Expression of a gene in seed scutellum is antagonistically regulated by abscisic acid and gibberellic acid signaling pathways to control seed dormancy.
Abstract
Seed dormancy is a natural phenomenon in plants. It ensures that seeds complete the grain-filling stage before germination and prevents germination in unsuitable ecological conditions. In this study, we determined the previously unknown function of the rice (Oryza sativa) gene GERMIN-LIKE PROTEIN 2-1 (OsGLP2-1) in seed dormancy. Using artificial microRNA and CRISPR/CAS9 approaches, suppression of OsGLP2-1 expression in rice resulted in the release of dormancy in immature seeds. Conversely, overexpression of OsGLP2-1 driven by the OsGLP2-1 native promoter led to greater seed dormancy. Seed scutellum-specific expression of OsGLP2-1 was increased by exogenous abscisic acid, but decreased with gibberellic acid treatment. We provide evidence that OsGLP2-1 is antagonistically controlled at the transcriptional level by ABA INSENSITIVE5 and GAMYB transcription factors. We conclude that OsGLP2-1 acts as a buffer, maintaining appropriate equilibrium for the regulation of primary dormancy during seed development in rice.
A proper level of seed dormancy is important for crop cultivars. Seeds that show weak dormancy are susceptible to preharvest sprouting, which causes substantial loss in grain yield and quality. In contrast, strong dormancy inhibits germination of the seed. Published investigations provide evidence that the seed dormancy or germination developmental outcomes are mainly determined by the cooperative action of abscisic acid (ABA)- and gibberellic acid (GA)-associated signal pathways (Fang and Chu, 2008; Fang et al., 2008; Holdsworth et al., 2008a; Shu et al., 2013; Abraham et al., 2016; Wang et al., 2018). Both ABA and GA are important plant growth regulators. ABA mainly promotes seed dormancy, inhibits root growth, and facilitates abiotic stress tolerance, whereas, in contrast, GA initiates seed germination and stimulates plant growth. Other phytohormones, such as ethylene, indoleacetic acid, strigolactone, and brassinosteroid (Yang et al., 2011; Toh et al., 2012; Wang et al., 2013; Zhao et al., 2019), as well as external environmental factors such as temperature and light intensity, and internal developmental signals such as nitric oxide and reactive oxygen species, also play roles in the processes of seed dormancy or germination through cooperation with ABA or GA (Alboresi et al., 2005; Liu et al., 2010; Toorop et al., 2012). In addition, epigenetic mechanisms (histone modification, DNA methylation, and chromatin remodeling) also regulate seed dormancy (Gao and Ayele, 2014).
ABA biosynthetic and catabolic pathways have been elucidated by 18O labeling experiments, molecular genetic analysis of auxotrophs, and biochemical studies (Nambara and Marion-Poll, 2005). The ABA signal transduction model was established based on studies of the mechanism of the ABA receptors of the REGULATORY COMPONENTS OF ABA RECEPTOR/PYRABACTIN RESISTANCE1/PYL1-LIKE protein family (Zhu, 2016). Binding of ABA to these receptors leads to inactivation of type 2C protein phosphatases, such as ABA INSENSITIVE1 (ABI1) and ABI2. This in turn activates SUC NONFERMENTING 1-RELATED PROTEIN KINASES, which regulates activation of downstream transcription factors through phosphorylation (Fujii et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Santiago et al., 2009; Sheard and Zheng, 2009; Raghavendra et al., 2010). ABI5 (a basic region/Leu-zipper protein) and ABI3 (a B3 domain transcription factor) are two key ABA-responsive transcription factors that regulate downstream target genes by binding to the ABA-responsive element (ABRE) of ACGTG in the promoters of the genes (Gao and Ayele, 2014).
Gibberellins regulate a variety of growth and developmental programs in plants, including cell differentiation and elongation, reproductive development, and germination. Gibberellins are often considered to exert antagonistic roles to ABA to control seed germination. The major effect of GA is degradation of the DELLA protein, allowing the GAMYB transcription factor to activate the α-amylase genes during seed germination (Gubler et al., 1995; Gubler et al., 1999). The α-amylase in the aleurone layer catalyzes hydrolysis of starch to Glc, thus providing energy for seed germination. In addition to its role in germination, GA is involved in regulation of seed storage protein genes during seed development (Diaz et al., 2002). The exact molecular target of GA on seed dormancy during seed development is unclear.
The transition from seed dormancy to germination is a dynamic process. Freshly harvested mature water-permeable dormant seeds show primary dormancy, which has been induced through the involvement of ABA during seed maturation on the mother plant (Hilhorst, 1995; Miyoshi et al., 1999; Manz et al., 2005). During the period of after-ripening, seed dormancy is gradually relieved (Holdsworth et al., 2008a). As one of the most important ABA-responsive transcription factors, ABI5 plays a crucial role in seed dormancy via the ABA signaling pathway. However, in contrast with germination-related genes, only a small number of dormancy-related genes (such as EARLY MET-LABELED6 and MOTHER OF FT AND TFL1 [MFT]) regulated by ABI5 and ABI3/VIVIPAROUS1 have been identified and functionally studied (Hattori et al., 1995; Lopez-Molina et al., 2002; Xi et al., 2010; Nakamura et al., 2011). Previous studies also show that ABA content and expression of ABA metabolic genes and signal-related genes exhibit no apparent differences between whole imbibing dormant and after-ripened wheat (Triticum aestivum) seeds (Liu et al., 2013), which suggests that wheat seed dormancy and after-ripening may be controlled by separate genetic pathways.
Seed constituents are also associated with dormancy. The embryo and/or a seed coat component (such as the testa or endosperm) are reportedly associated with seed dormancy (Hilhorst, 1995; Finch-Savage and Leubner-Metzger, 2006; Müller et al., 2006; Gu et al., 2015). Assuming to be the primary determinant of seed dormancy, the endosperm surrounding the embryo inhibits germination in intact Arabidopsis (Arabidopsis thaliana) seeds (Bethke et al., 2007). During seed germination, ABA delays endosperm weakening and rupture, whereas GA, acting as an early embryo signal, promotes this process (Müller et al., 2006). The qSD7-1 and qSD12 quantitative trait loci were determined to be associated with maternal and offspring tissue-imposed dormancies, respectively (Gu et al., 2010). However, the mechanisms by which the embryo and endosperm coordinately control seed dormancy, especially during seed development, are not well understood.
The scutellum, which is part of the seed located between the embryo and endosperm, functions as conductive tissue between the endosperm and embryo. Several important metabolic events occur in the scutellum. In germinating wheat seeds, following degradation of endosperm starch, the resulting hexoses are converted to Suc in the scutellum and eventually transported to the embryo axis (Aoki et al., 2006; Asakura et al., 2007). In the germinating grain, the storage proteins in the endosperm are hydrolyzed into peptides and free amino acids, which was subsequently taken by the scutellum (Salmenkallio and Sopanen, 1989). Above all, the scutellum is a prospective site for the control of seed germination/dormancy mediated by the ABA/GA-response pathways (Asakura et al., 2007).
Germin-like proteins (GLPs) are a group of proteins that show a similar amino acid sequence to that of wheat germin, which are expressed during seed germination. In plants GLPs constitute a multigene family (Kim and Triplett, 2004), and perform a variety of biological functions in plant development, stress response, and pathogen resistance (Berna and Bernier, 1997; Membré et al., 1997; Liu et al., 2016). The promoter sequences of GLPs are responsive to environmental stresses and growth factors (Breen and Bellgard, 2010; Mahmood et al., 2010; Li et al., 2016). Among isolated GLPs, a portion of the proteins exhibit oxalate oxidase or superoxide dismutase activity. However, the function of most GLPs remains unknown.
Although the germins were first isolated from germinating wheat embryos since 1980s, and an increasing number of germins and GLPs were identified in the defense functions, so far, only one case (ZmGLP) is related to germination vigor of maize (Zea mays) seed (Lapik and Kaufman, 2003; Fu et al., 2016). ZmGLP helps to keep seed germination vigor, but its mechanism is unclear. In this study we show that a rice (Oryza sativa) germin-like protein, which is designated OsGLP2-1 according to a recent nomenclature (Li et al., 2016), played a key role in dormancy regulation of developing seeds. OsGLP2-1 is located on chromosome 2 (LOC_Os02g29000) and is expressed exclusively in the seed scutellum and minor veins of the leaf. Unlike other isolated GLPs, which are usually localized in the extracellular matrix, OsGLP2-1 was localized in the cytosol and showed rapid movement within the cell. When compared with the control, suppression of OsGLP2-1 in rice led to dormancy release and displayed a higher frequency of germination, whereas overexpression of OsGLP2-1 resulted in deep dormancy. Transcription of OsGLP2-1 was directly regulated by ABI5 and GAMYB through binding to the corresponding cis-regulatory elements in the promoter. This work shows that OsGLP2-1 is dual-targeted by ABA and GA signals involved in seed dormancy in opposite ways. Elucidation of the function of OsGLP2-1 in dormancy of developing seeds will help to clarify the mechanism of seed dormancy coregulated by ABA and GA.
RESULTS
Expression Pattern of OsGLP2-1
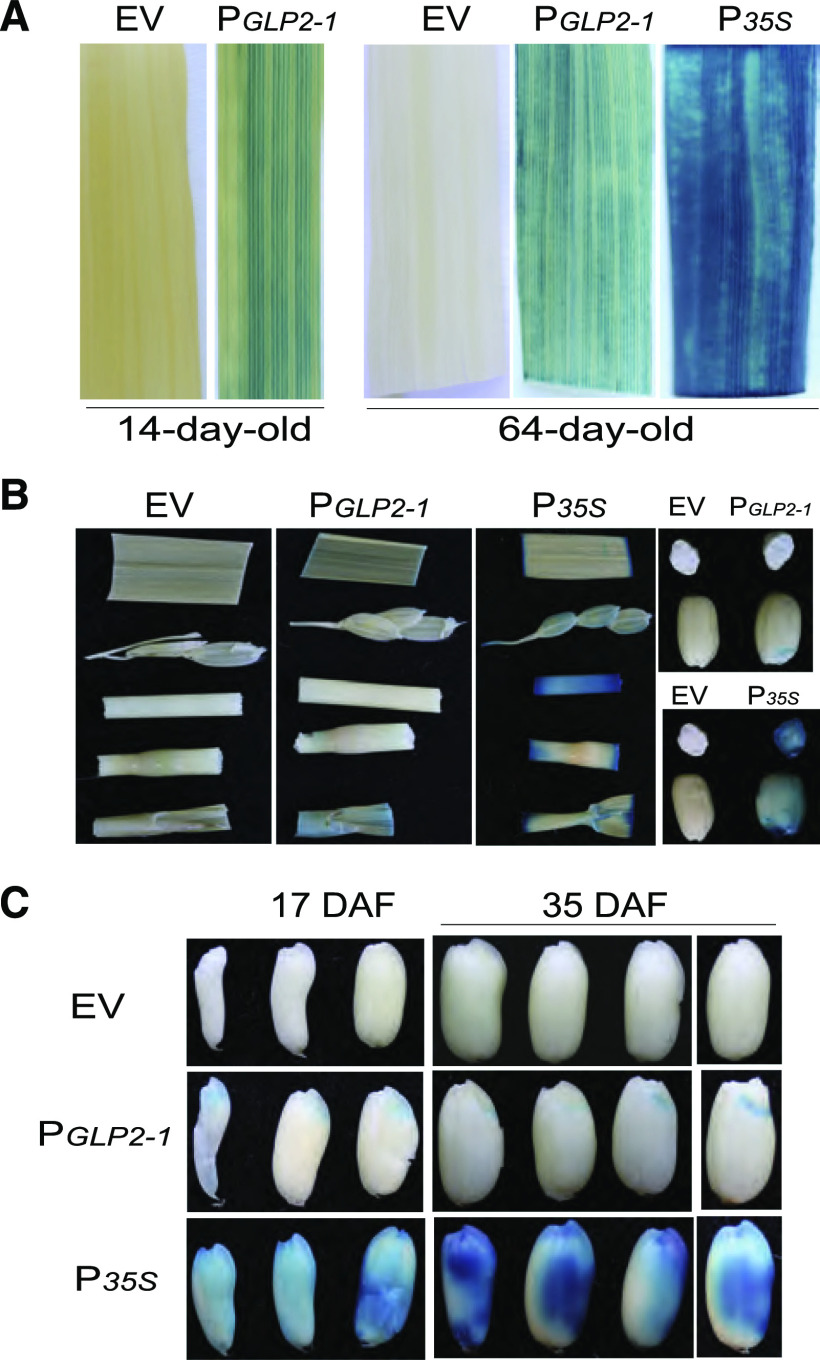
OsGLP2-1 (LOC_Os02g29000) encodes a germin-like protein of 216 amino acids containing a single cupin domain that comprises two conserved amino acid sequence motifs to form a β-barrel structure (Dunwell et al., 2004). To determine the expression pattern of OsGLP2-1, the promoter fragment of OsGLP2-1 (PGLP2-1) spanning from position −2,086 to −1 relative to the translation initiation site was amplified and cloned. The reporter expression vector PGLP2-1:GUS, in which the gene GUS was driven by PGLP2-1, was constructed and transformed into rice callus. Staining with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide of tissues from PGLP2-1:GUS rice plants showed intense GUS signal predominantly in the minor veins of the leaf in the vegetative stage (Fig. 1A). In the reproductive stage, only weak GUS signal was observed in the leaf sheath, and no signal was detected in the other organs examined except the seed scutellum (tissue located between the embryo and endosperm), which showed strong expression of GUS, forming a blue crescent in both immature and mature seeds (Fig. 1, B and C; Supplemental Fig. S1A). These results indicate that a potential role of OsGLP2-1 played in developing rice seed and seed dormancy.
Figure 1.
Expression pattern of PGLP2-1:GUS in transgenic rice plants at different growth stages. A, Specific expression in minor veins of the leaf in the vegetative phase. Leaves from 14-d-old seedlings: left, negative control (empty vector transformant [EV]) lacking signal; right, PGLP2-1:GUS leaf with strong blue GUS signal in minor veins. Leaves from 64-d-old plant: left, negative control; center, PGLP2-1:GUS leaf showing staining in the minor veins; right, CaMV 35S promoter (P35S) driving constitutive expression of GUS in the leaf. B, GUS staining in organs from mature plant, including leaf, spikelet, stem, internode, leaf sheath, and seed. Among these organs of PGLP2-1:GUS plants, light signal was observed in the leaf sheath and strong blue signal displayed specifically in the scutellum. C, GUS staining of developing seeds of PGLP2-1:GUS transformant. Seeds were sampled at different growth stages (17 and 35 DAF). Scutellum-specific expression pattern was exhibited by all developing seeds. EV seed was used as negative control, and P35S showing constitutive GUS expression was used as positive control.
OsGLP2-1 Positively Regulated the Dormancy of Developing Seed
To check the involvement of OsGLP2-1 in seed dormancy regulation, a complementary DNA fragment (737 bp) harboring the open reading frame (ORF) of the gene was amplified and cloned from rice ssp. japonica ‘Nipponbare’. Cauliflower mosaic virus (CaMV) 35S promoter-driven OsGLP2-1 ectopic overexpression (OE-GLP2-1) and suppression (amiR-GLP2-1) transgenic rice plants (Supplemental Fig. S2) as well as empty vector-transformed plants were generated. The homozygous lines for each type of transgenic rice plants were selected, and their seed dormancy phenotypes were compared using T3 seeds. The T3 immature seeds of OE-GLP2-1 and amiR-GLP2-1 (35 and 45 d after flowering [DAF]) exhibited marked different dormancy. When compared with the seed dormancy of empty vector control, the T3 seeds (35 DAF) of the majority of amiR-GLP2-1 independent lines (10/16) displayed dormancy releasing phenotype and a high germination percentage, and the germinated seeds developed into seedlings subsequently (Supplemental Fig. S3A; Supplemental Table S1). Contrary to the dormancy-breaking phenotype of amiR-GLP2-1 seeds, the sign of deepening dormancy was exhibited in OE-GLP2-1 seeds. Given that OsGLP2-1 is specifically expressed in scutellum, we further created the OsGLP2-1 overexpression transgenic rice lines driven by its native promoter (PGLP2-1:HA-GLP2-1-eGFP). The enhanced dormancy of PGLP2-1:HA-GLP2-1-eGFP seeds was consistently observed. The germination percentages of PGLP2-1:HA-GLP2-1-eGFP independent lines were significantly lower than that of empty vector control (Table 1). The contradictory dormancy phenotypes of amiR-GLP2-1 and PGLP2-1:HA-GLP2-1-eGFP seeds suggested that OsGLP2-1 played an important role in the maintenance of primary dormancy of the developing seed and that changed expression levels of OsGLP2-1 led to alteration of seed dormancy state.
Table 1. Comparison of the transgenic seeds (30 DAF) germination rate.
Empty vector seeds were used as a negative control. Values shown are means ± SE of triplicate measurements. Asterisks indicate significant difference at *P = 0.05 and **P = 0.01, respectively, in SPSS one-way ANOVA analysis with post hoc test. N, Number of seeds.
| Line No. | N | Germination Percentage | ||
|---|---|---|---|---|
| 9 d | 11 d | 13 d | ||
| % | ||||
| Empty vector | 30, 30, 30 | 34.97 ± 3.40 | 38.86 ± 2.51 | 38.86 ± 2.51 |
| PGLP2-1:HA-GLP2-1-eGFP | ||||
| PGLP-17 | 11, 10, 10 | 13.03 ± 11.88* | 13.03 ± 11.89** | 13.03 ± 11.89** |
| PGLP-22 | 12, 10, 10 | 0.00 ± 0.00** | 0.00 ± 0.00** | 0.00 ± 0.00** |
| PGLP-26 | 14, 12, 11 | 3.03 ± 3.71** | 3.03 ± 3.71** | 3.03 ± 3.71** |
| PGLP-29 | 8, 10, 10 | 10.83 ± 1.02** | 10.83 ± 1.02** | 10.83 ± 1.02** |
| PGLP-31 | 12, 12, 12 | 16.67 ± 5.89* | 16.67 ± 5.89* | 16.67 ± 5.89* |
| P35S-ABI5 | ||||
| AB5-44 | 10, 12, 10 | 5.56 ± 6.80** | 5.56 ± 6.80** | 5.56 ± 6.80** |
| AB5-6 | 12, 10, 12 | 0.00 ± 0.00** | 2.78 ± 3.40** | 2.78 ± 3.40** |
| AB5-13 | 14, 14, 16 | 0.00 ± 0.00** | 0.00 ± 0.00 ** | 0.00 ± 0.00 ** |
| P35S-GAMYB | ||||
| GAB6-5 | 10, 10, 10 | 96.67 ± 4.08** | 96.67 ± 4.08** | 96.67 ± 4.08** |
| GAB7-2 | 12, 10, 10 | 83.89 ± 25.97** | 83.89 ± 25.97** | 87.22 ± 26.29** |
| GAB8-11 | 12, 10, 10 | 93.89 ± 3.79** | 97.22 ± 3.40** | 97.22 ± 3.40** |
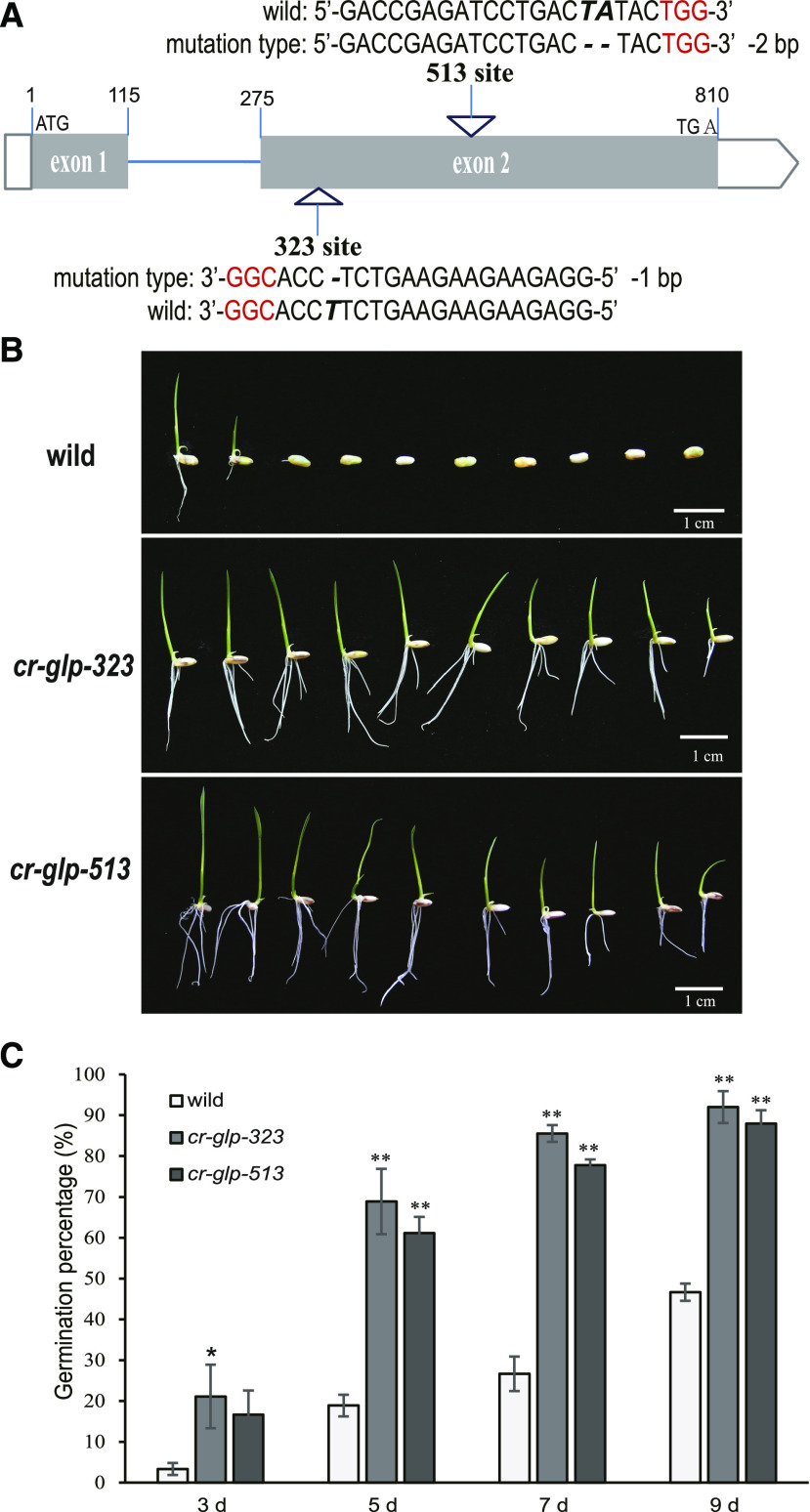
To further demonstrate the loss-of-function of OsGLP2-1 in rice, two knock-out lines (osglp2-1 323 and 513) were created by CRISPR-CAS9 system (Fig. 2A). The 20 DAF dehusked seeds of transfer DNA (T-DNA)-free homozygous osglp2-1 mutant were used for comparison of seed dormancy. When compared with the wild-type seed, seeds of both osglp2-1 lines 323 and 513 exhibited dormancy-breaking phenotype, with significantly high germination percentage (Fig. 2, B and C). Together with the enhanced dormancy of PGLP2-1:HA-GLP2-1-eGFP seeds, these results proved the positively regulation of seed dormancy by OsGLP2-1.
Figure 2.
Dormancy-breaking phenotype exhibited on the rice CRISPR-osglp2-1-seeds. A, Schematic diagram of CRISPR/CAS9-mediated targeted mutagenesis in OsGLP2-1. The ORF consists of two exons indicated as black boxes. The mutated sites (323 and 513) in 20-bp single guide RNA targeting sequences and protospacer adjacent motif are shown in bold italic letter and in red letter, respectively. B, and C, Comparisons of wild type and CRISPR-osglp2-1 developing seeds dormancy state. B, Dormancy-breaking phenotype exhibited on the CRISPR-osglp2-1-seeds (7 d after sowing). C, Compared with the low germination rate of wild-type seeds at 3, 5, 7, and 9 d after sowing, two lines of T-DNA–free homozygous CRISPR-osglp2-1-seeds, cr-glp-323 and cr-glp-513, displayed significantly higher germination percent, respectively. The surface sterilized 20 DAF-dehulling seeds were sown in MS medium, 15 seeds for each sample, and 6 replicates were performed. Error bars = SE. Asterisks indicate significant difference at *P = 0.05 and **P = 0.01 in SPSS one-way ANOVA analysis with post hoc test.
Altered Expression of OsGLP2-1 in Response to ABA and GA
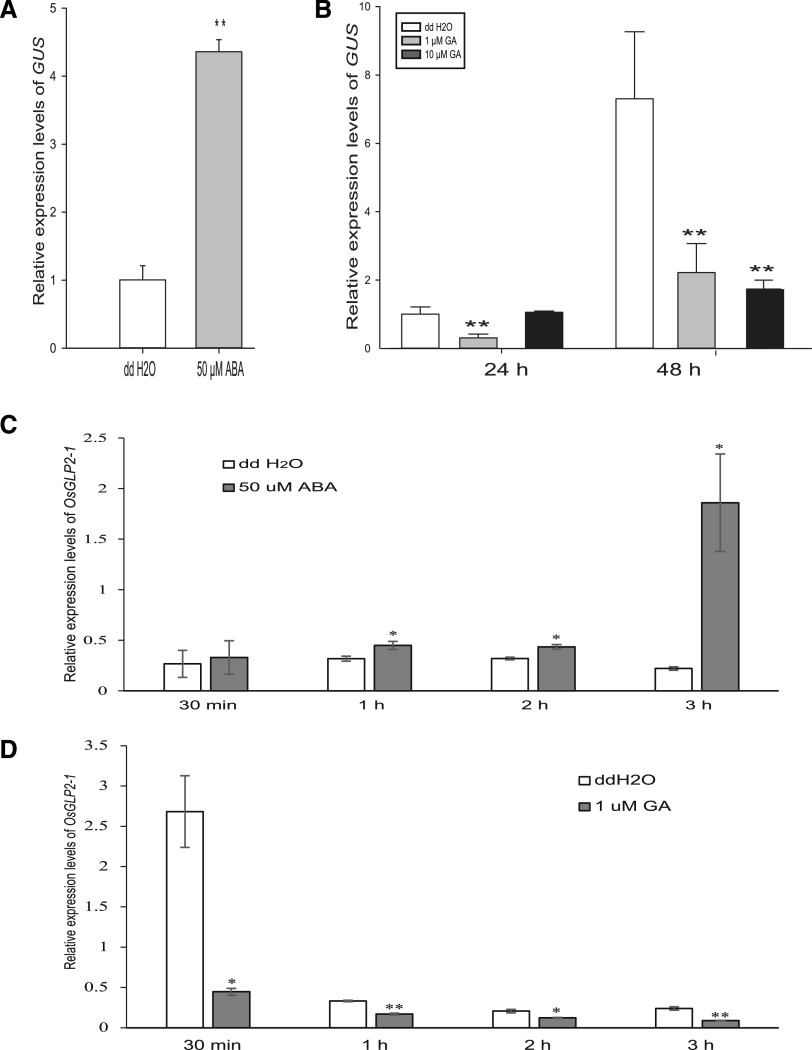
It is well known that seed dormancy is mainly associated with ABA and GA. Our results showed that OsGLP2-1 functions in control of seed dormancy, suggesting if there is a regulatory relationship between OsGLP2-1 and ABA and GA. To verify this, OsGLP2-1 expression in response to exogenous ABA and GA treatment was examined and compared in seed halves of PGLP2-1:GUS rice containing the embryo and scutellum collected 30 DAF. The expression level of GUS reporter gene was measured by reverse transcription-quantitative PCR (RT-qPCR). When compared with the relative expression level in the untreated control, GUS expression driven by PGLP2-1 was significantly increased by 50 μm ABA treatment, but was markedly decreased by 1 and 10 μm GA treatments (Fig. 3, A and B). In addition to use of the GUS reporter (PGLP2-1:GUS transgenic lines), we also detected OsGLP2-1 induction/suppression in response to ABA/GA in wild-type japonica cv Nipponbare seeds to see the response of OsGLP2-1 to ABA and GA. The results were consistent with that using PGLP2-1:GUS transgenic lines: the expression of OsGLP2-1 was induced by ABA, while suppressed by GA in wild-type developing seed. When compared with the expression level of OsGLP2-1 in untreated control seeds, significantly activated expression of OsGLP2-1 was displayed in 50 μm ABA treatment at 30 min, 1 to 3 h, whereas notable inhibited OsGLP2-1-expressing was shown in 1 μm GA treatment (Fig. 3, C and D). These results showed that the expression of OsGLP2-1 was involved in both of ABA and GA signaling pathways.
Figure 3.
Response of the expression of OsGLP2-1 to ABA and GA. A and B, Expression of GUS driven by PGLP2-1 was stimulated by exogenous ABA and suppressed by GA. C and D, Altered expression of OsGLP2-1 in response to ABA and GA in wild-type seeds. The induction/suppression of OsGLP2-1 expression was detected after ABA/GA treated within 3 h, respectively. The PGLP2-1:GUS or wild-type rice embryo including the scutellum was freshly cut from dehusked 30 DAF seeds, and immediately treated with exogenous 50 μm ABA (A and C), 1 or 10 μm GA (B and D) for different time points in a growth chamber (25°C, 12-h light/12-h dark). The samples were ground in liquid nitrogen for further monitoring of GUS or OsGLP2-1 expression by RT-qPCR. Collection of 15 embryos for each sample and three replicates were performed. Error bars = SE (n = 3). Asterisks indicate significant difference at *P = 0.05 and **P = 0.01 in SPSS independent-sample t test (A, C, and D) and one-way ANOVA analysis with post hoc test (B), respectively.
The roles of ABA and GA in seed dormancy and germination are well known. To further explore the interactions of OsGLP2-1 with these hormones in their controlled physiological processes, T3 seeds were collected at 45 DAF and cultured on one-half strength Murashige and Skoog (MS) medium supplemented with 50 μm ABA or 10 μm GA3. The germination of amiR-GLP2-1 seeds was repressed by 50 μm ABA treatment (Supplemental Figs. S3 and S4A; Supplemental Table S2). This result showed that exogenous ABA can effectively weaken the dormancy-breaking phenotype of amiR-GLP2-1. In the presence of 10 μm GA, germination of OE-GLP2-1 seeds was stimulated but did not fully complemented the low germination potential of the seeds (Supplemental Fig. S4B). Another seed dormancy comparison assay performed on T4 immature seeds of OE-GLP2-1 and amiR-GLP2-1 produced similar results (data not shown). These results showed that exogenous ABA and GA can effectively weaken and almost offset the dormancy phenotype of amiR-GLP2-1 and OE-GLP2-1, respectively, and further confirmed that the regulation of seed dormancy by OsGLP2-1 was closely related to ABA and GA pathways.
Expression of OsGLP2-1 Was Promoted by ABI5 But Suppressed by GAMYB
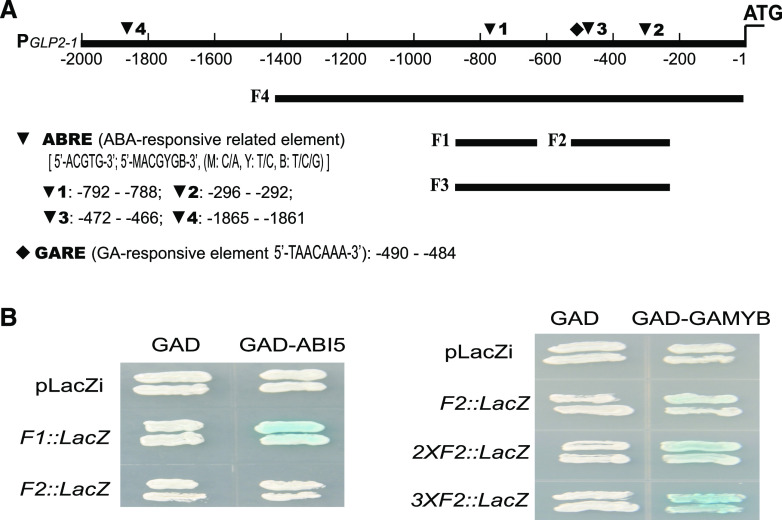
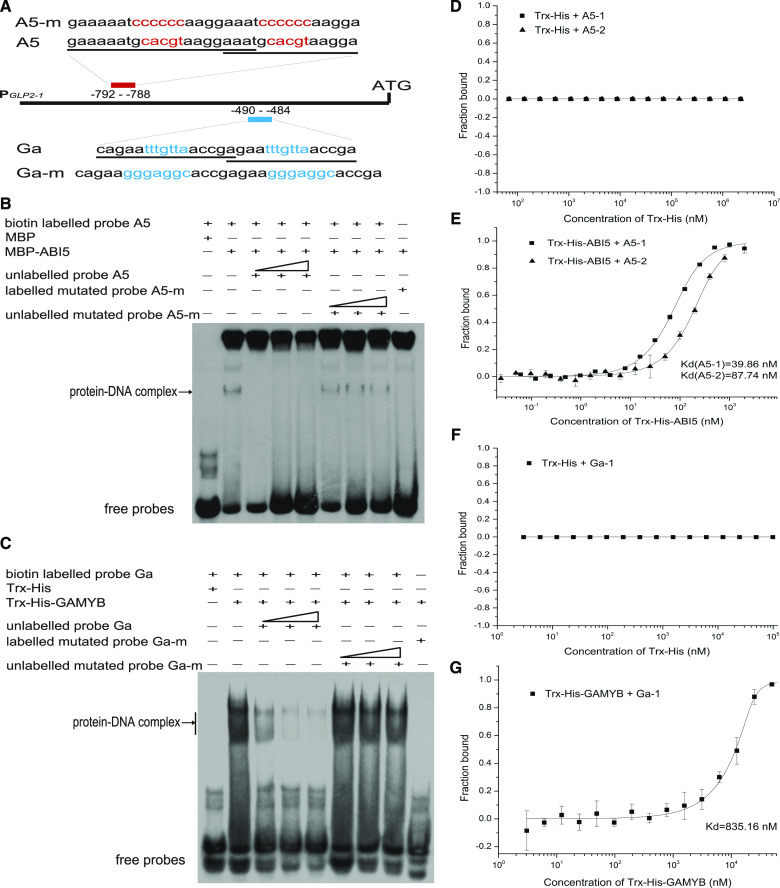
To explore the regulatory mechanism of OsGLP2-1 expression, the 2,086-bp OsGLP2-1 promoter sequence was analyzed using the plant cis-acting regulatory DNA elements database (Higo et al., 1999). The OsGLP2-1 promoter contained four potential ABRE cis-elements located at sites −1865 to −1861, −792 to −788, and −296 to −292 for 5′-ACGTG-3′ and −472 to −466 for 5′-MACGYGB-3′ (M: C/A, Y:T/C, B:T/C/G) upstream of the translation start site for OsGLP2-1. The GA response complex usually includes three sequence motifs: a GA-responsive element (GARE; TAACAAA-box) for binding to GAMYB; a pyrimidine box (5′-CCTTTT-3′) for binding with a DOF (DNA-binding One Zinc Finger) family transcription factor; and a 5′-TATCCAC-3′ element for an unknown transcription factor. The OsGLP2-1 promoter sequence contained the above three elements located respectively at −941 to −935, −490 to −484, and −62 to −57 bp upstream of the translation start site. The presence of these cis-element motifs suggested that OsGLP2-1 might be regulated by ABA- and GA-response-related transcription factors.
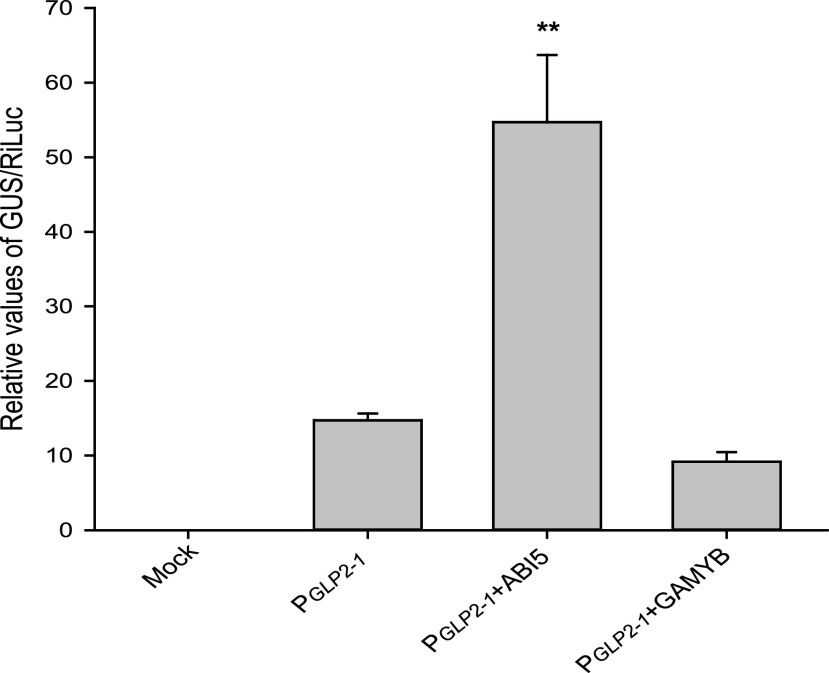
To determine whether transcription of OsGLP2-1 was directly regulated by transcription factors involved in the ABA and GA signaling pathways, the main transcription factors that mediate seed dormancy regulation in the ABA/GA signal pathways, i.e. ABI5 and ABL1, the DOF family transcription factors, and GAMYB, were tested in a preliminary agroinfiltration experiment (Fig. 4; Supplemental Fig. S5). Overexpression vectors for each transcription factor were constructed, and each was coexpressed with the PGLP2-1:GUS expression vector in Nicotiana benthamiana leaves. The relative activity of GUS was measured. Among the transcription factors tested, ABI5 significantly increased the expression of GUS controlled by PGLP2-1, whereas GAMYB exerted a negative impact (Fig. 4). These results suggested that the response of OsGLP2-1 to ABA and GA was directly mediated by ABI5 and GAMYB, respectively.
Figure 4.
Effect of ABI5 and GAMYB on activity of GUS driven by PGLP2-1. Effector construct: ABI5 (P35S:ABI5) and GAMYB (P35S:GAMYB); Reporter construct (PGLP2-1:GUS); Internal control (PNOS:RiLuc). The means of GUS/RiLuc ratios were obtained from three replicates of five pooled N. benthamiana leaves. Error bars = SE (n = 3). Mock, leaf sample infiltrated with MMA solution. RiLUC was used as an internal control in the agroinfiltration system. Asterisks indicates sgnificant difference at **P = 0.01 in SPSS independent-sample t test. ABI5: LOC_Os01g64000; GAMYB: LOC_Os01g59660.
ABI5 and GAMYB Coordinately Regulate OsGLP2-1 Expression through Binding to the Respective ABRE and GARE Elements
The agroinfiltration results implied that ABI5 and GAMYB may directly regulate OsGLP2-1 expression. To verify the physical interaction of the PGLP2-1 promoter with the ABI5 and GAMYB transcription factors, a yeast one-hybrid assay was performed (Fig. 5). When compared with the negative control (pLacZi+GAD), among the tested PGLP2-1, F1, F2, F3, and F4 promoter fragments (Fig. 5A), ABI5 directly bound to the F1 segment with an ABRE core cis-element (ACGT) and induced expression of LacZ. The GAMYB transcription factor bound to the F2 fragment and activated expression of the reporter gene, and the interaction was enhanced with multiple copies of F2 (Fig. 5B). These results are compatible with the presence of a GAMYB-binding GARE in the F2 fragment. Thus, ABI5 and GAMYB were indicated to function as transacting factors for the ABRE and GARE cis-elements, respectively, to regulate OsGLP2-1 expression.
Figure 5.
Binding activity of ABI5 and GAMYB to PGLP2-1 detected by yeast one-hybrid assay. A, Schematic diagram of the potential cis-elements in PGLP2-1 and promoter fragments. F1: −932 to −637 (295 bp); F2: −507 to −237 (270 bp); F3: −932 to −237 (696 bp); F4: −1419 to −1 (1419 bp); PGLP2-1: −2086 to −1 (2086 bp). B, Compared with the negative controls (empty vector GAD with pLacZi; F1::LacZ, or F2::LacZ, respectively, and GAD-ABI5 with pLacZi), ABI5 directly bound to the PGLP2-1 promoter F1 fragment and activated expression of the LacZ reporter gene. GAMYB directly bound to the F2 fragment and induced LacZ expression; moreover, the signal increased progressively with 2 or 3 GARE tandemly repeated in 2×F2::LacZ and 3×F2::LacZ, respectively. The combinations of GAD with pLacZi, F2::LacZ, 2×F2::LacZ, or 3×F2::LacZ; and GAD-GAMYB with pLacZi were used as negative controls. These binding interactions of PGLP2-1 with GAMYB and ABI5 were confirmed in at least three repeated experiments.
An electrophoresis mobility shift assay (EMSA) was conducted to confirm the results of the yeast one-hybrid assay. The assay demonstrated the DNA-binding specificity of the MBP-ABI5 fusion protein to the biotin-labeled ABRE cis-element in the OsGLP2-1 promoter (Fig. 6, A and B). The Trx-His-GAMYB fusion protein specifically bound to the GARE cis-element in the OsGLP2-1 promoter (Fig. 6, A and C). The shift bands displayed gradients of intensity from low to high amounts of fusion proteins (Supplemental Fig. S6), and the binding signals were gradually suppressed by the addition of unlabeled competitive probe (Fig. 6C). With mutated probes, no competitive ability was observed and the binding ability was completely abolished (Fig. 6, B and C). It was worth noting that a more complicated pattern displayed in the binding of ABI5 with its cis-element maybe caused by the protein stability or other factors. Moreover, the freshly purified Trx-His-ABI5 or Trx-His-GAMYB fusion protein expressed in Escherichia coli, together with their corresponding fragments (A5-1, A5-2 and Ga-1) in GLP2-1 promoter, respectively, were further served to a microscale thermophoresis (MST) assay, the direct bonding characteristic was found between Trx-His-ABI5 fusion protein with A5-1 or A5-2 fragment and the Trx-His-GAMYB with Ga-1 fragment. The dissociation constant (Kd) between Trx-His-ABI5 and A5-1, A5-2 was 39.86 nm and 87.74 nm, respectively; and the Kd between Trx-His-GAMYB and Ga-1 was 835.16 nm. The negative control protein, Trx-His, did not interact with these fragments (Fig. 6, d–g). These results together with the yeast one-hybrid and EMSA results proved that the OsGLP2-1 promoter was directly targeted by the ABI5 and GAMYB transcription factors through binding to the corresponding ABRE and GARE cis-elements, respectively, in the PGLP2-1 promoter.
Figure 6.
Physical interaction of ABI5 and GAMYB to the ABRE and GARE in PGLP2-1. A to C, EMSA results displayed that the transcription factors (ABI5 and GAMYB) could directly recognize and bind to PGLP2-1 fragments containing the ABRE or GARE, respectively. A, The probes used in EMSA were designed based on the predicted binding sequence of ABRE (red letters) and GARE (blue letters) in the F1 and F2 fragments, respectively, together with their flanking nucleotides (underlined), two overlapped binding sites were synthesized. B and C, Binding activity of ABI5 and GAMYB with their corresponding probes. The fusion proteins MBP-ABI5 and Trx-His-GAMYB specifically bound to the corresponding probes and resulted in the retardant band, whereas no binding activity was observed with the corresponding negative controls of MBP and Trx-His. The unlabeled cold probes (10, 50, 100×) gradually competed for the corresponding binding activity, whereas the mutated biotin-labeled probes (10, 50, 100×) completely abolished the protein-DNA interaction, and the mutated cold-probes showed no competitive activities. D to G, MST analysis showing that the specially binding activity of Trx-His-ABI5 with OsGLP2-1 promoter fragments of A5-1, A5-2 (E), and Trx-His-GAMYB with Ga-1 (G), respectively. The negative control, Trx-His, did not interact with these fragments (D and F). D and E, MST measurements of the interaction between ABI5 and A5-1, A5-2 fragments. F and G, MST assay testing the interaction between GAMYB and Ga-1 fragment. D to G data are average ± sd, n = 3. The data were analyzed with the software of MO. Affinity Analysis v2.2.4.
Dynamic Subcellular Localization of OsGLP2-1
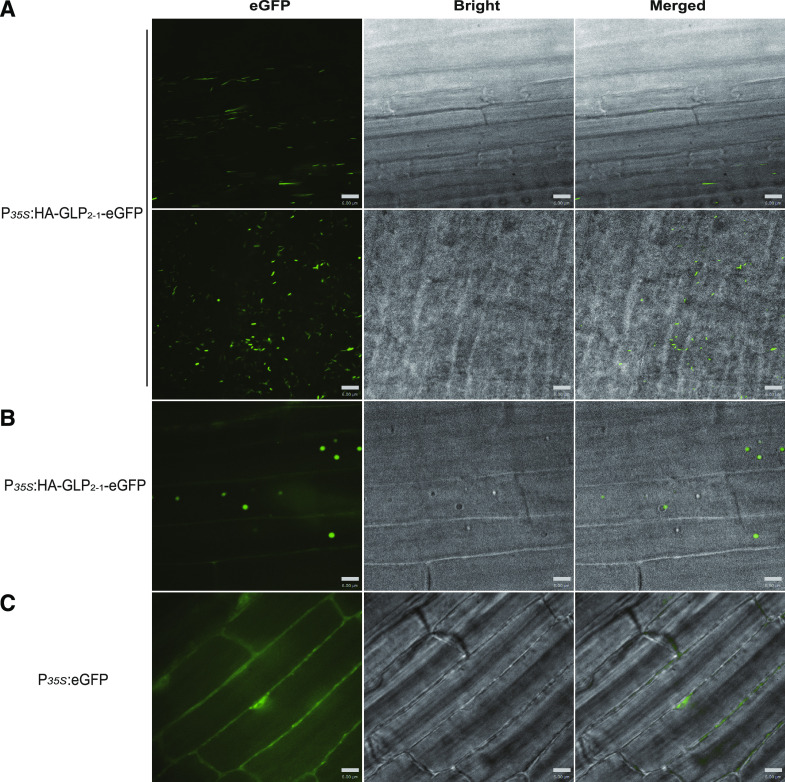
Knowledge of the subcellular localization of OsGLP2-1 may aid in understanding its biological function in rice. Thus the OsGLP2-1-eGFP fusion protein constructs, P35S:HA-GLP2-1-eGFP and PGLP2-1:HA-GLP2-1-eGFP, were generated, and transient expression of the constructs in the leaf of N. benthamiana was performed. The control (P35S:eGFP) showed constitutively expressed green fluorescent signal both in the cytosol and nucleus. The HA-GLP2-1-eGFP fusion protein driven by the PGLP2-1 native promoter or CaMV 35S (P35S) promoter exhibited single or bundles of mobile needle-shaped green fluorescent signal in the cytosol, and no signal was observed in the nucleus (Supplemental Fig. S7A). This result differed from previous reports that GLPs are usually associated with the cell wall (Li et al., 2016; Liu et al., 2016). To confirm this result, stable P35S:HA-GLP2-1-eGFP and PGLP2-1:HA-GLP2-1-eGFP transgenic Arabidopsis ecotype Columbia (Col-0) plants were generated, for which identical results were observed (Supplemental Fig. S7B). To verify this dynamic subcellular localization feature, homozygous lines of P35S:HA-GLP2-1-eGFP transgenic rice plant were further generated, and the 7-d-old seedling was used for subcellular localization observation. The result showed that, in addition to the mobile needle-shaped green fluorescence signal as shown in Arabidopsis and N. benthamiana, the moving round signal was also observed in the cytosol (Fig. 7; Supplemental Movies S1 and S2). These results demonstrated that OsGLP2-1 is a highly dynamic protein, and the specificity of OsGLP2-1-eGFP subcellular localization will help to reveal the molecular mechanism of OsGLP2-1 in rice.
Figure 7.
Subcellular localization of OsGLP2-1-eGFP in transgenic rice seedling. In 7-d-old rice seedling, the control P35S:eGFP leaves showed green fluorescence throughout the cytoplasm and nucleus (C); in contrast, green fluorescence from P35S: HA-GLP2-1-eGFP leaves exhibited moving needle-shaped signals in the cytosol, and no signal was observed in the nucleus (A). Moreover, the mobile round green fluorescent signal was also observed in P35S:HA-GLP2-1-eGFP seedling root (B). These dynamic green fluorescent signals of OsGLP2-1-eGFP in rice were confirmed in at least three independent experiments. Bars = 6 μm.
DISCUSSION
It is generally accepted that dormancy is induced by ABA during seed development on the mother plant. After maturation of the seed, germination is preceded by a decline in ABA content and the hormonal balance between ABA and GAs acts as an integrator of environmental cues to maintain dormancy or activate germination. ABI5 and GAMYB are the key positive transcription regulators of ABA and GA signaling, respectively. The expression patterns of ABI5 and GAMYB in rice show some similarities, for example, a low level of expression in organs during vegetative growth, but a relatively high level in the reproductive organs (Kaneko et al., 2004; Zou et al., 2008). In rice seeds, GAMYB is expressed specifically in the aleurone layer and scutellum. In the aleurone layer, GAMYB positively regulates α-amylase activity (Kaneko et al., 2004), but its function in the scutellum is unclear. MFT regulates seed germination via the ABA and GA signaling pathways in Arabidopsis, and its expression is directly regulated by ABI3, ABI5, and DELLA (Xi et al., 2010). A wheat homolog of MFT is also associated with seed dormancy and is exclusively expressed in the scutellum and coleorhiza (Nakamura et al., 2011). The overlapping scutellum expression pattern of GAMYB and ABI5-targeting genes (such as MFT) indicates that the possible cross-regulation of GA and ABA signals via GAMYB/ABI5 in the scutellum may be important for control of seed dormancy and germination.
GLPs are a group of proteins wide-spreading among plants. In rice, the GLP protein family contains 43 members. Although GLPs show a similar sequence to that of wheat germin, which is highly expressed during seed germination, GLPs show a variety of expression patterns and functions. For example, GLPs play important roles in plant disease resistance, including in rice (Manosalva et al., 2009; Davidson et al., 2010). Besides the similarity in protein structure, an additional common feature of germin and GLPs is their extracellular localization (Li et al., 2016; Liu et al., 2016). OsGLP2-1 is specifically expressed in the seed scutellum and minor veins of the leaf, together with the presence of ABRE and GARE binding motifs in the promoter; these results imply that interaction between ABI5 and GAMYB may be involved in the regulation of OsGLP2-1.
The present results indicated that OsGLP2-1 is involved in the ABA and GA signaling regulation of seed primary dormancy, and is directly regulated by the ABI5 and GAMYB transcription factors. Genetic analysis confirmed that the expression level of OsGLP2-1 in rice was positively correlated with premature seed dormancy. The expression of OsGLP2-1 is induced by ABI5 and suppressed by GAMYB, which is in accord with the process of seed development. During seed development, high quantities of ABA are synthesized to induce and maintain seed dormancy. With maturation of the seeds, ABA is degraded and dormancy is released by GAs. The OsGLP2-1 promoter contains both ABI5 and GAMYB binding motifs (ABRE and GARE). Furthermore, in a comparison of the binding activity of GAMYB and ABI5 to the PGLP2-1 promoter GAMYB showed relatively stronger specific binding activity in the in vitro assay. In the transient expression assay ABI5 displayed high transcription activity with the complete PGLP2-1 promoter, whereas in the yeast one-hybrid and EMSA assays ABI5 showed relatively weak binding activity. In yeast one-hybrid assay, ABI5 directly bound to the F1 segment with an ABRE core cis-element (ACGT), but the F2 fragment, which contained an ABRE and an ABRE-like element, failed to be activated by ABI5, and the ABRE flanking sequence, i.e. coupling elements previously reported to affect the binding activity in ABA responsive pathway (Hobo et al., 1999; Niu et al., 2002). Based on these results, we speculate there is competition balance between ABI5 and GAMYB in the regulation of OsGLP2-1 expression. Further investigation will help to explain how the two antagonistic signals ABA and GA, in a tissue-specific manner, coregulate the target genes to determine seed dormancy and germination during seed developing.
The signal for seed dormancy (or germination inhibitors) generally originates from the seed coat, endosperm, and embryo. It is necessary to investigate on which particular seed components OsGLP2-1 acts. Given that the dormancy phenotype of dehusked immature seeds is altered, a function for OsGLP2-1 in the seed coat is excluded. Comparison of the dormancy of isolated immature embryos and intact seeds showed that when compared with the vector control, only the intact OsGLP2-1 transgenic seeds exhibited significantly changed seed dormancy, whereas no significant variation was observed for the isolated embryos and the embryo-containing half-seed (Supplemental Fig. S8). These findings indicated that the control of dormancy by OsGLP2-1 may be associated with inhibition by the endosperm, but the mechanism requires further investigation.
Germin-like proteins are usually expressed during seed germination. To visualize the expression pattern of PGLP2-1:GUS on germinating seeds and seedlings, dry or germinating seeds and 7-d-old seedling were subjected to GUS staining. An identical scutellum-specific pattern of GUS expression was detected in the dry seeds and germinating seeds (Supplemental Figs. S1, A and B, S9B). After germination, the GUS signal changed to the vascular tissue of the seedling (Supplemental Figs. S1C and S9C). It is important to note that when compared with altered dormancy phenotypes of PGLP2-1:HA-GLP2-1-eGFP, amiR-GLP2-1, and osglp2-1 knock-out seeds displayed during developing-seed-stage, there was no obvious germination variation observed in after-ripened seeds. These results suggested the main function of OsGLP2-1 in dormancy-maintenance for developing seed. The PGLP2-1:GUS staining in germination seed indicated the expression of OsGLP2-1, but its potential role played in seed germination may relative weak. As previously reported, enhanced ABA content in imbibition seed failed to inhibit seed germination (Holdsworth et al., 2008b). Alternative possibility for the remaining expression of OsGLP2-1 during germination may act like seed-stored mRNAs. Some RNAs stored from seed maturation still existed in germinated seed. It is also found that posttranscriptional modification and posttranslational regulation mechanisms are involved in after-ripened seed germination (Holdsworth et al., 2008b; Gao et al., 2013). Collectively, seed development and maturation were dominated by the action of ABA, whereas seed germination was associated with multiple mechanisms.
Besides its function in seed dormancy, OsGLP2-1 might have additional functions in the plant. As we described above, during seedling and vegetative growth, OsGLP2-1 is expressed preferentially in minor veins of the leaf, and the subcellular localization differs from that of germins and all other investigated GLPs. OsGLP2-1 is localized within the cell, and the OsGLP2-1-eGFP fusion protein resembles a short needle or round object and shows rapid movement in the cytosol. Based on these results, we speculate that OsGLP2-1 might be a component of a transporter in the cell for nutrient uptake and transport in seed development, germination, seedling development, and plant growth, but much work is required to test this hypothesis.
Taken together, the present findings show that OsGLP2-1, a previously uncharacterized protein, is responsible for maintaining primary dormancy during seed developing stage, specifically in the scutellum. The transcription factors ABI5 and GAMYB, components of the ABA and GA signaling pathways, coregulate the expression of OsGLP2-1, and thereby maintain the dormancy of the developing seed. Further investigations, in particular on the subcellular localization of OsGLP2-1 and its interacting proteins, are needed to elucidate the molecular mechanism underlying its biological function in plants.
MATERIALS AND METHODS
Seed Germination and Treatment
Rice (Oryza sativa) ssp. japonica ‘Nipponbare’ seeds were used. The dehusked immature seeds were first surface-sterilized with 75% (v/v) ethanol for 2 min and then 30% (v/v) NaClO for 30 min, rinsed with double-distilled water, and sowed on MS salt mixture (phyto Technology Laboratories, catalog [cat.] no. M524) solid media, supplemented with either 50 μm ABA (Sigma, cat. no. A1049), or 1 or 10 μm GA (Sigma, cat. no. G7645). Germination was assessed at 3, 5, and 7 d after sowing. For comparison of embryo germination, two treatments were applied: (1) the embryo was isolated from 25 DAF dehusked seeds and cultured on one-half strength MS medium; and (2) immature seeds lacking the testa were cut into two halves either containing or lacking the embryo; only the embryo-containing half was used for germination. The seeds were cultured in a growth chamber at 25°C with a 12-h-light/12-hr-dark photoperiod.
Constructs
Rice genomic DNA was extracted from rice leaf with cetyltrimethylammonium bromide method (Murray and Thompson, 1980). The OsGLP2-1 promoter (PGLP2-1; 2,086 bp) was amplified using the promoter-specific primers C2-Pro-5-H3 and C2-pro3-Bgl (Supplemental Table S3). The PGLP2-1 PCR fragment was excised with HindIII and BglII, then cloned into the binary plasmid pCAMBIA-1300-SGN (digested with HindIII/BamHI) to generate the recombinant plasmid PGLP2-1:GUS containing the GUS report gene driven by PGLP2-1.
To generate the construct for overexpression of OsGLP2-1, the primer pair cup2Sxba and cup2RSac was used to amplify the cDNA containing the OsGLP2-1 ORF (OsGLP2-1-ORF). The OsGLP2-1-suppression construct was produced using an artificial microRNA method with the backbone sequence of OsmiR159a (Sunkar et al., 2005; Zhang et al., 2012). Based on the precursor sequence of OsmiR159a, the replacement specific target sequence of 5′-CTCAGTTCTGAAGTTTCTAAA-3′, spanning the second exon and 3′ untranslated region of the OsGLP2-1-cDNA, was amplified with the primer pair C2-5aS-Xba and C2-5aR-Sac. This fragment (amiR-GLP2-1) and the OsGLP2-1-ORF fragment were separately cloned into the plant expression vector pCAMBIA-1300-D4S with XbaI/SacI digestion to yield OsGLP2-1 overexpression or suppression vectors driven by the CaMV 35S promoter. The resulting constructs were designated OE-GLP2-1 and amiR-GLP2-1, respectively.
To generate the constructs for subcellular localization of OsGLP2-1-eGFP, the GLP2-1-ORF fragment was amplified with the primer pair HA-C2-SBam and C2-R-Sma to yield a 680-bp product, in which the HA-tag was fused at the N terminus of OsGLP2-1. The product was first digested with SmaI/BamHI, cloned into the 130-GFP plasmid to create the plasmid P35S:HA-GLP2-1-eGFP, then further digested with BamHI/HindIII; the PGLP2-1 promoter fragment (BglII/HindIII) was then inserted. Subsequently, the CaMV 35S promoter was substituted with PGLP2-1. The new construct was designated PGLP2-1:HA-GLP2-1-eGFP, in which the HA-GLP2-1-eGFP fusion protein was driven by the PGLP2-1 native promoter. Finally, the recombinant constructs P35S:HA-GLP2-1-eGFP and PGLP2-1:HA-GLP2-1-eGFP were used for observation of subcellular localization via transient and stable expression in the plant.
CRISPR-CAS9-Induced Mutation in the OsGLP2-1 Gene
The osglp2-1 mutant was developed using CRISPR/CAS9-based editing of OsGLP2-1 in rice by Biogle in 2016. Two targets in the second exon of OsGLP2-1 (cr-glp-323: 5′-GGAGAAGAAGAAGTCTTCCA-3′; cr-glp-513: 5′-GACCGAGATCCTGACTATAC-3′; Fig. 2A) were selected and the targeting specificity of target sequences (including protospacer adjacent motif) was confirmed using a BLAST search against the rice genome (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Hsu et al., 2013). The designed targeting sequences were synthesized and annealed to form the oligo adaptors, which were further inserted into pBGK032 vector digested by Bsa I, to produce CRISPR/CAS9 plasmids. Transformation of rice was carried out via Agrobacterium tumefaciens strain EHA105. The mutated osglp2-1 plant was identified by sequencing of PCR product with primers of C2-Cri-S/C2-Cri-R flanking the OsGLP2-1 genome. In order to obtain the T-DNA free osglp2-1 mutant, the crosses between wild-type plant and the T1 homozygous mutated plant were further performed. From F2 population plants, T-DNA–free osglp2-1 homozygous mutant was selected based on the absence of hygromycin band, while the presence of the osglp2-1 mutated site in a genomic DNA PCR detection.
Plant Stable Transformation
The plant expression constructs were introduced into A. tumefaciens strain EHA105 and transformed into rice via A.-mediated transformation (Hiei et al., 1994), and stable Arabidopsis (Arabidopsis thaliana; ecotype Col-0) transgenic plants were obtained by floral-dip transformation (Clough and Bent, 1998). For each construct, about 30 independent T0 transformants were obtained, and further screened by germinating seeds from T2 lines with 50 μg/mL hygromycin. The hygromycin-resistant homozygous seeds were used for further experiments. Seeds from homozygous lines transformed with the empty vector pCAMBIA 1300, or the construct harboring the GUS (or eGFP) gene driven by the CaMV 35S promoter, were used as a negative and positive control, respectively.
RT-qPCR
Freshly harvested immature rice seeds were ground to powder with the SPEX SamplePrep Geno/Grinder in liquid nitrogen. Total RNA was extracted following the method previously reported (Wang et al., 2012), and further treated with TURBO DNase according to the TURBO DNA-free Kit (Applied Biosystems, AM1907). cDNAs were produced using the Invitrogen superscript III First Strand Synthesis System (cat. no. 18080-051). RT-qPCR was performed with the SYBR Green Real-time PCR Master Mix (TOYOBO, QPK-201) to determine the relative expression level of OsGLP2-1 or GUS, using the primers listed in Supplemental Table S3. Rice ACTIN was used as an internal control.
GUS Staining
Staining of GUS activity was performed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as described previously (Gallagher, 1992). The GUS assay was first carried out with leaves from 14-d-old PGLP2-1:GUS transgenic rice seedlings, and with various organs (leaf, spikelet, internode, stem, leaf sheath, and seed) at the reproductive stage (25 DAF). Subsequently, the tissue-specific expression of GUS in minor veins of the leaf and in the scutellum with T3 plant were intensively examined. The assay was conducted on leaves at various growth stages (42, 52, 64, and 86 d after sowing), and on freshly harvested seeds at different development stages (17 and 35 DAF). To visualize the expression pattern of PGLP2-1:GUS during seed germination, dry or germinating seeds and 7-d-old seedling were also subjected to GUS staining. Briefly, dry seeds were imbibed in double-distilled water and incubated in a growth chamber at 25°C with a 12-h-light/12-hr-dark photoperiod. The germinated seeds (after 3 d of imbibition) stripped of the glume and 7-d-old seedlings were collected separately for GUS staining.
Transient Expression of OsGLP2-1 in Nicotiana benthamiana
To investigate the effect of transcription factors on the expression of OsGLP2-1, a transient expression assay was conducted in N. benthamiana. According to the potential cis-elements predicted by the plant cis-acting regulatory DNA elements database, the candidate genes of upstream of OsGLP2-1 were cloned. The cDNAs containing the ORF of different transcription factors were amplified from rice with appropriate primers (Supplemental Table S3), and cloned into the plant expression vector 1300-DAS downstream of the CaMV 35S promoter. For the reporter construct, the OsGLP2-1 promoter fragment (HindIII/BglII) was cloned into the plant vector 1300-intGUS (HindIII/BamHI) to generate the plasmid 130-PGLP2-1:intGUS in which PGLP2-1 drives the GUS report gene (intGUS). The construct 121-PNOS-Renilla luciferase (RiLuc) was used as an internal control. To avoid contamination of the reporter protein expressed from Agrobacterium, the reporter genes of both intGUS and RiLuc ORFs contained an intron. The plasmids were introduced into A. tumefaciens strain GV3101 for agroinfiltration.
Agroinfiltration of N. benthamiana was performed in accordance with a previous procedure with some modifications (Li et al., 2013). The freshly cultured single clones of GV3101 carrying the different plasmids were independently grown at 28°C overnight and then diluted by 1% (v/v) to continuous cultured until OD600 = 1.0, resuspended in MMA buffer (10 mm MgCl2, 10 mm MES, and 100 μm acetosyringone) to wash one time, resuspended in MMA, and then adjusted to OD600 = 1.5. The reporter, effector, and 121-PNOS-RiLuc constructs were mixed (2:3:1). Mixture of 130-PGLP2-1:intGUS and 121-PNOS-RiLuc (2:1) without the effector construct was used as a negative control, and the MMA buffer was used as a mock control. After agroinfiltration, the N. benthamiana plants were cultured in a greenhouse for 3 d; then leaves were sampled for RiLuc and GUS detection.
Yeast One-Hybrid Assay
Binding of the OsGLP2-1 promoter to the transcription factors ABI5 and GAMYB was detected with a yeast one-hybrid assay. PGLP2-1 fragments (F1: −932 to −637 [295 bp]; F2: −507 to −237 [270 bp]; F3: −932 to −237 [696 bp]; and F4: −1419 to −1 [1419 bp]) with predicted cis-elements were amplified by PCR with specific primers (Supplemental Table S3). The fragments were cloned into the pLacZi vector (Clontech) to generate the constructs pLacZi-PGLP2-1, −F1, −F2, −F3, and −F4, respectively. To confirm binding activity, duplicated fragments of 2×F1, 3×F1, and 2×F2, 3×F2 were constructed. The ORF fragments of the ABI5 and GAMYB effectors were separately cloned into the pGAD424 vector (Clontech). The pLacZi-Fn::LacZ series reporter gene plasmids and pGAD424-ABI5 or pGAD424-GAMYB constructs were cotransformed into Saccharomyces cerevisiae strain EGY48. The transformants were selected on synthetic complete medium lacking Ura and Leu.
EMSA
The MBP-ABI5 fusion protein was expressed in the pMAL-p2X vector (BioLabs, cat. no. E8000S) and purified in accordance with the manufacturer’s instructions. The specific primer pair AB5-S-Bam and AB5-R-H3 was used to amplify the ABI5-ORF fragment, which was cloned into the pMAL-p2X vector (BamHI/HindIII). The Trx-His-GAMYB fusion protein was constructed in the pET32a (+) expression vector via BamHI/HindIII digestion. Escherichia coli strain BL21 (DE3) pLysS cell harboring the fusion constructs was induced by 0.2 mm isopropylthio-β-galactoside at 16°C overnight (about 18 h) at 150 rpm. Cells expressing the ABI5 or GAMYB fusion protein were collected and lysed with a cell disrupter (JNBIO, JN-mini) at 1000 bar on ice. The recombinant proteins MBP-ABI5 and Trx-His-GAMYB were purified with Amylose Resin (BioLabs, cat. no. E8021L) and Ni Sepharose excel (GE Healthcare, cat. no. 17-3712-02), respectively, and then applied to the EMSA using the Chemiluminescent Nucleic Acid Detection Module Kit (Prod. no. 89880). The GARE- or ABRE-containing probes were separately labeled with biotin by Thermo Fisher Scientific (Fig. 6). The unlabeled probe was used as a competitive probe, and in the mutated probe the corresponding cis-element was changed. To confirm the binding activity of ABI5 and GAMYB to the corresponding cis-elements of GARE and ABRE in the PGLP2-1 promoter, different amounts of MBP-ABI5 (0, 0.3, 0.5, 1.0, and 1.5 μg) and Trx-His-GAMYB (0.05, 0.1, 0.3, 0.5, and 1.0 μg) fusion proteins were separately subjected to the EMSA. The expressed protein of MBP and Trx-His from the corresponding empty vector of pMAL-p2X and pET-32a (+), respectively, was used as a negative control.
Microscale Thermophoresis Assay
Protein-DNA binding experiments were performed with a Monolith NT.115 instrument (NanoTemper Technologies; www.nanotemper-technologies.com). The Trx-His-ABI5 fusion protein expressed from pET32a (+) in E. coli was further induced as described above. The recombinant proteins of Trx-His-ABI5 and Trx-His-GAMYB were purified using imidazole-gradient purification in Ni-NTA agarose (Qiagen). The OsGLP2-1 promoter fragments (A5-1: 5′-CCGGAGAAAAATGCACGTAAGGAAATAATAAT-3′ located at −805 to −774; A5-2: 5′-GCAGGCAGGCCACAACACGTTTATTCAGACTT-3′ at −311 to −280; and Ga-1: 5′-ACAGAATTTGTTAACCGAACGACGAACGCGTAATGCATA-3′ at −496 to −458, the underlined bases indicate ABRE or GARE element) were, respectively, synthesized and labeled with 5-carboxyfluorescein (FAM) at the 5′ end (Thermo Fisher Scientific). The labeled FAM-A5-1, FAM-A5-2, or FAM-Ga-1 fragment with 16 titrations of Trx-His, Trx-His-ABI5, or Trx-His-GAMYB proteins were loaded onto standard treated silicon capillaries (MO-K002 Monolith NT.115; NanoTemper Technologies), and fluorescence was measured. The experiment was accompanied by a step-by-step gradient dilution process. The optimized measuring parameters of instrument were as follows: light emitting diode power 20%; MST power 20% for Trx-His-ABI5 and 80% for Trx-His-GAMYB; fluorescence before time, 5 s; MST on time, 30 s; fluorescence after time, 5 s; delay time, 25 s. Data were analyzed using software MO. Affinity Analysis v2.2.4 (Nano Temper Technologies).
Confocal Microscopy
Leaf samples from the transient N. benthamiana, seedling of stable transgenic Arabidopsis, and rice plants were observed with confocal microscope (Leica TCS SP8 and UltraView Vox).
Statistical Analyses
Statistical analysis was performed using independent-sample t test in Statistical Package for Social Science (SPSS) for two groups comparison and one-way ANOVA post hoc tests for more than two groups comparison.
Accession Numbers
Rice genomic sequence data from this article can be found in the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu) under the following accession numbers: OsGLP2-1 (LOC_Os02g29000), ABI5 (LOC_Os01g64000), GAMYB (LOC_Os01g59660), ABL1 (LOC_Os6g10880), DOF2 (LOC_Os02g15350), DOF7 (LOC_Os07g32510), and DOF9 (LOC_Os09g29960).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GUS staining of PGLP2-1:GUS transformant during seed germination.
Supplemental Figure S2. Transcript levels of OsGLP2-1 in the leaf of OE-GLP2-1 and amiR-GLP2-1 rice detected by RT-qPCR.
Supplemental Figure S3. Dormancy phenotype of immature seeds of amiR-GLP2-1 rice at 9 d after sowing.
Supplemental Figure S4. Germination rate of OE-GLP2-1 and amiR-GLP2-1 seeds with hormone treatment.
Supplemental Figure S5. Influence of the transcription factors on the relative GUS activities (GUS/RiLuc) controlled by PGLP2-1.
Supplemental Figure S6. Protein concentration gradient assay in EMSA for MBP-ABI5 and Trx-His-GAMYB recombinant proteins.
Supplemental Figure S7. Subcellular localization of OsGLP2-1-eGFP.
Supplemental Figure S8. Percentage germination of OE-GLP2-1 and amiR-GLP2-1 rice seeds on one-half strength MS solid medium.
Supplemental Figure S9. GUS staining of developing seed, embryo and etiolated seedling of PGLP2-1:GUS transformant.
Supplemental Table S1. Percentage germination of amiR-GLP2-1 transgenic seeds (35 DAF) on MS medium.
Supplemental Table S2. Percentage germination of amiR-GLP2-1 transgenic seeds (35 DAF) on MS medium supplemented with 50 μm ABA.
Supplemental Table S3. Primers used in this study.
Supplemental Movie S1. The dynamic movement of the needle-shape green fluorescence in P35S:HA-GLP2-1-eGFP rice seedling leaf.
Supplemental Movie S2. The mobile round green fluorescent signal of GLP2-1-eGFP in 3-d-old seedling root.
Acknowledgments
We are grateful to Zhaosheng Kong, Dr Lei Su, and Yao Wu (Institute of Microbiology, Chinese Academy of Sciences) for technical assistance with confocal microscopy and microscale thermophoresis assay; Fen Tan (Institute of Microbiology, Chinese Academy of Sciences) for rice transformation; and Hangzhou Biogle for CRISPR-osglp2-1 construction and transformation.
Footnotes
This work was supported by grants from the National Transgenic Science and Technology Program (grant no. 2016ZX08010–002) and the National Natural Science Foundation of China (grant no. 91217304).
References
- Abraham Z, Iglesias-Fernández R, Martínez M, Rubio-Somoza I, Díaz I, Carbonero P, Vicente-Carbajosa J(2016) A developmental switch of gene expression in the barley seed mediated by HvVP1 (Viviparous-1) and HvGAMYB interactions. Plant Physiol 170: 2146–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN(2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28: 500–512 [DOI] [PubMed] [Google Scholar]
- Aoki N, Scofield GN, Wang XD, Offler CE, Patrick JW, Furbank RT(2006) Pathway of sugar transport in germinating wheat seeds. Plant Physiol 141: 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T, Hirose S, Asatsuma S, Nanjo Y, Nakaizumi T, Itoh K, Hori H, Komatsu S, Mitsui T(2007) Proteomic characterization of tissue expansion of rice scutellum stimulated by abscisic acid. Biosci Biotechnol Biochem 71: 1260–1268 [DOI] [PubMed] [Google Scholar]
- Berna A, Bernier F(1997) Regulated expression of a wheat germin gene in tobacco: Oxalate oxidase activity and apoplastic localization of the heterologous protein. Plant Mol Biol 33: 417–429 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung YY, Still DW, Jones RL(2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143: 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen J, Bellgard M(2010) Germin-like proteins (GLPs) in cereal genomes: Gene clustering and dynamic roles in plant defence. Funct Integr Genomics 10: 463–476 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Manosalva PM, Snelling J, Bruce M, Leung H, Leach JE(2010) Rice germin-like proteins: allelic diversity and relationships to early stress responses. Rice (N Y) 3: 43–55 [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martínez M, Isabel-La Moneda I, Carbonero P(2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 29: 453–464 [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S(2004) Cupins: The most functionally diverse protein superfamily? Phytochemistry 65: 7–17 [DOI] [PubMed] [Google Scholar]
- Fang J, Chai C, Qian Q, Li C, Tang J, Sun L, Huang Z, Guo X, Sun C, Liu M, et al. (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chu C(2008) Abscisic acid and the pre-harvest sprouting in cereals. Plant Signal Behav 3: 1046–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G(2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Fu ZY, Zhao Z, Qin YT, Xu MM, Chen YQ, Tang JH(2016) Functional marker related to germination vigor of maize seed. Mol Breed 36: 159 [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK(2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR.(1992) GUS protocols: using the GUS gene as a reporter of gene expression. Academic Press, San Diego [Google Scholar]
- Gao F, Ayele BT(2014) Functional genomics of seed dormancy in wheat: Advances and prospects. Front Plant Sci 5: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Rampitsch C, Chitnis VR, Humphreys GD, Jordan MC, Ayele BT(2013) Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotechnol J 11: 921–932 [DOI] [PubMed] [Google Scholar]
- Gu XY, Liu T, Feng J, Suttle JC, Gibbons J(2010) The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol Biol 73: 97–104 [DOI] [PubMed] [Google Scholar]
- Gu XY, Zhang JF, Ye H, Zhang LH, Feng JH(2015) Genotyping of endosperms to determine seed dormancy genes regulating germination through embryonic, endospermic, or maternal tissues in rice. G3 (Bethesda) 5: 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV(1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV(1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna S(1995) Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: Analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J 7: 913–925 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T(1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T(1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM.(1995) A critical update on seed dormancy. 1. Primary dormancy. Seed Sci Res 5: 61–73 [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T(1999) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19: 679–689 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ(2008a) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D(2008b) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13: 7–13 [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA(2004) Cotton fiber germin-like protein. I. Molecular cloning and gene expression. Planta 218: 516–524 [DOI] [PubMed] [Google Scholar]
- Lapik YR, Kaufman LS(2003) The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein alpha-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15: 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J(2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xu X, Chen C, Shen Z(2016) Genome-wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis. Int J Mol Sci 17: 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Gao F, Kanno Y, Jordan MC, Kamiya Y, Seo M, Ayele BT(2013) Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS One 8: e56570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yang J, Yan S, Zhang S, Zhao J, Wang W, Yang T, Wang X, Mao X, Dong J, et al. (2016) The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol Biol 92: 411–423 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ye N, Liu R, Chen M, Zhang J(2010) H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J Exp Bot 61: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH(2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Mahmood T, Nazar N, Abbasi BH, Naqvi SMS(2010) Comparative analysis of regulatory elements in different germin-like protein gene promoters. Afr J Biotechnol 9: 1871–1881 [Google Scholar]
- Manosalva PM, Davidson RM, Liu B, Zhu X, Hulbert SH, Leung H, Leach JE(2009) A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol 149: 286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz B, Müller K, Kucera B, Volke F, Leubner-Metzger G(2005) Water uptake and distribution in germinating tobacco seeds investigated in vivo by nuclear magnetic resonance imaging. Plant Physiol 138: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membré N, Berna A, Neutelings G, David A, David H, Staiger D, Sáez Vásquez J, Raynal M, Delseny M, Bernier F(1997) cDNA sequence, genomic organization and differential expression of three Arabidopsis genes for germin/oxalate oxidase-like proteins. Plant Mol Biol 35: 459–469 [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Nakata E, Nagato Y, Hattori T(1999) Differential in situ expression of three ABA-regulated genes of rice, RAB16A, REG2 and OSBZ8, during seed development. Plant Cell Physiol 40: 443–447 [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G(2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF(1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, et al. (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23: 3215–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A(2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ(2002) Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell 14: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E(2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Salmenkallio M, Sopanen T(1989) Amino acid and peptide uptake in the scutella of germinating grains of barley, wheat, rice, and maize. Plant Physiol 89: 1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA(2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Zheng N(2009) Plant biology: Signal advance for abscisic acid. Nature 462: 575–576 [DOI] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q(2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Girke T, Jain PK, Zhu JK(2005) Cloning and characterization of microRNAs from rice. Plant Cell 17: 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y(2012) Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol 53: 107–117 [DOI] [PubMed] [Google Scholar]
- Toorop PE, Cuerva RC, Begg GS, Locardi B, Squire GR, Iannetta PPM(2012) Co-adaptation of seed dormancy and flowering time in the arable weed Capsella bursa-pastoris (shepherd’s purse). Ann Bot 109: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang G, Zhang X, Wang F, Song R(2012) Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem Anal 23: 159–163 [DOI] [PubMed] [Google Scholar]
- Wang M, Li W, Fang C, Xu F, Liu Y, Wang Z, Yang R, Zhang M, Liu S, Lu S, et al. (2018) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 50: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Wang Z, Cao H, Sun Y, Li X, Chen F, Carles A, Li Y, Ding M, Zhang C, Deng X, et al. (2013) Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25: 149–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H(2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22: 1733–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang YN, Xue LJ, Zou MJ, Liu JY, Chen F, Xue HW(2011) Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol 156: 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Yan YS, Wang LN, Yang K, Xiao N, Liu YF, Fu YP, Sun ZX, Fang RX, Chen XY(2012) A novel rice gene, NRR responds to macronutrient deficiency and regulates root growth. Mol Plant 5: 63–72 [DOI] [PubMed] [Google Scholar]
- Zhao X, Dou L, Gong Z, Wang X, Mao T(2019) BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol 221: 908–918 [DOI] [PubMed] [Google Scholar]
- Zhu JK.(2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F(2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66: 675–683 [DOI] [PubMed] [Google Scholar]