Abstract
Background
UBE2O, as a member of the ubiquitin-conjugating enzyme family, is abnormally expressed and exhibits abnormal functions in human malignancies. However, the function of UBE2O in head and neck squamous cell carcinoma (HNSCC) remains unknown. Therefore, our study aims to investigate the role of UBE2O in HNSCC progression and the underlying mechanisms.
Methods
The expression of UBE2O in HNSCC patients was investigated with data from the Cancer Genome Atlas (TCGA) and from a separate primary tumor cohort. The function of UBE2O in HNSCC cells was studied by cell viability assay, colony formation assay, wound healing assay, and cell migration and invasion chamber assay. The effect of UBE2O on tumor growth in vivo was determined in a subcutaneous xenograft model of HNSCC.
Results
TCGA data showed that UBE2O mRNA expression was dramatically increased in HNSCC tissues and that patients with high expression of UBE2O transcripts had a worse survival prognosis than patients with low expression of UBE2O transcripts. Gain-of-function and loss-of-function analyses revealed that oncogenic UBE2O enhanced the proliferation, migration and invasion of HNSCC cells in vitro. Further, mechanistic analysis revealed that UBE2O induced the epithelial–mesenchymal transition (EMT) phenotype and also potentiated TGF-β1-induced EMT, and thus leading to an enhanced capacity of migration and invasion in HNSCC. Finally, xenograft models showed that UBE2O knockout obviously inhibited the occurrence of EMT, angiogenesis and tumor growth in HNSCC in vivo.
Conclusion
Our study indicates that UBE2O acts as an oncogene to promote the malignant progression and EMT of HNSCC.
Keywords: head and neck squamous cell carcinoma, ubiquitin-conjugating enzyme e2o, epithelial–mesenchymal transition, migration, invasion
Introduction
Head and neck squamous cell carcinoma (HNSCC) refers to SCC of the upper aerodigestive tract, including the nasal cavities, paranasal sinuses, oral cavity, pharynx and larynx; HNSCC is the sixth most common cancer with an annual incidence of more than 600,000 cases worldwide.1,2 Despite tremendous improvements in surgery, radiochemotherapy and immunotherapy over the past few decades, the overall 5-year survival rate for patients with HNSCC still remains unsatisfactory.3,4 Local recurrence and distant metastasis are the main causes of the frustrating prognosis in patients with HNSCC.5,6 Therefore, it is urgent and imperative to advance our understanding of the molecular mechanisms associated with HNSCC progression; advanced understanding will make substantial contributions to the development of novel therapeutic strategies and improve patient survival outcomes.
Ubiquitin-conjugating enzyme E2O (UBE2O), also known as E2-230K, is a relatively large ubiquitin-conjugating enzyme of the E2 family,7 which has been proposed to play important roles in erythroid differentiation, such as remodeling of the proteome.8 The role of UBE2O in the ubiquitin-proteasome protein degradation system has also been gradually determined in recent years. For example, UBE2O mediates ubiquitination without a separate ubiquitin ligase and target orphans of multiprotein complexes for degradation.9 UBE2O also mediates the monoubiquitination of SMAD6 in bone morphogenetic protein signaling10 and leads to the polyubiquitination of BAP1 in chromatin-associated processes.11 Interestingly, UBE2O is located on the 17q25.1 chromosome in a region that is amplified in a variety of cancers, strongly indicating the potential role of UBE2O in tumorigenesis.7,12-14 Vila et al validated that UBE2O promoted tumor initiation and progression by targeting AMPK-α2 for degradation,15 but in myeloma, UBE2O exerted tumor suppressive roles by modulating the stability of the protein c-Maf.16 Therefore, the controversial and complicated functions of UBE2O in cancer are still poorly understood, and there have been no reports related to the role of UBE2O in HNSCC tumorigenesis.
Here, the Cancer Genome Atlas (TCGA) data were initially utilized to determine that UBE2O transcripts were remarkably increased in HNSCC tissues, and UBE2O overexpression predicted a worse prognosis in patients with HNSCC. Subsequent in vitro and in vivo evidence demonstrated that UBE2O, as an oncogene, obviously promoted the proliferation, migration and invasion of HNSCC. Mechanistically, UBE2O induced the occurrence of epithelial–mesenchymal transition (EMT) and potentiated TGFβ1-induced EMT in HNSCC. UBE2O knockout obviously inhibited the occurrence of EMT, angiogenesis and tumor growth in HNSCC in vivo. These results indicate that UBE2O acts as an oncogene in HNSCC progression and EMT induction.
Materials and Methods
The Cancer Genome Atlas (TCGA) Data Analysis
Publicly available human HNSCC datasets from TCGA were analyzed. The mRNA expression data of UBE2O (Illumina Hiseq RNAseq V2, RSEM normalized) were downloaded from the TCGA data portal (https://cancergenome.nih.gov/). The prognosis analysis was conducted in The Human Protein Atlas (http://www.proteinatlas.org/).
Patient Cohort
Paraffin-embedded samples from 187 patients with HNSCC were obtained from Xiangya Hospital, and 81 samples of paired adjacent noncancerous epithelial tissues were included. The pathological tumor-node-metastasis (TNM) stage was determined according to the 7th American Joint Committee on Cancer staging system. The patients’ clinical parameters, such as age, sex, smoking history, alcohol consumption, T stage, clinical stage and lymph node metastasis, were recorded. The study was approved by the Research Ethics Committee of Central South University, Changsha, China. Written informed consent was obtained from all the patients, and the study was conducted in accordance with the Declaration of Helsinki.
Immunohistochemistry and Evaluation
Immunohistochemical staining was performed as described previously.6,17 Briefly, antigen retrieval was carried out in 10 mmol/L citrate buffer (pH 6.0) for 15 min at 100°C in a microwave oven. The endogenous peroxidase activity was blocked by 3% hydrogen peroxide for 10 min at room temperature. The slides were then incubated overnight at 4°C with rabbit UBE2O polyclonal antibody (Abcam, Cambridge, MA), followed by horseradish peroxidase-labeled goat anti-rabbit IgG. The immunoreactivity was visualized with 3′,3′-diaminobenzidine and counterstained with Mayer’s hematoxylin. The negative control slides were incubated with normal goat serum instead of the primary antibody under the same experimental conditions. The final staining score of UBE2O-positive staining was obtained by the sum of the percentage of positive cells (<10%=1, 11–50%=2, 51–80%=3 and ≥81%=4) and staining intensity (0 points=no intensity, 1 point=weak intensity, 2 points=moderate intensity and 3 points=strong intensity). The tumors were classified into two groups, namely, the high UBE2O expression (scored as 5–7) and low UBE2O expression (scored as 0–4) groups, based on the final staining score.6,17
Cell Cultures and Reagents
The human HNSCC cell line Tu686 was kindly provided by Dr. Zhuo Chen (Emory University Winship Cancer Institute, Atlanta, GA, USA), which was approved by the Research Ethics Committee of Central South University. Human Fadu cells were purchased from ATCC. HNSCC cells were cultured in Dulbecco’s modified Eagle’s media/F12 medium (1:1) supplemented with 10% fetal bovine serum (Gibco BRL, Gaithersburg, MD, USA) and incubated at 37°C with saturated humidity and 5% CO2. Cells in an exponential growth state were used for the following experiments. TGF-β1 was obtained from Sigma-Aldrich Co. (St Louis, MO, USA). To induce EMT, the cells were seeded on the previous day, starved in serum-free medium overnight and treated with 10 ng/mL TGF-β1.
Construction of the CRISPR/Cas9 System Targeting UBE2O
Two short guide RNAs (sgRNA; sgRNA1 and 2) targeting exon 3 of the human UBE2O gene (NM_022066.4) were selected and synthesized to make lenti-UBE2O-sgRNA-Cas9 constructs.18 In brief, 25-bp DNA oligos containing the 20 bp target sequence and BsmbI sticky ends were annealed and inserted into the lentiCRISPR-v2 plasmid (Addgene: 52,961) digested with BsmbI. The DNA sequences for generating sgRNA1 and sgRNA2 are as follows: sgRNA1: 5ʹ-CATCTATCCCGTCAACAGCA-3ʹ; sgRNA2: 5ʹ-GACTGTGCCGTCAAGCTCAT-3ʹ. For the control plasmid, no sgRNA sequence was inserted into the construct. The sequencing-verified plasmids were then used to package the lentivirus with package vectors. The UBE2O-sgRNA-edited cells and control cells were selected with 6–8 μg/mL puromycin. A serial dilution method in a 96-cell plate was used to select the potential colony for the knockout efficiency test. After monoclonal expansion, the cells were stored at −80°C and Western blotting assays were used to verify the knockout efficiency of UBE2O in HNSCC cells.
Transfection of UBE2O cDNA
HNSCC parent or UBE2O knockout (KO) cells were transfected with UBE2O cDNA (Origene Technologies Inc., Rockville, MD, USA) according to the manufacturers’ protocol. To determine the efficiency of cDNA, UBE2O expression was assessed by Western blotting assays.
Western Blotting Analyses
Western blotting was conducted as previously described.19,20 Briefly, the total proteins were extracted from the cells with RIPA lysis buffer, separated in 8–12% SDS-PAGE gels, and transferred onto PVDF (polyvinylidene fluoride) membranes (Millipore, Billerica, MA, USA). After blocking with 5% nonfat milk at room temperature for 1 hour, the membranes were incubated with the relevant primary antibodies followed by an appropriate secondary antibody. The primary antibodies in the current study included anti-UBE2O (1:800, Abcam), anti-E-cadherin, anti-Vimentin, anti-N-cadherin, anti-Snail, anti-Zeb1 and anti-Zeb2 (1:800, Cell Signaling Technology, Danvers, MA, USA). Anti-β-Actin (1:1000, Beyotime, Shanghai, China) was used as a loading control. The blots were visualized using ECL reagents (Pierce, Rockford, IL, USA) and a chemiluminescence imaging system (Bio-Rad, Richmond, CA, USA).
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analyses
Total RNA of the cells and tissue samples was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The detection of target mRNAs was carried out with the High Capacity cDNA Reverse Transcription kit and SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Quantification of gene expression was conducted using the 2−ΔΔCT method and normalized to GAPDH. The primer sequences used for PCR are listed in Supplementary Table S1.
Cell Viability Assays
Tu686 or Fadu cells after UBE2O modulation were seeded into 6-well plates for cell counting at different time points, or into 96-well plates for proliferation assays with the Cell Counting Kit-8 (CCK-8, Dojindo, Japan).
Colony Formation Assays
For the colony formation assay, the cells were seeded in 6-well plates (1 × 103 cells per well) and incubated for 2 weeks. The colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet. All the experiments were repeated three times.
Wound Healing Assays
The cell migration activity was examined by wound-healing assay. Briefly, the cells were seeded in 6-well plates and incubated to almost total confluence. The cell monolayer was disrupted with a 10 μL plastic pipette tip. Then, the cells were cultured in serum-free medium. The initial gap length (0 hours) and the residual gap length at 24 or 60 hours after scratching were photographed under an inverted microscope. The experiments were performed in triplicate.
Cell Migration and Invasion Chamber Assays
The cell migration and invasion chamber assays were performed with the Corning Transwell migration chambers and Corning BioCoat Matrigel Invasion chambers (Corning Inc., Corning, NY, USA) according to the manufacturer’s instructions. Briefly, 1×105 cells were seeded into the upper chamber in serum-free medium. After incubation for 24 hours, nonmigrating or invading cells on the surface of the upper chamber were removed with a cotton-tipped swab. The migrated or invaded cells on the lower side were fixed and stained. The cells were photographed under a microscope and counted with ImageJ software. Each experiment was repeated for three times.
Tumor Xenografts
Four-week-old female nude mice were obtained from Hunan SJA Laboratory Animal Co., Ltd. (Changsha, Hunan, China) and maintained under specific pathogen-free conditions with the approval of the Institutional Animal Care and Use Committee of Xiangya Hospital, Central South University. The animal experiments were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals. Tumor xenografts were induced by subcutaneous injection 2×106 Tu686 control (Ctrl) or UBE2O KO cells suspended in 200 μL PBS into the flanks of nude mice. The tumors were measured with an external caliper, and the volume was calculated as length×width2/2. When the mice were sacrificed, the xenograft tumors were removed from the different groups, and the final weight of each tumor was measured. The tumor samples were then subjected to Western blotting and qRT-PCR assays.
Statistical Analysis
All the data shown are representative of at least three independent experiments and are presented as the mean ± standard deviation (SD). Positive staining rates of the UBE2O protein in the HNSCC tissues and adjacent noncancerous tissues were compared by χ2 test, which was also used to analyze the association between the UBE2O protein levels and the clinicopathological parameters. Statistical significance was determined with a two-sided unpaired Student’s t-test (for equal variances) or Welch’s corrected t-test (for unequal variances). P < 0.05 was considered statistically significant.
Results
UBE2O Expression Is Elevated in HNSCC Samples
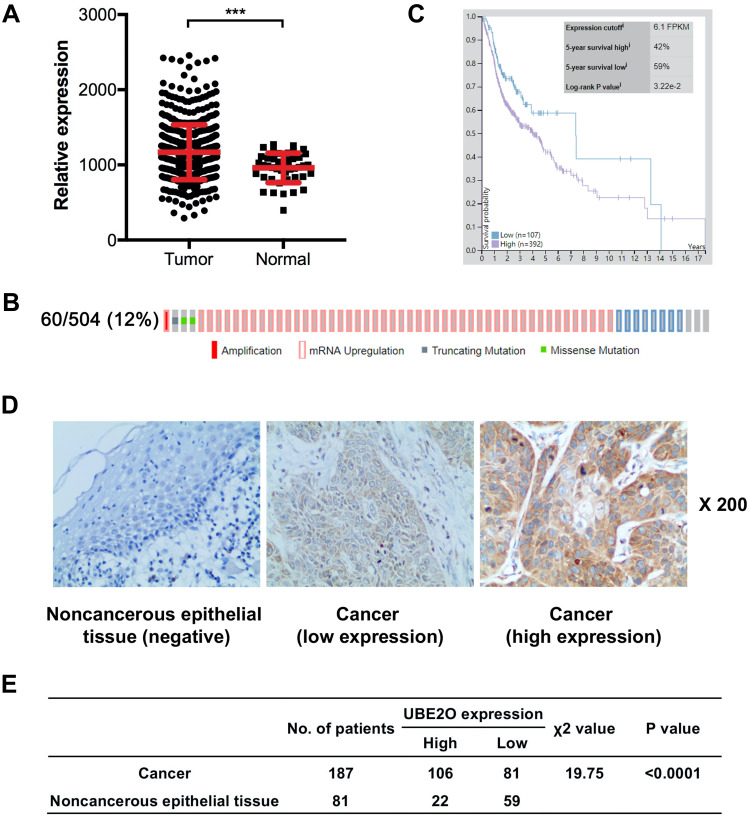
In our study, UBE2O expression was initially investigated using public TCGA RNA-sequencing data, in which both HNSCC and adjacent noncancerous samples were analyzed. As a result, these data showed that UBE2O transcripts were higher in the HNSCC samples than in the adjacent noncancerous samples (Figure 1A). With regard to the location of the tumors, compared with the adjacent noncancerous tissues, the expression of UBE2O mRNA was also upregulated in all the HNSCC tissues derived from different regions, such as the hypopharynx, larynx, oropharynx and oral cavity, but there were no significant differences in UBE2O mRNA expression among these HNSCC tissues derived from the different regions (Supplementary Figure S1). Genetic alterations of UBE2O, including gene amplification, mRNA upregulation, truncation and missense mutations, were detected in 12% (60/504) of the HNSCC tissues (Figure 1B). Kaplan-Meier analysis revealed that the 5-year survival rate of patients with high expression of UBE2O transcripts was obviously lower than that of patients with low expression (P < 0.05; Figure 1C). To further validate the results predicted online, a patient cohort including 187 HNSCC tissues and 81 paired adjacent noncancerous epithelial tissues was used to quantify the UBE2O protein and its clinical significance; this examination revealed that UBE2O protein was dramatically elevated in the HNSCC samples (P < 0.001; Figure 1D and E). However, the protein level of UBE2O failed to be positively associated with the clinical parameters in patients with HNSCC (all P > 0.05; Supplementary Table S2). Collectively, these clinical data suggest that UBE2O overexpression may be associated with the progressive behaviors in patients with HNSCC.
Figure 1.
UBE2O expression is elevated in HNSCC samples. (A) UBE2O transcripts in HNSCC tissues and corresponding noncancerous adjacent tissues from the public TCGA dataset were analyzed. (B) Frequencies of UBE2O alterations, including amplification, mRNA upregulation, truncation and missense mutations, from the HNSCC TCGA database were analyzed. (C) Survival prognosis of patients with HNSCC based on UBE2O transcript level. (D) Representative images of UBE2O protein expression in HNSCC tissues and adjacent noncancerous epithelial tissues determined by immunohistochemistry. Original magnification: 200×. (E) Expression of UBE2O protein in HNSCC tissues and adjacent noncancerous epithelial tissues (***P < 0.001).
UBE2O Promotes the Progression of HNSCC Cells
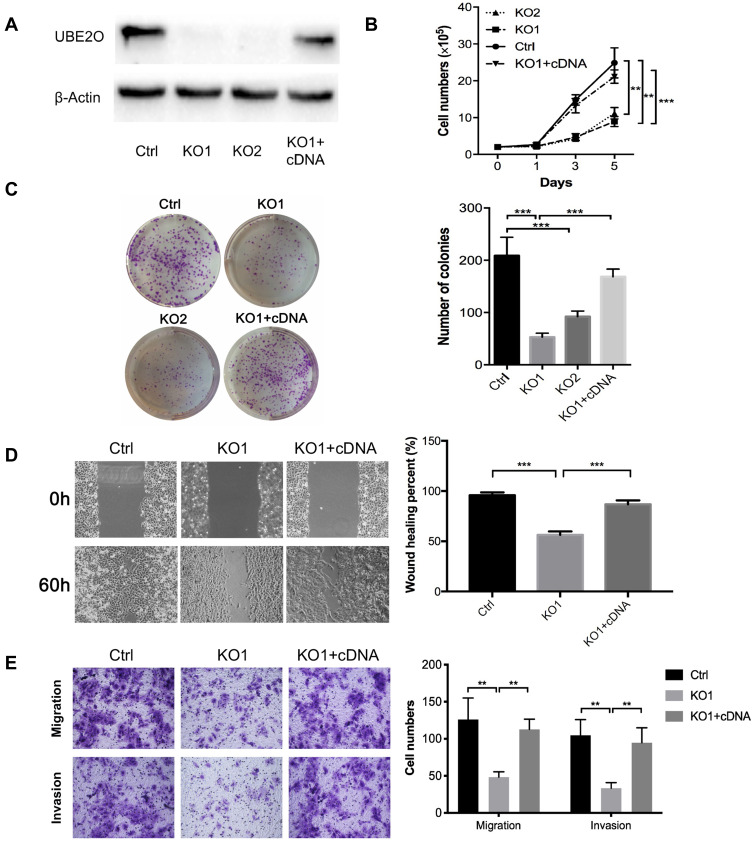
To investigate the role of UBE2O in HNSCC proliferation, endogenic UBE2O was knocked out in HNSCC Tu686 cells with the CRISPR/Cas9 system, and single colonies that exhibited efficient knockout were selected and expanded for subsequent experiments. Then, UBE2O was overexpressed by transfecting cDNA into the confirmed UBE2O knockout (KO1) cells (Figure 2A). Western blotting assays confirmed that UBE2O was successfully knocked out and overexpressed in the UBE2O KO1 and cDNA groups, respectively (Figure 2A). Consequently, CCK-8 assays showed that compared with the control group, the proliferation in the UBE2O knockout (KO1 and KO2) group was effectively inhibited, and the KO1 cells presented a higher inhibitory efficiency; additionally, the restored expression of UBE2O in the KO1 cells correspondingly enhanced its capacity of proliferation (P < 0.05; Figure 2B). Colony formation assays showed that colonies were greatly decreased in the UBE2O KO cells and increased in the UBE2O-overexpressing cells (P < 0.001, Figure 2C). Therefore, these data indicate that UBE2O enhances the proliferation of HNSCC cells in vitro.
Figure 2.
UBE2O promotes the progression of HNSCC cells. (A) Western blotting assays confirmed that UBE2O was successfully knocked out and overexpressed in Tu686 cells. (B) Cells in each group were counted at different time points. (C) Colony formation assays indicated that the number of colonies was significantly decreased in the UBE2O knockout cells and that the number of colonies was increased in the UBE2O-overexpressing cells. (D) Cell migration in different groups was examined by wound healing experiments in Tu686 cells. (E) Cell migration and invasion were examined using Transwell invasion assays in Tu686 cells. The results represent the average of three independent experiments ± standard deviation (S.D.) (**P < 0.01, ***P < 0.001).
To clarify whether UBE2O affects the metastatic capacities of HNSCC cells, we examined the impact of UBE2O on cell migration and invasion. As a result, wound healing assays showed that the cell migratory ability was significantly inhibited when UBE2O was knocked out and strengthened when UBE2O was upregulated (P < 0.001; Figure 2D). Transwell chamber assays demonstrated that the number of migratory and invading cells was reduced in the UBE2O KO group and correspondingly increased in the UBE2O cDNA group (P < 0.01; Figure 2E). Therefore, these results demonstrate that UBE2O enhances the migration and invasion of HNSCC cells in vitro.
UBE2O Knockout Impairs the TGF-β1-Induced EMT Process
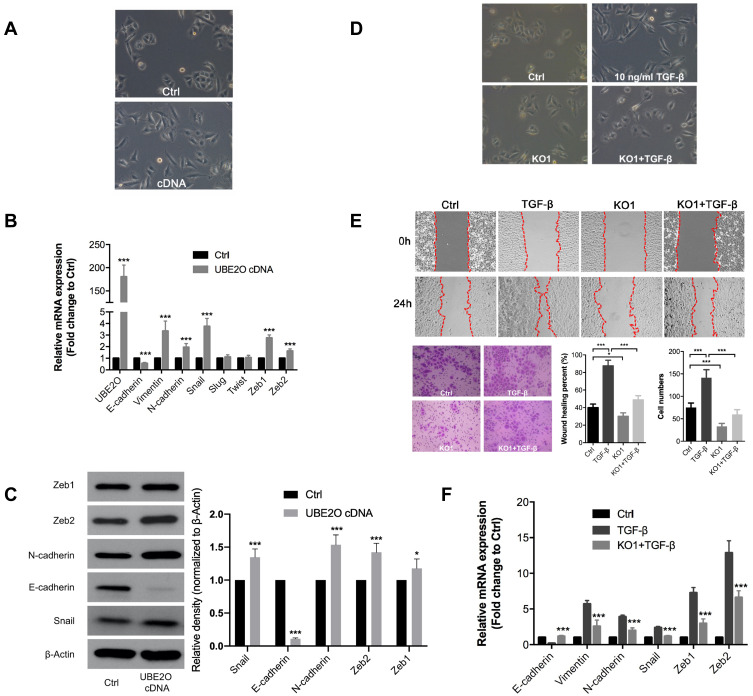
EMT is always considered a classic mechanism of cancer invasion and metastasis,21 and we found that following UBE2O overexpression in Tu686 cells, the cells underwent canonical EMT morphological changes with the loss of an adherent phenotype, which was reflected by decreased cell-to-cell contacts and a fibroblast-like appearance (Figure 3A). In addition, the mRNA levels of the epithelial marker E-cadherin decreased, while the levels of mesenchymal markers (Vimentin and N-cadherin) and EMT transcription factors including Snail, Zeb1 and Zeb2, increased (P < 0.001, Figure 3B). Correspondingly, the protein level of E-cadherin was decreased, while the protein levels of N-cadherin, Snail, Zeb1 and Zeb2 were upregulated (Figure 3C). These data suggest that EMT might be involved in UBE2O-mediated invasion and metastasis of HNSCC cells.
Figure 3.
UBE2O knockout impairs the TGF-β1-induced EMT process. (A) Overexpression of UBE2O induced EMT-related morphological changes in Tu686 cells. PCR and Western blotting assays were performed to evaluate the expression of EMT markers at the mRNA (B) and protein (C) levels following UBE2O overexpression. (D) Knockout of UBE2O attenuated TGF-β1-induced EMT morphological changes. (E) Knockout of UBE2O impaired TGF-β1-induced changes in cell migratory and invasive ability. (F) Knockout of UBE2O attenuated TGF-β1-induced mRNA changes at the mRNA level changes (*P < 0.05, ***P < 0.001).
TGF-β1 is a well-known and potent inducer of EMT in numerous human cancers, including HNSCC.22,23 To further confirm whether UBE2O affected HNSCC EMT, we treated parent Tu686 cells and the UBE2O KO1 cells with 10 ng/mL TGF-β1. Our data showed that UBE2O KO attenuated the TGF-β1-induced morphological changes in EMT (Figure 3D). In addition, the wound healing and transwell invasion assays showed that UBE2O KO1 mitigated the migratory and invasive ability enhanced by TGF-β1 (Figure 3E). Consistent with alterations in morphology and invasion, at the molecular level, the qRT-PCR data clearly revealed that UBE2O KO1 significantly reversed the TGF-β1-induced mRNA changes in EMT markers, including E-cadherin, Vimentin, N-cadherin, Snail, Zeb1 and Zeb2 (Figure 3F). Taken together, these results indicate that UBE2O induces EMT and increases the TGF-β1-mediated EMT, which may in turn lead to enhanced invasion and metastasis of HNSCC.
UBE2O Enhances the Proliferation and Invasion of HNSCC Fadu Cells
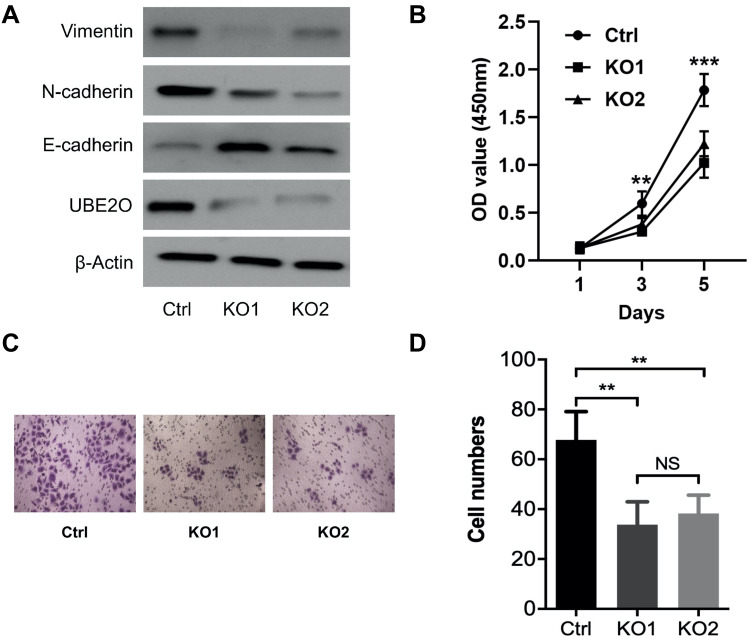
All the above data indicate that UBE2O accelerates the malignant progression of HNSCC through the occurrence of EMT. However, to exclude the possibility that the effect of UBE2O on HNSCC was Tu686 cell-specific, another HNSCC cell line, Fadu, was used to verify the main results as above. With the successful knockout of UBE2O (Figure 4A), the in vitro proliferation of Fadu cells was similarly inhibited (P < 0.05; Figure 4B). In addition, as expected, UBE2O KO also led to decreased invasive capacity of the Fadu cells in the Transwell Matrigel chamber assays (P < 0.05; Figure 4C and D), and this observation was accompanied by increased E-cadherin expression and decreased Vimentin and N-cadherin expression at the protein level (Figure 4A). Together, these data are consistent with the data derived from the Tu686 cells, which further supports the oncogenic role of UBE2O in the regulation of proliferation and invasion of HNSCC in vitro.
Figure 4.
UBE2O enhances the proliferation and invasion of HNSCC Fadu cells. (A) Western blotting assays were used to quantify the protein levels following UBE2O knockout. (B) CCK-8 assays were used to evaluate the proliferation ability of the Fadu cells in each group. (C) Representative images of Transwell invasion assays. (D) Quantitative results of Transwell invasion assays (NS, not significant; **P < 0.01; ***P < 0.001).
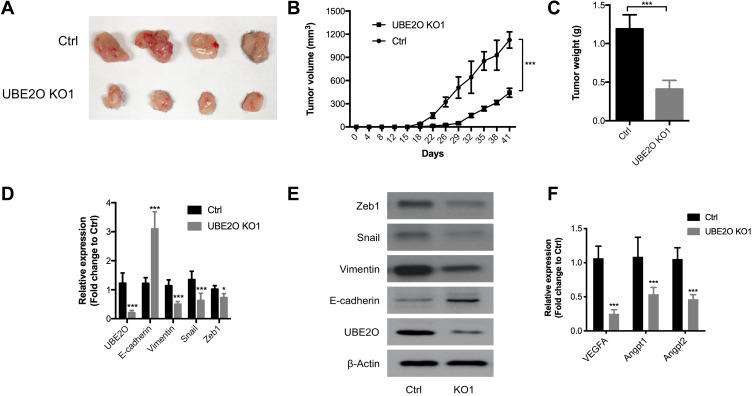
UBE2O Promotes HNSCC Tumor Growth in vivo
To further validate the oncogenic role of UBE2O in HNSCC in vivo, HNSCC Tu686 cells were used to establish a subcutaneous xenograft model in nude mice. As shown in Figure 5A and B, compared with the control group, tumor growth in the UBE2O KO group was dramatically decreased. The weight of the tumors in the UBE2O KO group was also significantly decreased compared with that in the control group (P < 0.001; Figure 5C). In addition, in the UBE2O KO xenografts, the mRNA levels of E-cadherin increased, while the expression levels of UBE2O, Vimentin, Snail and Zeb1 all correspondingly decreased (Figure 5D), which was also confirmed by Western blotting assays (Figure 5E). As EMT is associated with cancer angiogenesis,24 we found that in the UBE2O KO xenografts, the mRNA levels of proangiogenic factors, including VEGFA, Angpt1 and Angpt2, all decreased, suggesting that UBE2O may also be involved in cancer angiogenesis (Figure 5F). All these results demonstrate that UBE2O induces the occurrence of EMT and finally contributes to tumor growth and invasion of HNSCC in vivo.
Figure 5.
UBE2O promotes HNSCC tumor growth in vivo. (A and B) The UBE2O KO tumor xenografts grew substantially more slowly than those in the control group. (C) The average weight of the tumors in the UBE2O KO group was significantly lighter than that in the control group. (D) UBE2O affected the mRNA levels of EMT biomarkers in the xenograft tumors. (E) UBE2O affected the protein levels of EMT biomarkers in the xenograft tumors. (F) UBE2O affected the mRNA levels of angiogenesis biomarkers in the xenograft tumors (*P < 0.05, ***P < 0.001).
Discussion
Elucidating the underlying molecular mechanisms associated with HNSCC progression is beneficial for designing promising therapeutic strategies for patients with HNSCC. The ubiquitin-proteasome system, as an important posttranslational modification system that facilitates the degradation of target proteins, is involved in the malignant behaviors of distinct human cancers, including HNSCC, by the functional modulation of potential oncogenes, tumor suppressors and diverse signaling pathways.25–29 In the ubiquitin-proteasome system, ubiquitin-conjugating enzyme E2 accepts ubiquitin from enzyme E1 and transfers the activated ubiquitin molecules to E3 or directly mediates the ubiquitination of its substrate. Therefore, enzyme E2 is critical for protein ubiquitination and stability and acts as an important regulator in tumor initiation and progression.30,31 For instance, UBE2C is overexpressed in diverse human cancers and promotes malignant tumor progression through the activation of multiple oncogenic signaling pathways, including Wnt/β‑catenin, Erk and PI3K/Akt pathways.32–34 However, the function of UBE2O still remains unknown.
UBE2O was first purified from rabbit reticulocytes and considered an E2 enzyme due to its specific and conserved ubiquitin-conjugating domain.7,35 Initially, based on the data from TCGA and our verifying patient cohort, the transcript and protein expression of UBE2O was obviously increased in HNSCC tissues compared to the corresponding adjacent epithelial tissues, and patients with high UBE2O transcripts had a worse survival prognosis. However, we failed to confirm the positive links between the UBE2O protein and the clinical parameters. Our negative result in HNSCC was inconsistent with the data in breast cancer samples, in which UBE2O overexpression indicated a high risk of metastasis and poor prognosis.36 The discordant prognostic capacity of the UBE2O mRNA and protein may be partially explained by the complicated posttranslational regulation of UBE2O itself in HNSCC, which also suggests that the clinical significance of the UBE2O protein varies greatly in distinct cancer types. Therefore, investigations of clinical samples of other human cancers will further expand and determine the clinical significance of UBE2O.
Recently, although definite physiological ubiquitination substrates of UBE2O, such as BMAL1 and BAP1, have been identified,11,37 UBE2O was also found to decrease the level of the TRAF6 protein in a catalytic activity-independent manner, rather than to induce its polyubiquitin.38 These data indicated the complicated functions of UBE2O in distinct biopathological processes. In regard to cancer, only limited reports revealed that UBE2O could target AMPK-α2 and c-Maf for ubiquitination, which consequently led to the activation of downstream signaling pathways, such as the mTOR/HIF1α and MYC pathways and the apoptotic pathway.15,16,36 To the best of our knowledge, this study is the first to demonstrate that UBE2O functions as an oncogene to promote the proliferation, migration and invasion of HNSCC cells. However, whether the oncogenic role of UBE2O in HNSCC depends on its E2 activity and the identity of its potential downstream substrates remain to be further determined.
EMT is a highly regulated process in which cancer epithelial cells are reprogrammed into mesenchymal cells, and this transition is accompanied by enhanced capabilities of migration, invasion and resistance to diverse therapeutic approaches.21,39 Cancer cells that undergo EMT display decreased expression of the epithelial marker E-cadherin and increased expression of mesenchymal markers, such as Vimentin, N-cadherin and key transcription factors.39 EMT is obviously stimulated by developmental signaling pathways, among which the TGF-β pathway is always considered to be a primary inducer of EMT via a Smad-dependent mechanism.23 In response to TGF-β1, Smad2/3 are phosphorylated, form a trimer with Smad4 and then translocate into the cell nucleus, which in turn starts the transcription of target genes responsible for EMT. On the other hand, Smad6/7 are inhibitory Smads that act as a negative feedback loop in the Smad-dependent signaling pathway.23,40 Recently, the ubiquitin-conjugating enzyme E2 was also found to participate in the regulation of the EMT process in several human cancers. In gastric and prostate cancers, ectopic expression of UBE2T could promote cell proliferation and metastasis by inducing EMT.41 In addition, UBE2C knockdown impeded EMT by repressing the phosphorylation of the hub oncogenic gene Aurora-A in gastric adenocarcinoma.32 In this study, our data revealed that UBE2O could potentiate TGF-β1-induced EMT, leading to enhanced proliferation, motility and metastatic capacity in HNSCC cells. Interestingly, Zhang et al reported that UBE2O promoted the monoubiquitination of inhibitory Smad6 and therefore facilitated adipogenesis induced by BMP7;10 these findings indicated that UBE2O could skew the balance of the Smad-dependent signaling pathways toward a stimulatory status via the inactivation of inhibitory Smads, including Smad6/7. Considering that both TGF-β1 and BMP7 are potent stimulators for the cancer EMT process via the activation of the Smad-dependent signaling pathway, UBE2O may also enhance TGF-β1-induced EMT via the inactivation of inhibitory Smads; however, this possibility needs to be verified in the future.
Angiogenesis is a crucial element for cancer malignant proliferation, local invasion and systemic dissemination; therefore, it plays a crucial role in cancer metastasis.42 A number of anti-angiogenic agents have been approved by the FDA and are being applied in clinical management to fight against blood vessel formation in cancer; these agents have promising prospects.43 Our study clearly indicated that UBE2O knockout significantly decreased the expression of proangiogenic factors, including Angpt1, Angpt2 and VEGFA, all of which are important stimulators of the proliferation, migration, adhesion and spreading of blood vessel endothelial cells and the subsequent acceleration of angiogenesis in solid cancers.44 Although our study lacked direct evidence to clarify how UBE2O regulated the expression of these angiogenic factors and how UBE2O mediated the crosstalk between cancer cells and endothelial cells, these data clearly indicated that the targeted inhibition of UBE2O could be a promising strategy to suppress cancer angiogenesis and therefore block cancer metastasis.
In conclusion, our current study demonstrated that UBE2O could function as an oncogene to promote the proliferation, migration and invasion of HNSCC cells involving EMT. Therefore, UBE2O may be a promising therapeutic target for patients with HNSCC. Further detailed studies are required to determine the exact molecular mechanisms of UBE2O-mediated carcinogenesis and whether these results can be extended to other solid human cancers.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 81974424, 81874133, 81773243 and 81772903), the Natural Science Foundation of Hunan Province (Nos. 2019JJ40481, 2019JJ50944 and 2018JJ2630), the Huxiang Young Talent Project (No. 2018RS3024) and the Fundamental Research Funds for the Central South University (No. 1053320182447).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.She L, Qin Y, Wang J, et al. Tumor-associated macrophages derived CCL18 promotes metastasis in squamous cell carcinoma of the head and neck. Cancer Cell Int. 2018;18:120. doi: 10.1186/s12935-018-0620-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen EE, LaMonte SJ, Erb NL, et al. American cancer society head and neck cancer survivorship care guideline. CA Cancer J Clin. 2016;66(3):203–239. doi: 10.3322/caac.21343 [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Guo T, Xu G, et al. Characterization of alternative splicing events in HPV-negative head and neck squamous cell carcinoma identifies an oncogenic DOCK5 variant. Clin Cancer Res. 2018;24(20):5123–5132. doi: 10.1158/1078-0432.CCR-18-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duprez F, Berwouts D, De Neve W, et al. Distant metastases in head and neck cancer. Head Neck. 2017;39(9):1733–1743. doi: 10.1002/hed.24687 [DOI] [PubMed] [Google Scholar]
- 6.Huang D, Qiu Y, Li G, et al. KDM5B overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck. J Cancer. 2018;9(1):198–204. doi: 10.7150/jca.22145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokota T, Nagai H, Harada H, et al. Identification, tissue expression, and chromosomal position of a novel gene encoding human ubiquitin-conjugating enzyme E2-230k. Gene. 2001;267(1):95–100. doi: 10.1016/S0378-1119(01)00407-3 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen AT, Prado MA, Schmidt PJ, et al. UBE2O remodels the proteome during terminal erythroid differentiation. Science. 2017;357:6350. doi: 10.1126/science.aan0218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagitani K, Juszkiewicz S, Hegde RS. UBE2O is a quality control factor for orphans of multiprotein complexes. Science. 2017;357(6350):472–475. doi: 10.1126/science.aan0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Zhang J, Bauer A, et al. Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2-230K-mediated monoubiquitination of SMAD6. EMBO J. 2013;32(7):996–1007. doi: 10.1038/emboj.2013.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mashtalir N, Daou S, Barbour H, et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell. 2014;54(3):392–406. doi: 10.1016/j.molcel.2014.03.002 [DOI] [PubMed] [Google Scholar]
- 12.Lin M, Smith LT, Smiraglia DJ, et al. DNA copy number gains in head and neck squamous cell carcinoma. Oncogene. 2006;25(9):1424–1433. doi: 10.1038/sj.onc.1209166 [DOI] [PubMed] [Google Scholar]
- 13.Briffa R, Um I, Faratian D, et al. Multi-scale genomic, transcriptomic and proteomic analysis of colorectal cancer cell lines to identify novel biomarkers. PLoS One. 2015;10(12):e0144708. doi: 10.1371/journal.pone.0144708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toffoli S, Bar I, Abdel-Sater F, et al. Identification by array comparative genomic hybridization of a new amplicon on chromosome 17q highly recurrent in BRCA1 mutated triple negative breast cancer. Breast Cancer Res. 2014;16(6):466. doi: 10.1186/s13058-014-0466-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vila IK, Yao Y, Kim G, et al. A UBE2O-AMPKalpha2 axis that promotes tumor initiation and progression offers opportunities for therapy. Cancer Cell. 2017;31(2):208–224. doi: 10.1016/j.ccell.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Zhang Z, Li J, et al. The ubiquitin-conjugating enzyme UBE2O modulates c-Maf stability and induces myeloma cell apoptosis. J Hematol Oncol. 2017;10(1):132. doi: 10.1186/s13045-017-0499-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing Q, Li G, Chen X, et al. Wnt3a promotes radioresistance via autophagy in squamous cell carcinoma of the head and neck. J Cell Mol Med. 2019;23(7):4711–4722. doi: 10.1111/jcmm.14394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Li G, Yang N, et al. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer Cell Int. 2017;17:2. doi: 10.1186/s12935-016-0372-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Li G, Ren S, et al. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol Lett. 2017;13(4):2631–2636. doi: 10.3892/ol.2017.5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11(1):28–39. doi: 10.1002/1878-0261.12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu C, Liu Y, Huang D, et al. TGF-beta1 mediates epithelial to mesenchymal transition via the TGF-beta/Smad pathway in squamous cell carcinoma of the head and neck. Oncol Rep. 2011;25(6):1581–1587. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi: 10.1038/cr.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardali E, Ten Dijke P. Transforming growth factor-beta signaling and tumor angiogenesis. Front Biosci. 2009;14:4848–4861. doi: 10.2741/3573 [DOI] [PubMed] [Google Scholar]
- 25.Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16(7):634–648. doi: 10.1080/15384101.2017.1288326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YJ, Wu H, Shen XZ. The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett. 2016;379(2):245–252. doi: 10.1016/j.canlet.2015.06.023 [DOI] [PubMed] [Google Scholar]
- 27.Sahasrabuddhe AA, Elenitoba-Johnson KS. Role of the ubiquitin proteasome system in hematologic malignancies. Immunol Rev. 2015;263(1):224–239. doi: 10.1111/imr.12236 [DOI] [PubMed] [Google Scholar]
- 28.Ding F, Xiao H, Wang M, Xie X, Hu F. The role of the ubiquitin-proteasome pathway in cancer development and treatment. Front Biosci. 2014;19:886–895. doi: 10.2741/4254 [DOI] [PubMed] [Google Scholar]
- 29.Voutsadakis IA. Ubiquitination and the ubiquitin - proteasome system in the pathogenesis and treatment of squamous head and neck carcinoma. Anticancer Res. 2013;33(9):3527–3541. [PubMed] [Google Scholar]
- 30.van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24(4):981–993. doi: 10.1096/fj.09-136259 [DOI] [PubMed] [Google Scholar]
- 31.Alpi AF, Chaugule V, Walden H. Mechanism and disease association of E2-conjugating enzymes: lessons from UBE2T and UBE2L3. Biochem J. 2016;473(20):3401–3419. doi: 10.1042/BCJ20160028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Song Y, Liu X, et al. UBE2C induces EMT through Wnt/β‑catenin and PI3K/Akt signaling pathways by regulating phosphorylation levels of Aurora-A. Int J Oncol. 2017;50(4):1116–1126. doi: 10.3892/ijo.2017.3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Liu P, Wang J, et al. Ubiquitin-conjugating enzyme E2C regulates apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway. Med Oncol. 2015;32(5):149. doi: 10.1007/s12032-015-0609-8 [DOI] [PubMed] [Google Scholar]
- 34.Xie C, Powell C, Yao M, Wu J, Dong Q. Ubiquitin-conjugating enzyme E2C: a potential cancer biomarker. Int J Biochem Cell Biol. 2014;47:113–117. doi: 10.1016/j.biocel.2013.11.023 [DOI] [PubMed] [Google Scholar]
- 35.Klemperer NS, Berleth ES, Pickart CM. A novel, arsenite-sensitive E2 of the ubiquitin pathway: purification and properties. Biochemistry. 1989;28(14):6035–6041. doi: 10.1021/bi00440a047 [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Ma F, Liu C, et al. UBE2O promotes the proliferation, EMT and stemness properties of breast cancer cells through the UBE2O/AMPKalpha2/mTORC1-MYC positive feedback loop. Cell Death Dis. 2020;11(1):10. doi: 10.1038/s41419-019-2194-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Yang J, Zhang Y, et al. Ubiquitin-conjugating enzyme UBE2O regulates cellular clock function by promoting the degradation of the transcription factor BMAL1. J Biol Chem. 2018;293(29):11296–11309. doi: 10.1074/jbc.RA117.001432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Zhang J, Zhang L, van Dam H, Ten Dijke P. UBE2O negatively regulates TRAF6-mediated NF-kappaB activation by inhibiting TRAF6 polyubiquitination. Cell Res. 2013;23(3):366–377. doi: 10.1038/cr.2013.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh M, Yelle N, Venugopal C, Singh SK. EMT: mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 40.Katsuno Y, Lamouille S, Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25(1):76–84. doi: 10.1097/CCO.0b013e32835b6371 [DOI] [PubMed] [Google Scholar]
- 41.Luo C, Yao Y, Yu Z, et al. UBE2T knockdown inhibits gastric cancer progression. Oncotarget. 2017;8(20):32639–32654. doi: 10.18632/oncotarget.15947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielenberg DR, Zetter BR. The contribution of angiogenesis to the process of metastasis. Cancer J. 2015;21(4):267–273. doi: 10.1097/PPO.0000000000000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasudev NS, Reynolds AR. Anti-angiogenic therapy for cancer: current progress, unresolved questions and future directions. Angiogenesis. 2014;17(3):471–494. doi: 10.1007/s10456-014-9420-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8(6):464–478. doi: 10.1038/nrm2183 [DOI] [PubMed] [Google Scholar]