Abstract
Phagocytosis, or ‘cell eating’, is a eukaryote-specific process where particulate matter is engulfed via invaginations of the plasma membrane. The origin of phagocytosis has been central to discussions on eukaryogenesis for decades, where it is argued as being either a prerequisite for, or consequence of, the acquisition of the ancestral mitochondrion. Recently, genomic and cytological evidence has increasingly supported the view that the pre-mitochondrial host cell—a bona fide archaeon branching within the ‘Asgard’ archaea—was incapable of phagocytosis and used alternative mechanisms to incorporate the alphaproteobacterial ancestor of mitochondria. Indeed, the diversity and variability of proteins associated with phagosomes across the eukaryotic tree suggest that phagocytosis, as seen in a variety of extant eukaryotes, may have evolved independently several times within the eukaryotic crown-group. Since phagocytosis is critical to the functioning of modern marine food webs (without it, there would be no microbial loop or animal life), multiple late origins of phagocytosis could help explain why many of the ecological and evolutionary innovations of the Neoproterozoic Era (e.g. the advent of eukaryotic biomineralization, the ‘Rise of Algae’ and the origin of animals) happened when they did.
Keywords: eukaryogenesis, Asgard archaea, phagocytosis, eukaryovory, Rise of Algae, Neoproterozoic
1. Introduction
The origin of eukaryotes is one of the most daunting and captivating problems in all biology, and was one of the foremost events in the history of life on Earth. One of the primary traits separating eukaryotes from bacteria and archaea is the ability of eukaryotes to internalize entire prey cells via phagocytosis (see glossary for explanation of terms) [1]. The origin of phagocytosis in eukaryotes set the stage for modern food webs, as phagotrophic protists serve as one of the primary bridges connecting the ‘microbial loop’ with classical, animal-containing food chains, and are key regulators of carbon remineralization, nutrient retention, and primary productivity in marine, freshwater, and soil ecosystems [2–4]. Phagocytosis was also a prerequisite for the origin of animal multicellularity, where it is involved in nutrition, embryogenesis, tissue remodelling and immunity [5–8]. Recent palaeontological, geochemical and molecular clock evidence has suggested that various forms of eukaryotic predation may have helped drive certain global environmental and ecological changes during the Neoproterozoic Era (1000–541 million years ago, or Ma)—from the proliferation of marine algae to the origin of animals and other multicellular clades. Why eukaryotic predation became ecologically widespread at this time in Earth history remains unclear, and raises the question of when phagocytosis itself evolved—a question central to the origin of eukaryotes. Recent discoveries of novel archaeal lineages closely related to eukaryotes, equipped with proteins essential to phagocytosis and previously thought to be unique to eukaryotes, have revitalized debates on when phagocytosis first evolved. These two active and ongoing discussions—the origin of phagocytosis in the context of eukaryogenesis, and the role of eukaryotic predation in the Neoproterozoic Earth system—have largely occurred independently of one another in the literature. In this paper, I review both of these topics and argue that when and how phagocytosis first evolved in the context of eukaryogenesis can greatly inform our understanding of the earliest geologic evidence for eukaryotic predation—and vice versa.
2. The earliest evidence for eukaryotic predation
2.1. Palaeontological and molecular evidence
The oldest evidence of eukaryotic predation in the rock record arguably comes from the earliest fossil algae [9]. If the primary endosymbiotic origin of plastids from cyanobacteria in the ancestors of the Archaeplastida—the eukaryotic supergroup containing glaucophytes, red algae and green algae [10]—occurred via phagocytosis, as the morphology and phagocytic behaviour of certain early-branching green algae arguably suggests [11,12], then fossil Archaeplastida would serve as indirect evidence for bacterivory. Likewise, fossil taxa belonging to lineages that acquired plastids from algae via secondary or tertiary endosymbiosis—e.g. stramenopiles and diatoms, respectively [13]—would serve as indirect evidence for eukaryovory [14]. The only other known example of primary plastid acquisition directly from cyanobacteria (and not from algae)—Paulinella chromatophora [15]—likely occurred via phagocytosis [16], but only 140–90 Ma [17], and therefore cannot be considered among the earliest evidence for bacterivory in the rock record. The oldest widely accepted evidence for primary-plastid-containing algae in the fossil record is Bangiomorpha pubescens [18,19], a likely fossil red alga first appearing in the ca 1.05 billion-year-old (Ga) Angmaat Formation of northeastern Canada [20,21]. Older, more equivocal candidates for the earliest archaeplastid fossils exist, namely putative crown-group red algae from the ca 1.6 Ga Chitrakoot Formation of central India [22], and potential stem-group red or green algae (or stem-archaeplastids) from the 1.56 Ga Gaoyuzhuang Formation of North China [23]. If these older taxa indeed represent fossil Archaeplastida, then the oldest indirect evidence for bacterivory could be pushed back over 500 Myr to 1.60–1.56 Ga. If these older fossil taxa instead represent photosynthetic stem-group eukaryotes, which has also been proposed [21,23], then they could still potentially serve as indirect evidence for bacterivory, although determining plastid capture via phagocytosis (and by extension bacterivory) in these unknown stem-lineages would be difficult to determine [12]. Recent molecular clock estimates for the last common ancestor (LCA) of Archaeplastida yield 95% credibility intervals of 2.12–1.69 Ga [24] and 1.67–1.12 Ga [25], while analyses including the most recent age constraints on B. pubescens as fossil calibration points suggest that primary plastid acquisition in the Archaeplastida occurred by 1.37–1.14 Ga [21]. Given these uncertainties, it remains reasonable that the earliest evidence for crown-group Archaeplastida dates to ca 1.05 Ga in the rock record, with molecular clock estimates for the first primary origin of plastids dating to ca 1.25 Ga [21]. This fossil and molecular evidence serves as indirect evidence for bacterivory, insofar as the ancestral plastid in stem-group Archaeplastida was truly acquired via phagocytosis by a wall-less host—an assumption that may not have been the case [1,12,26]. Lastly, with respect to symbiont capture more generally, while some consider phagocytosis a prerequisite to the origin of mitochondria [27,28]—implying bacterivory among primitively amitochondriate stem-group eukaryotes necessarily predated crown-group eukaryotes—this scenario is critically examined in the following section.
Outside of fossil algae, the oldest fossil evidence for eukaryotic predation currently comes from the ca 1150–900 Ma lower Shaler Supergroup in Arctic Canada in the form of ovoid and circulation perforations preserved in the organic walls of diverse eukaryotic microfossils [29]. These perforations, which broadly resemble the holes made by modern myzocytotic and protoplast-feeding predators as they puncture and piece their prey [30,31], are interpreted as direct evidence for eukaryovory [29,32]. Similar perforations are also found in walls of younger organic-walled eukaryotic microfossils (the vase-shaped microfossils, or VSMs) from the 780 to 740 Ma Chuar Group, Grand Canyon, Arizona, USA [30,31], which are also widely interpreted as the direct result of eukaryovory [9,29,32–34]. These VSMs—and others from comparably aged assemblages ca 789–729 Ma [35,36]—are interpreted as representing the oldest fossil evidence for arcellinid testate amoebae [37,38], which are abundant and diverse in modern freshwater and soil habitats, where they are largely bacterivorous and eukaryovorous [39–41]. Therefore, the VSMs themselves—with or without perforations—also serve as evidence for bacterivory and eukaryovory by ca 789 Ma [9,30,33,34,37,38]. The oldest evidence for biologically controlled eukaryotic biomineralization—the ca 810-million-year-old apatitic scale microfossils (ASMs) from the Fifteenmile Group in Yukon, Canada—has also been interpreted as indirect evidence for eukaryovory in the early Neoproterozoic [32,42]. This conclusion is based on the reasoning that these apatitic scales may have been selected for their deterrence of piercing and/or ingestion by predatory protists [43], analogous in function to the silica frustules and chitinous threads of modern diatoms [44–47]. Together, fossil evidence suggests that something like protoplast feeding or myzocytosis was an active feeding strategy by ca 1150–900 Ma [29], that a modern bacterivorous and eukaryovorous lineage (i.e. the arcellinid testate amoebae) had diverged by ca 789–759 Ma [37,38], and that armour potentially adapted to deter or resist eukaryovory had evolved by ca 810 Ma [42]—collectively serving as the oldest evidence for eukaryotic predation in the fossil record [32] (figure 1).
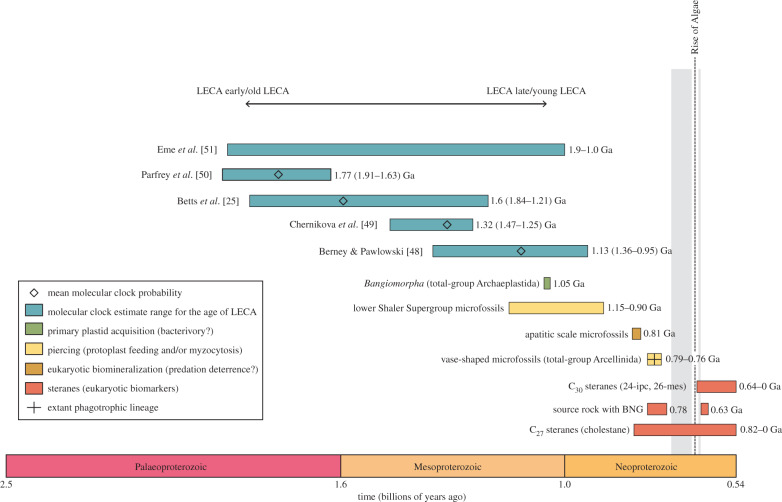
Figure 1.
Timeline of palaeontological, organic geochemical and molecular clock evidence regarding the age of crown-group eukaryotes and the earliest evidence for eukaryotic predation. The Sturtian (717–660 Ma) and the Marinoan (640–635 Ma) glaciations are each shown as vertical grey bars. For the molecular clock estimates, the bars display the highest credibility intervals (95%) for the age of LECA, with the diamonds indicating the mean age estimates from each analysis. For Berney & Pawlowski [48], these values were obtained from fig. 1 (node 1). For Chernikova et al. [49], these values were obtained from the first set of values reported in table 2, where Bangiomorpha—then dated to 1.2 Ga—was used a fossil calibration constraint. The values from Parfrey et al. [50] come from fig. 2, listed in table S1 under analysis ‘a’, while the values from Betts et al. [25] come from fig. 3. The range reported for Eme et al. [51] covers a variety of analyses exploring the impact of using different tree topologies, fossil calibration constraints and substitution models on estimating the age of LECA—hence no reported mean estimate for this overall range. Given that modern type phagocytosis may not have facilitated the capture of the ancestral plastid [1,26], Bangiomorpha may or may not serve as indirect evidence for bacterivory—hence the question mark in the key. While the lower Shaler Supergroup microfossils and the vase-shaped microfossils both preserve perforations likely sourced from eukaryotic predators, it is unclear if these punctures resulted from protoplast feeding, myzocytosis, or some other form of predatory piercing. While the apatitic scale microfossils are reasonably interpreted as having been adapted to deter predation, the relatively indirect nature of this evidence, as discussed in the main text, is reflected by the question mark in the key.
The appearance of VSMs and ASMs in the fossil record broadly correlates with an apparent increase in the taxonomic richness of eukaryotic microfossils ca 800–750 Ma [33,34]—although this trend may represent a sampling artefact [52]. Molecular clock estimates have also suggested that the major eukaryotic clades began diversifying ca 800 Ma [33,50]. As the VSMs and ASMs have both been interpreted as some of the earliest evidence for eukaryovory, either the origin or expansion of eukaryovory has been invoked as an ecological mechanism for this Neoproterozoic diversification of eukaryotes [9,32–34]. Indeed, Stanley originally proposed that the origin of bacterivory (the crossing of the ‘heterotroph barrier’), quickly followed by the origin of eukaryovory, during the late Proterozoic (2.5–0.541 Ga) actively promoted diversification at multiple trophic levels through the introduction of novel predator–prey interactions [53]. Out of this diversification, animals and macroalgae emerged and perpetuated these dynamics on larger spatial scales—with animals stimulating the diversity of macroalgae via grazing—thereby setting the stage for the Phanerozoic (0.541–0 Ga). Similarly, the origin or expansion of eukaryovory in the Neoproterozoic has been invoked to explain this apparent increase in eukaryotic microfossil richness, as well as the origin of animals and other multicellular eukaryotic clades. Indeed, it has been argued that the unicellular ancestors of these multicellular groups may have escaped predatory engulfment by increasing their size via coloniality and simple multicellularity [33,34,54]. Support for this ecological driver for the origin of animal multicellularity comes, in part, from recent molecular clock estimates placing the origin of crown-group animals near 800 Ma [55–58], coincident with the early Neoproterozoic diversification of eukaryotes and the appearance of VSMs and ASMs in the rock record [33,34]. Further support for the role of eukaryovory during the Neoproterozoic diversification of eukaryotes comes from the reconstruction of the ancestral feeding mode of many major eukaryotic clades, which, coupled with molecular clock estimates [50,51,59], suggests that many ancestrally eukaryovorous groups, like the foraminifera and ciliates, diverged in the early-to-mid Neoproterozoic [34]. Overall, ecological theory, the reconstruction of ancestral feeding modes across the eukaryotic tree, and molecular clock analyses have all been used to argue that eukaryovory drove eukaryotic diversification, including multiple origins of eukaryotic multicellularity, in the early Neoproterozoic, explaining the correlation between the appearance of VSMs and ASMs in the rock record and the increase in the taxonomic richness of eukaryotic microfossils ca 800 Ma [34].
2.2. Organic geochemical evidence
In addition to the fossil and molecular records, the organic geochemical (biomarker) record has been used to argue for the presence and importance of eukaryotic predation in the mid-Neoproterozoic. The oldest currently recognized biomarkers of eukaryotic origin (i.e. steranes, the geologically stable forms of eukaryotic sterols) date to 820–720 Ma, predominantly in the form of cholestane (C27) with traces of ergostane (C28) and no detectable stigmastane (C29) [60]. This 100:0:0% distribution of the three most abundant steranes in the rock record (cholestane, ergostane and stigmastane, respectively) has been interpreted to represent a primarily heterotrophic eukaryotic source with potential—and perhaps major [61]—contributions from red algae (rhodophytes, belonging to the Archaeplastida supergroup) [60,62]. Rocks from this time interval also preserve the C28 sterane, 26-methylcholestane (or cryostane), which has been interpreted as being derived from 26-methylsterols potentially protective against membranolytic toxins released by other eukaryotes either as a method of, or defence against, predation [63]. While this interpretation further emphasizes the role of eukaryovory at this time—especially considering that cryostane is recovered from the Chuar Group, where the VSMs were originally discovered—any interpretation on the origin of cryostane remains speculative, as its natural precursor sterols remain unidentified in modern taxa [64–66]. The shift to more modern sterane distributions, as well as higher ratios between steranes and hopanes (biomarkers derived from bacterial bacteriohopanepolyols), apparently occurred 659–645 Ma, between the two Snowball Earth events of the Cryogenian Period (720–635 Ma), signalling a proliferation of planktonic marine archaeplastids, particularly chlorophytes, known as the ‘Rise of Algae’ [60,62]. While the mechanisms underlying this transition remain uncertain, bacterivory on cyanobacteria has been invoked to explain how the incumbency of cyanobacteria in marine ecosystems could have been broken for the first time, actively promoting a greater relative abundance of planktonic algae [60,67], similarly to Stanley's ecological model for the Proterozoic–Phanerozoic transition [53].
More recently, additional biomarker evidence has been used to argue for the role of bacterivory in promoting algal proliferation in the Ediacaran (635–541 Ma). Immediately following the Marinoan glaciation (649–635 Ma), sterane distributions suggest a return to algal-lean, cyanobacterial-dominated ecosystems before shifting again to algal-rich conditions—the ‘Ediacaran Rise of Algae’ [60,68]. The recently described biomarker 25,28-bisnorgammacerane (BNG)—abundant in the post-Marinoan cap dolostones of the Araras Group in Brazil—has been interpreted to reflect the microbially degraded remains of ciliate biomass from redox-stratified, bacterially dominated ecosystems [68]. The decrease and ultimate disappearance of BNG correlated with increasing sterane/hopane ratios in the earliest Ediacaran potentially reflects active bacterivory by ciliates [68], a lineage of predatory protists belonging to the Alveolata supergroup [10]. Selective predation on bacteria by ciliates may have decreased cyanobacterial concentrations, reduced water column turbidity, and increased nutrient availability, overall resulting in a proliferation of marine planktonic algae and the breaking of the self-sustaining, cyanobacterially dominated ecosystems of the earliest Ediacaran [68]. This argument has been reiterated by the controversial interpretation of 24-isopropylcholestane (24-ipc) and 26-methylstigmastane (26-mes)—two C30 steranes that appear in rock record by 640 Ma and are generally considered the remains of demosponges [64,69,70]—as the remains of Rhizaria [66], a eukaryotic supergroup containing primarily heterotrophic (i.e. bacterivorous and eukaryovorous) amoebae [10]. This particular (and, again, controversial) interpretation of 24-ipc and 26-mes from the Cryogenian and Ediacaran has also been used to argue for the importance of eukaryotic predation in promoting algal proliferation through the selective feeding on bacteria by rhizarians [66]. Taken together, the abundance and distribution of BNG, 24-ipc, and 26-mes in Neoproterozoic rocks have been used to argue for the presence and ecological importance of both ciliates and rhizarians (two phagotrophic lineages belonging to the SAR clade) during the ecological transition to more algal-rich conditions in the late Cryogenian and early Ediacaran [66,68] (figure 1).
2.3. Eukaryotic predation and the Neoproterozoic Earth system
Eukaryotic predation has been invoked to explain a number of events in the Neoproterozoic, from the Rise of Algae to the origin of animals [33,34,60,66–68]. At the same time, the last eukaryotic common ancestor (LECA) is generally thought to have arisen by 1.6 Ga [14,50], and is commonly reconstructed as a bacterivorous flagellate [71,72]. If both of these conclusions are true, then an 800 Myr gap exists between the inferred origin of bacterivory and the appearance of definitive geological evidence for eukaryotic predation. What could possibly explain this gap? Oxygen limitation has been invoked to explain the gap between bacterivory (ca 1.9–1.6 Ga) and eukaryovory (ca 800 Ma) [34,50]—comparable to the observed relationship between oxygen concentration and animal-on-animal predation in modern marine oxygen minimum zones [73]. Similarly, phosphate limitation and low bacterial prey densities have been invoked to explain why bacterivory did not become ecologically ‘sustained’ until the Cryogenian [60]. Are these two arguments compatible with one another, or two separate explanations inspired by two different datasets? That is, why would eukaryovory take off ca 800 Ma in response to oxygenation before bacterivory took off ca 659–645 Ma in response to enhanced nutrient levels? Especially when eukaryovory demonstrably occurs under anoxia [74], and bacterivory demonstrably persists in oligotrophic settings [75]. The timing and mechanisms of these two different narratives disagree with one another, and arguably fail to work on their own. Meanwhile, other narratives very clearly leave the mid-Neoproterozoic proliferation of bacterivory a mystery, attributing it to neither oxygenation nor nutrients [67], while others seem to overlook this temporal gap altogether, invoking an unexplained expansion of bacterivory in the Cryogenian and Ediacaran [66,68]. These uncertainties and discrepancies raise important questions—namely, are we confident phagocytosis (and bacterivory) evolved before 1.6 Ga and that such a long temporal gap, spanning well over half a billion years, indeed exists and needs explaining? Answering these questions depends on dating LECA and determining its phagocytic capacities.
3. Phagocytosis and eukaryogenesis
3.1. Models of eukaryogenesis
The distinction between eukaryotic cells from the cells of bacteria and archaea has been called ‘the greatest single evolutionary discontinuity to be found in the present-day living world’ [76]. One of the major traits separating eukaryotes from bacteria and archaea is the widespread ability of eukaryotes to phagocytose. There is only one described example of a bacterium—the planctomycete ‘Candidatus Uab amorphum’—engaging in behaviour approaching phagocytosis [77]. However, planctomycetes possess a definitely Gram-negative cellular organization, incompatible with true endocytic invaginations of the outer membrane [78]. Meanwhile, behaviour resembling phagocytosis is currently unobserved in archaea [1]. The origin of phagocytosis itself is generally treated as either a prerequisite for, or consequence of, the acquisition of the ancestral mitochondrion (in what are known as ‘mitochondria-late’ and ‘mitochondria-early’ models of eukaryogenesis, respectively) [1,79]. The timing, or relative ordering, of mitochondrial acquisition (i.e. early versus late, first versus last) is often coupled (although not necessarily so) to the mechanism of mitochondrial acquisition, as well as the affinity and nature of the host cell [80]. As such, mitochondria-late scenarios are also called ‘phagotrophic’ models, where an ostensibly eukaryotic (or ‘protoeukaryotic’, neither archaeal nor bacterial) host cell, already equipped with phagocytic machinery and other eukaryote-specific traits (acquired ‘autogenously’, through point mutation and natural selection prior to the origin of mitochondria), engulfed the ancestral mitochondrion via phagocytosis [1,71,80,81]. By contrast, mitochondria-early scenarios are also called ‘syntrophic’ models, where at least two kinds of prokaryotic cells (generally an archaeal host and a bacterial symbiont, the ancestral mitochondrion), metabolically dependent on one another via anaerobic syntrophy, became integrated into one cell (hence the additional label of ‘fusion’ models) with many, if not all, defining eukaryotic traits, like phagocytosis, evolving afterward (‘endosymbiotically’, as a result of mitochondrial acquisition) [1,71,80–82]. While virtually all researchers agree that mitochondria descend from free-living α-proteobacteria, and that LECA definitively possessed mitochondria (from which mitosomes and hydrogenosomes descend), the precise nature and affinity of the host cell and the relative timing of the major events of eukaryogenesis prior to LECA remain active areas of research [71,80,83,84]. Namely, was the host cell already capable of phagocytosis, even at a rudimentary level, and did this ability mediate the acquisition of the ancestral mitochondrion? Or was the host cell strictly non-phagocytic, with phagocytosis evolving sometime after (and as a result of) mitochondrial acquisition?
3.2. Archaea and phagocytosis
In recent years, phylogenetic analyses have increasingly supported what is called the ‘two primary domain’ (2D) scenario for the tree of life [85–93]. In this scenario, the eukaryotic nuclear lineage (i.e. the lineage belonging to the host cell that acquired the α-proteobacterial symbiont) branches within the Archaea, relegating Eukarya to the status of a ‘secondary’ domain, formed as a merger between Bacteria and Archaea, the two ‘primary’ domains [87]. The 2D scenario is also labelled the ‘Eocyte’ tree [94,95], based on the predicted sister-group relationship between the Eukarya and the ‘Eocyta’— or ‘eocytes’, since renamed the Crenarchaeota [96]—originally based on the comparative analysis of ribosome structures [97]. By contrast, the ‘three primary domain’ scenario for the universal tree of life predicts that Archaea and Eukarya are two distinct, monophyletic groups, sister to one another, with Bacteria on the opposite side of the root [96].
This recent proliferation of 2D (or eocyte) phylogenies has bolstered support for fusion models of eukaryogenesis, involving the integration of a bona fide archaeal host with the α-proteobacterial symbiont [82,94]. While a 2D tree of life certainly represents a major prediction of syntrophic models of eukaryogenesis [98,99], it importantly does not falsify a phagotrophic origin of mitochondria [80,100]. For example, the recovery of 2D phylogenies supporting a eukaryotic origin from within the archaeal ‘TACK superphylum’ (comprising the Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota), as well as the discovery of actin and tubulin (two major and essential components of the eukaryotic cytoskeleton) homologues within the TACK [26,101,102], led to the formulation of the ‘phagocytosing archaeon theory’ (PhAT) [103]. In PhAT, the host cell is envisioned as a secondarily wall-less archaeon, likely representing a TACK lineage, with an actin-based cytoskeleton sufficiently ‘complex’ for phagocytosis, through which the host cell internalized the ancestral mitochondrion [103]. In this sense, PhAT represents both a fusion model [94] and a mitochondria-late model [79], involving a bona fide archaeon autogenously evolving phagocytic abilities prior to the origin of mitochondria. In other words, PhAT involves a phagocytosing archaeon serving as the ‘primitive phagocyte’ or ‘protoeukaryote’ of other phagotrophic models [28], despite being a fusion model, and is therefore predicated upon the plausibility of an archaeal lineage capable of phagocytosis.
Shortly after the proposal of PhAT, metagenomic data revealed that a new lineage of archaea—originally identified though environmental 16S rRNA genes from marine sediments and categorized as the Deep Sea Archaeal Group [104,105]—branches more closely to eukaryotes than any other known archaeal lineage [106]. The renamed ‘Lokiarchaeota’—which branch deeply within the TACK superphylum—form a clade with eukaryotes, and contain, in the composite Lokiarchaeum genome, a number of proteins previously thought to be exclusive to eukaryotes, namely actin and GTPases, which are essential to phagocytosis. These observations lead to the prediction that the Lokiarchaeota might possess dynamic, actin-based cytoskeletons and the capacity for endo- and/or phagocytosis [106,107], consistent with PhAT [103]. These conclusions were reinforced by the subsequent metagenomic discovery of the Thor-, Odin- and Heimdallarchaeota—lineages closely related to the Lokiarchaeota, together forming the ‘Asgard superphylum’, the archaeal lineages most closely related to (and paraphyletic to) the eukaryotic nuclear lineage [108,109].
In 2020, descriptions of the first cultured representative of the Asgard archaea were published [110]. This archaeon—‘Candidatus Prometheoarchaeum syntrophicum’ strain MK-D1—forms membrane supported protrusions with unique branching patterns, consistent with PhAT [103] and earlier models like it [26]. However, MK-D1 is apparently unable to phagocytose, as it is only 500 nm in diameter, and lacks the proteins and means of energy production arguably required to evolve and perform phagocytosis [111,112]. Indeed, while the Asgard archaea, and the TACK archaea more generally, feature certain proteins that are both critical to phagocytosis and currently unobserved in other archaea and bacteria, they lack many of the phagocytosis-related proteins truly specific to eukaryotes, as well as the phagocytosis-related proteins most likely sourced to eukaryotes from bacteria [112]. Therefore, while actin and tubulin are necessary for phagocytosis, they are insufficient, suggesting that the archaeal host cell was incapable of phagocytosis [110,112]. Given this conclusion, Imachi et al. [110] propose an alternative fusion model of eukaryogenesis—the entangle-engulf-endogenize (E3) model—in which the ancestral mitochondrion was not internalized by the archaeal host via phagocytosis (or any other endocytosis-like behaviour), but through interactions between the symbiont and extracellular structures projected out by the host cell into the surrounding environment—similar to the ‘inside-out’ model proposed by Baum & Baum [113].
While bacteria and archaea are apparently unable to invaginate their outer membranes in a truly endocytic manner [1,78], they are able to form and secrete outer membrane vesicles derived from membrane-supported protrusions, or blebs, that form away from the cytosol [114]. Tube-like projections and other surface appendages—likely involved in a range of processes, from nutrient exchange to genetic transfer—are also exhibited by a number of archaea [115–117]. With respect to the archaeal host cell, such protrusions would have increased its surface area to volume ratio, and may have enhanced physical contact and metabolite exchange with the α-proteobacterial symbiont [110,113]. A combination of protrusion formation (entangling) and blebbing (engulfing), as depicted in the E3 model, may have therefore been involved in surrounding and ultimately encapsulating the ancestral mitochondrion. While the acquisition of the bacterial symbiont is often envisioned to have occurred via invaginations of the host's plasma membrane—what have been called ‘outside-in’ models—a scenario involving protrusions and blebbing, like E3, serves as the inverse—an ‘inside-out’ model of eukaryogenesis [113]. Inside-out models have the advantage of being based on behaviours readily observable in archaea (i.e. protrusion formation and blebbing), while outside-in models, like PhAT, rely on behaviours never before described in archaea (i.e. phagocytosis). Overall, despite the presence of actin, tubulin and other proteins essential to phagocytosis in the archaeal lineages most closely related to eukaryotes, there is currently no evidence for phagocytosis (or endocytosis more generally) in archaea [1].
3.3. Last eukaryotic common ancestor and phagocytosis
If the archaeal host cell, prior to the acquisition of mitochondria, could not phagocytose, then when in eukaryotic evolution did phagocytosis evolve? LECA is generally reconstructed as a phagotrophic flagellate, suggesting that phagocytosis necessarily evolved along the eukaryotic stem-lineage before the origin of the eukaryotic crown-group [71,72]. However, there are alternatives to, and modifications of, this scenario. Firstly, LECA has also been reconstructed as an osmotroph [34,118], depending on an opisthokont rooting of the eukaryotic tree [50,119]. Although there is currently no agreed upon topology for the tree of eukaryotes [10], an opisthokont rooting, if ultimately supported, would potentially suggest that LECA was an osmotroph, and perhaps an obligate (i.e. non-phagocytic) osmotroph, and that phagocytosis evolved independently in virtually every eukaryotic supergroup. Indeed, even primarily non-phagocytic clades, like Archaeplastida and Fungi, contain phagocytosing representatives [11,120], suggesting either a secondarily loss of phagocytosis in these groups, or an independent origination within certain lineages [121]. An alternative scenario to both of these reconstructions (phagotrophic LECA and a strictly osmotrophic LECA) is the sort of intermediary scenario proposed by Yutin et al. [26]. Similar to PhAT (yet published four years earlier), this fusion model of eukaryogenesis suggests that the fundamental actin-based machinery underlying phagocytosis, present and conserved across the eukaryotic tree, was ultimately inherited from the eukaryotic host cell, a bona fide archaeon [26]. However, while these generic components were present in LECA, they were only fully elaborated upon within the eukaryotic crown-group, in multiple, independent origins of ‘full-fledged’ or ‘modern-type’ phagocytosis. This conclusion is based on the diversity and distribution of phagosome-associated proteins across different eukaryotic lineages, suggesting that LECA was perhaps unable to perform phagocytosis as it is currently expressed in modern amoeba, ciliates, and other sampled phagotrophs. For instance, while actin, tubulin, and numerous actin-binding proteins are apparently universal to eukaryotes, forming the core phagocytic machinery almost certainly present in LECA, other proteins involved in phagocytosis, namely receptor proteins, are poorly conserved with no universal examples extending back to LECA [26]. This apparent lack of conservation contrasts with the results of similar efforts to reconstruct the evolutionary origins of other eukaryotic systems, such as the nuclear pore complex, which is more confidently reconstructed as being present in LECA in more or less its modern form [122–125]. Together, these results suggest that while LECA was almost certainly able to engage in endocytosis as is expressed in many modern eukaryotes, the same cannot be said of phagocytosis, which arguably does not extend to LECA in any of its modern expressions. In this case, LECA may have, therefore, been primarily dependent on endocytic osmotrophy (i.e. pinocytosis) for nutrition, and perhaps only performed phagocytosis incidentally, if at all.
The earliest forms of phagocytosis, even if inefficient compared to those of modern phagotrophs, would have still offered a significant selective advantage at a time in Earth history when no other cells were capable of engulfing one another [71]. Therefore, LECA may have engaged in phagocytosis, but perhaps unreliably or inconsistently compared to modern bacterivores and eukaryovores. Indeed, among modern eukaryotes, many parasitic and phagotrophic lineages, including animals, have lost the biosynthetic capacity for many essential amino acids, which they instead obtain from their hosts and prey [71,126–128]. The observation that these amino acid biosynthesis pathways are conserved across the eukaryotic lineages that retain them, such as primarily non-phagotrophic clades like fungi [71], potentially suggests that LECA itself was not a dedicated or ‘advanced’ phagotroph, otherwise these pathways would have been lost along the eukaryotic stem-lineage. In other words, while LECA may have been mechanistically capable of a rudimentary form of particle capture via endocytosis (i.e. the beginnings of phagocytosis), it may not have been a true phagotroph primarily reliant on phagocytosis for nutrition—instead it may have relied on osmotrophy via pinocytosis. Overall, as a sort of intermediate scenario between a ‘truly’ phagocytosing LECA and an obligately or strictly osmotrophic (non-phagocytic) LECA, LECA instead may have exhibited a sort of rudimentary, yet selectively advantageous, form of phagocytosis that was independently elaborated upon (and completely lost) in various eukaryotic lineages [26]. If this was the case, the multiple, relatively ‘late’ (i.e. post-LECA) origins of ‘true’ phagotrophy may help explain the apparent temporal gap separating the inferred origin of crown-group eukaryotes from the earliest evidence of eukaryotic predation in the rock record, as described in the previous section. However, defining this gap also depends on constraining when LECA originated.
4. Scenarios for last eukaryotic common ancestor
The age of LECA is only very broadly constrained (figure 1). Both fossil data and molecular clock estimates suggest an origin of the eukaryotic crown-group sometime between 2.0 and 1.0 Ga [14,25,48–51,129,130]. While total-group eukaryotic fossils likely extend back to the Paleoproterozoic Era (2.5–1.6 Ga), the oldest widely accepted crown-group eukaryotic fossils date from ca 1.05 Ga [18,19,21] to ca 789–759 Ma [37,38]. Likewise, the oldest eukaryotic steranes date to ca 820 to 720 Ma [60]. A conservative reading of these combined records (biomarker, fossil, and molecular) might suggest an origin of LECA after 1.2 Ga or so [129,131], with all older total-group eukaryotic fossils necessarily belonging to eukaryotic stem-lineages [48]. Overall, we could then put the origin of LECA into two camps or possibilities: ‘LECA-early’ (ca 1.8–1.6 Ga) and ‘LECA-late’ (less than 1.2 Ga)—or alternatively ‘old LECA’ and ‘young LECA’ [49] (figure 1 and table 1). Likewise, as described in the previous section, LECA may have been a phagotroph (in the modern sense), a strict (non-phagocytosing) osmotroph, or perhaps an ‘inefficient’ or ‘primitive’ phagotroph [26]. The eukaryovory hypothesis for the Neoproterozoic diversification of eukaryotes described above [33,34] presumes an early, phagotrophic LECA (1.9–1.6 Ga) [50] (table 1, Scenario 1), implying a temporal gap of 450–900 Myr between the origin of bacterivory and the origin (or proliferation) of eukaryovory (using the ca 1150–900 Ma Shaler Supergroup microfossils as the oldest direct fossil evidence for eukaryovory) [29]. This considerable gap is then explained by invoking low oxygen availability [34]—although see Mills & Canfield [6] for a response to this mechanism. Alternatively, if LECA dates to ca 1.2 Ga, and was capable only of ‘rudimentary’ phagocytosis (i.e. endocytic osmotrophy, or pinocytosis, with only incidental phagocytosis) (table 1, Scenario 4), then the temporal gap between LECA and the oldest fossil evidence for eukaryovory would be only 50–300 Myr, and could be explained by the relatively late (post-LECA) origin of phagocytosis itself. This particular scenario (table 1, Scenario 4) could be falsified if predatory perforations (comparable to those seen in the Shaler Supergroup microfossils and the VSMs) and/or other signs of eukaryotic predation are discovered in the fossil record prior to ca 1.2 Ga. Such findings would then imply that specialized, modern forms of phagocytosis (e.g. protoplast feeding) evolved prior to LECA, if LECA is determined to have evolved ca 1.2 Ga or later (table 1, Scenario 2). Alternatively, if LECA evolved earlier in time (greater than 1.2 Ga), these fossils could still be compatible with a post-LECA origin of phagocytosis, depending on the age of the fossils and the estimated age of LECA. For instance, if new predatory perforations from the Mesoproterozoic (1.6–1.0 Ga) are discovered, but LECA is understood to have originated in the Palaeoproterozoic (LECA-early), then these new fossils would still be consistent with a post-LECA origin of phagocytosis (table 1, Scenario 3). In this case, falsifying a post-LECA origin of phagocytosis would probably have to rely on the comparative analysis of neontological data [26,112]. Overall, before invoking environmental factors, such as nutrient and oxygen limitation, to explain the temporal gap separating LECA from the proliferation of eukaryotic predators in the Neoproterozoic [34,60], the age and phagocytic capacities of LECA first need to be determined (table 1).
Table 1.
The uncertainties surrounding LECA's age (figure 1) and phagocytic abilities suggest at least four different scenarios for when and how phagocytosis evolved. Phagocytosis here refers strictly to its modern form, which may or not extend back to LECA, as discussed in the main text.
| LECA-early (>1.6 Ga) | LECA-late (<1.2 Ga) | |
|---|---|---|
| pre-LECA phagocytosis | (1) phagocytosis is ancestral to crown-group eukaryotes, which originated by the end Palaeoproterozoic | (2) phagocytosis is ancestral to crown-group eukaryotes, which originated towards the end Mesoproterozoic |
| post-LECA phagocytosis | (3) phagocytosis evolved independently within crown-group eukaryotes, which originated by the end Palaeoproterozoic | (4) phagocytosis evolved independently within crown-group eukaryotes, which originated towards the end Mesoproterozoic |
5. Conclusion
Crown-group eukaryotes are generally thought to have emerged over 1.6 Ga from an ancestrally bacterivorous state. On the other hand, palaeontological and organic geochemical evidence suggests that eukaryotic predation (both bacterivory and eukaryovory) became ecologically widespread in the early-to-mid Neoproterozoic (figure 1). This apparent temporal gap—spanning over a half a billion years—remains difficult to explain. As one potential solution, a late origin of the eukaryotic crown-group (less than 1.2 Ga), coupled with multiple late, post-LECA origins of ‘modern’ phagocytosis (i.e. phagocytosis as it is currently expressed in many extant eukaryotic lineages, such as amoebae, ciliates and animals), could dramatically reduce the duration of this temporal gap, leading to a scenario remarkably similar to that predicted by Stanley in 1973 [53]. Indeed, the eukaryotic host appears to have been a bona fide archaeon, a major prediction of mitochondria-early hypotheses based on anaerobic syntrophy [98], even if an archaeal host does not definitively falsify mitochondria-late scenarios [80,100]. While this archaeal host likely sourced the major components of the eukaryotic cytoskeleton, essential to phagocytosis, it was arguably unable to phagocytose by itself prior to the origin of mitochondria [1,110,112]. This ‘bottom-up’ approach to eukaryogenesis, therefore, arguably supports mitochondria-early scenarios, suggesting that the eukaryotic host cell was incapable of phagocytosis, and used phagocytosis-independent mechanisms (capture facilitated by protrusions and blebs that formed away from the host cytosol, rather than endocytic invaginations) to ultimately encapsulate the ancestral mitochondrion [110,113]. At the same time, ‘top-down’ approaches, based on the diversity and distribution of phagosome-associated proteins across the eukaryotic tree, suggest that while LECA almost certainly possessed the basic cytoskeletal machinery underlying endocytosis, phagocytosis as it is currently exhibited by various extant eukaryotic clades potentially evolved independently multiple times within the eukaryotic crown-group [26]. As no unequivocally crown-group eukaryotic fossils, or biomarkers of eukaryotic origin, currently pre-date 1.05 Ga, a relatively late origin of LECA (less than 1.2 Ga) [129,131], coupled with a relatively long eukaryotic stem-lineage dating back to 1.6 Ga or greater [25], further suggests that modern-type phagocytosis may have only evolved toward the end of the Mesoproterozoic Era. These predictions suggest a dramatically reduced temporal gap between the evolutionary origin of phagocytosis and the earliest signs of phagocytosis in the rock record, consistent with Stanley's prediction of a late-Proterozoic crossing of the ‘heterotroph barrier’—a key prerequisite to the origin of modern ecosystems [53].
Acknowledgements
The author wishes to gratefully acknowledge the helpful comments of Erik Sperling, as well as influential conversations with Morgan Gaia, Sriram Garg, Patrick Keeling and Bill Martin. The author also acknowledges helpful exchanges with Brian Leander and Sebastian Hess concerning the terminology of different eukaryotic feeding modes. The manuscript benefitted from the insightful comments of three anonymous reviewers.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
This work was supported by the Agouron Institute Geobiology Postdoctoral Fellowship programme.
Glossary of terms
- Bacterivory
(also bacteriotrophy) a predatory mode whereby organisms (generally microbial eukaryotes, but also bacteria like Bdellovibrio and Micavibrio) obtain and ingest nutrients directly from bacterial prey [132,133], irrespective of the bacterial carbon source or metabolism [3]. While a variety of bacteria prey upon other bacteria for nutrition [134], for the purposes of this effort, bacterivory refers to the predation of bacteria by microbial eukaryotes via phagocytosis, unless otherwise noted.
- Crown-group
a clade consisting of the last common ancestor plus all of its descendants, living or extinct [135,136].
- Endocytosis
the eukaryote-specific process in which cells internalize foreign materials and molecules via invaginations of the plasma membrane that pinch off from the cell surface to form intracellular vesicles within the cytosol [137,138].
- Eukaryogenesis
the origin of the eukaryotic cell—the major evolutionary transition from FECA to LECA [84,109].
- Eukaryophagy
eukaryovory in which microbial eukaryotes phagocytose entire, and generally ‘large’, eukaryotic prey cells for nutrition [14,33,139]. Eukaryophagy arguably contrasts with myzocytosis, where myzocytosis is categorized as a form of non-phagocytic endocytosis [140–142].
- Eukaryotic predation
any kind of eukaryotic predatory behaviour, regardless of prey (bacterivory or eukaryovory), or feeding mechanism (phagocytosis or myzocytosis).
- Eukaryovory
(also eukaryotrophy) a predatory mode whereby organisms (generally microbial eukaryotes) obtain and ingest nutrients from ‘large’ microbial eukaryotic prey [132,133], whether through eukaryophagy or myzocytosis.
- FECA
the first eukaryotic common ancestor, marked by the divergence of total-group Eukarya from its sister-lineage.
- LECA
the last eukaryotic common ancestor (the most recent common ancestor of all living eukaryotes).
- Myzocytosis
(cell sucking) a predatory mode and form of endocytosis in which microbial eukaryotes pierce the cortex of prey cells to draw out the cytoplasmic contents, either entirely or partially [133]. This predatory style is often very explicitly contrasted with phagocytosis in that only the cytosol—not the plasma membrane or the entire organism—is ingested via vesicular uptake [140–142], although some taxa, such as the euglenid Peranema trichophorum, can perform both phagocytosis and myzocytosis [143]. Myzocytosis is sometimes treated as specific to the Myzozoa—the clade encompassing the Apicomplexa, chrompodellids, and dinoflagellates [142,144].
- Osmotrophy
a feeding mechanism in which an organism uptakes dissolved nutrients and metabolites via osmosis, active transport or pinocytosis [145]. Osmotrophy very explicitly contrasts with phagocytosis [146], yet overlaps with endocytosis through pinocytosis.
- Phagocytosis
(cell eating) the form of endocytosis in which ‘large’ (greater than or equal to 0.5 µm) particles (traditionally those visible by light microscopy and generally thought of as entire cells) are captured (e.g. via pseudopodia) and internalized while excluding most, if not all, of the surrounding extracellular fluid [1,114,138,140,147].
- Phagotrophy
the nutritional mode whereby ‘large’ food particles, such as entire prey cells, are ingested via phagocytosis [133]. While phagotrophy includes bacterivory and eukaryophagy, it arguably excludes myzocytosis [140].
- Pinocytosis
(cell drinking) the form of endocytosis in which extracellular fluid, small particles, soluble macromolecules, and low-molecular-weight solutes are internalized via vesicular uptake [138]. Pinocytosis was coined as a contrast, and analogue, to phagocytosis [148], and classifies under osmotrophy as a mechanism for dissolved nutrient uptake [145].
- Protoplast feeding
a predatory mode whereby microbial eukaryotes, namely vampyrellid amoebae (Vampyrellidae, Rhizaria) and the Viridiraptoridae (Filosa, Rhizaria), locally dissolve the cell wall of their prey—primarily algae, as well as fungal spores and hyphae [149]—to phagocytose the entire protoplast without engulfing the entire cell [150].
- Stem-group
a paraphyletic group of all extinct taxa that diverged before the last common ancestor of any particular crown-group, but after the split from its most closely related sister-group [135,136]. Note that a ‘stem-group’ placement only refers to a particular set of nodes—dinosaurs, for instance, are stem-group birds, but are also crown-group amniotes and tetrapods.
- Total-group
the combined stem-group and crown-group of any particular clade [135,136].
References
- 1.Martin WF, Tielens AGM, Mentel M, Garg SG, Gould SB. 2017. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol. Mol. Biol. Rev. 81, e00008-17 ( 10.1128/MMBR.00008-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. ( 10.3354/meps010257) [DOI] [Google Scholar]
- 3.Sherr EB, Sherr BF. 1994. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb. Ecol. 28, 223–235. ( 10.1007/BF00166812) [DOI] [PubMed] [Google Scholar]
- 4.Fenchel T. 2008. The microbial loop–25 years later. J. Exp. Mar. Biol. Ecol. 366, 99–103. ( 10.1016/j.jembe.2008.07.013) [DOI] [Google Scholar]
- 5.Desjardins M, Houde M, Gagnon E. 2005. Phagocytosis: the convoluted way from nutrition to adaptive immunity. Immunol. Rev. 207, 158–165. ( 10.1111/j.0105-2896.2005.00319.x) [DOI] [PubMed] [Google Scholar]
- 6.Mills DB, Canfield DE. 2017. A trophic framework for animal origins. Geobiology 15, 197–210. ( 10.1111/gbi.12216) [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T. 2017. Origin of animal multicellularity: precursors, causes, consequences—the choanoflagellate/sponge transition, neurogenesis and the Cambrian explosion. Phil. Trans. R. Soc. B 372, 20150476 ( 10.1098/rstb.2015.0476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills DB, Francis WR, Canfield DE. 2018. Animal origins and the Tonian Earth system. Emerg. Top. Life Sci. 2, 289–298. ( 10.1042/ETLS20170160) [DOI] [PubMed] [Google Scholar]
- 9.Porter S. 2011. The rise of predators. Geology 39, 607–608. ( 10.1130/focus062011.1) [DOI] [Google Scholar]
- 10.Keeling PJ, Burki F. 2019. Progress towards the tree of eukaryotes. Curr. Biol. 29, R808–R817. ( 10.1016/j.cub.2019.07.031) [DOI] [PubMed] [Google Scholar]
- 11.Maruyama S, Kim E. 2013. A modern descendant of early green algal phagotrophs. Curr. Biol. 23, 1081–1084. ( 10.1016/j.cub.2013.04.063) [DOI] [PubMed] [Google Scholar]
- 12.Nowack ECM, Weber APM. 2018. Genomics-informed insights into endosymbiotic organelle evolution in photosynthetic eukaryotes. Annu. Rev. Plant Biol. 69, 51–84. ( 10.1146/annurev-arplant-042817-040209) [DOI] [PubMed] [Google Scholar]
- 13.Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Phil. Trans. R. Soc. B 365, 729–748. ( 10.1098/rstb.2009.0103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butterfield NJ. 2015. Early evolution of the Eukaryota. Palaeontology 58, 5–17. ( 10.1111/pala.12139) [DOI] [Google Scholar]
- 15.Marin B, Nowack ECM, Melkonian M. 2005. A plastid in the making: evidence for a second primary endosymbiosis. Protist 156, 425–432. ( 10.1016/j.protis.2005.09.001) [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya D, Price DC, Yoon HS, Yang EC, Poulton NJ, Andersen RA, Das SP. 2012. Single cell genome analysis supports a link between phagotrophy and primary plastid endosymbiosis. Sci. Rep. 2, 356 ( 10.1038/srep00356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaye L, Valadez-Cano C, Pérez-Zamorano B. 2016. How really ancient is Paulinella chromatophora? PLoS Curr. 8 ( 10.1371/currents.tol.e68a099364bb1a1e129a17b4e06b0c6b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butterfield NJ, Knoll AH, Swett K. 1990. A bangiophyte red alga from the Proterozoic of arctic Canada. Science 250, 104–107. ( 10.1126/science.11538072) [DOI] [PubMed] [Google Scholar]
- 19.Butterfield NJ. 2000. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26, 386–404. () [DOI] [Google Scholar]
- 20.Knoll AH, Wörndle S, Kah LC. 2013. Covariance of microfossil assemblages and microbialite textures across an upper Mesoproterozoic carbonate platform. Palaios 28, 453–470. ( 10.2110/palo.2013.p13-005r) [DOI] [Google Scholar]
- 21.Gibson TM, et al. 2018. Precise age of Bangiomorpha pubescens dates the origin of eukaryotic photosynthesis. Geology 46, 135–138. ( 10.1130/G39829.1) [DOI] [Google Scholar]
- 22.Bengtson S, Sallstedt T, Belivanova V, Whitehouse M. 2017. Three-dimensional preservation of cellular and subcellular structures suggests 1.6 billion-year-old crown-group red algae. PLoS Biol. 15, e2000735 ( 10.1371/journal.pbio.2000735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu S, Zhu M, Knoll AH, Yin Z, Zhao F, Sun S, Qu Y, Shi M, Liu H. 2016. Decimetre-scale multicellular eukaryotes from the 1.56-billion-year-old Gaoyuzhuang Formation in North China. Nat. Commun. 7, 11500 ( 10.1038/ncomms11500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Baracaldo P, Raven JA, Pisani D, Knoll AH. 2017. Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc. Natl Acad. Sci. USA 114, E7737–E7745. ( 10.1073/pnas.1620089114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betts HC, Puttick MN, Clark JW, Williams TA, Donoghue PCJ, Pisani D. 2018. Integrated genomic and fossil evidence illuminates life's early evolution and eukaryote origin. Nat. Ecol. Evol. 2, 1556–1562. ( 10.1038/s41559-018-0644-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yutin N, Wolf MY, Wolf YI, Koonin EV. 2009. The origins of phagocytosis and eukaryogenesis. Biol. Direct 4, 9 ( 10.1186/1745-6150-4-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354. ( 10.1099/00207713-52-2-297) [DOI] [PubMed] [Google Scholar]
- 28.de Duve C. 2007. The origin of eukaryotes: a reappraisal. Nat. Rev. Genet. 8, 395–403. ( 10.1038/nrg2071) [DOI] [PubMed] [Google Scholar]
- 29.Loron CC, Rainbird RH, Turner EC, Greenman JW, Javaux EJ. 2018. Implications of selective predation on the macroevolution of eukaryotes: evidence from Arctic Canada. Emerg. Top. Life Sci. 2, 247–255. ( 10.1042/ETLS20170153) [DOI] [PubMed] [Google Scholar]
- 30.Porter SM, Meisterfeld R, Knoll AH. 2003. Vase-shaped microfossils from the Neoproterozoic Chuar Group, Grand Canyon: a classification guided by modern testate amoebae. J. Paleontol. 77, 409–429. () [DOI] [Google Scholar]
- 31.Porter SM. 2016. Tiny vampires in ancient seas: evidence for predation via perforation in fossils from the 780–740 million-year-old Chuar Group, Grand Canyon, USA. Proc. R. Soc. B 283, 20160221 ( 10.1098/rspb.2016.0221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen PA, Riedman LA. 2018. It's a protist-eat-protist world: recalcitrance, predation, and evolution in the Tonian–Cryogenian ocean. Emerg. Topics Life Sci. 2, 173–180. ( 10.1042/ETLS20170145) [DOI] [PubMed] [Google Scholar]
- 33.Knoll AH. 2014. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb. Perspect. Biol. 6, a016121 ( 10.1101/cshperspect.a016121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoll AH, Lahr DJ. 2016. Fossils, feeding, and the evolution of complex multicellularity. In Multicellularity, origins and evolution (eds Niklas KJ, Newman SA), pp. 3–16. Cambridge, MA: MIT Press. [Google Scholar]
- 35.Strauss JV, Rooney AD, Macdonald FA, Brandon AD, Knoll AH. 2014. 740 Ma vase-shaped microfossils from Yukon, Canada: implications for Neoproterozoic chronology and biostratigraphy. Geology 42, 659–662. ( 10.1130/G35736.1) [DOI] [Google Scholar]
- 36.Riedman LA, Porter SM, Calver CR. 2018. Vase-shaped microfossil biostratigraphy with new data from Tasmania, Svalbard, Greenland, Sweden and the Yukon. Precambrian Res. 319, 19–36. ( 10.1016/j.precamres.2017.09.019) [DOI] [Google Scholar]
- 37.Lahr DJG, et al. 2019. Phylogenomics and morphological reconstruction of arcellinida testate amoebae highlight diversity of microbial eukaryotes in the Neoproterozoic. Curr. Biol. 29, 991–1001.e1003. ( 10.1016/j.cub.2019.01.078) [DOI] [PubMed] [Google Scholar]
- 38.Porter SM, Riedman LA. 2019. Evolution: ancient fossilized amoebae find their home in the tree. Curr. Biol. 29, R212–R215. ( 10.1016/j.cub.2019.02.003) [DOI] [PubMed] [Google Scholar]
- 39.Nikolaev SI, Mitchell EAD, Petrov NB, Berney C, Fahrni J, Pawlowski J. 2005. The testate lobose amoebae (order Arcellinida Kent, 1880) finally find their home within Amoebozoa. Protist 156, 191–202. ( 10.1016/j.protis.2005.03.002) [DOI] [PubMed] [Google Scholar]
- 40.Gomaa F, Todorov M, Heger TJ, Mitchell EAD, Lara E. 2012. SSU rRNA phylogeny of Arcellinida (Amoebozoa) reveals that the largest Arcellinid genus, Difflugia Leclerc 1815, is not monophyletic. Protist 163, 389–399. ( 10.1016/j.protis.2011.12.001) [DOI] [PubMed] [Google Scholar]
- 41.Fiz-Palacios O, Leander BS, Heger TJ. 2014. Old lineages in a new ecosystem: diversification of arcellinid amoebae (Amoebozoa) and peatland mosses. PLoS ONE 9, e95238 ( 10.1371/journal.pone.0095238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen PA, Strauss JV, Rooney AD, Sharma M, Tosca N. 2017. Controlled hydroxyapatite biomineralization in an 810 million-year-old unicellular eukaryote. Sci. Adv. 3, e1700095 ( 10.1126/sciadv.1700095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smetacek V. 2001. A watery arms race. Nature 411, 745 ( 10.1038/35081210) [DOI] [PubMed] [Google Scholar]
- 44.Verity PG, Villareal TA. 1986. The relative food value of diatoms, dinoflagellates, flagellates, and cyanobacteria for tintinnid ciliates. Archiv für Protistenkunde 131, 71–84. ( 10.1016/S0003-9365(86)80064-1) [DOI] [Google Scholar]
- 45.Hamm CE, Merkel R, Springer O, Jurkojc P, Maier C, Prechtel K, Smetacek V. 2003. Architecture and material properties of diatom shells provide effective mechanical protection. Nature 421, 841–843. ( 10.1038/nature01416) [DOI] [PubMed] [Google Scholar]
- 46.Tillmann U. 2004. Interactions between planktonic microalgae and protozoan grazers. J. Eukaryot. Microbiol. 51, 156–168. ( 10.1111/j.1550-7408.2004.tb00540.x) [DOI] [PubMed] [Google Scholar]
- 47.Hamm C, Smetacek V. 2007. Armor: why, when, and how. In Evolution of primary producers in the sea (eds Falkowski PG, Knoll AH), pp. 311–332. London, UK: Academic Press; ( 10.1016/B978-012370518-1/50015-1) [DOI] [Google Scholar]
- 48.Berney C, Pawlowski J. 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. R. Soc. B 273, 1867–1872. ( 10.1098/rspb.2006.3537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chernikova D, Motamedi S, Csürös M, Koonin EV, Rogozin IB. 2011. A late origin of the extant eukaryotic diversity: divergence time estimates using rare genomic changes. Biol. Direct 6, 26 ( 10.1186/1745-6150-6-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl Acad. Sci. USA 108, 13 624–13 629. ( 10.1073/pnas.1110633108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eme L, Sharpe SC, Brown MW, Roger AJ. 2014. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb. Perspect. Biol. 6, a016139 ( 10.1101/cshperspect.a016139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole DB, Mills DB, Erwin DH, Sperling EA, Porter SM, Reinhard CT, Planavsky NJ. 2020. On the co-evolution of surface oxygen levels and animals. Geobiology 319, 55 ( 10.1111/gbi.12382) [DOI] [PubMed] [Google Scholar]
- 53.Stanley SM. 1973. An ecological theory for the sudden origin of multicellular life in the late Precambrian. Proc. Natl Acad. Sci. USA 70, 1486–1489. ( 10.1073/pnas.70.5.1486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Cortona A, et al. 2020. Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds. Proc. Natl Acad. Sci. USA 117, 2551–2559. ( 10.1073/pnas.1910060117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288. ( 10.1093/bioinformatics/btp368) [DOI] [PubMed] [Google Scholar]
- 56.Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. 2011. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334, 1091–1097. ( 10.1126/science.1206375) [DOI] [PubMed] [Google Scholar]
- 57.dos Reis M, Thawornwattana Y, Angelis K, Telford MJ, Donoghue PC, Yang Z. 2015. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr. Biol. 25, 2939–2950. ( 10.1016/j.cub.2015.09.066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dohrmann M, Wörheide G. 2017. Dating early animal evolution using phylogenomic data. Sci. Rep. 7, 3599 ( 10.1038/s41598-017-03791-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groussin M, Pawlowski J, Yang Z. 2011. Bayesian relaxed clock estimation of divergence times in foraminifera. Mol. Phylogenet. Evol. 61, 157–166. ( 10.1016/j.ympev.2011.06.008) [DOI] [PubMed] [Google Scholar]
- 60.Brocks JJ, Jarrett AJM, Sirantoine E, Hallmann C, Hoshino Y, Liyanage T. 2017. The rise of algae in Cryogenian oceans and the emergence of animals. Nature 548, 578–581. ( 10.1038/nature23457) [DOI] [PubMed] [Google Scholar]
- 61.Zumberge JA, Rocher D, Love GD. 2019. Free and kerogen-bound biomarkers from late Tonian sedimentary rocks record abundant eukaryotes in mid-Neoproterozoic marine communities. Geobiology 115, 246 ( 10.1111/gbi.12378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brocks JJ. 2018. The transition from a cyanobacterial to algal world and the emergence of animals. Emerg. Topics Life Sci. 2, 181–190. ( 10.1042/ETLS20180039) [DOI] [PubMed] [Google Scholar]
- 63.Brocks JJ, Jarrett AJM, Sirantoine E, Kenig F, Moczydłowska M, Porter S, Hope J. 2016. Early sponges and toxic protists: possible sources of cryostane, an age diagnostic biomarker antedating Sturtian Snowball Earth. Geobiology 14, 129–149. ( 10.1111/gbi.12165) [DOI] [PubMed] [Google Scholar]
- 64.Zumberge JA, Love GD, Cárdenas P, Sperling EA, Gunasekera S, Rohrssen M, Grosjean E, Grotzinger JP, Summons RE. 2018. Demosponge steroid biomarker 26-methylstigmastane provides evidence for Neoproterozoic animals. Nat. Ecol. Evol. 2, 1709–1714. ( 10.1038/s41559-018-0676-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sperling EA, Stockey RG. 2018. The temporal and environmental context of early animal evolution: considering all the ingredients of an ‘Explosion’. Integr. Comp. Biol. 58, 605–622. ( 10.1093/icb/icy088) [DOI] [PubMed] [Google Scholar]
- 66.Nettersheim BJ, et al. 2019. Putative sponge biomarkers in unicellular Rhizaria question an early rise of animals. Nat. Ecol. Evol. 3, 577–581. ( 10.1038/s41559-019-0806-5) [DOI] [PubMed] [Google Scholar]
- 67.Lenton TM, Daines SJ. 2018. The effects of marine eukaryote evolution on phosphorus, carbon and oxygen cycling across the Proterozoic–Phanerozoic transition. Emerg. Topics Life Sci. 2, 267–278. ( 10.1042/ETLS20170156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Maldegem LM, et al. 2019. Bisnorgammacerane traces predatory pressure and the persistent rise of algal ecosystems after Snowball Earth. Nat. Commun. 10, 476 ( 10.1038/s41467-019-08306-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love GD, et al. 2009. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457, 718–721. ( 10.1038/nature07673) [DOI] [PubMed] [Google Scholar]
- 70.Gold DA, Grabenstatter J, de Mendoza A, Riesgo A, Ruiz-Trillo I, Summons RE. 2016. Sterol and genomic analyses validate the sponge biomarker hypothesis. Proc. Natl Acad. Sci. USA 113, 2684–2689. ( 10.1073/pnas.1512614113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC. 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit. Rev. Biochem. Mol. Biol. 48, 373–396. ( 10.3109/10409238.2013.821444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knoll AH, Lahr DJG. 2016. Fossils, feeding, and the evolution of complex multicellularity. In Multicellularity, origins and evolution. The Vienna Series in Theoretical Biology, pp. 1–16. Boston, MA: Massachusetts Institute of Technology. [Google Scholar]
- 73.Sperling EA, Frieder CA, Raman AV, Girguis PR, Levin LA, Knoll AH. 2013. Oxygen, ecology, and the Cambrian radiation of animals. Proc. Natl Acad. Sci. USA 110, 13 446–13 451. ( 10.1073/pnas.1312778110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fenchel T, Finlay BJ. 1995. Ecology and evolution in anoxic worlds. Oxford, UK: Oxford University Press. [Google Scholar]
- 75.Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ, Zubkov MV. 2012. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc. Natl Acad. Sci. USA 109, 5756–5760. ( 10.1073/pnas.1118179109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanier R, Douderoff M, Adelberg E. 1963. The microbial world, 2nd edn Englewood Cliffs, NJ: Prentice-Hall Inc. [Google Scholar]
- 77.Shiratori T, Suzuki S, Kakizawa Y, Ishida K-I. 2019. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat. Commun. 10, 5529 ( 10.1038/s41467-019-13499-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boedeker C, et al. 2017. Determining the bacterial cell biology of planctomycetes. Nat. Commun. 8, 14853 ( 10.1038/ncomms14853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roger AJ, Muñoz-Gómez SA, Kamikawa R. 2017. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192. ( 10.1016/j.cub.2017.09.015) [DOI] [PubMed] [Google Scholar]
- 80.Poole AM, Gribaldo S. 2014. Eukaryotic origins: how and when was the mitochondrion acquired? Cold Spring Harb. Perspect. Biol. 6, a015990 ( 10.1101/cshperspect.a015990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O'Malley MA. 2010. The first eukaryote cell: an unfinished history of contestation. Stud. Hist. Phil. Biol. Biomed. Sci. 41, 212–224. ( 10.1016/j.shpsc.2010.07.010) [DOI] [PubMed] [Google Scholar]
- 82.Forterre P. 2011. A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res. Microbiol. 162, 77–91. ( 10.1016/j.resmic.2010.10.005) [DOI] [PubMed] [Google Scholar]
- 83.Müller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. MMBR 76, 444–495. ( 10.1128/MMBR.05024-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dacks JB, Field MC, Buick R, Eme L, Gribaldo S, Roger AJ, Brochier-Armanet C, Devos DP. 2016. The changing view of eukaryogenesis—fossils, cells, lineages and how they all come together. J. Cell Sci. 129, 3695–3703. ( 10.1242/jcs.178566) [DOI] [PubMed] [Google Scholar]
- 85.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. 2008. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA 105, 20 356–20 361. ( 10.1073/pnas.0810647105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Foster PG, Cox CJ, Embley TM. 2009. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Phil. Trans. R. Soc. B 364, 2197–2207. ( 10.1098/rstb.2009.0034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. 2010. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat. Rev. Microbiol. 8, 743–752. ( 10.1038/nrmicro2426) [DOI] [PubMed] [Google Scholar]
- 88.Guy L, Ettema TJG. 2011. The archaeal ‘TACK' superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587. ( 10.1016/j.tim.2011.09.002) [DOI] [PubMed] [Google Scholar]
- 89.Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM. 2012. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc. R. Soc. B 279, 4870–4879. ( 10.1098/rspb.2012.1795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams TA, Foster PG, Cox CJ, Embley TM. 2013. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236. ( 10.1038/nature12779) [DOI] [PubMed] [Google Scholar]
- 91.Guy L, Saw JH, Ettema TJG. 2014. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 6, a016022 ( 10.1101/cshperspect.a016022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McInerney JO, O'Connell MJ, Pisani D. 2014. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat. Rev. Microbiol. 12, 449–455. ( 10.1038/nrmicro3271) [DOI] [PubMed] [Google Scholar]
- 93.Raymann K, Brochier-Armanet C, Gribaldo S. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl Acad. Sci. USA 112, 6670–6675. ( 10.1073/pnas.1420858112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Forterre P. 2013. The common ancestor of Archaea and Eukarya was not an archaeon. Archaea 2013, 1–18. ( 10.1155/2013/372396) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gaia M, Da Cunha V, Forterre P. 2018. The Tree of Life. In Molecular mechanisms of microbial evolution (ed. Rampelotto PH.), pp. 55–99. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 96.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579. ( 10.1073/pnas.87.12.4576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lake JA, Henderson E, Oakes M, Clark MW. 1984. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc. Natl Acad. Sci. USA 81, 3786–3790. ( 10.1073/pnas.81.12.3786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41. ( 10.1038/32096) [DOI] [PubMed] [Google Scholar]
- 99.Martin WF, Garg S, Zimorski V. 2015. Endosymbiotic theories for eukaryote origin. Phil. Trans. R. Soc. B 370, 20140330 ( 10.1098/rstb.2014.0330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Poole AM, Neumann N. 2011. Reconciling an archaeal origin of eukaryotes with engulfment: a biologically plausible update of the Eocyte hypothesis. Res. Microbiol. 162, 71–76. ( 10.1016/j.resmic.2010.10.002) [DOI] [PubMed] [Google Scholar]
- 101.Ettema TJG, Lindås A-C, Bernander R. 2011. An actin-based cytoskeleton in archaea. Mol. Microbiol. 80, 1052–1061. ( 10.1111/j.1365-2958.2011.07635.x) [DOI] [PubMed] [Google Scholar]
- 102.Yutin N, Koonin EV. 2012. Archaeal origin of tubulin. Biol. Direct 7, 10 ( 10.1186/1745-6150-7-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martijn J, Ettema TJG. 2013. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem. Soc. Trans. 41, 451–457. ( 10.1042/BST20120292) [DOI] [PubMed] [Google Scholar]
- 104.Jorgensen SL, et al. 2012. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl Acad. Sci. USA 109, E2846–E2855. ( 10.1073/pnas.1207574109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jørgensen SL, Thorseth IH, Pedersen RB, Baumberger T, Schleper C. 2013. Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge. Front. Microbiol. 4, 299 ( 10.3389/fmicb.2013.00299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spang A, et al. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179. ( 10.1038/nature14447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Embley TM, Williams TA. 2015. Steps on the road to eukaryotes. Nature 521, 169–170. ( 10.1038/nature14522) [DOI] [PubMed] [Google Scholar]
- 108.Zaremba-Niedzwiedzka K, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358. ( 10.1038/nature21031) [DOI] [PubMed] [Google Scholar]
- 109.Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. 2018. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 16, 120 ( 10.1038/nrmicro.2017.154) [DOI] [PubMed] [Google Scholar]
- 110.Imachi H, et al. 2020. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577, 519–525. ( 10.1038/s41586-019-1916-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467, 929–934. ( 10.1038/nature09486) [DOI] [PubMed] [Google Scholar]
- 112.Burns JA, Pittis AA, Kim E. 2018. Gene-based predictive models of trophic modes suggest Asgard archaea are not phagocytotic. Nat. Ecol. Evol. 2, 751 ( 10.1038/s41559-018-0520-8) [DOI] [PubMed] [Google Scholar]
- 113.Baum DA, Baum B. 2014. An inside-out origin for the eukaryotic cell. BMC Biol. 12, 76 ( 10.1186/s12915-014-0076-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gould SB, Garg SG, Martin WF. 2016. Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol. 24, 525–534. ( 10.1016/j.tim.2016.03.005) [DOI] [PubMed] [Google Scholar]
- 115.Rosenshine I, Tchelet R, Mevarech M. 1989. The mechanism of DNA transfer in the mating system of an archaebacterium. Science 245, 1387–1389. ( 10.1126/science.2818746) [DOI] [PubMed] [Google Scholar]
- 116.Albers S-V, Meyer BH. 2011. The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426. ( 10.1038/nrmicro2576) [DOI] [PubMed] [Google Scholar]
- 117.Marguet E, Gaudin M, Gauliard E, Fourquaux I, du Plouy lB, Matsui S, Forterre I. 2013. Membrane vesicles, nanopods and/or nanotubes produced by hyperthermophilic archaea of the genus Thermococcus. Biochem. Soc. Trans. 41, 436–442. ( 10.1042/BST20120293) [DOI] [PubMed] [Google Scholar]
- 118.Martin W, Rotte C, Hoffmeister M, Theissen U, Gelius-Dietrich G, Ahr S, Henze K. 2003. Early cell evolution, eukaryotes, anoxia, sulfide, oxygen, fungi first (?), and a tree of genomes revisited. IUBMB Life 55, 193–204. ( 10.1080/1521654031000141231) [DOI] [PubMed] [Google Scholar]
- 119.Katz LA, Grant JR, Parfrey LW, Burleigh JG. 2012. Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst. Biol. 61, 653–660. ( 10.1093/sysbio/sys026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Powell MJ. 1984. Fine structure of the unwalled thallus of Rozella polyphagi in its host Polyphagus euglenae. Mycologia 76, 1039–1048. ( 10.1080/00275514.1984.12023948) [DOI] [Google Scholar]
- 121.Raven JA. 2013. Cells inside cells: symbiosis and continuing phagotrophy. Curr. Biol. 23, R530–R531. ( 10.1016/j.cub.2013.05.006) [DOI] [PubMed] [Google Scholar]
- 122.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2, e380 ( 10.1371/journal.pbio.0020380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mans BJ, Anantharaman V, Aravind L, Koonin EV. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3, 1612–1637. ( 10.4161/cc.3.12.1345) [DOI] [PubMed] [Google Scholar]
- 124.Bapteste E, Charlebois RL, MacLeod D, Brochier C. 2005. The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol. 6, R85 ( 10.1186/gb-2005-6-10-r85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neumann N, Lundin D, Poole AM. 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 5, e13241 ( 10.1371/journal.pone.0013241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Payne SH, Loomis WF. 2006. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot. Cell 5, 272–276. ( 10.1128/EC.5.2.272-276.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guedes RLM, Prosdocimi F, Fernandes GR, Moura LK, Ribeiro HAL, Ortega JM. 2011. Amino acids biosynthesis and nitrogen assimilation pathways: a great genomic deletion during eukaryotes evolution. BMC Genomics 12(Suppl. 4), S2 ( 10.1186/1471-2164-12-S4-S2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Richter DJ, Fozouni P, Eisen MB, King N. 2018. Gene family innovation, conservation and loss on the animal stem lineage. Elife 7, e38726 ( 10.7554/eLife.34226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cavalier-Smith T. 2013. Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur. J. Protistol. 49, 115–178. ( 10.1016/j.ejop.2012.06.001) [DOI] [PubMed] [Google Scholar]
- 130.Javaux EJ, Lepot K. 2018. The Paleoproterozoic fossil record: implications for the evolution of the biosphere during Earth's middle-age. Earth-Sci. Rev. 176, 68–86. ( 10.1016/j.earscirev.2017.10.001) [DOI] [Google Scholar]
- 131.Porter SM. 2020. Insights into eukaryogenesis from the fossil record. Interface Focus 10, 20190105 ( 10.1098/rsfs.2019.0105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leander BS, Triemer RE, Farmer MA. 2001. Character evolution in heterotrophic euglenids. Eur. J. Protistol. 37, 337–356. ( 10.1078/0932-4739-00842) [DOI] [Google Scholar]
- 133.Leander BS. 2004. Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 12, 251–258. ( 10.1016/j.tim.2004.04.001) [DOI] [PubMed] [Google Scholar]
- 134.Jurkevitch E. 2007. Predatory behaviors in bacteria—diversity and transitions. Microbe-Am. Soc. Microbiol. 2, 67 ( 10.1128/microbe.2.67.1) [DOI] [Google Scholar]
- 135.Jefferies RPS. 1979. The origin of chordates—a methodological essay. In The origin of major invertebrate groups (ed. House MR.), pp. 443–477. London, UK: Academic Press. [Google Scholar]
- 136.Budd GE, Jensen S. 2000. A critical reappraisal of the fossil record of the bilaterian phyla. Biol. Rev. 75, 253–295. ( 10.1017/S000632310000548X) [DOI] [PubMed] [Google Scholar]
- 137.Goldstein JL, Anderson RG, Brown MS. 1979. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 279, 679–685. ( 10.1038/279679a0) [DOI] [PubMed] [Google Scholar]
- 138.Silverstein SC, Steinman RM, Cohn ZA. 1977. Endocytosis. Annu. Rev. Biochem. 46, 669–722. ( 10.1146/annurev.bi.46.070177.003321) [DOI] [PubMed] [Google Scholar]
- 139.Ratti S, Knoll AH, Giordano M. 2013. Grazers and phytoplankton growth in the oceans: an experimental and evolutionary perspective. PLoS ONE 8, e77349 ( 10.1371/journal.pone.0077349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schnepf E, Deichgräber G. 1984. ‘Myzocytosis’, a kind of endocytosis with implications to compartmentation in endosymbiosis. Naturwissenschaften 71, 218–219. ( 10.1007/BF00490442) [DOI] [Google Scholar]
- 141.Larsen J. 1988. An ultrastructural study of Amphidinium poecilochroum (Dinophyceae), a phagotrophic dinoflagellate feeding on small species of cryptophytes. Phycologia 27, 366–377. ( 10.2216/i0031-8884-27-3-366.1) [DOI] [Google Scholar]
- 142.Cavalier-Smith T. 2018. Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 255, 297–357. ( 10.1007/s00709-017-1147-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Triemer RE. 1997. Feeding in Peranema trichophorum revisited (Euglenophyta). J. Phycol. 33, 649–654. ( 10.1111/j.0022-3646.1997.00649.x) [DOI] [Google Scholar]
- 144.Rueckert S, Pipaliya SV, Dacks JB. 2019. Evolution: parallel paths to parasitism in the apicomplexa. Curr. Biol. 29, R836–R839. ( 10.1016/j.cub.2019.07.047) [DOI] [PubMed] [Google Scholar]
- 145.Margulis L, McKhann HI, Olendzenski L. 1990. Illustrated glossary of protoctista Boston, MA: Jones & Bartlett. [Google Scholar]
- 146.Richards TA, Talbot NJ. 2018. Osmotrophy. Curr. Biol. 28, R1179–R1180. ( 10.1016/j.cub.2018.07.069) [DOI] [PubMed] [Google Scholar]
- 147.Flannagan RS, Jaumouillé V, Grinstein S. 2012. The cell biology of phagocytosis. Annu. Rev. Pathol. 7, 61–98. ( 10.1146/annurev-pathol-011811-132445) [DOI] [PubMed] [Google Scholar]
- 148.Holter H. 1959. Pinocytosis. Int. Rev. Cytol. 8, 481–504. ( 10.1016/s0074-7696(08)62738-2) [DOI] [PubMed] [Google Scholar]
- 149.Berney C, Romac S, Mahé F, Santini S, Siano R, Bass D. 2013. Vampires in the oceans: predatory cercozoan amoebae in marine habitats. ISME J. 7, 2387–2399. ( 10.1038/ismej.2013.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hess S. 2017. Hunting for agile prey: trophic specialisation in leptophryid amoebae (Vampyrellida, Rhizaria) revealed by two novel predators of planktonic algae. FEMS Microbiol. Ecol. 93, fix104 ( 10.1093/femsec/fix104) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.