Matrix-assisted laser desorption/ionization-mass spectrometry imaging reveals that biosynthesis and recycling of the plant defense compound dhurrin are key events during sorghum grain germination.
Abstract
Dhurrin is the most abundant cyanogenic glucoside found in sorghum (Sorghum bicolor) where it plays a key role in chemical defense by releasing toxic hydrogen cyanide upon tissue disruption. Besides this well-established function, there is strong evidence that dhurrin plays additional roles, e.g. as a transport and storage form of nitrogen, released via endogenous recycling pathways. However, knowledge about how, when and why dhurrin is endogenously metabolized is limited. We combined targeted metabolite profiling with matrix-assisted laser desorption/ionization-mass spectrometry imaging to investigate accumulation of dhurrin, its recycling products and key general metabolites in four different sorghum lines during 72 h of grain imbibition, germination and early seedling development, as well as the spatial distribution of these metabolites in two of the lines. Little or no dhurrin or recycling products were present in the dry grain, but their de novo biosynthesis started immediately after water uptake. Dhurrin accumulation increased rapidly within the first 24 h in parallel with an increase in free amino acids, a key event in seed germination. The trajectories and final concentrations of dhurrin, the recycling products and free amino acids reached within the experimental period were dependent on genotype. Matrix-assisted laser desorption/ionization-mass spectrometry imaging demonstrated that dhurrin primarily accumulated in the germinating embryo, confirming its function in protecting the emerging tissue against herbivory. The dhurrin recycling products, however, were mainly located in the scutellum and/or pericarp/seed coat region, suggesting unknown key functions in germination.
Cyanogenic glucosides (CNglcs) are nitrogenous defense compounds that are widely distributed in the plant kingdom (Gleadow and Møller, 2014). They are of particular interest due to their presence in several crops, including the economically important cereals barley (Hordeum vulgare; Nielsen et al., 2002), wheat (Triticum aestivum; Erb et al., 1981; Jones, 1998), and sorghum (Sorghum bicolor; Kojima et al., 1979; Poulton, 1990), and because they often accumulate in high amounts in a wide range of plants (Kojima et al., 1979; Swain and Poulton, 1994; Crush and Caradus, 1995; Gleadow and Woodrow, 2002; Busk and Møller, 2002; Forslund et al., 2003; Jørgensen et al., 2005; Gleadow et al., 2008; Sánchez-Pérez et al., 2008). The CNglcs exert their function as defense compounds by releasing hydrogen cyanide (HCN), which is toxic to many living organisms and potentially auto-toxic to the plant (Lechtenberg, 2011). The HCN release is caused by β-glycosidase catalyzed hydrolysis of the CNglcs, initiated by tissue disruption, as for example caused by chewing herbivores (Morant et al., 2008). Besides this well-established function, CNglcs serve roles as storage and transport forms of reduced nitrogen to be reincorporated in general metabolism upon demand (Selmar, 1993; Swain and Poulton, 1994; Pičmanová et al., 2015; Nielsen et al., 2016; Del Cueto et al., 2017; Bjarnholt et al., 2018; Blomstedt et al., 2018; Schmidt et al., 2018). Although all plants possess a detoxification and nitrogen recovery pathway for HCN produced as a by-product of ethylene biosynthesis (Matilla, 2000; Piotrowski, 2008; Iqbal et al., 2013), nitrogen recovery from CNglcs proceeds via pathways that avoid release of the auto-toxic HCN. One such pathway has been elucidated (Bjarnholt et al., 2018), whereas the existence of others is documented by the coherent series of CNglc-derived metabolites identified in several cyanogenic plants. These include CNglcs decorated with extra sugars or acyl groups, or amide and carboxylic acid derivatives (Li et al., 2013; Pičmanová et al., 2015; Blomstedt et al., 2016; Del Cueto et al., 2017; Bjarnholt et al., 2018; Bøgeskov Schmidt et al., 2018). A number of studies have indicated that CNglcs and possibly their metabolites are intimately involved in modulation of plant growth, development, and stress responses (Jørgensen et al., 2005; Møller, 2010; Blomstedt et al., 2012; Del Cueto et al., 2017; Ionescu et al., 2017).
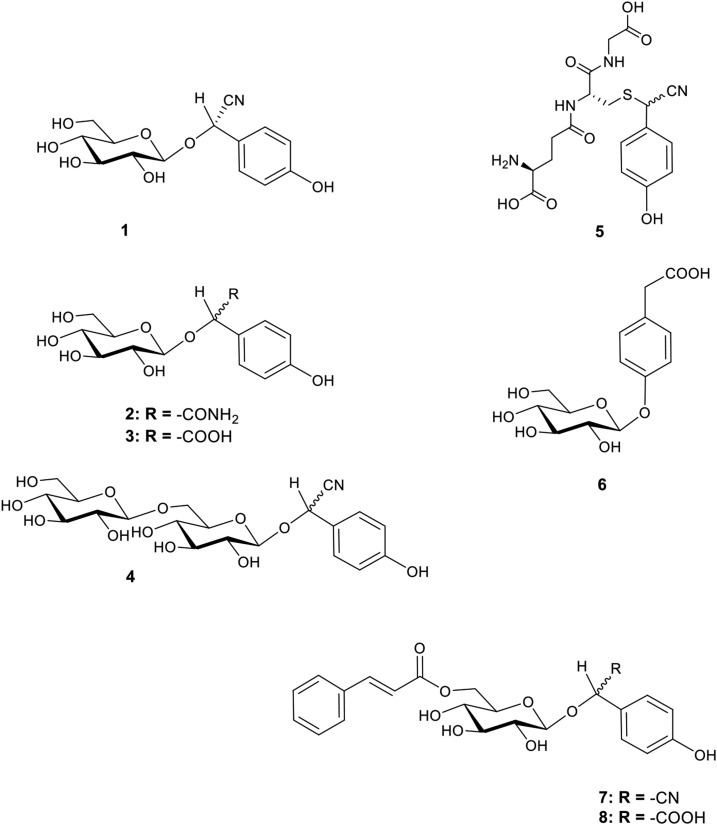
Sorghum contains the Tyr derived CNglc dhurrin and the dhurrin derived metabolites shown in Figure 1 (Pičmanová et al., 2015; Blomstedt et al., 2016; Bjarnholt et al., 2018). The biosynthetic pathway and the enzymes catalyzing dhurrin biosynthesis are well known (Gleadow and Møller, 2014), and the genes and enzymes forming a dhurrin recycling pathway have also been identified (Bjarnholt et al., 2018). This pathway proceeds via an intermediate glutathione conjugate (Fig. 1, 5) to form the end products ammonia and p-hydroxyphenylacetic acid. The latter compound exhibits hormonal activity in algae (Fries, 1976, 1977), and it is stored as the glucoside p-glucosyloxyphenylacetic acid in sorghum (Fig. 1, 6; Pičmanová et al., 2015; Blomstedt et al., 2016; Bjarnholt et al., 2018).The dhurrin derived carboxylic acid p-hydroxymandelic acid has antifungal and allelopathic activity (Kope et al., 1991; Yamamoto et al., 1999; Gu et al., 2008) and is likewise stored as a glucoside (dhurrin acid; Fig. 1, 3) in high concentrations in sorghum (Pičmanová et al., 2015; Blomstedt et al., 2016). The sorghum grain contains little or no dhurrin (Tipples and Norris, 1983; Panasiuk and Bills, 1984; Veličković et al., 2014; Nielsen et al., 2016), but dhurrin is rapidly biosynthesized at the early stages of plant development where it accumulates in high amounts to protect the newly formed tissues (Halkier and Møller, 1989; Busk and Møller, 2002). Interestingly, dhurrin recycling occurs concurrently with dhurrin de novo biosynthesis at the early stage of sorghum seedling development (Adewusi, 1990; Busk and Møller 2002; Bjarnholt et al., 2018). At that stage, nitrogen recovery may not be the main purpose of dhurrin recycling. Previous studies did not investigate if dhurrin biosynthesis and recycling takes place already during seed germination.
Figure 1.
Structures of dhurrin and its identified recycling products found in sorghum. (1) Dhurrin, (2) dhurrin amide, (3) dhurrin acid, (4) dhurrin glucosides (representative structure, exact linkage positions unknown), (5) GS-pOHPACN, (6) pGPAAc, (7) caffeoyl-dhurrin, and (8) caffeyol-dhurrin acid.
The seeds, formally called caryopsis, of Poaceae are characterized by a typical tissue compartmentalization, which depends on their taxonomical clade. Grains of cereals such as sorghum are constituted mainly by a starch and protein filled endosperm and a dormant embryo composed of the embryonic axis and scutellum (Fig. 2; Dante et al., 2014). Seed germination and initiation of central metabolism is induced by imbibition and hormone-controlled remobilization of resources from the endosperm storage tissue (Yamaguchi, 2008; Sponsel, 2016). Hydrolytic enzymes produced in the aleurone layer surrounding the endosperm are released to degrade the storage polymers. The released general metabolites are then transported via the scutellum into the developing embryo as a source of energy and building blocks (Dante et al., 2014; Rosental et al., 2014) ensuring rapid growth of the vulnerable young seedling. Channeling of resources toward formation of specialized metabolites would thus entail that these play a role in germination or early growth of the seedling. In germinating barley seeds, hordatines have been found to accumulate in the embryonic axis, presumably for the purpose of defending the tissue against pests and diseases at this vulnerable growth stage (Gorzolka et al., 2016). Plant hormones and specialized metabolites are biosynthetically and structurally closely related, and several specialized metabolites are suspected of playing a role in hormonal signaling. The production of hydrolytic enzymes excreted by the aleurone and scutellum layers is triggered by gibberellins, hormones produced in the scutellum and transported to the aleurone cells (reviewed by Yamaguchi, 2008; Sponsel, 2016). In wheat for example, the phenolic specialized metabolite coumarin has been shown to induce production of hydrolytic enzymes in aleurone cells (Saleh and Abu El-Soud, 2015). Tissue-wise localization of compounds during early germination and seedling development may, therefore, lend clues to their significance and biological activity.
Figure 2.
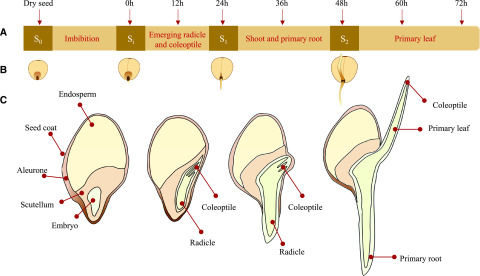
Sorghum germination process; embryo and seedling development and sampling time points. A, Timeline of imbibition, germination, and collection of samples. Sorghum grain were imbibed in water for 12 h and germinated for 72 h. Samples were collected every 12 h at the desired time points, as indicated with arrows. All eight time points were subjected to metabolite profiling. From the eight time points, four developmental stages (S) were selected for analysis by MSI and named: S0, dry seed; Si, after 12 h of imbibition; S1, emerged radicle and coleoptile; and S2, developed shoot and primary root. B, Graphical representation of the stages collected for MSI analyses. For the cvBTx623 line these stages matched the indicated time points (for tcd1 see Fig. 3). C, Schematic of longitudinal sections of the four developmental stages selected for MSI analysis, showing detailed morphological features and main compartments.
In this study, we investigate the spatial distribution of dhurrin and its derivatives in the germinating sorghum grain using mass spectrometry imaging (MSI). This technique enables determination of the in situ localization of metabolites based on their molecular masses. A number of different methods are available, but the most popular is matrix assisted laser desorption ionization (MALDI)-MSI (Bjarnholt et al., 2014; Boughton et al., 2016). This method allows the detection of a wide variety of metabolites and offers the possibility of very high spatial resolution (so far demonstrated down to 5 µm in plant tissue analyses; Bjarnholt et al., 2014; Korte et al., 2015; Boughton et al., 2016). Coupling the MALDI ion source with mass analyzers that provide high mass accuracy (<2 ppm), such as the Orbitrap, provides identification of compounds based on their accurate mass. Here, we applied highly sensitive liquid chromatography (LC)-triple quadrupole mass spectrometry (MS) analyses in combination with MALDI-Orbitrap-MSI to study the emergence and spatial distribution of dhurrin and its derivatives compared with selected general metabolites. To gain more insight into the significance of dhurrin biosynthesis and its endogenous recycling pathways, we monitored the changes in accumulation of dhurrin and its derived metabolites during the early stages of the germination process, from the dry seed until the protrusion and early growth of the coleoptile where dhurrin biosynthesis is at its highest rate (Busk and Møller, 2002), in four varieties of sorghum, one of which lacked the capacity to synthesize dhurrin (Blomstedt et al., 2012).
RESULTS
To explore the formation and recycling of dhurrin in the course of grain imbibition and germination, four different genotypes of sorghum were analyzed. The wild types used were a commercial breeding line (wild-type parent; Blomstedt et al., 2012) and the cultivar BTx623 for which the genome sequence has been reported (www.phytozome.org). The mutant line tcd1 (totally cyanide deficient) obtained by ethyl methanesulfonate mutagenesis does not produce dhurrin because it carries a mutation in CYP79A1 rendering it inactive (Blomstedt et al., 2012). The tcd1 line was generated in the commercial breeding line (Blomstedt et al., 2012) and has now been backcrossed twice into the cv BTx623. This was included as a unique reference system to confirm the identities of dhurrin and its derivatives detected in the wild type, and to explore their metabolic relationship and possible significance in the germination process. A sibling from the last tcd1×BTx623 cross named TCD1, which does not contain the mutation, was analyzed as an additional control. Grains and seedlings from all four genotypes were collected at specific time points (Fig. 2A) following imbibition and germination in the dark. All samples were subjected to quantitative metabolite analysis by LC-MS. Selected cv BTx623 and tcd1 samples were analyzed by MALDI-MSI to monitor the spatio-temporal developments in accumulation of interesting analytes at four stages of germination (Fig. 2, B and C). The grains were imbibed for 12 h, and the levels of dhurrin and its recycling products were quantified in the whole-grain extracts at 0, 12, 24, 36, 48, 60, and 72 h after imbibition (AI), as well as in samples from the mature, unimbibed grain.
Accumulation of Dhurrin and its Recycling Products during Germination and Seedling Formation
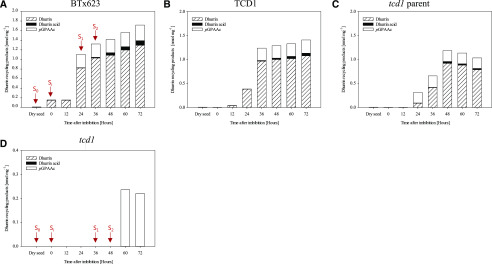
Analyses of the dry unimbibed grain of the three dhurrin producing sorghum lines confirmed the previous observation from other genotypes that dhurrin is absent in the dry grain (Tipples and Norris, 1983; Panasiuk and Bills, 1984; Veličković et al., 2014; Nielsen et al., 2016). Previously, the earliest reported presence of dhurrin was in shoots after a total of 44 h of imbibition and germination in the dark (Halkier and Møller, 1989). In this study of germinating grain and the germinated grain including emerging shoots and roots, dhurrin reached detectable levels already at 0 h AI in cv BTx623 and at 12 and 24 h AI, respectively, in the TCD1 and wild-type lines (Fig. 3, A–C). Dhurrin levels were generally found to be higher in cv BTx623 compared with other dhurrin producing lines and continued to increase in cv BTx623 at the end of the experiment, whereas it was more stagnant in the two other lines. At the end of the experiment, at 72 h AI, the dhurrin concentration in cv BTx623 and TCD1 was ∼1.3 nmol mg−1 (se = 0.08) and 1.1 nmol mg−1 (se = 0.04), respectively. In the tcd1 parent line dhurrin content peaked at 48 h AI (0.9 nmol mg−1, se = 0.08).
Figure 3.
Levels of dhurrin and its recycling products during germination and early seedling development in four sorghum lines. Absolute quantification of dhurrin, dhurrin acid, and pGPAAc in wild types is shown. cv BTx623 (A), TCD1 (B), tcd1 parent line (C), and mutant line tcd1 (D); note the different scale compared with A to C. For tcd1, pGPAAc was the only detected dhurrin related compound, and this was only detected at the last two time points (60–72 h AI). GS-pOHPACN was detected in the wild types, however, in minute concentrations, which are therefore shown separately in Supplemental Figure S1. Data for semiquantification of dhurrin glucosides are shown in Supplemental Figure S2. Arrows in A and D indicate the time points where the two lines analyzed by MSI had reached the different developmental stages S0 to S2. Values are mean of n = 3 biological replicate for each time point. Exact values and the calculated ses are listed in Supplemental Table S1.
The targeted analysis also documented the presence of previously reported dhurrin derivatives, namely the dhurrin glucosides (Fig. 1, 4), dhurrin acid (Fig. 1, 3), p-hydroxyphenyl(S-glutathione)acetonitrile (GS-pOHPACN; Fig. 1, 5), and p-glucosyloxyphenylacetic acid (pGPAAc; Fig. 1, 6). Other products and intermediates of the proposed recycling pathways were not detected (Fig. 1, 2, 7, and 8). With a few specific exceptions discussed below, the accumulation patterns of the detected metabolites matched their expected formation from dhurrin, i.e. in most cases they were present in detectable levels at one or two time points after the appearance of dhurrin, and the concentrations were either increasing or leveling out toward the end of the experiment, similar to those of dhurrin.
An overall trend of proportionality between the level of dhurrin and the recycling products was observed, i.e. the tcd1 parent samples exhibited the lowest levels of dhurrin and also of recycling products compared with the other wild-type lines. Average values and ses for all compounds/time points and lines are presented in the Supplemental Table S1. The dhurrin derivative present at the highest concentration was pGPAAc. The timing of the appearance of pGPAAc was slightly shifted between the lines, and in the tcd1 parent line the concentration of pGPAAc was already at its maximum level at the first time point where dhurrin was detected (Fig. 3C). After the initial increase in concentration all dhurrin-producing lines maintained a stable level at 0.3 nmol mg−1 throughout the experimental period. The same accumulation pattern was observed for GS-pOHPACN, although the concentration of this compound was much lower reaching 0.006, 0.002, and 0.0011 nmol mg−1 in cv BTx623, TCD1, and tcd1 parent line, respectively (Supplemental Fig. S1). Such a pattern is in accordance with the position of GS-pOHPACN as an intermediate in the recycling pathway of dhurrin to pGPAAc (Bjarnholt et al., 2018). In all three wild types, dhurrin acid was present as the second highest accumulated recycling product, reaching ∼0.1, 0.06, and 0.03 nmol mg−1 in cv BTx623, TCD1, and parent, respectively. Although at barely detectable levels (between 0.005 and 0.01 nmol mg−1), dhurrin acid was also detected in dry mature seeds. Strikingly, the mutant line tcd1 accumulated pGPAAc to roughly the same level as the wild-type and sibling lines (Fig. 3D), but with a distinct time delay of approximately 24 h. This was unexpected, as tcd1 in the original parent background was previously reported to not accumulate pGPAAc during plant development (from 7 to 46 d after sowing; Blomstedt et al., 2016; Bjarnholt et al., 2018). In this study, we did detect trace amounts of dhurrin in one replicate from each of two growth stages, indicating that it may in fact be produced in the mutant line at levels below the detection limit. However, at this stage it cannot be concluded whether dhurrin was actually present and could be a precursor for the detected pGPAAc.
Accumulation of Free Amino Acids
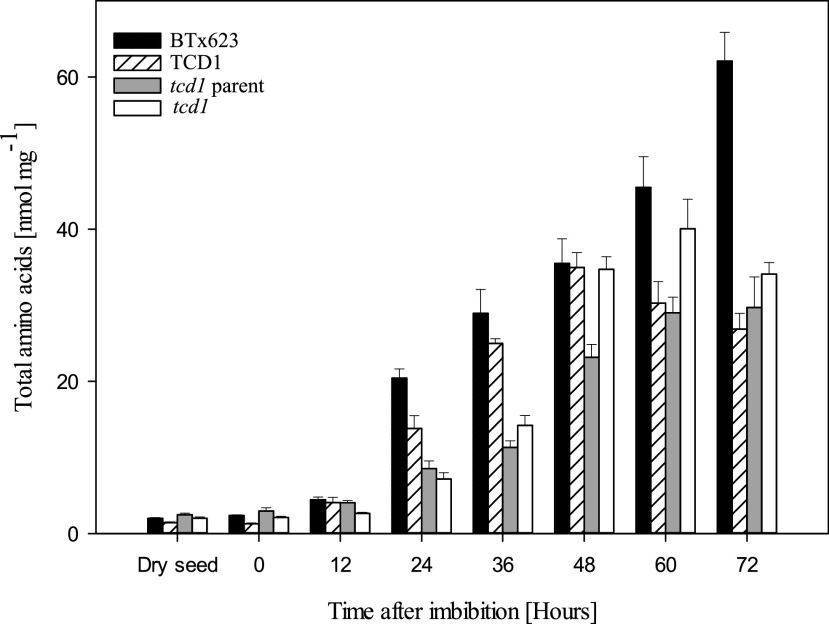
To investigate the timing of the onset of dhurrin biosynthesis in more detail, we compared it with one of the very early events in general metabolism during seed germination: the increase in concentration of free amino acids, caused by hydrolysis of storage proteins or de novo biosynthesis, to feed the need of the developing embryo. Dhurrin is biosynthesized from the amino acid Tyr, and furthermore, the recycling of dhurrin into dhurrin acid and pGPAAc leads to release of ammonia, which is toxic in high concentrations and therefore efficiently assimilated into amino acid metabolism via incorporation into Glu and Gln. It can therefore be envisioned that the ratio or concentration of some or all free amino acids may be affected in the mutant line compared with the dhurrin producing control lines. Figure 4 displays the development in the total sum of free amino acids in the four different lines. It should be noted that Gly could not be detected with the applied method. However, its concentration is expected to be relatively small (Feenstra et al., 2017). Differences were observed in the concentrations between the individual amino acids (Supplemental Fig. S3), but the overall concentration trajectories over time as well as between the sorghum lines were roughly identical to that displayed for the total sum. The concentrations of individual free amino acids in the dry grain varied from below 0.01 nmol mg−1 for Trp to around 1 nmol mg−1 for Asn. In all lines, the concentrations of free amino acids increased immediately upon imbibition, but initially at a slow rate. A dramatic increase was observed at 24 to 48 h AI, with roughly the same differences between the wild-type control lines as was observed for dhurrin accumulation. As exemplified by the plot of the total free amino acid content in Figure 4, the levels were generally highest in cv BTx623 and consistently lower in the parent line. The sibling TCD1 line was quite similar to cv BTx623 until 48 h AI. After 48 h, the concentrations of free amino acids in the parent and TCD1 lines leveled out or even decreased, whereas they were still increasing in the cv BTx623 line. These trajectories and line-dependent differences completely matched the dhurrin trajectories (Fig. 3). Differences in concentrations of free amino acids in germinating seeds were previously shown to depend on the genetic background (Feenstra et al., 2017). As previously mentioned, the tcd1 line was originally generated in the genetic background of a near isogenic commercial breeding line, and the tcd1 and TCD1 lines used in the current study are the F2 generation of the tcd1×BTx623 cross. Consequently, these two lines have a mixed parent×BTx623 genetic background, which is presumably what is reflected by the somewhat intermediate position of TCD1 between cv BTx623 and the mutant parent line with regard to concentrations of dhurrin and free amino acids. Interestingly, the free amino acid levels were different in tcd1 compared with its sibling TCD1, as illustrated for the total sum in Figure 4. The tcd1 line seemed to be delayed with respect to amino acids accumulation until ∼48 h AI, after which the levels were often higher in tcd1 compared with TCD1 and consequently also the parent line. With only three biological replicates to measure changes in such a dynamic pool of metabolites undergoing rapid change, the SEs were relatively high. However, Gln, Pro, and Tyr stood out, as the apparent levels of these amino acids reached higher or similar levels in tcd1 than in cv BTx623 at one or more of the time points 48 to 72 h AI (Supplemental Fig. S3). Almost all other amino acids were higher in cv BTx623 than in all other lines at most time points, with the most pronounced differences being found for Asp, Asn, His, Ile, Met, Ser, Thr, and Val. Levels of Ala, Arg, and Lys appeared somewhat unaffected or differently affected by the genetic background, and the remaining amino acids followed the trends of the total sum, but with moderate differences between the lines (Supplemental Fig. S3).
Figure 4.
Total free amino acids content during germination and early seedling development in four sorghum lines. The bars show the total sum of all free proteinaceous amino acids, except Gly, which could not be quantified with the applied method but is usually present in low amounts. Data for individual amino acids are shown in Supplemental Figure S3. Values are mean of n = 3 replicates for each time point. The error bars represent the se within the biological replicates.
Spatial Distribution of General and Specialized Metabolites
The stages selected for MSI analyses were chosen by a combination of the metabolite analyses and development of particular anatomical features. The four stages were (1) S0, the dry grain representing a base line; (2) Si, the imbibed grain, which was the first time point where dhurrin was detected in the wild type; (3) S1, where the radicle and early shoot emerged and dhurrin recycling products were first detected; and (4) S2, the stage where the primary root was fully developed and the coleoptile, containing the primary leaf, completely protruded from the embryonic axis of the embryo. For cv BTx623 these stages corresponded to the time points shown in Figure 2. The development of tcd1 was slightly delayed compared with cv BTx623, and therefore the seeds selected for MSI analysis from these lines were chosen based on the developmental stage rather than time (time points shown in Fig. 3D). After the first 36 h AI the mutant line was delayed by ∼12 h compared to the wild type. At this stage the radicle and the coleoptile containing the primary leaves were fully emerged in cv BTx623, whereas in tcd1 that was delayed until 48 h AI. For each stage and line, three seeds (including roots and seedlings when present) were sectioned longitudinally at 10 µm thickness, placed on precooled glass slides, and immediately freeze-dried, using the method reported by Kawamoto and Kawamoto (2014), with the few modifications reported by Gorzolka et al. (2016), Boughton and Thinagaran (2018), Sarabia et al. (2018), and Schmidt et al. (2018). Three consecutive sections were selected and subjected to respectively fluorescence microscopy and MSI analyses in both positive and negative mode to allow detection of an extensive range of metabolites.
Dhurrin and its recycling products and a number of general metabolites were detected in positive mode using 2,5-dihydroxybenzoic acid (DHB) as matrix. Amino acids were detected in negative mode using 1,5-diaminonaphthalene (DAN) as matrix on a different section. To minimize the crystal size and obtain uniform matrix deposition and thereby minimize delocalization of analytes, sublimation was used for matrix deposition (Hankin et al., 2007). The sections were analyzed with 30 µm pixel size, which provided sufficient spatial resolution for matching the morphological features observed in the fluorescence microscopy images while maintaining a reasonable acquisition time (∼4–6 h per sample). During data processing to produce the images, each ion was normalized to the total ion count (TIC) in each pixel, making up for some of the variation between analyses. Many factors, including sample handling, matrix deposition, and plant-tissue background can cause differences in ionization efficiencies for individual metabolites across the sample. Potentially this may result in images of metabolite distributions that do not provide a true representation of the sample constituents. To validate our results, we therefore compared with previous data obtained using different approaches.
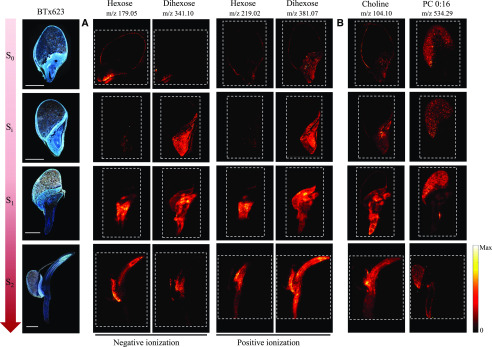
During the germination process, breakdown of the starchy endosperm into mono- and disaccharides provides an important source of energy for the developing embryo (Lopes and Larkins, 1993; Aoki et al., 2006; Leonova et al., 2010; Sánchez-Linares et al., 2012; Yan et al., 2014; Feenstra et al., 2017). Quantification of mono- and disaccharides of microdissected germinating cereal seeds from maize (Zea mays) and oat (Avena sativa) showed that monohexose (Hex) and dihexose (Hex2) accumulate in the embryo and scutellum (Leonova et al., 2010; Feenstra et al., 2017). The same distribution was observed in the MALDI-MSI analyses of the germinating sorghum seeds (Fig. 5A). Hex2 was lowly abundant in unimbibed samples, but its signal greatly increased in the embryo and scutellum already after imbibition (Si). As the germination proceeded and the axis protruded, Hex2 was concentrated more extensively in the emerging radicle and coleoptile (S2; Fig. 5A). Accumulation of Hex occurred after the protrusion of the radicle from the embryo (S1), where it was mainly found in the differentiation/elongation zone of the emerging radicle. At the last stage analyzed (S2), the Hex signal was highly concentrated in the coleoptile and slightly less abundant in the radicle. At the S0 stage, Hex and Hex2 were detected in the pericarp/aleurone layers (the two layers could not be distinguished with the lateral resolution applied in the MSI analyses). This is consistent with observation of MALDI-MSI in germinating maize kernels (Feenstra et al., 2017).
Figure 5.
Distribution and compartmentalization of general metabolites during germination and early seedling development of the cv BTx623 wild-type line. Three consecutive cryo-sections were imaged: The optical images on the far left (cv BTx623) were acquired with fluorescence microscopy, and the remaining images using MALDI-MSI in both negative and positive ion mode, using respectively DAN and DHB as matrix at 30 µm spatial resolution. Developmental stages are as in Figure 2. The dashed lines define the outlines of the recorded MS images. A, Comparative distribution of hexose and dihexose using the two ionization modes. In positive mode, compounds were detected as potassium adducts ([M+K]+), and in negative mode as deprotonated ions ([M-H]−). B, Specific tissue compartmentalization of choline [M+H]+ and phosphocholine (PC 0:16) [M+K]+. Scale bar = 1 mm. Signals for all MSI images are normalized to TIC on each pixel, and maximum values for generating images are listed in Supplemental Figure S4.
It is important to note that the compounds are detected based on their accurate mass and corresponding sum formulae. In the scutellum and developing embryo Hex and Hex2 are the major general metabolites Glc and Suc, as demonstrated in the microdissection experiments (Leonova et al., 2010; Feenstra et al., 2017); however, it is possible that Hex and Hex2 in the layer(s) encasing the seeds are different saccharides. For Hex and Hex2, the potassium adducts [C6H12O6+K]+ and [C12H22O11+K]+ gave the strongest signals in positive mode and were therefore used to map the distribution of these compounds. This was in accordance with the maize kernel study using a similar MALDI-MSI method (Feenstra et al., 2017), which has also been shown to provide a good signal quality from Hex2 in negative mode (Korte et al., 2015). Based on this knowledge, the efficiency and consistency of our ionization methods were compared by analyzing the distribution of Hex and Hex2 as deprotonated species ([C6H12O6-H]− and [C12H22O62-H]−) detected in negative mode (Fig. 5A). At all stages analyzed, the distribution of [Hex+K]+ and [Hex2+K]+ versus [Hex-H]− and [Hex2-H]− were as identical as can be expected for different sections, demonstrating the efficiency of both ionization methods.
Choline is an important plant constituent as a precursor in the formation of many metabolites including acetylcholine and phosphatidylcholine (PC; Miura and Shin, 1984). In chick pea (Cicer arietinum) seedlings, choline is highly abundant in the cotyledon and growing part of the embryo including the radicle (Ahmad and Karim, 1951), whereas PC is preferentially stored in the endosperm together with many other seed oil reserves (Fincher, 1989; Leonova et al., 2010). The same tissue-specific distributions were demonstrated for germinating barley seeds using MALDI analysis, showing strong accumulation of PC in the endosperm, whereas choline was detected in the scutellum+embryo and the aleurone layer (Gorzolka et al., 2016). Similar to Gorzolka et al. (2016), we detected choline as protonated species and PC (16:0) as a potassium adduct ([C5H13NO1+H]+ and [C24H50NO7+K]+, respectively) in positive mode. These analyses demonstrated that our MALDI-MSI method afforded efficient ionization across all examined germination stages and tissues, including the endosperm (Fig. 5B). Nearly identical results were obtained using the tcd1 mutant line (Supplemental Fig. S5), demonstrating similar ionization efficiencies and basic metabolite distributions in the two genotypes and corroborating the robustness of the methods.
Localization of Dhurrin and its Recycling Products during Germination and Seedling Formation
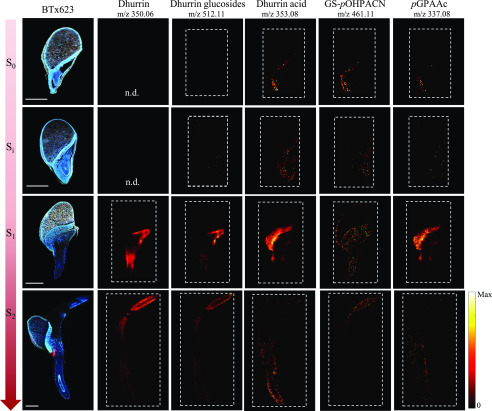
Dhurrin and its recycling products were detected as potassium or sodium adducts in positive mode. The ions providing the most intense signal were selected for generation of the images. Dhurrin was most consistently detected as a potassium adduct ([C14H17NO7+K]+), and only in a few individual replicates also detected as its sodium adduct with much lower signal intensity. Therefore, the potassium adduct was used to generate the images. Dhurrin glucosides were the only recycling products detected as a potassium adduct [M+K]+, whereas dhurrin acid, GS-pOHPACN, and pGPAAc were exclusively detected as their sodium adducts ([M+Na]+). Supplemental Table S3 provides a list of accurate masses applied to generate the images. The intensities varied between replicates, but the distributions were the same. The most clear results were selected for presentation in Figure 6, and the replicate results can be viewed in Supplemental Figure S7. In the metabolic extract analysis, dhurrin was first detected after imbibition (Si) in the BTx623 line, and the recycling products were first detected after axis protrusion (S1). In the MSI analyses, these compounds were all detected in the dry seeds as well as after imbibition (Fig. 3; Supplemental Fig. S7, S0 + Si), but the signal intensities were minute at these early stages, and the compounds not detected in all replicates. This explains why the very same constituents were not detected in the total extracts that were analyzed by LC-MS, due to the dilution effect. The weak signals make it difficult to assess the exact localization of the compounds, but it appears that dhurrin and its glucosides were present in both scutellum and embryo of the dry seeds. The three dhurrin glucosides have the same sum formula and thus identical exact mass and could therefore not be differentiated in the MSI analysis. The distribution of the ions corresponding to the dhurrin glucosides consistently followed the same distribution pattern as dhurrin until the end of the experiment. On the other hand, the dhurrin recycling products dhurrin acid and pGPAAc mainly accumulated in the scutellum and other tissues enclosing the embryo. After water uptake, this difference was much more pronounced than at S0, as the signal for dhurrin and the dhurrin glucosides were clearly much more intense in the embryo than in the scutellum, whereas the recycling products seemed to be almost exclusively located in the scutellum (Fig. 6, Si). As the germination proceeds, the radicle is the first part of the seedling to visibly emerge from the seed coat. Subsequently, the coleoptile containing the plumule (primordial leaf) protrudes from the embryonic axis, and this marks the end of germination. In cv BTx623, this developmental stage, S1, was reached at 24 h AI. At this stage the dhurrin and dhurrin glucosides appeared to be more concentrated in the differentiation/elongation zone of the emerging radicle and in the area protecting the plumule. The remaining compounds were still mainly found in the scutellum.
Figure 6.
Distribution and compartmentalization of dhurrin and dhurrin recycling products during germination and early seedling development of the cv BTx623 wild-type line. Procedures, settings, and overall layout are as in Figure 5. All MS images were acquired in positive mode. Dhurrin was most consistently detected as a potassium adduct [M+K]+, and therefore the accurate mass of dhurrin potassium adducts (m/z 350.0636) was used to generate the images. Dhurrin glucosides were the only recycling products detected as a potassium adduct [M+K]+, whereas dhurrin acid, GS-pOHPACN, and pGPAAc were detected as their sodium adducts [M+Na]+. MS images of tcd1 samples were blank for dhurrin (see Supplemental Fig. S6) and its derivatives. All MS images were normalized to the TIC in each pixel, and maximum values for generating images are listed in Supplemental Figure S4. n.d., Not determined. Scale bars = 1 mm.
At the last time point, S2, all compounds were exclusively detected in the newly formed root and coleoptile tissue. In particular, dhurrin was detected at very high intensity in the tip of the fully emerged coleoptile, consistent with a previous study (Halkier and Møller, 1989). The dhurrin derivatives were weakly detected at this stage and there were variations between replicates (Supplemental Fig. S7), making it difficult to determine whether the compounds were uniformly distributed or not. The strongest signal was found for the dhurrin glucosides, whereas dhurrin acid, pGPAAc and GS-pOHPACN were barely detectable. The distribution of GS-pOHPACN diverged from the remaining compounds, as it was generally more broadly distributed. At S0 it was detected at highest intensity in the embryo, whereas at Si it was equally intense in embryo and scutellum. In addition, in some replicates it was detected in the pericarp/aleurone layer at Si, S1, and possibly S2. As such, GS-pOHPACN and pGPAAc, which are respectively intermediate and product of the same metabolic pathway, displayed only partially overlapping distributions.
At all imaged stages, MSI signals corresponding to dhurrin and its recycling products were absent in the tcd1 mutant, at the applied mass tolerances (Supplemental Table S3) and default abundance threshold of MSiReader (Robichaud et al., 2013). This is visualized for dhurrin in Supplemental Figure S6A, showing average mass spectra of cv BTx623 and tcd1 at S1. This is difficult to visualize for earlier stages, where dhurrin and its derivatives were only detected in scattered pixels in cv BTx623 samples, making the average spectra uninformative. For dhurrin the most abundant isotope of the [M+K]+ ion is that containing a single 13C, which has a theoretical abundance of 15.1%. In the three pixels with highest dhurrin abundance in the S0 sample shown in Supplemental Figure S7, we were able to detect this isotope (mass-to-charge ratio [m/z] 351.0670) at, respectively, 8%, 9%, and 15% of the monoisotopic [dhurrin+K]+ peak (Supplemental Fig. S6C). Given the very low absolute peak intensities, which affects the accuracy of the measured ratios, and that this m/z value was absent from other pixels and tcd1 samples, this further supports the presence of low quantities of dhurrin and its derivatives in the BTx623 line at S0 and Si. The second most abundant isotope is [dhurrin+41K]+; however, at a theoretical abundance of 7.2%, this m/z value was not detectable at the S0 and Si stages. At S1 and S2, the MSI findings were corroborated by the LC-MS based metabolite analyses of the tcd1 mutant (Fig. 3D). Taken together, the results support that the detected ions in BTx623 do in fact correspond to the proposed compounds. pGPAAc was detected at later time points in the LC-MS analyses of tcd1, but unfortunately the size of the protruded axis (root and coleoptile) at those stages of seedling growth caused embedding and sectioning to be impossible and therefore the distribution of this compound could not be explored by MSI.
Distribution of Amino Acids
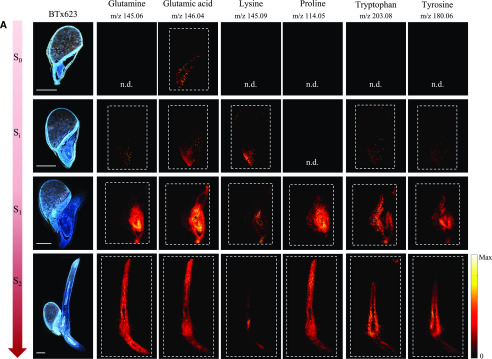
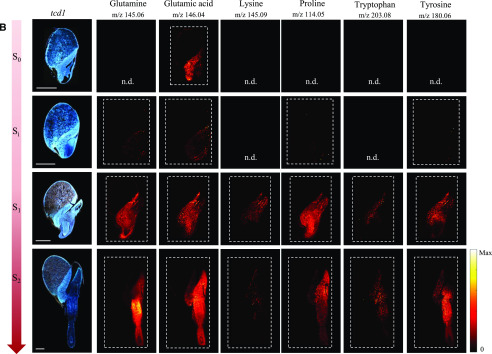
In the germinating maize kernels, only three proteinaceous amino acids were reliably detected by MALDI-MSI (Feenstra et al., 2017). In our study, all amino acids except Ala and Gly were readily detected as deprotonated species in negative mode [M-H]−. However, the relative signal intensities in the MS images did not completely reflect the concentrations determined in the LC-MS analyses. This serves to emphasize the well-known fact that the ionization efficiency in MS imaging depends on the type of applied matrix, the tissue matrix background, and in particular the physicochemical properties of the analytes. Figure 7 shows results for a selected subset of amino acids with a full data set provided in the supplementary data (Supplemental Figs. S8 and S9). Although MSI is not a quantitative technique, the overall trends in intensity of amino acid ions in images from samples of increasing age did match the overall trends found in the analysis of free amino acids in extracts. In cv BTx623 as well as in tcd1, there was a sharp increase in signal intensity from Si to S1 for most amino acid ions, reflecting the sharp increase in amino acid concentrations detected in extracts of whole samples at 24 to 36 h AI. Asp, Glu, and Asn were present in high concentrations at all stages, and genotypes analyzed by LC-MS and these amino acids were also detected at all stages in the MSI analyses of cv BTx623 and tcd1 (Fig. 7; Supplemental Figs. S8 and S9). In general, small, neutral amino acids such as Val, Met, Leu/Ile (which have identical sum formulae and cannot be differentiated in the MSI analyses), Ser, Pro, and Thr were only clearly detectable by MSI at the later growth stages, regardless of their concentration in the extract analysis. Conversely, Lys and Trp were found in low concentrations at all growth stages, but were nevertheless in some cases detectable in the MSI analysis already at stage Si.
Figure 7.
Distribution of a selected subset of amino acids during germination and early seedling development in the cv BTx623 wild-type line and the mutant line tcd1. Procedures, settings, and overall layout as in Figure 5. A, cv BTx623. B, tcd1. Amino acids were all detected as deprotonated species, [M-H]−, in negative ionization mode. Scale bars = 1 mm. A full data set of distributions of all amino acids in one replicate of BTx623 and in all analyzed replicates for the selected subset of amino acids in cv BTx623 and the mutant line tcd1 are provided in Supplemental Figures. S8 and S9). All MS images were normalized to the TIC in each pixel, and maximum values for generating images are listed in Supplemental Figure S4.
No free amino acids were detected in the endosperm, indicating that the rate of hydrolytic release of free amino acids from the storage proteins was lower than the transport of the amino acids toward the developing tissues. At the active growth stages from Si onward, where the amino acids are also synthesized de novo, the presence of amino acids was detected both in the scutellum and embryo. At these early stages of germination, the free amino acids are required for biosynthesis of protein and metabolites, presumably in both tissues, but eventually only the embryo will be active. This was reflected by the MSI results with a tendency toward the scutellar amino acid signals decreasing and the embryonic axis localized amino acid signals increasing from Si to S2. Asp, Glu, Gln, and Trp were in some cases also detected in the aleurone/pericarp layer at stage S0 and/or Si. The inconsistency in detection between stages (Supplemental Fig. S8) and replicates (Supplemental Fig. S9) may be exacerbated by the low concentrations present. At the last imaged growth stage, some amino acids displayed distinct localizations in the newly formed tissues. For example, the highly abundant Glu was almost detected everywhere, whereas Lys was mainly detected in the elongation zones of both root and shoot where Trp, on the other hand, was specifically not detected (Fig. 7). We observed no striking differences for tcd1 compared with cv BTx623, as illustrated for a subset of amino acids in Figure 7B and Supplemental Figure S9.
DISCUSSION
The spatio-temporal investigation of the accumulation of dhurrin and its recycling products in the germinating sorghum grain demonstrates that initiation of dhurrin biosynthesis and even the endogenous metabolism of dhurrin are very early events in seed germination and seedling development, initiated already during imbibition. The in situ localization of dhurrin metabolites showed that dhurrin and related cyanogenic di-glucosides accumulate in the newly formed tissues, protecting the vulnerable developing embryo. On the other hand, dhurrin derivatives accumulate mainly in the scutellum or are distributed across embryo, scutellum, and aleurone/pericarp layer, indicating key functions in the germination process.
Imbibition of the dry seeds is the starting point of germination. In the model plant Arabidopsis (Arabidopsis thaliana) metabolite abundances undergo substantial changes after the first few hours of imbibition (Rajjou and Debeaujon, 2008; Howell et al., 2009). The metabolic activity generates the essential building blocks for plant growth, energy production, and protein biosynthesis. In accordance with previous studies, we found only low concentrations of free amino acids in the dry sorghum grain (Bewley et al., 2013; Feenstra et al., 2017). The concentration of total and most of the individual free amino acids increased slowly at the first few time points after imbibition, and then increased dramatically at 24 h AI in cv BTx623 and slightly later or less steeply in the other control lines (Fig. 4; Supplemental Fig. S3). The trajectories of dhurrin concentration were exactly the same (Fig. 3), showing that dhurrin biosynthesis was switched on as early as the key event of amino acids release from the endosperm storage proteins. This suggests that dhurrin is of primary importance in the early phase of germination. Dhurrin localization at the tissue level as determined by MALDI-MSI furthermore points toward the presumed primary functionality of the metabolite. The presence of dhurrin in the differentiation/elongation zone of the emerging radicle and in the area protecting the plumule confirms the position of dhurrin as a key player in the sorghum defense system. Specialized metabolites that provide defense against general biotic stresses are commonly found in peripheral or key tissues where they can provide most benefit to the plant as a first line of chemical defense (Bellotti and Riis, 1994; Shroff et al., 2008; Ha et al., 2012; Bøgeskov Schmidt et al., 2018). In germinating barley, the antifungal hordatines were found to accumulate predominantly in the outer layer of the shoot (Gorzolka et al., 2016), similar to dhurrin accumulation in the outer layer of the cotyledon. However, the concurrent onset of the dhurrin recycling processes early in the development is indicative of other roles in the germinating grain.
The dhurrin glucosides were detected in the metabolite extracts as soon as dhurrin levels increased at 24 h AI. In the MS images of cv BTx623, they were colocalized with dhurrin. The biological functions of these di-glucosides are unknown, but as their aglycone is the dhurrin cyanohydrin, they will release HCN following possible hydrolysis resulting in the removal of both Glc residues. The cyanogenic di-glucosides found in cassava (Manihot esculenta) and the rubber plant (Hevea brasiliensis) have been proposed to be transport forms protected against cleavage by the monoglucosidase that hydrolyses the cyanogenic monoglucosides (Selmar, 1993). By contrast, in almonds (Prunus dulcis, syn. Prunus amygdalis), the cyanogenic di-glucoside amygdalin accumulates in the kernels and is responsible for the toxic cyanide release in the bitter seeds following the serial action of two glucosidases (Sánchez-Pérez et al., 2008, 2009, 2012; Del Cueto et al., 2017).
Dhurrin-6ˈ-glucoside was originally isolated from guttation droplets of sorghum leaves (Selmar et al., 1996), indicating a role in transport for this compound. The dhurrin glucosides may serve different functions in different tissues, e.g. transport forms in aerial tissues and defense purposes in roots where dhurrin is lowly abundant or absent. Determination of the distribution of the individual di-glucosides would require dissection and extract analysis where the isobaric compounds can be differentiated by retention time and their linkage types be determined by ion mobility MS (Pičmanová et al., 2015) or by NMR spectrometry of the isolated compounds.
The remaining dhurrin recycling products detected in the extracts of germinating seeds from the dhurrin producing lines either emerged at the same time as the dhurrin concentration increased (pGPAAc) or shortly after (GS-pOHPACN), or were detected at all time points, but found to increase shortly after the increase in dhurrin concentration (dhurrin acid). This early emergence or increase likewise indicate important roles of these compounds at the early stages of plant life. In fact, all derivatives were detected already at Si in the MS images, supporting their importance for the germinating seed or early seedling formation. Despite their apparent metabolic relation to dhurrin, the two major derivatives, dhurrin acid and pGPAAc, were apparently absent from the growing tissues until after germination at S2. This would suggest that before this stage, supply of reduced nitrogen to the growing tissues is not the main purpose of the pathways for conversion of dhurrin into these acids. The presence of dhurrin acid in the dry seed and its localization around the embryonic axis after imbibition at Si supports that it functions as a storage form of an antifungal agent (Kope et al., 1991), protecting the seed and the early embryo stage. In cassava, the main cyanogenic glucoside is linamarin, and its dhurrin acid analog, linamarin acid, was found to be localized to the stem and in the vascular tissue of the cassava tuber (Bøgeskov Schmidt et al., 2018). The presence in the vascular tissue proposes that the glucosylated CNglc-derived carboxylic acids may be transport forms of their potentially bio-active aglucones.
p-Hydroxyphenylacetic acid, the aglucone of pGPAAc, has been shown to exert growth promoting effects and auxin-like activities in algae (Fries, 1976, 1977). The compound has been isolated from plants, algae, and associated microbes, and its nonhydroxylated analog phenylacetic acid is well-established as an auxin-like plant hormone (Koepfli et al., 1938; Ferro et al., 2010; Simon and Petrášek, 2011). Glucosylation is a well-known mechanism used to control auxin homeostasis (Korasick et al., 2013) and the activity of gibberellins, both key hormones controlling seed germination (Yamaguchi, 2008). The observed localization of pGPAAc in the scutellum, the tissue that is considered the main source of gibberellin production in germinating seeds, renders it tempting to suggest that p-hydroxyphenylacetic acid has an equally important role in sorghum seed germination. Assignment of an important function to p-hydroxyphenylacetic acid is supported by the fact that the tcd1 mutant was shown to produce pGPAAc, which after germination accumulated at wild-type levels (Fig. 3D). The presence of pGPAAc in tcd1 was surprising in the light of previous reports where pGPAAc was found to be absent or present in trace amounts in vegetative tissues of tcd1 (Bjarnholt et al., 2018) and a second mutated line, tcd2, which is mutated in the last enzyme of dhurrin biosynthesis (Blomstedt et al., 2016). In the present study, we did detect traces of dhurrin in a few replicates of tcd1 extracts, and this may be what fuels the production of pGPAAc in the germinating seeds and/or young seedling tissues of this line, with the low dhurrin supply explaining the delayed accumulation. On the other hand, the eventual accumulation of wild-type levels of pGPAAc suggests that this compound is produced from a completely different pathway, which remains to be elucidated.
GS-pOHPACN is an intermediate in the production of pGPAAc from dhurrin (Bjarnholt et al., 2018). As such, it was convincing to find it localized to both embryo and scutellum, where its precursor, dhurrin, and its downstream product, pGPAAc, accumulated. At S1, the accumulation of precursor and product appeared almost completely separated between the two tissues. The presence of GS-pOHPACN in both tissues could therefore suggest that it is produced from dhurrin in the embryo and subsequently transported to the scutellum for further processing into pGPAAc. Compartmentalization between the different tissues of germinating seeds has also been suggested for the gibberellin biosynthetic pathway in Arabidopsis (Urbanova and Leubner-Metzger, 2016). However, this does not explain the presence of GS-pOHPACN in the aleurone/pericarp layer. In the aleurone tissue, the small molecules present can be expected to be mainly signaling compounds or products resulting from the cell wall degradation that happens during germination. It can be hypothesized that GS-pOHPACN is a transport molecule of the bio-active nonglucosylated p-hydroxyphenylacetic acid or even of free pOHPACN (p-hydroxyphenylacetonitrile), which may have an undescribed bio-activity. p-Hydroxyphenylacetic acid was not detected in the extract analyses, maybe because the locally produced and aleurone localized p-hydroxyphenylacetic acid were diluted too much in the total extracts. Conversion of GS-pOHPACN to p-hydroxyphenylacetic acid requires the action of one of the two lambda GSTs (glutathione S-transferases) SbGSTL1 or SbGSTL2, followed by the nitrilase heterodimer SbNIT4A/B2. The transcripts of these enzymes colocalized at the cellular level in sorghum seedling leaves (Bjarnholt et al., 2018) and were coexpressed in developing sorghum seeds (Nielsen et al., 2016). Transcriptome analyses of the individual tissues in the germinating sorghum seed, such as recently carried out for barley (Betts et al., 2017), could help identify source and sink tissues of GS-pOHPACN and lend further clues as to the nature of its biological function.
The overall pattern of free amino acid accumulation showed that the siblings tcd1 and TCD1 were somewhat intermediate between their parent wild-type lines. The tcd1 mutant displayed a delay in accumulation of free amino acids compared with its sibling. This delay was mirrored by the delay in germination in tcd1, as germination of TCD1 and the parent line were not significantly different from cv BTx623. Amino acids have been shown to influence the germination process. In Arabidopsis, de novo biosynthesis of Met was described to be of primary importance for seed germination and seedling growth (Gallardo et al., 2002). In other plants, such as barley and sugar beet (Beta vulgaris var. altissima), enzymes related to Met metabolic pathways dramatically increase in the first hours after imbibition before radical emergence (Soeda et al., 2005; Catusse et al., 2008). In cv BTx623 and TCD1, the concentration of Met increased much more steeply than in tcd1, in which comparable levels were reached with a delay of 24 to 36 h. It is possible that an imbalance in the free amino acids caused by possible over accumulation of Tyr in the absence of the dhurrin biosynthesis could influence the germination rate. Indeed, Tyr as well as Pro and Gln accumulated to higher levels in tcd1 compared with its sibling and even cv BTx623 after germination. Like other aromatic amino acids, Tyr is produced in the plastids and exported into the cytosol as precursor for synthesis of other metabolites. Degradation of Tyr via a pathway initiated by Tyr amino transferase (Tyr-AT) and transamination of α-ketoglutarate into Glu (Hildebrandt et al., 2015; Schenck and Maeda, 2018) avoids the risk of overaccumulation. Glu did not accumulate in the tcd1 mutant relative to the wild type, but as Glu is a precursor for Pro and Gln, it is likely that the observed increase in levels of the latter two in tcd1 is a consequence of Tyr overaccumulation and degradation. Tyr may also be directed into synthesis of other metabolites such as tyramine, tocopherols, Vitamin E, and numerous phenolic compounds such as the flavonoids. We did not target these compounds in our analyses, but it is also possible that an imbalance in accumulation of such metabolites may have caused the perturbation of the germination rate.
Our investigations of the MALDI-MSI method showed that the applied matrices worked similarly and well across tissue types, genotypes, and growth stages for a range of general metabolites with different physicochemical properties (Fig. 5). Our MSI results for hexoses and amino acids were in accordance with other studies using both MALDI-MSI and LC-MS analyses of microdissected tissues (Figs. 5 and 7; Leonova et al., 2010; Gorzolka et al., 2016; Feenstra et al., 2017). Although MSI is not a quantitative technique, the overall trends in intensity of the amino acid ions in samples taken at different stages of the germination process (Fig. 4) matched the overall trends found in the analyses of extracts (Supplemental Fig. S3). Amino acids are poorly ionizable compounds, and because of their amphiprotic nature they have been reported to be hardly detectable by MALDI-MSI (Toue et al., 2014; Feenstra et al., 2017). Despite this, we visualized the distribution of 18 out of the 20 free proteinaceous amino acids, many with highly intense signals. The very high selectivity of the Orbitrap operating at 140,000 mass resolving power (applying a bin width of 0.01 Th in image generation) has likely improved detection compared with experiments on low-resolution MALDI-time of flight systems (Toue et al., 2014), but this does not explain the difference between the current study and the work of Feenstra et al. (2017). Factors such as sample preparation, mounting technique (carboxymethylcellulose, or CMC, versus gelatin), matrix (DAN vs. DHB), and sample drying (freeze drying versus desiccation) as well as differences in plant background matrices might have a significant impact on signal intensity and therefore on the successful detection of metabolites of interest. Some discrepancies were indeed observed between the LC-MS and the MALDI-MSI analyses of dhurrin and dhurrin-derived metabolites. At the early stages of germination, the discrepancies might be explained by the more localized accumulation, allowing compounds to be more easily detected by MSI than in the total seed extracts. However, at the last point of seedling development investigated, where the radicle has evolved into the primary root and the coleoptile was fully emerged from the seed coat, the concentrations of dhurrin and its recycling products in the dhurrin producing wild-type lines were just as high as at the previous growth stage, but only weakly or barely detected by MALDI-MSI. This may be explained by the substantial change in the plant background during the rapid transition from the seed to a seedling, which may have affected the ionization efficiency of the compounds. Ion suppression caused by matrix background is a common problem in MS-based studies.
This study demonstrates the power of combining LC/MS and MALDI-MSI to gain insight into important biological processes. The results have demonstrated an apparent importance of dhurrin and its recycling processes at the earliest stages of sorghum grain germination and seedling formation and unveiled the high complexity of this metabolic grid and compartmentalizations. These insights point toward specific physiological functions of dhurrin and its recycling products.
MATERIALS AND METHODS
Plant Material
Seeds of four different sorghum (Sorghum bicolor) lines, namely ‘BTx623’, TCD1, tcd1 parent, and tcd1 mutant, were imbibed for 12 h in the dark at room temperature and germinated on moist filter paper in petri dishes at 28°C for 3 d. Samples were collected every 12 h at eight time points, dry grain and 0, 12, 24, 36, 48, 60, and 72 h, with 0 h corresponding to those taken immediately after 12 h of imbibition. At each time point, three seeds combined as one biological replicate (for a total of nine seeds for each time point) were immediately quenched in liquid nitrogen (N2) and used for metabolite quantification by LC-MS. For in situ visualization of metabolites in cv BTx623 and tcd1, seeds were collected at four stages based on the development of particular anatomical features, taking into account the different germination rates observed between the lines. To cover biological variation between individual seeds and technical variation between analyses, each time point was analyzed by MSI in triplicates for cv BTx623 and duplicates for tcd1.
Sample Preparation for Metabolite Profiling by LC-MS
Frozen plant material was ground in liquid nitrogen using mortar and pestle and extracted in 85% (v/v) aqueous methanol with 0.5% (v/v) formic acid (80°C, 5 min) in screw-lid microtubes (Sarstedt) and immediately cooled on ice. The extracts were transferred to screw cap glass vials and stored at −20°C until further pretreatment. For analyses, samples were diluted 5 times (1:5 [v/v]) in water and filtered through a membrane filter (0.45 μm; Merck Millipore) by centrifugation (2000 g, 5 min). After filtration, samples were stored at 4°C until analysis.
Quantification of Dhurrin and Related Metabolites by LC-3Q-MS
The diluted extracts were directly analyzed by LC-MS. Chromatography was performed on an Advance UHPLC system (Bruker). Separation was achieved on a Zorbax Eclipse XDB-C18 column (100 × 3.0 mm, 1.8 µm, 100 Å; Agilent Technologies), using a gradient of A = MilliQ grade water with 0.05% (v/v) formic acid and B = 100% methanol . The elution profile was 0 to 0.03 min, 2% B; 0.03 to 0.9 min, 2% to 15% B; 0.9 to 1.4 min, 15% to 60% B; 1.4 to 3.3 min, 60% to 100% B; 3.3 to 3.9 min, 100%; 3.9 to 4.0 min, 100% to 2% B; and 4.0 to 5.0 min 2% B. The mobile phase flow rate was 500 µL min−1, and column temperature was maintained at 40°C. The liquid chromatography was coupled to an EVOQ Elite TripleQuad mass spectrometer (Bruker) equipped with an electrospray ion source (ESI) operated in combined positive and negative ionization mode. Instrument parameters were optimized by infusion experiments with pure standards. The ion spray voltage was maintained at 5000 and −4000 V in positive and negative ionization mode, respectively. Cone temperature was set to 300°C and cone gas to 20 psi. Heated probe temperature was set to 200°C and probe gas flow to 50 psi. Nebulizing gas was set to 60 psi and collision gas to 1.6 mTorr. Nitrogen was used as probe and nebulizing gas and argon as collision gas. Active exhaust was constantly on. Multiple reaction monitoring (MRM) was used to monitor analyte molecular ion to fragment ion transitions: MRM transitions were chosen based on direct infusion experiments. Detailed values for mass transitions can be found in Supplemental Table S2A. Both Q1 and Q3 quadrupoles were maintained at unit resolution. Alternatively, a modified MRM setting was used to select and filter the parent ion in both Q1 and Q3 while not applying any collision energy in Q2. This variation of selected ion monitoring (SIM) was called SIM2 and increased the signal strength by ∼3.5- to 15-fold and improved signal-to-noise ratio up to 10-fold compared with normal SIM where only Q1 is used for ion selection. Also for SIM2, both Q1 and Q3 were kept at unit resolution. Bruker MS Workstation software (Version 8.2, Bruker) was used for data acquisition and processing. Linearity in ionization efficiencies was verified by analyzing dilution series of standard mixtures. Reference compounds for dhurrin, dhurrin amide, dhurrin acid, pGPAAc, p-hydroxyphenylacetic acid, p-hydroxyacetonitrile, and GS-pOHPACN were used for absolute quantification by using 12 points external calibration curves in a range of concentrations from 0.001 to 50 µm.
Quantification of Amino Acids by LC-3Q-MS
For the absolute quantification of free amino acids, the diluted sample was 1:10 mixed with 13C, 15N-labeled amino acids (Algal amino acids 13C, 15N, Isotec) at a concentration of 10 µg mL−1. Diluted samples were filtered (Durapore 0.22 µm polyvinylidene difluoride filters; Merck Millipore) and used directly for LC-MS analysis. The analysis was performed as described in Mirza et al. (2016), with changes as detailed in the rest of this section. Briefly, chromatography was performed on an Advance UHPLC system (Bruker). Separation was achieved on a Zorbax Eclipse XDB-C18 column (100 × 3.0 mm, 1.8 µm; Agilent Technologies). Formic acid (0.05% [v/v]) in water and acetonitrile (supplied with 0.05% [v/v] formic acid) were used as mobile phases A and B, respectively. The elution profile was 0 to 1.2 min, 3% B; 1.2 to 4.3 min, 3% to 65% B; 4.3 to 4.4 min 65% to 100% B; 4.4 to 4.9 min, 100% B; 4.9 to 5.0 min, 100% to 3% B; and 5.0 to 6.0 min, 3% B. Mobile phase flow rate was 500 µL min−1, and column temperature was maintained at 40°C. The liquid chromatography was coupled to an EVOQ Elite TripleQuad mass spectrometer (Bruker) equipped with an ESI. Instrument parameters were optimized by infusion experiments with pure standards. The ion spray voltage was maintained at 3000 V in positive ion mode. Cone temperature was set to 300°C and cone gas flow to 20 psi. Heated probe temperature was set to 400°C and probe gas flow set to 50 psi. Nebulizing gas was set to 60 psi and collision gas to 1.6 mTorr. Nitrogen was used as both cone gas and nebulizing gas and argon as collision gas. MRM was used to monitor analyte molecular ion → fragment ion transitions: MRMs for amino acids were chosen as described in Jander et al. (2004), with additions from Docimo et al. (2012), for Arg and Lys. Both Q1 and Q3 quadrupoles were maintained at unit resolution. Bruker MS Workstation software (version 8.2.1, Bruker) was used for data acquisition and processing. Individual amino acids in the sample were quantified by the respective 13C, 15N-labeled amino acid internal standard, except for Trp, and Asn: Trp was quantified using 13C, 15N-Phe applying a response factor of 0.42, Asn was quantified using 13C, 15N-Asp applying a response factor of 1.0 (Docimo et al., 2012). Further details for transitions and collision energies can be found in Supplemental Table S2B.
Detection of Dhurrin Glucosides by LC-Quadrupole Time-Of-Flight-MS
Full scan analysis of sorghum extracts was performed using a Thermo Scientific Dionex Ultimate 3000 RS (ThermoFisher Scientific) coupled with a Bruker compact quadrupole-quadrupole time of flight MS (Bruker Daltonics) equipped with an ESI source. For data acquisition and processing, Compass DataAnalysis software (version 4.3, Bruker Daltonics) was used. Then 8 µL of samples were injected on a LC Kinetex XB-C18 column (150 × 2.1 mm, 1.7 µm, Phenomenex), and compound separation was achieved using a gradient of A = MilliQ grade water with 0.05% (v/v) formic acid and B = acetonitrile with 0.05% (v/v) formic acid. The elution profile was 0 to 5 min, 2% B; 4 to 9 min, 30% B; 9 to 14 min, 100% B; 14 to 21 min, 100% to 2% B; 21 to 28 min, 2% B. The mobile phase flow rate was 300 µL min−1, and column temperature was maintained at 40°C. The full-scan mass spectra were obtained within a range of m/z 50 to1.300 with the instrument operated in the positive ion mode. The spray voltage was set at 4000 V with the maximum dry temperature set at 250°C at the pressure of 1.2 Bar. The up-front collision-induced dissociation was set to off, and the optimized collision energy was set at 7eV in the collision cell. The quadrupole mass filter was set with low mass and with the ion energy of 4 eV. Extracted ion chromatograms for specific [M+H]+ adduct ions were used to detect dhurrin glucosides. Their fragmentation patterns, accurate masses, and retention times were used to identify peaks according to Pičmanová et al. (2015).
MALDI-MSI Sample Preparation and Fluorescence Microscopy
Samples were prepared according to the Kawamoto method (Kawamoto, 2003; Kawamoto and Kawamoto, 2014) with slight modifications. Frozen samples were embedded into a 2% (w/v) aqueous medium of CMC into a cryo-mold. The mold containing the embedded sample was submerged into a coolant solution of hexane/dry ice for ∼5 min. When completely frozen, CMC blocks were removed from the mold and stored at −80°C until cryo-sectioning. Frozen samples were transferred to a Leica CM3050S cryostat (Leica Microsystems) at −30°C to thermally equilibrate for 1 h. The embedded samples were fixed on the sample holder of the cryo-microtome using optimum cutting temperature gel (Tissue-Tek), sectioned at 10 µm thickness, collected with cryofilm II C9 (Section Lab), which was attached to prechilled glass slides with double-sided adhesive carbon tape (Agar scientific), and finally freeze-dried under vacuum at 1.0 mbar overnight. Three consecutive sections were selected for each sample; one was used for fluorescence microscopy analysis inspection and the others were coated with, respectively, DHB and DAN by sublimation, as described by Hankin et al. (2007). All the fluorescence images were captured on a Leica DMR HC (Leica Microsystems) wide-field fluorescence microscope, using a filter cube with a 340 to 380 nm excitation filter, 400 nm beam splitter, and 420 nm long-pass emission filter. Autofluorescence exposure times varied were based on the stage of the grain and the tissue composition (i.e. dry grain or germinated grain with protruded axes) but generally were set around between 600 and 800 ms. FIJI ImageJ (Schindelin et al., 2012) was used for any additional image preprocessing, such as cropping, adding the scale bar, or other relevant operations.
MALDI-MSI Analyses
MALDI-MSI analysis was performed on a Thermo QExactive Orbitrap mass spectrometer (Thermo Scientific) equipped with the AP-SMALDI10 ion source (TransMIT). The AP-SMALDI10 ion source was equipped with a MNL100 nitrogen laser (LTB Lasertechnik) with 337 μm wavelength and 60 Hz frequency, operated using 30 laser pulses per pixel. Spectra were acquired in positive and negative ion mode (scan range m/z 70–700) at mass resolving power 140,000 at m/z 200. A matrix peak was used as lock-mass for internal mass calibration, ensuring a mass accuracy of 2 ppm or better. The acquired raw files were converted to imzML files using an imzML converter (Schramm and al., 2012), and images were generated using MSiReader v.1.01 (MATLAB; Robichaud et al., 2013) using a bin width of 0.01 Th and the following default parameters: MaxColorScale: 100,000, NormalizeOption: TIC, NormScale: 1 NormCutoff: 1, Minimum Abundance Threshold: 0.001. All MS images were normalized to the TIC in each pixel, and the maximum values for generating images are listed in Supplemental Figure S4. The same color scale was used regardless of genotype or germination stage. Metabolites were identified by accurate mass, and the complete list of m/z values and the mass tolerance applied for each is available in supplementary information (Supplemental Table S3).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. The concentrations of dhurrin and dhurrin recycling products in four different lines.
Supplemental Table S2. Transitions and response factors used for MRM for analysis by LC-3Q-MS.
Supplemental Table S3. Compounds visualized by MALDI-MSI.
Supplemental Figure S1. Concentration of GS-pOHPACN during germination and early seedling development in sorghum wild types.
Supplemental Figure S2. Dhurrin glucosides content during germination and early seedling development in sorghum wild types.
Supplemental Figure S3. Concentrations of individual amino during germination and early seedling development in four sorghum lines.
Supplemental Figure S4. Maximum values for generating images.
Supplemental Figure S5. Distribution and compartmentalization of general metabolites in tcd1.
Supplemental Figure S6. Comparison of average raw mass spectra of two selected samples of cv BTx623 and tcd1 at stage S1.
Supplemental Figure S7. All replicates of images in Figure 6.
Supplemental Figure S8. Distribution of all detected amino acids during germination in cv BTx623 by MALDI-MSI.
Supplemental Figure S9. Distribution of a subset of amino acids across biological replicates.
Acknowledgments
We thank Alexander Schulz (University of Copenhagen) for expert advice about fluorescence microscopy, Dr. Camilla Knudsen (University of Copenhagen) for helping out with Figure 2 graphics, and Dr. David Pattison (University of Copenhagen) for assistance with LC-quadrupole time of flight-MS.
Footnotes
This work was supported by the VELUX Foundation (grant nos. VKR023054 and 19151 to L.M., M.S.M., and N.B.), the European Research Council (ERC; advanced grant no. 323034), the Danish National Research Foundation (Danmarks Grundforskningsfond; grant no. DNRF99 to C.C.), the Department of Industry, Innovation, Science, Research and Tertiary Education, Australian Government, Centre of Excellence for Environmental Decisions, Australian Research Council (grant nos. LP0774941, LP100100434, and DP130101049 to R.M.G. for the development and characterization of the tcd1 mutants); and Det Frie Forskningsråd (Danish Council for Independent Research; grant no. DFF–4002–00391, in partnership with Pacific Seeds Pty Ltd). Support from the Carlsberg Foundation for MALDI-MSI instrumentation.
Articles can be viewed without a subscription.
References
- Adewusi SRA.(1990) Turnover of dhurrin in green sorghum seedlings. Plant Physiol 94: 1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad BK, Karim MA(1951) Biosynthesis of choline in the seedling of the chick-pea (Cicer arietinum). Biochem J 55: 817–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Scofield GN, Wang XD, Offler CE, Patrick JW, Furbank RT(2006) Pathway of sugar transport in germinating wheat seeds. Plant Physiol 141: 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellotti AC, Riis L(1994) Cassava cyanogenic potential and resistance to pests and diseases. Acta Hortic 141–152 [Google Scholar]
- Betts NS, Berkowitz O, Liu R, Collins HM, Skadhauge B, Dockter C, Burton RA, Whelan J, Fincher GB(2017) Isolation of tissues and preservation of RNA from intact, germinated barley grain. Plant J 91: 754–765 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Bradford KJ, Hilhors HWM, Nonogaki H(2013) Seeds: Physiology of Development, Germination, and Dormancy. Springer-Verlag, New York [Google Scholar]
- Bjarnholt N, Li B, D’Alvise J, Janfelt C(2014) Mass spectrometry imaging of plant metabolites--principles and possibilities. Nat Prod Rep 31: 818–837 [DOI] [PubMed] [Google Scholar]
- Bjarnholt N, Neilson EHJ, Crocoll C, Jørgensen K, Motawia MS, Olsen CE, Dixon DP, Edwards R, Møller BL(2018) Glutathione transferases catalyze recycling of auto-toxic cyanogenic glucosides in sorghum. Plant J 94: 1109–1125 [DOI] [PubMed] [Google Scholar]
- Blomstedt CK, Gleadow RM, O’Donnell N, Naur P, Jensen K, Laursen T, Olsen CE, Stuart P, Hamill JD, Møller BL, et al. (2012) A combined biochemical screen and TILLING approach identifies mutations in Sorghum bicolor L. Moench resulting in acyanogenic forage production. Plant Biotechnol J 10: 54–66 [DOI] [PubMed] [Google Scholar]
- Blomstedt CK, O’Donnell NH, Bjarnholt N, Neale AD, Hamill JD, Møller BL, Gleadow RM(2016) Metabolic consequences of knocking out UGT85B1, the gene encoding the glucosyltransferase required for synthesis of dhurrin in Sorghum bicolor (L. Moench). Plant Cell Physiol 57: 373–386 [DOI] [PubMed] [Google Scholar]
- Blomstedt CK, Rosati VC, Lindberg Møller B, Gleadow R(2018) Counting the costs: Nitrogen partitioning in Sorghum mutants. Funct Plant Biol 45: 705–718 [DOI] [PubMed] [Google Scholar]
- Bøgeskov Schmidt F, Heskes AM, Thinagaran D, Lindberg Møller B, Jørgensen K, Boughton BA(2018) Mass spectrometry based imaging of labile glucosides in plants. Front Plant Sci 9: 892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton BA, Thinagaran D(2018) Mass spectrometry imaging (MSI) for plant metabolomics. Methods Mol Biol 1778: 241–252 [DOI] [PubMed] [Google Scholar]
- Boughton BA, Thinagaran D, Sarabia D, Bacic A, Roessner U(2016) Mass spectrometry imaging for plant biology: A review. Phytochem Rev 15: 445–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Møller BL(2002) Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol 129: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catusse J, Strub JM, Job C, Van Dorsselaer A, Job D(2008) Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc Natl Acad Sci USA 105: 10262–10267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crush JR, Caradus JR(1995) Cyanogenesis potential and iodine concentration in white clover (Trifolium repens L.) cultivars. New Zeal Agric Res 38: 309–316 [Google Scholar]
- Dante RA, Larkins BA, Sabelli PA(2014) Cell cycle control and seed development. Front Plant Sci 5: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Cueto J, Ionescu IA, Pičmanová M, Gericke O, Motawia MS, Olsen CE, Campoy JA, Dicenta F, Møller BL, Sánchez-Pérez R(2017) Cyanogenic glucosides and derivatives in almond and sweet cherry flower buds from dormancy to flowering. Front Plant Sci 8: 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docimo T, Reichelt M, Schneider B, Kai M, Kunert G, Gershenzon J, D’Auria JC(2012) The first step in the biosynthesis of cocaine in Erythroxylum coca: The characterization of arginine and ornithine decarboxylases. Plant Mol Biol 78: 599–615 [DOI] [PubMed] [Google Scholar]
- Erb N, Zinsmeister HD, Nahrstedt A(1981) The cyanogenic glycosides of triticum, secale and sorghum. Planta Med 41: 84–89 [DOI] [PubMed] [Google Scholar]
- Feenstra AD, Alexander LE, Song Z, Korte AR, Yandeau-Nelson MD, Nikolau BJ, Lee YJ(2017) Spatial mapping and profiling of metabolite distributions during germination. Plant Physiol 174: 2532–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro N, Bredow T, Jacobsen HJ, Reinard T(2010) Route to novel auxin: Auxin chemical space toward biological correlation carriers. Chem Rev 110: 4690–4708 [DOI] [PubMed] [Google Scholar]
- Fincher GB.(1989) Molecular and cellular biology associated with endosperm mobilization in germinating cereal grain. Annu Rev Plant Physiol Plant Mol Biol 40: 305–351 [Google Scholar]
- Forslund K, Morant M, Jørgensen B, Olsen CE, Asamizu E, Sato S, Tabata S, Bak S(2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131: 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries L.(1977) Growth regulating effects of phenylacetic acid and p-hydroxyphenylacetic acid on Fucus spiralis L. (Phaeophyceae, Fucales) in axenic culture. Phycologia 16: 451–455 [Google Scholar]
- Fries L.(1976) p-Hydroxyphenylacetic acid and other phenolic compounds as growth stimulators of the red alga Porphyra tenera. Plant Sci Lett 6: 299–307 [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D(2002) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Gleadow RM, Haburjak J, Dunn JE, Conn ME, Conn EE(2008) Frequency and distribution of cyanogenic glycosides in Eucalyptus L’Hérit. Phytochemistry 69: 1870–1874 [DOI] [PubMed] [Google Scholar]
- Gleadow RM, Møller BL(2014) Cyanogenic glycosides: Synthesis, physiology, and phenotypic plasticity. Annu Rev Plant Biol 65: 155–185 [DOI] [PubMed] [Google Scholar]
- Gleadow RM, Woodrow IE(2002) Constraints on effectiveness of cyanogenic glycosides in herbivore defense. J Chem Ecol 28: 1301–1313 [DOI] [PubMed] [Google Scholar]
- Gorzolka K, Kölling J, Nattkemper TW, Niehaus K(2016) Spatio-temporal metabolite profiling of the barley germination process by MALDI-MS Imaging. PLoS One 11: e0150208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li HB, Kong CH(2008) Allelopathic potential of barnyard grass on rice and soil microbes in paddy. Allelopathy J 21: 389–396 [Google Scholar]
- Ha M, Kwak JH, Kim Y, Zee OP(2012) Direct analysis for the distribution of toxic glycoalkaloids in potato tuber tissue using matrix-assisted laser desorption/ionization mass spectrometric imaging. Food Chem 133: 1155–1162 [Google Scholar]
- Halkier BA, Møller BL(1989) Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial purification of the enzyme system involved. Plant Physiol 90: 1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin JA, Barkley RM, Murphy RC(2007) Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom 18: 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun HP(2015) Amino acid catabolism in plants. Mol Plant 8: 1563–1579 [DOI] [PubMed] [Google Scholar]
- Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J(2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149: 961–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu IA, López-Ortega G, Burow M, Bayo-Canha A, Junge A, Gericke O, Møller BL, Sánchez-Pérez R(2017) Transcriptome and metabolite changes during hydrogen cyanamide-induced floral bud break in sweet cherry. Front Plant Sci 8: 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N, Trivellini A, Masood A, Ferrante A, Khan NA(2013) Current understanding on ethylene signaling in plants: The influence of nutrient availability. Plant Physiol Biochem 73: 128–138 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu S, Li L, Last RL(2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39: 465–475 [DOI] [PubMed] [Google Scholar]
- Jones DA.(1998) Why are so many food plants cyanogenic? Phytochemistry 47: 155–162 [DOI] [PubMed] [Google Scholar]
- Jørgensen K, Bak S, Kamp-Busk P, Sørensen C, Olsen CE, Puonti-Kaerlas J, Møller BL(2005) Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol 139: 363–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Kawamoto K(2014) Preparation of thin frozen sections from nonfixed and undecalcified hard tissues using Kawamot’s film method (2012). Methods Mol Biol 1130: 149–164 [DOI] [PubMed] [Google Scholar]
- Kawamoto T.(2003) Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol 66: 123–143 [DOI] [PubMed] [Google Scholar]
- Koepfli JB, Thimann KV, Went FW(1938) Phytohormones: Structure and physiological activity. J Biol Chem 122: 763–780 [Google Scholar]
- Kojima M, Poulton JE, Thayer SS, Conn EE(1979) Tissue distributions of dhurrin and of enzymes involved in its metabolism in leaves of Sorghum bicolor. Plant Physiol 63: 1022–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kope HH, Tsantrizos YS, Fortin JA, Ogilvie KK(1991) p-Hydroxybenzoylformic acid and (R)-(-)-p-hydroxymandelic acid, two antifungal compounds isolated from the liquid culture of the ectomycorrhizal fungus Pisolithus arhizus. Can J Microbiol 37: 258–264 [DOI] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC(2013) Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte AR, Yandeau-Nelson MD, Nikolau BJ, Lee YJ(2015) Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal Bioanal Chem 407: 2301–2309 [DOI] [PubMed] [Google Scholar]
- Lechtenberg M.(2011) Cyanogenesis in higher plants and animals In eLS. John Wiley & Sons, Ltd, Chichester, United Kingdom [Google Scholar]
- Leonova S, Grimberg A, Marttila S, Stymne S, Carlsson AS(2010) Mobilization of lipid reserves during germination of oat (Avena sativa L.), a cereal rich in endosperm oil. J Exp Bot 61: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Knudsen C, Hansen NK, Jørgensen K, Kannangara R, Bak S, Takos A, Rook F, Hansen SH, Møller BL, Janfelt C, Bjarnholt N(2013) Visualizing metabolite distribution and enzymatic conversion in plant tissues by desorption electrospray ionization mass spectrometry imaging. Plant J 74: 1059–1071 [DOI] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA(1993) Endosperm origin, development, and function. Plant Cell 5: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla AJ.(2000) Ethylene in seed formation and germination. Seed Sci Res 10: 111–126 [Google Scholar]
- Mirza N, Crocoll C, Erik Olsen C, Ann Halkier B(2016) Engineering of methionine chain elongation part of glucoraphanin pathway in E. coli. Metab Eng 35: 31–37 [DOI] [PubMed] [Google Scholar]
- Miura GA, Shin TM(1984) Cholinergic constituents in plants: Characterization and distribution of acetylcholine and choline. Physiol Plant 61: 417–421 [Google Scholar]
- Møller BL.(2010) Plant science. Dynamic metabolons. Science 330: 1328–1329 [DOI] [PubMed] [Google Scholar]
- Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchez-Pérez R, Møller BL, Bak S(2008) β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Nielsen KA, Olsen CE, Pontoppidan K, Møller BL(2002) Leucine-derived cyano glucosides in barley. Plant Physiol 129: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LJ, Stuart P, Pičmanová M, Rasmussen S, Olsen CE, Harholt J, Møller BL, Bjarnholt N(2016) Dhurrin metabolism in the developing grain of Sorghum bicolor (L.) Moench investigated by metabolite profiling and novel clustering analyses of time-resolved transcriptomic data. BMC Genomics 17: 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasiuk O, Bills D(1984) Cyanide content of sorghum sprouts. J Food Sci 49: 791–793 [Google Scholar]
- Pičmanová M, Neilson EH, Motawia MS, Olsen CE, Agerbirk N, Gray CJ, Flitsch S, Meier S, Silvestro D, Jørgensen K, et al. (2015) A recycling pathway for cyanogenic glycosides evidenced by the comparative metabolic profiling in three cyanogenic plant species. Biochem J 469: 375–389 [DOI] [PubMed] [Google Scholar]
- Piotrowski M.(2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69: 2655–2667 [DOI] [PubMed] [Google Scholar]
- Poulton JE.(1990) Cyanogenesis in plants. Plant Physiol 94: 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Debeaujon I(2008) Seed longevity: Survival and maintenance of high germination ability of dry seeds. C R Biol 331: 796–805 [DOI] [PubMed] [Google Scholar]
- Robichaud G, Garrard KP, Barry JA, Muddiman DC(2013) MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom 24: 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosental L, Nonogaki H, Fait A(2014) Activation and regulation of primary metabolism during seed germination. Seed Sci Res 24: 1–15 [Google Scholar]
- Saleh AM, Abu El-Soud W(2015) Evidence for “gibberellin-like” activity of coumarin. S Afr J Bot 100: 51–57 [Google Scholar]
- Sánchez-Linares L, Gavilanes-Ruíz M, Díaz-Pontones D, Guzmán-Chávez F, Calzada-Alejo V, Zurita-Villegas V, Luna-Loaiza V, Moreno-Sánchez R, Bernal-Lugo I, Sánchez-Nieto S(2012) Early carbon mobilization and radicle protrusion in maize germination. J Exp Bot 63: 4513–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Belmonte FS, Borch J, Dicenta F, Møller BL, Jørgensen K(2012) Prunasin hydrolases during fruit development in sweet and bitter almonds. Plant Physiol 158: 1916–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Jørgensen K, Motawia MS, Dicenta F, Møller BL(2009) Tissue and cellular localization of individual β-glycosidases using a substrate-specific sugar reducing assay. Plant J 60: 894–906 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pérez R, Jørgensen K, Olsen CE, Dicenta F, Møller BL(2008) Bitterness in almonds. Plant Physiol 146: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabia LD, Boughton BA, Rupasinghe T, van de Meene AML, Callahan DL, Hill CB, Roessner U(2018) High-mass-resolution MALDI mass spectrometry imaging reveals detailed spatial distribution of metabolites and lipids in roots of barley seedlings in response to salinity stress. Metabolomics 14: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck CA, Maeda HA(2018) Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry 149: 82–102 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt FB, Cho SK, Olsen CE, Yang SW, Møller BL, Jørgensen K(2018) Diurnal regulation of cyanogenic glucoside biosynthesis and endogenous turnover in cassava. Plant Direct 2: e00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm T, Hester Z, Klinkert I, Both JP, Heeren RMA, Brunelle A, Laprévote O, Desbenoit N, Robbe MF, Stoeckli M, Spengler B, Römpp A(2012) imzML--a common data format for the flexible exchange and processing of mass spectrometry imaging data. J Proteomics 75: 5106–5110 [DOI] [PubMed] [Google Scholar]
- Selmar D.(1993) Transport of cyanogenic glucosides: Linustatin uptake by Hevea cotyledons. Planta 191: 191–199 [Google Scholar]
- Selmar D, Irandoost Z, Wray V(1996) Dhurrin-6′-glucoside, a cyanogenic diglucoside from Sorghum bicolor. Phytochemistry 43: 569–572 [DOI] [PubMed] [Google Scholar]
- Shroff R, Vergara F, Muck A, Svatos A, Gershenzon J(2008) Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc Natl Acad Sci USA 105: 6196–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Petrášek J(2011) Why plants need more than one type of auxin. Plant Sci 180: 454–460 [DOI] [PubMed] [Google Scholar]
- Soeda Y, Konings MCJM, Vorst O, Van Houwelingen AMML, Stoopen GM, Maliepaard CA, Kodde J, Bino RJ, Groot SPC, Van der Geest AHM(2005) Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiol 137: 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel VM.(2016) Signal achievements in gibberellin research: The second half-century. Annu Plant Rev 49: 1–36 [Google Scholar]
- Swain E, Poulton JE(1994) Utilization of amygdalin during seedling development of prunus serotina. Plant Physiol 106: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipples KH, Norris FW(1983) Polyphenol changes in sorghum grain during malting. Nov L J Sci Food Agric 31: 1295–1299 [Google Scholar]
- Toue S, Sugiura Y, Kubo A, Ohmura M, Karakawa S, Mizukoshi T, Yoneda J, Miyano H, Noguchi Y, Kobayashi T, Kabe Y, Suematsu M(2014) Microscopic imaging mass spectrometry assisted by on-tissue chemical derivatization for visualizing multiple amino acids in human colon cancer xenografts. Proteomics 14: 810–819 [DOI] [PubMed] [Google Scholar]
- Urbanova T, Leubner-Metzger G(2016) Gibberellins and seed germination. Annu Plant Rev 49: 253–284 [Google Scholar]
- Veličković D, Ropartz D, Guillon F, Saulnier L, Rogniaux H(2014) New insights into the structural and spatial variability of cell-wall polysaccharides during wheat grain development, as revealed through MALDI mass spectrometry imaging. J Exp Bot 65: 2079–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S.(2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Yokotani-Tomita K, Kosemura S, Yamamura S, Yamada K, Hasegawa K(1999) Allelopathic substance exuded from a serious weed, germinating barnyard grass (Echinochloa crus-galli L.), roots. J Plant Growth Regul 18: 65–67 [DOI] [PubMed] [Google Scholar]
- Yan D, Duermeyer L, Leoveanu C, Nambara E(2014) The functions of the endosperm during seed germination. Plant Cell Physiol 55: 1521–1533 [DOI] [PubMed] [Google Scholar]