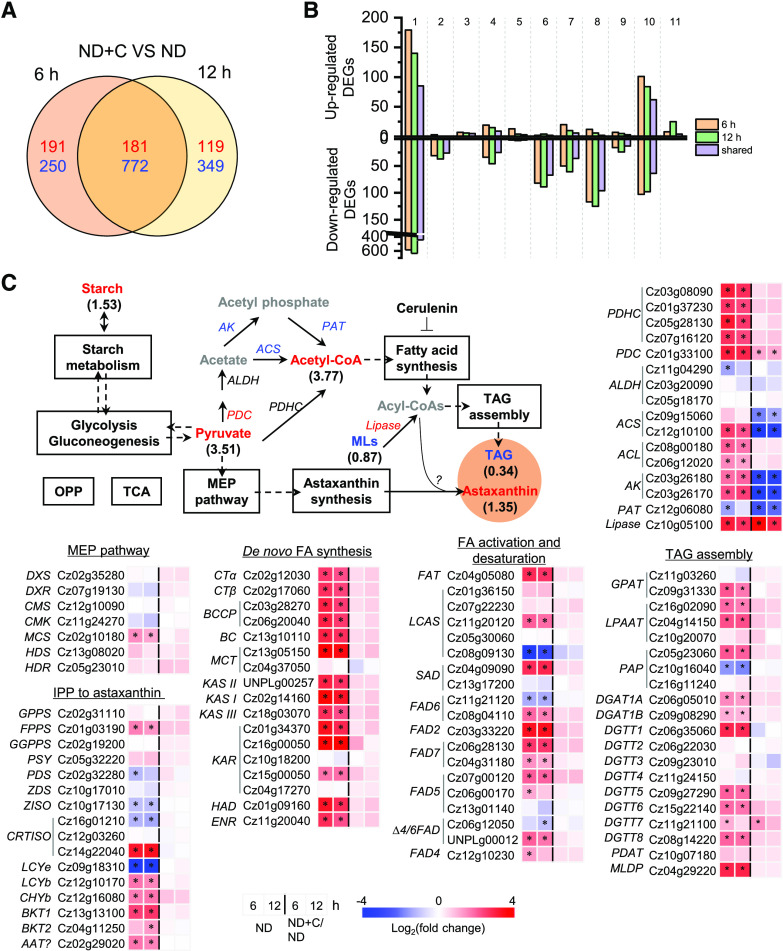
To elucidate the transcriptional regulation underlying augmented astaxanthin biosynthesis in response to ND, comparative RNA sequencing (RNA-seq) analysis was performed for algal cells induced by ND for 6 and 12 h with or without cerulenin. The presence of cerulenin led to 372 up-regulated and 1,022 down-regulated differentially expressed genes (DEGs) for the 6-h cultures and 300 up-regulated and 1,121 down-regulated DEGs for the 12-h cultures, with 181 up-regulated and 772 down-regulated DEGs shared by both time points (Fig. 7A). The shared DEGs were manually categorized into 11 functional groups (Fig. 7B). Metabolism represented the largest known functional category for up-regulated DEGs, over half of which were oxidoreductases such as reductase and dehydrogenase (Supplemental Data Set S1). By contrast, the largest known functional category for down-regulated DEGs was DNA, RNA, and gene expression, followed by cell structure and component and metabolism (Fig. 7B).
Figure 7.
Comparative transcriptomic analysis of ND-induced C. zofingiensis cells with and without cerulenin. A, Venn diagram of shared DEGs between 6 and 12 h of ND. The values in red indicate the number of up-regulated DEGs, whereas the values in blue designate the number of down-regulated DEGs in the presence of cerulenin compared with no cerulenin. B, Functional distribution of DEGs in up- and down-regulated categories, as follows: 1, function unknown; 2, cell cycle; 3, stress and cell death; 4, transport; 5, energy; 6, cell structure and component; 7, protein synthesis, modification, folding; and turnover; 8, DNA, RNA, and gene expression; 9, signaling; 10, metabolism; 11, photosynthesis. C, Transcriptional regulation of certain carbon metabolism pathways. Genes (italic) that are differentially expressed are shown in red (up-regulated) and blue (down-regulated). Compounds (roman) are shown in different colors: red, increased; blue, decreased; black, not determined or nonchanged. The pathways boxed in black indicate nonregulation. Values in parentheses designate fold changes of compound variation (48 h). The heat map shows log2 (fold change) values of transcripts. ACS, Acetyl-CoA synthetase; AK, acetate kinase; FA, fatty acid; MLs, membrane lipids; OPP, oxidative pentose phosphate pathway; PAT, phosphate acetyltransferase; PDC, pyruvate decarboxylase; TCA, tricarboxylic acid cycle. See Supplemental Data Set S2 for more details of the RNA-seq data.
Although cerulenin resulted in a decrease in lipids and increases in acetyl-CoA, pyruvate, starch, and astaxanthin, no significant changes occurred at the transcriptional level for most carbon metabolic pathways, including carotenogenesis for astaxanthin biosynthesis and lipogenesis for TAG biosynthesis (Fig. 7C; Supplemental Data Set S2). Therefore, transcriptional regulation might not be involved in cerulenin treatment-associated astaxanthin enhancement. Notably, acetyl-CoA biosynthesis pathways were severely attenuated by down-regulating acetyl-CoA synthetase, acetate kinase, and phosphate acetyltransferase genes, which may be due to the product feedback inhibition of acetyl-CoA buildup caused by cerulenin treatment (Fig. 7C).
DISCUSSION
C. zofingiensis Utilizes an Alternative Route for Astaxanthin Biosynthesis
In H. pluvialis, due to the efficient conversion of canthaxanthin to astaxanthin catalyzed by CHYb, only a trace amount of canthaxanthin accumulates (Lemoine and Schoefs, 2010; Shah et al., 2016). By contrast, C. zofingiensis CHYb may have no or low activity in hydroxylating canthaxanthin to astaxanthin, leading to the buildup of canthaxanthin that accounts for approximately 30% of the secondary carotenoids (Fig. 3A). On the other hand, adonixanthin, the intermediate of astaxanthin synthesis from zeaxanthin ketolation that is typically not found in H. pluvialis, is present in C. zofingiensis and accumulates under astaxanthin-induction conditions (Fig. 4A). Moreover, impairment of BKT activity abolishes astaxanthin biosynthesis while boosting zeaxanthin accumulation (Wang and Chen, 2008; Roth et al., 2017; Huang et al., 2018). These findings suggest that C. zofingiensis BKT is active in converting zeaxanthin to astaxanthin, although not at high efficiency. As a further study, our in vitro results provide direct evidence that, in C. zofingiensis, BKT is able to convert zeaxanthin to astaxanthin whereas CHYb has no activity in ketolating canthaxanthin for astaxanthin formation (Fig. 3B). Thus, C. zofingiensis employs a route alternative to H. pluvialis for astaxanthin synthesis, namely, hydroxylation of β-carotene to zeaxanthin mediated by CHYb and subsequent ketolation of zeaxanthin to astaxanthin mediated by BKT.
The functional difference of BKT and CHYb between C. zofingiensis and H. pluvialis may be attributed to the amino acid variance in proteins. Previous studies have identified several C. zofingiensis mutants deficient in astaxanthin synthesis (Roth et al., 2017; Huang et al., 2018; Ye and Huang, 2019). Among these, 15 mutants harbor a single amino acid substitution in CzBKT1 distributed across 12 positions of the protein (Supplemental Fig. S11), suggesting that these amino acid residues are essential for the function of CzBKT1. When compared with H. pluvialis BKT (HpBKT), CzBKT1 possesses three amino acid differences at the above-mentioned 12 positions: Arg-51, Pro-99, and Ser-251 (Supplemental Fig. S11). Notably, at the corresponding position 51 in CzBKT, C. reinhardtii BKT (CrBKT), which shows activity in converting zeaxanthin to astaxanthin in Escherichia coli systems (Zhong et al., 2011), harbors the same Arg residue (Supplemental Fig. S11). Moreover, the substitution of Arg-51 in CzBKT with Lys, the same residue at the corresponding position in HpBKT (Supplemental Fig. S11), abolishes astaxanthin production in C. zofingiensis (Ye and Huang, 2019). In this context, Arg-51 may render CzBKT1 different from HpBKT in the function of ketolating zeaxanthin for astaxanthin synthesis.
On the other hand, overexpressing CrBKT in C. reinhardtii, Arabidopsis (Arabidopsis thaliana), and tomato (Solanum lycopersicum) leads to canthaxanthin accumulation (Zhong et al., 2011; Huang et al., 2013; Perozeni et al., 2020), indicating that the endogenous CHYbs in these organisms may resemble CzCHYb and have no or low activity in hydroxylating canthaxanthin for astaxanthin synthesis. Therefore, the amino acid residues at the position where CzCHYb, CrCHYb, AtCHYb, and SlCHYb are the same but differ from HpCHYb (boxed in red in Supplemental Fig. S12) may contribute to the function of HpCHYb in converting canthaxanthin to astaxanthin. These assumptions, however, need experimental verification in future studies.
C. zofingiensis showed a rapid decline of β-carotene under astaxanthin-induction conditions and thereafter maintained a basal level (Fig. 4A). Nevertheless, this alga accumulates a certain amount of canthaxanthin and adonixanthin in addition to astaxanthin (Fig. 4A; Supplemental Fig. S13). In the presence of norflurazon, which blocks de novo carotenoid synthesis, the sum of the decreases in β-carotene, β-cryptoxanthin, and zeaxanthin is close to the sum of the increases in astaxanthin, canthaxanthin, adonixanthin, and echinenone in C. zofingiensis (Supplemental Table S3). This differs from H. pluvialis, in which the β-carotene decrease is comparable to the astaxanthin increase (Schoefs et al., 2001), and reflects the difference in astaxanthin biosynthetic routes between the two algae. Manipulating the pathways for efficient transformation of both canthaxanthin and zeaxanthin to astaxanthin may represent a feasible approach to improve astaxanthin synthesis in C. zofingiensis. HpCHYb and CrBKT are potential gene candidates for such a purpose, which have been evaluated in land plants and algae (Zhong et al., 2011; Huang et al., 2013; Zhu et al., 2018; Perozeni et al., 2020).
Astaxanthin Biosynthesis and Accumulation Are Independent of de Novo Fatty Acid Synthesis in C. zofingiensis
Astaxanthin and TAG share and may compete for carbon precursors in algae (Fig. 1). The astaxanthin increase caused by fatty acid synthesis inhibition in C. zofingiensis (Fig. 6A), however, is unlikely due to the shunt of carbon flux from fatty acids but from the transformation of other carotenoids, as the TC showed little change whereas β-carotene and canthaxanthin decreased (Table 1). Considering that starch increased considerably upon cerulenin treatment (Fig. 7C), carbon flux is likely rerouted from storage lipids to starch as a carbon sink.
Astaxanthin accumulation responds contrastingly to cerulenin treatment in C. zofingiensis and H. pluvialis, despite that fatty acids required for astaxanthin esterification are severely reduced in both algae (Fig. 6; Supplemental Fig. S10; Zhekisheva et al., 2005; Chen et al., 2015). One possibility is that C. zofingiensis synthesizes considerably lower astaxanthin than H. pluvialis (Liu et al., 2014) and thus needs fewer fatty acids for astaxanthin esterification and less TAG-filled LDs for astaxanthin storage. Consistent with this, the astaxanthin/TFA and astaxanthin/TAG ratios are lower in C. zofingiensis than in H. pluvialis (Fig. 6E; Supplemental Fig. S10). Nevertheless, when treated with cerulenin, both ratios increased considerably in C. zofingiensis and exceeded the values in H. pluvialis (Fig. 6E; Supplemental Fig. S10), indicating that fatty acids or TAG might not be limiting factors for astaxanthin biosynthesis and accumulation in C. zofingiensis.
Although the fatty acid level does not restrict astaxanthin accumulation in C. zofingiensis, the fatty acid profile may determine astaxanthin ester fatty acid composition. This is supported by the fact that many fatty acids of both astaxanthin ester and TFA follow the same trend in their relative abundance upon cerulenin treatment: a decrease in C18:1 accompanied by increases in PUFAs such as C16:3 and C18:3 (Supplemental Table S2; Supplemental Fig. S14). This can be explained by the fact that C18:1 is on the one hand severely impaired in its replenishment via the inhibition of de novo fatty acid synthesis by cerulenin and on the other hand subjected to fatty acid desaturases for further desaturation. The enhancement of astaxanthin ester, particularly diester, requires a supply of additional fatty acids, which may come from the recycling of membrane lipids. In corroboration, cerulenin treatment attenuated the glycolipids MGDG and DGDG, particularly MGDG (Supplemental Fig. S9). This is reasonable, as MGDG and DGDG in green algae including C. zofingiensis are mainly composed of PUFAs (Supplemental Fig. S9; Li-Beisson et al., 2015; Liu et al., 2016a), which can be recycled to meet the need of astaxanthin diester synthesis under cerulenin treatment (Supplemental Fig. S14).
Astaxanthin Esterification in Algae Involves Unknown Enzymes Yet To Be Identified
The formation of astaxanthin ester from free astaxanthin involves an acyltransferase that may transfer an acyl moiety from acyl-CoA and/or acyl-containing lipids to the hydroxyl end groups of astaxanthin. It has been proposed that DGATs are candidate enzymes responsible for astaxanthin esterification in algae (Chen et al., 2015). However, neither in vitro assay using the endoplasmic reticulum fraction (where DGAT enzymes reside) of C. zofingiensis cells (Supplemental Fig. S15) nor heterologous expression of DGAT genes from C. zofingiensis in the free astaxanthin-producing yeast strain (Zhou et al., 2015; Supplemental Fig. S16) supported the function of DGATs in astaxanthin esterification. Roth et al. (2017) proposed that AAT, a long-chain-alcohol O-fatty-acyltransferase up-regulated considerably under astaxanthin-producing conditions (Fig. 5E), might be involved in the esterification of astaxanthin in C. zofingiensis. Heterologous expression of this gene in the free astaxanthin-producing yeast strain also failed to produce astaxanthin ester (Supplemental Fig. S16). In this context, the genuine acyltransferase responsible for astaxanthin esterification has yet to be identified. This may be addressed by the functional screening of more putative acyltransferases, particularly those up-regulated ones under astaxanthin accumulation conditions (Supplemental Data Set S3).
MATERIALS AND METHODS
Algal Strain and Maintenance
The green microalga Chromochloris zofingiensis (ATCC 30412) was obtained from the American Type Culture Collection. This alga was maintained on an agar plate of modified BG-11 medium containing 0.6 g L–1 NaNO3. Ten milliliters of liquid medium was inoculated with cells from an agar plate, and the alga was grown aerobically in a 100-mL flask at 25°C for 6 d with orbital shaking at 150 rpm and illuminated with continuous light of 30 µmol photons m–2 s–1 (cool-white fluorescent tube light, from the top). The cells were then inoculated at 10% (v/v) into glass columns provided with illumination of 70 µmol photons m–2 s–1 (cool-white fluorescent tube light, from one side) and aeration of 1.5% (v/v) CO2-enriched air, grown to late exponential phase, and used as seed cultures for subsequent experiments.
Growth Conditions for the Induction of Astaxanthin and TAG in C. zofingiensis
The seed cultures were harvested, washed with nitrogen-deficient medium, and suspended in nitrogen-deficient medium at a cell density of 0.5 g L–1. The cultures were grown in 250-mL glass columns aerated with 1.5% (v/v) CO2-enriched air and illuminated with 70 µmol photons m–2 s–1 for ND conditions. For HL illumination, 350 µmol photons m–2 s–1 was provided with (HL conditions) or without (ND+HL conditions) supplementation of 0.6 g L–1 NaNO3. For HG, the cultures were supplemented with 10 g L–1 Glc and grown in the dark with orbital shaking at 150 rpm.
For the inhibition treatments under ND conditions, five inhibitors were employed: mevinolin (Sigma-Aldrich; PHR1285), fosmidomycin (Sigma-Aldrich; FR-900098), cerulenin (Sigma-Aldrich; 219557), norflurazon (Sigma-Aldrich; 34364), and DPA (Sigma-Aldrich; 242586). All five inhibitors were dissolved in methanol for stock solution preparation and added to the C. zofingiensis cultures at the onset of induction of ND, HL, or ND+HL to give working concentrations of 25, 50, and 100 µm for mevinolin and fosmidomycin; 5, 10, and 20 µm for cerulenin; 25, 50, and 100 nm for norflurazon; and 10, 20, and 40 µm for DPA. The control culture without inhibitor was supplemented with methanol at a concentration of 0.1% (v/v), the same as that in the treatments.
For the feeding of free fatty acids, C16:1, C18:1, C18:2, and C18:3n3 were delivered from ethanol stocks to C. zofingiensis cultures upon the induction of ND in the presence of 10 µm cerulenin, giving rise to the working concentrations of 25, 50, and 100 µm for C16:1, C18:2, or C18:3n3 and 50, 100, and 200 µm for C18:1. The ethanol concentration in the cultures with or without fatty acids was 0.1% (v/v).
Microscopic Observation of Algal Cells
C. zofingiensis cells were stained with the fluorescent probe BODIPY 505/515 (Invitrogen; D3921) with a final concentration of 1 µg mL–1 (from a stock of 1 mg mL–1 in dimethyl sulfoxide) and observed using an Olympus BX51 fluorescence microscope equipped with a DP72 digital camera.
Transmission electron microscopy was used to observe the subcellular structure of C. zofingiensis. Fresh samples were fixed with 4% (v/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde at 4°C for 12 h. The supernatant was removed and mixed with 5 µL of egg albumin in a 15-mL centrifuge tube. The collected algal cells were then incubated with 0.5% (v/v) osmic acid in 0.1 m phosphate-buffered saline (pH 7.4) at 4°C for 1 h followed by dehydration for 10 min with ethanol. Cell samples were aggregated within an aggregator (37°C for 12 h, 45°C for 12 h, and 60°C for 24 h). After drying, thin sections were stained with uranyl acetate for 5 min and observed with a JEOL 1230 microscope.
LD Isolation from ND-Treated C. zofingiensis Cells
Isolation and purification of LDs from C. zofingiensis were performed as described by Yoneda et al. (2016) with some modifications. In brief, algal cells were harvested by centrifugation at 5,000g for 5 min at room temperature and rinsed with washing buffer (0.2% [w/v] NaCl and 10 mm Tris-HCl, pH 7.6). The harvested cells were resuspended in Suc buffer (0.25 m Suc, 10 mm Tris-HCl, pH 7.6, and 1× protease inhibitor cocktail [EMD Millipore; 539131]) and lysed by passing through a French press (Spectronics Instruments) at an internal pressure of 15,000 p.s.i. The cell homogenate was centrifuged at 50,000g for 5 min at 4°C to remove unbroken cells and other organelles as a pellet. LDs floating on the surface layer of the supernatant were transferred to a new tube and loaded gently on top with one-fourth volume of 2.5 m Suc solution for another centrifugation (50,000g for 20 min at 4°C). LDs from the top fraction of the solution were collected into 10 1.5-mL tubes, followed by the addition of 1 mL of weak detergent buffer (0.2% [v/v] Triton X-100 and 10 mm Tris-HCl, pH 7.6), incubation on ice for 10 min, and centrifugation (20,000g for 10 min at 4°C) to remove the buffer. This washing process was repeated three times. The remaining LDs in the tubes were gathered into one tube and then recentrifuged at 20,000g for 10 min at 4°C to discard as much buffer solution as possible. After the detergent treatment, the LDs were washed three times with 10 mm Tris-HCl (pH 7.6) for immediate use or stored at –80°C.
In Vitro Assay for CzCHY and CzBKT
The open reading frames of CzCHY and CzBKT were cloned into the expression vector pYES2-CT (Invitrogen; V825120). The resulting vectors, once confirmed by restriction digestion and sequencing, were transformed into the Saccharomyces cerevisiae strain H1246 (∆dga1∆lro1∆are1∆are2; Sandager et al., 2001) by using the S. c. EasyComp Transformation Kit (Invitrogen; K505001). Transformants were selected on SC-uracil medium, and single colonies, confirmed by PCR, were grown in SC-uracil medium with 2% (w/v) raffinose for 24 h. Cells were harvested by centrifugation at 1,500g for 5 min and resuspended in SC-uracil medium with 2% (w/v) Gal with an initial OD600 of 0.4. After incubation at 30°C for 15 h, cells were washed twice and resuspended in 50 mm Tris-HCl buffer (pH 7.5, containing 1 mm dithiothreitol, 5% glycerol, and 1× protease inhibitor cocktail) to an OD600 of approximately 100 and then lysed by passing twice through a French pressure cell (Spectronics Instruments) at an internal pressure of 15,000 p.s.i. DNase was added to the broken cell extract at a concentration of 10 μg mL−1, and the mixture was incubated on ice for 15 min. Cell debris was removed from the suspension by centrifugation at 10,000g for 10 min at 4°C, and the supernatant was centrifuged further at 100,000g for 1 h at 4°C. The resulting microsomal membrane pellets were resuspended in microsomal storage buffer (50 mm Tris-HCl, pH 7.5, and 10% [v/v] glycerol) to give a protein concentration of 10 µg µL−1 for immediate use or stored at −80°C. Protein concentration was determined using a protein assay kit (Bio-Rad).
The in vitro assay was conducted according to the previously described procedures (Fraser et al., 1997, 1998) with minor modifications. Two micrograms of the substrates (β-carotene [Sigma-Aldrich; C4582], zeaxanthin [CSNpharm; CSN20552], and canthaxanthin [CSNpharm; CSN21545]; 1 mg mL−1 in chloroform) were added to the reaction mixture (200 µL; containing 50 mm Tris-HCl buffer [pH 7.5], 50 µg of microsome protein, 0.5 mm dithiothreitol, 0.25 mm FeSO4, 2.5 mm ascorbic acid, 0.25 mm 2-oxoglutarate, 1 mm ATP, and 0.05% [w/v] deoxycholate) and incubated at 30°C with vigorous shaking for 6 h. For the treatment of fatty acid inhibitor, cerulenin was added at a concentration of 5, 10, or 20 µm. The reaction was terminated by 2 mL of chloroform:methanol mixture (2:1, v/v), and the phase separation was achieved by the addition of 0.8 mL of 1% (w/v) KCl. The lower chloroform phase was transferred to a glass vial, dried under nitrogen gas, and dissolved in acetone for HPLC analysis.
RNA Isolation and RT-qPCR
RNA was isolated from aliquots of about 108 cells harvested under different conditions using the TRI Reagent (Invitrogen; AM9738) according to the manufacturer’s instructions. The concentration of total RNA was determined spectrophotometrically at 260 nm by NanoDrop 2000c (Thermo Scientific), and the quality was checked by electrophoresis. Total RNA (1 μg) extracted from different samples was reverse transcribed to cDNA by using a SuperScript III First-Strand Synthesis System (Invitrogen; 18080051) primed with oligo(dT) according to the manufacturer’s instructions. RT-qPCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems) in the presence of SYBR Green PCR Master Mix (Applied Biosystems; 4309155). Primer sequences used for RT-qPCR are listed in Supplemental Table S3. The mRNA expression level was normalized using the actin gene (Cz05g19150) as the internal control.
RNA-Seq and DEG Analysis
Total RNA (10 μg) extracted from each algal sample (0, 6, and 12 h of ND and 6 and 12 h of ND supplemented with 10 µm cerulenin; in triplicate) was treated by DNase I (Invitrogen; 18047019), and then mRNA was purified with Sera-mag Magnetic Oligo(dT) Beads (GE Healthcare; 38152103011150). The transcriptome libraries were constructed using the NEBNext mRNA Library Prep Reagent Set (New England Biolabs; E6100) and sequenced for two 150-bp runs (paired end) using a HiSeq 2500 sequencing system (Illumina) by Sangon Biotech. Reads were aligned to the C. zofingiensis genome (https://phytozome-next.jgi.doe.gov/info/Czofingiensis_v5_2_3_2) with TopHat (version 2.0.4; Trapnell et al., 2012). The RNA-seq data can be accessed via the accession number GSE113802 in the Gene Expression Omnibus.
For each of the RNA-seq data sets, gene expression was measured as the numbers of aligned reads to annotated genes by Cufflinks (version 2.0.4) and normalized to fragments per kilobase million values. DEGs between two conditions were defined as followed: the fragments per kilobase million value of at least one condition was no less than 1 and gene expression showed at least a 2-fold change with the false discovery rate-adjusted P < 0.05. The genes in each cluster were manually categorized into 11 groups according to Liu et al. (2019).
Extraction of Pigments and Lipids from C. zofingiensis
Cell samples were harvested and freeze-dried on a DW3 freeze-drier (Heto Dry Winner). Cells were disrupted by homogenization with liquid nitrogen and extracted by a solvent mixture of chloroform:methanol:0.75% (w/v) KCl solution (2:1:0.75, v/v). The lower chloroform phase containing pigments and lipids was dried under nitrogen gas and dissolved in acetone or chloroform for immediate use or stored at −20°C.
Pigment Identification and Quantification
The extracts in acetone were filtered through a 0.22-μm Millipore organic membrane. Twenty microliters of each extract was separated by HPLC on a Waters Spherisorb 5-μm, ODS2 4.6, 250-mm analytical column with a Waters 2695 HPLC system. Pigments were eluted at a flow rate of 1.2 mL min–1 with a linear gradient from 100% solvent A (acetonitrile:methanol:0.1 m Tris-HCl [pH 8], 84:2:14, v/v) to 100% solvent B (methanol:ethyl acetate, 68:32, v/v) over a 15-min period, followed by 10 min of solvent B. Individual carotenoids were identified by their absorption spectra, and their typical retention times were compared with standard samples of pure carotenoids.
For TLC separation of carotenoids, extracts were loaded on a TLC silica gel 60 plate (EMD Millipore) using hexane:tert-butyl methyl ether:acetic acid (80:2:2, v/v) as the development system. For recovering astaxanthin ester, the carotenoid extracts were subjected to two sequential TLC separation processes, namely, developed first with hexane:tert-butyl methyl ether:acetic acid (80:2:2, v/v) and then with butyl acetate:hexane (40:60, v/v).
Lipid and Fatty Acid Analysis
Neutral lipids and polar lipids were separated on a TLC plate using hexane:tert-butyl methyl ether:acetic acid (80:2:2, v/v) and chloroform:methanol:acetic acid:water (25:4:0.7:0.3, v/v) as the development system, respectively (Liu et al., 2019).
For lipid visualization by charring, the TLC plate was sprayed uniformly with 8% (w/v) H3PO4 containing 10% (w/v) copper (II) sulfate pentahydrate, air dried, and baked at 180°C for 3 min. For quantification, individual lipids on TLC plates were visualized with iodine vapor, recovered, transesterified, and analyzed by the GC-MS method as described below.
Fatty acid methyl esters (FAMEs) were prepared by direct transmethylation of recovered lipids and astaxanthin ester according to our previously described procedures (Liu et al., 2016b). The FAMES were separated by GC-MS using the Agilent 7890 capillary gas chromatograph equipped with a 5975 C mass spectrometry detector and a HP-88 capillary column (60 m × 0.25 mm; Agilent Technologies). The temperature program consisted of an initial hold at 100°C for 5 min, ramping at 3.5°C min−1 to 240°C, and a final hold at 240°C for 5 min. The injector was kept as 250°C with an injection volume of 2 μL in splitless mode. The flow rate of the carrier gas (helium) was 1.5 mL min−1, and the ionization energy was 70 eV (electron impact, full scan mode). The individual FAME in the samples was quantified by using a FAME standard (Sigma-Aldrich; 1269119).
Accession Numbers
The RNA-seq data from this article are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under the accession number GSE113802. The gene accession numbers are retrieved from the C. zofingiensis genome database (https://phytozome-next.jgi.doe.gov/info/Czofingiensis_v5_2_3_2). The names and accession numbers of the major genes/proteins mentioned in this article are detailed in Supplemental Table S4 and Supplemental Data Set S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of MEP and MVA pathways in C. zofingiensis.
Supplemental Figure S2. Quality evaluation of the isolated LD fraction.
Supplemental Figure S3. Correlation between astaxanthin and TAG and between astaxanthin and OA in C. zofingiensis cells under different conditions.
Supplemental Figure S4. Relative abundance of fatty acids of astaxanthin ester and TAG in C. zofingiensis.
Supplemental Figure S5. Plotting of astaxanthin content and carotenogenic gene expression and of TAG content and TAG synthesis-related gene expression in C. zofingiensis cells under different conditions of ND, HL, ND+HL, and HG.
Supplemental Figure S6. Effects of norflurazon, DPA, and cerulenin, without or with FFA feeding, on C. zofingiensis growth under ND conditions.
Supplemental Figure S7. Effects of carotenogenesis inhibitors on astaxanthin and TAG synthesis under ND conditions.
Supplemental Figure S8. TLC analysis of astaxanthin and TAG extracted from C. zofingiensis cells treated by cerulenin under different growth conditions of ND, HL, and ND+HL.
Supplemental Figure S9. Effects of cerulenin on polar lipids in C. zofingiensis under ND conditions.
Supplemental Figure S10. Effects of cerulenin on astaxanthin accumulation in H. pluvialis under ND conditions.
Supplemental Figure S11. Protein sequence alignment of BKTs from algae.
Supplemental Figure S12. Protein sequence alignment of CHYbs from algae and land plants.
Supplemental Figure S13. Change of carotenoids in C. zofingiensis under ND conditions with different inhibitors.
Supplemental Figure S14. Effects of cerulenin on astaxanthin diester fatty acid profile in C. zofingiensis under ND conditions.
Supplemental Figure S15. Effects of DGAT inhibitors on the in vitro astaxanthin transesterification activity of C. zofingiensis endoplasmic reticulum fraction.
Supplemental Figure S16. HPLC analysis of carotenoid extract from the free astaxanthin-producing yeast strain YPP7(−) expressing C. zofingiensis acyltransferase genes.
Supplemental Table S1. Fatty acid profiles as affected by carotenogenesis inhibitors.
Supplemental Table S2. Fatty acid profiles as affected by cerulenin.
Supplemental Table S3. Effects of different inhibitors on the change of carotenoids in C. zofingiensis.
Supplemental Table S4. Primer sequences of selected genes used in RT-qPCR experiments.
Supplemental Data Set S1. Details of DEGs revealed by RNA-seq.
Supplemental Data Set S2. RNA-seq data for genes involved in selected biological pathways.
Supplemental Data Set S3. RNA-seq data for genes encoding putative acyltransferases.
ACKNOWLEDGMENTS
We thank Hongwei Yu at Zhejiang University for providing the astaxanthin-producing yeast strain.
Footnotes
This work was supported by the National Key Research Program of China (grant no. 2018YFA0902500), the National Youth Thousand Talents Program of China, and Peking University CCUS project supported by BHP Billiton.
References
- Ambati RR, Phang SM, Ravi S, Aswathanarayana RG(2014) Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications. A review. Mar Drugs 12: 128–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar E, Rise M, Vishkautsan M, Arad S(1995) Pigments and structural changes in Chlorella zofingiensis upon light and nitrogen stress. J Plant Physiol 146: 527–534 [Google Scholar]
- Boussiba S.(2000) Carotenogenesis in the green alga Haematococcus pluvialis: Cellular physiology and stress response. Physiol Plant 108: 111–117 [Google Scholar]
- Chen G, Wang B, Han D, Sommerfeld M, Lu Y, Chen F, Hu Q(2015) Molecular mechanisms of the coordination between astaxanthin and fatty acid biosynthesis in Haematococcus pluvialis (Chlorophyceae). Plant J 81: 95–107 [DOI] [PubMed] [Google Scholar]
- Cordero BF, Couso I, León R, Rodríguez H, Vargas MA(2011) Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl Microbiol Biotechnol 91: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero BF, Couso I, Leon R, Rodriguez H, Vargas MA(2012) Isolation and characterization of a lycopene ε-cyclase gene of Chlorella (Chromochloris) zofingiensis: Regulation of the carotenogenic pathway by nitrogen and light. Mar Drugs 10: 2069–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero BF, Obraztsova I, Martin L, Couso I, Leon R, Vargas MA, Rodriguez H(2010) Isolation and characterization of a lycopene beta-cyclase gene from the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta). J Phycol 46: 1229–1238 [Google Scholar]
- Couso I, Cordero BF, Vargas M, Rodríguez H(2012) Efficient heterologous transformation of Chlamydomonas reinhardtii npq2 mutant with the zeaxanthin epoxidase gene isolated and characterized from Chlorella zofingiensis. Mar Drugs 10: 1955–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX Jr., Gantt E(2011) Elucidation of the pathway to astaxanthin in the flowers of Adonis aestivalis. Plant Cell 23: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo JA, Rodríguez H, Moreno J, Vargas MA, Rivas J, Guerrero MG(2004) Accumulation of astaxanthin and lutein in Chlorella zofingiensis (Chlorophyta). Appl Microbiol Biotechnol 64: 848–854 [DOI] [PubMed] [Google Scholar]
- Dose J, Matsugo S, Yokokawa H, Koshida Y, Okazaki S, Seidel U, Eggersdorfer M, Rimbach G, Esatbeyoglu T(2016) Free radical scavenging and cellular antioxidant properties of astaxanthin. Int J Mol Sci 17: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N, Wang C, Liu X, Zhao X, Liu Y, Liu X, Du Y, Zhang Z, Zhang H(2019) De novo synthesis of astaxanthin: From organisms to genes. Trends Food Sci Technol 92: 162–171 [Google Scholar]
- Fraser PD, Miura Y, Misawa N(1997) In vitro characterization of astaxanthin biosynthetic enzymes. J Biol Chem 272: 6128–6135 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Shimada H, Misawa N(1998) Enzymic confirmation of reactions involved in routes to astaxanthin formation, elucidated using a direct substrate in vitro assay. Eur J Biochem 252: 229–236 [DOI] [PubMed] [Google Scholar]
- Fucikova K, Lewis AL(2012) Intersection of Chlorella, Muriella and Bracteacoccus: Resurrecting the genus Chromochloris Kol et Chodat (Chlorophyceae, Chlorophyta). Fottea 12: 83–93 [Google Scholar]
- Galasso C, Orefice I, Pellone P, Cirino P, Miele R, Ianora A, Brunet C, Sansone C(2018) On the neuroprotective role of astaxanthin: New perspectives? Mar Drugs 16: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald K, Hagen C(2001) Beta-carotene is the intermediate exported from the chloroplast during accumulation of secondary carotenoids in Haematococcus pluvialis. J Appl Phycol 13: 89–93 [Google Scholar]
- Grünewald K, Hirschberg J, Hagen C(2001) Ketocarotenoid biosynthesis outside of plastids in the unicellular green alga Haematococcus pluvialis. J Biol Chem 276: 6023–6029 [DOI] [PubMed] [Google Scholar]
- Guerin M, Huntley ME, Olaizola M(2003) Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol 21: 210–216 [DOI] [PubMed] [Google Scholar]
- Gwak Y, Hwang YS, Wang B, Kim M, Jeong J, Lee CG, Hu Q, Han D, Jin E(2014) Comparative analyses of lipidomes and transcriptomes reveal a concerted action of multiple defensive systems against photooxidative stress in Haematococcus pluvialis. J Exp Bot 65: 4317–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen C, Grunewald K(2000) Fosmidomycin as an inhibitor of the non-mevalonate terpenoid pathway depresses synthesis of secondary carotenoids in flagellates of the green alga Haematococcus pluvialis. J Appl Bot 74: 137–140 [Google Scholar]
- Harker M, Young AJ(1995) Inhibition of astaxanthin synthesis in the green alga, Haematococcus pluvialis. Eur J Phycol 30: 179–187 [Google Scholar]
- Holtin K, Kuehnle M, Rehbein J, Schuler P, Nicholson G, Albert K (2009) Determination of astaxanthin and astaxanthin esters in the microalgae Haematococcus pluvialis by LC-(APCI)MS and characterization of predominant carotenoid isomers by NMR spectroscopy. Anal Bioanal Chem 395: 1613–1622 [DOI] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A(2008) Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Huang J, Liu J, Li Y, Chen F(2008) Isolation and characterization of the phytoene desaturase gene as a potential selective marker for genetic engineering of the astaxanthin-producing green alga Chlorella zofingiensis (Chlorophyta). J Phycol 44: 684–690 [DOI] [PubMed] [Google Scholar]
- Huang JC, Wang Y, Sandmann G, Chen F(2006) Isolation and characterization of a carotenoid oxygenase gene from Chlorella zofingiensis (Chlorophyta). Appl Microbiol Biotechnol 71: 473–479 [DOI] [PubMed] [Google Scholar]
- Huang JC, Zhong YJ, Liu J, Sandmann G, Chen F(2013) Metabolic engineering of tomato for high-yield production of astaxanthin. Metab Eng 17: 59–67 [DOI] [PubMed] [Google Scholar]
- Huang L, Gao B, Wu M, Wang F, Zhang C(2019) Comparative transcriptome analysis of a long-time span two-step culture process reveals a potential mechanism for astaxanthin and biomass hyper-accumulation in Haematococcus pluvialis JNU35. Biotechnol Biofuels 12: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Lin Y, He M, Gong Y, Huang J(2018) Induced high-yield production of zeaxanthin, lutein, and β-carotene by a mutant of Chlorella zofingiensis. J Agric Food Chem 66: 891–897 [DOI] [PubMed] [Google Scholar]
- Huang W, Ye J, Zhang J, Lin Y, He M, Huang J(2016) Transcriptome analysis of Chlorella zofingiensis to identify genes and their expressions involved in astaxanthin and triacylglycerol biosynthesis. Algal Res 17: 236–243 [Google Scholar]
- Ip PF, Chen F(2005) Production of astaxanthin by the green microalga Chlorella zofingiensis in the dark. Process Biochem 40: 733–738 [Google Scholar]
- Ip PF, Wong KH, Chen F(2004) Enhanced production of astaxanthin by the green microalga Chlorella zofingiensis in mixotrophic culture. Process Biochem 39: 1761–1766 [Google Scholar]
- Kopecky J, Schoefs B, Loest K, Stys D, Pulz O(2000) Microalgae as a source for secondary carotenoid production: A screening study. Algol Stud 98: 153–168 [Google Scholar]
- Lemoine Y, Rmiki NE, Creach A, Rachidi J, Schoefs B(2008) Cytoplasmic accumulation of astaxanthin by the green alga Haematococcus pluvialis (flotow) In Schoefs B, ed, Plant Cell Organelles: Selected Topics. Research Signpost, Kerala, India, pp 251–284 [Google Scholar]
- Lemoine Y, Schoefs B(2010) Secondary ketocarotenoid astaxanthin biosynthesis in algae: A multifunctional response to stress. Photosynth Res 106: 155–177 [DOI] [PubMed] [Google Scholar]
- Li Y, Huang J, Sandmann G, Chen F(2009) High-light and sodium chloride stress differentially regulate the biosynthesis of astaxanthin in Chlorella zofingiensis (Chlorophyceae). J Phycol 45: 635–641 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Beisson F, Riekhof W(2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82: 504–522 [DOI] [PubMed] [Google Scholar]
- Liu J, Han D, Yoon K, Hu Q, Li Y(2016a) Characterization of type 2 diacylglycerol acyltransferases in Chlamydomonas reinhardtii reveals their distinct substrate specificities and functions in triacylglycerol biosynthesis. Plant J 86: 3–19 [DOI] [PubMed] [Google Scholar]
- Liu J, Huang J, Fan KW, Jiang Y, Zhong Y, Sun Z, Chen F(2010a) Production potential of Chlorella zofingiensis as a feedstock for biodiesel. Bioresour Technol 101: 8658–8663 [DOI] [PubMed] [Google Scholar]
- Liu J, Mao X, Zhou W, Guarnieri MT(2016b) Simultaneous production of triacylglycerol and high-value carotenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour Technol 214: 319–327 [DOI] [PubMed] [Google Scholar]
- Liu J, Sun Z, Gerken H, Liu Z, Jiang Y, Chen F(2014) Chlorella zofingiensis as an alternative microalgal producer of astaxanthin: Biology and industrial potential. Mar Drugs 12: 3487–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Z, Mao X, Gerken H, Wang X, Yang W(2019) Multiomics analysis reveals a distinct mechanism of oleaginousness in the emerging model alga Chromochloris zofingiensis. Plant J 98: 1060–1077 [DOI] [PubMed] [Google Scholar]
- Liu J, Sun Z, Zhong Y, Gerken H, Huang J, Chen F(2013) Utilization of cane molasses towards cost-saving astaxanthin production by a Chlorella zofingiensis mutant. J Appl Phycol 25: 1447–1456 [Google Scholar]
- Liu J, Zhong Y, Sun Z, Huang J, Jiang Y, Sandmann G, Chen F(2010b) One amino acid substitution in phytoene desaturase makes Chlorella zofingiensis resistant to norflurazon and enhances the biosynthesis of astaxanthin. Planta 232: 61–67 [DOI] [PubMed] [Google Scholar]
- Lohr M, Im CS, Grossman AR(2005) Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol 138: 490–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Wu T, Kou Y, Shi Y, Zhang Y, Liu J(2019) Characterization of type I and type II diacylglycerol acyltransferases from the emerging model alga Chlorella zofingiensis reveals their functional complementarity and engineering potential. Biotechnol Biofuels 12: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Zhang Y, Wang X, Liu J(2020) Novel insights into salinity-induced lipogenesis and carotenogenesis in the oleaginous astaxanthin-producing alga Chromochloris zofingiensis: A multi-omics study. Biotechnol Biofuels 13: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F, Lu D, Li Y, Zeng M(2006) Characterization of astaxanthin esters in Haematococcus pluvialis by liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. Anal Biochem 352: 176–181 [DOI] [PubMed] [Google Scholar]
- Mulders KJM, Janssen JH, Martens DE, Wijffels RH, Lamers PP(2014) Effect of biomass concentration on secondary carotenoids and triacylglycerol (TAG) accumulation in nitrogen-depleted Chlorella zofingiensis. Algal Res 6, Part A: 8–16 [Google Scholar]
- Mulders KM, Weesepoel Y, Bodenes P, Lamers P, Vincken JP, Martens D, Gruppen H, Wijffels R(2015) Nitrogen-depleted Chlorella zofingiensis produces astaxanthin, ketolutein and their fatty acid esters: A carotenoid metabolism study. J Appl Phycol 27: 125–140 [Google Scholar]
- Nguyen HM, Baudet M, Cuiné S, Adriano JM, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, et al. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: With focus on proteins involved in lipid metabolism. Proteomics 11: 4266–4273 [DOI] [PubMed] [Google Scholar]
- Orosa M, Valero JF, Herrero C, Abalde J(2001) Comparison of the accumulation of astaxanthin in Haematococcus pluvialis and other green microalgae under N-starvation and high light conditions. Biotechnol Lett 23: 1079–1085 [Google Scholar]
- Peled E, Leu S, Zarka A, Weiss M, Pick U, Khozin-Goldberg I, Boussiba S(2011) Isolation of a novel oil globule protein from the green alga Haematococcus pluvialis (Chlorophyceae). Lipids 46: 851–861 [DOI] [PubMed] [Google Scholar]
- Perozeni F, Cazzaniga S, Baier T, Zanoni F, Zoccatelli G, Lauersen KJ, Wobbe L, Ballottari M (2020) Turning a green alga red: Engineering astaxanthin biosynthesis by intragenic pseudogene revival in Chlamydomonas reinhardtii. Plant Biotechnol J doi:10.1111/pbi.13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar R, Inoue R, Shiraishi H, Katsuda T, Katoh S(2008) High efficiency production of astaxanthin by autotrophic cultivation of Haematococcus pluvialis in a bubble column photobioreactor. Biochem Eng J 39: 575–580 [Google Scholar]
- Rise M, Cohen E, Vishkautsan M, Cojocaru M, Gottlieb HE, Arad SM(1994) Accumulation of secondary carotenoids in Chlorella zofingiensis. J Plant Physiol 144: 287–292 [Google Scholar]
- Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, Limon MC, Meléndez-Martínez AJ, Olmedilla-Alonso B, Palou A, et al. (2018) A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res 70: 62–93 [DOI] [PubMed] [Google Scholar]
- Roth MS, Cokus SJ, Gallaher SD, Walter A, Lopez D, Erickson E, Endelman B, Westcott D, Larabell CA, Merchant SS, et al. (2017) Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc Natl Acad Sci USA 114: E4296–E4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, Gallaher SD, Westcott DJ, Iwai M, Louie KB, Mueller M, Walter A, Foflonker F, Bowen BP, Ataii NN, et al. (2019) Regulation of oxygenic photosynthesis during trophic transitions in the green alga Chromochloris zofingiensis. Plant Cell 31: 579–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S (2001) Storage lipid synthesis is non-essential in yeast. J Biol Chem 277: 6478–6482 [DOI] [PubMed] [Google Scholar]
- Sandmann G, Römer S, Fraser PD(2006) Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab Eng 8: 291–302 [DOI] [PubMed] [Google Scholar]
- Schmidt I, Schewe H, Gassel S, Jin C, Buckingham J, Hümbelin M, Sandmann G, Schrader J(2011) Biotechnological production of astaxanthin with Phaffia rhodozyma/Xanthophyllomyces dendrorhous. Appl Microbiol Biotechnol 89: 555–571 [DOI] [PubMed] [Google Scholar]
- Schoefs B, Rmiki N, Rachadi J, Lemoine Y(2001) Astaxanthin accumulation in Haematococcus requires a cytochrome P450 hydroxylase and an active synthesis of fatty acids. FEBS Lett 500: 125–128 [DOI] [PubMed] [Google Scholar]
- Shah MMR, Liang Y, Cheng JJ, Daroch M(2016) Astaxanthin-producing green microalga Haematococcus pluvialis: From single cell to high value commercial products. Front Plant Sci 7: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovchenko A, Neverov K(2017) Carotenogenic response in photosynthetic organisms: A colorful story. Photosynth Res 133: 31–47 [DOI] [PubMed] [Google Scholar]
- Sun Z, Zhang Y, Sun LP, Liu J(2019) Light elicits astaxanthin biosynthesis and accumulation in the fermented ultrahigh-density Chlorella zofingiensis. J Agric Food Chem 67: 5579–5586 [DOI] [PubMed] [Google Scholar]
- Tang Y, Rosenberg JN, Bohutskyi P, Yu G, Betenbaugh MJ, Wang F(2016) Microalgae as a feedstock for biofuel precursors and value-added products: Green fuels and golden opportunities. BioResources 11: 2850–2885 [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L(2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W(2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64: 665–700 [DOI] [PubMed] [Google Scholar]
- Wang X, Wei H, Mao X, Liu J(2019) Proteomics analysis of lipid droplets from the oleaginous alga Chromochloris zofingiensis reveals novel proteins for lipid metabolism. Genomics Proteomics Bioinformatics 17: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen T(2008) The biosynthetic pathway of carotenoids in the astaxanthin-producing green alga Chlorella zofingiensis. World J Microbiol Biotechnol 24: 2927–2932 [Google Scholar]
- Ye Y, Huang JC(2019) Defining the biosynthesis of ketocarotenoids in Chromochloris zofingiensis. Plant Divers 42: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda K, Yoshida M, Suzuki I, Watanabe MM(2016) Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol 57: 397–406 [DOI] [PubMed] [Google Scholar]
- Yoneda K, Yoshida M, Suzuki I, Watanabe MM (2016) Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol 57: 397–406 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shi M, Mao X, Kou Y, Liu J(2019) Time-resolved carotenoid profiling and transcriptomic analysis reveal mechanism of carotenogenesis for astaxanthin synthesis in the oleaginous green alga Chromochloris zofingiensis. Biotechnol Biofuels 12: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhekisheva M, Zarka A, Khozin-Goldberg I, Cohen Z, Boussiba S(2005) Inhibition of astaxanthin synthesis under high irradiance does not abolish triacylglycerol accumulation in the green alga Haematococcus pluvialis (Chlorophyceae). J Phycol 41: 819–826 [Google Scholar]
- Zhong YJ, Huang JC, Liu J, Li Y, Jiang Y, Xu ZF, Sandmann G, Chen F(2011) Functional characterization of various algal carotenoid ketolases reveals that ketolating zeaxanthin efficiently is essential for high production of astaxanthin in transgenic Arabidopsis. J Exp Bot 62: 3659–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Ye L, Xie W, Lv X, Yu H(2015) Highly efficient biosynthesis of astaxanthin in Saccharomyces cerevisiae by integration and tuning of algal crtZ and bkt. Appl Microbiol Biotechnol 99: 8419–8428 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zeng D, Yu S, Cui C, Li J, Li H, Chen J, Zhang R, Zhao X, Chen L, et al. (2018) From golden rice to aSTARice: Bioengineering astaxanthin biosynthesis in rice endosperm. Mol Plant 11: 1440–1448 [DOI] [PubMed] [Google Scholar]