Transgenesis dissection of the functional roles of maize Glossy2 and Glossy2-like genes in cuticular lipid deposition reveals both common and distinct functions.
Abstract
Plant epidermal cells express unique molecular machinery that juxtapose the assembly of intracellular lipid components and the unique extracellular cuticular lipids that are unidirectionally secreted to plant surfaces. In maize (Zea mays), mutations at the glossy2 (gl2) locus affect the deposition of extracellular cuticular lipids. Sequence-based genome scanning identified a new Gl2 homolog in the maize genome, namely Gl2-like. Both the Gl2-like and Gl2 genes are members of the BAHD superfamily of acyltransferases, with close sequence similarity to the Arabidopsis (Arabidopsis thaliana) CER2 gene. Transgenic experiments demonstrated that Gl2-like and Gl2 functionally complement the Arabidopsis cer2 mutation, with differential influences on the cuticular lipids and the lipidome of the plant, particularly affecting the longer alkyl chain acyl lipids, especially at the 32-carbon chain length. Site-directed mutagenesis of the putative BAHD catalytic HXXXDX-motif indicated that Gl2-like requires this catalytic capability to fully complement the cer2 function, but Gl2 can accomplish complementation without the need for this catalytic motif. These findings demonstrate that Gl2 and Gl2-like overlap in their cuticular lipid function, but have evolutionarily diverged to acquire nonoverlapping functions.
Extracellular lipids are constituents of a protective hydrophobic structure, the cuticle, which covers aerial organs of all terrestrial plants. The cuticle plays important roles in many plant-environment interactions, including controlling water status (Kerstiens, 1996; Riederer, 2006), responding to abiotic stresses (Long et al., 2003), interacting with biotic pathogens (Kolattukudy, 1985; Jenks et al., 1994), and defining organ boundaries during development (Yephremov et al., 1999; Sieber et al., 2000; Kurdyukov et al., 2006). The extracellular cuticular lipids are chemically and physically arranged in distinct layers (Kolattukudy, 1965; Jellings and Leech, 1982; Jetter et al., 2000), with the epicuticular lipids being primarily linear alkyl chains, including very-long chain fatty acids (VLCFAs), hydrocarbons, ketones, primary and secondary alcohols, aldehydes, and wax esters; in addition, nonalkyl, terpene-type specialized metabolites are also components of the epicuticle (Martin and Juniper, 1970; Kolattukudy, 1976; Tulloch, 1976). These lipids are deposited on and embedded within a lipophilic cutin polymer matrix (a polymer of esterified ω−hydroxy and epoxy-C16 and C18 fatty acids, glycerol, and α,ω−αdicarboxylic acids; Kolattukudy, 2001; Heredia, 2003; Bonaventure et al., 2004; Franke et al., 2005; Pollard et al., 2008).
The molecular aspects of cuticular lipid biogenesis have been greatly facilitated by forward genetic approaches that use eceriferum (cer) mutants of Arabidopsis (Arabidopsis thaliana; Koornneef et al., 1989), glossy (gl) mutants of maize (Zea mays; Schnable et al., 1994) and tomato (Solanum lycopersicum; Vogg et al., 2004; Leide et al., 2007), and wax crystal-sparse leaf (wsl) mutants of rice (Oryza sativa; Yu et al., 2008; Wang et al., 2017). Results of these studies have been primarily interpreted in the context of a metabolic model that had been proposed from earlier physiological/biochemical studies (Bianchi et al., 1985; Post-Beittenmiller, 1996). At the core of this metabolic model is the endoplasmic reticulum-associated process of fatty acid elongation, which feeds two fatty acid modification pathways: a reductive pathway that generates fatty aldehydes, primary alcohols, and wax esters; and a decarbonylative pathway that converts the common, aldehyde-intermediate to hydrocarbons and ultimately ketones and secondary alcohols.
In maize, both fatty acid modification pathways are differentially expressed among different organs. Specifically, in juvenile leaves of maize seedlings, the reductive pathway predominates, and the expression of this metabolic network is under the control of the juvenile-to-adult phase transition, mediated by the transcription factor Gl15 (Moose and Sisco, 1996). By contrast, the fatty acid elongation-decarbonylative pathway is primarily expressed in silks (Perera et al., 2010; Loneman et al., 2017; Dennison et al., 2019). The gl mutations that have been characterized since the early-1900s have been identified via phenotypic screens of seedling leaves (Hayes and Brewbaker, 1928), which therefore primarily affect the fatty acid elongation-reductive pathway.
Glossy2 (Gl2) is exemplary of such a product of forward genetics, initially identified in 1928 as a mutant that causes “beading” of water droplets on seedling leaves, a common phenotype of all gl mutants (Hayes and Brewbaker, 1928). The initial characterization of the cuticular lipid profiles of gl2 (Bianchi et al., 1975) suggested deficiencies in lipids that are derived from the fatty acid elongation-reductive pathway, specifically a potential block in the fatty acid elongation process between chain lengths of 30 and 32 carbon atoms. The homology of Gl2 to the Arabidopsis CER2 gene became apparent when both loci were molecularly defined (Tacke et al., 1995; Xia et al., 1996); the two genes encode proteins that share ∼60% sequence similarity. The cer2 mutation causes the bright-green appearance of Arabidopsis stems (Koornneef et al., 1989), due to an underlying decrease in the products of the fatty acid elongation-decarboxylative pathway, with an apparent block in the elongation of C26 or C28 fatty acids to C30 fatty acid (McNevin et al., 1993; Jenks et al., 1995).
At the time of initial identification, these two proteins (GL2 and CER2) defined new sequences (Tacke et al., 1995; Xia et al., 1996), which ultimately became archetypal of the BAHD class of enzymes (St-Pierre and De Luca, 2000; D’Auria, 2006). BAHD enzymes are acyl-CoA-dependent acyltransferases that catalyze the acylation of alcohols or amine groups, forming ester and amide bonds in the assembly of a large number of specialized metabolites (St-Pierre and De Luca, 2000; D’Auria, 2006). These enzymes have been phylogenetically categorized into five clades (Clades I–V), with GL2 and CER2 being members of Clade II (D’Auria, 2006). The specific biochemical functions of the Gl2/CER2-containing Clade II BAHD enzymes remain unclear. More recently, yeast reconstitution experiments suggest that CER2 and CER2-LIKE proteins (Haslam et al., 2012; Haslam et al., 2015) and the rice homolog OsCER2 (Wang et al., 2017) associate with the fatty acid elongase (FAE) system, possibly by an interaction with the CER6-encoded FAE component, to alter the acyl-chain length profile of the system. However, the role of the BAHD acyltransferase catalytic capability of these proteins in this functionality is unclear.
In this study, we expanded the Gl2 characterization by identifying a Gl2-like gene in the maize genome that is a homolog of Gl2, which suggests that these two genes are products of an evolutionary gene duplication event that may have provided a template for potential neofunctionalization (Renny-Byfield and Wendel, 2014). Specifically, we used a transgenic strategy to evaluate and compare the functional ability of Gl2 and Gl2-like with genetically complement the Arabidopsis cer2 mutant. The resulting transgenic lines were used to evaluate the effects of these genetic manipulations on the cuticular lipid profiles and also on the broader lipid metabolic network (i.e. cutin and cellular lipidome). These experiments demonstrate that both the maize Gl2 and Gl2-like genes can complement the Arabidopsis cer2 mutant; however, the two genes have functionally drifted, with Gl2 expressing additional capabilities as compared with Gl2-like and CER2.
RESULTS
Computational Identification and Characterization of the Gl2-like Gene
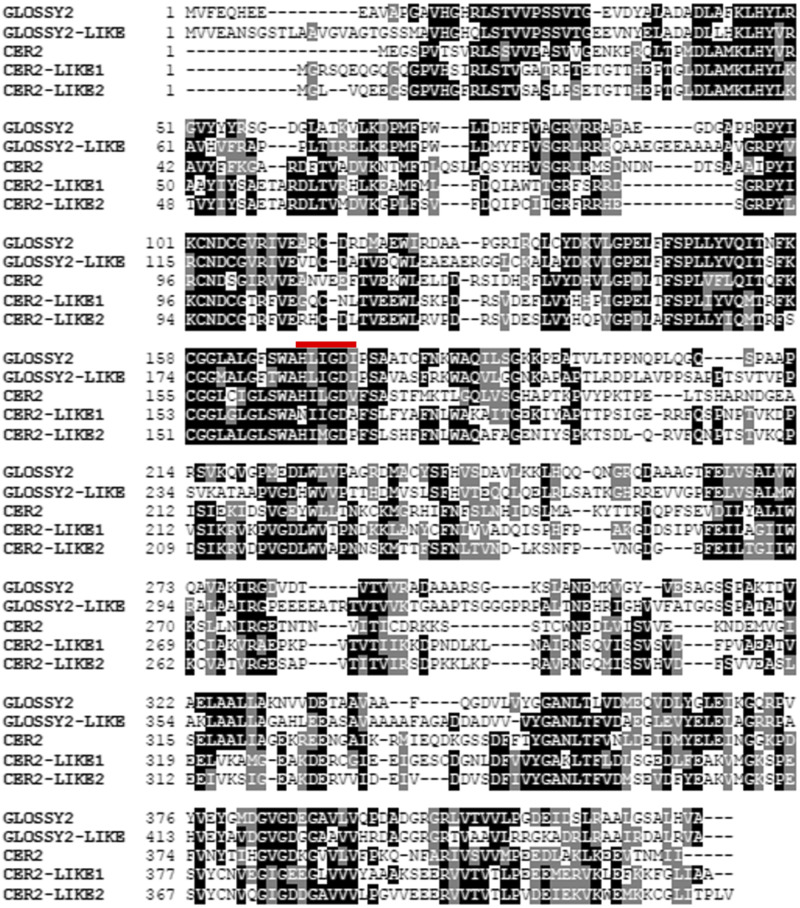
The phylogenetic diversity of the Gl2 (GRMZM2G098239; Zm00001d002353) gene was explored by sequence similarity searches of both the Arabidopsis and maize genomes (Altschul et al., 1997; Alonso et al., 2003; Andorf et al., 2016). This revealed that Gl2 shares high sequence similarity with an uncharacterized maize gene, which we term Gl2-like (GRMZM2G315767; Zm00001d024317); this gene also shares similarity with three Arabidopsis genes, CER2 (At4g24510), CER2-LIKE1 (At4g13840), and CER2-LIKE2 (At3g23840; Fig. 1; Table 1; Xia et al., 1996; Pascal et al., 2013). The GL2 and GL2-LIKE proteins share 50% sequence identity with each other, and ∼30% sequence identities with the CER2-family of proteins. This sequence identity includes the conserved HXXXDX, acyl-CoA-dependent acyl transferase domain, which is typical of the BAHD enzyme family (D’Auria, 2006).
Figure 1.
Amino acid sequence comparison of GL2 homologs. The maize GLOSSY2 (GRMZM2G098239; Zm00001d002353) and GLOSSY2-LIKE (GRMZM2G315767; Zm00001d024317) sequences are compared with the Arabidopsis CER2 (At4g24510), CER2-LIKE1 (At4g13840), and CER2-LIKE2 (At3g23840) sequences using Clustal O (1.2.4) and BOXSHADE (v3.21). Black shading indicates identical residues, and gray shading indicates similar residues. The conserved -HXXXDX- acyl-CoA-dependent acyltransferase catalytic-domain of BAHD enzymes is indicated with a red line.
Table 1. Conservation in the amino acid sequences of Z. mays GLOSSY2 homologs, CER2 homologs, and rice OsCER2.
The digits above the 100values are percent identity, and the digits below the 100-values are percent similarity.
| Identity/similarity | GLOSSY2 | GLOSSY2-LIKE | CER2 | CER2-LIKE1 | CER2-LIKE2 | OsCER2 |
|---|---|---|---|---|---|---|
| GLOSSY2 | 100 | 50 | 34 | 35 | 38 | 73 |
| GLOSSY2-LIKE | 63 | 100 | 32 | 33 | 34 | 51 |
| CER2 | 54 | 49 | 100 | 35 | 36 | 35 |
| CER2-LIKE1 | 51 | 48 | 53 | 100 | 64 | 35 |
| CER2-LIKE2 | 53 | 49 | 53 | 75 | 100 | 38 |
| OsCER2 | 81 | 63 | 53 | 52 | 54 | 100 |
Prior characterizations of the expression of the Gl2 gene indicate that it is highly expressed in young seedling leaves of maize (Tacke et al., 1995), and this is consistent with the role of this gene in cuticular lipid biosynthesis, as suggested with the glossy phenotype that is presented by mutant alleles (Hayes and Brewbaker, 1928). More extensive global transcriptomic analyses confirmed the high levels of Gl2 expression in seedling leaves, but also indicated that this gene is also highly expressed in tassels, endosperm, and silks (Supplemental Fig. S1; Supplemental Table S1). The particularly high levels of expression in silks is consistent with prior findings that this tissue deposits large quantities of cuticular lipids (Sekhon et al., 2011, 2012; Stelpflug et al., 2016; Portwood et al., 2019). The same RNA-sequencing dataset indicated that the expression pattern of the Gl2-like gene is distinct from that of Gl2, but more significantly, in most tissues examined, Gl2-like is expressed at lower levels than Gl2, by a factor of 5- to 10-fold (Supplemental Fig. S1).
Transgenic Expression of GL2 and GL2-LIKE Modifies the eceriferum Phenotype of the Arabidopsis cer2 Mutant Stems
The functionality of the Gl2 and Gl2-like genes was evaluated by their transgenic ability to complement the cer2 mutant of Arabidopsis. These effects were interpreted in the context of the effect of removing such functionality in the homologous host, i.e. the maize gl2 mutant (note a mutation at the gl2-like locus is not currently available). The gl2 mutation primarily affected the accumulation of the major components of the cuticular lipids on maize seedling leaves, reducing the levels of C32 primary alcohol and C32 aldehyde, which were associated with only a partial compensatory increase in C28 primary alcohol and aldehyde (Supplemental Fig. S2; Supplemental Table S2).
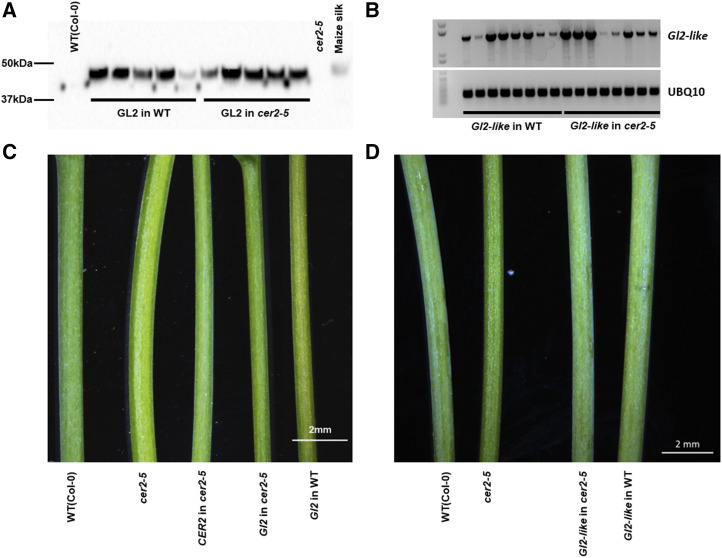
The open reading frame coding for the GL2 and GL2-LIKE proteins were expressed in Arabidopsis under the transcriptional control of the constitutive 35S promoter in homozygous lines that carried either wild-type or mutant cer2-5 alleles. Expression of the Gl2 and Gl2-like transgenes was confirmed at the protein level by Western blot analysis (Fig. 2A) of extracts using a GL2 antibody, and at the mRNA level by reverse transcription (RT)-PCR analysis of RNA isolated from these plants (Fig. 2B), respectively. As expected, the GL2 antibody detected a 46 kD polypeptide band in extracts from maize silk tissues, and this protein band was also detected in extracts prepared from Arabidopsis transgenic lines expressing the Gl2 transgene (i.e. genotype: Gl2 in wild type and Gl2 in cer2-5); this protein band was absent from the control, nontransgenic Arabidopsis plants. Gl2-like expression was detected via RT-PCR analysis with RNA-template preparations made from Arabidopsis plants carrying the Gl2-like transgene (in the wild-type and cer2-5 mutant lines), and this transcript was undetectable in the nontransgenic wild-type and cer2-5 mutant control plants (Fig. 2B).
Figure 2.
Transgenic expression of Gl2 and Gl2-like in Arabidopsis. A, Western blot analysis of the 46-kD GL2 protein transgenically expressed in either wild-type (WT) or cer2-5 mutant plants. Protein extract sample from maize silks serves as the positive control; protein extracts from the stems of wild-type Arabidopsis and cer2-5 mutant plants serve as negative controls. B, RT-PCR analysis of the Gl2-like mRNA heterologously expressed in either wild-type or cer2-5 mutant plants. Ubiquitin mRNA (At4g05320) is used as the internal control. C, Stem phenotypes of nontransgenic Arabidopsis wild-type and cer2-5 mutant, and transgenic lines expressing either maize Gl2 or CER2 in wild-type and cer2-5 mutant backgrounds.. D, Stem phenotypes of nontransgenic Arabidopsis wild-type and cer2-5 mutant, and transgenic lines expressing the maize Gl2-like transgene in either wild-type or cer2-5 mutant backgrounds.
Typical of cer mutants, the stems of the cer2-5 mutant show the eceriferum phenotype, presenting bright green stems (Koornneef et al., 1989), rather than the dull green appearance of the wild-type plants (Fig. 2, C and D). This phenotype is associated with epicuticle-deficiency and is indicative of changes in the cuticular surface lipid composition (Koornneef et al., 1989). As with the transgenic expression of the CER2 protein (Xia et al., 1996), the transgenic expression of the GL2-LIKE protein in the cer2-5 mutant background fully restores the stem eceriferum phenotype to the dull green, wild-type appearance (Fig. 2D). By contrast, the transgenic expression of the GL2 protein in the cer2-5 mutant only partially restored this phenotype to the wild-type appearance (Fig. 2C). As control experiments, we transgenically expressed the Gl2 or Gl2-like transgenes in wild-type Arabidopsis plants, and this did not alter the phenotype of the stems (i.e. they retained the dull green, wild-type appearance; Fig. 2, C and D).
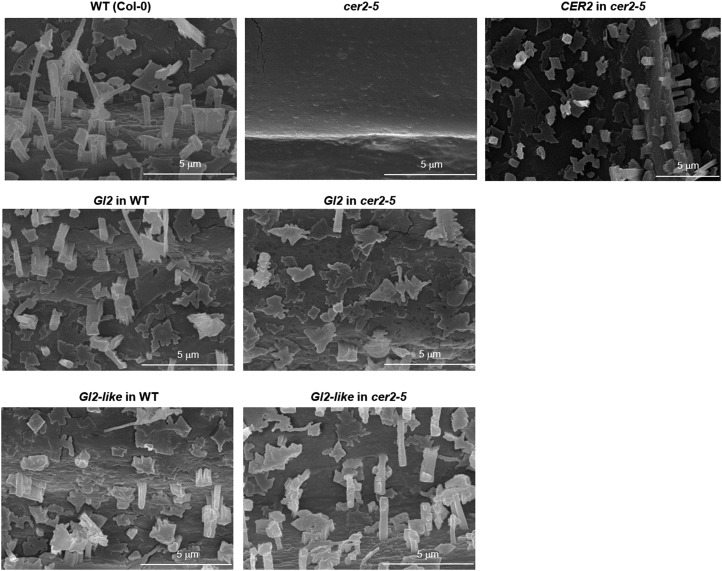
As was expected from prior studies (Xia et al., 1996), scanning electron microscopic (SEM) examination of the stems identified the crystalloid structures of the epicuticle, and these were absent from the surfaces of Arabidopsis cer2-5 mutant stems, but the transgenic expression of CER2 in this mutant background induced their reappearance, reinstating similar structures to those of wild-type plants (Fig. 3). The parallel SEM examination of the stem-surfaces of plants expressing the Gl2-like transgene in either the wild-type or cer2-5 mutant background indicated that the epicuticle was similar to that of wild-type plants (Fig. 3). By contrast, the crystalloids on those plants expressing the Gl2 transgene, in either the wild-type or cer2-5 mutant background, occurred at a lower density, and the structures of these crystalloids were more irregular, flattened, and have a thinner flake-like appearance than the wild-type control (Fig. 3).
Figure 3.
Arabidopsis stem epicuticular crystalloids. Scanning electron micrographs (10,000× magnification) of stem surfaces of nontransgenic Arabidopsis wild-type (WT) or cer2-5 mutant plants, compared with stem surfaces of transgenic plants expressing Gl2, Gl2-like, or CER2 transgenes in either wild-type or cer2-5 mutant plants.
Transgenic Expression of GL2 and GL2-LIKE Rescues the Cuticular Lipid-Deficiency Chemotype of the cer2 Mutant
Figure 4, Supplemental Figures S3 and S4, and Supplemental Table S2 present the data concerning the accumulation of extracellular cuticular lipids extracted from stems of the different genotypes developed in this study. The extractable cuticular lipid load on stems of the cer2-5 mutant was about half of the wild-type stems (Supplemental Fig. S3). As with the transgenic expression of CER2, the transgenic expression of either the maize Gl2 or the Gl2-like gene in this mutant increased the cuticular lipid load by about twofold, to near wild-type levels. The transgenic expression of Gl2-like in the wild-type background also increased the total cuticular lipid load by ∼15% compared with wild type (statistically significant by t test, P-value < 0.05), whereas the expression of the Gl2 transgene had no such effect on the total cuticular lipid load of the wild-type stems.
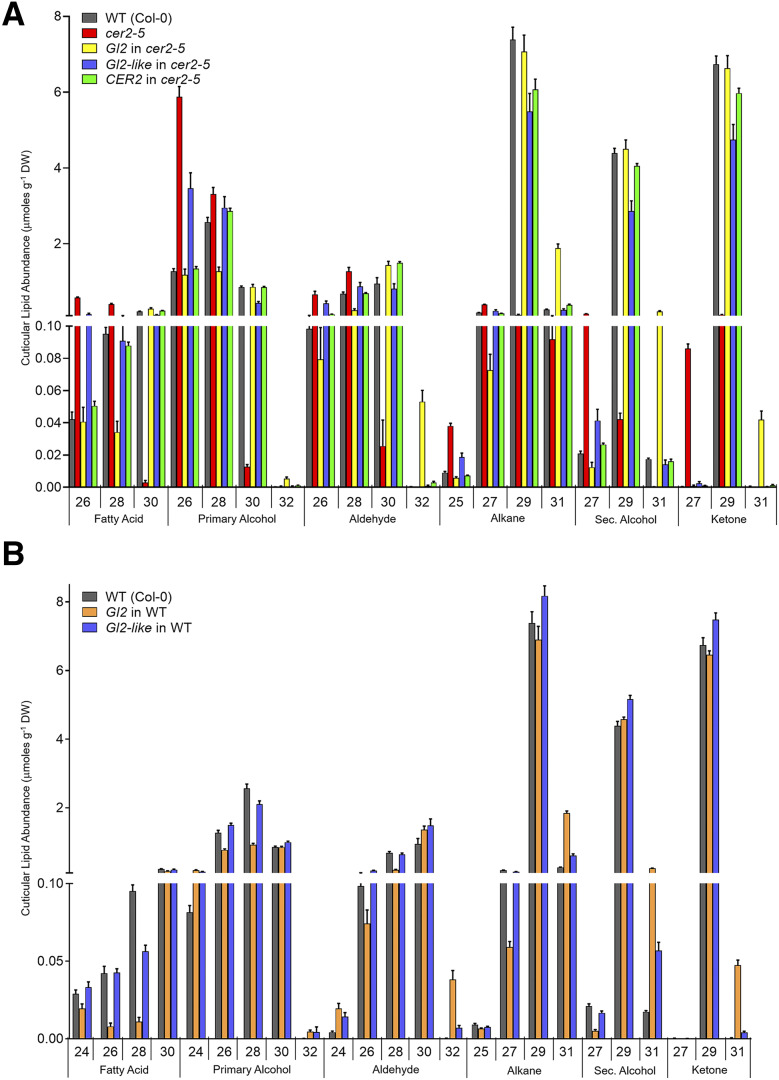
Figure 4.
Effect of transgenic expression of Gl2 and Gl2-like on the extracellular cuticular lipid profiles of Arabidopsis stems. A, Transgenic expression of Gl2, Gl2-like, or CER2 in the cer2-5 mutant background, as compared with the profiles in the nontransgenic wild-type (WT; Col-0) and cer2-5 controls. B, Transgenic expression of Gl2 and Gl2-like in the wild-type background. All numeric and statistical data, including the C20–C24 carbon chain-length minor components, can be found in Supplemental Table S2. The data represent means ± se of 10 to 15 replicates. DW, Dry weight.
Figure 4 shows the compositional changes in the cuticular lipid profiles of these stems; these data are focused on alkyl chain-lengths of 26-carbons and longer, whereas the effect on the accumulation of the shorter length alkyl chains was negligible (these latter data are included in Supplemental Figs. S3 and S4). The cer2-5 mutation primarily affected the accumulation of the major components of the cuticular lipids, which were nearly eliminated in the mutant; these constituents are C30 fatty acid alkyl derivatives, specifically C29 alkane, C29 secondary alcohol, and C29 ketone. These decreases in accumulation were associated with a partial compensatory increase in shorter chain fatty acid derivatives, predominantly the C26 primary alcohol. The transgenic expression of either Gl2, Gl2-like, or the CER2 transgenes in the cer2-5 mutant background resulted in a cuticular lipid compositional profile that was near identical to the wild-type profile in terms of the major cuticular lipid components. Thus, as compared with the cer2-5 mutant, these transgenic lines showed increased accumulation of the C29 alkane, C29 secondary alcohol, and C29 ketone. However, the effect on the less abundant cuticular lipids was different among the three transgenes. Thus, like the CER2 transgene, the Gl2 transgene decreased the accumulation of the C26 components (i.e. primary alcohols) to wild-type levels, but the Gl2-like transgene was not capable of this effect, and the C26 primary alcohol remained at elevated levels as in the cer2 mutant (Fig. 4A).
In addition to the transgenic compensatory effects on the cuticular lipid profiles, the expression of the two maize transgenes in the cer2-5 mutant induced new changes to the profiles that are not normally present in wild-type controls. In particular, Gl2 expression in cer2-5 mutant induced the formation of longer alkyl chains; namely derivatives of the C32 fatty acid, which includes C31 alkane, C31 secondary alcohol, and C31 ketone. These novel metabolites were also observed when the Gl2 transgene was expressed in the wild-type background (Fig. 4). These latter novel components were not manifest by the transgenic expression of either the CER2 or the Gl2-like genes in either the wild-type or the cer2-5 mutant (Fig. 4).
Therefore, collectively these biochemical changes establish that both the GL2 and GL2-LIKE proteins are functional homologs of CER2 and fully complement the biochemical deficiency associated with the cer2 mutation. However, the two maize genes are not equivalent in how they affect cuticular lipid profiles; the Gl2 transgene induced additional capabilities by enabling the production of even longer chain constituents than normal (up to 32-carbon fatty acids, and their alkyl derivatives) and Gl2-like was incapable of reversing the effect on the accumulation of 26-carbon atom constituents.
Mutations in the Putative BAHD Catalytic Domain Demonstrate Differences in the in Vivo Functionality Between GL2 and GL2-LIKE
The maize GL2 (Tacke et al., 1995) and the homologous Arabidopsis CER2 (Negruk et al., 1996; Xia et al., 1996) proteins initially defined the BAHD family of enzymes that catalyze acyltransferase reactions (D’Auria, 2006). A common structural feature is important to the catalytic function of BAHD enzymes, which has been identified through structural analysis and site-directed mutagenesis studies (Ma et al., 2005; D’Auria, 2006; Unno et al., 2007; Garvey et al., 2008). This is the HXXXDX-motif, which contains the catalytic His-residue that is responsible for deprotonating the alcohol or amine acyl acceptor-substrate, and is thus crucial to the acyltransferase catalytic mechanism. As with other biochemically characterized BAHD enzymes, this motif is similarly positioned in the primary sequences of GL2 and GL2-LIKE proteins (Fig. 1).
Interaction between potential substrates and this BAHD catalytic domain was computationally explored with structural models of GL2 and GL2-LIKE proteins, generated by Phyre2 (Kelley et al., 2015). With use of these structural models, the 3DLigandSite algorithm (Kelley and Sternberg, 2009) identified the Sorghum hydroxycinnamoyl transferase as the best BAHD structural template for GL2 and GL2-LIKE proteins. Using the experimentally determined structure of the substrate-enzyme complex of hydroxycinnamoyl transferase, we identified that in addition to the potential catalytic His residue, the last “X” residue of the HXXXDX-motif of GL2 and GL2-LIKE (i.e. Ile-174 and Ile-190, respectively) may be sufficiently close to a potential substrate to directly interact. Furthermore, the Ile residue at this position is rare among BAHD homologs (occurs <1% of 1,085 sequences that we analyzed), and residues with considerably smaller side chains are the most prevalent residues at this position (∼70% are Gly and ∼20% are Ala residues). By contrast, the middle 3 “X” residues are more conserved as hydrophobic amino acids.
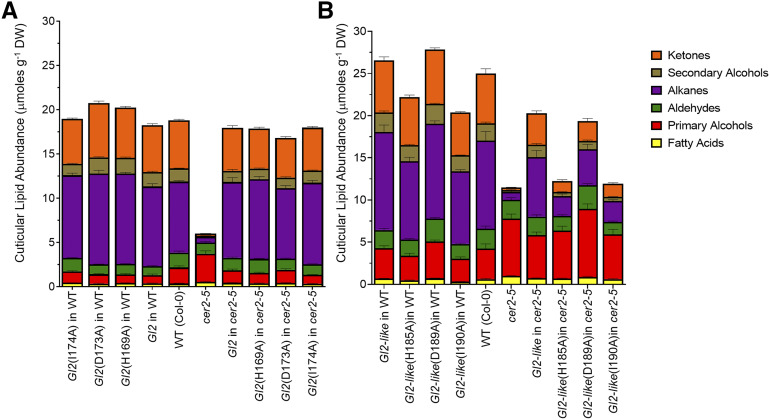
Therefore, site-directed mutagenesis was used to experimentally explore the functional role of the H, D, and final “X” residues in the HXXXDX-domain of the GL2 and GL2-LIKE proteins. Specifically, with each protein we generated three point-mutants by substituting Ala for each of the three critical residues in this domain [i.e., GL2(H169A), GL2(D173A), GL2(I174A), and GL2-LIKE(H185A), GL2-LIKE(D189A) and GL2-LIKE(I190A)]. Each of these mutants were transgenically expressed in the cer2-5 mutant and wild-type Arabidopsis plants and the stem cuticular lipid profiles were analyzed to assess the ability of the mutant proteins to complement the cer2-5 mutation (Fig. 5; Supplemental Table S3).
Figure 5.
The role of the HXXXDX, BAHD-defining catalytic motif in supporting in planta functionality of GL2 and GL2-LIKE proteins. Transgenes were either the wild-type (WT) Gl2 (A) or Gl2-like (B), or the indicated point mutants in the HXXXDX catalytic-motif. Each gene was expressed in Arabidopsis wild-type or cer2-5 mutant genetic backgrounds. All numeric data and statistical analysis can be found in Supplemental Table S3. The data represent means ± se of 10 to 40 replicates. DW, Dry weight.
All three Gl2 point mutants did not affect the ability of these transgenes to complement the cer2-5 chemotype; either the expression of the wild-type Gl2 transgene or any of the three Gl2 point mutants, which should have destroyed the BAHD catalytic capability, were still capable of reversing the reduction in epicuticular lipid accumulation that is characteristic of cer2-5 (Fig. 5A). By contrast, the H185A, and I190A point mutants of Gl2-like transgene could not fully complement the cer2-5 chemotype, and reverse the cuticular lipid load on these stems (Fig. 5B). These results demonstrate that the HXXXDX BAHD catalytic motif is not required for the ability of the GL2 protein to functionally replace the CER2 function; however, this motif is required for the ability of the GL2-LIKE protein to fully replace the CER2 function.
The Gl2 and Gl2-like Transgenes Alter the VLCFA and hydroxy-VLCFA Components of the Cutin and Cellular Lipidome Profiles
Because the genetic lesion that determines extracellular lipid traits occurs in the context of intracellular lipid metabolic processes, we profiled and compared the intracellular lipidomic pools in the stems of the different genotypes generated in this study. Furthermore, as potential BAHD enzymes, GL2 and/or GL2-LIKE may be involved in the assembly of the ester bonds, which are prevalent in the assembly of the cutin polymer. For these reasons, we also evaluated the effect of these genetic manipulations on the cutin monomers of the isolated cutin prepared from stems.
As with the cuticular lipid analyses described above, these comparisons were interpreted in the context of the effect of removing the gl2 functionality in maize. Thus, lipidomics analyses indicated that mutation at the gl2 locus in maize affects the VLCFA pool associated with the intracellular lipidome. These alterations were primarily associated with elongation of C26 and C28 fatty acids, leading to higher accumulation of these precursors (metabolites 60, 164, and 165; Supplemental Fig S5; Supplemental Table S4) and decreasing the level of C32 fatty acid (i.e. metabolite 167; Supplemental Fig. S5; Supplemental Table S4).
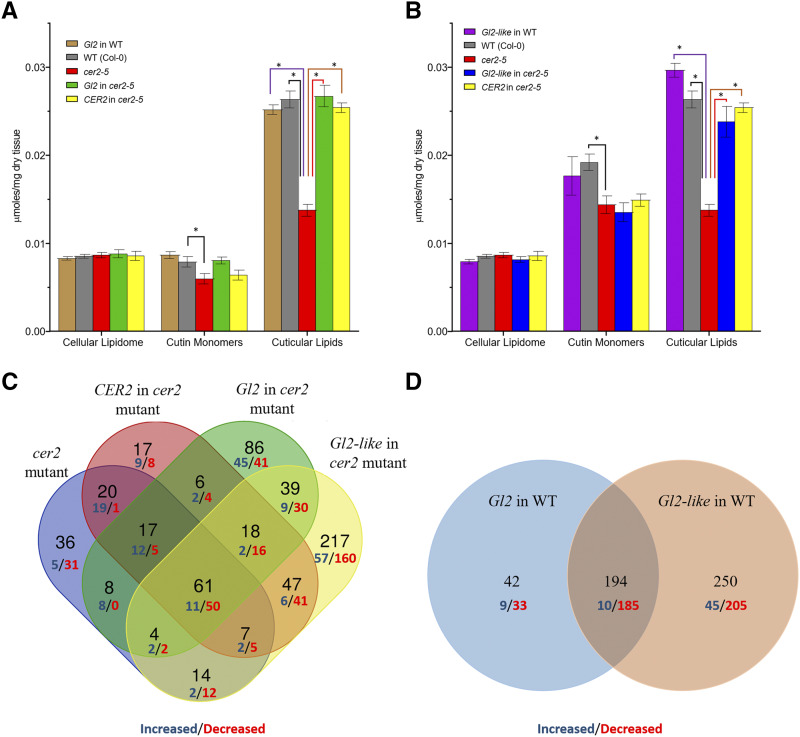
Figure 6 illustrates the effect of the transgenic expression of Gl2 (Fig. 6A) and Gl2-like (Fig. 6B) on the accumulation of the three lipid classes, the cellular lipidome, cutin monomers, and extractable cuticular lipids. These data indicate that quantitatively, the major effect of the cer2 mutation was in halving the total accumulation of the extractable cuticular lipids, with minimal or no effect on the total accumulation of the cellular lipidome or cutin monomers. Moreover, the transgenic expression of Gl2 or Gl2-like reversed the effect of the cer2 mutation, and returned cuticular lipid content to wild-type levels, and had no quantitative effect on the total accumulation of the other lipid components that were evaluated (Fig. 6, A and B).
Figure 6.
Effect of Gl2 and Gl2-like transgenes on accumulation of Arabidopsis stem lipids. Comparison of the cellular lipidome, cutin monomer, and cuticular lipid content of nontransgenic wild-type (WT) and cer2-5 mutant stems and transgenic plants expressing Gl2 or CER2 (A) and Gl2-like or CER2 (B). The data represent means ± se of 4 to 15 replicates. Asterisks indicate genetic manipulations that result in statistically significant changes as evaluated by Student’s t test (P-value < 0.05). Venn-diagram representation of changes in the accumulation of analytes identified in the cellular lipidome of stems in response to the transgenic expression of Gl2, Gl2-like, and CER2 in the cer2-5 mutant background (C) or in a wild-type background (D). Each subset identifies the number of lipid analytes whose abundance is either increased (blue digits) or decreased (red digits) in the indicated genetic background. The digital data can be found in Supplemental Table S4. The altered accumulation levels are statistically significant based on evaluation by Student’s t test (P-value < 0.05).
However, these transgenic manipulations had compositional effects on these lipids, and we focus the following text on the effect on VLCFA and derivative components. Although the cutin polymer is thought not to have VLCFA components, cutin preparations often contain VLCFA derivatives (i.e. 2-hydroxy-VLCFAs), which are probably associated with sphingolipids that copurify with cutin (Molina et al., 2006). Thus, regardless of their complex-lipid origins, because these VLCFAs are FAE-generated products, we evaluated the effect of Gl2 and Gl2-like transgenic expression on their abundance.
Specifically, in the cer2-5 mutant there were reductions in accumulation of the C22- and C24-fatty acids and their 2-hydroxy-derivatives, and an increase in the C26 fatty acid and the corresponding 2-hydroxy-derivative that was associated with the cutin preparations (i.e. metabolites 163, 165, 167, 176, 178, and 181; Supplemental Fig. S6A; Supplemental Table S4). Transgenic expression of Gl2 in the cer2-5 mutant reversed these alterations in VLCFA components, and in some cases further enhanced their accumulation to double or quadruple the levels that occur in the wild-type controls (i.e. metabolites 163, 165, 176, 178, and 179; Supplemental Fig. S6C). By contrast, the transgenic expression of Gl2-like caused only minor alterations in the total levels of cutin monomers (Fig. 6B) and of the VLCFA components associated with the cutin preparations, and this was irrespective of whether Gl2-like is expressed in a wild-type or cer2-5 mutant background (Supplemental Fig. S6, E and F; Supplemental Table S4).
Insights into the effect of transgenic expression of Gl2 and Gl2-like on the cellular lipidome was provided by the analysis of lipid extracts via a standardized LC-qTOF analytical platform (Okazaki and Saito, 2018). These analyses determined the relative abundance of 1372 lipid analytes, 153 of which were chemically identified. The latter include phosphoglycerolipids (29), glycolipids (35), isoprenoids (24), acylated glycolipids (8), sterol lipids (12), storage lipids (33), and free fatty acids (12; Supplemental Table S4). Among these chemically defined lipids, the most striking alterations were the changes in the accumulation of the free fatty acids (i.e. metabolites 148 to 153; Supplemental Fig. S6, A–F; Supplemental Table S4). The cer2-5 mutation reduced the accumulation of C30 fatty acid (metabolite 150), resulting in the higher accumulation of the precursor fatty acids of 26 and 28 carbon chain lengths (i.e. metabolites 148 and 149; Supplemental Fig. S6A; Supplemental Table S4). The transgenic expression of maize Gl2 and Gl2-like in this mutant reversed these cer2-5-induced effects (Supplemental Fig. S6, C and E; Supplemental Table S4). Moreover, Gl2 has additional capabilities, inducing the increased accumulation of C32 fatty acid (i.e. metabolite 151; Supplemental Fig. S6, C and D; Supplemental Table S4). This latter effect correlates with the increased levels of C31 alkyl derivatives that were detected in the epicuticular lipid profiles of wild-type and cer2-5 mutant stems in the same transgenic lines (Fig. 4). These findings suggest that both Gl2 and Gl2-like have the ability to affect the terminal elongation processes of fatty acids to C30 and C32 chain-lengths, which are negatively impacted by the cer2 mutation.
Another alteration in the lipidome that is consistent with the shared functionality between CER2, Gl2, and Gl2-like in fatty acid elongation is the alteration in the accumulation pattern of the glycosylceramides that uses a 2-hydroxy C26 fatty acid building block (i.e. metabolites 58 and 59; Supplemental Fig. S6A; Supplemental Table S4). The accumulation of these metabolites is doubled in the cer2-5 mutant, and this effect is reversed by the transgenic expression of either Gl2 or Gl2-like (Supplemental Fig. S6, C and E; Supplemental Table S4). An additional similar genetic modulation, but not associated with fatty acid elongation, is the accumulation of the minor sterol esters, that is, β-sitosterol esterified with linoleic or linolenic acid (i.e. metabolites 106 and 107; Supplemental Fig. S6, A, C, and E; Supplemental Table S4). When compared with the wild-type, the accumulation of these two sterol esters was increased by between 10- and 25-fold in the cer2-5 mutant, and their levels were decreased to normal when Gl2 was transgenically expressed in the mutant, whereas the transgenic expression of Gl2-like did not have this latter effect (Supplemental Fig. S6, C and E; Supplemental Table S4).
These lipidomics analyses also quantified changes in the accumulation of 1,219 analytes that are chemically undefined (Supplemental Table S4). On a mole basis, these chemically undefined lipids account for ∼15% of all the lipids that were collectively profiled in the wild-type and in the cer2-5 mutant tissue. These data would be more informative of Gl2 or Gl2-like functions once their chemical identities are determined; however, we evaluated their relative accumulation patterns as molecular markers to gain insights into the relative functionalities of the two maize genes.
The accumulation of approximately half of these chemically undefined lipid analytes were unaffected by the cer2-associated genetic manipulations. The Venn-diagram shown in Figure 6C illustrates that 35% of these analytes decreased in accumulation in response to these genetic manipulations, whereas only 15% showed increased accumulation. As molecular markers, the changes in the accumulation of these unknown metabolites associated with each transgenic event (i.e. Gl2 or Gl2-like expression) did not parallel the genetic complementation displayed by CER2 transexpression. For example, there were 156 analytes (13% of detected analytes) whose accumulation was similarly altered by the expression of either CER2, Gl2, or Gl2-like, whereas there were 17, 86, and 217 analytes (collectively 26% of the detected analytes) that were uniquely altered by the expression of each individual transgene (Fig. 6C). Moreover, the expression of the two maize homologs in a wild-type background induced changes in approximately 300 analytes, and only two-thirds of these were common to both transgenic lines (Fig. 6D). Therefore, although the two maize homologs can functionally complement the cuticular lipid phenotype of the cer2 mutation, the two maize genes also express unique attributes that are not equivalent to each other or to the CER2 gene.
DISCUSSION
The extracellular cuticular lipids that coat the outer surfaces of the aerial organs of terrestrial plants (Fernandes et al., 1964; Kolattukudy, 1980) are specifically produced by the epidermal cell layer of these organs. Because epidermal cells account for only about 10% of the cellular population of these aerial organs (Jellings and Leech, 1982), elucidating the molecular and biochemical mechanisms that regulate their biogenesis is confounded by the other 90% of the cell population that is not involved in these processes. This technical barrier has been partially overcome by using forward genetic approaches to identify and characterize genes involved in extracellular cuticular lipid accumulation (Kunst and Samuels, 2009; Shaheenuzzamn et al., 2019). This strategy has been successful in the isolation of causative genes that generate the cuticular lipid phenotypes (gl in maize and eceriferum in Arabidopsis). Yet in many cases, the exact mechanisms by which these gene products affect cuticular lipid deposition remain poorly understood.
Exemplary of such molecular characterization of cuticular lipid genes is the Gl2 gene of maize and its Arabidopsis homolog CER2 (Tacke et al., 1995; Xia et al., 1996). In this study, we specifically implemented a transgenic strategy to characterize the functional interrelationship between the maize Gl2 gene, the homologous Gl2-like gene, and the Arabidopsis homolog CER2. We identified the maize Gl2-like gene by the shared 63% sequence similarity with Gl2. Although Gl2 is known to be involved in cuticular lipid biosynthesis (Hayes and Brewbaker, 1928; Bianchi et al., 1975; Tacke et al., 1995), Gl2-like may have overlapping but distinct roles in this pathway. The transgenic experiments conducted in this study utilized Arabidopsis as the vehicle for these evaluations, testing for the ability of each maize gene to compensate for the missing function associated with the homologous Arabidopsis CER2 gene; in parallel, we also evaluated the effect of each transgenic event in the wild-type background.
The Ability of Gl2 and Gl2-like to Contribute to Extracellular Cuticular Lipid Deposition
The functionality of Gl2 in cuticular lipid biosynthesis was initially identified by the genetic observations that mutations at this locus eliminate the deposition of these constituents on the surfaces of juvenile seedling leaves (Hayes and Brewbaker, 1928; Bianchi et al., 1975). The identification of Gl2-like is solely based on sequence similarity to the GL2 and CER2 proteins, without any functional data to support its role in cuticular lipid biosynthesis. Mutations at the cer2 locus block the normal accumulation of cuticular lipids, by apparently blocking the conversion of C26 and/or C28 fatty acid to C30 fatty acid (McNevin et al., 1993; Jenks et al., 1995).
Our transgenic studies established the functionality of Gl2-like in cuticular lipid biosynthesis. Specifically, as with the transgenic expression of CER2 in a cer2 mutant, the transgenic expression of either Gl2 or Gl2-like in the same mutant, restores the normal accumulation of cuticular lipids, and restores the ability to convert the C26/C28 fatty acids to C30 fatty acid. Therefore, both Gl2 and Gl2-like are functional homologs of CER2. However, there are differences induced by the transgenic expression of Gl2 as compared to Gl2-like, which suggest that the two gene products have acquired distinct functionalities since the probable gene duplication event that gave rise to the two homologs. These differences between Gl2 and Gl2-like are revealed as differences in the (1) ability to complement the eceriferum phenotype on Arabidopsis stems; (2) epicuticular crystalloid morphologies; (3) total cuticular lipid loads and compositions; and (4) intracellular lipidomes in the cer2-5 lines complemented by each maize gene.
In these comparisons, the cer2 plants that were genetically complemented with the Gl2-like transgene presented traits that are more like the wild-type Arabidopsis plants than the Gl2-complemented plants. Moreover, although the transgenic expression of either the CER2 gene or Gl2-like in the wild-type background did not induce any new cuticular lipid components, the parallel experiments conducted with the transgenic expression of Gl2 resulted in plants that express new cuticular lipid components that are not associated with the wild-type Arabidopsis host (i.e. the ability to support the generation of C32 fatty acids and alkyl derivatives). Collectively, these observations are interpreted to indicate that although both maize Gl2 and Gl2-like genes express a functionality that can replace the CER2 function of Arabidopsis, the Gl2-like gene more completely and accurately recapitulates the CER2 functionality, and Gl2 encodes additional functionalities that are beyond those of Gl2-like and CER2 genes. This neofunctionalization that probably arose following the gene duplication event that generated the two maize paralogs enables Gl2 to induce the production of C32 alkyl-chains in Arabidopsis is consistent with the fact that the maize cuticular lipids are predominantly alkyl chains derived from C32 fatty acids. Similar neofunctionalization of the Arabidopsis CER2 gene family can be inferred with CER2-LIKE1 facilitating the production of 32-carbon alkyl-chains (Pascal et al., 2013).
Relationship between BAHD Catalytic Activity and the Ability of Gl2 and Gl2-like to Support In Planta Epicuticular Lipid Deposition
Although not initially recognized (Tacke et al., 1995; Xia et al., 1996), CER2 and GL2 proteins proved to be archetypal of the BAHD-family of acyltransferases (St-Pierre and De Luca, 2000; D’Auria, 2006). This catalytic ability of the BAHD enzymes was demonstrated by the biochemical characterization of a number of enzymes that catalyze acyltransferase reactions in the biosynthesis of diverse specialized metabolites. The BAHD enzymes are recognizable by the conservation of two primary sequence motifs: (1) the HXXXDX catalytic domain and (2) the DFGWG domain that appears to stabilize the substrate-enzyme complex (St-Pierre and De Luca, 2000; D’Auria, 2006). Although both domains have been reported to be essential for enzyme functionality, the latter domain is not fully conserved among all characterized BAHD enzymes (Suzuki et al., 2003; Bayer et al., 2004; Ma et al., 2005; Unno et al., 2007).
The neofunctionalization that was enabled by the gene duplication that gave rise to Gl2 and Gl2-like appears to be associated with the potential catalytic capabilities of this BAHD-defining catalytic domain. The role of the HXXXDX domain to support cuticular lipid deposition by GL2 and GL2-LIKE were tested in planta by directed mutagenesis. Point mutants that are predicted to disrupt BAHD catalytic activity (i.e. the His, Asp and terminal “X” residue of the HXXXDX domain) did not affect the ability of GL2 to support cuticular lipid deposition, whereas these mutations partially disrupted the ability of GL2-LIKE to support this metabolic outcome. These findings indicate that although the GL2 protein does not require BAHD catalytic activity to support cuticular lipid deposition, this domain is required for GL2-LIKE to fully achieve the same function.
The GL2 results agree with studies previously conducted with CER2, which demonstrated that the BAHD-defining HXXXDX domain is not needed to complement the cer2-associated cuticular lipid deficiency (Haslam et al., 2012). Moreover, this study also demonstrates that the yeast coexpression of CER2 with a 3-ketoacyl-CoA synthase (KCS) component of the plant FAE system results in the ability of the yeast strain to produce longer-chain fatty acids than normal, up to 28 and 30 carbon chain lengths; yeast strains normally only produce C26 VLCFA. Moreover, this latter capability does not require an intact HXXXDX catalytic domain. These results have been interpreted to indicate that CER2 noncatalytically interacts with the FAE system for VLCFA biosynthesis, and alters the product profile of the FAE system. Similar conclusions have been reached with the characterization of Arabidopsis CER2 homologs, such as CER2-LIKE1 and CER2-LIKE2, where the former lacks the His residue in the predicted HXXXDX motif (Haslam et al., 2012, 2015).
In contrast with CER2 and Gl2, Gl2-like requires the intact, and presumably functional, BAHD-defining HXXXDX catalytic domain to fully complement the in planta cer2 function. Moreover, a functional HXXXDX, BAHD-catalytic domain appears to be required by the rice CER2 homolog WSL4, which facilitates the interaction with KCS (Wang et al., 2017) to affect fatty acid elongation beyond C24 chain-lengths. These characterizations raise a number of questions concerning the ability of these proteins to support cuticular lipid deposition and the function of other Clade II BAHD proteins: (1) Is GL2-LIKE (and possibly the rice homolog, OsCER2) a bifunctional protein, one mediated by the HXXXDX catalytic function, and the second that is independent of this catalytic function? (2) Do other members of the Clade-II BAHD family possess an acyltransferase catalytic activity? (3) What are the two substrates (the acyl-donor and acyl-acceptor substrates) of the GL2-LIKE acyltransferase enzyme? Answering these questions will probably require the development of a direct biochemical assay that goes beyond the studies conducted to date; these latter studies primarily rely on correlations between genetic modifications and metabolic outcomes.
The Role of Gl2 and Gl2-like in Supporting the Accumulation of VLCFA Components of Intracellular Lipid Pools
Mutations in the cer2 locus appear to block the fatty acid elongation process from C26 or C28 to C30 fatty acids (McNevin et al., 1993; Jenks et al., 1995). Based on our current understanding of the mechanism of fatty acid elongation, it is mechanistically unclear how specific iterations of the elongation cycle (i.e. from C26 or C28 to C30) can be distinguished from any of the other six iterations that elongate a C18 fatty acid to a C30 fatty acid. Recent studies with the Arabidopsis CER2 and rice homologs, have proposed the possibility of a physical interaction between the condensing enzyme of the FAE complex (specifically KCS6) and CER2 (Haslam et al., 2015; Wang et al., 2017) and this interaction may affect a single iteration of the elongation cycle.
Based on the cuticular lipid profiles in transgenic Arabidopsis lines, we surmise that as with CER2, Gl2-like can support the ability of elongating fatty acids from C28 to C30. Although Gl2 also shares this capability, it can additionally contribute to the elongation of C30 to C32 fatty acids. Thus, we hypothesize that Gl2-like and Gl2 probably have overlapping functions in maize, where the former affects fatty acid elongation from C26/C28 to C30, and the latter the elongation from C26/C28 to C32.
The primary genetic lesion that determines these extracellular cuticular lipid traits occur in the intracellular lipid metabolic processes that underlie the origins of the cuticular lipids. These processes are primarily associated with the intracellular membranes (ER and possibly plasma membrane), which house the biochemical reactions associated with VLCFA biosynthesis and down-stream reactions that generate the other alkyl derivatives of the cuticle (Bernard and Joubès, 2013). By profiling and comparing the intracellular lipidomic pool and the cutin matrix of the stems of the different genotypes generated in this study, we evaluated the extent to which VLCFA metabolism is juxtaposed with other lipid metabolic pathways (e.g. phospholipids, sphingolipids, and storage lipids). These comparisons indicate that genetic manipulations associated with mutating the cer2 function and replacing it with either Gl2 or Gl2-like affects cuticular lipid profiles and also has pronounced effects on those lipids that utilize VLCFA building blocks (Li-Beisson et al., 2010; Bernard and Joubès, 2013). In addition, CER2, Gl2, and Gl2-like expression altered the accumulation of several unidentified intracellular lipid molecules. Therefore, these combined data imply that these genes may have regulatory roles in controlling the FAE complex.
Based on the lipidomics analyses and the HXXXDX-mutagenesis studies, it is possible to hypothesize that the acyltransferase reaction of GL2-LIKE may be part of the regulatory mechanism that modulates the FAE complex, particularly the terminal elongation cycle. However, it is unclear how an acyltransferase reaction mechanism can affect one of several FAE reaction cycles that utilize iterations of Claisen condensation-reduction-dehydration-reduction reaction mechanisms.
MATERIAL AND METHODS
Plant Material and Growth Conditions
Transfer DNA mutant line SALK_084443C (cer2-5; At4g24510) in the Col-0 background was obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). This transfer-DNA insertion disrupts the second exon of CER2. This cer2-5 mutant stock and the wild-type Col-0 stock were used in all the experiments described herein. Seeds were sown in LC1 Sunshine Mix (Sun Gro Horticulture) and treated at 4°C for 5 d to break seed dormancy. Seedlings were transferred to individual pots and grown to maturity under constant growth conditions in a regulated growth room at 22°C under continuous illumination (2568 Lux or photosynthetic photon flux density of 100 μmol of photons m−2 s−1). Plants were watered once a week, and the irradiance, temperature, and relative humidity were monitored using an Onset Computer Corporation Hobo monitor U12-012 (www.onsetcomp.com). Biochemical and microscopic analyses were conducted on stem tissues of Arabidopsis plants of different genotypes, harvested when the primary flower bolt was 35 to 40 cm in height. Flowers, cauline leaves, and siliques were removed, and stem tissue was used for analysis.
Maize (Zea mays) gl2 mutant seeds (gl2-Salamini; Maize Genetics COOP Stock Center catalog no. 208H; maizecoop.cropsci.uiuc.edu) were outcrossed to inbred B73, and the resulting heterozygous F1 seeds were selfed and backcrossed to the inbred B73. The BC1 seeds were planted in soil, and grown in a climate-controlled greenhouse under a diurnal cycle of 16-h light and 8-h dark at 27°C and 24°C, respectively, maintaining 30% relative humidity. The gl2 mutant seedlings were identified from the segregating progeny by their water beading phenotype, and nonbeading sibling seedlings were used as wild-type controls. Five individual maize seedling plants, at the 3 to 5 leaf stage, were pooled to generate a single replicate, and a total of 4 to 5 replicates were used for cuticular lipid and lipidomic analyses.
Molecular Cloning
The full-length Gl2 (GRMZM2G098239; Zm00001d002353) and Gl2-like (GRMZM2G315767; Zm00001d024317) open read frames were codon-optimized for expression in Arabidopsis (Arabidopsis thaliana) with GeneOptimizer (GeneArt, LifeTechnologies) and OptimumGene (GenScript; www.genscript.com), respectively, and these sequences were chemically synthesized (GenScript; GeneArt, LifeTechnologies). The Gl2 sequence was cloned into pMA-RQ and pDONR221 entry vectors by Life Technologies Corporation. Gl2-like sequence was initially obtained from GenScript as a pUC57-clone and was subcloned into pENTR/D-TOPO entry vector (Invitrogen). Further subcloning of Gl2 and Gl2-like sequences for plant expression experiments were performed using LR Clonase II Enzyme Mix (Invitrogen), using pEarleyGate100 vector (Earley et al., 2006), which controls expression of the transgene with the CaMV 35S promoter. The resultant recombinant vectors (p35S::Gl2 and p35S::Gl2-like) were introduced into Agrobacterium tumefaciens (strain C58C1) by electroporation (Sambrook et al., 1989). For site-directed mutagenesis experiments, mutation of the His-, Asp-, and Ile- residues of the HXXXDX motif of GL2 and GL2-LIKE were generated using QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies). The GL2 protein was also expressed in Escherichia coli BL21AI strain (Invitrogen; www.thermofisher.com) using the pDEST17 expression vector (Invitrogen). The authenticity of all recombinant plasmids was confirmed by DNA sequencing.
Plant Transformation and Selection
Gl2 and Gl2-like transgenes were transformed into wild-type Arabidopsis, Col-0 ecotype, and the homozygous cer2-5 mutant line via an A. tumefaciens-mediated floral-dip protocol, adapted from Clough and Bent (1999). Briefly, inflorescence bolts were submerged for 20 s in an infiltration medium containing A. tumefaciens (strain C58C1) carrying either p35S:Gl2 or p35S::Gl2-like. The infiltration medium consisted of 2% (w/v) Suc and 0.02% (v/v) Vac-in-stuff Silwet l-77 (Lehle Seeds). Plants were then returned to a 22°C growth chamber under continuous illumination until seed set. Seeds from transformed plants were collected and germinated in soil. Transformed seedlings were initially identified as being resistant to BASTA herbicide (i.e. glufosinate), applied at a dilution of 1:1000. The herbicide-resistant plants were propagated and selfed to the T2 generation. Plants carrying the Gl2 or Gl2-like transgene were molecularly confirmed with gene-specific primers by PCR analysis using DNA-templates isolated from individual herbicide resistant plants. RNA expression of Gl2-like was confirmed by RT-PCR, and ubiquitin mRNA (At4g05320) was used as the internal control; GL2 protein expression was evaluated by Western blot assays. Three replicate lines from each of three independent transformation events were maintained and used for all further experiments.
Western Blot Analysis
Protein extracts were prepared by homogenizing plant leaf tissue with a buffer consisting of 62.5 mm Tris-HCl, pH 6.8; 30% (v/v) glycerol; 10% (w/v) SDS; and 10% (v/v) 2-mercaptoethanol. Samples were vortexed for 5 min, boiled for 10 min, and centrifuged at 13,000g for 2 min. The clarified supernatant protein extracts were subjected to SDS-PAGE, and proteins were transferred to a nitrocellulose membrane according to manufacturer’s instructions (Bio-RAD). Protein blots were first probed with GL2-specific antibody, recovered from ascites fluid recovered from GL2 challenged mice (www.biotech.iastate.edu/Hybridoma; 1:1,000 dilution), and then with horseradish peroxidase-linked antimouse IgG antibody (Bio-Rad; 1:3,000 dilution). The antigen-antibody complexes were detected using the Pierce ECL chemi-luminescent detection system (Thermo Scientific) and visualized on the ChemiDoc XRS+ gel documentation system (Bio-Rad).
Stereomicroscopy
The eceriferum phenotype was visualized from the central 1-cm segment of the stems using a Zeiss macro-Zoom microscope (Zeiss Axio Zoom V16) with ZEN2 software (Carl Zeiss.).
Scanning Electron Microscopy
The 1-cm stem segments were mounted on aluminum stubs with double-sided carbon tape, dried in a desiccator and sputter-coated (www.tedpella.com) with a Cressington HR208 sputter coater with platinum for 90 s at 40 milliamps, depositing a 10-nm coating. The segments were examined at 10 kV with a Hitachi SU4800 field emission SEM (www.hitachi-hightech.com), and images were digitally captured in TIFF format.
Extraction and Analysis of Stem Extracellular Cuticular Lipids
Extracellular cuticular lipids were extracted from isolated tissue by immersing the stems for 10 s in 10 mL chloroform (HPLC-grade Fisher Chemicals), containing 5 µg hexacosane (Sigma-Aldrich) as an internal standard. The stems were then flash frozen using liquid nitrogen and lyophilized (Labconco FreeZone Benchtop Freeze Dry System), and the weight of the dry biomass was determined. A similar extraction procedure was used to extract cuticular lipids from maize seedling, collected at 3 to 5 leaf stage, using 60 μg hexacosane as the internal standard.
The chloroform was removed from the extracellular cuticular lipid extracts by evaporation under a stream of N2 gas, and the dried lipids were silylated using a protocol based on that of Wood et al. (2001) and Hannoufa et al. (1993). Specifically, the dried extracts were dissolved in 0.2 mL acetonitrile, and silylation was performed by the addition of 0.05 mL of N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% (v/v) trimethylchlorosilane (TMCS; Sigma-Aldrich), and incubated at 65°C for 30 min. The samples were cooled, dried under a stream of N2 gas, and resuspended in chloroform. One microliter of the derivatized sample was injected into gas chromatography-mass spectrometry (GC-MS) or GC-flame ionization detector.
GC-MS analysis was conducted with an Agilent 7890A GC, equipped with a HP-5ms capillary column (30 m × 0.25 mm × 0.25 μm; Agilent), interfaced to an Agilent 5975C quadrapole mass spectrometer. Chromatography was conducted with helium gas, at a flow-rate of 1.0 mL min−1, and an inlet temperature at 280°C. The column oven temperature was initially held at 120°C, ramped at 10°C/min to 260°C and held at this temperature for 10 min, and then ramped at 5°C/min to 320°C and held there for 4 min. EI-MS ionization energy was set to 70 eV, and the interface temperature was 280°C. Resulting chromatograms and mass-spectra were deconvoluted and queried against an in-house Mass-Spectral library and the NIST 14 Mass Spectral Library using the NIST AMDIS software (Stein, 1999).
GC-flame ionization detector analysis was conducted with an Agilent 6890 GC, equipped with a DB-1 MS capillary column (15 m × 0.25 mm × 0.25 μm, Agilent 122-0112). Chromatography was conducted with helium gas, at a flow-rate of 1.2 mL min−1, and an inlet temperature at 280°C. The column oven temperature was initially held at 80°C, ramped at 15°C/min to 220°C, ramped at 7.5°C/min to 310°C, and finally ramped at 20°C/min to 340°C and held for 5 min. The Agilent ChemStation software was used for peak alignments, and parallel GC-MS analysis was used to chemically identify eluting peaks.
The relative abundance of extracellular lipids per milligram dry weight of plant material was calculated based on the ion-strength of the internal standard. Statistical significance was determined using Student’s t test.
Extraction and Analysis of Stem Cutin Monomers
With use of procedures described by Li-Beisson et al. (2010), cutin was extracted from 50 to 100 mg of freshly harvested tissue, spiked with 10 μg heptadecanoic acid as an internal quantitation standard. The monomer components were converted to methyl esters by acid catalysis and analyzed by GC-MS methods as described by Li-Beisson et al. (2013).
Extraction and Analysis of Stem Cellular Lipidome
Intracellular lipids were extracted from 5–8 mg dry Arabidopsis stem tissues or dried maize seedling leaf tissues that had previously been extracted for epicuticular lipids. The samples were analyzed using a Waters Xevo G2 Q-TOF MS equipped with a Waters ACQUITY UPLC system as previously reported by Okazaki and Saito (2018). Briefly, flash-frozen plant tissue was lyophilized and pulverized to fine powder using a Mixer Mill 301 (Retsch). An extraction solvent containing a mixture of methyl tert-butyl ether and methanol (3:1, v/v), spiked with 1 μm 1,2-didecanoyl-sn-glycero-3-phosphocholine (Sigma-Aldrich) as internal standard, was added to the tissue, at a rate of 160 μL per mg dry tissue, and the mixture was thoroughly vortexed. Water was added (50 μL per mg tissue), and the mixture was thoroughly mixed for 10 min at room temperature using a sample tube mixer. After a 10-min incubation on ice, the mixture was centrifuged at 3,000g at 4°C for 10 min. Then 160 μL of the supernatant was collected and evaporated to dryness in a SpeedVac. The residue was dissolved in 200 μL of ethanol, vortexed for 10 min at room temperature, and centrifuged at 10,000g at 4°C for 10 min. The supernatant (180 μL) was used immediately for lipid analysis. Lipid analysis was conducted using liquid chromatography quadrupole time-of-flight mass spectrometer (HPLC, Waters Acquity UPLC system; MS, Waters Xevo G2 Qtof), as reported by Okazaki and Saito (2018).
The relative abundance of the cellular lipidome was calculated using abundances detected in positive ion mode for all lipid categories (including unknowns) with the exception of fatty acids, which were detected in negative ion mode. Statistical significance was determined using Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: GRMZM2G098239 and Zm00001d002353 (GL2);GRMZM2G315767 and Zm00001d024317 (GL2-LIKE);At4g24510 (AtCER2); At4g13840 (AtCER2-LIKE1); At3g23840 (AtCER2-LIKE2); and Os04g0611200 (OsCER2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gl2 and Gl2-like gene expression patterns.
Supplemental Figure S2. Effect of the gl2 mutation on the extracellular cuticular lipid profiles of seedling leaves of maize at the 3 to 5 leaf stage.
Supplemental Figure S3. Effect of transgenic expression of Gl2, Gl2-like or CER2 on the minor constituents of the extracellular cuticular lipid profiles of stems of the Arabidopsis cer2-5 mutant plants, as compared to the nontransgenic wild-type and cer2-5 mutant stems.
Supplemental Figure S4. Extracellular cuticular lipid composition of Arabidopsis stems from the indicated genotypes.
Supplemental Figure S5. Effect of the gl2 mutation on the cellular lipidome of seedling leaves of maize at the 3 to 5 leaf stage.
Supplemental Figure S6. Effect of transgenic expression of Gl2, Gl2-like, or CER2 on the stem cellular lipidome.
Supplemental Table S1. Tissue collection data for RNA-seq analysis from maize inbred B73.
Supplemental Table S2. Cuticular lipid abundance of Arabidopsis stems and maize leaves.
Supplemental Table S3. Cuticular lipid abundance of Arabidopsis stems expressing mutated Gl2 and Gl2-like transgenes.
Supplemental Table S4. Lipidome and cutin data.
Acknowledgments
The authors acknowledge Dr. Harry T. Horner, Tracey Stewart, and Randell Den Adel for helpful discussion and for use of equipment at Roy J. Carver High Resolution Microscopy Facility (Iowa State University); Drs. Zhihong Song, Ann Perera, and Lucas Showman of the WM Keck Metabolomics Research Laboratory (Iowa State University) for assistance in metabolic analyses; Bri Vidrine (Iowa State University) for providing maize glossy2 mutant seed, and propagating this genetic stock; and the Hybridoma Facility (Iowa State University) for characterization of GLOSSY2 antisera.
Footnotes
This work was supported by the State of Iowa, through Iowa State University Center for Metabolic Biology (grant no. IOS–1139489 to B.J.N.), the National Science Foundation (NSF; grant no. EEC–0813570 to B.J.N.), and the National Science Foundation-EAPSI (grant no. OISE–1614020 to L.E.A.).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ(1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorf CM, Cannon EK, Portwood JL II, Gardiner JM, Harper LC, Schaeffer ML, Braun BL, Campbell DA, Vinnakota AG, Sribalusu VV, et al. (2016) MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res 44(D1): D1195–D1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A, Ma X, Stöckigt J(2004) Acetyltransfer in natural product biosynthesis—functional cloning and molecular analysis of vinorine synthase. Bioorg Med Chem 12: 2787–2795 [DOI] [PubMed] [Google Scholar]
- Bernard A, Joubès J(2013) Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Bianchi G, Avato P, Salamini F(1985) Biosynthetic pathways of epicuticular wax of maize as assessed by mutation, light, plant age and inhibitor studies. Maydica (Italy) 30: 179–198 [Google Scholar]
- Bianchi G, Avato P, Salamini F(1975) Glossy mutants of maize. VI. Chemical constituents of glossy-2 epicuticular waxes. Maydica (Italy) 20: 165–173 [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M(2004) Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J 40: 920–930 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1999) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- D’Auria JC.(2006) Acyltransferases in plants: A good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dennison T, Qin W, Loneman DM, Condon SGF, Lauter N, Nikolau BJ, Yandeau-Nelson MD(2019) Genetic and environmental variation impact the cuticular hydrocarbon metabolome on the stigmatic surfaces of maize. BMC Plant Biol 19: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS(2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fernandes AMS, Baker EA, Martin JT(1964) Studies on plant cuticle. Ann Appl Biol 53: 43–58 [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L(2005) Apoplastic polyesters in Arabidopsis surface tissues--a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Garvey GS, Mccormick SP, Rayment I(2008) Structural and functional characterization of the TRI101 trichothecene 3-o-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: Kinetic insights to combating fusarium head blight. J Biol Chem 283: 1660–1669 [DOI] [PubMed] [Google Scholar]
- Hannoufa A, McNevin J, Lemieux B(1993) Epicuticular waxes of eceriferum mutants of Arabidopsis thaliana. Phytochemistry 33: 851–855 [Google Scholar]
- Haslam TM, Haslam R, Thoraval D, Pascal S, Delude C, Domergue F, Fernández AM, Beaudoin F, Napier JA, Kunst L, Joubès J(2015) ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol 167: 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam TM, Mañas-Fernández A, Zhao L, Kunst L(2012) Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol 160: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes HK, Brewbaker HE(1928) Glossy seedlings in maize. Am Nat 62: 228–235 [Google Scholar]
- Heredia A.(2003) Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim Biophys Acta 1620: 1–7 [DOI] [PubMed] [Google Scholar]
- Jellings AJ, Leech RM(1982) The importance of quantitative anatomy in the interpretation of whole leaf biochemistry in species of triticum, hordeum and avena. New Phytol 92: 39–48 [Google Scholar]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EN(1994) Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) moench. Plant Physiol 105: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Tuttle HA, Eigenbrode SD, Feldmann KA(1995) Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol 108: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R, Schäffer S, Riederer M(2000) Leaf cuticular waxes are arranged in chemically and mechanically distinct layers: Evidence from Prunus laurocerasus L. Plant Cell Environ 23: 619–628 [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE(2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE(2009) Protein structure prediction on the Web: A case study using the Phyre server 4. Nat Protoc 4: 363–71 [DOI] [PubMed] [Google Scholar]
- Kerstiens G.(1996) Cuticular water permeability and its physiological significance. J Exp Bot 47: 1813–1832 [Google Scholar]
- Kolattukudy PE.(1985) Enzymatic penetration of the plant cuticle by fungal pathogens. Annu Rev Phytopathol 23: 223–250 [Google Scholar]
- Kolattukudy PE.(1965) Biosynthesis of wax in Brassica oleracea. Biochemistry 4: 1844–1855 [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE.(2001) Polyesters in higher plants In Babel W, and Steinbüchel AA, eds, Biopolyesters Advances in Biochemical Engineering/Biotechnology, Vol 71 Springer, Berlin, Heidelberg: [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE.(1976) Chemistry and Biochemistry of Natural Waxes. Elsevier, Amsterdam [Google Scholar]
- Kolattukudy PE.(1980) Cutin, suberin, and waxes In Stumpf PK, ed, Lipids: Structure and Function—A Comprehensive Treaty, Vol Vol 4 Academic Press, New York, pp 571–645 [Google Scholar]
- Koornneef M, Hanhart CJ, Thiel F(1989) A genetic and phenotypic description of Eceriferum (cer) mutants in Arabidopsis thaliana. J Hered 80: 118–122 [Google Scholar]
- Kunst L, Samuels L(2009) Plant cuticles shine: Advances in wax biosynthesis and export. Curr Opin Plant Biol 12: 721–727 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, Bär S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Métraux J-P, Yephremov A(2006) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Reussing K, Riederer M, Vogg G(2007) The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: Effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol 144: 1667–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson M, Arondel V, Bates P, Baud S, Bird D, Debono A, Durrett T T, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi:10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. Arabidopsis Book 8: e0133, doi:10.1199/tab.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loneman DM, Peddicord L, Al-Rashid A, Nikolau BJ, Lauter N, Yandeau-Nelson MD(2017) A robust and efficient method for the extraction of plant extracellular surface lipids as applied to the analysis of silks and seedling leaves of maize. PLoS One 12: e0180850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long LM, Patel HP, Cory WC, Stapleton AE(2003) The maize epicuticular wax layer provides UV protection. Funct Plant Biol 30: 75–81 [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J(2005) Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem 280: 13576–13583 [DOI] [PubMed] [Google Scholar]
- Martin JT, and Juniper BE, eds; (1970) The Cuticles of Plants. Edward Arnold (Publishers) Ltd., London and New York [Google Scholar]
- McNevin JP, Woodward W, Hannoufa A, Feldmann KA, Lemieux B(1993) Isolation and characterization of eceriferum (cer) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome 36: 610–618 [DOI] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M(2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67: 2597–2610 [DOI] [PubMed] [Google Scholar]
- Moose SP, Sisco PH(1996) Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10: 3018–3027 [DOI] [PubMed] [Google Scholar]
- Negruk V, Yang P, Subramanian M, McNevin J, Lemieux B(1996) Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J 8: 137–145 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Saito K(2018) Plant lipidomics using UPLC-QTOF-MS. Methods Mol Biol 1778: 157–169 [DOI] [PubMed] [Google Scholar]
- Pascal S, Bernard A, Sorel M, Pervent M, Vile D, Haslam RP, Napier JA, Lessire R, Domergue F, Joubès J(2013) The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J 73: 733–746 [DOI] [PubMed] [Google Scholar]
- Perera MADN, Qin W, Yandeau-Nelson M, Fan L, Dixon P, Nikolau BJ(2010) Biological origins of normal-chain hydrocarbons: a pathway model based on cuticular wax analyses of maize silks. Plant J 64: 618–632 [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB(2008) Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Portwood JL II, Woodhouse MR, Cannon EK, Gardiner JM, Harper LC, Schaeffer ML, Walsh JR, Sen TZ, Cho KT, Schott DA, et al. (2019) MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Res 47: D1146–D1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D.(1996) Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 405–430 [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Wendel JF(2014) Doubling down on genomes: Polyploidy and crop plants. Am J Bot 101: 1711–1725 [DOI] [PubMed] [Google Scholar]
- Riederer M.(2006) Thermodynamics of the water permeability of plant cuticles: Characterization of the polar pathway. J Exp Bot 57: 2937–2942 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T(1989) Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- Schnable P, Stinard P, Wen T-J, Heinen S, Weber D, Schneer- man M, Zhang L, Hansen J, Nikolau B(1994) The genetics of cuticular wax biosynthesis. Maydica 39: 279–287 [Google Scholar]
- Sekhon RS, Childs KL, Santoro N, Foster CE, Buell CR, de Leon N, Kaeppler SM(2012) Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol 159: 1730–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM(2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Shaheenuzzamn M, Liu T, Shi S, Wu H, Wang Z(2019) Research advances on cuticular waxes biosynthesis in crops: A review. Int J Agric Biol 21: 911–921 [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Métraux JP, Nawrath C(2000) Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12: 721–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V(2000) Chapter nine evolution of acyltransferase genes: Origin and diversification fo the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv Phytochem 34: 285–315 [Google Scholar]
- Stein SEE.(1999) An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J Am Soc Mass Spectrom 10: 770–781 [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM(2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Nakayama T, Nishino T(2003) Proposed mechanism and functional amino acid residues of malonyl-CoA:anthocyanin 5-O-glucoside-6′’’-O-malonyltransferase from flowers of Salvia splendens, a member of the versatile plant acyltransferase family. Biochemistry 42: 1764–1771 [DOI] [PubMed] [Google Scholar]
- Tacke E, Korfhage C, Michel D, Maddaloni M, Motto M, Lanzini S, Salamini F, Döring H-P(1995) Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J 8: 907–917 [DOI] [PubMed] [Google Scholar]
- Tulloch A.(1976) Chemistry of waxes of higher plants In Chemistry and Biochemistry of Natural Waxes. Elsevier, Amsterdam, pp 235–287 [Google Scholar]
- Unno H, Ichimaida F, Suzuki H, Takahashi S, Tanaka Y, Saito A, Nishino T, Kusunoki M, Nakayama T(2007) Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J Biol Chem 282: 15812–15822 [DOI] [PubMed] [Google Scholar]
- Vogg G, Fischer S, Leide J, Emmanuel E, Jetter R, Levy AA, Riederer M(2004) Tomato fruit cuticular waxes and their effects on transpiration barrier properties: Functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J Exp Bot 55: 1401–1410 [DOI] [PubMed] [Google Scholar]
- Wang X, Guan Y, Zhang D, Dong X, Tian L, Qu LQ(2017) A β-Ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol 173: 944–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KV, Bonham CC, Jenks MA(2001) The effect of water on the ion trap analysis of trimethylsilyl derivatives of long-chain fatty acids and alcohols. Rapid Commun Mass Spectrom 15: 873–877 [DOI] [PubMed] [Google Scholar]
- Xia Y, Nikolau BJ, Schnable PS(1996) Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 8: 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H(1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, He C(2008) Wax crystal-sparse Leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228: 675–685 [DOI] [PubMed] [Google Scholar]