Abstract
Amide Proton Transfer (APT) reports on contrast derived from the exchange of protons between amide groups and water. Commonly, APT contrast is quantified by asymmetry analysis, providing an ensemble contrast of both amide proton concentration and exchange rate. An alternative is to sample the off-resonant spectrum and fit an exchange model, permitting the APT effect to be quantified, correcting automatically for confounding effects of spillover, field inhomogeneity and magnetization transfer (MT). Additionally, it should permit amide concentration and exchange rate to be independently quantified. Here a Bayesian method is applied to this problem allowing pertinent prior information to be specified. A three-pool model was used incorporating water protons, amide protons and MT effect. The method is demonstrated in simulations, creatine phantoms with varying pH and in vivo (n=7). The Bayesian model-based approach was able to quantify the APT effect accurately (RMSE < 2%) even when subject to confounding field variation and MT effect, unlike traditional asymmetry analysis. The in vivo results gave approximate APT concentration (relative to water) and exchange rate values of 3x10-3 and 15 s-1. A degree of correlation was observed between these parameter making the latter difficult to quantify with absolute accuracy, suggesting that more optimal sampling strategies might be required.
Keywords: Amide Proton Transfer, Chemical Exchange Saturation Transfer, Magnetization Transfer
1. Introduction
Amide Proton Transfer (APT) imaging is a form of Chemical Exchange Saturation Transfer (CEST) imaging. CEST permits the imaging of endogenous or exogenous protons that are in exchange with the bulk water pool, for example with amide groups, via selective saturation using radiofrequency (RF) irradiation (1–3). Various CEST agents may be employed to detect a range of physiological parameters and metabolites, however APT uniquely exploits endogenous amide protons on the peptide backbone and has been shown to be sensitive to changes in amide concentration in brain tumors (4–6) and also to pH-dependent exchange rates in animal models of ischemia (7).
The CEST effect is frequently quantified using the Magnetization Transfer Ratio, MTR=1-Ssat/S0, comparing signals with (Ssat) saturation of the exchangeable protons to that without any applied radio frequency saturation (S0). If the resonance frequency of the solute is sufficiently close to that of bulk water then the effects of indirect saturation of the bulk water, so called ‘spillover’, will also be present. This is often corrected for by calculating an asymmetry ratio using a further measurement symmetrically placed on the opposite side of the water resonance:
| [1] |
For pure APT, this asymmetry arises from proton exchange and not magnetization transfer (MT), thus this quantity is often referred to as the proton transfer ratio, PTR, and this will be adopted here. The size of the PTR is related to the concentration of the solute group protons, their accessibility to water, and the rate of exchange of these protons with bulk water. For endogenous peptide amide protons, the exchange is base-catalysed for pH values above 5 (4). A relationship between rate constant and pH in rat brain has been shown [4] assuming that the amide concentration was known, allowing interpretation of APT contrast in terms of pH.
Another potential source of error is that PTR is very sensitive to shifts in the resonant frequency of water from the expected value caused by inhomogeneity of the B0 field (8,9). MT effects also influence the shape of the z-spectrum in vivo. In theory, asymmetry analysis will correct for MT effects in a similar manner to the ‘spillover’ effect. However, this assumes that the MT effect is symmetric around the water center frequency, which has been shown not to be true in many tissues (10).
Acquisition of a sampled z-spectrum permits analysis by fitting of a model of the CEST process as given by the Bloch equations modified for exchange: often referred to as the Bloch-McConnell equations (11,12). The CEST effect is modeled with one pool representing bulk water and further pools for each labile proton source. The amide proton concentration and exchange rate appear as separate parameters within the model and thus might be individually estimated from sampled z-spectrum data by model fitting. A generally cited disadvantage of the model-fitting approach, especially for APT, is that a large number of parameters within the model need to be defined a priori, the majority of which are known only to a certain degree of accuracy and are subject to inter- and intra-subject variability. However, a model-fitting strategy has the advantage of directly incorporating spillover effects and can correct for shifts in the water center frequency by letting this parameter be determined from the data. MT effects can also be included by the addition of an extra pool within the model (13). However, these corrections all require the fitting of an even greater number of parameters from limited data leading to an increasing risk of over-fitting and thus inaccurate results.
Here we investigate the use of a model-based analysis for the quantification of the CEST effect in APT MRI using a sampled z-spectrum. We also investigate the separate quantification of amide proton concentration and exchange rate. To mitigate the risks of over fitting and to incorporate uncertainty in the model parameters we employ a Bayesian model-fitting method (14). This method has previously been applied with success to the similarly SNR limited case of arterial spin labeling (ASL) quantification (14) and Bayesian methods are increasingly being applied to MRI analyses (15). The Bayesian model fitting method permits prior information about each parameter to be incorporated into the analysis that reflects the expected variability or uncertainty of its value. We explore the accuracy of model-based analysis of CEST data in simulation, in vitro and in vivo.
2. Theory
The z-spectrum can be described using the Bloch-McConnell equations with two pools: one representing water protons and one representing amide protons. A third pool may be added to model MT effects between semisolid protons and water and further pools potentially being included to model other sources of labile protons. In a general multi-pool system the magnetization in pool i in exchange with pool(s) j is given in (13,16). Chemical exchange between pools is given by the rate constant kij. Each pool is described by its resonance frequency, ωi, the equilibrium magnetization related to the concentration of protons in the pool, M0i, and the the T1 and T2 relaxation time constants, T1i and T2i. Generally the magnetization of other pools is defined relative to the main water pool, M0i = rM0i*M0w, thus rM0i is a measure of the proton concentration in pool i relative to water. A general matrix formulation of the multi-pool exchange model is given in the Appendix.
By performing the analysis using the full model we hypothesized that it should be possible to quantify the amide proton exchange rate whilst also accounting for amide proton concentration and B0 field inhomogeneity. For the latter the water resonance frequency is included as a variable within the model fitting. However, to employ the full model for the z-spectrum a relatively large number of parameters need to be either defined or estimated from the data. In typical non-linear fitting algorithms, allowing a large number of parameters to vary increases the risks of over fitting of the data and thus providing parameter estimates that reflect the random noise more than the underlying signal of interest. The alternative is to make strict assumptions about the value of many of the parameters in the model, however this may potentially bias the results if these assumptions are wrong. In practice many of the parameters within the APT model are known to fall within a relatively restricted range (e.g., Table 1), even if their precise value in any individual is not known a priori.
Table 1. Model parameters with prior values – expressed as the mean and standard deviation (SD) of a normal distribution, i ∈ [w,APT,MT]. Those without SD values were taken to be known exactly a priori.
| Water Pool | APT Pool | MT Pool | |||||
|---|---|---|---|---|---|---|---|
| Parameter | Mean | SD | Mean | SD | Mean | SD | |
| in vivo | |||||||
| M0i | 0 | 106 | - | - | - | - | |
| rM0i (M0i/M0w) | - | - | 0 | 0.01 | 0 | 0.01 | |
| ki,w | s-1 | - | - | 20B | N/A | 40C | N/A |
| log(ki,w) A | - | - | 3.0 | 1.0 | 3.7 | 1.0 | |
| T1 | s | 1.3E | 0.15 | 0.77 | 0.15 | 1.0H | 0.15 |
| T2 | ms | 50E | 10 | 10 | 2 | 0.2F | 0.04 |
| ωi | ppm | 0 | 0.1 | 3.5G | 0.1 | -2.41D | 0.1 |
| in vitro (agarose gel 3% plus creatine) | |||||||
| M0i | 0 | 106 | - | - | - | - | |
| rM0i (M0i/M0w) | - | - | 0 | 0.01 | 0 | 0.01 | |
| ki,w | s-1 | - | - | 50 | N/A | 48J | N/A |
| log(ki,w) A | - | - | 3.9 | 1.0 | 3.87 | 1.0 | |
| T1 | s | 3I | 0.15 | 1I | 0.15 | 1.0J | 0.15 |
| T2 | ms | 60I | 12 | 8.5I | 1.7 | 15x10-6 J | 3x10-6 |
| ωi | ppm | 0 | 0.1 | 1.9 | 0.1 | 0 | 0.1 |
The log value of the proton exchange rate constant was estimated during model fitting, thus the prior was defined on the log value.
Approximate value expected in tissue based on rat brain study (4).
Using the asymmetry found by (10).
Based on (33) assuming a Lorentzian lineshape (see Discussion).
(4).
T1 for MT protons has a minimal influence on the z-spectrum and is this generally assumed to be approximately equal to 1 s.
(39)
(20).
An alternative to conventional nonlinear model fitting is to take a Bayesian approach. The measured signal is comprised of an underlying signal plus noise (assuming an additive noise process):
| [7] |
where f is the model for the signal parameterized by parameters θ and e is the additive noise component. The noise can be assumed to be random and thus can be described by a probability distribution. Hence, the likelihood of measuring any data value given a set of parameter values can be written. If white (Gaussian) noise is assumed the likelihood becomes:
| [8] |
where σ is the standard deviation of the white noise process. The outcome from a model-fitting procedure is an estimate of the parameter values that were responsible for generating the data observed. This is an ‘inversion’ of the likelihood in equation 8, which can be achieved via Bayes theorem:
| [9] |
where Pr(θ|S) is the posterior probability of the parameters given the data and Pr(θ)is the prior distribution of the model parameters. From the posterior distribution, the expected parameter values can be obtained by taking the mean, the uncertainty or confidence intervals calculated from the standard deviation. It is also possible to marginalize over model parameters by integration to leave the distribution for only the parameters of interest. Marginalization enables one to account for the uncertainty introduced in the parameters of interest by the ‘nuisance’ parameters over which marginalization has been performed.
The use of a Bayesian algorithm permits prior information to be incorporated into the analysis through the prior probability distribution term in equation 9. For parameters that are to be explicitly estimated from the data, the use of a prior probability distribution provides a ‘soft’ constraint on the value; thus, reducing the risk of over-fitting. For other parameters, such as T1 and T2 values, whose values are known but for which some observable variation is expected, the variability can be captured by the prior, allowing the uncertainty to be reflected in the final parameter estimates.
Using the results of a model-based analysis a further quantitative asymmetry measure may be calculated, PTR*. This measure uses the parameters from the model-fitting to generate an ideal 2-pool, water plus APT z-spectrum and compare it to an ideal 1-pool model of water (to remove direct saturation effects):
| [10] |
Thus PTR* provides a single quantitative measure of the CEST effect (combining both proton exchange rate and amide concentration contributions) without the B0 field inhomogeneity and MT effects.
3. Methods
3.1. Simulation study
A simulation study was conducted to examine the quantification of the APT effect using a Bayesian model-based approach including the separate estimation of proton concentration and exchange rate. Four scenarios were considered that were designed to match either the widely used creatine in vitro model of APT (8) or in vivo APT measurement:
Two pool, water plus creatine (at 1.9 ppm relative to water).
Three pool, water, creatine and symmetric MT pool to match the MT effects expected from agarose gel.
Two pool, water plus amide (at 3.5 ppm relative to water).
Three pool, water, amide and asymmetric MT pool to match the asymmetric effects observed in vivo (10).
Data were generated using the Bloch-McConnell equations with amide concentration and proton exchange rate both varying over a 5 x 5 matrix: rM0APT ranged from 0.001 to 0.003 in 0.0005 steps, log(kAPT,w) ranged from 2.5 to 4.5 in 0.5 steps (12.2, 20.1, 33.1, 54.6, 90.0 s-1) encompassing exchange rates expected for pH around the range 6 to 7 (4). Data were also generated for each combination with offset in the water center frequency in the range -0.5 to 0.5 ppm in 0.25 ppm steps. The water pool magnetization was set to 100 and the other parameters to the mean values given in Table 1. Continuous saturation was assumed with a magnitude of 0.55 μT and a saturation time of 2 s to match the in vitro and in vivo data acquisitions.
For each combination of amide concentration, exchange rate and water center frequency offset, 100 datasets were generated to which white noise was added with a signal-to-noise ratio (defined as the ratio of noise standard deviation to unsaturated water magnetization) of 100:1. The data were analyzed using a Bayesian model-based method using a 3-pool model and the appropriate prior parameters in Table 1. PTR was calculated using equation (1) from a spline interpolation of the CEST spectrum, PTR* was calculated from the estimated parameters from the model-based analysis using equation (10). The root-mean-square-error against the true values were calculated for PTR, PTR*, kAPT,w and rM0APT across the 100 instances. Additionally, the mean parameter estimate error was also calculated as a measure of bias in the estimates seen across the 100 instances.
3.2. Phantom Study
An in vitro study was performed to test the ability of the Bayesian model-based method to fit to CEST data and to examine the accuracy with which amide proton concentration and exchange rate could be quantified. Tissue-like phantoms were prepared according to (8) using an agarose gel with the addition of creatine (Sigma Aldrich, St. Louis, MO, USA). Briefly, creatine was added to deionized water to reach concentrations of 75, 100 and 125 mM. Once the creatine had fully dissolved, agarose was added to comprise 3 % of the mixed solution and then heated to boiling. Thereafter, the mixed solution was maintained at 50 °C and titrated to pH values of 5.3, 5.6, 6 and 6.4 before being transferred to 2 ml tubes. In order to minimize the magnetic field inhomogeneity, a plastic container was used to house all the tubes and filled with the same agarose gel. The sample was left to solidify at room temperature prior to the MRI experiment.
Data were acquired at 4.7 T in a Varian DirectDrive™ spectrometer (Varian Inc. subsidiary of Agilent Technologies, Santa Clara, CA, USA) using a CEST sequence developed in house with a field of view of 80 mm x 80 mm, matrix size of 64 x 64, slice thickness of 1 mm, acquisition bandwidth of 250 kHz, echo time (TE) of 20 ms and repetition time (TR) of 15 s. Saturation was achieved using a series of Gaussian pulses of duration 20 ms with effective flip angle 184°, and with 20 ms spacing for a total saturation of 10 s. After the saturation, single-slice spin-echo (SE) echo planar imaging (EPI) readout was used. Data were acquired for saturation frequency offsets from -3.8 to 3.8 ppm (-760 to 760 Hz for 4.7 T) with 0.1 ppm (20 Hz) increment plus a reference image at a saturation frequency of 20 ppm (4000 Hz), a total of 78 volumes being acquired in 39 min 32 s.
Two analyses were performed using the Bayesian model-based approach, in the first both kAPT,w and rM0APT were estimated from the data, in the second rM0APT was fixed using values derived from the literature for the concentrations of creatine used (17,18) (75 mM -> 0.0019, 100 mM ->0.0027, 125 mM -> 0.0034).
As a comparison WASSR data were also acquired from -0.75 to 0.75 ppm At intervals of 0.025 ppm (-150 to 150 Hz, increments of 5 Hz) using a series of 20 ms Gaussian pulses with an effective flip angle 31° total duration 2 s. The WASSR data were analyzed using the method of (9) to produce an estimate of the shift in the water center frequency for comparison with that derived from the model fitting. In addition a B0 map was also generated from the phase of gradient echo image acquisitions with TEs of 2.11, 3.17, 4.23, 5.29 and 6.35 ms (19) using a least squares fit.
PTR was calculated using equation (1) from a spline interpolation of the CEST spectrum, including the correction for the water center frequency derived from the WASSR data. PTR* was calculated, as for the simulated data, using the model estimates.
3.3. In vivo study
To test the method in vivo, data were acquired from healthy subjects (n=7, range 22-35 years) using a Siemens Verio 3T system (Siemens Healthcare, Erlangen, Germany), under a technical development protocol agreed with local ethics and institutional committees, using a CEST sequence developed in-house. Single slice imaging was performed approximately mid brain transverse using a single-shot spin-echo EPI readout with TR of 4 s, TE 26 ms, matrix 80 x 80, FOV = 240 x 240 mm, slice thickness = 5 mm. Saturation was achieved using a series of Gaussian pulses of duration 20 ms with effective flip angle = 184°, and with 20 ms spacing for a total pulse train duration of 2 s. Data were acquired for saturation frequency offsets from -4.5 to 4.5 ppm with 0.3 ppm increment plus a reference image with no saturation, a total of 32 volumes being acquired in 2 min 55 s. In all subjects WASSR data were also acquired using the same sequence, but using a single 50 ms Gaussian pulse with effective flip angle 90° and offset range -0.5 to 0.5 ppm with a 0.03 ppm increment.
Data were analyzed using the Bayesian model-based approach and images of the parameter values produced. As for the in vitro data PTR and PTR* images were produced along with WASSR images of the water center frequency shift.
3.4. Bayesian model-fitting
A Bayesian data fitting algorithm that is the probabilistic analogue of the more traditional non-linear least squares was employed (14). The algorithm was implemented in custom written C++ code and is available as a tool within the FMRIB Software Library (www.fmrib.ox.ac.uk/fsl).
The parameters included within the model are listed in Table 1. The equilibrium magnetization of the water pool was inferred directly from the data, with magnetization of the APT and MT pools being estimated relative to this, rM0APT and rM0MT respectively. The log-transformed value for exchange rates for both APT and MT pools were inferred by the fitting algorithm as indicated in Table 1. This was done to make the model more linear with respect to these parameters to improve algorithmic convergence. Additionally, the logarithm of the amide proton exchange rate is linearly related to pH (4), making this a more useful parameter to estimate directly. Voxelwise variation in the water resonant frequency was inferred relative to the Larmor frequency at the specified field strength, this parameter being reparameterised as an offset in the frequency quantified in ppm. The saturation pulse train used in the phantom experiments and in vivo acquisition was equated for analysis to an equivalent average power continuous saturation B1 value of 0.55 μT using the calculation presented in (20). This value is slightly lower than the optimal B1 value of 0.63 μT that would be predicted by the relationship in (21).
The prior on each parameter was defined by a normal distribution with the values of mean and standard deviation given in Table 1. Values for the magnetization of water and the magnetization ratios for APT and MT were intentionally non-informative, this being reflected in the large standard deviation chosen. The mean values for these priors were set to zero to reflect the ‘default’ condition that there is no signal arising from these pools in the case of pure noise. The mean values for the other parameters were derived from the literature and were chosen to be appropriate for the field strength used. The variability of these parameters, reflecting both variability reported in the literature and natural variability within the brain, was incorporated by setting the standard deviation of the prior. The water resonant frequency offset parameter was initialized using the frequency at which the minimum in the z-spectrum occurred.
4. Results
4.1. Simulation study
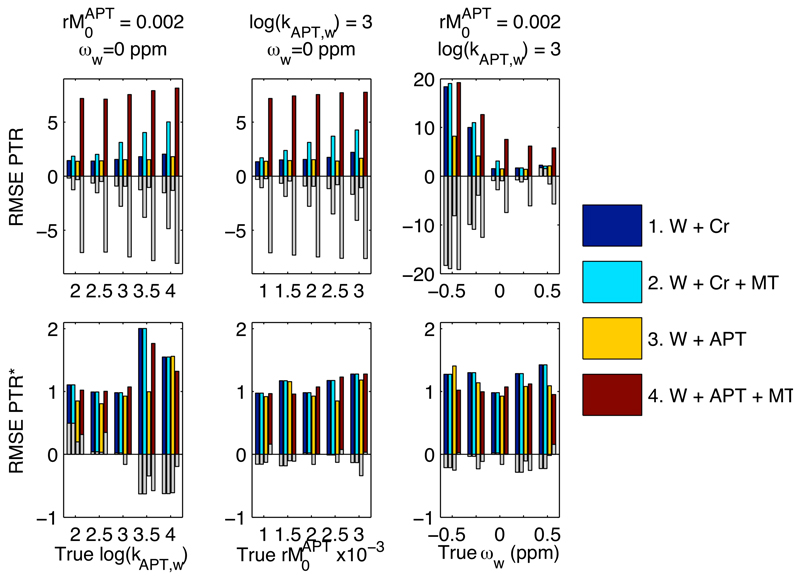
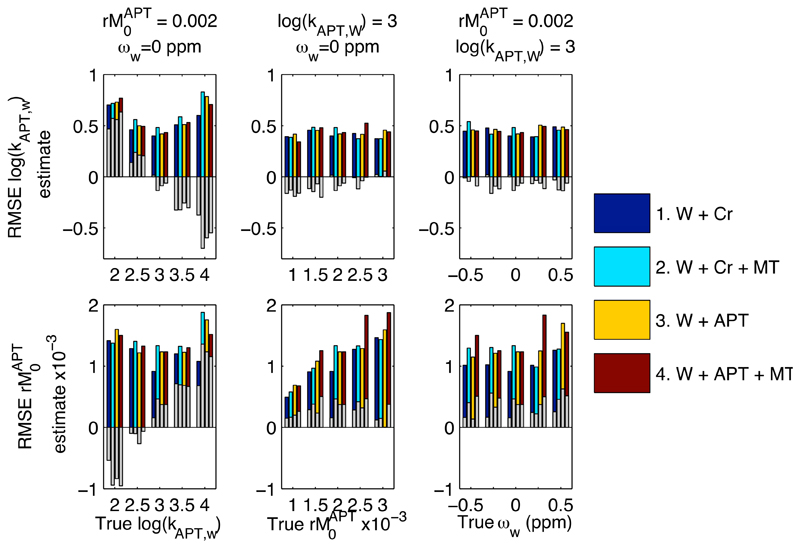
Figure 1 shows a RMSE and mean error values over the 100 instances for PTR and PTR* from the simulation study. Figure 2 shows the errors for kAPT,w and rM0APT as estimated by the model-based analysis. Three observations can be made from these results: firstly, that whilst the RMSE errors for both PTR and PTR* were the same for both of the two pool examples, the error was substantially increased in PTR once a third, MT, pool was included. This arose primarily though a bias in the estimates as demonstrated by the large mean error, due to changes in the z-spectrum not accounted for in the PTR calculation. The error was larger for the in vivo equivalent 3-pool simulation due to the asymmetry in the MT effect. For PTR* there was only a small increase in the error when the MT component was included, consistent with the extra variability introduced by having more parameters in the 3-pool model. Secondly, whilst the errors in PTR showed a substantial dependence on the shift in the water center frequency, ωw, those for PTR*, kAPT,w and rM0APT were broadly consistent for all shift values, indicating that the model-based method was correcting for this confound as expected. Thirdly, the errors in kAPT,w and rM0APT were seen to vary as the true kAPT,w value varied, whereas the errors would be expected to remain constant if unbiased estimation was being achieved. This same effect was also seen, to a lesser degree, for variations in the true value of rM0APT. The mean error indicated that this was a compensatory effect, such that underestimation of kAPT,w would be matched by overestimation of rM0APT and vice versa. Despite this under- or overestimation the PTR* measure retained its accuracy.
Figure 1.
RMSE and mean error for PTR and PTR* (%) over the 100 instances within the experiments of the simulation study. The coloured bars show RMSE, with the superimposed grey bars give the mean error (a measure of the bias in the estimates over the 100 noisy instances). PTR errors were substantially increased by inclusion of MT effects and shifts in the water center frequency.
Figure 2.
RMSE and mean error for kAPT,w and rM0APT over the 100 instances within the experiments of the simulation study. The coloured bars show RMSE, with the superimposed grey bars give the mean error (a measure of the bias in the estimates over the 100 noisy instances). Errors were largely insensitive to the inclusion of MT effects and shifts in the water center frequency. Underestimation of kAPT,w for some experiments was compensated for by overestimation of rM0APT and vice versa.
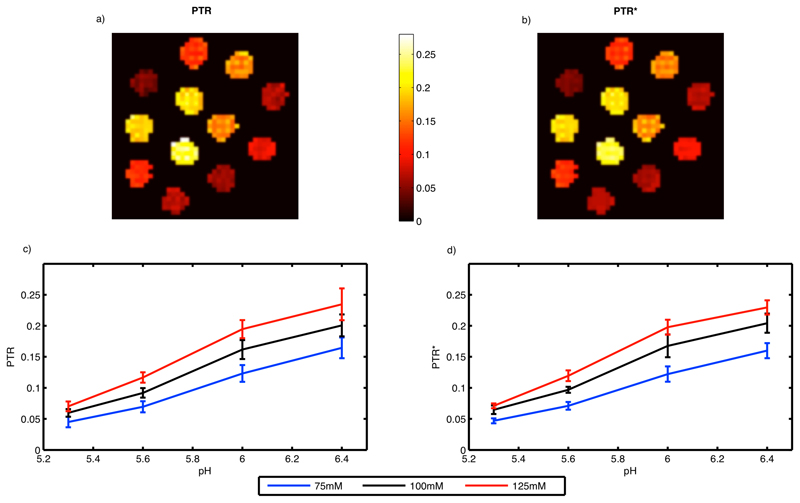
4.2. Phantom study
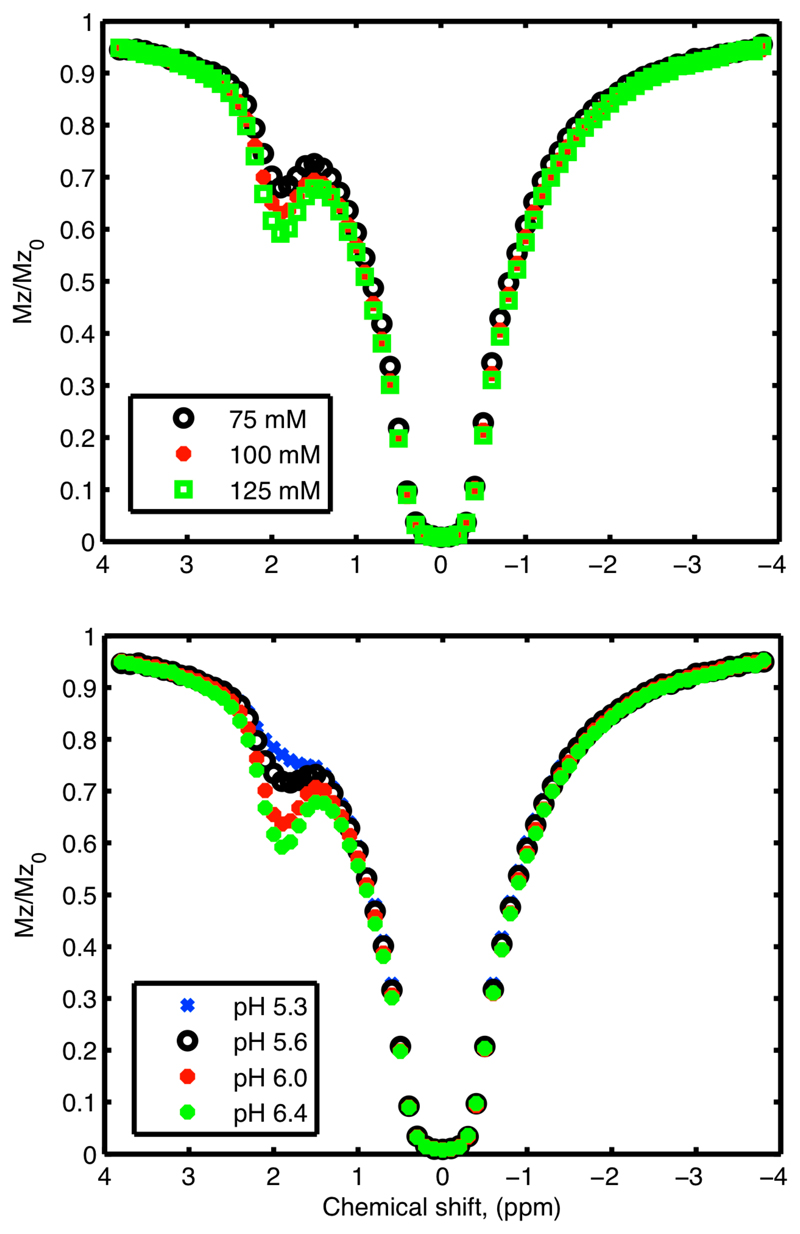
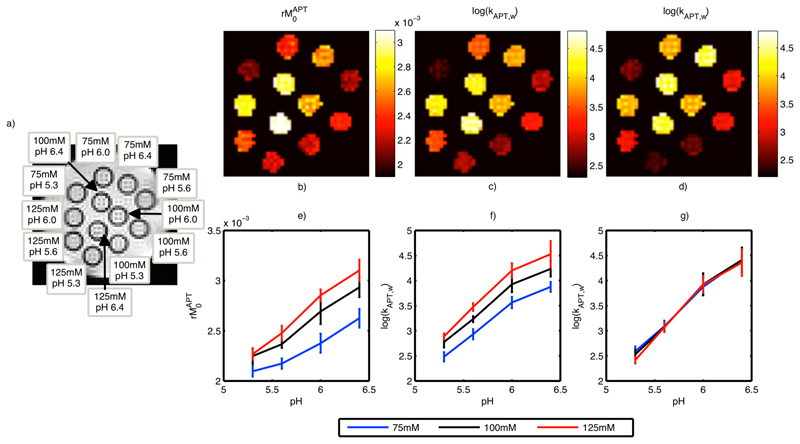
Figure 3 shows the measured z-spectra at a pH of 6.4 for different creatine concentrations (upper) and different pH values at 125 mM creatine (lower). Figure 4 shows the estimated kAPT,w and rM0APT images along with the equilibrium magnetization image labeled with the contents of each sample. Increases of rM0APT with increased creatine concentration and increasing kAPT,w with pH were visible in the images (b, c and d). Figure 4 also shows the mean and standard deviation within the samples of the estimated kAPT,w and rM0APT (e, f and g). Increases in rM0APT with creatine concentration and kAPT,w with pH were approximately linear, but it was clear that the rM0APT estimates were not independent of pH and the kAPT,w estimates were not independent of creatine concentration. The largest biases in the results appeared to occur for samples with low creatine concentration (where the CEST effect will be smallest). Figure 4 also shows the kAPT,w estimates for the APT pool when the model fitting analysis was performed using fixed rM0APT values, in this case the estimated kAPT,w was almost identical for all creatine concentration values.
Figure 3.
Example z-spectra from the phantom study for all three creatine concentrations at a pH of 6.4 (top) and all pH values at a creatine concentration of 125 mM (bottom).
Figure 4.
a) Equilibrium magnetization image from the in vitro study labeled with the creatine concentration and pH in each sample, b) and c) estimated rM0APT and kAPT,w from the model-based analysis method, d) estimated kAPT,w when the rM0APT was fixed using literature values e) mean estimated rM0APT vs. pH, f) mean estimated kAPT,w vs. pH within each sample from the in vitro study, g) mean estimated kAPT,w vs. pH when the rM0APT was fixed using literature values.
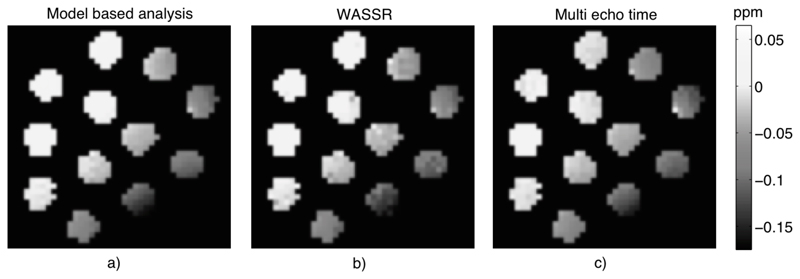
Figure 5 shows the PTR images along with their mean and standard deviation values within each sample. Both methods for calculating PTR produced visibly very similar results in the in vitro study. Figure 6 shows the estimated water resonance offset images using the three methods considered. The spatial distribution was similar across all three methods. The estimated water shift from the model-based analysis matched closely with the WASSR image and estimates from multiple echo time data.
Figure 5.
a) PTR and b) PTR* images from the in vitro study.
Figure 6.
Water resonant frequency offset images from the in vitro study estimated using a) model fitting the z-spectrum, b) WASSR data and maximum asymmetry method, c) multiple echo time data.
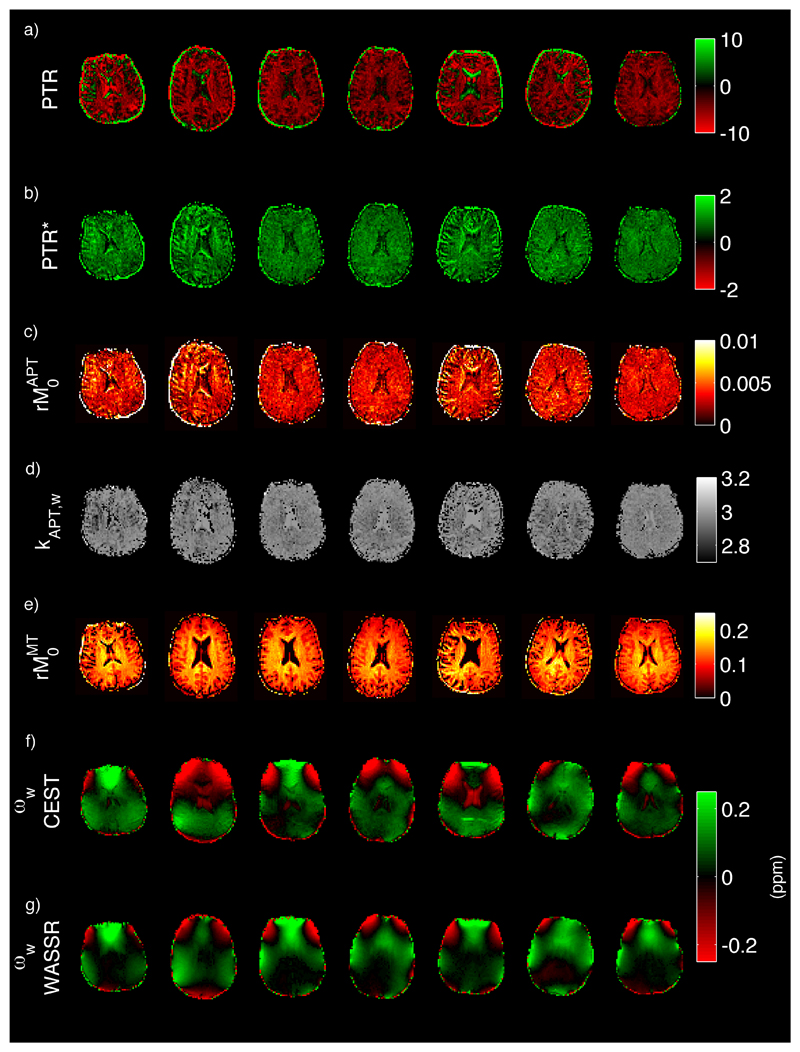
4.3. In vivo study
Figure 7 shows the images from the 7 healthy volunteers. Images of kAPT,w and rM0APT fitted using the model were relatively homogeneous across the brain. Likewise PTR* was relatively homogenous. PTR was negative and showed greater spatial variability than PTR*, which was positive throughout the brain. The model fitting estimated a substantial MT pool component within the z-spectrum, the relative magnetization of this pool appearing higher in regions of greater white matter prevalence.
Figure 7.
Images from the healthy subject data: a) PTR calculated from the data, b) PTR* using the parameters from the model fit to calculate the spectrum, c), d) and e) rM0APT, kAPT,w and rM0MT from the model fitting respectively, f) water resonant frequency offset from the model fitting, g) water center frequency offset as calculated for the WASSR data using the asymmetry method of (9).
Table 2 gives the group mean and standard deviation for concentrations in the APT and MT pools, and kAPT,w within the grey and white matter masks. There was general similarity between the water center frequency offset images from the model-based analysis and the WASSR images. However, in a number of subjects differences between the two images were present, for example, Subjects 2 and 5.
Table 2. Group mean and standard deviation within grey and white matter for APT concentration and rate constant, and MT concentration estimates.
| Parameter | Grey matter | White matter | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| rM0APT | 0.0031 | 0.0002 | 0.0033 | 0.0001 |
| log(kAPT,w) | 2.69 | 0.12 | 2.88 | 0.06 |
| rM0MT | 0.087 | 0.004 | 0.117 | 0.006 |
5. Discussion
In this work the analysis of APT z-spectra using a multi-pool model has been examined. Such an approach has been employed previously on CEST data (13,16), but it has generally been regarded as unsuitable for in vivo APT images given the small CEST contrast arising from endogenous amide protons, the close proximity of the amide resonant frequency to water and the large number of parameters whose precise value is not known a priori. Here a Bayesian analysis method has been adopted that offers robust model fitting in the face of such issues. In contrast to the non-linear least squares approach applied in these previous examples, the Bayesian method permits prior information on the parameters to be included in the form of probability distributions reducing the risk of over-fitting from having many free parameters in the model, whilst permitting some natural variation in these parameters to be inferred from the data. The Bayesian method employed is effectively a probabilistic version of the non-linear least-squares algorithm and thus has similar computational performance (14).
The majority of CEST analysis is performed using asymmetry of the z-spectrum, which in its simplest forms requires only two samples from the spectrum. However, correction for effects, such as a shift in the water center frequency, require the collection of supplementary data either of samples from the z-spectrum followed by some form of interpolation (22) or from another MRI method that can provide an estimate of the effect on the spectrum (8) (9). An advantage of a model-based approach is that confounding effects can be corrected for directly as part of the analysis via their inclusion as parameters in the model fit. The use of an extra spectrum concentrated around the water center frequency has been proposed in the form of the WASSR method. These data were also acquired here and comparisons of the water center frequency offset images in Figure 6 and those in Figure 7 demonstrated consistency in most subjects, indicating that it is possible to estimate the offset directly from the main z-spectrum. In principle where WASSR data is available this could be incorporated directly into the model-based analysis to improve upon the offset estimation.
MT is recognized as a confound for in vivo APT imaging, this was evident in the in vivo data acquired for this study. For example, the PTR images were negative throughout the brain indicating that the z-spectrum on the reference (-3.5 ppm) side exhibited more saturation than at the amide proton offset (+3.5 ppm). This result is consistent with the substantial asymmetry introduced into the spectrum by MT effect as predicted by the findings of (10). To account for MT effects a third pool representing the semi-solid/bound molecules was included in the analysis. The resulting PTR* measure was thus positive as would be expected from the APT effect alone. In practice the limited range of the frequency offsets sampled might make the absolute quantification of the MT effect inaccurate from these data. The sampling frequencies used did appear sufficient to correct for the confounding effect of MT on the spectrum as demonstrated with the simulated data.
The relatively spatially consistent nature of the images of the rate constant in Figure 7 is also consistent with the expected spatial homogeneity of pH in the healthy brain. Thus the application of the proposed analysis on typical z-spectrum APT data in healthy controls indicates that it is feasible to attempt estimations of both amide proton concentration and exchange rate in vivo. The ‘noisy’ appearance of the images, however, implies that the small size of the APT effect is difficult to detect accurately on a voxelwise basis. A potential limitation of APT methods to quantify pH in vivo, and in particular pH changes arising from ischemia, is the degree of correlation between amide concentration and exchange rate. As illustrated by the in vitro study it is difficult to unambiguously quantify both parameters independently from a single sampled z-spectrum. The estimated rM0APT values from the in vivo data in Table 2 were higher than those reported previously, for example 72 mM from WEX experiments (23). It is feasible that this is an example of overestimation of rM0APT with a matching underestimation of kAPT,w, as seen in the simulations. The estimated value of kAPT,w in Table 2 is of the order of 13 s-1 which might be lower than expected. Using the calibration in (4) this would correspond to a pH around 6.8, which is lower than the physiologically expected value, although caution should be exercised in taking that calibration and applying it to humans. In clinical practice it may be reasonable to assume that either one of rM0APT or kAPT,w is approximately fixed, for example that amide concentration varies minimally during the acute phase of an ischemic episode. It should be noted, however, that in tumors it is believed that both amide concentration and pH will vary from healthy tissue (5). Since the correlation between estimates of amide concentration and exchange rate is compensatory, an alternative is to use the PTR* measure to quantify the APT effect. This measure mirrors the widely used PTR, thus it provides a single summary measure of both exchange rate and concentration, with the advantage that PTR* benefits from the MT effect and water shift correction available from the model-based analysis.
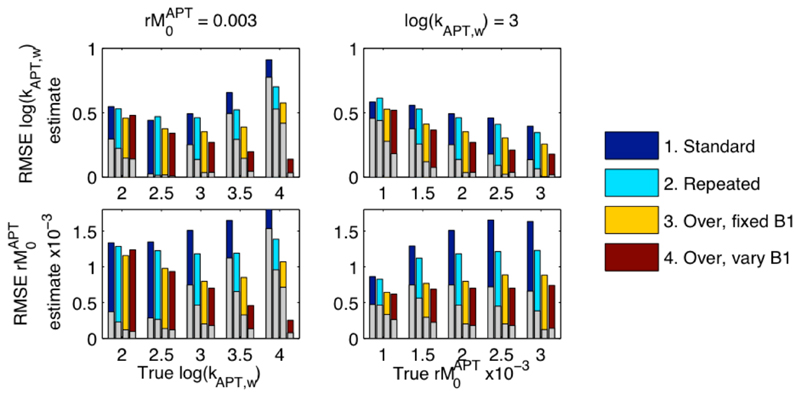
The use of measurements at multiple B1 values has been proposed to offer improved separation of amide concentration and exchange rate (24,25). The acquisition of data predominantly around the amide proton resonant frequency also appears to be advantageous, reflecting the fact that most of the information on these parameters will be derived from data in that region. These were both investigated using a further simulation study. Using the same parameters as the in vivo simulations (2-pool), four sampling strategies were considered:
B1 of 0.6 μT with offsets -4.5 to 4.5 ppm with 0.3 ppm increment, plus one image without saturation. Total 32 samples.
Strategy 1 with the spectrum from -4.5 to 4.5 repeated. Total 63 samples.
Strategy 1 plus spectrum from 2.8 to 4.2 repeated a further 6 times. Total 62 samples.
Strategy 1 plus spectrum from 2.8 to 4.2 repeated a further 6 times each at a different B1 value: 0.3, 0.4, 0.5, 0.7, 0.8, 0.9 μT. Total 62 samples.
Thus each sampling strategy contained a standard multiple saturation frequency acquisition, strategies 2 through 4 representing approaches to acquisition resulting in an approximate doubling of the number of samples: (i) by repeating the standard acquisition, (ii) by five-fold oversampling around the amide frequency, and (iii) by oversampling around the amide frequency using different B1 values.
Figure 8 shows the RMSE from the analysis of this simulated data, RMSE was generally lower when twice the number of samples were available and improved further if the extra samples were concentrated around the amide proton resonance. The use of multiple B1 values with concentrated sampling was seen to provide the lowest RMSE for practically all combinations of concentration and rate constant. The largest reductions in RMSE were seen for high concentration ratios and exchange rate constant value, with minimal improvements when these parameters were small. Like RMSE the bias was substantially reduced by more optimal placement of the samples and especially by the use of multiple B1 levels. The acquisition strategies investigated here were empirically chosen, it is likely that more optimally designed strategies could offer further advantages (26,27).
Figure 8.
Results from the simulation study. RMSE for estimating rM0APT and kAPT,w, when data were simulated with the ‘true’ values shown, for the four different sampling strategies. Superimposed on the bars is the absolute value of the mean error in the parameter estimate indicating the bias in the results toward under or over estimation. Both the RMSE and bias was reduced by placing more samples around the amide resonance and use of multiple B1 levels.
A disadvantage of a model-based analysis is the time taken in processing the data. A relatively rapid probabilistic fitting method has been employed here. However, processing of the single slice APT data required of the order of an hour of time on a standard desktop computer. The primary reason for this was the computational cost associated with the matrix exponential (see Appendix), which has to be computed for every single saturation frequency on each iteration of the algorithm. A faster solution would be obtained if the uncertainties associated with the T1, T2 and pool resonant frequencies were taken to be fixed, reducing the number of parameters in the inference. Far faster implementations should be feasible if analytic approximations to the model solution were adopted, such as those proposed by (17) (28), assuming that the inaccuracies introduced were acceptably accounted for.
The model used here assumes that the pulsed saturation strategy employed for the in vitro and in vivo data can be acceptably approximated by an equivalent continuous saturation of the same total duration with an equivalent B1 value. We have adopted the average power approximation here, which has been proposed as a better approximation than the more widely used average field value (29). Simulations (not shown) suggest that the continuous approximation is not a perfect match to the effect of pulsed saturation. This could be accounted for in the analysis by fully modeling the pulse train, for example using a discretized approximation that has been used by others (30). Currently the disadvantage of this approach is the increase in computational complexity due to the increased number of calculations associated with the discrete steps in the saturation pulse approximation. Comparisons of model fitting using both pulsed and discrete saturation in phantom studies suggest that the impact on the estimation of parameters from the model is minimal (31).
A 3-pool model was adopted to account for APT and MT contributions to the signal. It is likely that in vivo there will be other populations of exchangeable protons that will also contribute within the range of offset frequencies considered. For example the detection of myo-inositol has been proposed in the brain (19) and of glycogen in the liver (32). These components could be included by extending the model to extra pools as is commonly carried out for multi-site PARACEST agents.
In theory the modeling of MT as a pool within the Bloch-McConnell equations is equivalent, in the steady state, to assuming a Lorentzian lineshape. It has been proposed that MT effects are better modeled using a super-Lorentzian lineshape accounting for the fact that the pool is semi-solid, for example (33). This cannot be directly applied to the model used here, since the analysis deliberately accounts for imaging prior to reaching steady state saturation. The results of (33) (particularly their Fig. 1) indicate that within the range of saturation frequencies and B1 values employed here a Lorentzian fit is still reasonable. However, it is likely that the asymmetric component modeled here as an MT effect is in fact not a pure MT effect. (34) has observed this phenomenon at 7T and with a higher spectral resolution than used here; they attributed it to Nuclear Overhauser Effects (NOE) (3). NOE would appear to explain why a substantially asymmetric component is observed within the relatively narrow frequency range employed in CEST imaging (as compared to traditional MT imaging). As a model for NOE the single pool used here would serve only as a first order approximation, since NOE is likely to be composed of many exchange interactions of protons with differing offset frequencies.
6. Conclusions
Analysis of APT sampled z-spectra using the Bloch-McConnell model and a Bayesian fitting algorithm has been demonstrated. This permitted the APT effect to be quantified in vivo, even in the presence of asymmetries in the spectrum arising from MT or NOE contributions. Studies on tissue mimicking phantoms have shown that absolute independent quantification of amide concentration and exchange rate might not always be possible using a single sampled z-spectrum. To achieve such separation specialized sampling schedules incorporating multiple saturation field strengths may be required.
Supplementary Material
7. Acknowledgements
MAC was employed by The Centre of Excellence in Personalised Healthcare funded by the Wellcome Trust and EPSRC under grant number WT088877/Z/09/Z. YT is funded by Qualcomm Scholarship from Qualcomm Inc. NRS and AK are funded by Cancer Research UK, grant number C5255/A12678.
References
- 1.Zhou J, van Zijl PCM. Chemical exchange saturation transfer imaging and spectroscopy. Prog Nucl Mag Res Sp. 2006;48(2–3):109–36. [Google Scholar]
- 2.Sherry AD, Woods M. Chemical Exchange Saturation Transfer Contrast Agents for Magnetic Resonance Imaging. Annu Rev Biomed Eng. 2008 Aug 1;10(1):391–411. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Zijl PCM, Yadav NN. Chemical exchange saturation transfer (CEST): What is in a name and what isn't? Magn Reson Med. 2011 Apr 1;65(4):927–48. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Payen J-F, Wilson D, Traystman R, van Zijl P. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Medicine. 2003;9(8):1085–90. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 5.Jones C, Schlosser M, van Zijl P, Pomper M, Golay X, Zhou J. Amide proton transfer imaging of human brain tumors at 3T. Magn Reson Med. 2006 Sep 1;56(3):585–92. doi: 10.1002/mrm.20989. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, et al. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nature Medicine. 2011;17(1):130–4. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun P, Zhou J, Sun W, Huang J, van Zijl P. Detection of the ischemic penumbra using pH-weighted MRI. J Cereb Blood Flow Metab. 2007;27(6):1129–36. doi: 10.1038/sj.jcbfm.9600424. [DOI] [PubMed] [Google Scholar]
- 8.Sun P, Farrar C, Sorensen A. Correction for artifacts induced by B-0 and B-1 field inhomogeneities in pH-Sensitive chemical exchange saturation transfer (CEST) Imaging. Magn Reson Med. 2007 Dec 1;58(6):1207–15. doi: 10.1002/mrm.21398. [DOI] [PubMed] [Google Scholar]
- 9.Kim M, Gillen J, Landman B, Zhou J, van Zijl P. Water Saturation Shift Referencing (WASSR) for Chemical Exchange Saturation Transfer (CEST) Experiments. Magn Reson Med. 2009;61(6):1441–50. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua J, Jones C, Blakeley J, Smith S, van Zijl P, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med. 2007;58:786–93. doi: 10.1002/mrm.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell H. Reaction Rates by Nuclear Magnetic Resonance. J Chem Phys. 1958;28(3):430–1. [Google Scholar]
- 12.Woessner D. Nuclear Transfer Effects in Nuclear Magnetic Resonance Pulse Experiments. J Chem Phys. 1961;35(1) 41–&. [Google Scholar]
- 13.Li A, Hudson R, Barrett J, Jones C, Pasternak S, Bartha R. Four-Pool Modeling of Proton Exchange Processes in Biological Systems in the Presence of MRI-Paramagnetic Chemical Exchange Saturation Transfer (PARACEST) Agents. Magn Reson Med. 2008 Nov 1;60(5):1197–206. doi: 10.1002/mrm.21752. [DOI] [PubMed] [Google Scholar]
- 14.Chappell M, Groves A, Whitcher B, Woolrich M. Variational Bayesian Inference for a Nonlinear Forward Model. IEEE Transactions on Signal Processing. 2009;57(1):223–36. [Google Scholar]
- 15.Woolrich M, Jbabdi S, Patenaude B, Chappell MA, Makni S, Behrens T, et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage. 2009;45(1, Supplement 1):S173–86. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 16.Woessner D, Zhang S, Merritt M, Sherry A. Numerical solution of the Bloch equations provides insights into the optimum design of PARACEST agents for MRI. Magn Reson Med. 2005 Apr 1;53(4):790–9. doi: 10.1002/mrm.20408. [DOI] [PubMed] [Google Scholar]
- 17.Sun P, Sorensen A. Imaging pH using the chemical exchange saturation transfer (CEST) MRI: Correction of concomitant RF irradiation effects to quantify CEST MRI for chemical exchange rate and pH. Magn Reson Med. 2008 Aug 1;60(2):390–7. doi: 10.1002/mrm.21653. [DOI] [PubMed] [Google Scholar]
- 18.Sun P, Benner T, Kumar A, Sorensen A. Investigation of optimizing and translating pH-sensitive pulsed-chemical exchange saturation transfer (CEST) imaging to a 3T clinical scanner. Magn Reson Med. 2008 Oct 1;60(4):834–41. doi: 10.1002/mrm.21714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haris M, Cai K, Singh A, Hariharan H, Reddy R. In vivo mapping of brain myo-inositol. NeuroImage. 2011;54(3):2079–85. doi: 10.1016/j.neuroimage.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zu Z, Li K, Janve VA, Does MD, Gochberg DF. Optimizing pulsed-chemical exchange saturation transfer imaging sequences. Magn Reson Med. 2011 Oct;66(4):1100–8. doi: 10.1002/mrm.22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun P, van Zijl P, Zhou J. Optimization of the irradiation power in chemical exchange dependent saturation transfer experiments. Journal of Magnetic Resonance. 2005;175(2):193–200. doi: 10.1016/j.jmr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Stancanello J, Terreno E, Castelli D, Cabella C, Uggeri F, Aime S. Development and validation of a smoothing-splines-based correction method for improving the analysis of CEST-MR images. Contrast Media and Molecular Imaging. 2008 Jul 1;3(4):136–49. doi: 10.1002/cmmi.240. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Wilson DA, Sun PZ, Klaus JA, van Zijl PCM. Quantitative description of proton exchange processes between water and endogenous and exogenous agents for WEX, CEST, and APT experiments. Magn Reson Med. 2004 May 1;51(5):945–52. doi: 10.1002/mrm.20048. [DOI] [PubMed] [Google Scholar]
- 24.Sun P. Simultaneous determination of labile proton concentration and exchange rate utilizing optimal RF power: Radio frequency power (RFP) dependence of chemical exchange saturation transfer (CEST) MRI. Journal of Magnetic Resonance. 2010;202:155–61. doi: 10.1016/j.jmr.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon M, Gilad A, Zhou J, Sun P, Bulte J, van Zijl P. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly-L-lysine and a starburst dendrimer. Magn Reson Med. 2006;55(4):836–47. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cercignani M, Alexander DC. Optimal acquisition schemes for in vivo quantitative magnetization transfer MRI. Magn Reson Med. 2006 Oct 1;56(4):803–10. doi: 10.1002/mrm.21003. [DOI] [PubMed] [Google Scholar]
- 27.Tee Y, Chappell M, Xie J, Payne S. CEST Sensitivity Functions Based Sampling Schedule. Proc Int Soc Magn Reson Med. 2011 May 11;:4491. [Google Scholar]
- 28.Zaiß M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson. 2011 Aug;211(2):149–55. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Zu Z, Janve VA, Li K, Does MD, Gore JC, Gochberg DF. Multi-angle ratiometric approach to measure chemical exchange in amide proton transfer imaging. Magn Reson Med. 2011 Dec 8; doi: 10.1002/mrm.23276. 10.1002–mrm.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun PZ, Wang E, Cheung JS, Zhang X, Benner T, Sorensen AG. Simulation and optimization of pulsed radio frequency irradiation scheme for chemical exchange saturation transfer (CEST) MRI-demonstration of pH-weighted pulsed-amide proton CEST MRI in an animal model of acute cerebral ischemia. Magn Reson Med. 2011 Oct;66(4):1042–8. doi: 10.1002/mrm.22894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tee Y, Khrapichev A, Sibson N, Payne S, Chappell M. Evaluating the Use of a Continuous Approximation for Model-Based Quantification of Pulsed Chemical Exchange Saturation Transfer (CEST) Journal of Magnetic Resonance. doi: 10.1016/j.jmr.2012.07.003. inpress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Zijl PCM, Jones CK, Ren J, Malloy CR, Sherry AD. MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST) Proc Natl Acad Sci USA. 2007 Mar 13;104(11):4359–64. doi: 10.1073/pnas.0700281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison R, Henkelman R. A model for Magnetization Transfer in Tissues. Magn Reson Med. 1995;33:475–82. doi: 10.1002/mrm.1910330404. [DOI] [PubMed] [Google Scholar]
- 34.Jones CK, Polders D, Hua J, Zhu H, Hoogduin HJ, Zhou J, et al. In Vivo Three-Dimensional Whole-Brain Pulsed Steady- AQ1 State Chemical Exchange Saturation Transfer at 7 T. Magn Reson Med. 2011 Aug 2;:1–18. doi: 10.1002/mrm.23141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sled J, Pike B. Quantiative Imaging of Magnetization Transfer Exchange and Relaxation Properties in vivo using MRI. Magn Reson Med. 2001;46:923–31. doi: 10.1002/mrm.1278. [DOI] [PubMed] [Google Scholar]
- 36.Chai J, Chen C, Chen J, Lee S-K, Yeung H. Estimation of in vivo Proton Intrinsic and Cross-Relaxation Rtes in Human Brain. Magn Reson Med. 1996;36:147–52. doi: 10.1002/mrm.1910360123. [DOI] [PubMed] [Google Scholar]
- 37.Deoni S. High-resolution T1 mapping of the brain at 3T with driven equilibrium single pulse observation of T1 with high-speed incorporation of RF field inhomogeneities (DESPOT1-HIFI) J Magn Reson Imaging. 2007;26(4):1106–11. doi: 10.1002/jmri.21130. [DOI] [PubMed] [Google Scholar]
- 38.Deoni S. Transverse relaxation time (T2) mapping in the brain with off-resonance correction using phase-cycled steady-state free precession imaging. J Magn Reson Imaging. 2009;30(2):411–7. doi: 10.1002/jmri.21849. [DOI] [PubMed] [Google Scholar]
- 39.Sun PZ, Murata Y, Lu J, Wang X, Lo EH, Gregory Sorensen A. Relaxation-compensated fast multislice amide proton transfer (APT) imaging of acute ischemic stroke. Magn Reson Med. 2008 May 1;59(5):1175–82. doi: 10.1002/mrm.21591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.