Abstract
Inhibiting the disease progression in KRAS-driven cancers after diagnosis has been a difficult task for clinicians to manage due to lack of effective intervention/preventive therapies. KRAS-driven cancers depend on sustained KRAS-signaling. Although developing inhibitors of KRAS-signaling has proven difficult in past, the quest for identifying newer agents has not stopped. Based on reports showing potential of terpenoid-chemicals of modulating signaling pathways downstream of KRAS, we asked if this chemical family has affinity of inhibiting KRAS function. Using crystal structure as a bait in silico, we identified 20 terpenoids for their KRAS protein-binding affinity. We next carried out biological validation of in silico data by employing in situ, in vitro, patient-derived explant ex-vivo and KPC transgenic mouse models. In this report, we provide a comprehensive analysis of a Lup-20(29)-en-3b-ol (Lupeol) as a KRAS-inhibitor. Using nucleotide-exchange, ITC, DSF, and immunoprecipitation assays, we show that Lupeol has a potential to reduce the GDP/GTP exchange of KRAS protein including mutant-KRASG12V. Lupeol treatment inhibited the KRAS-activation in KRAS-activated cell models (NIH-panel, colorectal, Lung, & PanIN) and patient-tumor explants ex-vivo. Lupeol reduced the 3-dimensional growth of KRAS-activated cells. The pharmacokinetic analysis showed the bioavailability of Lupeol after consumption via oral and intraperitoneal routes in animals. Tested under prevention settings, the Lupeol consumption inhibited the PanIN development in LSL-KRASG12D/Pdx-cre mice (PDAC progression model). These data suggest that the selected members of triterpene family (such as Lupeol) could be exploited as clinical agents for preventing the disease progression in KRAS-driven cancers however warrant further investigation.
Keywords: Colon cancer, Lung cancer, pancreatic cancer, KRASG12D, Transgenic mice
Introduction
RAS proteins are binary molecular switches that cycle between active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate (GDP)-bound states and the reaction is catalyzed by nucleotide exchange factor (GEF).1 KRAS mutations are frequently found at amino acid positions G12, G13 and Q61 that hamper intrinsic and GAP-mediated GTPase function.2 This leads to an accumulation of GTP-bound KRAS, the activated form of protein.2 The KRAS-GTP functions as a nucleotide-dependent switch for growth signaling pathways.2 According to the National Cancer Institute > 30 % of all human cancers – including 95 % of pancreatic cancer (PDAC), 45 % of colorectal cancer and 30% of lung cancer are driven by mutations of the RAS.3 The KRAS-GTP protein mediates its diverse growth-stimulating function through its direct interaction with effectors including Raf, PI3K and Ral.4 Although considered as a useful drug target, several attempts have failed to curb the activity of mutated KRAS.4 Furthermore, attempts to constrain the downstream-pathways of KRAS showed limited success due to the development of drug-resistance and complicated feedback mechanisms.5 Previously, the inhibitors of fernesylation were explored for blocking the function of KRAS protein however due to redeeming lipidation failed to perform as drugs under clinical settings.6
Plant-derived molecules formed the basis of several clinical anti-cancer drugs.7 These include Topotecan (similar to camptothecin), Vinca Alkaloid, Etoposide, Teniposide (Podophyllotoxin), Ingenol Mebutate and Emtansine.7 Previously, we showed that Lupeol triterpenoid inhibits the growth of KRAS-mutant cancer cell lines whereas sparing wild-type KRAS expressing cells.8 Based on this information, we asked if activated-KRAS has an affinity to the triterpene class of chemicals. We hypothesized that identifying novel KRAS-binding agents followed by validation in relevant biological models could lead to the development of new drugs or respective synthetic analogues to be used in KRAS-driven cancer prevention and therapy.
In the current study, we screened a library of triterpenoid class of molecules using an in silico approach and identified several top-hits which bind to the KRAS protein. We subsequently investigated triterpene Lup-20(29)-en-3b-ol (Lupeol) for its KRAS blocking activity using several RAS-activation assays and tested its efficacy in KRAS-driven cancer cell panels, human PDX ex-vivo and in KC transgenic mice.6
Materials and Methods
Antibodies
Pancreatic marker Kit (Catalogue #8679; anti-α-amylase, anti-keratin, anti-PLA-2GB), anti-pERK, anti-phos-AKT, anti-BRAF, anti-Bcl2, anti-Ki67 and anti-β-actin were purchased from Cell Signaling technology (Danvers, MA) whereas anti-KRAS-GTP antibody was procured from New East Biosciences (Malvern, PA).
Cell culture
Human normal pancreatic epithelial cell (HPNE) and KRAS-mutant cell (HCT116) were purchased from ATCC and cultured in RPMI medium. KRAS-activated premalignant pancreatic cells (PDE-Ras, PDE-st, PDE-KRAS/st) provided by Dr. Paul Campbell (Moffit Cancer Center, FL) were cultured in DMEM as described.11 The RAS-reagents group at National Institutes of Health provided the KRAS-active mouse fibroblasts (MEF) cell panel (KRAS4BG12D, KRAS4BG12V, KRAS4B-WT).
Screening of compounds for KRAS binding
The three-dimensional crystal structure of human KRAS (PDB code: 4EPV) having a resolution of 1.35 Å was analyzed by using Schrodinger-GLIDE docking program.9 Briefly, water molecules were removed, hydrogen atoms and charges were added using OPLS-2005 force field. Furthermore, loops and missing side chains were built using Prime-3.0 module. The hydrogen bonding network (Asp, Glu, and His hydroxyl containing residues) with minim-maxim RMSD of 0.30 Å was optimized. LigPrep module 3.1 was used to prepare chemical-ligands. Using OPLS-2005, the specific chirality/geometry was retained with the least energy conformations at biological pH 7.4. The ligand/inhibitor binding-site in the crystal complex was used for Glide docking at standard precision mode. The binding affinity of ligands with KRAS-protein were calculated using the MM-GBSA continuum-solvent model. Based on binding-affinity, Lupeol was selected for Induced Fit Docking (IFD) as discussed previously.9
Isothermal titration calorimetry (ITC)
ITC measurements were performed using Nano ITC-TA Instruments (New Castle, DE, USA). The recombinant proteins and compounds were prepared in same buffer (50 mM HEPES pH 6.8, 50 mM KCl, 5 mM MgCl2). A typical titration involved 14 injections of Lupeol (15 μL aliquots/injection) at 300 sec intervals, into the sample cell (volume ∼1.4 mL) containing KRAS protein. The heat of ligand-dilution in the buffer alone was subtracted from the titration data. The data were analyzed using Origin®5.0 software.
Differential scanning fluorimetry (DSF)
The KRAS protein samples were added to the thermal shift buffer, fluorescent dye orange (SYPRO) in water and aliquots were placed in of a 96 well-plate. The plate was centrifuged at 1500 rpm for 1 min and subsequently loaded into a theramocycler (Applied Biosystems 7500) to perform a melt curve experiment. The temperature was set to escalate on a continuous mode from 25–90°C The binding affinity of ligands against the human KRAS at a rate of 1.0 °C/s. The florescence was read at the excitation and emission wavelengths of 580±10 and 623±14, respectively.10
GTP/KRAS nucleotide association and exchange assays
The association of mant-GTP with recombinant KRAS protein was observed by fluorescence measurement over time on a BioteK fluorescence spectrometer (excitation 360 nm, emission 440 nm). Lupeol at the indicated amounts was incubated with 1 μM recombinant-KRAS protein and 200 μM mant-GTP in buffer (25 mM Tris (pH 7.5), 50 mM NaCl, and 1 mM DTT) at 25 °C. After 2.0 h of incubation, MgCl2 (final concentration 10 μM) was added. The protein was passed through NAP-5 column to remove free nucleotide. KRAS and mant-GTP alone were the positive control for the association, and the competition with 200-fold excess unlabeled GTP served as the negative control. The half-lives were determined using Prism software (single-exponential decay fit).
KRAS activation assay
GTP-bound KRAS levels were measured using a Raf-RAS-binding pull-down assay kit as per vendors protocol (Millipore, Mountain View, CA).11
Cell viability
The effect of Lupeol (5–30 μM) on the growth of normal cells (HPNE), KRAS activated-tumor cell lines (Ras/st PDE, Ras PDE, Kpp2, HCT-116) and KRAS-MEF panel (KRAS4BG12D, KRAS4BG12V, KRAS4BWT) was determined by MTT assay as described.12
[3H] thymidine uptake, prostatospheroids proliferation, apoptosis, immunoblotting, immunoprecipitation, immunohistochemical (IHC) and immunofluorescence analysis
All tests were performed as per our published method.12. All in vitro experiments used 48h Lupeol treatment (20 μM) except for prostatospheroids formation which used 12 days treatment protocol.
Lupeol pharmacokinetics in mice
Female C57BL/6 mice (8-weeks old) were used for pharmacokinetics studies. Blood samples from the mandibular vein were collected in lithium-heparin coated tubes (at 0, 0.25, 0.5, 1, 2, 4, 6, 12, and 24 h) after a single-dose administration of Lupeol (200 mg/kg) by oral or intraperitoneal routes. The samples were prepared as described in Supplementary data. The quantitative analysis of Lupeol in plasma samples was performed on a TSQ Quantum Ultra Mass Spectrometer (MS) coupled with a Waters Nano-Acquity capillary UPLC. Lanosterol was used as an internal control. The detailed MS/UPLC method for Lupeol is provided in Supplementary data. The PK parameters (Tmax, AUCinf, AUClast) were determined by using WinNonlin Version 5.3 from Pharsight (Mountain View, CA).
KRASG12D/Pdx-cre mouse model and Lupeol diet-supplementation
We generated KRASG12D/Pdx-cre mice which develop pancreatic intraepithelial neoplasia (PanIN). The breeding pair of animals was procured from the National Institutes of Health. The conditional mice were obtained by cross breeding of LsL-KRAS-G12D and Pdx-Cre animals as per the method reported by Bardeesy et al and Hingorani et al.13–14 The genotyping condition is provided in the supplementary data 1. A high-grade purified Lupeol-supplemented diet was prepared in the form of easy-to chew tablets (5 mg Lupeol/tablet; non-flavored; 1-tablet weight =3g) (Bio-Serve Inc., Flemington, NJ). The female KRASG12D/cre mice were fed with Lupeol-supplemented diet (200 mg/kg). We provided one-tablet per 20 g mouse and adjusted the number of tablets to the weight of mice to maintain the final dose (200 mg/kg). The non-flavored placebo tablets constitute of protein 21.3%, fat 4.3%, fiber 4.0%, ash 7.8%, moisture <10%, carbohydrate 54.0% and total calories of 3.39 kcal/gm. Food intake and body weight were measured regularly. The animals were euthanized as per IACUC guidelines and the entire pancreas was harvested for biochemical and immunohistochemical analyses.
Histology
For the grading of PanIN stage, we conducted the hematoxylin and eosin (H&E) staining. PanIN lesions were classified according to clinical-histopathologic criteria including total number of ductal lesions and their grade as recommended elsewhere.14
Patient tumor explants
The pancreatic tumors of patients (non-identifiable) were provided by the Cooperative Human Tissue Network (CHTN-East branch, Nashville) in accordance with IRB regulations. The tissues in culture media were received by the host laboratory within 12 h post-surgery. Briefly, tumors from two patients (n=2) were dissected under sterile conditions minced into 1mm3 pieces and placed (3 pieces per well) on pre-soaked, 1 cm3 dental sponges (Novartis Animal Health, Greensboro, NC) in a 12-well plate. After 12 h, the explants were treated with control (DMSO, 0.05%) or Lupeol (20 μM; for 10-days) were refreshed every 48 h. Explants were harvested and underwent H&E staining and IHC for morphology and KRAS-markers.
Statistical analyses
Student’s t test for independent analysis was applied to evaluate differences between the treated and untreated cells with respect to the expression of various proteins. ANOVA was used to study the significance between control and treated mice. A p-value of < 0.05 was considered statistically significant.
Results
Identifying KRAS-binding molecules
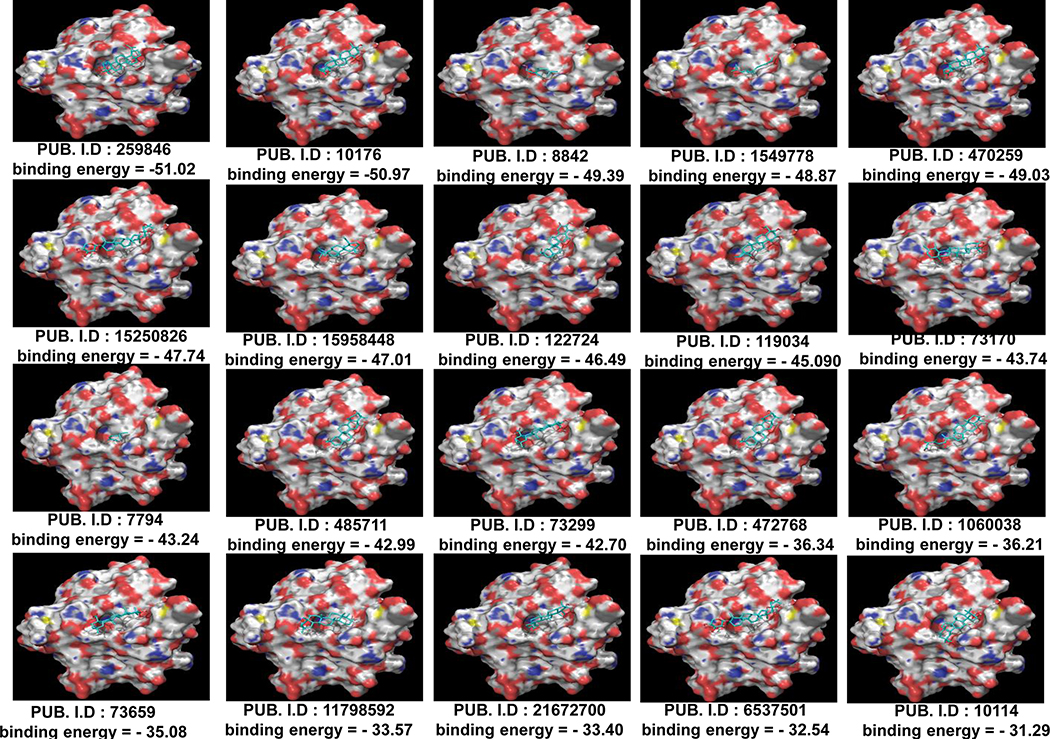
With a general-cutoff at −30.0 Kcal/mol energy, we shortlisted 21 molecules based on their binding potential to KRAS (Figure 1). Using high-stringency, the top molecules were further selected at a cut-off value of ˂ −45.0 Kcal/mol binding energy on the basis of which we identified Lupeol (−51.02 Kcal/mol), Cholanthrene (−50.97 Kcal/mol), Citronellol (−49.39 Kcal/mol), Arnidiol (−49.03 Kcal/mol), Geranylacetone (−48.87 Kcal/mol), Dehydroeburicoic acid (−47.74 Kcal/mol) and Celastrol (−46.49 Kcal/mol) (Figure 1). Lupeol (Lup-20(29)-en-3b-ol) is reported to be safe for consumption in animals. In the next series of in situ, in vitro and in vivo studies, we focused our attention on efficiency of Lupeol as a KRAS-inhibitor.
Figure 1: Screening of compounds for KRAS binding efficiency.
Figure 1 shows surface representations of Apo-KRAS (PDB# 4EPR) binding with small molecules as assessed by using a Glide software. Each surface representation is appended with PubChem ID and binding energy is represented in Kcal/mol.
Lupeol inhibits nucleotide association to KRAS protein
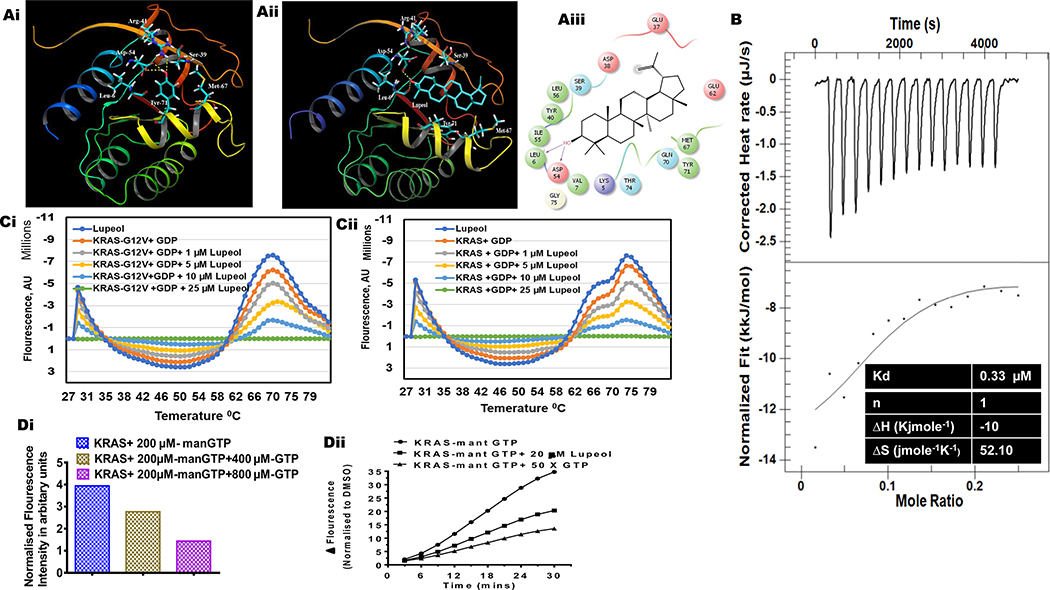
Molecular analysis of KRAS crystal structure (without a bound ligand) (PDB code: 4EPR) showed that Tyr-71 makes hydrogen bond interactions with Asp-54 and Arg-41 (Figure 2A). This hydrogen bonding network (crucial for KRAS activity) is disrupted upon ligand binding at a site situated between Switch I and Switch II regions (PDB code: 4EPV). The successful IFD execution of Lupeol with human KRAS generated multiple docking poses. The best pose chosen for further analysis exhibited multiple interactions with residues surrounding the hydrophobic pocket (Figure 2Ai). Further, Lupeol formed a hydrogen bond interaction with each of the residues Asp-54 and Leu-6 of KRAS (Figure 2Aii). In addition to hydrogen bond interactions, hydrophobic interactions were also observed. Total number of amino acid residues of KRAS that are involved in interactions with Lupeol are also presented (Figure 2Aiii). The Glide score, and binding affinity values (MM-GBSA values) calculated for Lupeol are −5.57 Kcal/mol and −92.93 Kcal/mol respectively. Furthermore, we found similarity in the binding pattern of Lupeol in the IFD KRAS-Lupeol docked complex with KRAS crystal co-complex structure (PDB code: 4EPV).
Figure 2: Effect of Lupeol on nucleotide binding and nucleotide exchange on KRAS.
(Ai) surface representation of residue interactions of Apo-KRAS (PDB code: 4EPR) (Aii) surface representation of KRAS-Lupeol docked complex ribbon representation of KRAS-Lupeol docked complex (Aiii) Ligand interaction diagram depicting KRAS amino acid residues involved in interaction with Lupeol. (B) Thermograms and binding isotherms show the interaction of Lupeol with recombinant KRAS protein (in terms of Enthalpy and Kd) as assessed by isothermal titration calorimetry (ITC). The top portion of each panel shows baseline corrected thermograms. The bottom portion shows the corresponding binding isotherms generated using nonlinear binding models. (Ci-ii) line graphs show the differential scanning fluorimetry (DSF) measurements for recombinant KRAS and KRAS-G12V loaded with GDP or GTP and Lupeol. (Di) The histogram shows the dose-dependent decrease in fluorescence intensity in presence of unlabeled GTP (yellow and purple bar). (Dii) line graphs shows effect of Lupeol on the association of mant-GTP with KRAS protein as assessed by fluorimetry. Data are representative of two biological replicates.
Lupeol-KRAS protein binding measured by ITC
We performed ITC to investigate the interactions between KRAS protein and Lupeol because ITC is the most powerful technique used to measure the energetics of drug-target interactions.15 The initial injection of Lupeol into solution resulted in the binding of Lupeol to KRAS protein and generation of maximal heat associated with the total enthalpy (Δ) of the interaction (Figure 2B). With subsequent injections, the amount of KRAS available for binding decreased which was indicated by the reduction of associated heat of interaction (Figure 2B). The heat of individual injections were integrated with respect to time and plotted against molar ratio of Lupeol (Figure 2B). When the resultant titration curve was fitted using one-site binding model, it yielded an association constant of 330 nm and enthalpy change (ΔH) of −10 Kcal mole−1 for a binding stoichiometry of n = 1. The entropy (ΔS) calculated were 52 jmole-1K-1 as shown in the inset of the (Figure 2B).
Lupeol interaction with KRAS and KRASG12V protein measured by DFS
The DSF is a method for measuring thermal stability that relies on a dye that becomes fluorescent when in contact with hydrophobic residues as proteins unfold upon heating.16 We used DSF technique to measure binding of Lupeol to KRAS or mutant-KRAS protein. Recombinant wild-type KRAS or mutant KRAS-G12V proteins were loaded with either GDP and incubated with Lupeol. Samples were incubated (at increasing temperatures) in the presence of a fluorescent dye that binds to hydrophobic surfaces exposed during thermal denaturation. We observed that Lupeol increases the amplitude of the thermal denaturation curve of wild-type KRAS and KRAS-G12V (loaded with GDP) suggesting binding of Lupeol to both wt-KRAS and KRAS-G12V protein (Figure 2Ci–ii). These data suggest that Lupeol interacts both with GDP-bound KRAS protein and might be blocking the formation of GTP-bound state by interfering with GDP-GTP exchange mechanism.
Effect of Lupeol on nucleotide exchange at KRAS protein
We performed florescence-based KRAS nucleotide exchange assay. As demonstrated by an increase in fluorescence, the addition of the fluorescent-GTP analog, mant-GTP (2′-/3′-O-(N′-methylanthraniloyl) guanosine-5′ O-triphosphate) exhibited an association with wild-type KRAS in a time-dependent fashion (Figure 2Di). We noted that addition of Lupeol (to KRAS/mant-GTP solution) caused an inhibition of mant-GTP association to KRAS protein (Figure 2Di). These data confirms that Lupeol blocks the GTP-GDP exchange at KRAS protein. The KRAS protein incubated with mant-GTP and excess unlabeled GTP (which completely blocked the mant-GTP association) served as the negative control (Figure 2Dii).
Lupeol inhibits the growth of KRAS-activated 2-dimensional and 3-dimensional models
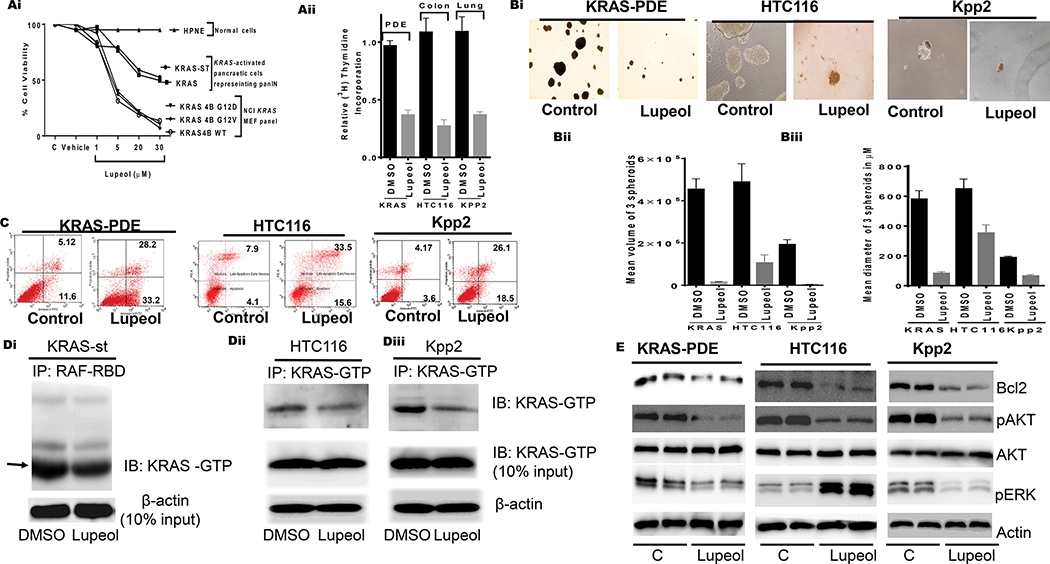
To determine the effect of Lupeol on the KRAS-activated monolayer or 2-dimensional (2D) cell cultures, we selected human pancreatic ductal-epithelial (PDE-KRAS and PDE-KRAS/st), mouse lung cancer (Kpp2), human Colon cancer (HCT116), and NCI-KRAS-active panel of cells. As a control, we used mouse embryonic fibroblast (MEF), and normal pancreatic (HPNE) cells. KRAS-expressing pancreatic and NCI cell panels were treated with Lupeol (1–50μM). At 48h post-treatment, cells were measured for growth by employing an MTT assay. Lupeol treatment inhibited the growth of KRAS-activated pancreatic neoplastic and NCI-KRAS cells in a dose-dependent manner while sparing normal cells (Figure 3Ai). Among NCI-KRAS-expressing cells, the prominent effect was observed in RPZ26198- KRASG12D and RPZ26425-KRAS-G12V cells (Figure 3Ai). An important observation is that Lupeol inhibited the growth of KRAS-mutant cells (HCT116-KRASG13D, KPP2-KRASG12D, RPZ26198-KRASG12D, RPZ26425-KRASG12V) at a lower dose (IC50 15–20μM) than wild-type KRAS-expressing cells (RPZ25854, RPZ26379; IC50 25–50 μM). Next, the [3H] thymidine uptake assay of cells showed that single treatment of Lupeol (20 μM) causes a reduction in the rate of proliferation of premalignant (PDE-KRAS and PDE-KRAS/st) and carcinoma (KPP2; HCT116) cells (Figure 3Aii).
Figure 3: Effect of Lupeol on growth proliferation, apoptosis, 3-dimensional spheroids, KRAS-GTP and KRAS downstream targets.
(Ai) Shows growth of normal, KRAS activated pancreatic cells representing PanIN and MEF having KRAS-G12D mutation. (Aiii) Histogram shows the rate of proliferation (3H[thymidine-uptake) of human pancreatic (KRAS-st), human colon (HTC116) and murine lung (KPP2). (Bi) microphotographs show the effect of Lupeol on prostatospheroids formation of KRAS-activated cells. (Bi-ii) Bar graphs show quantification spheroids in terms of volume and dimeter. (C) FACS analysis image shows the effect of Lupeol on Apoptosis in cells. (Di) immunoblot image shows KRAS-GTP level in pancreatic KRASst cells as assessed by RAF-RBD pull-down assay. (Dii-iii) immunoblot images show KRAS-GTP levels in colon and lung cells by KRAS-GTP-specific immunoprecipitation assays. (E) immunoblot image shows the KRAS-downstream targets in KRAS-activated cells.
Compared to 2D culture-models, 3-dimensional (3D) models closely mimics the microenvironment and growth pattern of cells within tumors. For this purpose, we allowed cells to form spheroids in a 3D Tumor-sphere Medium XF (PromoCell GmbH). After 4-days of spheroid formation in cells, the cultures were grouped into two: (i) control and (ii) Lupeol-treated of sub-lethal dose (10 μM). After 10–14 days of therapy, spheroids from control and the treated group were measured for size (an index of clonal proliferation). The control KRAS-activated pancreatic and colon cells formed spheroids of larger diameters (4×106 – 6 ×106 μm), whereas the Kpp2-spheroids exhibited smaller size (mean 2×106 μm). The long-term Lupeol therapy significantly (p<0.05) decreased the number of spheroid formations (Figure 3Bi–ii). We speculated that Lupeol-induced inhibition of clonal proliferation might be driving such clones to commit apoptosis. Next, KRAS/st, HCT116 and KPP2 cells were exposed to sub-lethal dose of Lupeol (10 μM) for 48 h and subjected to FACS analysis to measure apoptosis. An increase in a number of apoptotic cells was registered in pancreatic (23.02%), colon (3.6%) and Lung (5.4%) cells (Figure 3C).
Lupeol decreases active-KRAS protein (GTP-bound) levels in KRAS-activated cells
Mutations in the KRAS gene cause the accumulation of KRAS-GTP (activated form of KRAS) levels in premalignant and malignant cells.17–20 We asked if Lupeol-induced inhibition in proliferation is associated to the alterations in active protein (KRAS-GTP) levels in cells. We measured KRAS-GTP levels in cells treated with Lupeol (10 μM for 24 h) by using affinity pulldown (RAF/RBD) and KRAS-GTP specific immunoprecipitations assays. Lupeol treatment caused a reduction in GTP-bound active KRAS protein levels in pancreatic, colon and lung cancer cells (Figure 3Di–iii). Lupeol treatment did not cause any effect on total KRAS protein levels in cancer cells (Figure 3Di–iii) or normal cells (data not shown). These data validate the inhibitory potential of Lupeol as a KRAS-inhibitor in solution and in biological models.
Effect of Lupeol on KRAS-downstream pathways in KRAS-activated cells
Activation of KRAS triggers a cascade of molecular pathways, the majority of which are associated with the proliferation and therapy-resistance.3 The KRAS-induced pro-proliferative signal is relayed by Raf/Mek/Erk, PI3K/Pdk1/Akt and the Ral-guanine pathways in neoplastic cells.21–22 We next determined effect of Lupeol-treatment on the KRAS-downstream targets in cells by employing immunoblot assay. Lupeol treatment caused a marked reduction in phosphorylated-AKT and Bcl2 proteins in cells (Figure 3E). These data are significant because activated-Akt is promotes tumor progression whereas Bcl2-overexpression confers resistance to tumor cells.23 Whereas Lupeol caused a decrease in the phosphorylated-ERK levels in lung and colon cancer cells, an inverse case was observed in PDAC cells (Figure 3E). This data is consistent with studies which reported phosphorylation of ERK as a KRAS-independent event in PDAC models.24 Brandt et al showed cell type-dependent differential activation of ERK by oncogenic KRAS.25 Taken together, the data generated from in silico, in solution and cell-based biological assays firmly suggest the potential of Lupeol-triterpene as a strong KRAS-inhibitor.
Pharmacokinetics profile of Lupeol in vivo
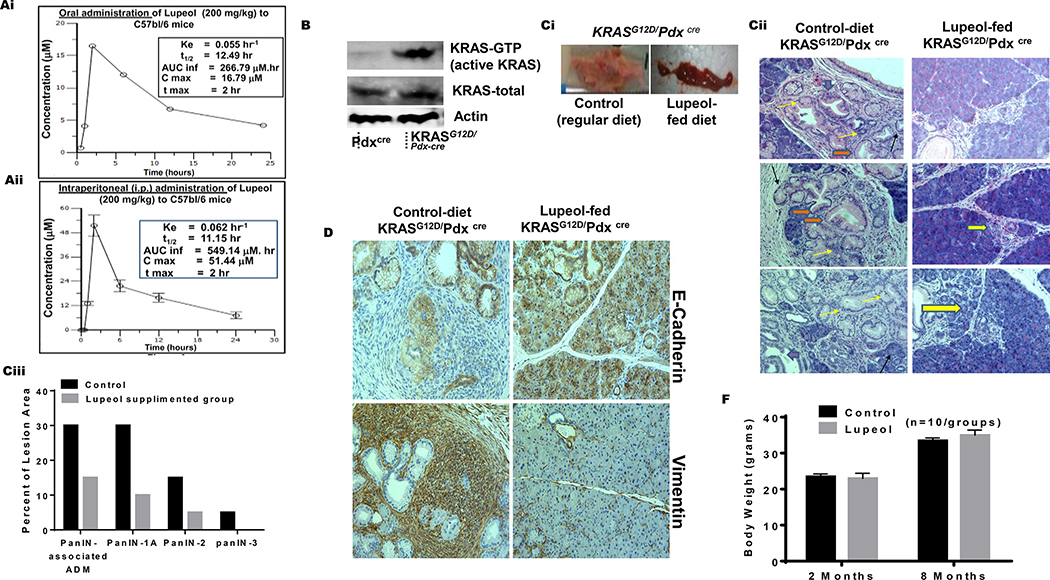
For quantification of Lupeol in mice plasma for in vivo pharmacokinetics, a sensitive and specific method with LC/MS/MS was developed as described under material and methods. The mean plasma concentration of Lupeol versus time was measured in the blood of mice collected at different time-points following a single-dose (200 mg/kg) of oral and intraperitoneal administration of Lupeol in two-independent experiments. The summarized pharmacokinetic parameters of Lupeol are presented in inset tables in Figures 4Ai–ii. Mice achieved a maximum plasma concentration of 16.79 μM and 51.44 μM at 2h for oral and intraperitoneal administration, respectively. The half-life (T1/2) of Lupeol was recorded as 12.49 h and 11.15 h following oral and intraperitoneal administration, respectively (Figure 4Ai–ii). The area under the curve from 0-inf (AUCInf) was estimated using a linear trapezoidal rule and equal to 266.79 μM and 549.14 μM for oral and intraperitoneal administrations, respectively (Figure 4Ai–ii).
Figure 4: Lupeol inhibits development of PanIN in KRASG12D/Pdxcre transgenic mice.
(Ai-ii) Graphs show 24 h pharmacokinetic profile of Lupeol in mice following 200 mg/kg oral and intraperitoneal administration. Inset in Figures Ai-ii shows pharmacokinetic profile (Cmax = Maximum plasma concentration; Tmax = Time to maximum concentration; AUClast = Area under the curve from time 0 to 24 hrs. AUCinf = Area under the curve from time 0 to infinity). (B) immunoblot image shows the pancreatic KRAS-GTP levels in Pdxcre and KRASG12D/Pdx-cre mice (Ci) Picto-micrographs show the pancreatic morphology in control and Lupeol-fed KRASG12D/Pdxcre mice. (Cii) Representative H&E-stained images of pancreatic acinar-to-ductal metaplasia from a placebo and Lupeol-fed KRASG12D/Pdx-cre mice at 8 months of age. (Ciii) Histogram shows the quantification of mPanIN 1A mPanIN 2A and mPanIN 3 lesions in KRASG12D/Pdxcre control and Lupeol-fed mice (D) Pictures show the EMT phenotype markers E-cadherin (epithelial) and vimentin (mesenchymal) in pancreatic tissues of control and Lupeol-fed mice. (F) Histogram shows the effect of Lupeol treatment on the body weight in mice.
Lupeol feeding inhibits PanIN development in KRASG12D /Pdx-cre mice
The development of PDAC starts from transformation of normal epithelium to carcinoma via a series of histologically well-defined precursor lesion formations termed as PanIN lesions. The activated-KRAS oncogene is a driver for the PanIN formation and multiple pathways are required for the progression of disease to carcinoma.14 Because Lupeol caused a significant inhibition in KRAS-activity in PDE-KRAS and PDE-KRAS-st in vitro models which mimics PanIN, we asked if similar effect could be achieved on PanIN development under in vivo conditions. We selected conditional KRASG12D/Pdx-cre transgenic model which exhibits the progression of PanIN stages, ranging from low-grade PanINs (1A and 1B) to high-grade PanINs (PanIN-2 and PanIN-3).13–14 We evaluated pancreatic tissues from Pdxcre and KRASG12D/Pdx-cre mice for KRAS-activity by using the KRAS-GTP-specific immunoblot assay. Consistent with expression of the KRASG12D allele, the pancreatic tissues of KRASG12D/Pdx-cre mice exhibited increased levels of KRAS-GTP levels (Figure 4B).
At 4th week post-birth, mice were genotyped for KRAS mutation. The KRASG12D/Pdx-cre mice are reported to develop PanIN or early stage PDAC between 24–28 weeks of age.14 The KRASG12D/Pdx-cre mice were divided into two groups: (1) Control group (fed with regular diet), and (ii) prevention group (fed with Lupeol-supplemented diet; 200mg/kg weight). After genotyping, we started Lupeol-feeding in mice till an age of 28 weeks (using a piggy-banking protocol). At this point, mice were sacrificed and viscera harvested for histopathological.
Histopathological analysis of pancreatic tissues showed different stages of mPanIN (mPanIN-1, mPanIN-2, and mPanIN-3) in control KRASG12D/Pdx-cre mice (Figure 4Ci–ii). Notably, Lupeol feeding in KRASG12D/Pdx-cre mice for 24 weeks showed significant (p<0.05) inhibition of mPanIN-1, mPanIN-2, and mPanIN-3 lesion development (Figure 4Cii). The histopathological analysis of the pancreatic tissue in Lupeol-fed mice showed normal area (or isolated acinar-to-ductal metaplasia). Metaplastic acinar-to-ductal metaplasia cells make luminal structures. Some metaplastic cells were noted to contain eosinophilic cytoplasmic granules (Figure 4Cii). PanIN-1A marked with a brown arrow are flat epithelial lesions composed of tall columnar cells with basally located nuclei and abundant supranuclear mucin. Most of the nuclei are small and round to oval in shape (Figure 4Cii). PanIN-2 shown with a yellow arrow as in Figure 4Cii are mucinous epithelial lesions, either flat or papillary and cytological, these lesions are having some nuclear abnormalities including loss of polarity, nuclear crowding, enlarged nuclei, pseudo-stratification and hyperchromatism. Occasionally we found PanIN-3 lesion as shown in Figure 4Cii with blue arrow. These lesions are usually papillary or micropapillary. The quantification of PanIN lesions (on histopathological basis) showed that KRASG12D/Pdx-cre mice develop mPanIN-1, (30%), mPanIN-2 (15%), and mPanIN-3 (5%) in control group (Figure 4Ciii). On the contrary, Lupeol-fed KRASG12D/Pdx-cre mice developed mPanIN1 and mPanIN2 in 10% and 5% animals, respectively (Figure 4Ciii). It is noteworthy that none of the Lupeol-fed mice exhibited mPanIN-3 lesions. Furthermore, 10% of pancreatic ducts in the control animals appeared normal, whereas 20% of pancreatic ducts were classified normal in the Lupeol-fed animals (Figure 4Ciii).
The epithelial-mesenchymal transition (EMT) is critical in the development of epithelial malignancies and accelerated by chronic inflammation in KRASG12D/Pdx-cre mice.26 The important molecular changes related to EMT are the loss of E-cadherin and gain of vimentin.26 Recent studies showed KRAS-mutation and E-Cadherin inactivation in KRAS-induced malignancies.27 We observed a marked loss of E-Cadherin and gain of vimentin protein expression in pancreatic tissues of 28-week KRASG12D/Pdx-cre mice (Figure 4D). We observed increased E-cadherin and decreased vimentin levels in the pancreatic tissues of Lupeol-fed KRASG12D/Pdx-cre mice (Figure 4D). Long-term Lupeol feeding did not cause any toxicity in mice. Control and Lupeol-fed mice showed similar weight gain as a function of age (Figure 4E).
Effect of Lupeol on the KRAS-downstream pathways in KRASG12D/Pdx-cre mice
We determined the effect of Lupeol-feeding on KRAS-GTP levels in KRASG12D/Pdx-cre mice using protein extracts of pancreatic tissues and employing the BRAF-RBD affinity pull-down assay. Lupeol-feeding was observed to reduce the KRAS-GTP protein levels in pancreatic tissues of KRASG12D/Pdx-cre mice (Figure 5Ai). We next validated the affinity-pull down data by conducting an immunoprecipitation assay of pancreatic-cell lysates by using anti-BRAF and anti-GTP-KRAS antibodies. As compared to control group, the Lupeol-fed mice showed reduced interaction of KRAS-GTP protein to BRAF protein pancreatic tissues (Figure 5Aii). These data strongly suggest that Lupeol has the potential of decreasing active KRAS (KRAS-GTP) levels under in vivo conditions.
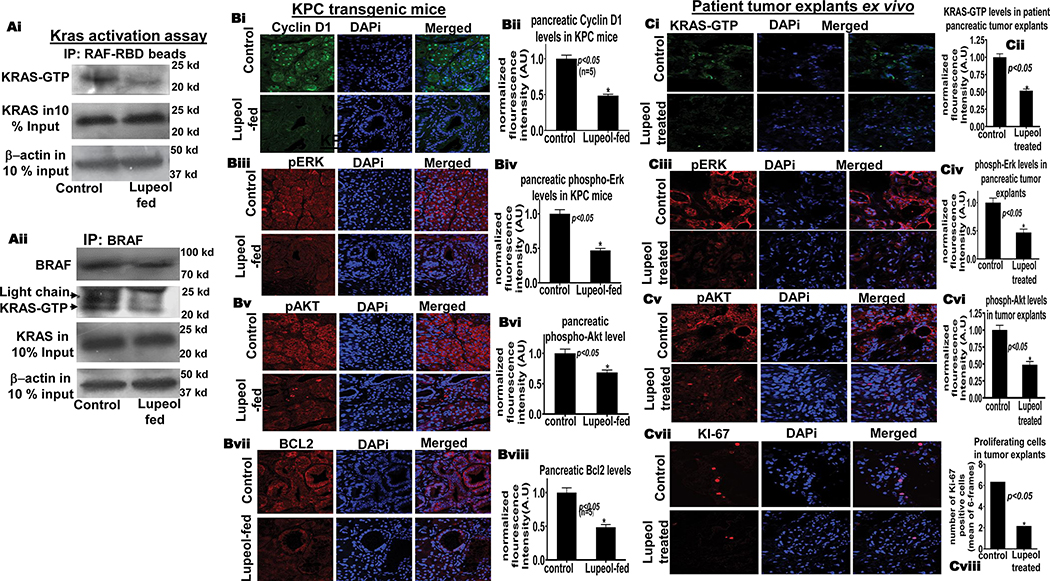
Figure 5. Lupeol reduces KRAS-GTP in KRASG12D/Pdxcre mice and patient tumor explants ex-vivo.
(Ai) Immunoblot image shows KRAS-GTP levels in pancreatic tissues KRASG12D/Pdx-cre mice as assessed of GST-Raf-RBD pull-down assay. (Aii) Immunoblot image shows KRAS-GTP levels in pancreatic tissues of control and Lupeol-fed mice by using BRAF-specific immunoprecipitation. (B) Immunofluorescence-microscopy images and respective histograms show the KRAS-GTP level and KRAS-downstream targets in control and Lupeol-fed mice. (C) Immunofluorescence-microscopy images and respective histograms show the (Ci-vi) KRAS-GTP level and KRAS-downstream targets (Cvii-viii) proliferation marker Ki67 staining in control and Lupeol-treated human pancreatic tumors (explant/PDX) ex vivo.
We next determined the inhibitory effects of Lupeol on downstream targets of KRAS signaling in pancreatic tissue of KRASG12D/Pdx-cre mice by employing the immunofluorescence-microscopy technique. The Lupeol-fed KRASG12D/Pdx-cre group exhibited reduced levels of phospho-ERK1/2, phospho-AKT, cyclin D1 and Bcl-2 proteins in pancreatic tissues than in control group (Figure 5B). Notably control mice exhibited enrichment of cyclin D1 in the nucleus, whereas Lupeol-fed mice exhibited decreased nuclear accumulation of cyclin D1. These data strengthens the notion that Lupeol is a strong KRAS-signaling inhibitor validated in vitro as well as in vivo.
Testing efficacy of Lupeol in patient-derived pancreatic explants ex vivo
After successful validation under in vitro and preclinical mouse models, our data strengthens the argument that Lupeol as a KRAS inhibitor warrants a testing in translational settings resembling close to the clinical condition. Patient tumor-derived xenografts or explants (PDX) resemble the disease very similar to the clinical condition and retain associated stromal components that play roles in the therapeutic sensitivities.28. Next, we utilized patient pancreatic tumors, cultured ex vivo (explants) to directly monitor the efficacy of Lupeol therapy. Pancreatic explants were treated with Lupeol (30 μM) for 10 days and subsequently analyzed for active KRAS protein and surrogate biomarkers of growth by using immunofluorescence-microscopy. When compared to control, the Lupeol-treated tumor-explants exhibited significantly (p<0.05) reduced KRAS-GTP protein levels (Figure 5Ci). Immunofluorescence analysis showed that tumor-explants treated with Lupeol exhibit significantly reduced p-Erk and p-AKT levels than control (Figure 5Cii–iii). As shown in pictographs and respective histogram, Lupeol treatment significantly reduced the KI-67 -positive (proliferation index) cells in tumor-explants (Figure 5Civ). To summarize, these data show that Lupeol is a strong and non-toxic KRAS-inhibitor with a translational potential to prevent and treat KRAS-driven malignancies.
Discussion
Mutations of RAS are associated with initiation, maintenance and poor prognosis of several types of cancers.29–32 The major stumbling block in the management of KRAS-driven cancers is the non-responsiveness to conventional therapies.32 This makes the development of anti-RAS therapies as a major health priority. KRAS protein functions as a molecular switch cycling between ON and OFF state to affect the intracellular signaling.30 Whereas the ON switch to the active state is promoted by guanine nucleotide exchange factors (GEFs), the OFF state is regulated by GTPase-activating proteins (GAPs).33 Oncogenic alleles of KRAS lose the capability of GAP-induced GTP hydrolysis resulting in increased GTP-bound KRAS protein levels which in turn trigger the pro-growth signaling pathways.30 According to Waters and Der et al, the adenosine triphosphate (ATP)-competitive inhibitors or small molecule GTP antagonists should provide a straightforward strategy to target mutant KRAS.31 However, with milli-molar GTP cellular concentrations and complex topography of RAS protein, competitive inhibitors of GTP faced enormous challenges and were deemed non-feasible.5, 14, 34. The alternative strategies to target KRAS pathway have been explored. These include inhibitors of (i) fernesyl transferase (FTIs) and (ii) KRAS processing enzymes, however these approaches failed in KRAS-mutant cancers.35–36. Other strategies include disruption of active state conformations of Switch-I (aa 30–38) and Switch-II (aa 59–76) regions that target (i) GEF interaction (ii) nucleotide-binding site and (iii) adjacent shallow surface pockets under the Switch II loop.15, 32, 37 Recently Janes et al showed a similar attempt for ARS-1620 compound as a KRAS activity disruptor between Switches I and II.38 Ganguly et al showed that SCH54292 and its water-soluble cognate bind to a region in between switch II and helix 4 of RAS-GDP.39 Andrographilides were shown to binding between Switch I and II of RAS protein however this chemical family binds to several proteins at different regions and is not specific to KRAS protein.40 Unfortunately several attempts to target unknown binding pockets on RAS protein resulted in dismal performance under in vitro and in vivo conditions.33 In this context this study is significant study as it investigated a series of small chemicals which showed high-affinity of binding to KRAS crustal structure and identified one terpenoid (Lupeol) that has potential to bind between Switch I and II regions of KRAS-protein. By employing different experimental platforms ranging from in situ to in vivo models, this chemical (Lupeol) was comprehensively validated as a KRAS-inhibitor. Sun et al reported that Tyr-71 is required to makes hydrogen bond interactions with Asp-54 and Arg-41 in the binding pocket of KRAS protein, in its Apo-state.41–43 We speculate that interaction of Lupeol in the binding pocket leads to conformational change of KRAS due to flipping away of Tyr-71 (thus leading to the disruption in the interaction between Tyr-71, Asp-54 and Arg-41 in Apo-KRAS). It is possible that due to disruption of Tyr-71 interactions by Lupeol, the orientation of Met-67 also flipped away.
The biggest concern for previously reported KRAS-inhibitors was their failure in physiological availability and systemic toxicity. However, this study confirms the safety, bioavailability and pharmacokinetic profile of Lupeol under physiological conditions. Previously several studies were performed to identify inhibitor GDP-GTP exchange as potential KRAS-inhibitors however majority of them were cell-specific and did not perform well as Pan-inhibitor of KRAS activity.33 This study validated the KRAS-inhibitor function of Lupeol in multiple human and murine KRAS-driven cancer models including PDAC, Colon and lung cancer suggesting utility of this small molecule as a future broad therapy for various KRAS-driven malignancies.
The concern regarding specificity of KRAS-inhibitors has proven to be detrimental in progress of such preclinical agents to clinical level.43 The high-affinity claim of Lupeol for activated-KRAS is strengthened on the basis of (i) the data from NIH-KRAS cell array (mutant KRAS or wild-type KRAS), (ii) PDE-KRAS-st cells and (iii) previous report where AsPC1 cells (mutant KRAS) responded more to Lupeol than BXPC3 (wild-type KRAS).44 This notion is supported by animal studies data showing a marked effect on KRAS-GTP levels in the pancreatic tissues of KRASG12D/Pdx-cre mice by long-term Lupeol-consumption. The translational significance of data from KRASG12D/Pdx-cre mice is strengthened by similar findings in patient tumor explants. Because Lupeol did not exhibit toxicity under in vivo conditions, therefore we foresee this inhibitor and its analogues as a safer future therapy for treating KRAS-driven cancers in humans.
Supplementary Material
Acknowledgment
This study was supported by an investigator award from American Institute of Cancer Research (AICR application#09A074) to author (MS). The author (MS) is supported by the US PHS grant (CA193739). We thank Neelofar Jan for providing help in animal studies. We thank the National Institute of Health-supported RAS-reagents group (Dr. Dominic Esposito, Frederick National Laboratory for Cancer Research & Leidos Biomedical Research, Inc.) for providing the KRAS-active MEF panel of cells.
Footnotes
Conflict of Interest statement:
Authors declare no conflict of interest in the current study. The manuscript has been approved by all authors. The funding statement has been provided under the acknowledgment section.
Data Sharing Statement:
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017;170(1):17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smit VT, Boot AJ, Smits AM, Fleuren GJ, Cornelisse CJ, Bos JL. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res. 1988;16(16):7773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCormick F Progress in targeting RAS with small molecule drugs. Biochem J. 2019;476(2):365–74. [DOI] [PubMed] [Google Scholar]
- 4.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111(5):817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jancik S, Drabek J, Radzioch D, Hajduch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;2010:150960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novotny CJ, Hamilton GL, McCormick F, Shokat KM. Farnesyltransferase-Mediated Delivery of a Covalent Inhibitor Overcomes Alternative Prenylation to Mislocalize K-Ras. ACS Chem Biol. 2017;12(7):1956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cragg GM, Newman DJ. Plants as a source of anti-cancer agents. J Ethnopharmacol. 2005;100(1–2):72–9. [DOI] [PubMed] [Google Scholar]
- 8.Saleem M, Kaur S, Kweon MH, Adhami VM, Afaq F, Mukhtar H. Lupeol, a fruit and vegetable based triterpene, induces apoptotic death of human pancreatic adenocarcinoma cells via inhibition of Ras signaling pathway. Carcinogenesis. 2005;26(11):1956–64. [DOI] [PubMed] [Google Scholar]
- 9.Sheikh IA. Stereoselectivity and the potential endocrine disrupting activity of di-(2-ethylhexyl)phthalate (DEHP) against human progesterone receptor: a computational perspective. J Appl Toxicol. 2016;36(5):741–7. [DOI] [PubMed] [Google Scholar]
- 10.Vivoli M, Novak HR, Littlechild JA, Harmer NJ. Determination of protein-ligand interactions using differential scanning fluorimetry. J Vis Exp. 2014(91):51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddique HR, Liao DJ, Mishra SK, Schuster T, Wang L, Matter B, et al. Epicatechin-rich cocoa polyphenol inhibits Kras-activated pancreatic ductal carcinoma cell growth in vitro and in a mouse model. Int J Cancer. 2012;131(7):1720–31. [DOI] [PubMed] [Google Scholar]
- 12.Ganaie AA, Beigh FH, Astone M, Ferrari MG, Maqbool R, Umbreen S, et al. BMI1 Drives Metastasis of Prostate Cancer in Caucasian and African-American Men and Is A Potential Therapeutic Target: Hypothesis Tested in Race-specific Models. Clin Cancer Res. 2018;24(24):6421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006; 103(15):5947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003. December;4(6):437–50. [DOI] [PubMed] [Google Scholar]
- 15.Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, et al. Multivalent Small-Molecule Pan-RAS Inhibitors. Cell. 2017;168(5):878–89 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Harrison RA, Li L, Zeng M, Gondi S, Scott D, et al. KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry. Structure. 2017;25(9):1442–8 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bournet B, Muscari F, Buscail C, Assenat E, Barthet M, Hammel P, et al. KRAS G12D Mutation Subtype Is A Prognostic Factor for Advanced Pancreatic Adenocarcinoma. Clin Transl Gastroenterol. 2016;7:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1(1):2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–83. [DOI] [PubMed] [Google Scholar]
- 20.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129(7):1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer. 2011;2(3):261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2(8):685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandt R, Sell T, Lüthen M, Uhlitz F, Klinger B, Riemer P, Giesecke-Thiel C, Schulze S, El-Shimy IA, Kunkel D, Fauler B, Mielke T, Mages N, Herrmann BG, Sers C, Blüthgen N, Morkel M. Cell type-dependent differential activation of ERK by oncogenic KRAS in colon cancer and intestinal epithelium. Nat Commun. 2019. July 2;10(1):2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Huang S, Sun YL. Epithelial-Mesenchymal Transition in Pancreatic Cancer: A Review. Biomed Res Int. 2017;2017:2646148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang B RAS signaling and anti-RAS therapy: lessons learned from genetically engineered mouse models, human cancer cells, and patient-related studies. Acta Biochim Biophys Sin (Shanghai). 2016;48(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra S, Moro CF, Del Chiaro M, Pouso S, Sebestyen A, Lohr M, et al. Ex vivo organotypic culture system of precision-cut slices of human pancreatic ductal adenocarcinoma. Sci Rep. 2019;9(1):2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu W, Sahin F, Iacobuzio-Donahue CA, Garcia-Carracedo D, Wang WM, Kuo CY, et al. Disruption of p16 and activation of Kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget. 2011;2(11):862–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39(2):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters AM, Der CJ. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med. 2018;8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickerson S, Joy ST, Arora PS, Bar-Sagi D. An orthosteric inhibitor of the RAS-SOS interaction. Enzymes. 2013;34 Pt. B:25–39. [DOI] [PubMed] [Google Scholar]
- 34.Burns MC, Sun Q, Daniels RN, Camper D, Kennedy JP, Phan J, et al. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc Natl Acad Sci U S A. 2014;111(9):3401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, et al. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Natl Acad Sci U S A. 2010;107(14):6471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H. Small-molecule modulation of Ras signaling. Nat Chem Biol. 2014;10(8):613–22. [DOI] [PubMed] [Google Scholar]
- 38.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 2018;172(3):578–89 e17. [DOI] [PubMed] [Google Scholar]
- 39.Ganguly AK, Pramanik BN, Huang EC, Liberles S, Heimark L, Liu YH, et al. Detection and structural characterization of ras oncoprotein-inhibitors complexes by electrospray mass spectrometry. Bioorg Med Chem. 1997;5(5):817–20. [DOI] [PubMed] [Google Scholar]
- 40.Hocker HJ, Cho KJ, Chen CY, Rambahal N, Sagineedu SR, Shaari K, et al. Andrographolide derivatives inhibit guanine nucleotide exchange and abrogate oncogenic Ras function. Proc Natl Acad Sci U S A. 2013;110(25):10201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter JJ, Anderson M, Blades K, Brassington C, Breeze AL, Chresta C, et al. Small molecule binding sites on the Ras:SOS complex can be exploited for inhibition of Ras activation. J Med Chem. 2015;58(5):2265–74. [DOI] [PubMed] [Google Scholar]
- 42.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012;109(14):5299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51(25):6140–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saleem M. Lupeol, a novel anti-inflammatory and anti-cancer dietary triterpene. Cancer Lett. 2009;285(2):109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.