Abstract
Cetuximab and panitumumab, as the highly effective antibodies targeting epidermal growth factor receptor (EGFR), have clinical activity in the patients with metastatic colorectal cancer (mCRC). These agents have good curative efficacy, but drug resistance also exists at the same time. The effects of KRAS, NRAS, and BRAF mutations and HER2 amplification on the treatment of refractory mCRC have been elucidated and the corresponding countermeasures have been put forward. However, the changes in EGFR and its ligands, the mutations or amplifications of PIK3CA, PTEN, TP53, MET, HER3, IRS2, FGFR1, and MAP2K1, the overexpression of insulin growth factor-1, the low expression of Bcl-2-interacting mediator of cell death, mismatch repair-deficient, and epigenetic instability may also lead to drug resistance in mCRC. Although the emergence of drug resistance has genetic or epigenetic heterogeneity, most of these molecular changes relating to it are focused on the key signaling pathways, such as the RAS/RAF/mitogen-activated protein kinase or phosphatidylinositol 3-kinase/Akt/mammalian target of the rapamycin pathway. Accordingly, numerous efforts to target these signaling pathways and develop the novel therapeutic regimens have been carried out. Herein, we have reviewed the underlying mechanisms of the resistance to anti-EGFR therapy and the possible implications in clinical practice.
Keywords: metastatic colorectal cancer, EGFR, drug resistance, cetuximab, panitumumab
Introduction
Colorectal cancer (CRC) is one of the most common tumors. Data from 185 countries have shown that >1.8 million new CRC cases and 881,000 deaths were estimated to have occurred in 2018 and the incidence and mortality rank third and second, respectively [1]. In addition, ∼20% of new cases are diagnosed as metastatic colorectal cancer (mCRC) at the time of initial visiting [2]. Fortunately, although the incidence and mortality of CRC in some underdeveloped areas are still increasing, they are declining in the whole world, especially in the developed countries [3]. At present, a limited number of targeted agents such as cetuximab, panitumumab, bevacizumab, aflibercept, and regorafenib have been shown to be active in the treatment of the patients with mCRC (hereinafter referred to as the patients or mCRCs). However, a number of the patients benefit a lot from the clinical application of these drugs [4–8].
Cetuximab is a chimeric (mouse/human) IgG-1 monoclonal antibody (mAb). As a targeted drug, cetuximab can competitively bind to epidermal growth factor receptor (EGFR) with the natural ligands such as EGF [9]. Its main pharmacological mechanism is to inhibit the phosphorylation of EGFR tyrosine kinase caused by the ligands binding and then block a series of reactions such as gene transcription and cell proliferation induced by the activation of the phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway and the RAS/RAF/mitogen-activated protein kinase (MAPK) pathway. Cetuximab can also mediate antibody-dependent cellular cytotoxicity and receptor internalization, so as to fully play the antitumor role [10–12]. The major side effects of cetuximab are skin toxicity and venous thromboembolic; skin rash may be related to the prognosis [13–17]. Panitumumab is a recombinant humanized IgG-2kappa mAb; both its pharmacological mechanisms and side effects are similar to those of cetuximab. In September 2006, panitumumab was approved by the US Food and Drug Administration as a single agent for the treatment of advanced CRC, but now it is mainly used in the combined-therapy regimens with other chemotherapeutic agents.
Although the use of cetuximab and panitumumab has greatly improved the overall survival (OS) and other clinical outcomes of mCRCs [4, 5], some patients cannot benefit from it. The discovery of the relationship between KRAS gene mutations and resistance to anti-EGFR mAbs has aroused the interest of a large number of scholars in the study of drug resistance. In this review, the molecular mechanisms of the resistance to anti-EGFR mAbs and the potential therapy regimens will be systematically elaborated on from three aspects: abnormal molecules in the EGFR pathway, abnormal activations between the paralleled pathways, and the other mechanisms.
Abnormal molecules in the EGFR pathway
Abnormalities of EGFR and its ligands
EGFR is one of the members of the ERBB/HER protein family that is composed of EGFR (HER1/ERBB1), HER2 (ERBB2/neu), HER3 (ERBB3), and HER4 (ERBB4). It consists of extracellular domains, transmembrane domains, and signal-transduction domains with tyrosine kinase activity. After binding to its ligands through the extracellular binding region, the proteins in the signal-transduction region phosphorylate and this finally leads to the intracellular cascade reactions that mainly promote cell proliferation [10, 18, 19]. In principle, the mutations of EGFR, the low expression of EGFR, and the changes in its ligands are detrimental to the efficacy of anti-EGFR therapy in some cancers. For example, in lung cancer, the EGFR gene copy number (GCN) or mutations (especially EGFRT790M) has been confirmed to be related to the resistance of molecule-targeted drugs [20, 21]; in the latest molecular-detection guideline of lung cancer, the importance of both EGFR GCN and mutations detection before the targeted treatment is reiterated [22]. Although some studies have pointed out that patients are all responsive to anti-EGFR mAbs regardless of the status of the EGFR [23–25], emerging evidence has shown that EGFR GCN and the acquired mutations of extracellular domains are related to the resistance to anti-EGFR mAbs in mCRCs [26–31], but EGFR-related detection in mCRCs has not been recommended at present [32]. Therefore, whether this indicator can be used as an efficacy predictor of anti-EGFR therapy in patients should be further investigated.
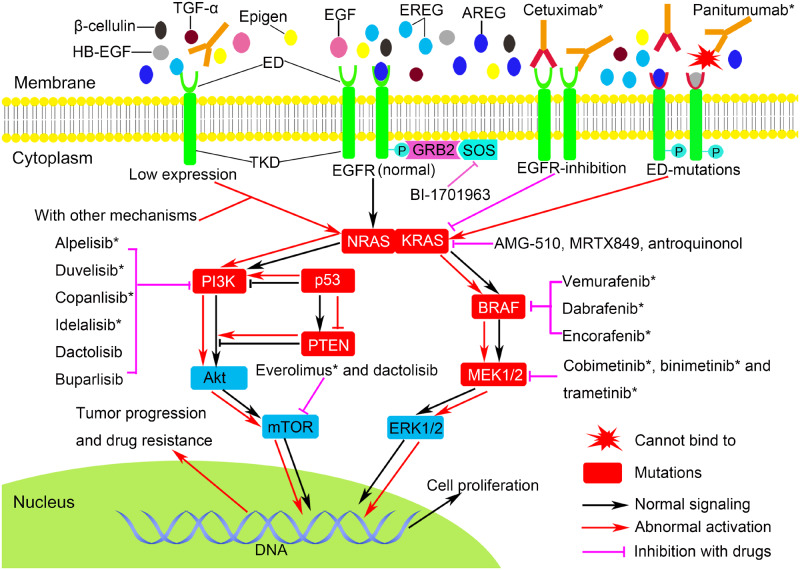
The ligands of EGFR mainly include EGF, transforming growth factor α (TGF-α), amphiregulin (AREG), epiregulin (EREG), epigen (EPGN), β-cellulin, and heparin-binding EGF (HB-EGF). As early as 2007, researchers had noticed that the expression levels of EREG and AREG are related to the efficacy of cetuximab in mCRCs [33]. Subsequently, accumulating studies have confirmed this finding; they have found that the higher the expression levels of these ligands, the better the observed efficacy of anti-EGFR mAbs in mCRCs can be, but this correlation is not consistent in patients with KRAS mutations [34]. In 2016, a randomized clinical trial of panitumumab plus irinotecan and ciclosporin in the treatment of advanced CRC has come out with a new ligand-expression model and pointed out that the expression levels of EREG and AREG are related to the therapeutic efficacy of panitumumab [35]. All of the above studies have suggested that the downregulation of EGFR ligands, especially EREG and AREG, may be one of the mechanisms of resistance to anti-EGFR mAbs. Therefore, the expression levels of EREG and AREG may be valuable markers for the choice of regimens in mCRCs, and it is worthy to be further optimized and then used for clinical guidance in order to reduce drug resistance and improve anti-EGFR mAbs efficacy (Figure 1).
Figure 1.
EGFR signaling and potential regimens in cetuximab- or panitumumab-resistant mCRC. In addition to NRAS/KRAS/BRAF mutations, low expression of EGFR/AREG/EREG, the mutations of extracellular domains of EGFR, PIK3CA, PTEN, TP53, and MEK1 are related to cetuximab or panitumumab resistance. The agents targeting BRAF and MEK1/2 have been approved for the subsequent therapy of advanced or metastatic CRC; some other targeted drugs such as PI3K, Mtor, and RAS inhibitors also deserve our attention, especially the KRAS G12C inhibitors AMG-510 and MRTX849. EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; HB-EGF, heparin-binding EGF; TGF-α, transforming growth factor α; AREG, amphiregulin; EREG, epiregulin; ED, extracellular domains; TKD, tyrosine kinase domains; GRB2, growth factor receptor bound-2; SOS, son of sevenless; MEK1/2, mitogen-activated protein kinase kinase; ERK1/2, extracellular-signal-regulated kinase; PI3K, phosphatidylinositol 3-kinase; mTOR, mammalian target of rapamycin; p, phosphorylation; * have been approved for markets.
Mutations of RAS
KRAS, HRAS, and NRAS are three major members of the RAS family. RAS as a proto-oncogene encodes the protein with GTPase activity, which plays a role of transducing and self-inactivating the signals in EGFR signaling. At present, the abnormalities of the RAS family have been reported to be related to a variety of tumors, and the mutations of KRAS and NRAS closely relate to CRC among them, but the relationship between CRC and the abnormality of HRAS is rarely reported [36]. According to the statistics, ∼37%–45% of CRCs harbor KRAS mutations in exon 2 (codons 12 or 13) and nearly 10% of CRCs harbor non-KRAS exon 2 RAS mutations that consist of non-exon 2 KRAS mutations (codons 59 or 61 mutations in exon 3, codons 147 or 117 mutations in exon 4) and NRAS mutations (codons 12 or 13 in exon 2 and codons 59 or 61 in exon 3) [5, 37–42].
Although studies have shown that the status of KRAS and NRAS is independent of the tumor staging [37, 43], since Lievre et al. [44] first reported that the mutations of KRAS can reduce the efficacy of anti-EGFR mAbs in 2006, emerging studies have confirmed that RAS mutations are the major causes of resistance to anti-EGFR therapy [5, 43, 45–50]. These pieces of evidence are mainly as follows: not only the resistance to anti-EGFR mAbs is related to KRAS mutations in exon 2 codons 12, but also the other mutant types of RAS such as codons 13 mutation in exon 2 and non-KRAS exon 2 RAS mutations can render the resistance to cetuximab or panitumumab in mCRCs; in addition to primary RAS mutations, there are many patients who harbor acquired RAS mutations in response to EGFR blockade. At present, the genotypes of KRAS and NRAS have become the main basis for choosing the chemotherapy regimen in mCRCs; anti-EGFR therapy in patients with KRAS and NRAS wild type (WT) has greatly improved their benefits. At the same time, in order to solve the problem of genes-detection sensitivity and specificity, not only should genes testing be performed only in the laboratories that are certified under the Clinical Laboratory Improvement Amendments of 1988 (CLIA-88), but also the latest National Comprehensive Cancer Network (NCCN) Colon/Rectal Cancer Panel and tumor-molecular-detection guidelines have recommended the next-generation sequencing (NGS) technology (Table 1) [32, 51].
Table 1.
Currently recommended molecular testing for colorectal cancer
| Biomarker | Test defect | Time | Technology | Recommendations | Evidence |
|---|---|---|---|---|---|
| KRAS/NRASa | Mutation | Workup for metastatic disease (suspected or proven) | NGSb, PCR | Avoid cetuximab or panitumumab treatment in mCRC patients who have tumors with KRAS and NRAS mutations (exons 2, 3, and 4 in both) | NCCN indicates lower-level, uniform acceptance (category 2A); but many believe classification is high-level, uniform acceptance |
| BRAFa | Mutation V600E | Workup for metastatic disease (suspected or proven) | NGS, PCRb | Cetucimab or panitumumab treatment is not recommended in mCRC patients harboring BRAFV600E mutation unless given with BRAF inhibitors with or without MEK inhibitors at the same time | NCCN indicates lower-level, uniform acceptance (category 2A); but many believe classification is high-level, uniform acceptance |
| HER2 | Amplification | Workup for metastatic or advanced disease (suspected or proven) | NGS, IHC, or FISH, need more evidence | Patients with HER2 amplification may be resistance to Cetuximab or panitumumab, trastuzumab plus (pertuzumab or lapatinib) regimen is an option for mCRC patients with HER2 amplified and RAS WT, but need more evidenced | NCCN indicates lower-level, wide acceptance (category 2B); but many believe classification is high-level, wide acceptance |
| NTRK | Gene fusion | Workup for metastatic disease (suspected or proven) | NGS, FISH, PCR | Larotrectinib is a treatment option for mCRC patients who are NTRK gene-fusion positive | NCCN indicates lower-level, uniform acceptance (category 2A); but many believe classification is high-level, uniform acceptance |
| MMR/MS | dMMRc/MSI-H | All patients with colon or rectal cancer | NGS, IHCc, PCR | Stage II MSI-H CRC patients may have a good prognosis but lack of efficacy of fluorouracil-based adjuvant therapy (nivolumab with or without ipilimumab) or pembrolizumab regimens are recommended for the patients with dMMR/MSI-H only | NCCN indicates lower-level, uniform acceptance (category 2A); but many believe classification is high-level, uniform acceptance |
NGS, next-generation sequencing; PCR, polymerase chain reaction; NCCN, National Comprehensive Cancer Network; IHC, immunohistochemistry; MMR, mismatch repair; dMMR, mismatch repair-deficient; MS, microsatellite; MSI-H, microsatellite highly instability; mCRC, metastatic colorectal cancer; FISH, fluorescence in situ hybridization; WT, wild-type.
KRAS and NRAS are determined alongside BRAF mutations.
Testing can be performed on primary and/or metastatic colorectal tissue specimens.
IHC refers to staining tumor tissue for protein expression of the four MMR genes known to be mutated in Lynch syndrome (MLH1, MSH2, MSH6, and PMS2).
If no previous treatment with HER2 inhibitor.
The structures of KRAS- and NRAS-mutant proteins are complex; although they have been known for a long time, the research and development of the related agents such as molecule-targeted drugs, mAbs, or vaccines are still quite difficult, and there is no RAS-targeted drug that has been successfully marketed so far. Therefore, the chemotherapy regimens with bevacizumab are still recommended in the guidelines for patients harboring RAS mutations. However, there have been several agents that have deserved our attention in recent years. In 2013, a paper showed that KRAS G12C could be targeted by a covalent compound that locks the mutant protein in its inactive GDP-bound state, which is supported by the discovery that KRAS G12C retains the highest residual intrinsic GTPase activity [52]. In addition, after the development and verification of the tool covalent compounds ARS-853 and ARS-1620, the first specific irreversible covalent inhibitors of KRAS-G12C in the clinic came from Amgen, AMG510, then from Mirati Therapeutics, MRTX849 (Figure 1). These two agents have been studying in clinic trials or in vitro and have been showing excellent antitumor efficacy that includes driving antitumor immunity [53–55]. Some clinic trials still aim to further confirm this evidence (NCT03600883 and NCT03785249). BI-1701963 is a non-specific inhibitor of KRAS; it blocks RAS signal transduction through inhibiting SOS1 selectively. The preclinical study has proved that BI-1701963 is effective for KRAS-mutant tumors. At present, China and other countries are participating in the global early clinical trial of this drug (NCT04111458). Antroquinol is also a non-specific inhibitor of RAS; it can indirectly inhibit the activation of RAS and RAS-related GTP-binding protein by inhibiting the activity of isoprenyl transferase and then promoting apoptosis. Preclinical studies have proved that antroquinol has a curative effect on RAS-mutant CRC [56, 57]. The trial of antroquinol as a first-line treatment of metastatic pancreatic cancer has been started (NCT03310632), but the research on mCRCs needs to be carried out. In addition, the vaccines designed for RAS-mutant peptide or immunotherapy with polyclonal T-cells for the patients with RAS mutations could also be a great breakthrough [58–61].
Mutations of BRAF
BRAF proto-oncogene belongs to the RAF genes family that includes two other members: CRAF and ARAF. As an important component of the RAS/RAF/MAPK pathway, BRAF mediates the combination of RAF and MAPK kinase (MAPKK/MEK1/2) in the signal transduction and regulates cell proliferation [62]. Statistics have shown that 30%–60% of melanoma, 30%–50% of thyroid cancer, and 5%–9% of CRC harbor BRAF mutations. The features of BRAF mutations in CRC are likely to occur in smokers and elderly and female patients whose primary tumor is mainly located in the right colon. Such patients commonly have high-grade cancers at diagnosis and a higher number of cancer-involved lymph nodes; they are more likely to be microsatellite instability-high (MSI-H) or mismatch repair-deficient (dMMR) and to progress rapidly [63].
BRAF V600 mutations are the most common mutations (about 80%). Among them, BRAFV600E accounts for >90% of BRAFV600 mutations, which refers to the T-to-A changing at nucleotide 1799 and a substitution of valine to glutamic acid [64]. The result of these changes is the rendering of BRAF protein to be an active monomer that is independent of the upstream signals, causing the synchronous and continuous activation of downstream pathways and promoting cell proliferation and differentiation. Some other rare BRAF-mutant types include mutant-mutant BRAF dimers that lead to high extracellular-signal-regulated kinase (ERK1/2) activation in a RAS-independent manner and mutant BRAF-wild-type CRAF dimers that amplify the signaling downstream of RAS. Many studies have proved that patients harboring BRAF mutations are resistant to cetuximab or panitumumab and have a generally poor prognosis [4, 5, 41, 65–67]. Therefore, BRAF status is a strong predictor for the predictive prognosis of CRC. In addition to RAS typing, high-sensitivity typing of the BRAF gene in mCRCs is equally important (Table 1) [32].
Different from the development of RAS inhibitors, many breakthroughs have been made in the development of BRAF inhibitors. At present, the approved BRAF monomers-targeted inhibitors include vemurafenib, dabrafenib, and encorafenib (LGX818) (Figure 1). They are mainly used in the treatment of melanoma, but the efficacy of single-agent therapy for mCRCs remains unclear [68], which is mainly related to the fact that, after inhibiting the MAPK pathway using BRAF inhibitors, EGFR can be activated by feedback in mCRCs, whereas melanoma only expresses low-level EGFR [69, 70]. However, the emergence of BRAF inhibitors makes the combined-therapy regimens possible. According to the latest NCCN Guidelines of Colon/Rectal Version, for the patients with BRAFV600E mutation, the first-line targeted-therapy regimens should include bevacizumab, while the subsequent options include targeting BRAF, MEK, and EGFR at the same time, such as dabrafenib plus trametinib and cetuximab or panitumumab, or encorafenib plus binimetinib (MEK162) and cetuximab or panitumumab. These two regimens have been proven to be effective and tolerable, which improves the benefits of the patients to some extent [71–73]; another multi-targeted-therapy regimen [MEK162 plus LGX818 and LEE011 (cyclin-dependent kinase (CDK) 4/CDK6 inhibitor)] is in a phase IB/II clinical trial (NCT01543698). In addition, as mentioned above, this type of patient situation is often accompanied by dMMR/MSI-H, so that anti-programmed death1 (PD-1)/anti-programmed death ligand 1 (PD-L1) and anti-cytotoxic T-lymphocyte antigen 4 (CTLA-4) immunotherapy may also have an ideal therapeutic effect [63, 74].
Mutations of PIK3CA, PTEN, and TP53
The KRAS/NRAS/BRAF status is a strong predictor of anti-EGFR mAbs efficacy in mCRCs, but more predictive factors are needed for the better selection of patients who will benefit highly from anti-EGFR mAbs. PIK3CA encodes p110alpha, a catalytic subunit of PI3K that mediates the PI3K/AKT pathway and promotes cell survival, whereas PTEN and TP53 are negative regulators of this pathway as the tumor-suppressor genes. PTEN plays a role as 3-phosphatase that dephosphorylates PIP3 or PIP2, and therefore inhibits the activation of PI3K and blocks this pathway [18, 75, 76]. TP53 encodes p53 protein, which acts not only as a guardian of the genome in normal cells, but also as an inhibitor of the oncogenes in cancer cells. Therefore, changes in PIK3CA could lead to the abnormal activation of the PI3K pathway, while the abnormalities of PTEN and TP53 genes could lead to the release of pathway inhibition. The final results could be that the regulation of the cells is out of control and abnormal proliferation of the cells occurs. The statistics have shown that the mutation rates of PIK3CA and PTEN in CRCs are ∼12% and ∼6%, respectively; mutations of PIK3CA are common in exon 9 and exon 20, and mainly occur in colon-cancer patients [77–79]. However, the frequency of TP53 mutations in CRCs is still unclear.
Increasing evidence has shown that the efficacy of anti-EGFR therapy will be greatly reduced in patients harboring the above mutations or expressing low PTEN and TP53 levels [80–87]. Although the differences in the clinical outcomes between patients with single exon 9 mutations of PIK3CA and patients with single exon 20 mutations of PIK3CA are not significant, when there are double mutations in patients, the clinical outcomes will be worse [85, 86]. Therefore, although there is no guideline to recommend using the genotypes of PIK3CA, PTEN, and TP53 to guide the choice of treatment regimens in CRCs, the role of these three genotypes or the expression levels of PTEN and p53 in anti-EGFR mAbs resistance should not be ignored.
The PI3K pathway has two major targets: PI3K and mTOR (Figure 1). At present, mTOR inhibitors such as everolimus are mainly used in the targeted therapy of advanced renal-cell carcinoma; they have not been recommended for the treatment of mCRC [88, 89]. Although they have shown curative efficacy in mCRCs, their safety and effectiveness still need to be further studied. At the same time, the study and development of PI3K inhibitors have achieved preliminary success: a number of PI3K inhibitors such as alpelisib (BYL719), duvelisib, copanlisib, and idelalisib have been approved for the targeted therapy of PIK3CA-mutant head and neck tumors, breast cancer, or lymphoma; besides, dactolisib (BEZ235, dual PI3K/mTOR inhibitor), PI-103, buparlisib (BKM120), and so on have been approved for the clinical trials of solid tumors. However, similarly to mTOR inhibitors, there are only a few studies that relate to mCRCs. A phase Ib dose-escalation study has shown that the combined treatment of encorafenib plus cetuximab and alpelisib is tolerable and provides promising clinical activity in the difficult-to-treat patient population with BRAF-mutant mCRC [90]. The results of some ongoing clinical trials that aim to combine PI3K or mTOR inhibitors with other targeted agents (NCT01719380, NCT01337765, and NCT01591421) to block the PI3K and MAPK pathways at the same time and eliminate drug resistance in mCRCs deserve our exploration. In addition, because PTEN mutations are also often accompanied by MSI-H [79], researchers are trying to use one PI3K inhibitor in combination with nivolumab to treat patients (NCT03711058, NCT03735628).
Abnormal activations between the parallel pathways
Amplifications or mutations of HER2 and HER3
HER2 and HER3 are two distinct members of the ERBB/HER protein family; they are both non-autonomous. HER2 (ERBB2/neu) does not bind with EGF-like ligands, but it acts as the preferred heterodimer partner of the other three members of the ERBB family [91]. Heterodimers such as EGFR/HER2 have higher affinity and specificity for the ligands [92]. Meanwhile, HER2 can bind more phosphotyrosine-binding protein than the other receptors [93]. In addition, the internalization process of dimers-containing HER2 will be slower and the receptors will return to the cell surface more effectively to receive the second stimulation [94, 95]. While the kinase activity of HER3 (ERBB3) is defective, it works by forming heterodimeric complexes with other receptors that are capable of generating potent cellular signals and then recruiting PI3K to six distinct sites in heterodimeric complexes, thus strongly activating PI3K in a RAS-independent manner [96]. Therefore, both HER2 and HER3 amplifications (also HER2 and HER3 overexpression) and mutations could increase the malignancy degree of the tumors through these mechanisms.
Although the amplification and mutations of HER2 are most common in breast cancer, research over the past decade has shown that 3%–5% of CRCs harbor primary overexpression of HER2 or HER2 mutations, and the prevalence is higher in RAS and BRAF WT CRCs (reported in about 5%–14%) (according to HERACLES criteria: immunohistochemistry 3+ or 2+ in >50% cells confirmed by fluorescence in situ hybridization) [97–100]. Meanwhile, 2% of patients harbor acquired mutations or amplification of HER2 gene after receiving anti-EGFR therapy, and their primary tumors mostly focus in the left colon and rectum and are RAS WT [101, 102]. However, the prevalence of HER3 amplification and mutations seems higher than that of HER2. According to the statistics, 5.7%–11% of CRCs harbor HER3 mutations [100, 103] and 51%–70% of CRCs show the overexpression of HER3 protein, but the evaluated criteria of HER3 overexpression are different [104, 105]. Since 2011, emerging studies have reported that the resistance to anti-EGFR mAbs is related to the primary or acquired amplification and mutations of HER2 [106–110], but the evidence does not support a prognostic role for HER2 overexpression [111]. As for HER3, although the study of the relationship between HER3 and anti-EGFR mAbs resistance is relatively later and smaller than that of HER2, much evidence has also revealed that abnormalities in HER3 could also reduce the efficacy of anti-EGFR therapy in the treatment of mCRCs [104, 112–114]. These findings reveal a novel mechanism of resistance to anti-EGFR mAbs: the abnormalities of HER2 and HER3. Therefore, HER2 has become a new target of mCRC genotyping and treatment up to now (Table 1). However, the diagnostic criteria for the overexpression of HER3, more authoritative evidence of the relationship between HER3 and the resistance to anti-EGFR mAbs, and HER3-targeted agents approved for markets are all required to render HER3 as a new genotyping and treatment target of mCRC.
HER2-targeted agents that have been approved for markets include the mAbs trastuzumab and pertuzumab, the inhibitors of ERBB family tyrosine kinase neratinib, afatinib, and pyrotinib, the conjugated agents trastuzumab-emtansine (T-DM1) and tratuzumab-hyaluronidase-oysk, and the dimerization inhibitors of EGFR/HER2 tyrosine kinase lapatinib (Table 2); they are mainly approved for the treatment of breast cancer. A number of multicenter, open phase II clinical trials have shown that trastuzumab plus lapatinib has good efficacy and tolerability in the treatment of patients who are HER2-positive [98, 115]; therefore, the latest NCCN guideline recommends that trastuzumab plus pertuzumab or lapatinib can be used in RAS WT mCRCs with HER2 amplification for the subsequent therapy. Although a randomized, open-label phase II trial of afatinib vs cetuximab in mCRCs has shown that the efficacy of afatinib is modest in patients with KRAS mutations [116], the use of tyrosine kinase inhibitors in mCRCs with or without HER2 mutations or amplification needs more evidence to support it [108]. The conjugated agents ado-trastuzumab-emtansine (T-DM1) and tratuzumab-hyaluronidase-oysk were approved for markets by the USA, Europe, and China for the treatment of HER2-positive breast cancer in 2019, but they have been rarely tried for mCRC. At present, many related clinical trials have been started (NCT03418558, NCT03225937, NCT03457896, NCT01919879, and NCT03843749). However, no HER3-targeted agent has been approved for markets. HER3-specific or bispecific inhibitors that have been approved for clinical trials in the past 5 years mainly include the following types: the humanized HER3 mAbs lumretuzumab, ISU104, and CDX-3379; the dual-action HER3/EGFR mAbs zenocutuzumab, duligotazumab (MEHD7945A); and the novel conjugated agent U3-1402 (Table 2); however, the efficacy of these agents in the trials is inconsistent and controversial [117–121]. In addition, both the efficacy and the tolerability of HER2/HER3-targeted drugs plus EGFR mAbs are also worthy of exploration.
Table 2.
Abnormal activations between the parallel pathways and the potential regimens
| Receptor/ligand | Functions | Change | Main targeted drugs (for market* or clinic trial) |
|---|---|---|---|
| HER2 | As the preferred heterodimeric partner of the other three ERBB members and amplifying the cascades | Amplification or mutations |
|
| HER3 | As the heterodimeric partner of the growth factor receptors and directly activating PI3K signaling in a RAS-independent way | Amplification or mutations |
|
| HGFR | Binding to HGF and activating multiple signal-transduction pathways, such as the MAPK pathway, the PI3K pathway, the NF-κB pathway, and the STAT3 pathway | Amplification |
|
| IGF1 | Binding to IGF1R and activating multiple growth-related signal-transduction pathways that include the MAPK pathway and the PI3K pathway | Amplification |
|
| IGF2 | Similar to IGF1 | Amplification |
HGF, hepatocyte growth factor; HGFR, hepatocyte growth factor receptor; PI3K, phosphatidylinositol 3-kinase; MAPK, mitogen-activated protein kinase; STAT3, transducer and activator of transcription 3; IGF, insulin-like growth factor; IGF1R, insulin-like growth factor 1 receptor; IR, insulin receptor; *have been approved for markets.
Amplification of MET
Hepatocyte growth factor receptor (HGFR, also known as MET tyrosine kinase receptor) is encoded by proto-oncogene MET [122], whereas its ligand HGF is produced by the surrounding fibroblasts and plays a biological effect by paracrine on the adjacent cells that express HGFR [19, 123]. The binding of HGF and HGFR leads to efficient activation of downstream signal-transduction pathways that include MAPK cascades, PI3K/Akt axis, the nuclear factor-κB inhibitor-α (IκBα)–nuclear factor-κB (NF-κB) complex, and the signal transducer and activator of transcription 3 (STAT3) [124]; besides, HGFR can also be coupled with HER3 to form a heterodimer under certain conditions [125]. Therefore, MET can not only participate in the occurrence of various morphological events both in the embryo and adulthood, but also drive the malignant progress or drug resistance of a variety of tumors, including CRC.
Currently, both preclinical and clinical studies have shown that MET abnormalities in CRC mainly manifest in gene amplification that can resist the inhibition effect of anti-EGFR mAbs on tumors and rarely in mutations [126–128]. Although the incidence of innate MET amplification in CRC patients is only 2%–4% [129, 130], as MET is similar to HER2, some CRCs harbor acquired MET amplification during the process of anti-EGFR therapy [128, 131, 132]. Hence, the failure of anti-EGFR therapy caused by the amplification of MET cannot be ignored.
At present, some non-specific inhibitors of MET tyrosine kinase such as crizotinib and cabozantinib have been approved for the treatment of other cancers, but all of the macromolecule or specific agents targeting HGFR/HGF are still at the clinical-trials stage. In recent years, the agents and relative clinical trials that deserve our special attention mainly include specific MET tyrosine kinase volitinib, conjugated drugs ABBV-399 and SHR-A1403, and dual-action EGFR/HGFR mAbs Sym-015, JNJ-61186372, LY3164530, and EMB01 (Table 2). Before that, many trials were related to the receptor mAb onartuzumab, the ligand mAb rilotumumab (AMG102), and the selective non-ATP competitive kinase inhibitor tivantinib (ARQ197). Both phase I and phase II clinical trials of those drugs have also shown great efficacy and tolerability in mCRCs [131, 133–135]. However, with many failures of those drugs in phase III clinical trials of other cancers [136–138], these drugs have not been approved for CRC yet in phase III clinical trials.
Overexpression of IGF1 and IGF2
Insulin-like growth factor 1 and 2 (IGF1 and IGF2) are quite important growth factors in the human body. IGF1 receptor (IGF1R) is the common receptor of IGF1 and IGF2. After binding with them, IGF1R can activate multiple signal-transduction pathways, including the RAS/RAF/MAPK and PI3K/AKT pathway. Therefore, the IGF1R and EGFR pathways have a close crosstalk and participate in cell proliferation, differentiation, angiogenesis, apoptosis, and other life processes together [139, 140]. Although the concentration of IGF2 in blood circulation is higher than that of IGF1, IGF1 plays a more important role in tumor progression than IGF2 because it is regulated by growth hormones, is not easily degraded, and has 15 times higher affinity with IGF1R [141, 142].
The evidence from clinical trials has shown that the overexpression of IGF1 can render the obvious resistance to anti-EGFR mAbs in mCRCs [104, 143], whereas patients with IGF2 overexpression only show lower sensitivity and a marginal response to anti-EGFR therapy [144]. However, few studies have reported the relationship between abnormal IGF1R and cancers. An in vitro study has shown that the regimen of anti-EGFR mAb ICR62 plus IGF1R tyrosine kinase inhibitor NVP-AEW541 can synergistically enhance the antitumor effect in some CRC cells [145]. Consequently, IGF1 and IGF1R could be two potential factors for anti-EGFR mAbs resistance.
Since IGF1 and IGF2 both act on IGF1R to play a physiological role, many researchers are focusing on targeting IGF1R. However, there are few breakthroughs in the development of IGF1R-targeted agents. Although an IGF1R mAb temprotumumab has been approved for the treatment of thyroid-associated ophthalmopathy, there is no trial of this agent for cancers. A randomized phase II clinical trial showed that the IGF1R inhibitor IMC-A12 has not shown the expected efficacy in vivo, either alone or combined with cetuximab [146]. A larger-sample-size II/III clinical trial suggested that an IGF1R inhibitor dalotuzumab (MK-0646) plus irinotecan and cetuximab regimen has good tolerability in patients, but there is no significant improvement in OS [147]. The other IGF1R inhibitor ganitumab plus panitumumab also did not improve the OS of mCRCs in a phase IB/II clinical study [133]. A phase I clinical trial confirmed that a dual-action inhibitor linsitinib (OSI-906) that targets IGF1R and insulin receptor (IR) at the same time has good antitumor activity in vivo [148]; unfortunately, before reaching the maximum tolerated dose, the OSI-906 plus mTOR inhibitor everolimus regimen has not manifested clinical efficacy when being used to treat refractory CRC [149]. At present, although it is feasible to inhibit IGF1R in theory, more clinical evidence and novel targeted agents are needed to support that IGF1R can be a therapeutic target after the emergence of resistance to anti-EGFR mAbs.
Other mechanisms
In addition to the primary or acquired changes in these common targets, some changes in genes or expression levels that have been rarely reported, unknown mutations, or epigenetic instability can also render the patients resistant to anti-EGFR therapy.
Some changes that have been rarely reported include NTRK gene fusion, IRS2 amplification or mutations, FGFR1 and MAP2K1 mutations, and the low expression of the Bcl-2-interacting mediator of cell death (Bim) [28, 150, 151]; the occurrence of unknown mutations mostly relates to dMMR. Previous studies have suggested that dMMR is associated with a good prognosis in early-stage disease and is rare in advanced patients, especially in sporadic CRC [152]. However, the latest research has pointed out that, under the pressure of anti-EGFR drugs, human CRC cells with mismatch repair proficient/microsatellite stable (pMMR/MSS) can manifest dMMR phenotype in a variety of ways, including downregulating the expression of mismatch repair (MMR) genes (MLH1, MLH2, and MLH6), homologous recombination genes (BBRCA2 and RAD51), and double-strand break repair gene EXO1, and gradually replacing high-fidelity DNA polymerase by low-fidelity DNA polymerases to participate in the DNA-replication process. Under the above integrated mechanisms, the genome of these cells begins to produce a large number of unknown adaptive mutations and MSI-H, which leads to these cells obtaining resistance to EGFR inhibitors. At the same time, the study also pointed out that the resistance to anti-EGFR agents in cancer cells will change from temporary to irreversible as the agents last longer [153], which put forward a new thinking on the choice of the targeted agents and the design of the treatment regimens.
Epigenetic instability includes aberrant DNA methylation, histone modification, chromosome remodeling, and non-coding RNA interference [154]; aberrant methylation phenotypes and microRNA (miRNA) interference are closely related to both the development and the drug resistance of CRC [155].
The data have shown that methylation events are more common than mutations in CRC patients [155, 156]. The whole-genome hypermethylation status of patients is significantly associated with the poor efficacy of anti-EGFR therapy, but many related mechanisms remain unclear [157]. At present, the study of cytosine-phosphate-guanine (CpG) island methylation phenotype (CIMP) is relatively extensive; however, due to lack of a unified definition standard, the prevalence of CIMP is temporarily unknown. Some analyses have shown that CIMP often accompanies BRAF mutations and relates to the low expression of MLH1, AREG, and EREG in CRCs, which may be a major mechanism by which patients are resistant to anti-EGFR mAbs [158, 159]. miRNA is a classification of endogenous small non-coding RNA that can regulate the expression of genes after transcription [160]. Although the study of miRNA interference with respect to resistance to anti-EGFR mAbs started relatively late, its role cannot be ignored due to the reversibility and activity of epigenetics that are similar to the interval drug resistance of mCRCs in clinic [161]. At present, miRNA-199a-5p/miRNA-375, miRNA-100/miRNA-125/miRNA-181a-5p, miRNA-425-5p, and miRNA-31-5p have been reported to be related to anti-EGFR therapy resistance in mCRCs [162–165] and the interference mechanisms of some miRNAs are relatively clear. MiRNA-199a-5p and miRNA-375 target PHLPP1, which indirectly activates the PI3K/AKT pathway [162], whereas miRNA-100/miRNA-125/miRNA-181a-5p regulates the Wnt/β-catenin-signaling pathway [163]. The above phenomena of epigenetic instability can interact with genetic mutations or amplification to mediate the resistance to anti-EGFR therapy in patients.
Discussion and future perspective
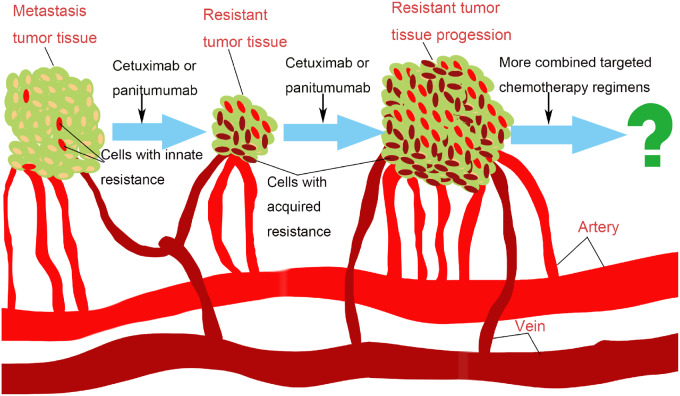
Anti-EGFR mAbs cetuximab and panitumumab were approved for the treatment of mCRC in 2004 and 2006, respectively; after years of clinical practice, they have been approved for the first-line treatment of KRAS/NRAS/BRAF WT mCRC. Although the OS, objective response rate, and progression-free survival of most patients with KRAS/NRAS/BRAF WT are significantly prolonged or increased after treatment with anti-EGFR mAbs, we have to face the reality that there are still some patients harboring the above genotypes who cannot obtain similar benefits, and even some patients who have experienced the clinical process of changing tumor inhibition to drug resistance and disease progression (Figure 2). This process is extremely complex and highly heterogeneous because there are both innate and acquired factors that contribute to the occurrence of drug resistance and, in addition to KRAS/NRAS/BRAF mutations, it is also related to a variety of molecular changes and genetic or epigenetic events. On the other hand, these mechanisms of drug resistance mainly manifest as EGFR-independent activations of the RAS/RAF/MAPK or PI3K/AKT pathway, which also provides some ideas for the solution to the anti-EGFR mAbs-resistance problem.
Figure 2.
The clinical process of changing tumor inhibition to drug resistance and tumor progression. This process is extremely complex and highly heterogeneous because there are both innate and acquired factors that contribute to the occurrence of drug resistance and the strategy of combined targeted chemotherapy deserves our attention.
At present, the principle of precise classification for treatment is being widely adopted to solve this problem. The NCCN Guidelines of Colon/Rectal Cancer Version have recommended the treatment regimens of KRAS/NRAS/BRAF mutations, HER2 amplifications, and dMMR/MSI (Table 1). Meanwhile, in order to obtain more accurate genotypes, the latest guidelines also have recommended using NGS technology for the genes detection of CRCs. However, with more agents that act on the resistance-related targets entering clinical trials or being approved for markets, the regimens of combined targeted chemotherapy could solve this problem under the premise of tolerable toxicity. Therefore, evaluation has now become the most important task (Figure 2), which should include the toxicity, the comparative efficacy, the best combination regimens, the timing of medication, and the appropriate efficacy evaluation biomarkers or index. We are looking forward to more positive results in order to improve the clinical efficacy of anti-EGFR therapy in mCRCs.
Authors' contributions
All authors contributed to the concept development, determined the search strategy, evaluated the results for inclusion, and provided critical review of the manuscript. Finally, all authors have read and approved the final manuscript.
Acknowledgements
None
Conflicts of interest
The authors declare that they have no competing interests.
Funding
This work was supported by the National Natural Science Foundation of China [81871994 and 81701834], the Guangdong Natural Science Foundation [2019B151502063], and the Guangdong Science and Technology Planning Program [20190202018].
References
- 1. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Ng K, Zhu AX.. Targeting the epidermal growth factor receptor in metastatic colorectal cancer. Crit Rev Oncol Hematol 2008;65:8–20. [DOI] [PubMed] [Google Scholar]
- 3. Arnold M, Sierra MS, Laversanne M. et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683–91. [DOI] [PubMed] [Google Scholar]
- 4. Maughan TS, Adams RA, Smith CG. et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet 2011;377:2103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douillard JY, Oliner KS, Siena S. et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023–34. [DOI] [PubMed] [Google Scholar]
- 6. Hurwitz H, Fehrenbacher L, Novotny W. et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004;350:2335–42. [DOI] [PubMed] [Google Scholar]
- 7. Van Cutsem E, Tabernero J, Lakomy R. et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol 2012;30:3499–506. [DOI] [PubMed] [Google Scholar]
- 8. Grothey A, Van Cutsem E, Sobrero A. et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013;381:303–12. [DOI] [PubMed] [Google Scholar]
- 9. Li S, Schmitz KR, Jeffrey PD. et al. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 2005;7:301–11. [DOI] [PubMed] [Google Scholar]
- 10. Ciardiello F, Tortora G.. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160–74. [DOI] [PubMed] [Google Scholar]
- 11. Kurai J, Chikumi H, Hashimoto K. et al. Antibody-dependent cellular cytotoxicity mediated by cetuximab against lung cancer cell lines. Clin Cancer Res 2007;13:1552–61. [DOI] [PubMed] [Google Scholar]
- 12. Tomas A, Futter CE, Eden ER.. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol 2014;24:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petrelli F, Borgonovo K, Barni S.. The predictive role of skin rash with cetuximab and panitumumab in colorectal cancer patients: a systematic review and meta-analysis of published trials. Target Oncol 2013;8:173–81. [DOI] [PubMed] [Google Scholar]
- 14. Stintzing S, Kapaun C, Laubender RP. et al. Prognostic value of cetuximab-related skin toxicity in metastatic colorectal cancer patients and its correlation with parameters of the epidermal growth factor receptor signal transduction pathway: results from a randomized trial of the GERMAN AIO CRC Study Group. Int J Cancer 2013;132:236–45. [DOI] [PubMed] [Google Scholar]
- 15. Van Cutsem E, Tejpar S, Vanbeckevoort D. et al. Intrapatient cetuximab dose escalation in metastatic colorectal cancer according to the grade of early skin reactions: the randomized EVEREST study. J Clin Oncol 2012;30:2861–8. [DOI] [PubMed] [Google Scholar]
- 16. Petrelli F, Cabiddu M, Borgonovo K. et al. Risk of venous and arterial thromboembolic events associated with anti-EGFR agents: a meta-analysis of randomized clinical trials. Ann Oncol 2012;23:1672–9. [DOI] [PubMed] [Google Scholar]
- 17. Zhang D, Ye J, Xu T. et al. Treatment related severe and fatal adverse events with cetuximab in colorectal cancer patients: a meta-analysis. J Chemother 2013;25:170–5. [DOI] [PubMed] [Google Scholar]
- 18. Citri A, Yarden Y.. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006;7:505–16. [DOI] [PubMed] [Google Scholar]
- 19. Chen WS, Lazar CS, Poenie M. et al. Requirement for intrinsic protein tyrosine kinase in the immediate and late actions of the EGF receptor. Nature 1987;328:820–3. [DOI] [PubMed] [Google Scholar]
- 20. Fukuoka M, Wu YL, Thongprasert S. et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866–74. [DOI] [PubMed] [Google Scholar]
- 21. Bonomi PD, Gandara D, Hirsch FR. et al. Predictive biomarkers for response to EGFR-directed monoclonal antibodies for advanced squamous cell lung cancer. Ann Oncol 2018;29:1701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindeman NI, Cagle PT, Aisner DL. et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323–58. [DOI] [PubMed] [Google Scholar]
- 23. Chung KY, Shia J, Kemeny NE. et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005;23:1803–10. [DOI] [PubMed] [Google Scholar]
- 24. Hecht JR, Mitchell E, Neubauer MA. et al. Lack of correlation between epidermal growth factor receptor status and response to panitumumab monotherapy in metastatic colorectal cancer. Clin Cancer Res 2010;16:2205–13. [DOI] [PubMed] [Google Scholar]
- 25. Cunningham D, Humblet Y, Siena S. et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337–45. [DOI] [PubMed] [Google Scholar]
- 26. Moroni M, Veronese S, Benvenuti S. et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 2005;6:279–86. [DOI] [PubMed] [Google Scholar]
- 27. Sartore-Bianchi A, Moroni M, Veronese S. et al. Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol 2007;25:3238–45. [DOI] [PubMed] [Google Scholar]
- 28. Bertotti A, Papp E, Jones S. et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015;526:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arena S, Bellosillo B, Siravegna G. et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res 2015;21:2157–66. [DOI] [PubMed] [Google Scholar]
- 30. Llovet P, Sastre J, Ortega JS. et al. Prognostic value of BRAF, PI3K, PTEN, EGFR copy number, amphiregulin and epiregulin status in patients with KRAS Codon 12 wild-type metastatic colorectal cancer receiving first-line chemotherapy with anti-EGFR therapy. Mol Diagn Ther 2015;19:397–408. [DOI] [PubMed] [Google Scholar]
- 31. Van Emburgh BO, Arena S, Siravegna G. et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun 2016;7:13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. El-Deiry WS, Goldberg RM, Lenz HJ. et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J Clin 2019;69:305–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khambata-Ford S, Garrett CR, Meropol NJ. et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007;25:3230–7. [DOI] [PubMed] [Google Scholar]
- 34. Jacobs B, De Roock W, Piessevaux H. et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol 2009;27:5068–74. [DOI] [PubMed] [Google Scholar]
- 35. Seligmann JF, Elliott F, Richman SD. et al. Combined epiregulin and amphiregulin expression levels as a predictive biomarker for panitumumab therapy benefit or lack of benefit in patients with RAS wild-type advanced colorectal cancer. JAMA Oncol 2016;2:633–42. [DOI] [PubMed] [Google Scholar]
- 36. Karnoub AE, Weinberg RA.. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol 2008;9:517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brink M, de Goeij AF, Weijenberg MP. et al. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis 2003;24:703–10. [DOI] [PubMed] [Google Scholar]
- 38. Peeters M, Price TJ, Cervantes A. et al. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol 2010;28:4706–13. [DOI] [PubMed] [Google Scholar]
- 39. Fernandez-Medarde A, Santos E.. Ras in cancer and developmental diseases. Genes Cancer 2011;2:344–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bos JL. ras oncogenes in human cancer: a review. Cancer Res 1989;49:4682–9. [PubMed] [Google Scholar]
- 41. Van Cutsem E, Kohne CH, Lang I. et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011–9. [DOI] [PubMed] [Google Scholar]
- 42. Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol 2015;33:1809–24. [DOI] [PubMed] [Google Scholar]
- 43. Schirripa M, Cremolini C, Loupakis F. et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer 2015;136:83–90. [DOI] [PubMed] [Google Scholar]
- 44. Lievre A, Bachet JB, Le Corre D. et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992–5. [DOI] [PubMed] [Google Scholar]
- 45. Peeters M, Oliner KS, Price TJ. et al. Analysis of KRAS/NRAS mutations in a phase III study of panitumumab with FOLFIRI compared with FOLFIRI alone as second-line treatment for metastatic colorectal cancer. Clin Cancer Res 2015;21:5469–79. [DOI] [PubMed] [Google Scholar]
- 46. Karapetis CS, Khambata-Ford S, Jonker DJ. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757–65. [DOI] [PubMed] [Google Scholar]
- 47. Van Cutsem E, Köhne CH, Hitre E. et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360:1408–17. [DOI] [PubMed] [Google Scholar]
- 48. Schirripa M, Loupakis F, Lonardi S. et al. Phase II study of single-agent cetuximab in KRAS G13D mutant metastatic colorectal cancer. Ann Oncol 2015;26:2503. [DOI] [PubMed] [Google Scholar]
- 49. Segelov E, Thavaneswaran S, Waring PM. et al. Response to cetuximab with or without irinotecan in patients with refractory metastatic colorectal cancer harboring the KRAS G13D mutation: Australasian Gastro-Intestinal Trials Group ICECREAM Study. J Clin Oncol 2016;34:2258–64. [DOI] [PubMed] [Google Scholar]
- 50. Cremolini C, Rossini D, Dell’Aquila E. et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol 2019;5:343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Linardou H, Dahabreh IJ, Kanaloupiti D. et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008;9:962–72. [DOI] [PubMed] [Google Scholar]
- 52. Ostrem JM, Peters U, Sos ML. et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503:548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hallin J, Engstrom LD, Hargis L. et al. The KRAS inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coelho MA, de Carné Trécesson S, Rana S. et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity 2017;47:1083–99.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Canon J, Rex K, Saiki AY. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 56. Ho CL, Wang JL, Lee CC. et al. Antroquinonol blocks Ras and Rho signaling via the inhibition of protein isoprenyltransferase activity in cancer cells. Biomed Pharmacother 2014;68:1007–14. [DOI] [PubMed] [Google Scholar]
- 57. Lin HC, Lin MH, Liao JH. et al. Antroquinonol, a ubiquinone derivative from the mushroom antrodia camphorata, inhibits colon cancer stem cell-like properties: insights into the molecular mechanism and inhibitory targets. J Agric Food Chem 2017;65:51–9. [DOI] [PubMed] [Google Scholar]
- 58. Toubaji A, Achtar M, Provenzano M. et al. Pilot study of mutant ras peptide-based vaccine as an adjuvant treatment in pancreatic and colorectal cancers. Cancer Immunol Immunother 2008;57:1413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carbone DP, Ciernik IF, Kelley MJ. et al. Immunization with mutant p53- and K-ras-derived peptides in cancer patients: immune response and clinical outcome. J Clin Oncol 2005;23:5099–107. [DOI] [PubMed] [Google Scholar]
- 60. Gjertsen MK, Bakka A, Breivik J. et al. Vaccination with mutant ras peptides and induction of T-cell responsiveness in pancreatic carcinoma patients carrying the corresponding RAS mutation. Lancet 1995;346:1399–400. [DOI] [PubMed] [Google Scholar]
- 61. Tran E, Robbins PF, Lu YC. et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375:2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wellbrock C, Karasarides M, Marais R.. The RAF proteins take centre stage. Nat Rev Mol Cell Biol 2004;5:875–85. [DOI] [PubMed] [Google Scholar]
- 63. Gonsalves WI, Mahoney MR, Sargent DJ. et al. Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/Alliance N0147. J Natl Cancer Inst 2014;106:dju106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Davies H, Bignell GR, Cox C. et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 65. Di Nicolantonio F, Martini M, Molinari F. et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705–12. [DOI] [PubMed] [Google Scholar]
- 66. Pietrantonio F, Petrelli F, Coinu A. et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 2015;51:587–94. [DOI] [PubMed] [Google Scholar]
- 67. Bokemeyer C, Van Cutsem E, Rougier P. et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer 2012;48:1466–75. [DOI] [PubMed] [Google Scholar]
- 68. Kopetz S, Desai J, Chan E. et al. Phase II pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol 2015;33:4032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prahallad A, Sun C, Huang S. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
- 70. Corcoran RB, Ebi H, Turke AB. et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Corcoran RB, Andre T, Atreya CE. et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov 2018;8:428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kopetz S, Grothey A, Yaeger R. et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 73. Van Cutsem E, Huijberts S, Grothey A. et al. Binimetinib, encorafenib, and cetuximab triplet therapy for patients with BRAF V600E-mutant metastatic colorectal cancer: safety lead-in results from the phase III BEACON Colorectal Cancer Study. J Clin Oncol 2019;37:1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Clancy C, Burke JP, Kalady MF. et al. BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: a systematic review and meta-analysis. Colorectal Dis 2013;15:e711–8. [DOI] [PubMed] [Google Scholar]
- 75. Malek M, Kielkowska A, Chessa T. et al. PTEN regulates PI(3,4)P2 signaling downstream of Class I PI3K. Mol Cell 2017;68:566–80.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Simpson L, Parsons R.. PTEN: life as a tumor suppressor. Exp Cell Res 2001;264:29–41. [DOI] [PubMed] [Google Scholar]
- 77. Shen Y, Wang J, Han X. et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling ofKRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One 2013;8:e81628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sukawa Y, Yamamoto H, Nosho K. et al. Alterations in the human epidermal growth factor receptor 2-phosphatidylinositol 3-kinase-v-Akt pathway in gastric cancer. World J Gastroenterol 2012;18:6577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Day FL, Jorissen RN, Lipton L. et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clin Cancer Res 2013;19:3285–96. [DOI] [PubMed] [Google Scholar]
- 80. Sartore-Bianchi A, Martini M, Molinari F. et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69:1851–7. [DOI] [PubMed] [Google Scholar]
- 81. Razis E, Pentheroudakis G, Rigakos G. et al. EGFR gene gain and PTEN protein expression are favorable prognostic factors in patients with KRAS wild-type metastatic colorectal cancer treated with cetuximab. J Cancer Res Clin Oncol 2014;140:737–48. [DOI] [PubMed] [Google Scholar]
- 82. Oden-Gangloff A, Di Fiore F, Bibeau F. et al. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br J Cancer 2009;100:1330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Di Bartolomeo M, Pietrantonio F, Perrone F. et al. Lack of KRAS, NRAS, BRAF and TP53 mutations improves outcome of elderly metastatic colorectal cancer patients treated with cetuximab, oxaliplatin and UFT. Target Oncol 2014;9:155–62. [DOI] [PubMed] [Google Scholar]
- 84. Huemer F, Thaler J, Piringer G. et al. Sidedness and TP53 mutations impact OS in anti-EGFR but not anti-VEGF treated mCRC—an analysis of the KRAS registry of the AGMT (Arbeitsgemeinschaft Medikamentöse Tumortherapie). BMC Cancer 2018;18:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Therkildsen C, Bergmann TK, Henrichsen-Schnack T. et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: a systematic review and meta-analysis. Acta Oncol 2014;53:852–64. [DOI] [PubMed] [Google Scholar]
- 86. Liao X, Morikawa T, Lochhead P. et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res 2012;18:2257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vitiello PP, Cardone C, Martini G. et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res 2019;38:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cai Z, Ke J, He X. et al. Significance of mTOR signaling and its inhibitor against cancer stem-like cells in colorectal cancer. Ann Surg Oncol 2014;21:179–88. [DOI] [PubMed] [Google Scholar]
- 89. Wolpin BM, Ng K, Zhu AX. et al. Multicenter phase II study of tivozanib (AV-951) and everolimus (RAD001) for patients with refractory, metastatic colorectal cancer. Oncologist 2013;18:377–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. van Geel R, Tabernero J, Elez E. et al. A phase Ib dose-escalation study of encorafenib and cetuximab with or without alpelisib in metastatic BRAF-mutant colorectal cancer. Cancer Discov 2017;7:610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tzahar E, Waterman H, Chen X. et al. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol 1996;16:5276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Worthylake R, Opresko LK, Wiley HS.. ErbB-2 amplification inhibits down-regulation and induces constitutive activation of both ErbB-2 and epidermal growth factor receptors. J Biol Chem 1999;274:8865–74. [DOI] [PubMed] [Google Scholar]
- 93. Jones RB, Gordus A, Krall JA. et al. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 2006;439:168–74. [DOI] [PubMed] [Google Scholar]
- 94. Baulida J, Kraus MH, Alimandi M. et al. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem 1996;271:5251–7. [DOI] [PubMed] [Google Scholar]
- 95. Lenferink AE, Pinkas-Kramarski R, van de Poll ML. et al. Differential endocytic routing of homo- and hetero-dimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. Embo j 1998;17:3385–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Black LE, Longo JF, Carroll SL.. Mechanisms of receptor tyrosine-protein kinase ErbB-3 (ERBB3) action in human neoplasia. Am J Pathol 2019;189:1898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cancer Genome Altas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sartore-Bianchi A, Trusolino L, Martino C. et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:738–46. [DOI] [PubMed] [Google Scholar]
- 99. Siena S, Sartore-Bianchi A, Marsoni S. et al. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann Oncol 2018;29:1108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Loree JM, Bailey AM, Johnson AM. et al. Molecular landscape of ERBB2/ERBB3 mutated colorectal cancer. J Natl Cancer Inst 2018;110:1409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. La Salvia A, Lopez-Gomez V, Garcia-Carbonero R.. HER2-targeted therapy: an emerging strategy in advanced colorectal cancer. Expert Opin Investig Drugs 2019;28:29–38. [DOI] [PubMed] [Google Scholar]
- 102. Mohan S, Heitzer E, Ulz P. et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 2014;10:e1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jaiswal BS, Kljavin NM, Stawiski EW. et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 2013;23:603–17. [DOI] [PubMed] [Google Scholar]
- 104. Scartozzi M, Giampieri R, Maccaroni E. et al. Analysis of HER-3, insulin growth factor-1, nuclear factor-kB and epidermal growth factor receptor gene copy number in the prediction of clinical outcome for K-RAS wild-type colorectal cancer patients receiving irinotecan-cetuximab. Ann Oncol 2012;23:1706–12. [DOI] [PubMed] [Google Scholar]
- 105. Lédel F, Hallström M, Ragnhammar P. et al. HER3 expression in patients with primary colorectal cancer and corresponding lymph node metastases related to clinical outcome. Eur J Cancer (Oxford, England: 1990) 2014;50:656–62. [DOI] [PubMed] [Google Scholar]
- 106. Ciardiello F, Normanno N.. HER2 signaling and resistance to the anti-EGFR monoclonal antibody cetuximab: a further step toward personalized medicine for patients with colorectal cancer. Cancer Discov 2011;1:472–4. [DOI] [PubMed] [Google Scholar]
- 107. Yonesaka K, Zejnullahu K, Okamoto I. et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med 2011;3:99ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kavuri SM, Jain N, Galimi F. et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 2015;5:832–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bertotti A, Migliardi G, Galimi F. et al. A molecularly annotated platform of patient-derived xenografts (‘xenopatients’) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1:508–23. [DOI] [PubMed] [Google Scholar]
- 110. Martin V, Landi L, Molinari F. et al. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013;108:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wu S-W, Ma C-C, Li W-h.. Does overexpression of HER-2 correlate with clinicopathological characteristics and prognosis in colorectal cancer? Evidence from a meta-analysis. Diagn Pathol 2015;10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Scartozzi M, Mandolesi A, Giampieri R. et al. The role of HER-3 expression in the prediction of clinical outcome for advanced colorectal cancer patients receiving irinotecan and cetuximab. Oncologist 2011;16:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Seligmann JF, Hatch AJ, Richman SD. et al. Association of tumor HER3 messenger RNA expression with panitumumab efficacy in advanced colorectal cancer. JAMA Oncol 2018;4:564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lièvre A, Ouine B, Canet J. et al. Protein biomarkers predictive for response to anti-EGFR treatment in RAS wild-type metastatic colorectal carcinoma. Br J Cancer 2017;117:1819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Meric-Bernstam F, Hurwitz H, Raghav KPS. et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol 2019;20:518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hickish T, Cassidy J, Propper D. et al. A randomised, open-label phase II trial of afatinib versus cetuximab in patients with metastatic colorectal cancer. Eur J Cancer (Oxford, England: 1990) 2014;50:3136–44. [DOI] [PubMed] [Google Scholar]
- 117. Koganemaru S, Kuboki Y, Koga Y. et al. U3-1402, a novel HER3-targeting antibody-drug conjugate, for the treatment of colorectal cancer. Mol Cancer Ther 2019;18:2043–50. [DOI] [PubMed] [Google Scholar]
- 118. Lieu CH, Hidalgo M, Berlin JD. et al. A phase Ib dose-escalation study of the safety, tolerability, and pharmacokinetics of cobimetinib and duligotuzumab in patients with previously treated locally advanced or metastatic cancers with mutant KRAS. Oncologist 2017;22:1024–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Meulendijks D, Jacob W, Martinez-Garcia M. et al. First-in-human phase I study of lumretuzumab, a glycoengineered humanized anti-HER3 monoclonal antibody, in patients with metastatic or advanced HER3-positive solid tumors. Clin Cancer Res 2016;22:877–85. [DOI] [PubMed] [Google Scholar]
- 120. Juric D, Dienstmann R, Cervantes A. et al. Safety and pharmacokinetics/pharmacodynamics of the first-in-class dual action HER3/EGFR antibody MEHD7945A in locally advanced or metastatic epithelial tumors. Clin Cancer Res 2015;21:2462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hill AG, Findlay MP, Burge ME. et al. Phase II study of the dual EGFR/HER3 inhibitor duligotuzumab (MEHD7945A) versus cetuximab in combination with FOLFIRI in second-line wild-type metastatic colorectal cancer. Clin Cancer Res 2018;24:2276–84. [DOI] [PubMed] [Google Scholar]
- 122. Bottaro DP, Rubin JS, Faletto DL. et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991;251:802–4. [DOI] [PubMed] [Google Scholar]
- 123. Stoker M, Gherardi E, Perryman M. et al. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature 1987;327:239–42. [DOI] [PubMed] [Google Scholar]
- 124. Trusolino L, Bertotti A, Comoglio PM.. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol 2010;11:834–48. [DOI] [PubMed] [Google Scholar]
- 125. Engelman JA, Zejnullahu K, Mitsudomi T. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 126. Luraghi P, Reato G, Cipriano E. et al. MET signaling in colon cancer stem-like cells blunts the therapeutic response to EGFR inhibitors. Cancer Res 2014;74:1857–69. [DOI] [PubMed] [Google Scholar]
- 127. Liska D, Chen CT, Bachleitner-Hofmann T. et al. HGF rescues colorectal cancer cells from EGFR inhibition via MET activation. Clin Cancer Res 2011;17:472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bardelli A, Corso S, Bertotti A. et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013;3:658–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Misale S, Di Nicolantonio F, Sartore-Bianchi A. et al. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 2014;4:1269–80. [DOI] [PubMed] [Google Scholar]
- 130. Zhang M, Li G, Sun X. et al. MET amplification, expression, and exon 14 mutations in colorectal adenocarcinoma. Hum Pathol 2018;77:108–15. [DOI] [PubMed] [Google Scholar]
- 131. Pietrantonio F, Oddo D, Gloghini A. et al. MET-driven resistance to dual EGFR and BRAF blockade may be overcome by switching from EGFR to MET inhibition in BRAF-mutated colorectal cancer. Cancer Discov 2016;6:963–71. [DOI] [PubMed] [Google Scholar]
- 132. Pietrantonio F, Vernieri C, Siravegna G. et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin Cancer Res 2017;23:2414–22. [DOI] [PubMed] [Google Scholar]
- 133. Van Cutsem E, Eng C, Nowara E. et al. Randomized phase Ib/II trial of rilotumumab or ganitumab with panitumumab versus panitumumab alone in patients with wild-type KRAS metastatic colorectal cancer. Clin Cancer Res 2014;20:4240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Eng C, Bessudo A, Hart LL. et al. A randomized, placebo-controlled, phase 1/2 study of tivantinib (ARQ 197) in combination with irinotecan and cetuximab in patients with metastatic colorectal cancer with wild-type KRAS who have received first-line systemic therapy. Int J Cancer 2016;139:177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bendell JC, Ervin TJ, Gallinson D. et al. Treatment rationale and study design for a randomized, double-blind, placebo-controlled phase II study evaluating onartuzumab (MetMAb) in combination with bevacizumab plus mFOLFOX-6 in patients with previously untreated metastatic colorectal cancer. Clin Colorectal Cancer 2013;12:218–22. [DOI] [PubMed] [Google Scholar]
- 136. Spigel DR, Edelman MJ, O’Byrne K. et al. Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non-small-cell lung cancer: METLung. J Clin Oncol 2017;35:412–20. [DOI] [PubMed] [Google Scholar]
- 137. Catenacci DVT, Tebbutt NC, Davidenko I. et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:1467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Rimassa L, Assenat E, Peck-Radosavljevic M. et al. Tivantinib for second-line treatment of MET-high, advanced hepatocellular carcinoma (METIV-HCC): a final analysis of a phase 3, randomised, placebo-controlled study. Lancet Oncol 2018;19:682–93. [DOI] [PubMed] [Google Scholar]
- 139. Furstenberger G, Senn HJ.. Insulin-like growth factors and cancer. Lancet Oncol 2002;3:298–302. [DOI] [PubMed] [Google Scholar]
- 140. Hu YP, Patil SB, Panasiewicz M. et al. Heterogeneity of receptor function in colon carcinoma cells determined by cross-talk between type I insulin-like growth factor receptor and epidermal growth factor receptor. Cancer Res 2008;68:8004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Jones JI, Clemmons DR.. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 1995;16:3–34. [DOI] [PubMed] [Google Scholar]
- 142. Rajaram S, Baylink DJ, Mohan S.. Insulin-like growth factor-binding proteins in serum and other biological fluids: regulation and functions. Endocr Rev 1997;18:801–31. [DOI] [PubMed] [Google Scholar]
- 143. Scartozzi M, Mandolesi A, Giampieri R. et al. Insulin-like growth factor 1 expression correlates with clinical outcome in K-RAS wild type colorectal cancer patients treated with cetuximab and irinotecan. Int J Cancer 2010;127:1941–7. [DOI] [PubMed] [Google Scholar]
- 144. Zanella ER, Galimi F, Sassi F. et al. IGF2 is an actionable target that identifies a distinct subpopulation of colorectal cancer patients with marginal response to anti-EGFR therapies. Sci Transl Med 2015;7:272ra12. [DOI] [PubMed] [Google Scholar]
- 145. Cunningham MP, Thomas H, Marks C. et al. Co-targeting the EGFR and IGF-IR with anti-EGFR monoclonal antibody ICR62 and the IGF-IR tyrosine kinase inhibitor NVP-AEW541 in colorectal cancer cells. Int J Oncol 2008;33:1107–13. [PubMed] [Google Scholar]
- 146. Reidy DL, Vakiani E, Fakih MG. et al. Randomized, phase II study of the insulin-like growth factor-1 receptor inhibitor IMC-A12, with or without cetuximab, in patients with cetuximab- or panitumumab-refractory metastatic colorectal cancer. J Clin Oncol 2010;28:4240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sclafani F, Kim TY, Cunningham D. et al. A randomized phase II/III study of dalotuzumab in combination with cetuximab and irinotecan in chemorefractory, KRAS wild-type, metastatic colorectal cancer. J Natl Cancer Inst 2015;107:djv258. [DOI] [PubMed] [Google Scholar]
- 148. Puzanov I, Lindsay CR, Goff L. et al. A phase I study of continuous oral dosing of OSI-906, a dual inhibitor of insulin-like growth factor-1 and insulin receptors, in patients with advanced solid tumors. Clin Cancer Res 2015;21:701–11. [DOI] [PubMed] [Google Scholar]
- 149. Bendell JC, Jones SF, Hart L. et al. A phase Ib study of linsitinib (OSI-906), a dual inhibitor of IGF-1R and IR tyrosine kinase, in combination with everolimus as treatment for patients with refractory metastatic colorectal cancer. Invest New Drugs 2015;33:187–93. [DOI] [PubMed] [Google Scholar]
- 150. Lian T, Li C, Wang H.. Trametinib in the treatment of multiple malignancies harboring MEK1 mutations. Cancer Treat Rev 2019;81:101907. [DOI] [PubMed] [Google Scholar]
- 151. Marzi L, Combes E, Vié N. et al. FOXO3a and the MAPK p38 are activated by cetuximab to induce cell death and inhibit cell proliferation and their expression predicts cetuximab efficacy in colorectal cancer. Br J Cancer 2016;115:1223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Koopman M, Kortman GA, Mekenkamp L. et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer 2009;100:266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Russo M, Crisafulli G, Sogari A, et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science2019;366:1473–80. [DOI] [PubMed] [Google Scholar]
- 154. Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148–59. [DOI] [PubMed] [Google Scholar]
- 155. Okugawa Y, Grady WM, Goel A.. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 2015;149:1204–25.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lao VV, Grady WM.. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol 2011;8:686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ouchi K, Takahashi S, Yamada Y. et al. DNA methylation status as a biomarker of anti-epidermal growth factor receptor treatment for metastatic colorectal cancer. Cancer Sci 2015;106:1722–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Weisenberger DJ, Siegmund KD, Campan M. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787–93. [DOI] [PubMed] [Google Scholar]
- 159. Lee MS, McGuffey EJ, Morris JS. et al. Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br J Cancer 2016;114:1352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 161. Salgia R, Kulkarni P.. The genetic/non-genetic duality of drug ‘resistance’ in cancer. Trends Cancer 2018;4:110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Mussnich P, Rosa R, Bianco R. et al. MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Exp Opin Ther Targets 2015;19:1017–26. [DOI] [PubMed] [Google Scholar]
- 163. Lu Y, Zhao X, Liu Q. et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med 2017;23:1331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Igarashi H, Kurihara H, Mitsuhashi K. et al. Association of MicroRNA-31-5p with clinical efficacy of anti-EGFR therapy in patients with metastatic colorectal cancer. Ann Surg Oncol 2015;22:2640–8. [DOI] [PubMed] [Google Scholar]
- 165. Angius A, Pira G, Scanu AM. et al. MicroRNA-425-5p expression affects BRAF/RAS/MAPK pathways in colorectal cancers. Int J Med Sci 2019;16:1480–91. [DOI] [PMC free article] [PubMed] [Google Scholar]