ABSTRACT
The red yeast Rhodosporidium toruloides naturally produces microbial lipids and carotenoids. In the past decade or so, many studies demonstrated R. toruloides as a promising platform for lipid production owing to its diverse substrate appetites, robust stress resistance and other favorable features. Also, significant progresses have been made in genome sequencing, multi-omic analysis and genome-scale modeling, thus illuminating the molecular basis behind its physiology, metabolism and response to environmental stresses. At the same time, genetic parts and tools are continuously being developed to manipulate this distinctive organism. Engineered R. toruloides strains are emerging for enhanced production of conventional lipids, functional lipids as well as other interesting metabolites. This review updates those progresses and highlights future directions for advanced biotechnological applications.
Keywords: chassis organism, genetic modification, microbial lipids, multi-omic analysis, Rhodosporidium toruloides
A review updates research progresses on the red yeast Rhodosporidium toruloidesand highlights future engineering directions.
INTRODUCTION
Lipids such as triacylglycerols and fatty acid derivatives are formerly sourced from plants and animal fats. When used as commodity, lipids are important feedstocks for oleochemicals and drop-in biofuels (d'Espaux et al. 2015; Jin et al. 2015; Leong et al. 2018). Compared with extraction from oil crops and plants, microbial production of lipids has many advantages including short production cycle, tailored processes and better accessibility to structural diversity. Therefore, intensive efforts have been made to design advanced strains for microbial production of lipid-based biofuels and chemicals (Yu et al. 2018; Zhou, Kerkhoven and Nielsen 2018; Yan and Pfleger 2020). In particular, Saccharomyces cerevisiae and Yarrowia lipolytica have been engineered to produce diverse oleochemicals. However, these yeasts remain ineffective in xylose utilization and inhibitor resistance, hence preventing large-scale production from lignocellulosic biomass (Ledesma-Amaro and Nicaud 2016; Spagnuolo et al. 2018).
Many natural oleaginous microorganisms intracellularly accumulate lipids under nutrient limitation conditions. In the past two decades, the red yeast R. toruloides has been used for lipid production from diverse feedstocks and with different bioprocess strategies (Li, Zhao and Bai 2007; Lin et al. 2010; Zhao et al. 2011; Xu et al. 2012; Shen et al. 2013; Dias et al. 2015; Fei et al. 2016). A successful pilot-scale process was demonstrated at 1000-L scale using sugarcane juice as substrate by R. toruloides DEBB 5533 for microbial lipid production, and a preliminary analysis showed its economic competitiveness with soybean oil in terms of biodiesel production (Soccol et al. 2017). Meanwhile, an early systems study on R. toruloides pioneered by Prof. Zhao's group at Dalian Institute of Chemical Physics has paved the way for further engineering of advanced strains (Zhu et al. 2012). Thus far, genetic tools including the CRISPR-Cas9 technology have been developed. Engineered R. toruloides strains have emerged for improved production profiles or increased product portfolios. Very recently, brief reviews on R. toruloides have also been published (Xu and Liu 2017; Park, Nicaud and Ledesma-Amaro 2018; Osorio-González et al. 2019a; Saini et al. 2020). Here, we present an extensive summary of research progresses on R. toruloides and offer discussions on the bottlenecks and perspectives to explore R. toruloides as a distinctive chassis for further engineering.
General background of R. toruloides
Rhodosporidium toruloides is a red heterothallic, dimorphism yeast, first isolated in 1922 from the air in Dalian, China and named as Torula rubescen (Banno 1967). It can exist either in the yeast form or as a mycelial form. In nature, R. toruloides is also found in pine wood pulp, soil, seawater, acid sewage and plant leaves (Buck and Andrews 1999; Gadanho and Sampaio 2005; Gadanho, Libkind and Sampaio 2006; Into et al. 2020). It is classified into the Sporidiobolaceae family, Sporidiobolales order, Microbotryomycetes class and Basidiomycota phylum. Some typical fungi in the Basidiomycota phylum include mushrooms, smuts and rusts. This classification makes R. toruloides unique, as many other yeasts for engineering studies such as Saccharomyces cerevisiae, Lypomyces starkeyi and Y. lypolytica are placed in the Ascomycota phylum. Because yeast species in the Pucciniomycotina of Basidiomycota phylum are polyphyletic, a recent phylogenetic classification proposes to rename Rhodosporidium as Rhodotorula (Wang et al. 2015). In this review, however, we used the name Rhodosporidium toruloides in order to better connecting with the majority of literature.
Rhodosporidium toruloides grows well at varying temperatures and pH (Gadanho, Libkind and Sampaio 2006). It uses diverse carbon sources as carbon and energy sources, for instance, monosaccharides including hexoses and pentoses; oligosaccharides such as sucrose, maltose, cellobiose, trehalose, raffinose and melezitose; alcohols such as ethanol, glycerol, mannitol and sorbitol; organic acids such as acetate, lactate, succinate, citrate and long-chain fatty acids as well as D-galacturonic acid (Yang et al. 2015; Xu and Liu 2017; Yaegashi et al. 2017; Jagtap and Rao 2018; Protzko et al. 2019). The robustness of R. toruloides in terms of resistance against biomass-derived inhibitors has been demonstrated (Hu et al. 2009; Zhao et al. 2012; Nogué et al. 2018). Thus, growing R. toruloides on biomass hydrolysates has been documented for production of lipids and carotenoids (Zhao et al. 2010; Matsakas et al. 2015; Singh et al. 2018; Dai et al. 2019; Osorio-González et al. 2019b; Bertacchi et al. 2020; Lopes, Bonturi and Miranda 2020). In terms of nitrogen sources, ammonium, nitrate, cadaverine, amino acids and small peptides are effective (Li, Zhao and Bai 2007; Wu et al. 2010; Li et al. 2020). To accumulate high amounts of lipids, oleaginous yeasts are usually cultivated under nitrogen-limited conditions (Evans and Ratledge 1984). Besides, limitation on other nutrients such as inorganic phosphate, sulfate and iron also facilitate lipid production by R. toruloides (Wu et al. 2010; Wu et al. 2011; Wang et al. 2018).
Systems biology of R. toruloides
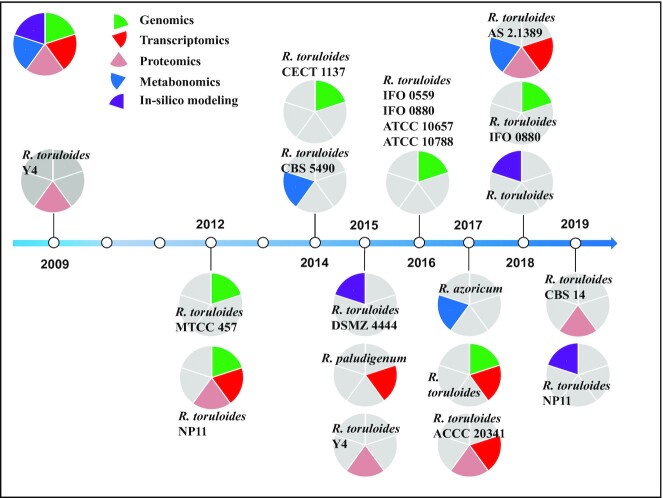
Genetic background and data in gene transcription, protein expression and basal metabolism are essential information for genetic modification and metabolic engineering. Significant progresses have been achieved in terms of understanding the genome, transcriptome, proteome and metabolome of R. toruloides, as summarized in Fig. 1 and Table S1 (Supporting Information).
Figure 1.
Milestones of systems biology studies in genus Rhodosporidium. The publication year, strain name and research approach are listed. Different colors are shown for genomics, proteomics, transcriptomics, metabolomics and in-silico modeling.
Genome sequencing
Genome sequence is key to modern molecular biology of microorganisms. A genome sequencing study of R. toruloides NP11 by Zhao and his coworkers revealed a genome of 20.2 Mb in size with a GC content of 61.9% containing 8171 protein-coding genes (Zhu et al. 2012). Ever since, draft genomes of several Rhodosporidium species have been described (Kumar et al. 2012; Morin et al. 2014; Zhang et al. 2016c; Tran et al. 2019). More recently, two Rhodosporidium haploids with A1 and A2 mating type isolated from the same diploid strain were sequenced for comparison and sequencing data showed very close genome size (20.75 and 21.49 Mbp), GC content (62.0% and 61.8%) and protein-coding genes (7730 and 7800) (Hu and Ji 2016). With aids of functional genomics and comparative genomics, the chromosomal structure, protein-coding genes, predicted proteins and functional RNA of Rhodosporidium species can be elucidated.
Transcriptomics
The combination of genomics and transcriptomics facilitated the discovery of introns in the genome of R. toruloides NP11, and a novel fatty acid synthase system, which participates in metabolic pathways absent in non-oleaginous yeasts (Zhu et al. 2012). In other studies, transcriptomic studies were used to investigate gene expression profiles and global responses of R. toruloides to various stress conditions or utilization of a broad range of carbon sources (Zhu et al. 2012; Lu et al. 2015; Qi et al. 2017; Coradetti et al. 2018; Wang et al. 2018; Protzko et al. 2019). These efforts have identified some regulatory and stress-resistant genes.
Proteomics
Proteomic studies of R. toruloides have been adopted to compare differences in protein expression at diverse conditions. This technique is vital in providing a tool for the identification of proteins involved in lipid accumulation (Liu et al. 2009). In addition, it also help in the study of xylose metabolism (Tiukova et al. 2019a), and screening for dynamic changes of relevant proteins associated with lipid droplets (Zhu et al. 2015). Elsewhere, proteomic studies of the conventional yeast S. cerevisiae and two oleaginous yeast strains, Cryptococcus albidus and R. toruloides, provided an overall understanding of the excessive lipid storage in oleaginous yeasts (Shi et al. 2013). However, unlike isolated proteomic or transcriptomic analysis, integrated assay of proteomes and transcriptomes offers more evidence to identity key genes and pathways (Zhu et al. 2012; Qi et al. 2017).
Metabolomics
More recently, differences in titers and key metabolites under different culture conditions were investigated via metabolomics, which revealed the relevance between glycerol metabolism and carotenoid production in R. toruloides (Shen et al. 2017). Also, using metabolomics principles, the inhibition of glucose on glycerol metabolism, along with the importance of oxygen supply to the microbial lipid composition and yield was clarified (Lee et al. 2014). The analysis of metabolomes coupled with transcriptomes and proteomes revealed a comprehensive molecular mechanism of lipid accumulation during phosphorus depletion and predicted target genes for strain engineering (Wang et al. 2018).
Metabolic network reconstruction and modeling
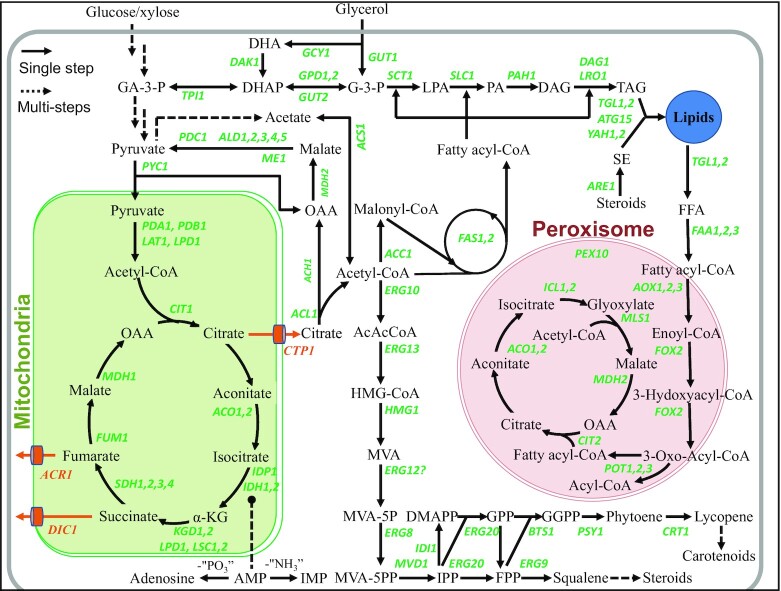
Available omic studies and the framework of the molecular basis of the lipid metabolism, physiology and responses of R. toruloides to environmental stresses have provided datasets for metabolic network reconstruction and in-silico modeling (Zhu et al. 2012; Bommareddy et al. 2015; Castaneda et al. 2018; Coradetti et al. 2018; Dinh et al. 2019; Tiukova et al. 2019b). The primary metabolic pathways in cytoplasm, mitochondria and peroxisome are shown in Fig. 2.
Figure 2.
The primary metabolic pathways of R. toruloides. Key genes involved in glycolysis, pentose phosphate pathway, TCA cycle in mitochondria, biosynthesis and degradation of fatty acids, triacylglycerols and phospholipids, isoprenoid biosynthesis, glyoxylate cycle pathway, and the β-oxidation pathways in peroxisomes are highlighted in green. Abbreviations: GA-3-P, glyceraldehyde 3-phosphate; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; G-3-P, glycerol-3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; SE, steryl ester; FFA, free fatty acid; OAA, oxaloacetate; AHG, α-ketoglutarate; AcAcCoA, aceto-acetyl-CoA; HMG-CoA, hydroxymethylglutaryl-CoA; MVA, mevalonate; MVA5P, mevalonate-5-phosphate; MVA5PP, mevalonate-5-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; AMP, adenosine monophosphate; IMP, inosine monophosphate.
According to the metabolism framework, acetyl-CoA is a key precursor of lipid synthesis, which mainly derives from the citric acid cleavage and pyruvate decarboxylation in the cytoplasm. Important genes involved in the conversion of acetyl-CoA into lipids, such as ACC1 (coding acetyl-CoA carboxylase), SCD1 (coding stearic CoA desaturase) and DGAT1 (coding diacylglycerol acyltransferase) are well characterized and presumed to catalyze the first, intermediate and final step, respectively, in triacylglycerol biosynthesis (Yang et al. 2012; Chen et al. 2014; Cui et al. 2016; Liu et al. 2016b). NADPH, a reducing power used for fatty acid acyl carrier protein synthesis, springs from the oxidative pentose phosphate pathway, and the POM cycle (pyruvate/oxaloacetate/malate cycle or transhydrogenase cycle) via cytosolic NADP+-dependent malic enzyme (Zhu et al. 2012; Coradetti et al. 2018). Acetyl-CoA can also be used to synthesize dimethylallyldiphosphate and its isomer 3-isopentenyl diphosphate—the common precursors of terpenoids, for instance, lycopene and β-carotene. Free fatty acids are also degraded in peroxisome to generate acetyl-CoA which could be converted into succinate by the glyoxylate cycle (Bommareddy et al. 2015; Castaneda et al. 2018).
In addition to the basic metabolic network, the effects of nitrogen and phosphorus depletion on lipid accumulation and relevant metabolism have been explored by system biology study (Zhu et al. 2012; Wang et al. 2018). Nitrogen or phosphorus limitations are linked to intracellular adenosine monophosphate (AMP) which converts to inosine monophosphate (IMP) through deamination or to adenosine (Ade) through dephosphorylation. These processes impair the activity of mitochondrial isocitrate dehydrogenase (Idh) owing to its high dependence on AMP. Low Idh activity is also implicated to influence the accumulation of mitochondrial citrate and its export to the cytoplasm, with further cleavage of citrate into OAA and acetyl-CoA for enhanced lipid production. In principle, nitrogen starvation promotes the up-regulation of genes involved in fatty acid synthesis and decomposition pathways. The pentose phosphate pathway and the transhydrogenase pathway increase NADPH supply and basically enhance the synthesis of triacylglycerols. Nitrogen starvation makes the cells prone to autophagy (Zhu et al. 2012) whereas the autophagy and RNA degradation pathways get activated in phosphorus exhaustion. Simultaneously, the expression of phosphatidate phosphatase (Pah1) and diacylglycerol acetyltransferase (Dag1) are significantly up-regulated while the β-oxidation pathway gets inhibited. These regulatory proteins and pathways contribute to the synthesis of triacylglycerol (Wang et al. 2018).
Recent studies on the genome, transcriptome, proteome and metabolome of R. toruloides are summarized in Table S1 (Supporting Information). It shows a trend of multi-omic analysis, because the results obtained from different omic layers can be reciprocally verified and provide more reliable conclusions. However, very few integrated studies of various omics and models are reported (Bommareddy et al. 2015; Castaneda et al. 2018). A curated version of a small-scale metabolic model for de novo lipid production by R. toruloides showed that the central nitrogen metabolism essential to predict the lipid metabolism at different culture conditions was unblocked (Castaneda et al. 2018). Moreover, the process of mass and charge balancing incorporated additional constraints on cell mass production. With the accumulation of omic data, as well as advances in technologies such as big data analysis and artificial intelligence, multi-omics research will pave the way and provide clues for genetic technology development, metabolic engineering, evolutionary engineering and process engineering.
Genetic tools development
Omics study promoted the discovery of biological elements such as replicons, promoters, terminators and selection markers for R. toruloides. However, there was no endogenous plasmid or autonomously replicating sequence and centromere sequence functional as a replication element being verified in R. toruloides.
Transformation methods
Genetic transformation was first reported with random integration of the phenylalanine ammonia lyase (PAL) gene into the chromosome of R. toruloides MS7013 through polyethylene glycol (PEG)-mediated protoplast transformation with a transformation efficiency of about 1000 transformants/μg DNA (Tully and Gilbert 1985). Because protoplast transformation efficiency is highly dependent on protoplast preparation and operation, subsequent studies sought to use Agrobacterium tumefaciens-mediated transformation (ATMT) (Liu et al. 2013). ATMT transformation is widely applicable, and the transformation efficiency varies from 70 to 1000 transformants per plate depending on the species and selection markers (Koh et al. 2014; Lin et al. 2014). Even though ATMT is easy to operate, its process is time-consuming as a single round preparation takes almost 2 weeks (Sun et al. 2017; Hooykaas et al. 2018). Moreover, as the intrinsic feature of ATMT transformation is random integration of the genome, it is essential to screen massive clones to identify those with superior phenotype and to diminish the risk of unexpected interference with endogenous genes (Wang et al. 2017).
Lithium acetate/PEG-mediated chemical transformation methods have also been evaluated for genetic transformation of R. toruloides DMKU3-TK16. In this technique, the transformation protocol is easy to master, even as it is currently hampered by the low efficiency of about 25 transformants/μg DNA (Tsai et al. 2017). More recently, electroporation has been adopted to transform R. toruloides (Liu et al. 2017), with a varied transformation efficiency of 40–1000 CFU/μg DNA depending on the strains.
The outcomes and features of different transformation methods applied in R. toruloides were compared in Table S2 (Supporting Information). It is clear that various methods have been attempted for random integration and site-specific deletion, with wide-ranging efficiency. Currently, ATMT is still the most reliable method, despite problems of T-DNA integration into the genome with a high probability of random insertion via non-homologous end joining (Koh et al. 2014; Sun et al. 2017; Wang et al. 2017). This method leads to a very low proportion of mutants generated from homologous recombination, thus necessitating the quest for the designing of some new methods to effectively filter out false-positive mutants. Previous research suggested that once a counter selection marker or a lethal gene such as herpes simplex virus thymidine kinase gene gets randomly integrated into chromosome by ATMT, the mutants get hardly grown in counter-screening medium (Khang et al. 2005). This phenomenon can be applied to filter ectopic transformants, while preserving the target mutants. Plenty of studies have illustrated strategies of transferring Ti plasmids harboring counter selection marker cassette locating outside homologous arms. This technique has proved helpful in the screening of positive mutants in other fungi including Magnaporthe grisea, Fusarium oxysporum and Verticillium dahliae (Khang et al. 2005; Tian et al. 2011; Jiang et al. 2013), which could be used to improve ATMT in Rhodosporidium species.
Generally, low transformation efficiency is one of the bottlenecks for all gene transfer methods. The highest transformation efficiency of R. toruloides is usually at the level of 103 transformants/μg DNA (or 105 cells), which makes the success of genetic manipulation much dependent on tools with higher transformation efficiency such as transposon-mediated mutations (Kumar 2016). It is important to note the difficulty to obtain a favorable mutant when the positive frequency is below 10−3. As a result, a lot of manpower and resources are committed to increasing transformants number, as the background ectopic mutants need to be filtered out by means of counter-screening and other methods. Besides (non-)homologous recombination of R. toruloides, there are also a few reports on the site-specific integration and the in-frame deletion of target genes free of scar (Koh et al. 2014; Sun et al. 2017; Sun et al. 2018).
Genetic parts
Like in other eukaryotes, a gene expression module in R. toruloides also follows a pattern of ‘promoter-functional gene-terminator’, in which the ‘functional gene’ includes selection markers, reporter genes and those coding for dedicated proteins. A number of toolsets such as promoters, terminators, selection markers and reporter genes have been verified functional in R. toruloides, as shown in Table S3 (Supporting Information). Endogenous constitutive and inducible promoters could be predicted from RNA sequencing data, as seen by a recent example of identification of 12 monodirectional and 8 bidirectional native promoters for R. toruloides (Nora et al. 2019).
Usually, the strength of promoters is characterized by the expression levels of reporter genes or resistance markers. For instance, the strength order of the constitutive promoters of PPGI, PPGK, PFBA, PTPI and PGPD in R. toruloides Y4 was determined through comparing the expression level of hygromycin resistance gene driven by these promoters (Wang et al. 2016b). In another study, the PGPD1 promoter of R. toruloides ATCC 10657 was identified and luciferase was used as a reporter gene to measure the strength of the 6 promoters regulating important genes involved in the lipid synthesis by R. toruloides (Liu et al. 2016a). Among them, the strength of the PLDP1 promoter is up to10-fold higher than that of the PGPD1 promoter, which has generally been considered as one of the strongest promoters in yeast. In addition, a PDAO1 promoter system that can be induced by D-amino acids was developed (Liu et al. 2015). Depending on the concentration of the inducer (such as D-alanine), the strength of the PDAO1 promoter can vary up to 10-fold, implying very good potential in gene expression regulation. Furthermore, inducible promoters PPHO89, PADH2 and PGAL1were identified, characterized and tightly regulated by phosphate and glucose concentration (Ma 2015). More recently, new inducible promoters such as PNAR1, PICL1, PCTR3 and PMET16 have been isolated which further enriched the repertoire of tunable genetic components (Johns, Love and Aves 2016). The promoter of RNA polymerase III was also identified that was used to express small RNA such as sgRNA (Jiao et al. 2019; Schultz, Cao and Zhao 2019).
In addition to the promoter from the genome, the virus 2A sequence is also effective to mediate the co-expression of multiple genes in R. toruloides (Jiao et al. 2018). Two different 2A sequences, porcine teschovirus-1 2A (P2A) and foot-and-mouth disease virus 2A (F2A) were evaluated as genetic elements to drive co-expression of the gene fused downstream the 2A sequence.
Terminators are specific DNA sequences within a few hundred base pairs in length and are needed to terminate gene transcription. As usual, terminators are located downstream of the gene stop codon, with feature sequences of (T-rich).TA (T) GT.(AT-rich)..TTT and TTTTTATA. Several endogenous terminators of genus Rhodosporidium, such as Thsp, Tgpd, etc., have been applied for gene expression termination (Jiao et al. 2018). Some heterogeneous terminators like T35s from Cauliflower mosaic virus and Tnos from A. tumefaciens have also been verified or reported functional in R. toruloides (Diaz et al. 2018).
Like promoters and terminators, selection markers and reporter genes also play crucial roles in genetic engineering. The available auxotrophic markers in genus Rhodosporidium are URA3 and LEU2, while that of antibiotics resistance include HYG, BLE, NAT and G418. Multiple rounds of integration experiments indicated that antibiotics resistance markers HYG, BLE and NAT could work independently in R. toruloides (Yang et al. 2008; Lin et al. 2014). Reporter genes such as luciferase- and green fluorescent protein (GFP)-coding genes have been applied in promoter characterization and strain screening (Liu et al. 2013; Liu et al. 2015; Johns, Love and Aves 2016; Diaz et al. 2018).
As summarized in Table S3 (Supporting Information), genetic parts are now largely in R. toruloides. Yet, some other elements such as nuclear localization sequence (NLS) and the application of other common reporter genes such as YFP and auxotrophic markers such as HIS remain to be demonstrated.
Targeted genome editing
RNA interference (RNAi) is a biological process in which RNA molecules inhibit gene expression or translation, by neutralizing targeted mRNA molecules. RNAi is one of the primary genetic tools that are independent of homologous recombination. It has been demonstrated that RNAi machinery is functional for gene-downregulation in R. toruloides (Liu et al. 2019). Inactivation of specific endogenous genes responsible for easy-screening phenotypes and genes overexpression by random integration has also been demonstrated (Koh et al. 2014; Sun et al. 2017). However, in-frame deletion or targeted integration process remains challenging.
One of the primary challenges of precise genetic modification is the low homologous recombination efficiency. When the knockout cassette of phytoene dehydrogenase gene CRT1 was introduced in R. toruloides NP11 through the ATMT method, only about 2% transformants were found with homologous recombination (Sun et al. 2017). Similarly, knocking out the gene CAR2 (encoding carotene cyclase) in the wild-type strain R. toruloides ATCC 10657 using the ATMT mediated homologous recombination gave a positive rate of 10.5% (Koh et al. 2014). In the Ku70-deficient strain R. toruloides ATCC 10657 △ku70, the targeted deletion frequency increased to 75.3% but the total number of transformants decreased to 14% of that of the wild-type strain. This suggested that the knockout of Ku70 reduces the ability of T-DNA to randomly insert into the chromosome, thereby excluding a large number of ectopic mutants. Thus, it seemed that Ku70 knockout decreased the screening workload with limited contribution to homologous recombination. Native homologous recombination factors or orthologous genetic parts such as RAD52 from S. cerevisiae are required to further improve homologous recombination in R. toruloides (van Attikum and Hooykaas 2003; Rolloos et al. 2014; Hooykaas et al. 2018).
As low homologous recombination efficiency and low transformation efficiency collectively limit the development of genetic tools based on homologous recombination, the application of other approaches especially the CRISPR-Cas system in Rhodosporidium becomes important. Very recently, three research groups independently developed the CRISPR-Cas9 system applicable in R. toruloides (Jiao et al. 2019; Otoupal et al. 2019; Schultz, Cao and Zhao 2019). These methods adopted different sgRNA transcription strategies to assist in Cas9 editing, hence achieving single or multi-genes editing via homologous recombination or NHEJ. However, the current technology is shrouded with several putative risks. First, both Cas9 and sgRNA cassettes are randomly integrated into the genome via ATMT or lithium chloride transformation, which may result in the inactivation of genes locating at the integration site. It could also be linked to the integrative and constitutive expression of sgRNA which result in off-target or toxicity of Cas9.
Since no replicable plasmid is available in R. toruloides, a gene of interest must be integrated into the chromosome for its function analysis. A classical Cre/loxP site-specific recombination system with the introduction of the Flp/FRT recombinase system was also applied to realize multiple rounds of random integration of heterogeneous genes (Sun 2017). Elsewhere, other recombinases like I-SceI have been adapted for the recovery of the G418 resistance marker in R. toruloides CECT 13085 (Fillet et al. 2016). In general, the application of recombinases implied that it is possible to achieve in-frame deletion and targeted insertion in the genome.
Currently, genetic parts, transformation methods and genetic tools remain to be enriched for complicated strain engineering of R. toruloides. Fortunately, many of those functional in other yeasts have been introduced successfully into genus Rhodosporidium (Lin et al. 2014; Johns, Love and Aves 2016; Wang et al. 2016b). Ultimately, mining, characterization and standardization of genetic toolsets will no doubt boost the emergence of novel gene-editing technologies which improve efficiency of strain engineering study.
Strain engineering
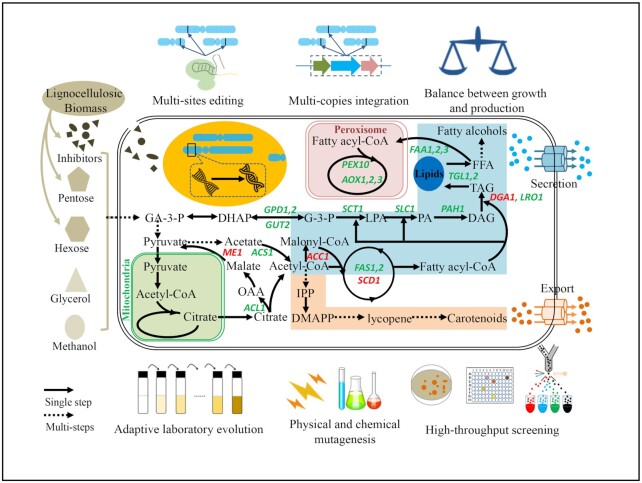
Increasingly enriched knowledge at systems level has been facilitating metabolic engineering of R. toruloides for diverse products including fatty acid derivatives, carotenoids, terpenoids and blue pigments. Similarly, efforts are devoted to expanding the substrate spectrum and stress resistance space (Fig. 3). A brief list of these examples is also shown in Table S4 (Supporting Information). In fact, the potential of R. toruloides as a microbial platform in biotechnology has also been reviewed recently (Xu and Liu 2017; Park, Nicaud and Ledesma-Amaro 2018; Saini et al. 2020).
Figure 3.
Strains engineering of genus Rhodosporidium as microbial platform. With the aid of rational and semi-rational approaches, including metabolic engineering, adaptive laboratory evolution, physical & chemical mutagenesis, high-throughput screening, Rhodosporidium is expected to be engineered to utilize lignocellulosic biomass, glycerol and methanol to produce and export fatty acid derivatives (highlighted in blue), terpenoids (highlighted in light orange). Key genes involved in fatty acid derivatives are highlighted in green and red (identified and characterized well in literature). Abbreviations: GA-3-P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; G-3-P, glycerol-3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; FFA, free fatty acid; OAA, oxaloacetate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate.
Engineering for fatty acid derivatives
Manipulation of important genes related to lipid biosynthesis has been successful in several aspects. Engineered R. toruloides strains were found with significantly increased lipid contents and yields under phosphorus-limited conditions upon expression of the key enzyme malic enzyme (Me) for improved NADPH supply (Wang et al. 2018). This is in-line with another work in which over-expression of Me in R. toruloides IFO 0880 boosted lipid production by 24% under nitrogen-limited conditions (Zhang et al. 2016b). In R. toruloides IFO 0880, random integration of genes encoding Acc1 and Dag1 led to increasing lipid content by 49% and lipid titer by 74% (Zhang et al. 2016c). Random integration of endogenous genes DGAT1 and SCD1 into R. toruloides CECT 13085 revealed an enhanced lipid yield and titer by 13% and 13%, respectively, while engineered strains produced about 39 g/L of lipid from lignocellulosic biomass hydrolysates (Diaz et al. 2018). As acetyl-CoA is a sole precursor to fatty acid biosynthesis, many studies have known to enhance its supply for improved production of fatty acid derivatives by in S. cerevisiae or Y. lipolytica (Xu et al. 2016; Zhou et al. 2016b; Qiao et al. 2017; Yu et al. 2018). In R. toruloides NP11, the introduction of phosphotransacetylase-coding gene from Bacillus subtilis to establish the conversion of phosphoacetate into acetyl-CoA led to increasing cell mass, lipid yield and titer by 26%, 11% and 54%, respectively (Yang et al. 2018).
Another scenario is to engineer advanced strains for the production of high-valued lipids. Previous studies demonstrated the composition of neutral lipids of R. toruloides as follows: palmitic acid 23%–30%, oleic acid (OA) 30%–37%, stearic acid 32%–37%, and linoleic acid 2%–4% (Papanikolaou and Aggelis 2011). To improve OA contents, R. toruloides strains were engineered by integrating an expression cassette for the Δ9-fatty acid desaturase from S. cerevisiae or the endogenous Δ9-fatty acid desaturase, leading to more than 5-fold higher OA contents, i.e. over 70% of the fatty acid composition of the total lipids (Wang et al. 2016a; Tsai et al. 2019). For linoleic acid production, an expression cassette for Δ12-fatty acid desaturase was integrated into the diploid strain R. toruloides AS 2.1389, thus achieving a titer of 1.3 g/L (Wang et al. 2016a). Random insertion of genes encoding Δ12 desaturase and ω-3 desaturase in combination with aldehyde dehydrogenase in-frame deletion afforded R. toruloides strains capable of producing lipids with α-linolenic acid content of ∼49% of total fatty acids (Fillet et al. 2016). More recently, genes encoding 3-ketoacyl-CoA synthase from different plants were genetically integrated into R. toruloides CECT 13085 and the recombinant strains produced very long-chain fatty acids, erucic acid and nervonic acid for the first time (Fillet et al. 2017). It was found that the titers of erucic acid and nervonic acid were stimulated upon increasing the copy numbers of the 3-ketoacyl-CoA synthase gene.
By overexpression of fatty acyl-CoA reductase from Marinobacter aquaeolei VT8 in R. toruloides CECT 13085, the engineered strain produced up to 8.0 g/L of long-chain fatty alcohols with 16–18 carbon atoms upon cultivation on sucrose for 75 h in a 7-L bioreactor (Fillet et al. 2015). By using nonionic surfactants during the culture process, fatty alcohol production was further enhanced by engineered strains (Liu et al. 2020).
Engineering for carotenoids and terpenoids
One key feature of R. toruloides is to accumulate both triacylglycerides and carotenoids (Buzzini et al. 2007; Singh et al. 2016; Singh et al. 2018; Bertacchi et al. 2020; Tran et al. 2020). Some efforts have been done by regulation of its endogenous enzymes to enhance carotenoids biosynthesis (Sun et al. 2017; Liu et al. 2018; Jiao et al. 2019). Attempts have also been tried to use Rhodosporidium as a production host for terpene-based biofuels by evaluation of 16 terpene synthases (TS) from plants, bacteria and fungi. Eight of these TS were found functional in R. toruloides and a total of nine different monoterpenes were obtained. The engineered strain produced either a single terpene compound or mixtures of others such as 1,8-cineole, sabinene, ocimene, pinene, limonene, and careen (Zhuang et al. 2019). Similarly, R. toruloides has been engineered for the production of other terpenoids including bisabolene, amorphadiene and ent-kaurene. When cultivated on the alkaline corn stover hydrolysate, bisabolene was produced by the engineered strain at a titer of over 680 mg/L; while on a glucose-only medium, it was at 521 mg/L (Yaegashi et al. 2017). Ionic liquids-rich hydrolysates were also feasible substrates for the R. toruloides strain to produce bisabolene, demonstrating a good compatibility with biomass pretreatment method (Sundstrom et al. 2018). Furthermore, bisabolene titers were improved to 2.2 g/L in a 20-L bioreactor using separation-free biomass hydrolysates as substrate (Pimienta et al. 2019). The diterpene ent-kaurene was produced in a 2-L bioreactor at a titer of 1.4 g/L from corn stover hydrolysates by engineered R. toruloides strain that expressed genes encoding the kaurene synthase from Gibberella fujikuroi and a mutated farnesyl diphosphate synthase from Gallus gallus for enhanced geranylgeranyl diphosphate supply (Geiselman et al. 2020).
Engineering for other metabolites
Indigoidine is a natural blue pigment produced by several bacteria including Streptomyces lavendulae and its biosynthetic gene cluster has been well established. This 3′,3′-bipyridyl pigment is formed through condensation of L-glutamine catalyzed by a non-ribosomal peptide synthetase (NRPS). Interestingly, by expressing the BpsA gene cluster from S. lavendulae in R. toruloides IFO 0880, the native metabolite pools of glutamine were converted into indigoidine. The engineered strain produced indigoidine at a titer of 2.9 g/L using a sorghum hydrolysate as carbon sources in a batch process and the highest titer of 86 g/L on glucose in a high-gravity fed-batch process within a 2-L bioreactor (Wehrs et al. 2019). This study further demonstrated the potential of R. toruloides for biotechnological applications.
It should also be noted that R. toruloides wild-type strains have been found as workhorses to produce sugar polyols (Jagtap and Rao 2018; Jagtap et al. 2019) and important enzymes.
Engineering with traditional approaches
It should be mentioned that traditional approaches such as physical or chemical mutagenesis and adaptive laboratory evolution have also been used for strain engineering. For example, UV mutagenesis, chemical mutagenesis along with atmospheric and room temperature plasma methods are used to mutate R. toruloides for enhanced lipid or carotenoid production (Qi et al. 2014; Zhang et al. 2016a; Guo et al. 2019; Tran et al. 2020). These approaches have been explored to identify strains with superior physiological features and then putative clues may be delineated via systems biology analysis for reverse metabolic engineering studies (Yamada, Kashihara and Ogino 2017).
Future directions
Apparently, R. toruloides has attracted major interests in academia and industry due to its unique physiological traits and vast imbedded potentials (Xu and Liu 2017; Park, Nicaud and Ledesma-Amaro 2018; Osorio-González et al. 2019a; Saini et al. 2020). Our knowledge has been substantially enriched in terms of the molecular basis of the genome, metabolism and physiology. Reliable toolsets are now accessible for strain engineering and systems biology studies. To facilitate more fruitful researches with this red yeast, several directions are worthwhile mentioning.
First of all, more efficient genetic tools should be developed. Albeit many genetic parts are available and genome editing methods have been documented very recently, the overall efficiency remains considerably lower to engineer R. toruloides than several other yeasts including S. cerevisiae and Y. lipolytica. This is largely due to the low efficiency of homologous recombination and transformation, and the absence of replicable plasmids for R. toruloides. In this regard, strains with much improved homologous recombination should be designed and evaluated. Of course, it is always valuable to characterize more genetic parts and devise more effective transformation protocols. Of special interests is to further customize the CRISPR-Cas9 system recently developed for targeted genome editing of R. toruloides.
Second, fundamentals on the systems biology of lipid production should be emphasized. Although lipid biology related to the health issues has been intensively studied at the systems biology level, efforts are limited to address those issues of truly biotechnology-relevant. As an excellent lipid producer, R. toruloides is a wonderful eukaryotic organism to understand the sophisticated regulatory circuits and networks related to lipid metabolism such as lipid droplet dynamics, lipid transportation and the relationships among lipid accumulation, autophagy and senescence. The analysis of existing and to-be-generated omics data with advanced bioinformatics tools will be fruitful in this subject. Another topic of great engineering interests is the mechanism regulating fatty acid biosynthesis and isoprenoid biosynthesis. Noting that acetyl-CoA is the central precursor to fatty acids and isoprenoids, both of them being produced by R. toruloides natively, the potential to direct acetyl-CoA for production of isoprenoids in high yields remains to be demonstrated.
Third, more efforts should be devoted to devising R. toruloides strains for high-value products. So far, limited success has been known for the synthesis of unsaturated fatty acids, fatty alcohols, carotenoids and blue pigments. Of special interests is to engineer the fatty acid biosynthesis machinery for designer chemicals as demonstrated recently in S. cerevisiae (Zhu et al. 2017). Although the main stream is to engineer R. toruloides for functional lipids, the potential has been documented to use this yeast for the production of non-ribosomal peptides and sugar alcohols. While the integration of key functional genes into the genome has been readily accessible to deliver new products, more studies should also consider pathway bypass, cofactor balance, regulatory genes and subcellular organelle engineering as seen in other yeasts (Zhou et al. 2016a; Qiao et al. 2017).
Lastly, it is almost always overlooked but in fact essential to engineer the physiology beneficial to the techno-economics of microbial lipid production. Some very important traits including but not limited to secretory lipid production, stress resistance to high temperatures, extreme pH and high osmotic pressure and expanded substrate scope to organic components found in typical biomass hydrolysates. For example, a membrane transporter was used to export lipid products from R. toruloides cells into the medium in a bi-phasic culture for improved product separation and yields (Lee et al. 2016). R. toruloides strains with improvements in these features are much more attractive for applications to use the real world low-cost feedstocks. Yet, this is very challenging as the desired strains are expected to result from systematic and global rewiring of cellular metabolism.
Together, R. toruloides is expected to subject rational genetic engineering and semi-rational approaches integrated with high-throughput screening for advanced strains in diverse biotechnology applications of great academic and economic value. Concurrently, our understanding on the fundamentals of physiology and biology of this yeast will be enriched.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Natural Science Foundation of China (Nos. 31870042 and 21721004), Natural Science Foundation of Jiangsu Province (No. BK20170037), Key Laboratory of Biomass Chemical Engineering of Ministry of Education, Zhejiang University (No. 2018BCE003) and Dalian Institute of Chemical Physics, CAS (Nos. DMTO201701 and DICP I201947).
Contributor Information
Zhiqiang Wen, School of Environmental and Biological Engineering, Nanjing University of Science & Technology, 200 Xiaolingwei St, Nanjing 210094, China.
Sufang Zhang, Laboratory of Biotechnology, Dalian Institute of Chemical Physics, CAS, 457 Zhongshan Rd, Dalian 116023, China.
Chuks Kenneth Odoh, Laboratory of Biotechnology, Dalian Institute of Chemical Physics, CAS, 457 Zhongshan Rd, Dalian 116023, China.
Mingjie Jin, School of Environmental and Biological Engineering, Nanjing University of Science & Technology, 200 Xiaolingwei St, Nanjing 210094, China.
Zongbao K Zhao, Laboratory of Biotechnology, Dalian Institute of Chemical Physics, CAS, 457 Zhongshan Rd, Dalian 116023, China; Dalian Key Laboratory of Energy Biotechnology, Dalian Institute of Chemical Physics, CAS, 457 Zhongshan Rd, Dalian 116023, China.
Conflict of interest
None declared.
REFERENCES
- Banno I. Studies on the sexuality of Rhodotorula. J Gen Appl Microbiol. 1967;13:169–96. [Google Scholar]
- Bertacchi S, Bettiga M, Porro Det al.. Camelina sativa meal hydrolysate as sustainable biomass for the production of carotenoids by Rhodosporidium toruloides. Biotechnol Biofuels. 2020;13:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy RR, Sabra W, Maheshwari Get al.. Metabolic network analysis and experimental study of lipid production in Rhodosporidium toruloides grown on single and mixed substrates. Microb Cell Fact. 2015;14:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck JW, Andrews JH. Attachment of the yeast Rhodosporidium toruloides is mediated by adhesives localized at sites of bud cell development. Appl Environ Microbiol. 1999;65:465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzini P, Innocenti M, Turchetti Bet al.. Carotenoid profiles of yeasts belonging to the genera Rhodotorula, Rhodosporidium, Sporobolomyces, and Sporidiobolus. Can J Microbiol. 2007;53:1024–31. [DOI] [PubMed] [Google Scholar]
- Castaneda MT, Nunez S, Garelli Fet al.. Comprehensive analysis of a metabolic model for lipid production in Rhodosporidium toruloides. J Biotechnol. 2018;280:11–8. [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu P, Liu Yet al.. Identification and characterization of a type-2 diacylglycerol acyltransferase (DGAT2) from Rhodosporidium diobovatum. Anton Leeuw Int J G. 2014;106:1127–37. [DOI] [PubMed] [Google Scholar]
- Coradetti ST, Pinel D, Geiselman GMet al.. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. eLife. 2018;7:e32110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, He S, Ji Xet al.. Identification and characterization of a novel bifunctional Delta(12)/Delta(15)-fatty acid desaturase gene from Rhodosporidium kratochvilovae. Biotechnol Lett. 2016;38:1155–64. [DOI] [PubMed] [Google Scholar]
- d'Espaux L, Mendez-Perez D, Li Ret al.. Synthetic biology for microbial production of lipid-based biofuels. Curr Opin Chem Biol. 2015;29:58–65. [DOI] [PubMed] [Google Scholar]
- Dai X, Shen H, Li Qet al.. Microbial lipid production from corn stover by the oleaginous yeast Rhodosporidium toruloides using the PreSSLP process. Energies. 2019;12:1053. [Google Scholar]
- Dias C, Sousa S, Caldeira Jet al.. New dual-stage pH control fed-batch cultivation strategy for the improvement of lipids and carotenoids production by the red yeast Rhodosporidium toruloides NCYC 921. Bioresour Technol. 2015;189:309–18. [DOI] [PubMed] [Google Scholar]
- Diaz T, Fillet S, Campoy Set al.. Combining evolutionary and metabolic engineering in Rhodosporidium toruloides for lipid production with non-detoxified wheat straw hydrolysates. Appl Microbiol Biotechnol. 2018;102:3287–300. [DOI] [PubMed] [Google Scholar]
- Dinh HV, Suthers PF, Chan SHJet al.. A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data. Metab Eng Commun. 2019;9:e00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CT, Ratledge C. Influence of nitrogen metabolism on lipid accumulation by Rhodosporidium toruloides CBS 14. Microbiology. 1984;130:1705–10. [Google Scholar]
- Fei Q, O'Brien M, Nelson Ret al.. Enhanced lipid production by Rhodosporidium toruloides using different fed-batch feeding strategies with lignocellulosic hydrolysate as the sole carbon source. Biotechnol Biofuels. 2016;9:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillet S, Gibert J, Suarez Bet al.. Fatty alcohols production by oleaginous yeast. J Ind Microbiol Biot. 2015;42:1463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillet S, Ronchel C, Callejo Cet al.. Engineering Rhodosporidium toruloides for the production of very long–chain monounsaturated fatty acid-rich oils. Appl Microbiol Biotechnol. 2017;101:7271–80. [DOI] [PubMed] [Google Scholar]
- Fillet SC, Gonzalez BS, Barreno MDCRet al.. Production of microbial oils with an elevated oleic acid content. WO2016185073A12016.
- Gadanho M, Libkind D, Sampaio JP. Yeast diversity in the extreme acidic environments of the Iberian Pyrite Belt. Microb Ecol. 2006;52:552–63. [DOI] [PubMed] [Google Scholar]
- Gadanho M, Sampaio JP. Occurrence and diversity of yeasts in the Mid-Atlantic ridge hydrothermal fields near the Azores Archipelago. Microb Ecol. 2005;50:408–17. [DOI] [PubMed] [Google Scholar]
- Geiselman GM, Zhuang X, Kirby Jet al.. Production of ent-kaurene from lignocellulosic hydrolysate in Rhodosporidium toruloides. Microb Cell Fact. 2020;19:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Chen S, Chen Get al.. Improvement of lipid production in oleaginous yeast Rhodosporidium toruloides by ultraviolet mutagenesis. Engineer Life Sci. 2019;19:548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooykaas PJJ, van Heusden GPH, Niu Xet al.. Agrobacterium-mediated transformation of yeast and fungi. Curr Top Microbiol Immunol. 2018;18:349–74. [DOI] [PubMed] [Google Scholar]
- Hu C, Zhao X, Zhao Jet al.. Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour Technol. 2009;100:4843–7. [DOI] [PubMed] [Google Scholar]
- Hu J, Ji L. Draft genome sequences of Rhodosporidium toruloides strains ATCC 10788 and ATCC 10657 with compatible mating types, Genome Announc. 2016;2:e00098–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Into P, Pontes A, Sampaio JPet al.. Yeast diversity associated with the phylloplane of corn plants cultivated in Thailand. Microorganisms. 2020;8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap SS, Bedekar AA, Liu J-Jet al.. Production of galactitol from galactose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Biotechnol Biofuels. 2019;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap SS, Rao CY. Production of D-arabitol from D-xylose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Appl Microbiol Biotechnol. 2018;102:143–51. [DOI] [PubMed] [Google Scholar]
- Jiang D, Zhu W, Wang Yet al.. Molecular tools for functional genomics in filamentous fungi: Recent advances and new strategies. Biotechnol Adv. 2013;31:1562–74. [DOI] [PubMed] [Google Scholar]
- Jiao X, Zhang Q, Zhang Set al.. Efficient co-expression of multiple enzymes from a single promoter mediated by virus 2A sequence in the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2018;18:foy086. [DOI] [PubMed] [Google Scholar]
- Jiao X, Zhang Y, Liu Xet al.. Developing a CRISPR/Cas9 system for genome editing in the basidiomycetous yeast Rhodosporidium toruloides. Biotechnol J. 2019;14:e1900036. [DOI] [PubMed] [Google Scholar]
- Jin M, Slininger PJ, Dien BSet al.. Microbial lipid-based lignocellulosic biorefinery: feasibility and challenges. Trends Biotechnol. 2015;33:43–54. [DOI] [PubMed] [Google Scholar]
- Johns AMB, Love J, Aves SJ. Four inducible promoters for controlled gene expression in the oleaginous yeast Rhodotorula toruloides. Front Microbiol. 2016;7:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang CH, Park SY, Lee YHet al.. A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet Biol. 2005;42:483–92. [DOI] [PubMed] [Google Scholar]
- Koh CMJ, Liu YB, Moehninsiet al.. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014;14:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. Multipurpose transposon-insertion libraries in yeast. Cold Spring Harb Protoc. 2016;2016. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kushwaha H, Bachhawat AKet al.. Genome sequence of the oleaginous red yeast Rhodosporidium toruloides MTCC 457. Eukaryot Cell. 2012;11:1083–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma-Amaro R, Nicaud JM. Metabolic engineering for expanding the substrate range of Yarrowia lipolytica. Trends Biotechnol. 2016;34:798–809. [DOI] [PubMed] [Google Scholar]
- Lee JJL, Chen LW, Cao Bet al.. Engineering Rhodosporidium toruloides with a membrane transporter facilitates production and separation of carotenoids and lipids in a bi-phasic culture. Appl Microbiol Biotechnol. 2016;100:869–77. [DOI] [PubMed] [Google Scholar]
- Lee JJL, Chen LW, Shi JHet al.. Metabolomic profiling of Rhodosporidium toruloides grown on glycerol for carotenoid production during different growth phases. J Agric Food Chem. 2014;62:10203–9. [DOI] [PubMed] [Google Scholar]
- Leong W-H, Lim J-W, Lam M-Ket al.. Third generation biofuels: A nutritional perspective in enhancing microbial lipid production. Renew Sust Energ Rev. 2018;91:950–61. [Google Scholar]
- Lin J, Shen H, Zhang Zet al.. Microbial lipid production by Rhodosporidium toruloides in a two-stage culture mode. Chin J Biotechnol. 2010;26:997–1002. [PubMed] [Google Scholar]
- Lin X, Wang Y, Zhang Set al.. Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2014;14:547–55. [DOI] [PubMed] [Google Scholar]
- Li Q, Kamal R, Wang Qet al.. Lipid production from amino acid wastes by the oleaginous yeast Rhodosporidium toruloides. Energies. 2020;13:1576. [Google Scholar]
- Liu D, Geiselman GM, Coradetti Set al.. Exploiting nonionic surfactants to enhance fatty alcohol production in Rhodosporidium toruloides. Biotechnol Bioeng. 2020;117:1418–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jiao X, Wang Yet al.. Fast and efficient genetic transformation of oleaginous yeast Rhodosporidium toruloides by using electroporation. FEMS Yeast Res. 2017;17:fox017. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhao X, Wang Fet al.. Comparative proteomic analysis of Rhodosporidium toruloides during lipid accumulation. Yeast. 2009;26:553–66. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Y, Liu Het al.. RNA interference in the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2019;19:foz031. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang M, Wang Tet al.. Two acetyl-CoA synthetase isoenzymes are encoded by distinct genes in marine yeast Rhodosporidium diobovatum. Biotechnol Lett. 2016a;38:417–23. [DOI] [PubMed] [Google Scholar]
- Liu YB, Koh CMJ, Ngoh STet al.. Engineering an efficient and tight D-amino acid-inducible gene expression system in Rhodosporidium/Rhodotorula species. Microb Cell Fact. 2015;14::170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YB, Koh CMJ, Sun LHet al.. Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides. Appl Microbiol Biotechnol. 2013;97:719–29. [DOI] [PubMed] [Google Scholar]
- Liu YB, Koh CMJ, Yap SAet al.. Identification of novel genes in the carotenogenic and oleaginous yeast Rhodotorula toruloides through genomewide insertional mutagenesis. BMC Microbiol. 2018;18:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YB, Yap SA, Koh CMJet al.. Developing a set of strong intronic promoters for robust metabolic engineering in oleaginous Rhodotorula (Rhodosporidium) yeast species. Microb Cell Fact. 2016b;15:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao Z, Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb Technol. 2007;41:312–7. [Google Scholar]
- Lopes HJS, Bonturi N, Miranda EA. Rhodotorula toruloides single cell oil production using Eucalyptus urograndis hemicellulose hydrolysate as a carbon source. Energies. 2020;13:795. [Google Scholar]
- Lu L, Wang J, Zhu Ret al.. Transcript profiling analysis of Rhodosporidium paludigenum-mediated signalling pathways and defense responses in mandarin orange. Food Chem. 2015;172:603–12. [DOI] [PubMed] [Google Scholar]
- Ma S. Inducible expression vector set for engineering in the oleaginous yeast Rhodosporidium toruloides. MA dissertation. China: Dalian Polytechnic University, 2015. [Google Scholar]
- Matsakas L, Nemailla Bonturi N, Miranda EAet al.. High concentrations of dried sorghum stalks as a biomass feedstock for single cell oil production by Rhodosporidium toruloides. Biotechnol Biofuels. 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin N, Calcas X, Devillers Het al.. Draft genome sequence of Rhodosporidium toruloides CECT1137, an oleaginous yeast of biotechnological interest. Genome Announc. 2014;2:e00641–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué VS, Black BA, Kruger JSet al.. Integrated diesel production from lignocellulosic sugars via oleaginous yeast. Green Chem. 2018;20:4349–65. [Google Scholar]
- Nora LC, Wehrs M, Kim Jet al.. A toolset of constitutive promoters for metabolic engineering of Rhodosporidium toruloides. Microb Cell Fact. 2019;18:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio-González CS, Hegde K, Brar SKet al.. Challenges in lipid production from lignocellulosic biomass using Rhodosporidium sp.; A look at the role of lignocellulosic inhibitors. Biofuel Bioprod Bior. 2019a;13:740–59. [Google Scholar]
- Osorio-González CS, Hegde K, Ferreira Pet al.. Lipid production in Rhodosporidium toruloides using C-6 and C-5 wood hydrolysate: A comparative study. Biomass Bioenerg. 2019b;130:105355. [Google Scholar]
- Otoupal PB, Ito M, Arkin APet al.. Multiplexed CRISPR-Cas9-based genome editing of Rhodosporidium toruloides. mSphere. 2019;4:e00099–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part II: technology and potential applications. Eur J Lipid Sci Tech. 2011;113:1052–73. [Google Scholar]
- Park YK, Nicaud JM, Ledesma-Amaro R. The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol. 2018;36:304–17. [DOI] [PubMed] [Google Scholar]
- Pimienta JAP, Papa G, Rodrigue Aet al.. Pilot-scale hydrothermal pretreatment and optimized saccharification enables bisabolene production from multiple feedstock. Green Chem. 2019;21:3152–64. [Google Scholar]
- Protzko RJ, Hach CA, Coradetti STet al.. Genomewide and enzymatic analysis reveals efficient D-galacturonic acid metabolism in the basidiomycete yeast Rhodosporidium toruloides. mSystems. 2019;4:e00389–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao K, Wasylenko TM, Zhou Ket al.. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol. 2017;35:173–7. [DOI] [PubMed] [Google Scholar]
- Qi F, Kitahara Y, Wang ZTet al.. Novel mutant strains of Rhodosporidium toruloides by plasma mutagenesis approach and their tolerance for inhibitors in lignocellulosic hydrolyzate. J Chem Technol Biotechnol. 2014;89:735–42. [Google Scholar]
- Qi F, Zhao XB, Kitahara Yet al.. Integrative transcriptomic and proteomic analysis of the mutant lignocellulosic hydrolyzate-tolerant Rhodosporidium toruloides. Eng Life Sci. 2017;17:249–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolloos M, Dohmen MH, Hooykaas PJet al.. Involvement of Rad52 in T‐DNA circle formation during A grobacterium tumefaciens‐mediated transformation of Saccharomyces cerevisiae. Mol Microbiol. 2014;91:1240–51. [DOI] [PubMed] [Google Scholar]
- Saini R, Hegde K, Brara SKet al.. Advanced biofuel production and road to commercialization: An insight into bioconversion potential of Rhodosporidium sp. Biomass Bioenerg. 2020;132:105439. [Google Scholar]
- Schultz JC, Cao M, Zhao H. Development of a CRISPR/Cas9 system for high efficiency multiplexed gene deletion in Rhodosporidium toruloides. Biotechnol Bioeng. 2019, 116:2103–9. [DOI] [PubMed] [Google Scholar]
- Shen H, Gong Z, Yang Xet al.. Kinetics of continuous cultivation of the oleaginous yeast Rhodosporidium toruloides. J Biotechnol. 2013;168:85–9. [DOI] [PubMed] [Google Scholar]
- Shen H, Zhang X, Gong Zet al.. Compositional profiles of Rhodosporidium toruloides cells under nutrient limitation. Appl Microbiol Biotechnol. 2017;101:3801–9. [DOI] [PubMed] [Google Scholar]
- Shi J, Feng H, Lee Jet al.. Comparative proteomics profile of lipid-cumulating oleaginous yeast: an iTRAQ-coupled 2-D LC-MS/MS analysis. PLoS One. 2013;8:e85532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Jawed A, Paul Det al.. Concomitant production of lipids and carotenoids in Rhodosporidium toruloides under osmotic stress using response surface methodology. Front Microbiol. 2016;7:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Sinha S, Bandyopadhyay KKet al.. Triauxic growth of an oleaginous red yeast Rhodosporidium toruloides on waste ‘extract’ for enhanced and concomitant lipid and β-carotene production. Microb Cell Fact. 2018;17:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soccol CR, Neto CJD, Soccol VTet al.. Pilot scale biodiesel production from microbial oil of Rhodosporidium toruloides DEBB 5533 using sugarcane juice: Performance in diesel engine and preliminary economic study. Bioresour Technol. 2017;223:259–68. [DOI] [PubMed] [Google Scholar]
- Spagnuolo M, Hussain MS, Gambill Let al.. Alternative substrate metabolism in Yarrowia lipolytica. Front Microbiol. 2018;9:1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom E, Yaegashi J, Yan Jet al.. Demonstrating a separation-free process coupling ionic liquid pretreatment, saccharification, and fermentation with Rhodosporidium toruloides to produce advanced biofuels. Green Chem. 2018;20:2870–9. [Google Scholar]
- Sun W, Yang X, Wang Xet al.. Developing a flippase-mediated maker recycling protocol for the oleaginous yeast Rhodosporidium toruloides. Biotechnol Lett. 2018;40:933–40. [DOI] [PubMed] [Google Scholar]
- Sun W, Yang X, Wang Xet al.. Homologous gene targeting of a carotenoids biosynthetic gene in Rhodosporidium toruloides by Agrobacterium-mediated transformation. Biotechnol Lett. 2017;39:1001–7. [DOI] [PubMed] [Google Scholar]
- Sun W. Agrobacterium-mediated genetic transformation-based genetic recombination system for Rhodosporidium toruloides. PhD dissertation. China: Dalian University of Technology, 2017. [Google Scholar]
- Tian L, Chen J, Wang Jet al.. High efficient gene knockout in Verticillium dahliae. Acta Mmicrobiol Sinica. 2011;51:906–13. [PubMed] [Google Scholar]
- Tiukova IA, Brandenburg J, Blomqvist Jet al.. Proteome analysis of xylose metabolism in Rhodotorula toruloides during lipid production. Biotechnol Biofuels. 2019a;12:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiukova IA, Prigent S, Nielsen Jet al.. Genome‐scale model of Rhodotorula toruloides metabolism. Biotechnol Bioeng. 2019b;116:3396–408. [DOI] [PubMed] [Google Scholar]
- Tran TN, Ngo D-H, Nguyen NTet al.. Draft genome sequence data of Rhodosporidium toruloides VN1, a strain capable of producing natural astaxanthin. Data Brief. 2019;26:104443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TN, Ngo D-H, Tran QTet al.. Enhancing astaxanthin biosynthesis by Rhodosporidium toruloides mutants and optimization of medium compositions using response surface methodology. Process. 2020;8:497. [Google Scholar]
- Tsai YY, Ohashi T, Kanazawa Tet al.. Development of a sufficient and effective procedure for transformation of an oleaginous yeast, Rhodosporidium toruloides DMKU3-TK16. Curr Gen. 2017;63:359–71. [DOI] [PubMed] [Google Scholar]
- Tsai YY, Ohashi T, Wu CCet al.. Delta-9 fatty acid desaturase overexpression enhanced lipid production and oleic acid content in Rhodosporidium toruloides for preferable yeast lipid production. J Biosci Bioeng. 2019;127:430–40. [DOI] [PubMed] [Google Scholar]
- Tully M, Gilbert HJ. Transformation of Rhodosporidium toruloides. Gene. 1985;36:235–40. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Hooykaas PJJ. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yurkov AM, Göker Met al.. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud Mycol. 2015;81:149–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen H, Tang Xet al.. Molecular tools for gene manipulation in filamentous fungi. Appl Microbiol Biotechnol. 2017;101:8063–75. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lin X, Zhang Set al.. Cloning and evaluation of different constitutive promoters in the oleaginous yeast Rhodosporidium toruloides. Yeast. 2016a;33:99–106. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang S, Potter Met al.. Overexpression of delta12-fatty acid desaturase in the oleaginous yeast Rhodosporidium toruloides for production of linoleic acid-rich lipids. Appl Biochem Biotechnol. 2016b;180:1497–507. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang S, Zhu Zet al.. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol Biofuels. 2018;11:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrs M, Gladden JM, Liu Yet al.. Sustainable bioproduction of the blue pigment indigoidine: Expanding the range of heterologous products in R. toruloides to include non-ribosomal peptides. Green Chem. 2019;21:3394–406. [Google Scholar]
- Wu S, Hu C, Jin Get al.. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour Technol. 2010;101:6124–9. [DOI] [PubMed] [Google Scholar]
- Wu S, Zhao X, Shen Het al.. Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour Technol. 2011;102:1803–7. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu D. Exploitation of genus Rhodosporidium for microbial lipid production. World J Microb Biot. 2017;33:54. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhao X, Wang Wet al.. Microbial conversion of biodiesel byproduct glycerol to triacylglycerols by oleaginous yeast Rhodosporidium toruloides and the individual effect of some impurities on lipid production. Biochem Eng J. 2012;65:30–6. [Google Scholar]
- Xu P, Qiao K, Ahn WSet al.. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. P Natl Acad Sci USA. 2016;113:10848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaegashi J, Kirby J, Ito Met al.. Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels. 2017;10:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Kashihara T, Ogino H. Improvement of lipid production by the oleaginous yeast Rhodosporidium toruloides through UV mutagenesis. World J Microb Biot. 2017;33:99. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang S, Tang Wet al.. Identification of the orotidine‐5′‐monophosphate decarboxylase gene of the oleaginous yeast Rhodosporidium toruloides. Yeast. 2008;25:623–30. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang S, Zhou YJet al.. Characterization of the mitochondrial NAD+-dependent isocitrate dehydrogenase of the oleaginous yeast Rhodosporidium toruloides. Appl Microbiol Biotechnol. 2012;94:1095–105. [DOI] [PubMed] [Google Scholar]
- Yang X, Jin G, Wang Yet al.. Lipid production on free fatty acids by oleaginous yeasts under non-growth conditions. Bioresour Technol. 2015;193:557–62. [DOI] [PubMed] [Google Scholar]
- Yang X, Sun W, Shen Het al.. Expression of phosphotransacetylase in Rhodosporidium toruloides leading to improved cell growth and lipid production. RSC Adv. 2018;8:24673–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Pfleger BF. Revisiting metabolic engineering strategies for microbial synthesis of oleochemicals. Metab Eng. 2020;58:35–46. [DOI] [PubMed] [Google Scholar]
- Yu T, Zhou YJ, Huang Met al.. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174:1549–58. [DOI] [PubMed] [Google Scholar]
- Zhang C, Shen H, Zhang Xet al.. Combined mutagenesis of Rhodosporidium toruloides for improved production of carotenoids and lipids. Biotechnol Lett. 2016a;38:1733–8. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ito M, Skerker JMet al.. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl Microbiol Biotechnol. 2016b;100:9393–405. [DOI] [PubMed] [Google Scholar]
- Zhang S, Skerker JM, Rutter CDet al.. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol Bioeng. 2016c;113:1056–66. [DOI] [PubMed] [Google Scholar]
- Zhao X, Hu C, Wu Set al.. Lipid production by Rhodosporidium toruloides Y4 using different substrate feeding strategies. J Ind Microbiol Biotechnol. 2011;38:627–32. [DOI] [PubMed] [Google Scholar]
- Zhao X, Peng F, De Wet al.. Effects of some inhibitors on the growth and lipid accumulation of oleaginous yeast Rhodosporidium toruloides and preparation of biodiesel by enzymatic transesterification of the lipid. Bioprocess Biosyst Eng. 2012;35:993–1004. [DOI] [PubMed] [Google Scholar]
- Zhao X, Wu S, Hu Cet al.. Lipid production from Jerusalem artichoke by Rhodosporidium toruloides Y4. J Ind Microbiol Biotechnol. 2010;37:581–5. [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Buijs NA, Zhu Zet al.. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J Am Chem Soc. 2016a;138:15368–77. [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Buijs NA, Zhu Zet al.. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016b;7:11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YJ, Kerkhoven EJ, Nielsen J. Barriers and opportunities in bio-based production of hydrocarbons. Nat Energy. 2018;3:925–35. [Google Scholar]
- Zhuang X, Kilian O, Monroe Eet al.. Monoterpene production by the carotenogenic yeast Rhodosporidium toruloides. Microb Cell Fact. 2019;18:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Ding Y, Gong Zet al.. Dynamics of the lipid droplet proteome of the oleaginous yeast Rhodosporidium toruloides. Eukaryot Cell. 2015;14:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang S, Liu Het al.. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun. 2012;3:1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhou YJ, Krivoruchko Aet al.. Expanding the product portfolio of fungal type I fatty acid synthases. Nat Chem Biol. 2017;13:360–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.