A rice receptor-like kinase induces immune responses in rice and Arabidopsis by kinase and guanylate cyclase activities, respectively.
Abstract
Plant pathogens secrete cell wall-degrading enzymes that degrade various components of the plant cell wall. Plants sense this cell wall damage as a mark of infection and induce immune responses. However, the plant functions that are involved in the elaboration of cell wall damage-induced immune responses remain poorly understood. Transcriptome analysis revealed that a rice (Oryza sativa) receptor-like kinase, WALL-ASSOCIATED KINASE-LIKE21 (OsWAKL21.2), is up-regulated following treatment with either Xanthomonas oryzae pv oryzae (a bacterial pathogen) or lipaseA/esterase (LipA; a cell wall-degrading enzyme of X. oryzae pv oryzae). Overexpression of OsWAKL21.2 in rice induces immune responses similar to those activated by LipA treatment. Down-regulation of OsWAKL21.2 attenuates LipA-mediated immune responses. Heterologous expression of OsWAKL21.2 in Arabidopsis (Arabidopsis thaliana) also activates plant immune responses. OsWAKL21.2 is a dual-activity kinase that has in vitro kinase and guanylate cyclase activities. Interestingly, kinase activity of OsWAKL21.2 is necessary to activate rice immune responses, whereas in Arabidopsis, OsWAKL21.2 guanylate cyclase activity activates these responses. Our study reveals a rice receptor kinase that activates immune responses in two different species via two different mechanisms.
The plant cell wall acts as a formidable barrier for pathogens. Plant pathogens secrete a battery of cell wall-degrading enzymes (CWDEs) to degrade different components of the plant cell wall (Albersheim and Anderson-Prouty, 1975; Hématy et al., 2009). CWDEs act as a double-edged sword for pathogens, as the activity of these enzymes leads to cell wall degradation, but this also releases cell wall degradation products that can elicit plant immune responses (Jha et al., 2007). Such host-derived molecules that can elicit immune responses are called damage-associated molecular patterns (DAMPs). External application of oligogalacturonides (OGs), xyloglucan oligomers, cellobiose, or cellotriose has been shown to induce immune responses in plants. These compounds might serve as DAMPs that are released upon the action of microbial hydrolytic enzymes on plant cell walls (Davis et al., 1986; de Azevedo Souza et al., 2017; Gust et al., 2017; Claverie et al., 2018). DAMPs also include molecules that are secreted by plants in response to the perception of danger. Such DAMPs include extracellular ATP and secreted peptides (e.g. plant elicitor peptide [Pep], systemin, etc.; Pearce et al., 1991; Huffaker et al., 2006; Chen et al., 2017). These DAMPs are sensed by membrane-localized receptor-like kinases and receptor-like proteins that activate defense response-associated signaling. Some known receptors of DAMPs are AtPEPR1/2 for Pep, AtDORN1 for extracellular ATP, SYR1 for systemins, and AtWAK1/2 for OGs (Brutus et al., 2010; Krol et al., 2010; Choi et al., 2014; Gust et al., 2017; Wang et al., 2018).
The wall-associated kinases (WAKs) constitute a unique class of receptor kinases that are known to be closely associated with the plant cell wall (Verica and He, 2002). WAKs are known to be involved in many physiological processes, including cell elongation, pollen development, and abiotic and biotic stress tolerance (Kohorn, 2016). Members of the WAK gene family have been known to interact with pectin and pectin degradation products (OGs). AtWAK1 and AtWAK2 have been reported to interact with pectin and OGs in vitro (Kohorn et al., 2006, 2009). Some proteins of the WAK gene family have also been known to be involved in immune responses in many plant species, such as Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays), and wheat (Triticum aestivum; He et al., 1998; Li et al., 2009; Hurni et al., 2015; Zuo et al., 2015; Harkenrider et al., 2016; Hu et al., 2017; Zhang et al., 2017b; Saintenac et al., 2018). In most cases, a receptor kinase or receptor-coreceptor complex recognizes the ligand and triggers phosphorylation events leading to the activation of MAP kinase signaling and its downstream targets (Meng and Zhang, 2013). However, some recent studies also indicate the presence of an alternate signaling system in plants, which is mediated by cyclic nucleotides such as cyclic guanosine monophosphate (cGMP) and cAMP (Gehring and Turek, 2017). cGMP is generated by guanylate cyclases (GCs), and most of the reported plant GCs are membrane-localized receptor kinases that also contain a functional GC motif inside the kinase domain (Gehring and Turek, 2017). Such kinases showing these dual activities are called moonlighting kinases (Wong et al., 2015). However, there have been some concerns regarding the presence of GC activities in plant receptor kinases, as they exhibit relatively less in vitro biochemical activity as compared with their counterparts in animals (Ashton, 2011; Bojar et al., 2014). Nevertheless, a growing number of reports indicate the importance of cyclic nucleotides and their synthases in plants (Gehring and Turek, 2017). In Arabidopsis, some receptor kinases, including a wall-associated kinase-like gene (AtWAKL10), are reported as moonlighting kinases (Meier et al., 2010). In a recent observation, Shen et al. (2019) indicated the role of a cGMP-responsive dual-activity protein kinase in GA signaling and salt-stress response in rice (Shen et al., 2019).
Rice serves as a staple food for more than half of the global population. Xanthomonas oryzae pv oryzae (Xoo) causes a serious bacterial blight disease of rice. CWDEs secreted by Xoo include cellulases, xylanases, and lipase/esterase (LipA; Rajeshwari et al., 2005; Jha et al., 2007). LipA is an important CWDE of Xoo, and deletion of the LipA gene results in a reduction in the virulence of Xoo in rice (Jha et al., 2007). Treatment of rice leaves with purified LipA leads to the activation of plant immune responses, including callose deposition, programmed cell death, and an enhanced tolerance toward Xoo (Aparna et al., 2009). Heat inactivation or mutation of the active-site residues of LipA abolishes its biochemical activity as well as its ability to induce immune responses in rice, indicating that the enzymatic activity of LipA is essential for the induction of the immune response (Jha et al., 2007; Aparna et al., 2009). However, rice functions involved in the perception and elaboration of LipA-induced immune responses remain to be identified.
In this study, transcriptome analysis was initially performed to identify gene expression changes that occur during LipA-induced immune responses in rice. An enhanced transcript level of a wall-associated kinase-like gene, OsWAKL21.2, was observed after treatment of rice leaves with either purified LipA or the pathogen Xoo but not after treatment with a LipA mutant of Xoo. Sequence alignment and biochemical studies indicated that OsWAKL21.2 is a dual-function receptor-like kinase that has in vitro kinase as well as GC activities. OsWAKL21.2 is a key component of signaling involved in LipA-induced immunity, as its down-regulation leads to attenuation of the LipA-induced immune response. Overexpression of OsWAKL21.2 in rice and its heterologous expression in Arabidopsis induce plant defense responses and confer enhanced tolerance to subsequent bacterial infection. However, we have observed that the mode of action of the protein is dissimilar in rice and Arabidopsis. Our results suggest that the kinase activity of OsWAKL21.2 is required to induce the immune response in rice, whereas its GC activity activates immune responses in Arabidopsis.
RESULTS
Expression of OsWAKL21.2 Is Enhanced after Treatment of Rice Leaves Either with LipA or Xoo
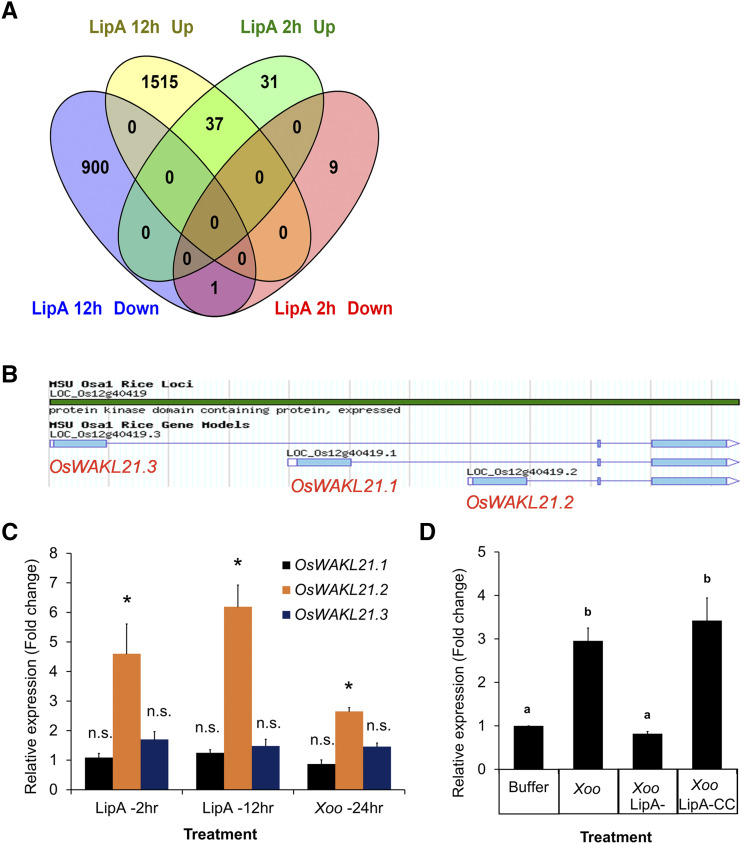
In order to identify rice functions that are potentially involved in early stages of LipA-induced immune responses, we performed transcriptome analysis of rice leaves after 30 min and 2 h of infiltration with LipA. After 30 min, no significantly altered gene expression was observed, whereas 78 genes (78 probes covering 74 unique genes) were differentially expressed (68 up and 10 down; fold change [FC] > 1.5-fold) after 2 h of LipA treatment (Supplemental Fig. S1A; Supplemental Table S1). These differentially expressed genes included those with potential roles in signaling, defense responses, or transcription/translation (Supplemental Fig. S1B). When compared with a previous microarray (Ranjan et al., 2015) performed after 12 h of LipA treatment, we observed that 38 of these 78 genes are differentially expressed (37 up and 1 down) at both time points (Fig. 1A; Supplemental Table S2). We compared these data with a publicly available microarray data set that was performed 24 h after treatment of rice leaves with various X. oryzae strains (Gene Expression Omnibus [GEO] accession no. GSE36272) and observed that some of these 38 genes were commonly up-regulated following X. oryzae treatment (Supplemental Table S3). The up-regulation of six of these commonly up-regulated genes was validated by reverse transcription quantitative PCR (RT-qPCR) after treatment of rice leaves with either Xoo or LipA (Supplemental Fig. S1C). Three of the 37 genes that were most commonly up-regulated after X. oryzae treatments include a putative wall-associated receptor kinase-like gene (OsWAKL21, LOC_Os12g40419), a putative ubiquitin ligase (OsPUB38, LOC_Os04g35680), and a putative Fru-bisphosphate aldolase (LOC_Os08g02700; Supplemental Table S3). In this study, we have focused on exploring the function of the wall-associated kinase OsWAKL21 in plant immune responses.
Figure 1.
Expression of OsWAKL21.2 is enhanced in rice leaves after treatment with either LipA or Xoo. A, Venn diagram indicating the number of genes that are differentially expressed after 2 and 12 h of LipA treatment. B, Three splice variants of OsWAKL21 as shown in the Rice-MSU database. C, RT-qPCR analysis of the expression of all three splice variants of OsWAKL21 after 2 and 12 h of LipA treatment and after 24 h of Xoo treatment in rice leaves. Asterisks represent significant differences in FC using Student’s t test with P < 0.05. n.s. indicates no significant difference in relative expression. D, RT-qPCR analysis of the expression of OsWAKL21.2 in rice leaves after 24 h of treatment with Xoo, LipA mutant of Xoo (Xoo LipA-), or LipA-complementing clone of Xoo (Xoo LipA-CC). Lowercase letters (a and b) above the bars indicate significant differences in FC using Student’s t test with P < 0.05. In C and D, 12- to 14-d-old leaves were infiltrated with either LipA (0.5 mg mL−1) or Xoo (OD of 1). Each bar represents the average value, and error bars denote se of at least three independent experiments. Relative expression was calculated in leaves treated with LipA or Xoo with respect to leaves treated with buffer. OsACTIN1 was used as an internal control for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method.
OsWAKL21 has three splice variants (OsWAKL21.1 [LOC_Os12g40419.1], OsWAKL21.2 [LOC_Os12g40419.2], and OsWAKL21.3 [LOC_Os12g40419.3]; Fig. 1B). RT-qPCR analyses indicated that the second splice variant (OsWAKL21.2) is mainly up-regulated in rice leaves after either LipA or Xoo treatment (Fig. 1C). Interestingly, treatment of rice leaves with the LipA mutant of Xoo did not enhance the expression of OsWAKL21.2, whereas introduction of a LipA-complementing clone into the LipA mutant restored its ability to enhance the expression of OsWAKL21.2 (Fig. 1D).
Overexpression of OsWAKL21.2 in Rice Induces Immune Responses
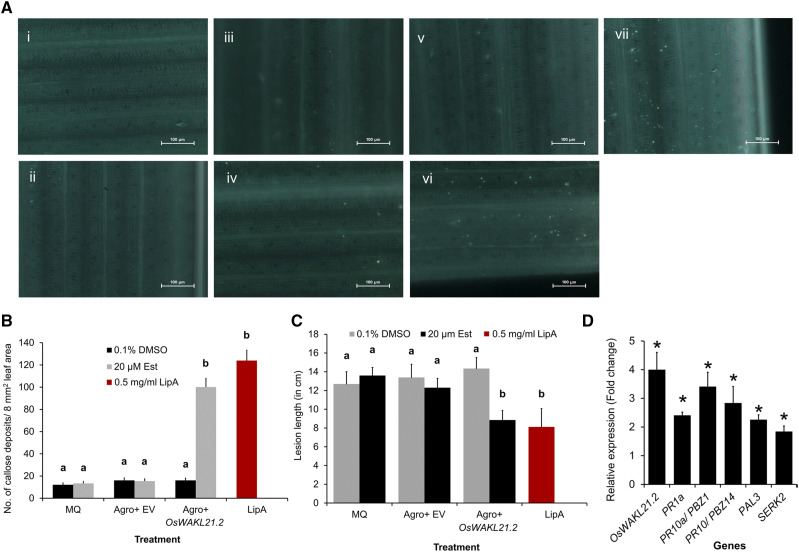
Treatment of rice leaves with LipA induces immune responses such as callose deposition, enhanced expression of defense-related genes, activation of the jasmonic acid (JA) pathway, and enhanced tolerance against subsequent Xoo infection (Jha et al., 2007; Ranjan et al., 2015). Agrobacterium tumefaciens-mediated transient overexpression of OsWAKL21.2 in young rice leaves significantly induced callose deposition, which is comparable to callose deposition induced by LipA treatment (Fig. 2, A and B). Transient overexpression of OsWAKL21.2 in rice leaves also enhanced tolerance against subsequent Xoo infection, leading to reduced lesion length caused by Xoo, which was also observed following treatment with LipA (Fig. 2C; Supplemental Fig. S2A). The overexpression of OsWAKL21.2 was confirmed by RT-qPCR and immunoblot analysis (Supplemental Fig. S2, B and C).
Figure 2.
Overexpression of OsWAKL21.2 in rice leaves induces plant immune responses. A, Callose deposition in rice leaves after treatment with various A. tumefaciens constructs or controls. The images shown are representative of one viewing area for each category. Numbered images are as follows: (i) 0.1% (v/v) dimethyl sulfoxide (DMSO); (ii) 20 µm Est; (iii) A. tumefaciens containing pMDC7 (empty vector [EV]) with 0.1% (v/v) DMSO; (iv) A. tumefaciens containing pMDC7 (empty vector) with 20 µm Est; (v) A. tumefaciens containing pMDC7::OsWAKL21.2 with 0.1% (v/v) DMSO; (vi) A. tumefaciens containing pMDC7::OsWAKL21.2 with 20 µm Est; and (vii) 0.5 mg mL−1 LipA. Images were taken at 20× magnification. Bars = 100 µm. B, Quantification of callose deposition in rice leaves after treatment with various A. tumefaciens constructs or controls as shown in A. Bar diagram shows the quantification of the number of callose deposits per area in rice leaves. Numbers of callose deposits in eight viewing areas (at 10× magnification) per leaf were considered. Each bar represents the average, and error bars represent se of 10 to 15 leaves per treatment in one set of experiments. Similar results were obtained in three independent experiments. C, Lesion length caused by Xoo in rice leaves when midveins of the leaves were previously treated with various A. tumefaciens constructs or controls. Midveins of rice leaves of 60-d-old plants were injected with MilliQ or water (MQ), LipA, or A. tumefaciens carrying empty vector or OsWAKL21.2 and also with (20 µm Est) or without (0.1% [v/v] DMSO) inducer. After 24 h, the leaves were pin-prick inoculated with Xoo 1 cm above the point of A. tumefaciens injection. Lesion length was measured after 10 d of infection with Xoo (Supplemental Fig. S2A). Each bar indicates the average, and error bars represent se of more than 20 leaves per treatment in one set of experiments. Similar results were obtained in three independent experiments. D, Relative expression of key defense-related genes after transient overexpression of OsWAKL21.2 in rice leaves. Each bar represents the average FC, and the error bars indicate se of three independent experiments (n = 12 in each experiment). For each gene, the transcript level of the uninduced condition (treatment with A. tumefaciens carrying OsWAKL21.2 with 0.1% [v/v] DMSO) was considered as 1 and was compared with the induced condition (treatment with A. tumefaciens carrying OsWAKL21.2 with 20 µm Est). OsACTIN1 was used as an internal control for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method. Asterisks represent significant difference in FC using Student’s t test with P < 0.05. In A and B, 12- to 14-d-old rice leaves were infiltrated with MQ, A. tumefaciens carrying empty vector, or vector containing OsWAKL21.2 and also with (20 µm Est) or without (0.1% [v/v] DMSO) inducer. In B and C, lowercase letters (a and b) above the bars indicate significant differences with P < 0.05 using one-way ANOVA followed by the Tukey-Kramer multiple comparison test. MQ treatment indicates control without any A. tumefaciens treatment. In A, B, and C, leaves treated with LipA were used as positive controls.
Plant immune responses are known to be modulated via the expression of defense-related genes. Therefore, we tested the expression of some key defense-related genes of rice after the transient overexpression of OsWAKL21.2 in midveinal regions of rice leaves. OsWAKL21.2 overexpression in rice enhanced the expression of three pathogenesis-related genes (OsPR1a, OsPR10/OsPBZ14, and OsPR10a/OsPBZ1), a somatic embryogenesis receptor kinase (OsSERK2), and a Phe ammonia lyase (OsPAL3; Fig. 2D). OsPR10, OsPR10a, OsSERK2, and OsPAL3 were also reported to be up-regulated in the microarray performed after 12 h of LipA treatment. We also tested the expression of 10 genes that are up-regulated following LipA/Xoo treatment (Supplemental Table S3) through microarray analysis and observed that seven of these 10 genes are also significantly up-regulated following overexpression of OsWAKL21.2 in rice leaves (Supplemental Fig. S2D). These results indicate that A. tumefaciens-mediated transient overexpression of OsWAKL21.2 in rice leaves induces immune responses that are at least in part similar to immune responses activated following LipA treatment.
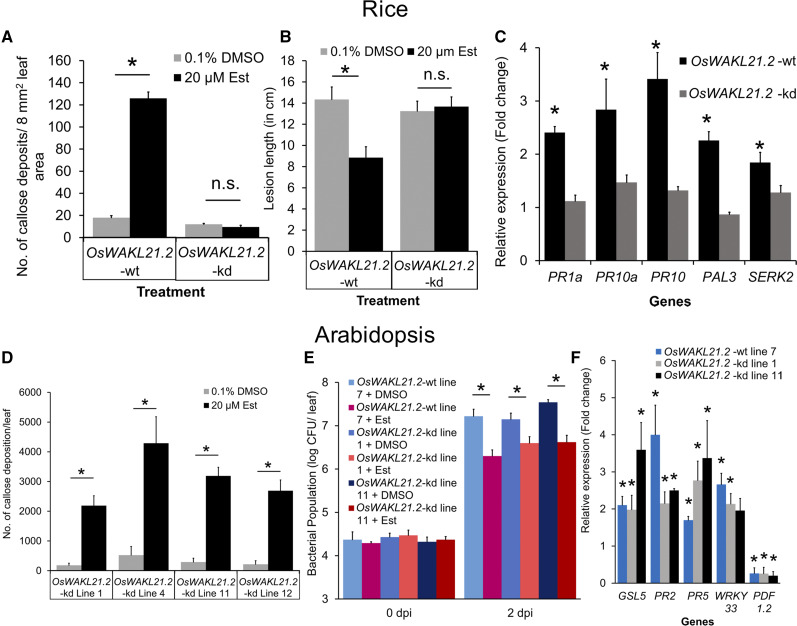
Transient Down-Regulation of OsWAKL21.2 Attenuates LipA-Induced Immune Responses in Rice
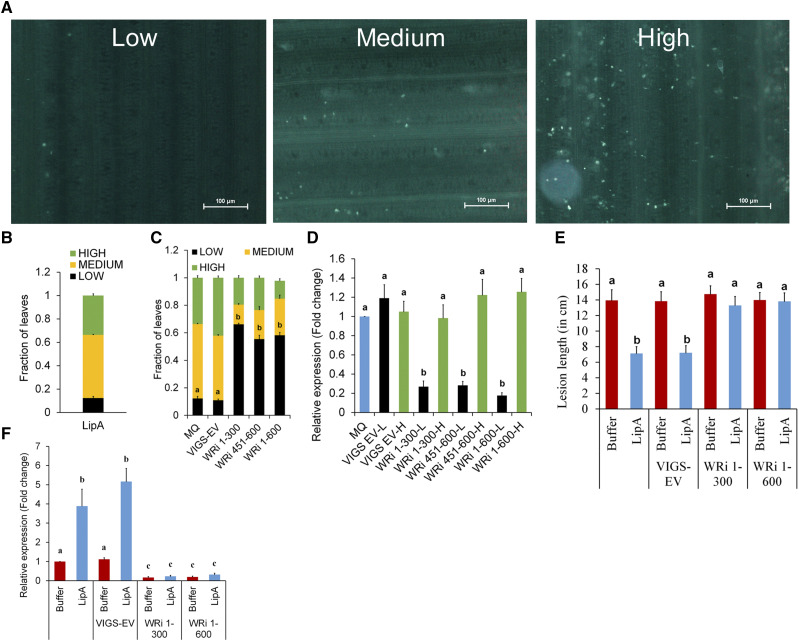
We subsequently assessed the effect of transient knockdown of OsWAKL21.2 by virus-induced gene silencing (VIGS) on LipA-induced immune responses. It was observed that the down-regulation was not retained by all leaves over time, which was also observed previously using this vector system (Kant and Dasgupta, 2017). So, an alternative approach was used for the assessment of callose deposition in RNA interference (RNAi) lines after LipA treatment (Supplemental Fig. S3B). We categorized the leaf samples into three classes based on the amount of callose deposition as low (less than 30 deposits per leaf), medium (approximately 30–80 deposits per leaf), or high (more than 80 deposits per leaf; Fig. 3A). Following LipA treatment, about 30% to 40% of the leaf samples showed high callose deposition, 10% to 15% showed low callose deposition, and the remainder (about 50%) showed a medium level of callose deposition (Fig. 3B). A similar ratio was observed if the seedlings were previously treated with VIGS-empty vector (Fig. 3C). The number of leaves showing low callose deposition significantly increased to more than 50% in WAKL-RNAi lines (WRi 1-300, WRi 451-600, and WRi 1-600 correspond to the fragments of OsWAKL21.2 that were used for down-regulation), whereas there was a reduction in the leaves that showed high or medium callose deposition (Fig. 3C). In RNAi lines, the leaves that exhibit low callose deposition following LipA treatment also exhibited significantly lower transcript/protein level of OsWAKL21.2, which was not observed in the leaves that showed high callose deposition (Fig. 3D; Supplemental Fig. S4A).
Figure 3.
Down-regulation of OsWAKL21.2 attenuates LipA-induced immune responses in rice. A, Categorization of the number of callose deposits in three different groups: low, medium, and high. The images shown are representative of one viewing area for each group. Eight such areas per leaf were viewed for categorization. Images were captured at 20× magnification. Bars = 100 µm. B, Fraction of leaves showing low, medium, or high callose deposition after LipA treatment. C, Fraction of leaves showing callose deposits after LipA infiltration that were previously treated with MilliQ or water (MQ; mock treatment) or A. tumefaciens containing VIGS-empty vector (EV) or WAK-RNAi constructs (OsWAKL21.2-RNAi 1-300 [WRi 1-300], OsWAKL21.2-RNAi 451-600 [WRi 451-600], or OsWAKL21.2-RNAi 1-600 [WRi 1-600]) in 12- to 14-d-old rice leaves. D, RT-qPCR analysis of OsWAKL21.2 transcript levels in leaves showing either low (L) or high (H) callose deposits. Each bar represents the average FC, and error bars indicate se observed in three biological replicates. For each sample, four to five leaves showing each callose phenotype were used for RNA isolation. The transcript level in mock (MQ)-treated leaves was considered as 1, and FC in A. tumefaciens-treated leaves was calculated with respect to it. E, Lesion length caused by Xoo in midveins of 60-d-old rice leaves that were pretreated with either buffer and LipA alone or along with A. tumefaciens strain WRi 1-300 or WRi 1-600. Each bar represents the average lesion length, and error bars show se of at least 20 leaves in one experiment. Similar results were obtained in three independent experiments. F, Expression levels of OsWAKL21.2 in rice leaves after 24 h of injection with either buffer and LipA alone or along with A. tumefaciens strain WRi 1-300 or WRi 1-600. Each bar represents the average of three independent experiments, n > 10 in each experiment. The transcript level of buffer-injected leaves was considered as 1, and FC in A. tumefaciens with buffer/LipA-treated leaves was calculated with respect to it. In B, C, D, and F, each bar represents the average and error bars denote the se of three different biological replicates. In B and C, each bar denotes the ratio of leaves showing each phenotype in at least 40 leaves. In C and E, lowercase letters (a and b) above the bars indicate significant differences with P < 0.05 using one-way ANOVA followed by the Tukey-Kramer multiple comparison test. In D and F, lowercase letters (a, b, and c) represent significant differences in FC using Student’s t test with P < 0.05. In D and F, OsACTIN1 was used as an internal control for RT-qPCR and the relative FC was calculated by using the 2−∆∆Ct method.
Since down-regulation of OsWAKL21.2 attenuated LipA-induced callose deposition, we decided to test its effect on LipA-induced tolerance toward Xoo. VIGS-mediated down-regulation of OsWAKL21.2 in rice midvein attenuates LipA-induced tolerance against subsequent Xoo infection (Fig. 3E; Supplemental Fig. S4B). RT-qPCR and immunoblot analysis using anti-OsWAKL21 antibodies indicated down-regulation of OsWAKL21.2 in the midveinal region following VIGS-mediated OsWAKL21.2 down-regulation (Fig. 3F; Supplemental Fig. S4C). There was a slight but usually nonsignificant reduction in transcript level of the other splice variants, and no significant difference was observed in transcript levels of other predicted off-target genes (Supplemental Fig. S5). This suggests that optimal expression of OsWAKL21.2 in rice leaves is required for LipA-induced tolerance against Xoo.
Heterologous Expression of OsWAKL21.2 in Transgenic Arabidopsis Lines Induces Plant Immune Responses
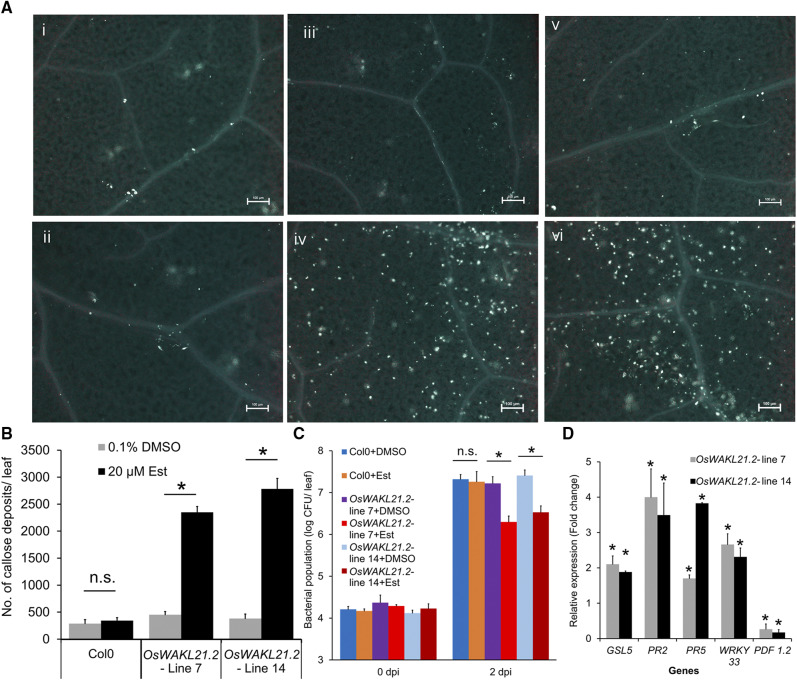
In order to determine whether the expression of OsWAKL21.2 activates immune responses in other plants, we generated stable Arabidopsis transgenic lines expressing OsWAKL21.2 under the control of a 17-β-estradiol (Est)-inducible promoter. Expression of OsWAKL21.2 in transgenic lines was examined after treatment with the inducer (Est) through RT-qPCR and immunoblotting (Supplemental Fig. S6, A and B). We observed that heterologous expression of OsWAKL21.2 in Arabidopsis results in enhanced callose deposition (Fig. 4, A and B) and enhanced tolerance against subsequent Pseudomonas syringae pv tomato DC3000 (Pst DC3000) infection (Fig. 4C). As there was a possibility that wounding caused during infiltration was eliciting defense responses, we tested callose deposition in leaves of transgenic Arabidopsis plants that had been sprayed with 20 µm Est. We observed a significant increase in callose deposition and OsWAKL21.2 transcript levels in transgenic Arabidopsis plants in which heterologous expression of OsWAKL21.2 was induced by spraying Est (Supplemental Fig. S6, C and D). In Arabidopsis, the salicylic acid (SA) and JA pathways are widely known to be involved in immune responses. We examined the expression of key genes linked to these two pathways in Arabidopsis transgenic lines. The heterologous expression of OsWAKL21.2 in Arabidopsis resulted in significant increases in the transcript levels of key SA pathway-related genes (AtPR2, AtPR5, and AtWRKY33) and AtGSL5, encoding a major callose synthase of Arabidopsis (Fig. 4D; Jacobs et al., 2003; Janda and Ruelland, 2015), whereas the transcript level of the key JA-responsive gene AtPDF1.2 was decreased. Overall, these data imply that heterologous expression of OsWAKL21.2 in Arabidopsis enhances callose deposition, enhances the expression of SA pathway-related genes, and, in addition, enhances tolerance against subsequent Pst DC3000 infection.
Figure 4.
Heterologous expression of OsWAKL21.2 in Arabidopsis induces immune responses. A, Callose deposition in leaves of wild-type Columbia (Col-0) or OsWAKL21.2 transgenic Arabidopsis lines following treatment with 20 µm Est (inducer) or 0.1% (v/v) DMSO (control). Numbered images are as follows: (i) Col-0 treated with DMSO; (ii) Col-0 treated with Est; (iii) OsWAKL21.2 transgenic line 7 treated with DMSO; (iv) OsWAKL21.2 transgenic line 7 treated with Est; (v) OsWAKL21.2 transgenic line 14 treated with DMSO; and (vi) OsWAKL21.2 transgenic line 14 treated with Est. Images were taken at 10× magnification. Bars = 100 µm. B, Quantification of the number of callose deposits in wild-type Col-0 and two different Arabidopsis OsWAKL21.2 transgenic lines after treatment with control or inducer. Leaves were treated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control). Each bar represents the average, and error bars represent se of three different leaves for each treatment in an experiment. C, Effects of heterologous expression of OsWAKL21.2 on the growth of Pst DC3000 after subsequent infection. Each bar represents the average, and error bars represent se of five leaves for each treatment in an experiment. CFU, Colony-forming units. D, Effects of heterologous expression of OsWAKL21.2 in transgenic Arabidopsis lines on the expression of SA or JA pathway-responsive genes. Expression in 0.1% (v/v) DMSO-treated leaves was considered as 1, and relative expression in 20 µm Est-treated leaves was calculated with respect to it. Each bar represents the average, and error bars represent se of three independent experiments for each line. For each sample, RNA was isolated from three leaves for every treatment. AtACTIN2 was used as an internal control for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method. Asterisks represent significant differences in FC using Student’s t test with P < 0.05. Transgenic or wild-type plant leaves were treated with 0.1% (v/v) DMSO (control) or 20 μm Est (inducer). Following 12 h of treatment, leaves were collected for callose deposition or transcript/protein analysis or were infected with Pst DC3000. Similar results were obtained in three independent experiments for A, B, and C. In B and C, asterisks represent significant differences when calculated using one-way ANOVA followed by the Tukey-Kramer multiple comparison test with P < 0.05. n.s. indicates no significant difference.
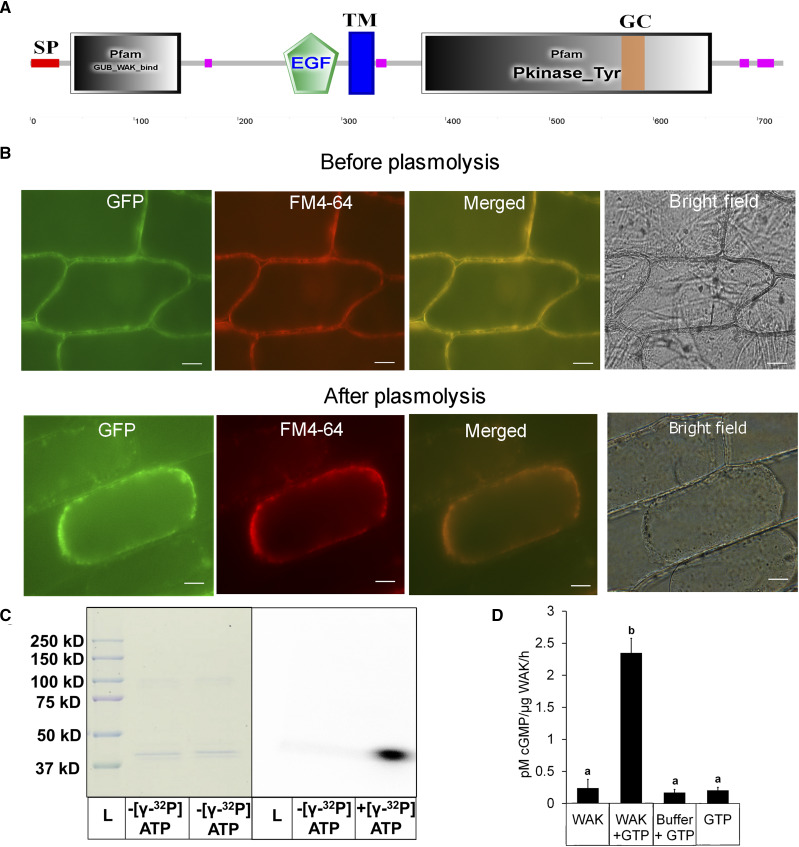
OsWAKL21.2 Is a Membrane-Localized Moonlighting Receptor Kinase That Has in Vitro Kinase and GC Activities
Sequence analysis of OsWAKL21.2 indicated that it is a receptor-like Ser/Thr kinase that accommodates an N-terminal extracellular galacturonan-binding domain, an epidermal growth factor-like repeat, and an intracellular C-terminal kinase domain, resembling other known wall-associated kinases (Fig. 5A). The analyses of OsWAKL21.2 also revealed the presence of a putative GC motif (residues 569–585) inside the kinase domain (Supplemental Fig. S7, A and B; Xu et al., 2018). Enhanced GFP (EGFP)-tagged recombinant OsWAKL21.2 (OsWAKL21.2:EGFP) colocalized with the cell membrane in onion (Allium cepa) epidermal cells and Nicotiana benthamiana leaf cells, indicating that it is a membrane-bound receptor (Fig. 5B; Supplemental Fig. S7D).
Figure 5.
Biochemical characterization and localization of OsWAKL21.2. A, Domain architecture of OsWAKL21.2 using the SMART tool (http://smart.embl-heidelberg.de/). EGF, Epidermal growth factor-like repeat; GC, GC motif; GUB, galacturonan-binding domain; Pkinase_Tyr, kinase domain; SP, signal peptide; TM, transmembrane region. Pink boxes represent low-complexity regions as indicated by the SMART tool. B, OsWAKL21.2:EGFP localizes on the cell membrane in onion peel after transient expression. OsWAKL21.2:EGFP was transiently transformed to onion peel cells using A. tumefaciens, and peels were visualized after 2 d using an epifluorescence microscope. Plasmolysis was done by incubating onion peel in 0.8 m mannitol for 30 min. The experiment was repeated three times, and similar results were obtained. FM4-64 staining was done by incubating in 10 μg mL−1 dye for 10 min. Images were captured at 60×. Bars = 10 μm. C, Kinase assay. The kinase domain of OsWAKL21 cloned and purified from E. coli shows autophosphorylation activity. Fifty micrograms of affinity-purified recombinant protein was used for assays with or without radiolabeled ATP. After 1 h, denatured samples were loaded onto a 10% SDS-PAGE gel. The gel was further subjected to autoradiography and Coomassie Brilliant Blue staining. The experiment was repeated three times, and similar results were obtained. D, GC assay. Fifty micrograms (in 50 μL) of affinity-purified recombinant protein was used for GC assay with or without GTP. After 1 h, 5 μL of the sample was directly used for cGMP quantification. GTP alone and GC buffer + GTP were used as controls. Each bar indicates the average, and error bars represent se of three independent experiments. Lowercase letters (a and b) above the bars indicate significant differences when analyzed using Student’s t test with P < 0.05.
The biochemical characterization was performed by cloning the intracellular kinase domain of OsWAKL21.2 (OsWAKL21376–725) with an N-terminal 6× His tag and expressing this in Escherichia coli. The purified cytoplasmic domain of OsWAKL21.2 showed autophosphorylation activity when incubated with [γ-32P]ATP, indicating that it is an active kinase (Fig. 5C). For analysis of GC activity, the same purified protein was incubated with GTP and cGMP was detected by qualitative and quantitative assays. cGMP was detected only when GTP was incubated with purified OsWAKL21376–725 (Fig. 5D; Supplemental Fig. S7C). The rate of cGMP synthesis was 2.1 ± 0.75 pm µg−1 protein h−1 (Fig. 5D), which is comparable to other known plant GCs such as AtPEPR1, AtPSKR1, and AtWAKL10 (Meier et al., 2010; Qi et al., 2010; Kwezi et al., 2011). These biochemical analyses strongly suggest that OsWAKL21.2 is a dual-function enzyme with both kinase and GC activities.
The Kinase Activity of OsWAKL21.2 Induces Immune Responses in Rice But Not in Arabidopsis
Considering that OsWAKL21.2 is a receptor kinase, we hypothesized that kinase activity of the protein would be required for the induction of immune responses. Based on homology with other plant receptor kinases, we mutated four active-site residues (Lys-407, Asp-504, Thr-542, and Thr-547) to Ala and generated a kinase-deficient mutant (OsWAKL21.2-kinase deficient or OsWAKL21.2-kd). The purified kinase domain of OsWAKL21.2-kd had almost no kinase activity but it retained GC activity (Supplemental Fig. S8, A–C). Furthermore, we observed that A. tumefaciens-mediated transient overexpression of the full-length OsWAKL21.2-kd in rice leaves did not induce rice immune responses such as callose deposition, enhanced tolerance against Xoo, or increased expression of key defense-related genes (Fig. 6, A–C; Supplemental Fig. S9, A and B). These results indicate that the kinase activity of OsWAKL21.2 is required for the induction of immune responses in rice.
Figure 6.
Kinase activity of OsWAKL21.2 is required for the induction of immune responses in rice but not in Arabidopsis. A, Quantification of callose deposition after transient overexpression of either wild-type (wt) OsWAKL21.2 or kinase-deficient mutant OsWAKL21.2-kd in rice leaves. Each bar represents the average, and error bars represent se of at least 12 leaves per treatment in an experiment. B, Lesion lengths after 10 d of Xoo pin-prick inoculation when OsWAKL21.2 or OsWAKL21.2-kd was transiently overexpressed prior to infection by Xoo. Each bar represents the average, and error bars represent se of lesion length in 20 to 30 leaves in an experiment. C, Relative expression of key defense-related genes after transient overexpression of either OsWAKL21.2 or OsWAKL21.2-kd in rice leaves. For each gene, the transcript level of the uninduced condition (treatment with A. tumefaciens carrying OsWAKL21.2 or OsWAKL21.2-kd with 0.1% [v/v] DMSO) was considered as 1 and was compared with the induced condition (treatment with A. tumefaciens carrying OsWAKL21.2 or OsWAKL21.2-kd with 20 µm Est). Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 12 in each experiment). D, Quantification of callose deposition in leaves of four different OsWAKL21.2-kd Arabidopsis transgenic lines (lines 1, 4, 11, and 12) treated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control). Each bar represents the average, and error bars represent se of three leaves in an experiment. E, Effects of heterologous expression of OsWAKL21.2-kd on the growth of Pst DC3000 after subsequent infection. Leaves of wild-type OsWAKL21.2 and two different OsWAKL21.2-kd Arabidopsis transgenic lines (lines 1 and 11) were infiltrated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control) and were subsequently inoculated with Pst DC3000 12 h post infiltration. Each bar represents the average, and error bars represent se of five leaves in each sample. CFU, Colony-forming units. F, Effects of heterologous expression of OsWAKL21.2-kd on the expression of key defense-related OsWAKL21.2-responsive genes in transgenic Arabidopsis lines. Expression in 0.1% (v/v) DMSO-treated leaves was considered as 1, and relative expression in 20 µm Est-treated leaves was calculated with respect to it. Each bar represents the average FC, and error bars indicate se in three independent experiments (n = 3 in each experiment). In C and F, OsACTIN1 and AtACTIN2 were used, respectively, as internal controls for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method. Similar results were obtained in three different experiments in A, B, D, and E. In A, B, D, and E, asterisks represent significant differences when calculated using one-way ANOVA followed by the Tukey-Kramer multiple comparison test with P < 0.05. In C and F, asterisks represent significant differences in FC using Student’s t test with P < 0.05. n.s. indicates no significant difference.
In order to further investigate the role of OsWAKL21.2 kinase activity in the induction of plant immune responses, we generated transgenic Arabidopsis lines expressing OsWAKL21.2-kd. Interestingly, we observed that the heterologous expression of OsWAKL21.2-kd in Arabidopsis caused an increase in callose deposition (Fig. 6D; Supplemental Fig. S9, C and D). Similar results were observed in four different transgenic lines. In Arabidopsis, the heterologous expression of OsWAKL21.2-kd resulted in enhanced tolerance toward Pst DC3000 and also changed the expression of defense-related genes in a similar pattern to OsWAKL21.2 (Fig. 6, E and F). As mentioned above, this mutant version did not induce immune responses in rice, indicating that the kinase activity of OsWAKL21.2 induces immune responses in rice but not in Arabidopsis.
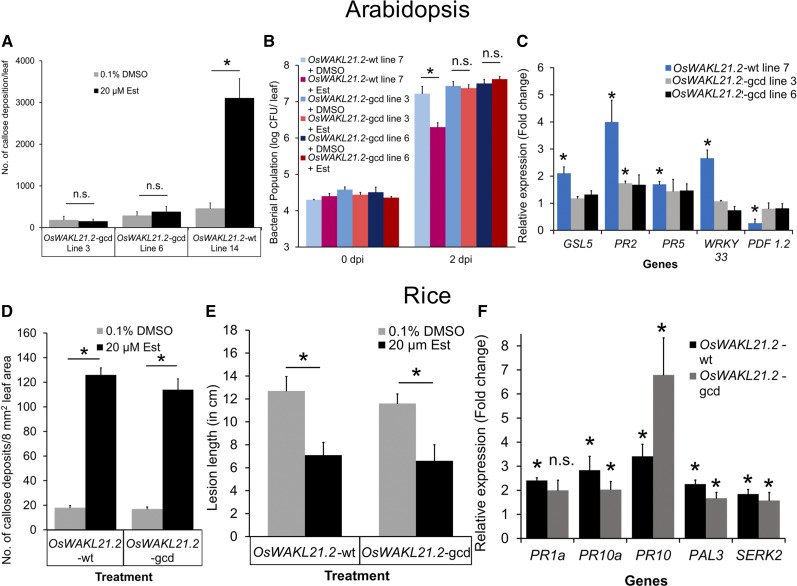
GC Activity of OsWAKL21.2 Induces Immune Responses in Arabidopsis But Not in Rice
Owing to the fact that the kinase-deficient mutant of OsWAKL21.2 retained the ability to induce immune responses in Arabidopsis, we decided to investigate whether the GC activity of OsWAKL21.2 had a role in the induction of immune responses in Arabidopsis. In order to test this hypothesis, we initially induced the expression of OsWAKL21.2 in Arabidopsis in the presence of the GC inhibitor LY83583 and observed that GC activity inhibition attenuates OsWAKL21.2- and OsWAKL21.2-kd-induced callose deposition in Arabidopsis (Supplemental Fig. S10). We then generated a mutant of OsWAKL21.2 that lacked GC activity (OsWAKL21.2-GC deficient or OsWAKL21.2-gcd) but retained its kinase activity (Supplemental Fig. S8, A–C; Ma et al., 2012). Heterologous expression of OsWAKL21.2-gcd in Arabidopsis did not induce callose deposition or lead to enhanced tolerance toward Pst DC3000 (Fig. 7, A and B; Supplemental Fig. S9, C and D).
Figure 7.
GC activity of OsWAKL21.2 is required for the induction of immune responses in Arabidopsis but not in rice. A, Quantification of callose deposition in leaves of two different Arabidopsis transgenic lines (lines 3 and 6) expressing GC-deficient OsWAKL21.2-gcd that were treated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control). Each bar represents the average, and error bars represent se of three leaves in an experiment. B, Effects of heterologous expression of OsWAKL21.2-gcd on the growth of Pst DC3000 after subsequent infection. Leaves of wild-type (wt) OsWAKL21.2 and two different OsWAKL21.2-gcd Arabidopsis transgenic lines (lines 3 and 6) were infiltrated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control) and were subsequently inoculated with Pst DC3000 12 h post infiltration. Each bar represents the average, and error bars represent se of five leaves in each sample. CFU, Colony-forming units. C, Effects of heterologous expression of OsWAKL21.2-gcd on the expression of key defense-related OsWAKL21.2-induced genes in transgenic Arabidopsis lines. Expression in 0.1% (v/v) DMSO-treated leaves was considered as 1, and relative expression in 20 µm Est-treated leaves was calculated with respect to it. Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 3 in each experiment). D, Quantification of callose deposition after transient overexpression of either wild-type OsWAKL21.2 or OsWAKL21.2-gcd in rice leaves. Each bar represents the average, and error bars represent se of at least 12 leaves per treatment in an experiment. E, Lesion lengths after 10 d of Xoo pin-prick inoculation when OsWAKL21.2 or OsWAKL21.2-gcd was transiently overexpressed prior to infection by Xoo. Each bar represents the average, and error bars represent se of lesion length in 20 to 30 leaves in an experiment. F, Relative expression of key defense-related genes after transient overexpression of either OsWAKL21.2 or OsWAKL21.2-gcd in rice leaves. For each gene, the transcript level under uninduced conditions (treatment with A. tumefaciens carrying OsWAKL21.2 or OsWAKL21.2-gcd with 0.1% [v/v] DMSO) was considered as 1 and was compared with that under the induced conditions (treatment with A. tumefaciens carrying OsWAKL21.2 or OsWAKL21.2-gcd with 20 µm Est). Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 12 in each experiment). In C and F, AtACTIN2 and OsACTIN1 were used, respectively, as internal controls for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method. Similar results were obtained in three different experiments in A, B, D, and E. In A, B, D, and E, asterisks represent significant differences when calculated using one-way ANOVA followed by the Tukey-Kramer multiple comparison test with P < 0.05. In C and F, asterisks represent significant differences in FC using Student’s t test with P < 0.05. n.s. indicates no significant difference.
Furthermore, OsWAKL21.2-gcd failed to significantly alter the expression of the majority of defense-related genes that are induced following heterologous expression of OsWAKL21.2 in Arabidopsis (Fig. 7C). Heterologous expression of OsWAKL21.2 in Arabidopsis leaves also enhanced the in planta cGMP level, which was not observed when OsWAKL21.2-gcd was expressed in transgenic Arabidopsis plants (Supplemental Fig. S11, A–C). However, transient overexpression of OsWAKL21.2-gcd induced immune responses in rice that were similar to the ones induced by wild-type OsWAKL21.2 (Fig. 7, D–F; Supplemental Fig. S9, A and B). These observations clearly indicate that the GC activity of OsWAKL21.2 induces immune responses in Arabidopsis but not in rice.
OsWAKL21.2 Possibly Induces the JA Pathway in Rice, Whereas It Activates the SA Pathway in Arabidopsis
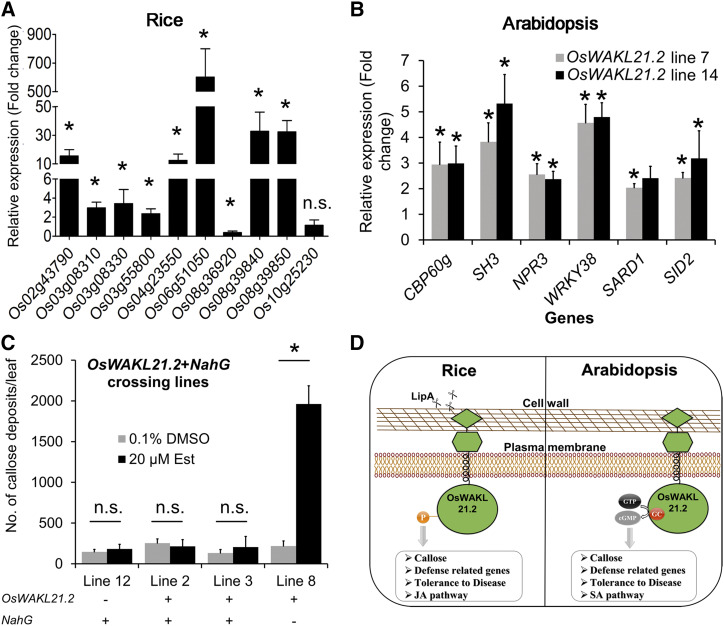
The results in this study indicate that kinase activity of OsWAKL21.2 is required to induce rice immune responses and that its GC activity is required for the induction of Arabidopsis immune responses. Our previous report indicated that the JA pathway is activated in rice leaves after treatment with LipA (Ranjan et al., 2015). We selected a subset of 10 genes that were previously predicted to be associated with the JA pathway in rice and were found to be up-regulated after 12 h of LipA infiltration (Ranjan et al., 2015). We tested the expression of these 10 genes and observed that eight out of 10 genes showed significant up-regulation after OsWAKL21.2 overexpression (Fig. 8A), whereas none of these showed significantly altered expression following overexpression of the kinase-deficient version of OsWAKL21.2 (OsWAKL21.2-kd) in rice (Supplemental Fig. S12A). We also assessed the expression of four key SA pathway-responsive WRKY genes in rice and found that overexpression of OsWAKL21.2 does not significantly alter the expression of any of these SA pathway-related genes (Supplemental Fig. S12B; Lan et al., 2013; Takatsuji, 2014). These results indicate that overexpression of OsWAKL21.2 in rice enhances the expression of JA pathway-related genes.
Figure 8.
OsWAKL21.2 induces the expression of JA pathway-related genes in rice, whereas it activates SA pathway-related genes in Arabidopsis. A, Relative expression of 10 JA pathway-related genes after transient overexpression of OsWAKL21.2 in rice leaves. These genes include three ZIM domain-containing proteins (LOC_Os03g08310, LOC_Os03g08330, and LOC_Os10g25230), two lipoxygenases (LOC_Os08g39840 and LOC_Os08g39850), one allene oxide synthase (LOC_Os03g55800), one basic helix-loop-helix transcription factor (LOC_Os04g23550), one ethylene-responsive transcription factor (LOC_Os02g43790), one chitinase (LOC_Os06g51050), and an AP2 domain-containing transcription factor (LOC_Os08g36920). For each gene, the transcript level under uninduced conditions (treatment with A. tumefaciens carrying WAK-wt [wild type] with 0.1% [v/v] DMSO) was considered as 1 and was compared with that under the induced conditions (treatment with A. tumefaciens carrying WAK-wt with 20 µm Est). Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 12 in each experiment). OsACTIN1 was used as an internal control. The relative FC was calculated by using the 2−∆∆Ct method. Asterisks represent significant differences in FC using Student’s t test with P < 0.05. n.s. indicates no significant difference in relative expression. B, Effects of heterologous expression of OsWAKL21.2 on the expression of SA pathway-related genes in transgenic Arabidopsis lines. Expression in 0.1% (v/v) DMSO-treated leaves was considered as 1, and relative expression in 20 µm Est-treated leaves was calculated with respect to it. Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 3 in each experiment). AtACTIN2 was used as an internal control for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method. Asterisks represent significant differences in FC using Student’s t test with P < 0.05. C, Quantification of callose deposits in Arabidopsis crossing lines expressing NahG and OsWAKL21.2 (lines 2 and 3) or either one of those (NahG only, line 12; OsWAKL21.2-wt only, line 8). Leaves were treated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control). Each bar represents the average, and error bars represent se of three leaves in an experiment. The asterisk represents a significant difference when calculated using one-way ANOVA followed by the Tukey-Kramer multiple comparison test with P < 0.05. n.s. indicates no significant difference. D, Model depicting the predicted role of OsWAKL21.2 in the induction of immune responses in rice and Arabidopsis. OsWAKL21.2 seems to be either directly or indirectly involved in the activation of immune responses following treatment with LipA. The kinase activity of OsWAKL21.2 is required for the induction of immune responses and the activation of the JA pathway in rice. On the other hand, in Arabidopsis, the GC activity of OsWAKL21.2 is required for the induction of immune responses that appear to be through the SA pathway.
The results described above (Fig. 4D) suggest that the expression of SA-related genes is enhanced after heterologous expression of OsWAKL21.2 in Arabidopsis. We further tested the expression of a few SA and JA pathway-related Arabidopsis genes (SA: AtSID2, AtCBP60g, AtSARD1, AtSH3, AtNPR3, and AtWRKY38; JA: AtAOS, AtCOI1, AtEEF1, AtJAZ1, AtJAZ10, AtJAR1, AtLOX2, AtPR2, and AtPR3) after heterologous expression of OsWAKL21.2 (Janda and Ruelland, 2015; Huang et al., 2016; Zhang et al., 2017a). We observed significantly enhanced expression of SA pathway-related genes, whereas the expression of most of the JA pathway-related genes was not significantly altered (Fig. 8B; Supplemental Fig. S12C). The JA biosynthetic gene AtCOI1 was significantly down-regulated, whereas a negative regulator of the JA pathway, AtJAZ1, was significantly up-regulated (Supplemental Fig. S12C). Heterologous expression of GC-deficient versions of OsWAKL21.2 (OsWAKL21.2-gcd) did not lead to significant changes in the expression of SA pathway-related genes in Arabidopsis (Supplemental Fig. S12D). In order to validate the role of the SA pathway in OsWAKL21.2-induced immune responses in Arabidopsis, we performed crosses between OsWAKL21.2 transgenic lines and NahG transgenic lines that do not accumulate SA (Delaney et al., 1994). Transgenic offspring lines that express both OsWAKL21.2 and NahG did not show enhanced callose deposition, whereas sister lines that expressed only OsWAKL21.2 showed enhanced callose deposition after treatment with Est (Fig. 8C; Supplemental Fig. S13). This observation indicates that OsWAKL21.2 induces immune responses in Arabidopsis via activation of the SA pathway.
DISCUSSION
CWDEs are important for the virulence of microbial plant pathogens. Xoo secretes several CWDEs such as cellulase A, cellobiosidase, and LipA, which are important for virulence. Treatment with these purified enzymes leads to the induction of rice immune responses (Jha et al., 2007). At least for the LipA protein, biochemical activity has been shown to be important for the induction of immune responses (Aparna et al., 2009). The molecular players involved in the perception and elaboration of LipA-induced immune responses in rice are yet to be identified. To discern the functions involved in LipA-induced immune responses, we performed transcriptome analyses at various time points following LipA treatment. Comparisons with microarray data available online indicated a handful of genes that are commonly up-regulated following LipA or Xoo treatment. One such gene was the second splice variant of a rice wall-associated kinase-like gene (OsWAKL21.2). The WAK genes are the only gene family known to recognize plant cell wall-derived DAMPs (Kohorn, 2016). Our study suggests that the expression of OsWAKL21.2 is enhanced after treatment of rice leaves with either LipA or Xoo but not after treatment with a LipA mutant of Xoo. This indicates that the increase in OsWAKL21.2 expression after Xoo treatment is specifically because of the presence of LipA in Xoo. We also observed that OsWAKL21.2 is a membrane-localized receptor kinase that has in vitro kinase and GC activities.
Down-regulation of some WAK gene family members in rice, such as OsWAK14, OsWAK91, OsWAK92, or Xa4-WAK, has been reported to enhance the susceptibility of rice plants toward subsequent infection (Delteil et al., 2016; Hu et al., 2017). We down-regulated the expression of OsWAKL21.2 in rice leaves using VIGS. Although down-regulation of OsWAKL21.2 did not alter susceptibility to Xoo, it attenuated LipA-induced tolerance to Xoo and callose deposition in rice, indicating that it is involved in the elaboration of LipA-induced immune responses. Since optimal expression of OsWAKL21.2 is essential for LipA-induced immune responses, it might be an upstream component in signaling activated following LipA treatment.
Treatment of rice leaves with LipA leads to callose deposition, activation of the JA pathway, enhanced expression of some defense-related genes, and enhanced tolerance against subsequent Xoo infection (Jha et al., 2007; Ranjan et al., 2015). Callose deposition is a hallmark of the immune response that is observed after treatment of the plant tissue with either CWDEs (including LipA) or DAMPs (Jha et al., 2007; Galletti et al., 2008). We also observed that the overexpression of OsWAKL21.2 in rice and its heterologous expression in Arabidopsis leaves lead to fortification of the cell wall in the form of callose deposition. Activation of the immune response leads to an increased tolerance toward subsequent infection in plants. We also observed that OsWAKL21.2-induced immune responses led to enhanced tolerance against subsequent bacterial infection in rice and Arabidopsis. Overexpression of several other WAKs, such as OsWAK1 (Li et al., 2009), OsWAK25 (Harkenrider et al., 2016), OsWAK14, OsWAK91, or OsWAK92 (Delteil et al., 2016), as well as AtWAK2 (Kohorn et al., 2009), AtWAK1 (Brutus et al., 2010), and TaWAKL4 (Saintenac et al., 2018), has been reported to enhance tolerance toward subsequent infections in different plant species. Overexpression of OsWAKL21.2 induces immune responses even in the absence of elicitor. Similar results have been reported after overexpression of OsWAK25 in rice (Harkenrider et al., 2016). We also observed a high number of callose deposits near the zone of infiltration of DMSO/Est, indicating that wounding could have an additive effect in total callose deposit density observed after infiltration. Moreover, we observed enhanced callose deposition after spray of Est in stable transgenic lines expressing OsWAKL21.2, indicating that wounding is not necessary for OsWAKL21.2-induced immune responses.
Immune responses are usually correlated with enhanced expression of defense-related genes. The overexpression of OsWAKL21.2 in the midveinal regions of rice leaves enhanced the expression of five defense-related and LipA-responsive genes. The key defense-related genes up-regulated by OsWAKL21.2 overexpression include OsPR1a (Park et al., 2008), OsPR10a (Bai et al., 2011), OsPR10 (Harkenrider et al., 2016), OsSERK2 (Chen et al., 2014), and OsPAL3 (Chen et al., 2018), which are well categorized as defense-related genes implicated in tolerance to Xoo. Interestingly, four of these five key defense genes (except OsPR1a) that are up-regulated by OsWAKL21.2 overexpression are also up-regulated after 12 h of LipA treatment (Ranjan et al., 2015). Overexpression of OsWAKL21.2 also enhances the expression of most of the tested LipA-responsive genes (seven of 10) and most of the tested JA pathway-related LipA-responsive genes (eight of 10), indicating that the overexpression of OsWAKL21.2 partially mimics LipA treatment conditions. These results establish that the overexpression of OsWAKL21.2 in rice induces immune responses that are similar to LipA treatment-induced immune responses.
Heterologous expression of OsWAKL21.2 in Arabidopsis leads to enhanced expression of SA-responsive genes such as AtPR2, AtPR5, and AtWRKY33 and down-regulation of the JA-responsive gene AtPDF1.2, indicating that OsWAKL21.2 likely activates the SA pathway in this species. We observed enhanced expression of several other SA biosynthesis-, regulation-, and response-related genes in Arabidopsis (AtSID2, AtSARD1, AtCBP60G, AtNPR3, AtWRKY33, AtWRKY38, and AtSH3) following OsWAKL21.2 heterologous expression (Janda and Ruelland, 2015), indicating up-regulation of the SA pathway. Expression analysis of most of the JA pathway-related genes was not significantly altered. We also found that transgenic plants expressing OsWAKL21.2 and NahG together did not show callose deposition, demonstrating that SA accumulation is required for the OsWAKL21.2-induced immune responses in Arabidopsis. These outcomes also explain the enhanced tolerance to Pst DC3000, as activation of the SA pathway in Arabidopsis leads to increased tolerance to Pst DC3000 (Xin and He, 2013). Activation of the SA pathway in Arabidopsis enhances the expression of the biotic stress-responsive callose synthase gene AtGSL5 (Dong et al., 2008), which was also up-regulated following heterologous expression of OsWAKL21.2. These results indicate that OsWAKL21.2, when expressed heterologously in Arabidopsis, acts as a defense gene and activates SA pathway-mediated immune responses. Some members of the WAK gene family in Arabidopsis, such as AtWAK1, AtWAK2, AtWAK3, AtWAK5, and AtWAKL10, are known as SA-responsive genes; treatment with SA leads to the enhanced expression of these genes, indicating correlation of the SA pathway and WAKs in Arabidopsis (He et al., 1998, 1999; Meier et al., 2010).
Ligand binding onto receptor kinases triggers phosphorylation that is further conveyed downstream via phosphorylation by/of kinases and their targets (Macho and Zipfel, 2014). A number of receptor kinases, such as AtBRI1, AtPSKR1, AtPEPR1, AtWAKL10, and HpPEPR1 (Hippeastrum hybridum PEPR1), are also known to possess dual enzymatic activity (i.e. they possess GC activity along with kinase activity; Meier et al., 2010; Ma et al., 2012; Świeżawska et al., 2015, 2017; Gehring and Turek, 2017). When compared with the animal GCs, plant GCs show very low in vitro GC activity, leading to a controversy about the actual existence and functional role of GC activity within plant receptor kinases (Ashton, 2011; Bojar et al., 2014). However, a growing number of reports confirm the existence of low GC activity in plant receptor kinases (Świeżawska et al., 2017; Wheeler et al., 2017). OsWAKL21.2 also possesses such a dual activity, which is comparable with other plant GCs. Treatment with a GC inhibitor and mutation of active-site residues of the GC motif showed that the GC activity of OsWAKL21.2 is required to induce immune responses in Arabidopsis but not in rice. GCs convert GTP to cGMP, which acts as a secondary signaling molecule (Gehring and Turek, 2017). Overexpression of AtBRI1, AtPSKR1, and AtPEPR1 (having GC activity) in Arabidopsis leads to a partial increase in cytoplasmic cGMP concentrations (Gehring and Turek, 2017), which was also observed following heterologous expression of OsWAKL21.2 in Arabidopsis. Some of the moonlighting kinases, such as AtPEPR1, AtBRI1, and AtPSKR1, are already known for their direct or modulatory role in Arabidopsis immune responses (Igarashi et al., 2012; Lozano-Durán and Zipfel, 2015). AtPEPR1 is a receptor of DAMP (Peps), and its GC activity is required for the activation of immune responses (Ma et al., 2012). AtWAKL10 has also been predicted as a defense-related gene that belongs to a similar gene family to OsWAKL21.2. These observations point toward the possible involvement of GCs in Arabidopsis immune responses. We have observed that, in rice, OsWAKL21.2 requires its kinase activity to induce immunity, whereas, in Arabidopsis, it requires its GC activity. Although the GC activity of OsWAKL21 is not essential for elaboration of the immune responses that we have studied in rice, it is possible that it might be involved in some other functions not studied here.
Treatment with CWDEs such as LipA leads to the induction of rice immune responses. The rice functions involved in the elaboration of LipA-induced immune responses remain to be identified. We have found that knockdown of the rice receptor kinase OsWAKL21.2 attenuates plant immune responses following LipA treatment. This suggests that OsWAKL21.2 could be involved in the elaboration of immune responses following LipA treatment. Overexpression of OsWAKL21.2 in plants induces immune responses and enhances tolerance toward hemibiotrophic pathogens. We observed that this receptor kinase is a moonlighting kinase that has in vitro GC activity along with kinase activity, making it one of the few moonlighting kinases known in plants. An interesting observation about OsWAKL21.2 is that for the induction of immune responses in rice, its kinase activity is required, but in Arabidopsis, its GC activity is needed. Figure 8D represents a mechanistic model of the role of OsWAKL21.2 in the induction of immune responses in rice and Arabidopsis. It should be noted that the analyses in rice reported in this article were performed by transient overexpression/down-regulation. These results need to be confirmed in stable overexpression and gene-edited knockdown lines. Future studies could also be aimed at identifying interacting partners of OsWAKL21.2 that are involved in the elaboration of immune responses. Furthermore, the possibility of using this gene to provide enhanced tolerance to bacterial pathogens in a variety of crops, including monocots and dicots, can be explored.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ssp. indica) variety Taichung Native 1, which is susceptible to Xanthomonas oryzae pv oryzae, was used for plant experiments. All rice experiments were performed in either a growth chamber (12-h/12-h day/night cycle) or a greenhouse at 28°C. Arabidopsis (Arabidopsis thaliana) ecotype Columbia and NahG lines were used for Arabidopsis experiments. Transgenic lines were generated using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected by adding hygromycin and/or kanamycin (NahG lines) to a final concentration of 20 or 50 µg mL−1, respectively. Plants were maintained in a growth chamber at 22°C/18°C day/night temperatures at about 70% humidity and with a 12-h/12-h day/night cycle. Leaves of 4- to 5-week-old plants that were in the rosette growth stage were used for experiments.
Bacterial Cultures
Xoo wild-type strain BXO43 (lab isolate) was used as a rice pathogen. The LipA mutant (BXO2001) of Xoo (BXO43) and its complemented strain (BXO2008) were also used in this study (Rajeshwari et al., 2005). Pseudomonas syringae pv tomato DC3000 was used as an Arabidopsis pathogen. Transient transformation in rice and floral dip of Arabidopsis were performed using Agrobacterium tumefaciens strain LBA4404. Escherichia coli BL21-AI was used for recombinant protein expression for biochemical assays.
LipA Purification from Xoo Culture Supernatant
Xoo BXO2008, a LipA-overproducing strain derived from BXO2001, was used for LipA overproduction and purification, and LipA was purified by the protocol described previously (Aparna et al., 2007). The purity and activity of the enzyme were tested by running it on an SDS-PAGE gel and testing on tributyrin-containing plates, respectively.
Microarray Analysis
The leaf treatment and microarray analysis were performed as described previously (Ranjan et al., 2015). RNA was isolated from 25 to 30 leaves after 30 min or 2 h of treatment either with LipA (0.5 mg mL−1) or buffer. Processed data and .cel files were also submitted to GEO-National Center for Biotechnology Information (accession no. GSE53940). RMA and PLIER16 algorithms were used for analysis, and probes showing significant differential expression (FC ≥ 1.5-fold and P < 0.05) in both analyses were considered as differentially expressed genes.
Vector Construction and Site-Directed Mutagenesis
Gateway cloning technology was used for cloning. OsWAKL21.2 was amplified using rice cDNA and cloned into pENTR/D-TOPO (Invitrogen). The gene was subcloned using the LR clonase reaction (Invitrogen) into pMDC7 plasmid (Curtis and Grossniklaus, 2003) for plant expression studies and in pH7FWG2 plasmid (Karimi et al., 2002) for localization experiments. In pMDC7, the target gene sequence is cloned downstream to the XVE promoter, which is Est inducible. A concentration of 20 μm Est (Sigma-Aldrich) was used in all overexpression studies as an inducer, whereas 0.1% (v/v) DMSO was used as a control (uninduced condition). The kinase domain OsWAKL21376–725 was cloned into bacterial expression vector pDEST17 (Invitrogen) and transformed into E. coli BL21-AI for recombinant protein expression. The constructs in pENTR/D-TOPO were used for site-directed mutagenesis (Zheng et al., 2004). The mutant versions were then transferred into the desired destination vectors using the LR clonase reaction. All the clones and mutations were confirmed using Sanger sequencing. All the plant expression constructs were introduced into A. tumefaciens strain LBA4404. LBA4404:XVEpro:OsWAKL21.2, LBA4404:XVEpro:OsWAKL21.2-kd, and LBA4404:XVEpro:OsWAKL21.2-gcd were used for transient transformation in rice and for the generation of Arabidopsis transgenic lines.
Callose Deposition Assay in Rice and Arabidopsis
For callose deposition assays in rice, 12- to 14-d-old leaves were used for A. tumefaciens-mediated transient transformation (Jha et al., 2010; Pillai et al., 2018). The suspension was infiltrated in the third rice leaf using a needleless 1-mL syringe with inducer (20 µm Est; Sigma-Aldrich) or control (0.1% [v/v] DMSO). Leaves collected for callose deposition were stained with Aniline Blue according to Millet et al. (2010). Callose deposition was visualized under blue light (excitation wavelength, 365 nm) using an ECLIPSE Ni-E epifluorescence microscope (Nikon) with 10× magnification. Eight images (∼1 mm2 each) were captured from each leaf from the zone of infiltration and proximal region. The number of callose deposits in all eight images for a leaf was added to get callose deposition per leaf (per 8 mm2). The average was calculated for 10 to 12 leaves for each treatment. Representative images were captured at 20× magnification for better indication of callose deposits.
For callose deposition in Arabidopsis transgenic plants, similar-sized rosette-stage leaves were infiltrated either with 100 µL of 0.1% (v/v) DMSO or 20 µm Est (inducer) using a needleless 1-mL syringe. After 12 h, leaves were collected and stained for callose deposition and then observed with the microscope as mentioned above for rice. Nearly 40 to 50 images per leaf were captured, and the number of callose deposits in each image was added to calculate the number of callose deposits in one leaf. For each sample, the average was calculated for three such leaves obtained from three separate plants.
Virulence Assay in Rice and Arabidopsis
Approximately 60-d-old Taichung Native 1 rice plants were used for infection of Xoo. For transient overexpression in rice midvein, 200 µL of actively growing A. tumefaciens (LBA4404) resuspended in 10 mm MES + 10 mm MgCl2 + 200 µm acetosyringone (final OD of 0.8; either with [20 µm Est] or without [0.1% {v/v} DMSO] inducer) was injected using a 1-mL syringe. After 24 h, about 1 cm above the A. tumefaciens injection site, the midveins of leaves were pin pricked with a needle touched to a fresh Xoo colony. Lesion length caused by Xoo was measured after 10 d of Xoo infection.
Pst DC3000 was used for infection in Arabidopsis leaves. Similar-sized leaves from five different rosette-stage plants were infiltrated with either 0.1% (v/v) DMSO or 20 µm Est. After 12 h, leaves were infected with actively growing culture of Pst DC3000 (diluted to OD of 0.02 in 10 mm MgCl2) by infiltration using a needleless 1-mL syringe. Leaves were harvested after 20 min (0 d post infiltration) or after 48 h (2 d post infiltration), crushed in 1 mL of autoclaved water, and dilutions were spread on Luria-Bertani medium plates containing 25 μg mL−1 rifampicin. Colony-forming units were calculated after incubation of plates at 28°C.
Down-Regulation of OsWAKL21.2 using VIGS
VIGS was used for A. tumefaciens-mediated transient down-regulation of OsWAKL21.2 in rice. Three RNAi constructs of different lengths from the unique 5′ end of OsWAKL21.2 were cloned in pRTBV-MVIGS (Supplemental Fig. S3A; Purkayastha et al., 2010). Down-regulation was performed according to a modified protocol mentioned previously (Purkayastha et al., 2010; Kant and Dasgupta, 2017). For callose deposition studies, newly germinated rice seedlings (1 d old) were dipped in activated A. tumefaciens culture (in 10 mm MES + 10 mm MgCl2 + 200 µm acetosyringone) for 24 h (Supplemental Fig. S3B). Ten days after A. tumefaciens treatment, the third leaf of each plant was infiltrated with LipA using a needleless syringe (0.5 mg mL−1; at least 40 leaves for each A. tumefaciens strain). After 16 h, a small piece (∼1.5 cm) of each leaf around the zone of infiltration was collected for callose deposition, whereas the rest of the leaf piece was stored for transcript/protein quantification. Each leaf was collected separately for callose and transcript/protein quantification and labeled. Callose deposition was observed as mentioned above for the callose deposition assay. The remainder of the four to five leaves that showed either low or high callose deposition were pooled, and RNA/protein was isolated from those pooled leaves for RT-qPCR or immunoblotting.
For virulence assay after down-regulation of OsWAKL21.2, midveins of 60-d-old rice plants were injected with 200 µL of A. tumefaciens containing the VIGS construct along with either buffer or LipA (0.5 mg mL−1; n > 40). After 24 h, midveins of 10 leaves were collected (3 cm each) for OsWAKL21.2 transcript/protein quantification, whereas the remaining 20 to 30 leaves were infected with a freshly growing colony of Xoo as mentioned above. Lesion length caused by Xoo was measured after 10 d of infection.
Purification of Recombinant Protein and in Vitro Biochemical Assays
The recombinant kinase domain of OsWAKL21.2, OsWAKL210.2376–725, with a 6× His tag was cloned, expressed, and purified from E. coli BL21-AI. Fifty micrograms of purified recombinant protein was used for kinase or GC assays in a 50-µL reaction. For kinase assays, the purified protein was incubated with 10 µCi of [γ-32P]ATP in kinase assay buffer (50 mm Tris [pH 7.5], 10 mm MgCl2, 2 mm MnCl2, 0.5 mm CaCl2, 1 mm DTT, and 20 mm ATP) for 1 h at room temperature (Li et al., 2009), run on a 10% SDS-PAGE gel, and the gel was subsequently exposed to a phosphoimager screen that was later scanned in a phosphoimager (Personal Molecular Imager; Bio-Rad) instrument.
GC assays were also performed from the same purified recombinant protein in GC assay buffer (50 mm Tris [pH 7.5], 2 mm MgCl2, 1 mm MnCl2, 0.5 mm CaCl2, and 0.2 mm NONOate [Sigma]) modified from the protocol described previously (Meier et al., 2010). The reaction was incubated at 37°C for either 1 or 12 h. The 1-h reaction was used for quantitative analysis, whereas 12-h reactions were used for qualitative analysis. cGMP produced after 1 h was quantified using a cGMP enzyme immunoassay kit (Sigma-Aldrich, catalog no. CG201) according to the manufacturer’s protocol, and the data were analyzed using the online tool Elisaanalysis. For qualitative analysis, the resultant product was blotted on a nitrocellulose membrane (Amersham, catalog no. RPN203E) and dried in the laminar hood with the UV light on for 1 h. The nucleotides were further cross-linked to the membrane in a UV transilluminator for 30 min. The membrane was blocked, washed, further incubated with anti-cGMP antibody (1:1,000; Sigma-Aldrich, catalog no. G4899), and processed as mentioned in the immunoblot section.
RNA Isolation and Gene Expression Analysis
For RT-qPCR, RNA was isolated by the protocol of Oñate-Sánchez and Vicente-Carbajosa (2008) with some modifications (Couto et al., 2015). For rice, 10 to 12 leaf pieces (or midvein pieces) were crushed together for each treatment unless mentioned otherwise. For Arabidopsis, three leaf pieces from separate plants were ground together for each treatment. cDNA was made from 5 µg of total RNA (RNA to cDNA EcoDry Premix [Oligo dT]; Clontech]) according to the manufacturer’s protocol. RT-qPCR was performed with diluted cDNA using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) in the ViiA 7 Real-Time PCR System (Applied Biosystems). Relative expression was calculated in enzyme- or Est-treated leaves with respect to mock/control (buffer or 0.1% [v/v] DMSO)-treated leaves. The FC was calculated using the 2−∆∆Ct method (Livak and Schmittgen, 2001). OsACTIN1 and AtACTIN2 were used as internal controls for rice and Arabidopsis, respectively. All the primers for RT-qPCR were designed using QuantPrime (Arvidsson et al., 2008; Supplemental Table S4).
Protein Isolation and Immunoblotting
For immunoblotting, total protein was isolated from 10 to 12 leaf pieces of rice or three leaves of Arabidopsis using the protocol described previously with minor modifications (Rohila et al., 2006). Twenty micrograms of total protein was loaded onto 10% SDS-PAGE gels for immunoblotting/Coomassie Brilliant Blue staining. The protein was transferred to a polyvinylidene difluoride membrane (Millipore) and processed for blotting. Anti-OsWAKL21376–725 antibodies were generated in rabbit in our institute animal house facility and used in a dilution of 1:100. Horseradish peroxidase-tagged anti-rabbit IgG secondary antibody (Abcam; dilution, 1:50,000) was used, and the blot was visualized in chemidoc (Vilber Lourmat).
Localization of OsWAKL21.2
The localization of OsWAKL21.2 was observed by transient transformation of onion (Allium cepa) peel cells and Nicotiana benthamiana leaves as described previously (Sun et al., 2007; Li, 2011). OsWAKL21.2 was cloned into Gateway-compatible vector pH7FWG2 (Karimi et al., 2002) and transformed in onion peel or N. benthamiana leaves by A. tumefaciens-mediated transient transformation. Plasmolysis was performed by incubating in 0.8 m mannitol for 30 min. Plasma membrane was stained by 10 μg mL−1 FM4-64. The GFP signal was visualized under a GFP filter using an ECLIPSE Ni-E epifluorescence microscope (Nikon), and images were captured at 60× magnification.
cGMP Quantification
cGMP was quantified in leaves of rosette-stage transgenic Arabidopsis plants by the method reported previously (Dubovskaya et al., 2011; Nan et al., 2014; Chen et al., 2018) with minor modifications. Six similar-sized leaves (total ∼200 mg) were collected from different plants for the untreated control. Three similar-sized leaves each from three different plants were infiltrated either with 0.1% (v/v) DMSO or 20 µm Est. Two leaves from each plant (total six leaves, ∼200 mg) were collected for cGMP quantification, whereas the third leaf was used for testing the expression of OsWAKL21.2. After 3 h of infiltration, leaves were collected and ground to a fine powder under liquid nitrogen. The powder was resuspended in 2 mL of ice-cold 6% (v/v) TCA and was collected in a 5-mL tube. After brief vortexing (10 s), tubes were centrifuged twice at 1,000g for 15 min at 4°C, and the supernatant was collected each time in 5-mL tubes. The aqueous supernatant was washed seven to eight times with water-saturated diethyl ether. The solvent was evaporated in a cold vacuum centrifuge at 4°C (SCANVAC; CoolSafe). cGMP was quantified in the extract using a cGMP enzyme immunoassay kit (Sigma-Aldrich, catalog no. CG201) according to the manufacturer’s protocol. Data were analyzed using the online tool Elisaanalysis.
Analyses of Publicly Available Transcriptome Data
Rice microarray data performed after X. oryzae treatment was obtained from GEO-National Center for Biotechnology Information (accession no. GSE36272). The .cel files were downloaded, analyzed, and processed using expression console (Affymetrix) using RMA-based normalization. The .chp files obtained after analysis were used in Transcriptome Analysis Console v3.0 software (Affymetrix) for relative expression analysis. Genes that show differential expression ≥ 1.5-fold with P < 0.05 were considered as differentially expressed.
Statistical Analysis
All experiments were independently performed at least three times. All data represented here indicate means ± se. The results of lesion length, callose deposition, and bacterial growth in colony-forming units were analyzed by one-way ANOVA (P < 0.05) followed by the Tukey-Kramer test. The results of RT-qPCR were analyzed by Student’s t test, and the genes that showed significantly altered expression (P < 0.05) between control and treated were considered as differentially expressed.
Accession Numbers
The PLIER16- and RMA-processed microarray data files generated and used in this experiment have been submitted to GEO (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE53940. Other publicly available microarray data used in our analysis were harvested from GEO under the accession numbers GSE49242 and GSE36272. The accession numbers of genes referred to in this study are provided in Supplemental Table S5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transcriptome profiling of rice leaves after treatment with LipA.
Supplemental Figure S2. Overexpression of OsWAKL21.2 induces rice immune responses.
Supplemental Figure S3. Methodology for down-regulation of OsWAKL21.2 in rice using VIGS.
Supplemental Figure S4. Transient down-regulation of OsWAKL21.2 in rice.
Supplemental Figure S5. VIGS-mediated transient down-regulation of OsWAKL21.2 does not have a significant effect on the expression of predicted off-target genes.
Supplemental Figure S6. RT-qPCR and immunoblot validation for heterologously expressing OsWAKL21.2 transgenic Arabidopsis plants.
Supplemental Figure S7. Biochemical characterization of OsWAKL21.2.
Supplemental Figure S8. Biochemical activities of the purified kinase domain of mutant versions of OsWAKL21.2.
Supplemental Figure S9. RT-qPCR and immunoblot validation of the expression of mutant versions of OsWAKL21.2 by transient transformation in rice and heterologous expression in Arabidopsis transgenic lines.
Supplemental Figure S10. Treatment with GC inhibitor attenuates OsWAKL21.2-induced callose deposition in transgenic Arabidopsis leaves.
Supplemental Figure S11. Heterologous expression of OsWAKL21.2 in Arabidopsis enhances the in planta cGMP level by its GC activity.
Supplemental Figure S12. Expression of JA pathway-related genes in rice and SA pathway-related genes in Arabidopsis following overexpression of mutant versions of OsWAKL21.2.
Supplemental Figure S13. Immunoblot validation of heterologous expression of OsWAKL21.2 in Arabidopsis transgenic lines generated after crossing with NahG lines.
Supplemental Table S1. List of probe sets that show differential expression after 2 h of LipA treatment.
Supplemental Table S2. List of differentially expressed genes after 2 and 12 h of LipA treatment.
Supplemental Table S3. Frequency of differentially expressed genes after LipA treatment in the microarray data performed after 24 h of X. oryzae treatment in GEO submission GSE36272.
Supplemental Table S4. List of primers used in this study.
Supplemental Table S5. Accession numbers of the genes mentioned in this study.
Acknowledgments
We thank Ramesh Palaparthi (Council of Scientific and Industrial Research, Centre for Cellular and Molecular Biology) for helping in analyzing the microarray data; Dr. Alok K. Sinha (Department of Biotechnology, National Institute of Plant Genome Research), Dr. Gopaljee Jha (Department of Biotechnology, National Institute of Plant Genome Research), and Dr. Puran Singh Sijwali (Council of Scientific and Industrial Research-Centre for Cellular and Molecular Biology) for their key suggestions in experiments; and Dr. Subhadeep Chatterjee (Department of Biotechnology, Centre for DNA Fingerprinting and Diagnostics) for providing NahG transgenic lines and the Pst DC3000 strain.
Footnotes
This work was supported by the 12th 5-year plan project, Plant-Microbe and Soil Interactions, of the Council of Scientific and Industrial Research (grant no. BSC0117) and a J.C. Bose fellowship to R.V.S. by the Department of Science and Technology, Ministry of Science and Technology (grant no. GAP0444).
References
- Albersheim P, Anderson-Prouty AJ(1975) Carbohydrates, proteins, cell surfaces, and the biochemistry of pathogenesis. Annu Rev Plant Physiol 26: 31–52 [Google Scholar]
- Aparna G, Chatterjee A, Jha G, Sonti RV, Sankaranarayanan R(2007) Crystallization and preliminary crystallographic studies of LipA, a secretory lipase/esterase from Xanthomonas oryzae pv. oryzae. Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 708–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparna G, Chatterjee A, Sonti RV, Sankaranarayanan R(2009) A cell wall-degrading esterase of Xanthomonas oryzae requires a unique substrate recognition module for pathogenesis on rice. Plant Cell 21: 1860–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B(2008) QuantPrime: A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton AR.(2011) Guanylyl cyclase activity in plants? Proc Natl Acad Sci USA 108: E96-NaN–E98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai W, Chern M, Ruan D, Canlas PE, Sze-To WH, Ronald PC(2011) Enhanced disease resistance and hypersensitivity to BTH by introduction of an NH1/OsNPR1 paralog. Plant Biotechnol J 9: 205–215 [DOI] [PubMed] [Google Scholar]
- Bojar D, Martinez J, Santiago J, Rybin V, Bayliss R, Hothorn M(2014) Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J 78: 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G(2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Cao Y, Li H, Kim D, Ahsan N, Thelen J, Stacey G(2017) Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun 8: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zuo S, Schwessinger B, Chern M, Canlas PE, Ruan D, Zhou X, Wang J, Daudi A, Petzold CJ, et al. (2014) An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant 7: 874–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Chen T, Sathe AP, He Y, Zhang X-B, Wu JL(2018) Identification of a novel semi-dominant spotted-leaf mutant with enhanced resistance to Xanthomonas oryzae pv. oryzae in rice. Int J Mol Sci 19: 3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G(2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Claverie J, Balacey S, Lemaître-Guillier C, Brulé D, Chiltz A, Granet L, Noirot E, Daire X, Darblade B, Héloir MC, et al. (2018) The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front Plant Sci 9: 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Couto D, Stransfeld L, Arruabarrena A, Zipfel C, Lozano-Durán R(2015) Broad application of a simple and affordable protocol for isolating plant RNA. BMC Res Notes 8: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U(2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KR, Darvill AG, Albersheim P, Dell A(1986) Host-pathogen interactions. XXIX. Oligogalacturonides released from sodium polypectate by endopolygalacturonic acid lyase are elicitors of phytoalexins in soybean. Plant Physiol 80: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Azevedo Souza C, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC(2017) Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol 173: 2383–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Delteil A, Gobbato E, Cayrol B, Estevan J, Michel-Romiti C, Dievart A, Kroj T, Morel JB(2016) Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol 16: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Chatterjee J, Kim S, Verma DPS(2008) Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229: 87–98 [DOI] [PubMed] [Google Scholar]
- Dubovskaya LV, Bakakina YS, Kolesneva EV, Sodel DL, McAinsh MR, Hetherington AM, Volotovski ID(2011) cGMP-dependent ABA-induced stomatal closure in the ABA-insensitive Arabidopsis mutant abi1-1. New Phytol 191: 57–69 [DOI] [PubMed] [Google Scholar]
- Galletti R, Denoux C, Gambetta S, Dewdney J, Ausubel FM, De Lorenzo G, Ferrari S(2008) The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring C, Turek IS(2017) Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front Plant Sci 8: 1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Pruitt R, Nürnberger T(2017) Sensing danger: Key to activating plant immunity. Trends Plant Sci 22: 779–791 [DOI] [PubMed] [Google Scholar]
- Harkenrider M, Sharma R, De Vleesschauwer D, Tsao L, Zhang X, Chern M, Canlas P, Zuo S, Ronald PC(2016) Overexpression of rice Wall-Associated Kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS ONE 11: e0147310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD(1999) A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol 39: 1189–1196 [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD(1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14: 55–63 [DOI] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S(2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Hu K, Cao J, Zhang J, Xia F, Ke Y, Zhang H, Xie W, Liu H, Cui Y, Cao Y, et al. (2017) Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat Plants 3: 17009. [DOI] [PubMed] [Google Scholar]
- Huang PY, Catinot J, Zimmerli L(2016) Ethylene response factors in Arabidopsis immunity. J Exp Bot 67: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA(2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni S, Scheuermann D, Krattinger SG, Kessel B, Wicker T, Herren G, Fitze MN, Breen J, Presterl T, Ouzunova M, et al. (2015) The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci USA 112: 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D, Tsuda K, Katagiri F(2012) The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J 71: 194–204 [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB(2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15: 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Ruelland E(2015) Magical mystery tour: Salicylic acid signalling. Environ Exp Bot 114: 117–128 [Google Scholar]
- Jha G, Patel HK, Dasgupta M, Palaparthi R, Sonti RV(2010) Transcriptional profiling of rice leaves undergoing a hypersensitive response like reaction induced by Xanthomonas oryzae pv. oryzae cellulase. Rice (N Y) 3: 1–21 [Google Scholar]
- Jha G, Rajeshwari R, Sonti RV(2007) Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol Plant Microbe Interact 20: 31–40 [DOI] [PubMed] [Google Scholar]
- Kant R, Dasgupta I(2017) Phenotyping of VIGS-mediated gene silencing in rice using a vector derived from a DNA virus. Plant Cell Rep 36: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A(2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kohorn BD.(2016) Cell wall-associated kinases and pectin perception. J Exp Bot 67: 489–494 [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P(2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J 60: 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Friedman HP, Fischer A, Byers N(2006) Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J Cell Sci 119: 2282–2290 [DOI] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L, Ruzvidzo O, Wheeler JI, Govender K, Iacuone S, Thompson PE, Gehring C, Irving HR(2011) The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J Biol Chem 286: 22580–22588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan A, Huang J, Zhao W, Peng Y, Chen Z, Kang D(2013) A salicylic acid-induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana. Plant Biol (Stuttg) 15: 452–461 [DOI] [PubMed] [Google Scholar]
- Li H, Zhou SY, Zhao WS, Su SC, Peng YL(2009) A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol 69: 337–346 [DOI] [PubMed] [Google Scholar]
- Li X.(2011) Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio Protoc 1: e95 [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R, Zipfel C(2015) Trade-off between growth and immunity: Role of brassinosteroids. Trends Plant Sci 20: 12–19 [DOI] [PubMed] [Google Scholar]
- Ma Y, Walker RK, Zhao Y, Berkowitz GA(2012) Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc Natl Acad Sci USA 109: 19852–19857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C(2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Meier S, Ruzvidzo O, Morse M, Donaldson L, Kwezi L, Gehring C(2010) The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE 5: e8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S(2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM(2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan W, Wang X, Yang L, Hu Y, Wei Y, Liang X, Mao L, Bi Y(2014) Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J Exp Bot 65: 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J(2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas PE, Ronald PC(2008) Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol 6: e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA(1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897 [DOI] [PubMed] [Google Scholar]
- Pillai SE, Kumar C, Patel HK, Sonti RV(2018) Overexpression of a cell wall damage induced transcription factor, OsWRKY42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BMC Plant Biol 18: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A, Mathur S, Verma V, Sharma S, Dasgupta I(2010) Virus-induced gene silencing in rice using a vector derived from a DNA virus. Planta 232: 1531–1540 [DOI] [PubMed] [Google Scholar]
- Qi Z, Verma R, Gehring C, Yamaguchi Y, Zhao Y, Ryan CA, Berkowitz GA(2010) Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc Natl Acad Sci USA 107: 21193–21198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeshwari R, Jha G, Sonti RV(2005) Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol Plant Microbe Interact 18: 830–837 [DOI] [PubMed] [Google Scholar]
- Ranjan A, Vadassery J, Patel HK, Pandey A, Palaparthi R, Mithöfer A, Sonti RV(2015) Upregulation of jasmonate biosynthesis and jasmonate-responsive genes in rice leaves in response to a bacterial pathogen mimic. Funct Integr Genomics 15: 363–373 [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Xu X, Gribskov M, Kanrar S, et al. (2006) Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J 46: 1–13 [DOI] [PubMed] [Google Scholar]
- Saintenac C, Lee WS, Cambon F, Rudd JJ, King RC, Marande W, Powers SJ, Bergès H, Phillips AL, Uauy C, et al. (2018) Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat Genet 50: 368–374 [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhan X, Yang P, Li J, Chen J, Tang B, Wang X, Hong Y(2019) Dual activities of plant cGMP-dependent protein kinase and its roles in gibberellin signaling and salt stress. Plant Cell 31: 3073–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Cao Z, Li Y, Zhao Y, Zhang H(2007) A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62: 529–532 [Google Scholar]
- Świeżawska B, Jaworski K, Duszyn M, Pawełek A, Szmidt-Jaworska A(2017) The Hippeastrum hybridum PepR1 gene (HpPepR1) encodes a functional guanylyl cyclase and is involved in early response to fungal infection. J Plant Physiol 216: 100–107 [DOI] [PubMed] [Google Scholar]
- Świeżawska B, Jaworski K, Szewczuk P, Pawełek A, Szmidt-Jaworska A(2015) Identification of a Hippeastrum hybridum guanylyl cyclase responsive to wounding and pathogen infection. J Plant Physiol 189: 77–86 [DOI] [PubMed] [Google Scholar]
- Takatsuji H.(2014) Development of disease-resistant rice using regulatory components of induced disease resistance. Front Plant Sci 5: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, He ZH(2002) The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol 129: 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithöfer A, Kalbacher H, Felix G(2018) The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat Plants 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Wheeler JI, Wong A, Marondedze C, Groen AJ, Kwezi L, Freihat L, Vyas J, Raji MA, Irving HR, Gehring C(2017) The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant J 91: 590–600 [DOI] [PubMed] [Google Scholar]
- Wong A, Gehring C, Irving HR(2015) Conserved functional motifs and homology modeling to predict hidden moonlighting functional sites. Front Bioeng Biotechnol 3: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, He SY(2013) Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Xu N, Fu D, Li S, Wang Y, Wong A(2018) GCPred: A web tool for guanylyl cyclase functional centre prediction from amino acid sequence. Bioinformatics 34: 2134–2135 [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY(2017a) Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot 68: 1371–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang B, Zuo W, Xing Y, Konlasuk S, Tan G, Zhang Q, Ye J, Xu M(2017b) Cytological and molecular characterization of ZmWAK-mediated head-smut resistance in maize. Mol Plant Microbe Interact 30: 455–465 [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL(2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA, et al. (2015) A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 47: 151–157 [DOI] [PubMed] [Google Scholar]