Abstract
Structural variants (SVs) rearrange large segments of DNA1 and can have profound consequences in evolution and human disease2,3. As national biobanks, disease-association studies, and clinical genetic testing have grown increasingly reliant on genome sequencing, population references such as the Genome Aggregation Database (gnomAD)4 have become integral in the interpretation of single-nucleotide variants (SNVs)5. However, there are no reference maps of SVs from high-coverage genome sequencing comparable to those for SNVs. Here we present a reference of sequence-resolved SVs constructed from 14,891 genomes across diverse global populations (54% non-European) in gnomAD. We discovered a rich and complex landscape of 433,371 SVs, from which we estimate that SVs are responsible for 25–29% of all rare protein-truncating events per genome. We found strong correlations between natural selection against damaging SNVs and rare SVs that disrupt or duplicate protein-coding sequence, which suggests that genes that are highly intolerant to loss-of-function are also sensitive to increased dosage6. We also uncovered modest selection against noncoding SVs in cis-regulatory elements, although selection against protein-truncating SVs was stronger than all noncoding effects. Finally, we identified very large (over one megabase), rare SVs in 3.9% of samples, and estimate that 0.13% of individuals may carry an SV that meets the existing criteria for clinically important incidental findings7. This SV resource is freely distributed via the gnomAD browser8 and will have broad utility in population genetics, disease-association studies, and diagnostic screening.
Subject terms: Genome informatics, Chromosome abnormality, Structural variation, Genomics, Mutation
A large empirical assessment of sequence-resolved structural variants from 14,891 genomes across diverse global populations in the Genome Aggregation Database (gnomAD) provides a reference map for disease-association studies, population genetics, and diagnostic screening.
Main
SVs are DNA rearrangements that involve at least 50 nucleotides1. By virtue of their size and abundance, SVs represent an important mutational force that shape genome evolution and function2,3, and contribute to germline and somatic diseases9–11. The profound effect of SVs is also attributable to the numerous mechanisms by which they can disrupt protein-coding genes and cis-regulatory architecture12. SVs can be grouped into mutational classes that include ‘unbalanced’ gains or losses of DNA (for example, copy-number variants, CNVs), and ‘balanced’ rearrangements that occur without corresponding dosage alterations (such as inversions and translocations)1 (Fig. 1a). Other common forms of SVs include mobile elements that insert themselves throughout the genome, and multiallelic CNVs (MCNVs) that can exist at high copy numbers1. More recently, exotic species of complex SVs have been discovered that involve two or more distinct SV signatures in a single mutational event interleaved on the same allele, and can range from CNV-flanked inversions to rare instances of localized chromosome shattering, such as chromothripsis13,14. The diversity of SVs in humans is therefore far greater than has been widely appreciated, as is their influence on genome structure and function.
Fig. 1. Properties of SVs across human populations.
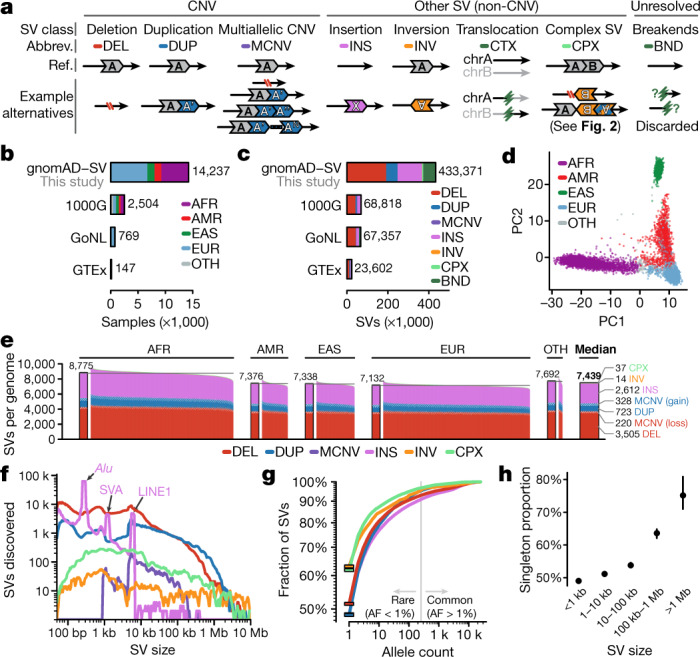
a, SV classes catalogued in this study. We also documented unresolved non-reference ‘breakends’ (BNDs), but they were excluded from all analyses as low-quality variants. b, After quality control, we analysed 14,237 samples across continental populations, including African/African American (AFR), Latino (AMR), East Asian (EAS), and European (EUR), or other populations (OTH). Three publicly available WGS-based SV datasets are provided for comparison (1000 Genomes Project (1000G), approximately 7× coverage; Genome of the Netherlands Project (GoNL), around 13× coverage; Genotype-Tissue Expression Project (GTEx), approximately 50× coverage)1,16,17. c, We discovered 433,371 SVs, and provide counts from previous studies for comparison1,16,17. d, A principal component (PC) analysis of genotypes for 15,395 common SVs separated samples along axes corresponding to genetic ancestry. e, The median genome contained 7,439 SVs. f, Most SVs were small. Expected Alu, SVA and LINE1 mobile element insertion peaks are marked at approximately 300 bp, 2.1 kb and 6 kb, respectively. g, Most SVs were rare (allele frequency (AF) < 1%), and 49.8% of SVs were singletons (solid bars). h, Allele frequencies were inversely correlated with SV size across all 335,470 resolved SVs in unrelated individuals. Values are mean and 95% confidence interval from 100-fold bootstrapping. Colour codes are consistent between a, c, e–h, and between b and d.
Although SVs alter more nucleotides per genome than SNVs and short insertion/deletion variants (indels; <50 bp)1, surprisingly little is known about their mutational spectra on a global scale. The largest published population study of SVs using whole-genome sequencing (WGS) remains the 1000 Genomes Project (n = 2,504; 7× sequence coverage)1, and the substantial technical challenges of SV discovery from WGS15 has led to non-uniform SV analyses across contemporary studies16–20. Moreover, short-read WGS is unable to capture a subset of SVs accessible to more expensive niche technologies, such as long-read WGS21. Owing to the combination of these challenges, SV references are dwarfed by contemporary resources for short variants, such as the Exome Aggregation Consortium (ExAC) and its successor, the Genome Aggregation Database (gnomAD), which have jointly analysed more than 140,000 individuals4,6. Publicly available resources such as ExAC and gnomAD have transformed many aspects of human genetics, including defining sets of genes constrained against damaging coding mutations6 and providing frequency filters for variant interpretation5. As short-read WGS is rapidly becoming the predominant technology in large-scale human disease studies, and will probably displace conventional methods for diagnostic screening, there is a mounting need for comparable references of SVs across global populations.
In this study, we developed gnomAD-SV, a sequence-resolved reference for SVs from 14,891 genomes. Our analyses revealed diverse mutational patterns among SVs, and principles of selection acting against reciprocal dosage changes in genes and noncoding cis-regulatory elements. From these analyses, we determined that SVs represent more than 25% of all rare protein-truncating events per genome, emphasizing the unrealized potential of routine SV detection in WGS studies. This SV reference has been integrated into the gnomAD browser (http://gnomad.broadinstitute.org) with no restrictions on reuse so that it can be mined for new insights into genome biology and applied as a resource to interpret SVs in diagnostic screening.
SV discovery and genotyping
We analysed WGS data for 14,891 samples (average coverage of 32×) aggregated from large-scale sequencing projects, of which 14,237 (95.6%) passed all quality thresholds, representing a general adult population depleted for severe Mendelian diseases (median age of 49 years) (Supplementary Table 1, Supplementary Figs. 1, 2). This cohort included 46.1% European, 34.9% African or African American, 9.2% East Asian, and 8.7% Latino samples, as well as 1.2% samples from admixed or other populations (Fig. 1). Following family-based analyses using 970 parent–child trios for quality assessments, we pruned all first-degree relatives from the cohort, retaining 12,653 unrelated genomes for subsequent analyses.
We discovered and genotyped SVs using a cloud-based, multi-algorithm pipeline for short-read WGS (Supplementary Fig. 3), which we prototyped in a study of 519 autism quartet families20. This pipeline integrated four orthogonal evidence types to capture SVs across the size and allele frequency spectra, including six classes of canonical SVs (Fig. 1a) and 11 subclasses of complex SVs22 (Fig. 2). We augmented this pipeline with new methods to account for the technical heterogeneity of aggregated datasets (Extended Data Fig. 1, Supplementary Figs. 4, 5), and discovered 433,371 SVs (Fig. 1c). After excluding low-quality SVs, which were predominantly (61.6%) composed of incompletely resolved breakpoint junctions (that is, ‘breakends’) that lack interpretable alternative allele structures for functional annotation and produce high false-discovery rates20 (Extended Data Fig. 2a), we retained 335,470 high-quality SVs for subsequent analyses (Supplementary Table 3). This final set of high-quality SVs corresponded to a median of 7,439 SVs per genome, or more than twice the number of variants per genome captured by previous WGS-based SV studies such as the 1000 Genomes Project (3,441 SVs per genome from approximately 7× coverage WGS), which underscores the benefits of high-coverage WGS and improved multi-algorithm ensemble methods for SV discovery.
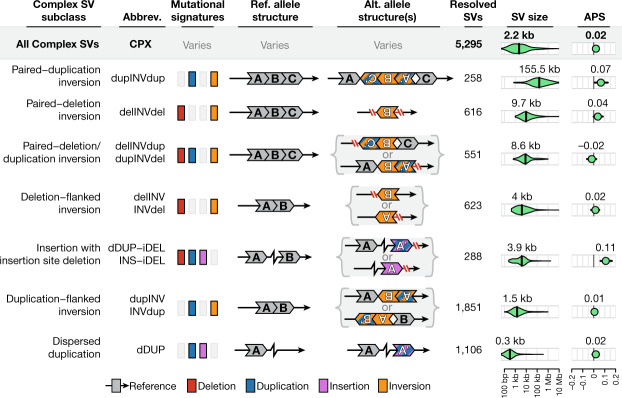
Fig. 2. Complex SVs are abundant in the human genome.
We resolved 5,295 complex SVs across 11 mutational subclasses, 73.7% of which involved at least one inversion. Each subclass is detailed here, including their mutational signatures, structures, abundance, density of SV sizes (vertical line indicates median size), and allele frequencies. Five pairs of subclasses have been collapsed into single rows due to mirrored or similar alternative allele structures (for example, delINV versus INVdel). Two complex SVs did not conform to any subclass (Extended Data Fig. 8).
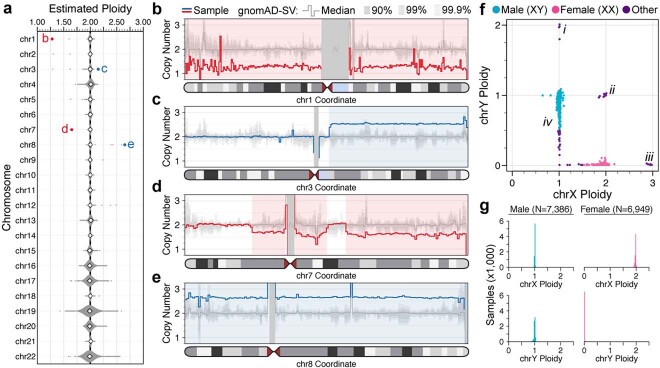
Extended Data Fig. 1. Detection of chromosome-scale dosage alterations.
We estimated ploidy (that is, whole-chromosome copy number) for all 24 chromosomes per sample. a, Distribution of autosome ploidy estimates across 14,378 samples passing initial data quality thresholds. White diamonds indicate medians. Individual points are outlier samples at least three standard deviations away from the cohort-wide mean. The outlier points marked in red and blue correspond to the samples highlighted in b–e. b–e, Samples with outlier autosome ploidy estimates typically contained somatic or mosaic chromosomal abnormalities, such as somatic aneuploidy of chromosome 1 (chr1) (b) or chromosome 8 (e), or large focal somatic or mosaic CNVs on chromosome 3 (c) and chromosome 7 (d). Each panel depicts copy-number estimates in 1-Mb bins for each rearranged sample in red or blue. Dark, medium and light-grey background shading indicates the range of copy number estimates for 90%, 99% and 99.9% of all gnomAD-SV samples, respectively, and the medium grey line indicates the median copy number estimate across all samples. Regions of unalignable N-masked bases >1 Mb in the reference genome are masked with grey rectangles. f, Sex chromosome ploidy estimates for all samples from a. We inferred karyotypic sex by clustering samples to their nearest integer ploidy for sex chromosomes. Several abnormal sex chromosome ploidies are marked, including XYY (i), XXY (ii), XXX (iii), and mosaic loss-of-Y (iv). g, Histogram representation of the data from f. Essentially all samples conformed to canonical sex chromosome ploidies.
Extended Data Fig. 2. Benchmarking the technical qualities of the gnomAD-SV callset.
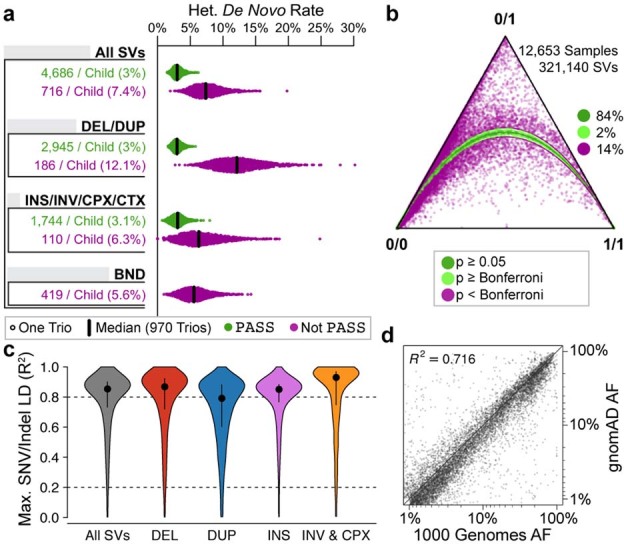
We evaluated the quality of gnomAD-SV with seven orthogonal analyses detailed in Supplementary Table 4, Supplementary Figs. 6–9 and Supplementary Note 1. Four core analyses are presented here. a, Apparent rates of de novo (that is, spontaneous) heterozygous SVs per child across 970 parent–child trios. Each point is a single trio, and vertical lines denote medians. Given the expected mutation rate of SVs accessible to short-read WGS1,20 (<1 true de novo SV per trio; see also Fig. 3a), effectively all de novo SVs represented a combination of false-positive genotypes in children and/or false-negative genotypes in parents. SVs passing all filters and included in the final gnomAD-SV callset (‘pass’) are shown in green. For comparison, variants that did not pass post hoc site-level filters (‘not pass’) are also shown in purple. b, Hardy–Weinberg equilibrium (HWE) metrics for all biallelic SVs localized to autosomes. Deviation from HWE was assessed using a chi-square goodness-of-fit test with one degree of freedom. Vertex labels reflect genotypes: 0/0 denotes homozygous reference; 0/1 denotes heterozygous; and 1/1 denotes homozygous alternate, with all sites shaded by chi-squared P value. c, Linkage disequilibrium between SVs and SNVs or indels for 23,706 common (allele frequency > 1%) SVs represented as cross-population maximum R2 values after excluding repetitive and low-complexity regions (see Supplementary Fig. 7). Points and vertical bars represent medians and interquartile ranges, respectively. d, Correlation of allele frequency (AF) for 37,907 common SVs captured by both the 1000 Genomes Project and gnomAD-SV1. Pearson’s correlation coefficient (R2) is provided.
Given that there are no gold-standard benchmarking procedures for SVs from WGS, we evaluated the technical qualities of gnomAD-SV using seven orthogonal approaches. These analyses are described in detail in Extended Data Figs. 2, 3, Supplementary Figs. 6–12, Supplementary Table 4 and Supplementary Note 1, but we highlight just a few here to demonstrate that gnomAD-SV conforms to many fundamental principles of population genetics, including Mendelian segregation, genotype distributions, and linkage disequilibrium. We found that the precision of gnomAD-SV was comparable to our previous study of 519 autism quartets that attained a 97% molecular validation rate for all de novo SV predictions20: in gnomAD, analyses of 970 parent–child trios indicated a median Mendelian violation rate of 3.8% and a heterozygous de novo rate of 3.0%. We also observed that 86% of SVs were in Hardy–Weinberg equilibrium, and common SVs were in strong linkage disequilibrium with nearby SNVs or indels (median peak R2 = 0.85). We performed extensive in silico confirmation of 19,316 SVs predicted from short-read WGS using matched long-read WGS from four samples21,23, finding a 94.0% confirmation rate with breakpoint-level read evidence, and revealing that 59.8% of breakpoint coordinates were accurate within a single nucleotide of the long-read data. These and other benchmarking approaches suggested that gnomAD-SV was sufficiently sensitive and specific to be used as a reference dataset for most applications in human genomics.
Extended Data Fig. 3. In silico confirmation of SVs in gnomAD-SV with long-read WGS.
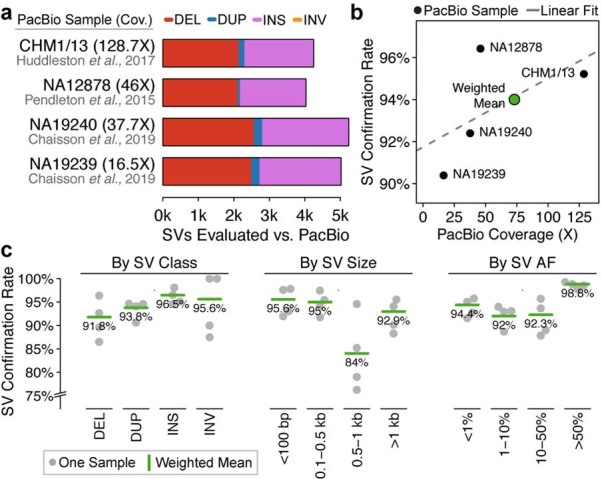
We used Pacific Biosciences (PacBio) long-read WGS data available for four samples in this study to perform in silico confirmation to estimate the positive predictive value and breakpoint accuracy for SVs in gnomAD-SV21,45,46 (Supplementary Fig. 10). a, Counts of SVs evaluated per sample in this analysis. SVs were restricted to those with breakpoint-level read support (that is, ‘split-read’ evidence, 92.8% of all SVs) and did not have breakpoints localized to annotated simple repeats or segmental duplications. b, An iterative local long-read WGS realignment algorithm, VaPoR47, was used to perform in silico confirmation of SVs predicted from short-read WGS in gnomAD-SV. As noted by the VaPoR developers47, the performance of this approach was sensitive to the sequencing depth of long-read WGS data. Therefore, the weighted mean of the four samples was used as a study-wide long-read WGS confirmation rate, weighting the confirmation rate of each sample based on the square root of its long-read WGS sequencing depth. c, Confirmation rates stratified by SV class, size and allele frequency. A mean of 4,829 SVs per sample were assessed. Horizontal green bars denote weighted means.
Population genetics and genome biology
The distribution of SVs across samples matched expectations based on human demographic history, with the top three components of genetic variance separating continental populations (Fig. 1d, Supplementary Fig. 13). African and African American samples exhibited the greatest genetic diversity and their common SVs were in weaker linkage disequilibrium with nearby short variants than Europeans, whereas East Asians featured the highest levels of homozygosity (Fig. 1e, Extended Data Fig. 4a–d, Supplementary Fig. 7). The mutational diversity of gnomAD-SV was extensive: we completely resolved 5,295 complex SVs across 11 mutational subclasses, of which 3,901 (73.7%) involved inverted segments (Fig. 2), confirming that inversion variation is predominantly composed of complex SVs rather than canonical inversions1,24. Across all SV classes, most SVs were small (median size of 331 bp) and rare (allele frequency < 1%; 92% of SVs), with half of all SVs (49.8%) appearing as ‘singletons’ (that is, only one allele observed across all samples) (Fig. 1f, g). Although the proportion of singletons varied by SV class, it was strongly dependent on SV size across all classes, which suggests that the amount of DNA rearranged is a key determinant of selection against most SVs (Fig. 1h, Extended Data Fig. 5a).
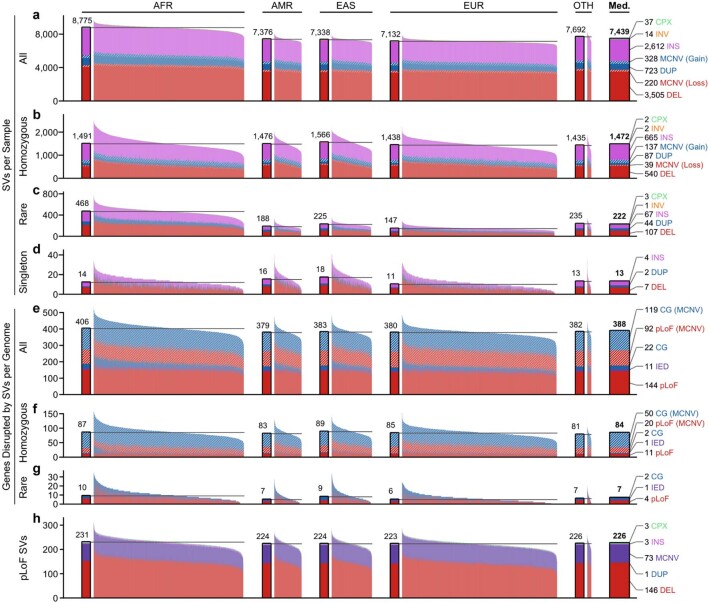
Extended Data Fig. 4. SVs contribute a substantial burden of rare, homozygous, and coding mutations per genome.
a–d, Counts of SVs per genome across a variety of parameters, corresponding to median counts of total SVs (a), homozygous SVs (b), rare SVs (c) and singleton SVs (d). Samples are grouped by population and coloured by SV types. The solid bar to the left of each population indicates the population median. e–g, Median counts of genes disrupted by SVs per genome when considering all SVs (including MCNVs) (e), homozygous SVs (including MCNVs) (f), and rare SVs (g). Colours correspond to predicted functional consequence. h, Counts of pLoF SVs per genome. For certain categories, such as genes disrupted by rare SVs per genome, a subset of samples (<5%) were enriched above the population average, as expected for individuals carrying large, rare CNVs predicted to cause the disruption of dozens or hundreds of genes (see Extended Data Fig. 1); for the purposes of visualization, the y axis for all panels has been restricted to a maximum of three interquartile ranges above the third quartile across all samples for each category.
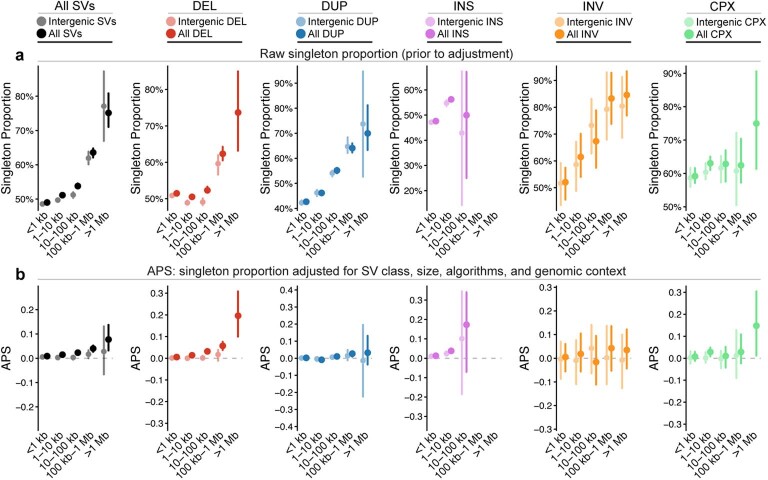
Extended Data Fig. 5. Rearrangement size is a primary determinant of allele frequency for most classes of SVs.
a, Proportion of singleton SVs in five SV size bins for each class of biallelic SVs considered in this study. Intergenic SVs (light colours; n = 206,954) exhibited reduced singleton proportions when compared to all SVs (dark colours; n = 335,470) of the same size and class. Bars reflect 95% confidence intervals from 100-fold bootstrapping. Categories with fewer than ten SVs are not shown. b, To account for the strong dependency of singleton proportion on SV size and class, we developed the APS metric, which normalizes singleton proportions using SV-specific technical and genomic covariates to permit comparisons of the frequency spectra across SV classes (see Supplementary Fig. 14). The same data as in a are shown, transformed onto the APS scale, which shows effectively no dependency on SV size for intergenic SVs. Bars reflect 95% confidence intervals from 100-fold bootstrapping. Residual deviation from APS = 0 is maintained when considering all SVs, owing to APS being intentionally calibrated to intergenic SVs as a proxy for neutral variation. Because larger SVs are more likely to be gene-disruptive, they upwardly bias the APS point estimates due to residual negative selection not captured by SV size alone. Counts of SVs per category for both a and b are listed in Supplementary Table 9.
Mutation rate estimates for SVs have remained elusive owing to limited sample sizes, poor resolution of conventional technologies, technical challenges of SV discovery, and use of cell line-derived DNA in population studies1,25. Here, we used the Watterson estimator26 to project a mean mutation rate of 0.29 de novo SVs (95% confidence interval 0.13–0.44) per generation in regions of the genome accessible to short-read WGS, or roughly one new SV every 2–8 live births, with mutation rates varying markedly by SV class (Fig. 3a). Although this imperfect method extrapolates from data pooled across unrelated individuals, we previously demonstrated comparable rates from molecularly validated observations in 519 quartet families20. Like mutation rates, the distribution of SVs throughout the genome was non-uniform, significantly correlated with repetitive sequence contexts, and was enriched near centromeres and telomeres23 (Supplementary Fig. 16). These trends were dependent on SV class, as biallelic deletions and duplications were predominantly enriched at telomeres, whereas MCNVs were enriched in centromeric segmental duplications (Fig. 3b–d). Given the reduced sensitivity of short-read WGS in repetitive sequences, this study certainly underestimates the true SV mutation rates; nevertheless, these analyses implicate several aspects of chromosomal context and SV class in determining SV mutation rates throughout the genome.
Fig. 3. Genome-wide mutational patterns of SVs.
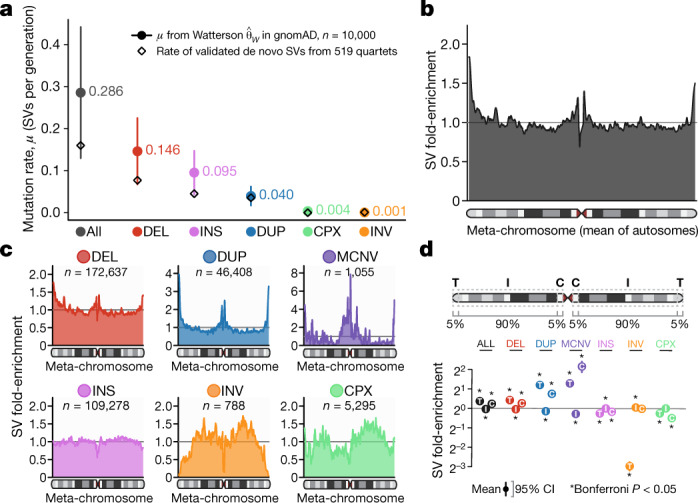
a, Mutation rates (μ) from the Watterson estimator for each SV class26. Bars represent 95% confidence intervals. Rates of molecularly validated de novo SVs from 519 quartet families are provided for comparison20. b, Smoothed enrichment of SVs per 100-kb window across the average of all autosomes normalized by chromosome arm length (a ‘meta-chromosome’) (Supplementary Fig. 16). c, The distribution of SVs along the meta-chromosome was dependent on variant class. d, SV enrichment by class and chromosomal position provided as mean and 95% confidence intervals (CI). C, centromeric; I, interstitial; T, telomeric. P values were computed using a two-sided t-test and were Bonferroni-adjusted for 21 comparisons. *P ≤ 2.38 × 10−3.
Dosage sensitivity of coding and noncoding loci
Owing to their size and mutational diversity, SVs can have varied consequences on protein-coding genes12 (Fig. 4a, Supplementary Fig. 17). In principle, any SV can result in predicted loss-of-function (pLoF), either by deleting coding nucleotides or altering open-reading frames. Coding duplications can result in copy-gain of entire genes, or of a subset of exons within a gene (referred to here as intragenic exonic duplication, or IED). The average genome in gnomAD-SV contained a mean of 179.8 genes altered by biallelic SVs (144.3 pLoF, 24.3 copy-gain, and 11.2 IED), of which 11.6 were predicted to be completely inactivated by homozygous pLoF (Fig. 4b, Extended Data Fig. 4e–h). When restricted to rare (allele frequency < 1%) SVs, we observed a mean of 10.2 altered genes per genome (5.5 pLoF, 3.4 copy-gain, and 1.3 IED). By comparison, a companion gnomAD paper estimated 122.4 pLoF short variants per genome, of which 16.3 were rare4. These analyses suggest that 29.4% of rare heterozygous gene inactivation events per individual are contributed by SVs, or conservatively 25.2% of pLoF events if we exclude IEDs given the context-dependence of their functional impact.
Fig. 4. Pervasive selection against SVs in genes mirrors coding short variants.
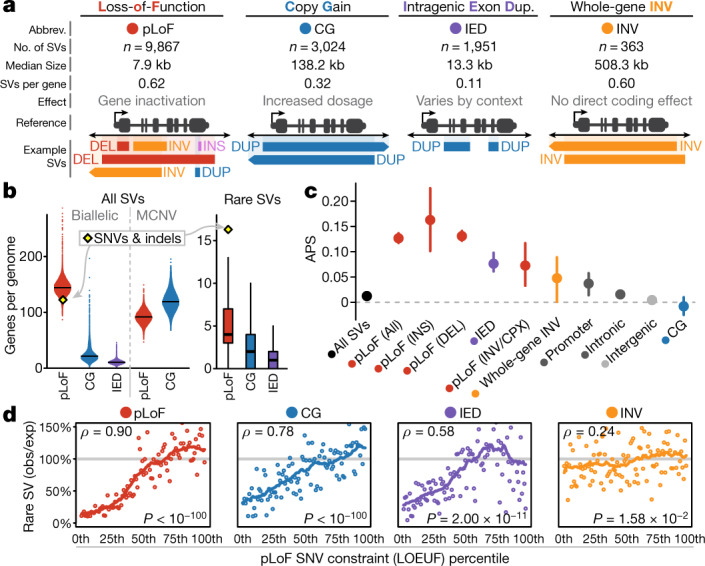
a, Four categories of gene-overlapping SVs, with counts of total SVs, median SV size, and mean SVs per gene in gnomAD-SV. b, Count of genes altered by SVs per genome. Horizontal lines indicate medians. Sample sizes per category listed in Supplementary Table 9. c, APS value for SVs overlapping genes. Bars indicate 100-fold bootstrapped 95% confidence intervals. SVs per category listed in Supplementary Table 9. d, Relationships of constraint against pLoF SNVs versus gene-overlapping SVs in 100 bins of around 175 genes each, ranked by SNV constraint4. Correlations were assessed with a two-sided Spearman correlation test. Solid lines represent 21-point rolling means. See Supplementary Fig. 19 for comparisons to missense constraint.
A fundamental question in human genetics is the degree to which natural selection acts on coding and noncoding loci. The proportion of singleton variants has been established as a proxy for strength of selection6; however, this metric is confounded for SVs given the strong correlation between allele frequency and SV size, among other factors. Therefore, we developed a new metric, adjusted proportion of singletons (APS), to account for SV class, size, genomic context, and other technical covariates (Extended Data Fig. 5, Supplementary Fig. 14). Under this normalized APS metric, a value of zero corresponds to a singleton proportion comparable to intergenic SVs, whereas values greater than zero reflect purifying selection, similar to the ‘mutability-adjusted proportion of singletons’ (MAPS) metric used for SNVs6. Applying this APS model revealed signals of pervasive selection against nearly all classes of SVs that overlap genes, including intronic SVs, whole-gene inversions, SVs in gene promoters, and deletions as small as a single exon (Fig. 4c, Extended Data Fig. 6, Supplementary Fig. 18). The one notable exception was copy-gain duplications, which showed no clear evidence of selection beyond what could already be explained by their sizes, which were vastly larger than non-copy-gain duplications (median copy-gain duplication size = 134.8 kb; median non-copy-gain duplication size = 2.7 kb; one-tailed Wilcoxon test, W = 1.18 × 108, P < 10−100). This result could have numerous explanations, but it is consistent with the known diverse evolutionary roles of gene duplication events, including positive selection reported in humans27,28.
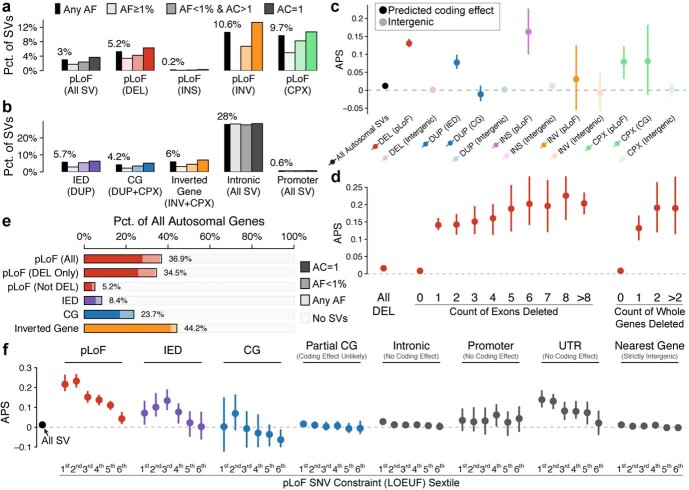
Extended Data Fig. 6. Most SVs within genes appear under negative selection.
a, Enrichments for pLoF consequences among rare and singleton SVs across SV classes. b, Enrichments for non-pLoF functional consequences among rare and singleton SVs across SV classes. c, Adjusted proportion of singletons across SV types and functional consequences. d, APS among deletions relative to count of exons and whole genes deleted. e, Fractions of all autosomal protein-coding genes with at least one SV across a variety of functional consequences. f, Relationship of APS and constraint against pLoF SNVs4. For this analysis, intronic, promoter and UTR SVs were required to have precise breakpoints (that is, have ‘split-read’ support) to protect against any cryptic overlap with coding sequence unable to be annotated due to imprecise breakpoints. For c, d and f, points and vertical bars represent 95% confidence intervals from 100-fold bootstrapping, respectively. Counts of SVs per category in c and d are provided in Supplementary Table 9. For d and f, deletions in highly repetitive or low-complexity sequence (≥30% coverage by annotated segmental duplications or simple repeats) were excluded.
Methods that quantify evolutionary constraint on a per-gene basis, such as the probability of intolerance to heterozygous pLoF variation (pLI)6 and the pLoF observed/expected upper fraction (LOEUF)4, have become core resources in human genetics. Nearly all existing metrics, including pLI and LOEUF, are derived from SNVs. Although previous studies have attempted to compute similar scores using large CNVs detected by microarray and exome sequencing29,30, or to correlate deletions with pLI18, no gene-level metrics comparable to LOEUF exist for SVs at WGS resolution. To gain insight into this problem, we built a model to estimate the depletion of rare SVs per gene compared to expectations based on gene length, genomic context, and the structure of exons and introns. This model is imperfect, as current sample sizes are too sparse to derive precise gene-level metrics of constraint from SVs. Nevertheless, we found strong concordance between the depletion of rare pLoF SVs and existing pLoF and missense SNV constraint metrics4 (pLoF Spearman correlation test, ρ = 0.90, P < 10−100) (Fig. 4d, Supplementary Fig. 19). Notably, a comparable positive correlation was also observed for copy-gain SVs and SNV constraint (pLoF Spearman correlation test, ρ = 0.78, P < 10−100), whereas a weaker yet significant correlation was detected for IEDs (pLoF Spearman correlation test, ρ = 0.58, P = 2.0 × 10−11). As orthogonal support for these trends, we identified an inverse correlation between APS and SNV constraint across all functional categories of SVs, which was consistent with our observed depletion of rare, functional SVs in constrained genes (Extended Data Fig. 6f). These comparisons confirm that selection against most classes of gene-altering SVs mirrors patterns observed for short variants18,30. They further suggest that SNV-derived constraint metrics such as LOEUF capture a general correspondence between haploinsufficiency and triplosensitivity for a large fraction of genes in the genome. It therefore appears that the most highly pLoF-constrained genes not only are sensitive to pLoF, but also are more likely to be intolerant to increased dosage and other functional alterations.
In contrast to the well-studied effects of coding variation, the effects of noncoding SVs on regulatory elements are largely unknown. There are a handful of examples of SVs with strong noncoding effects, although they are scarce in humans and model organisms31,32. In gnomAD-SV, we explored noncoding dosage sensitivity across 14 regulatory element classes, ranging from high-confidence experimentally validated enhancers to large databases of computationally predicted elements (Supplementary Table 5). We found that noncoding CNVs overlapping most element classes had increased proportions of singletons, although none exceeded the APS observed for pLoF SVs (Fig. 5a). In general, the effects of noncoding deletions appeared stronger than noncoding duplications, and CNVs predicted to delete or duplicate entire elements were under stronger selection than partial element disruption (Fig. 5b). We also observed that primary sequence conservation was correlated with selection against noncoding CNVs (Fig. 5c, d), which provides a foothold for future work on interpretation and functional effect prediction for noncoding SVs. Broadly, these results followed trends we observed for protein-coding SVs, which we interpreted as evidence for weak but widespread selection against CNVs altering most classes of annotated regulatory elements.
Fig. 5. Dosage sensitivity in the noncoding genome.
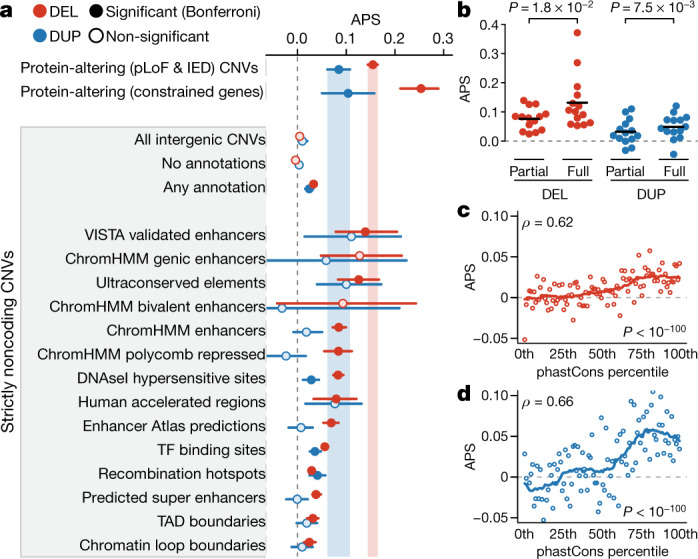
a, Strength of selection (APS) for noncoding CNVs overlapping 14 categories of noncoding elements (Supplementary Table 5). Bars reflect 95% confidence intervals from 100-fold bootstrapping. Each category was compared to neutral variation (APS = 0) using a one-tailed t-test. Categories surpassing Bonferroni-corrected significance for 32 comparisons are indicated with dark shaded points. SVs per category listed in Supplementary Table 9. DEL, deletion; DUP, duplication; TAD, topologically associating domain; TF, transcription factor. b, CNVs that completely covered elements (‘full’) had significantly higher average APS values than CNVs that only partially covered elements (‘partial’). P values calculated using a two-tailed paired two-sample t-test for the 14 categories from a. c, d, Spearman correlations between sequence conservation and APS for noncoding deletions (n = 143,353) (c) and duplications (n = 30,052) (d). Noncoding CNVs were sorted into 100-percentile bins based on the sum of the phastCons scores overlapped by the CNV. Correlations were assessed with a two-sided Spearman correlation test. Solid lines represent 21-point rolling means.
Trait association and clinical genetics
Most large-scale trait association studies have only considered SNVs in genome-wide association studies (GWAS). Taking advantage of the sample size and resolution of gnomAD-SV, we evaluated whether SNVs associated with human traits might be in linkage disequilibrium with SVs not directly genotyped in GWAS. We identified 15,634 common SVs (allele frequency >1%) in strong linkage disequilibrium (R2 ≥ 0.8) with at least one common short variant (Supplementary Fig. 7), 14.8% of which matched a reported association from the NHGRI-EBI GWAS catalogue or a recent analysis of 4,203 phenotypes in the UK Biobank33,34. Common SVs in linkage disequilibrium with GWAS variants were enriched for genic SVs across multiple functional categories (Supplementary Table 6), and included candidate SVs such as a deletion of a thyroid enhancer in the first intron of ATP6V0D1 at a hypothyroidism-associated locus34 (Extended Data Fig. 7). We also identified matches for previously proposed causal SVs tagged by common SNVs, including pLoF deletions of CFHR3 or CFHR1 in nephropathies and of LCE3B or LCE3C in psoriasis35,36. These results demonstrate the value of imputing SVs into GWAS, and for the eventual unification of short variants and SVs in all trait association studies. Given the potential value of this resource, we have released these linkage disequilibrium maps in Supplementary Table 7.
Extended Data Fig. 7. gnomAD-SV can augment disease association studies.
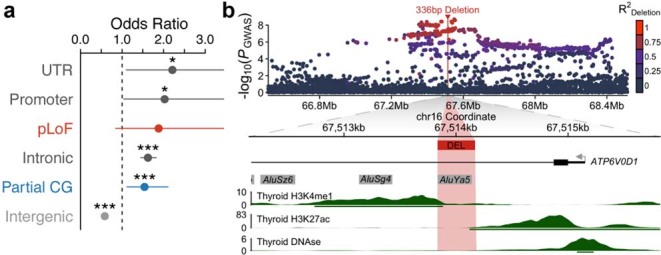
a, Functional enrichments of 2,307 common SVs in strong linkage disequilibrium (R2 ≥ 0.8) with an SNV associated with a trait or disease in the GWAS catalogue or the UK Biobank33,34. Points represent odds ratios of SVs being in strong linkage disequilibrium with at least one GWAS-significant SNV among all SVs in strong linkage disequilibrium with at least one SNV (total n = 15,634 SVs). Single and triple asterisks correspond to nominal (P < 0.05) and Bonferroni-corrected (P < 0.0083) significance thresholds from a two-sided Fisher’s exact test, respectively. Bars represent 95% confidence intervals. Test statistics, SV counts, and P values are provided in Supplementary Table 6. b, Example locus at 16q22.1, where we identified a 336-bp deletion in strong linkage disequilibrium with SNVs significantly associated with hypothyroidism in the UK Biobank34. Top, the GWAS signal among genotyped SNVs in the UK Biobank, coloured by strength of linkage disequilibrium (Pearson’s R2 value) with the 336-bp deletion identified in gnomAD-SV. Bottom, the local genomic context of this deletion, which overlaps an annotated intronic Alu element near (<1 kb) the first exon of a highly constrained, thyroid-expressed gene, ATP6V0D1. The deletion lies amidst histone mark peaks commonly found at active enhancers (H3K27ac and H3K4me1) based on publicly available chromatin data from adult thyroid samples, a phenotype-relevant tissue48. Human Alu elements are known to frequently act as enhancers, and the sentinel hypothyroidism SNV from the UK Biobank GWAS is a significant expression-modifying variant (that is, eQTL) for ATP6V0D1 and other nearby genes across many tissues, which indicates that the hypothyroidism risk haplotype modifies expression of ATP6V0D1 and/or other genes, potentially through the deletion of an intronic enhancer4,49.
As genomic medicine advances towards diagnostic screening at sequence resolution, computational methods for variant discovery from WGS and population references for interpretation will become indispensable. One category of disease-associated SVs, recurrent CNVs mediated by homologous segmental duplications known as genomic disorders, are particularly important because they collectively represent a common cause of developmental disorders37. Accurate detection of large, repeat-mediated CNVs is thus crucial for WGS-based diagnostic testing as chromosomal microarray is the recommended first-tier diagnostic screen at present for unexplained developmental disorders37. Using gnomAD-SV, we evaluated our ability to detect genomic disorders in WGS data by calculating CNV carrier frequencies for 49 genomic disorders across 10,047 unrelated samples with no known neuropsychiatric disease and found that CNV carrier frequencies in gnomAD-SV were consistent with those reported from chromosomal microarray in the UK Biobank38 (R2 = 0.669; Pearson correlation test, P = 7.38 × 10−13) (Fig. 6a, Supplementary Table 8, Supplementary Fig. 20). The frequencies of carriers of genomic disorders did not vary significantly among populations, with the exception of duplications of NPHP1 at 2q13, in which carrier frequencies in East Asian samples were up to 4.6-fold higher than in other populations, further highlighting the potential for variant interpretation to be confounded by the limited diversity of existing SV references (Supplementary Fig. 21).
Fig. 6. gnomAD-SV as a resource for clinical WGS interpretation.
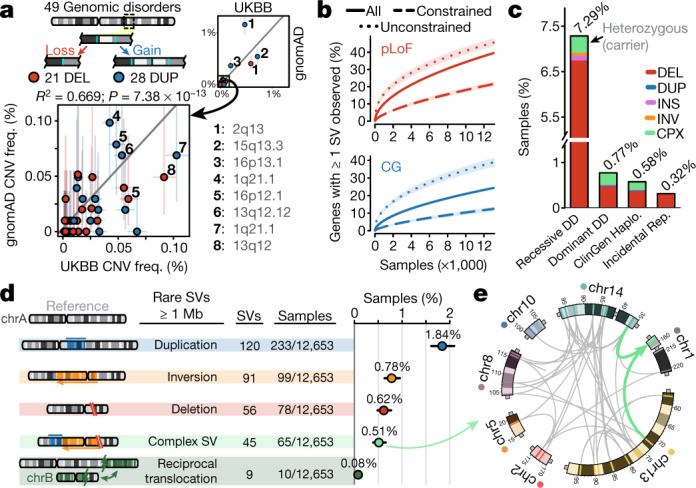
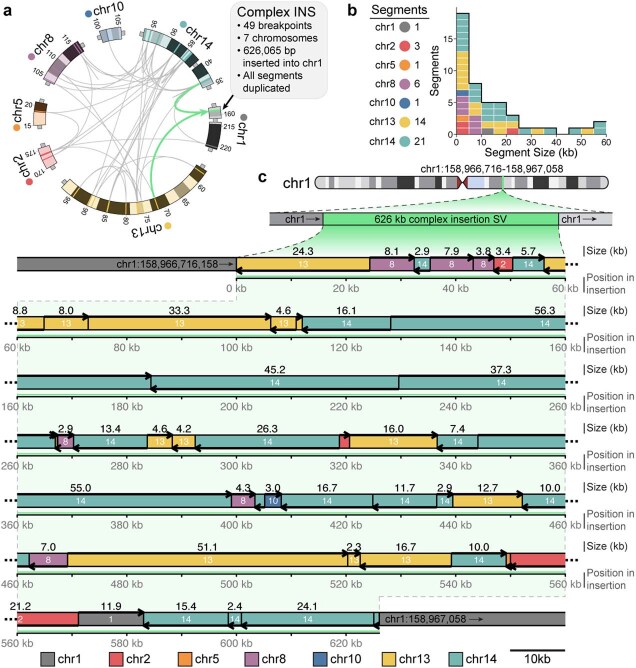
a, Comparison of carrier frequencies for 49 putatively disease-associated deletions (red) and duplications (blue) at genomic disorder loci between gnomAD-SV and microarray analyses in the UK Biobank (UKBB)38. Light bars indicate binomial 95% confidence intervals. Solid grey line represents linear best fit. b, At least one pLoF or copy-gain SV was detected in 36.9% and 23.7% of all autosomal genes, respectively. ‘Constrained’ and ‘unconstrained’ includes the least and most constrained 15% of all genes based on LOEUF4, respectively. c, Carrier rates for very rare (allele frequency < 0.1%) pLoF SVs in medically relevant genes across several gene lists7,39,44. SVs per category listed in Supplementary Table 9. d, Carrier rates for very large (≥1 Mb) rare autosomal SVs among 12,653 genomes. Bars represent binomial 95% confidence intervals. e, A complex SV involving at least 49 breakpoints and seven chromosomes (also see Extended Data Fig. 8). Teal arrows indicate insertion point into chromosome 1.
In the context of variant interpretation, the current gnomAD-SV resource will permit a screening threshold of allele frequencies less than 0.1% when matching on ancestry to the populations sampled here, and allele frequencies less than 0.004% globally. In the current release, we catalogued at least one pLoF or copy-gain variant for 36.9% and 23.7% of all autosomal genes, respectively, and 490 genes with at least one homozygous pLoF SV (Fig. 6b, Extended Data Fig. 6e, Supplementary Fig. 22). We also benchmarked carrier rates for several categories of clinically relevant variants in gnomAD-SV. First, 0.32% of samples carried a very rare (allele frequency < 0.1%) SV resulting in pLoF of a gene for which incidental findings are clinically actionable, nearly half of which (that is, 0.13% of all samples) would meet diagnostic criteria as pathogenic or likely pathogenic based upon the American College of Medical Genetics (ACMG) recommendations7 (Fig. 6c). Second, 7.22% of individuals were heterozygous carriers of rare pLoF SVs in known recessive developmental disorder genes39. Third, we estimated that 3.8% of the general population (95% confidence interval of 3.2–4.6%) carries at least one very large (≥1 Mb) rare autosomal SV, roughly half of which (45.2%) were balanced or complex (Fig. 6d). Among these was an example of localized chromosome shattering involving at least 49 breakpoints, yet resulting in largely balanced products, reminiscent of chromothripsis, in an adult with no known severe disease or DNA repair defect13,14,22 (Fig. 6e, Extended Data Fig. 8). Collectively, these analyses highlight the potential of gnomAD-SV and WGS-based SV methods to augment disease-association studies and clinical interpretation across a broad spectrum of variant classes and study designs.
Extended Data Fig. 8. An extremely complex SV involving 49 breakpoints and seven chromosomes.
A highly complex insertion rearrangement from gnomAD-SV in which 47 segments from six different chromosomes were duplicated and inserted into a single locus on chromosome 1, forming a 626,065 bp stretch of contiguous inserted sequence composed of shattered fragments. Given the involvement of multiple chromosomes, the signature of localized shattering, and the clustered breakpoints, we note that this rearrangement has several hallmarks of germline chromothripsis, which has been observed in healthy adults previously, albeit rarely22. However, unlike previous reports of germline chromothripsis, there are no apparent whole-chromosome translocations, and all segments were duplicated before being inserted in a compound manner into chromosome 1, potentially suggesting a replication-based repair mechanism. The exact origin of this rearrangement is unclear. a, Circos representation of all 49 breakpoints and seven chromosomes involved in this SV. Teal arrows indicate insertion point into chromosome 1. b, The median segment size was 8.4 kb. c, Linear representation of the rearranged inserted sequence. Colours correspond to chromosome of origin, and arrows indicate strandedness of the inserted sequence, relative to the GRCh37 reference.
Discussion
Human genetic research and clinical diagnostics are becoming increasingly invested in capturing the complete landscape of variation in individual genomes. Ambitious international initiatives to generate short-read WGS in many thousands of individuals from common disease cohorts have underwritten this goal40,41, and millions of genomes will be sequenced in the coming years from national biobanks42,43. A central challenge to these efforts will be the uniform analysis and interpretation of all variation accessible to WGS, particularly SVs, which are frequently invoked as a source of added value offered by WGS. Indeed, early WGS studies in cardiovascular disease and autism have been largely consistent in their analyses of short variants, but every study has differed in its analysis of SVs18–20,40,41. Thus, while ExAC and gnomAD have prompted remarkable advances in medical and population genetics for short variants, the same gains have not yet been realized for SVs. Although gnomAD-SV is not exhaustively comprehensive, it was derived from WGS methods and a reference genome that match those currently used in many research and clinical settings, which will help to facilitate the eventual standardization of SV discovery, analysis, and interpretation across studies.
Most foundational assumptions about human genetic variation were consistent between SVs and short variants in gnomAD, most notably that SVs segregate stably on haplotypes in the population and experience selection commensurate with their predicted biological consequences. This study also spotlights unique aspects of SVs, such as their remarkable mutational diversity, their varied functional effects on coding sequence, and the intense selection against large and complex SVs. Our analyses also demonstrate that gene-altering effects of SVs beyond pLoF are remarkably similar to the mutational constraints of SNVs, and that SNV constraint metrics are not specific to haploinsufficiency but underlie a general intolerance to alterations of both gene dosage and structure. Beyond genes, we uncovered widespread but modest selection against noncoding dosage alterations of many families of cis-regulatory elements. This study represents one of the largest empirical assessments of noncoding dosage sensitivity in humans, and underscores that: (1) few—if any—classes of noncoding cis-regulatory variants are likely to experience selection as strong as protein-truncating variants; (2) sequence conservation is unsurprisingly one of the strongest features associated with selection against noncoding SVs; and (3) current WGS sample sizes are vastly underpowered to identify individual constrained functional elements in the noncoding genome.
The value of the multi-algorithm ensemble approach and deep WGS is evident in the improved sensitivity of SV detection in gnomAD-SV. However, short-read WGS remains limited by comparison to emerging long-read technologies21. Given that short-read WGS is blind to a disproportionate fraction of repeat-mediated SVs and small insertions by comparison to long-read methods, this study certainly underestimates the true mutation rates within such hypermutable regions. Similarly, although our approach involves extensive methods to resolve complex SV alleles, some variants such as high-copy-state MCNVs often involve complicated haplotype configurations, and we expect that emerging de novo assembly and graph-based genome representations will greatly expand our knowledge of such SVs21,23. Nonetheless, 92.7% of all known autosomal protein-coding nucleotides are not localized to simple- or low-copy repeats, and therefore we expect that the catalogues of SVs accessible to short-read WGS across large populations like gnomAD-SV will capture a majority of the most interpretable gene-disrupting SVs in humans.
The scale of short-read WGS datasets currently in production has magnified the need for publicly available SV resources, and gnomAD-SV represents an initial effort to fill this void. Although these data remain insufficient to derive accurate estimates of gene-level constraint, sequence-specific mutation rates, and intolerance to noncoding SVs, they provide a step towards these goals and reinforce the value of data sharing and harmonized analyses of aggregated genomic data sets. These data have been made available without restrictions on reuse (https://gnomad.broadinstitute.org), and this resource will catalyse new discoveries in basic research while providing immediate clinical utility for the interpretation of rare structural rearrangements across human populations.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-020-2287-8.
Supplementary information
This file contains Supplementary Figures 1-24, Supplementary Notes, Supplementary Methods, and additional references.
Reviewer reports and authors' response from the peer review of this Article at Nature.
This file contains Supplementary Tables 1-9.
Acknowledgements
We thank the many individuals whose sequence data are aggregated in gnomAD for their contributions to research, and the users of gnomAD for their collaborative feedback. We are grateful to the families at the participating Simons Simplex Collection (SSC) sites, as well as the SSC principal investigators. We thank T. Hefferon of the NIH National Center for Biotechnology Information for his help hosting gnomAD-SV on dbVar. We have complied with all relevant ethical regulations. Research and contributing authors were supported by resources from the Broad Institute, the National Institutes of Health (NIH) (R01MH115957 to M.E.T., B.N. and D.G.M.; UM1HG008895 to M.J.D., B.N., S.G., E.S.L., S.K., M.E.T.; R01HD081256, P01GM061354, R01HD091797, R01HD096326, R01MH111776, R01HD099547 to M.E.T.; U01MH105669 to M.J.D., B.N. and M.E.T.; P50HD028138 to B.N. and M.E.T.; P01HD068250 to H.B.) and the Simons Foundation for Autism Research Initiative (SFARI #573206 to M.E.T.). R.L.C. was supported by NHGRI T32HG002295 and NSF GRFP #2017240332. H.B. was supported by NIDCR K99DE026824. A.V.K. was supported by NHGRI K08HG010155. M.E.T. was supported by Desmond and Ann Heathwood. L.C.F. was supported by the Swiss National Science Foundation (Advanced Postdoc.Mobility 177853). MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-00107, and UL1-TR-001420. MESA family is conducted and supported by the NHLBI in collaboration with MESA investigators. Support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258 and R01HL071259, by the National Center for Research Resources, grant UL1RR033176, and the National Center for Advancing Translational Sciences ULTR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Extended data figures and tables
Author contributions
R.L.C., H.B., K.J.K., X.Z., J.A., L.C.F, C.L., A.O’D.-L., E.V., H.J.L., J.I.R, M.J.D., D.G.M. and M.E.T. contributed to the writing of the manuscript and generation of figures. R.L.C., H.B., K.J.K., X.Z., L.C.F., C.L., L.D.G., H.W., E.V., J.F., M.J.D., E.B., D.G.M. and M.E.T. contributed to the analysis of data. R.L.C., H.B., X.Z., L.D.G., H.W., N.A.W., M.S., A.B., R.M., M.W., C.W., Y.H., T.B., T.S., M.R.S., E.V., J.F., V.R.-R., C.N., A.P., B.M.N., E.B., D.G.M. and M.E.T. developed tools and methods that enabled the scientific discoveries herein. R.L.C., H.B., K.J.K., X.Z., J.A., L.C.F., A.V.K., L.D.G., H.W., N.A.W., M.S., A.O’D.-L., A.B., R.M., G.T., K.M.L., C.S., N.G., C.C., L.M., K.D.T., H.J.L., S.S.R., W.P., Y.-D.I.C., J.I.R., C.N., A.P., E.L., S.G., B.M.N., S.K., M.J.D., E.B., D.G.M. and M.E.T. contributed to the production and quality control of the gnomAD dataset. All authors listed under The Genome Aggregation Database Consortium contributed to the generation of the primary data incorporated into the gnomAD resource. All authors reviewed the manuscript. R.L.C. and H.B. contributed equally to this study.
Data availability
All gnomAD-SV site-frequency data for appropriately consented samples (n = 10,847) have been distributed in VCF and BED format via the gnomAD browser (https://gnomad.broadinstitute.org/downloads/), as well as from NCBI dbVar under accession nstd166. Furthermore, these SVs have been integrated directly into the gnomAD browser8. The architecture of the gnomAD browser is described in the main gnomAD study4, as well as instructions for how to access and query the data hosted therein.
Code availability
The gnomAD-SV discovery pipeline is publicly available via a series of methods configured for the FireCloud/Terra platform (https://portal.firecloud.org/#methods) under the methods namespace ‘Talkowski-SV’. The svtk software package used extensively in the gnomAD-SV discovery pipeline is publicly available via GitHub (https://github.com/talkowski-lab/svtk). Most custom scripts used in the production and/or analysis of the gnomAD-SV dataset are publicly available via GitHub (https://github.com/talkowski-lab/gnomad-sv-pipeline). All code is made available under the MIT license, unless stated otherwise.
Competing interests
K.J.K. owns stock in Personalis. A.O’D.-L. has received honoraria from ARUP and Chan Zuckerberg Initiative. B.M.N. is a member of the scientific advisory board at Deep Genomics and consultant for Camp4 Therapeutics, Takeda Pharmaceutical, and Biogen. M.J.D. is a founder of Maze Therapeutics. D.G.M. is a founder with equity in Goldfinch Bio, and has received research support from AbbVie, Astellas, Biogen, BioMarin, Eisai, Merck, Pfizer, and Sanofi-Genzyme. M.E.T has received research support from Levo Therapeutics. All other authors declare no competing interests. S.K. is an employee of Verve Therapeutics, and holds equity in Verve Therapeutics, Maze Therapeutics, Catabasis, and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; he has served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, Novo Nordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Haug Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest, and Medscape; he reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetics risk predictor (20190017119).
Footnotes
Peer review information Nature thanks Don Conrad, Jan Korbel, Tobias Rausch and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Pamela Sklar
These authors contributed equally: Ryan L. Collins, Harrison Brand
Change history
2/3/2021
A Correction to this paper has been published: 10.1038/s41586-020-03176-6
Contributor Information
Michael E. Talkowski, Email: talkowsk@broadinstitute.org
Genome Aggregation Database Production Team:
Jessica Alföldi, Irina M. Armean, Eric Banks, Louis Bergelson, Kristian Cibulskis, Ryan L. Collins, Kristen M. Connolly, Miguel Covarrubias, Beryl Cummings, Mark J. Daly, Stacey Donnelly, Yossi Farjoun, Steven Ferriera, Laurent Francioli, Stacey Gabriel, Laura D. Gauthier, Jeff Gentry, Namrata Gupta, Thibault Jeandet, Diane Kaplan, Konrad J. Karczewski, Kristen M. Laricchia, Christopher Llanwarne, Eric V. Minikel, Ruchi Munshi, Benjamin M. Neale, Sam Novod, Anne H. O’Donnell-Luria, Nikelle Petrillo, Timothy Poterba, David Roazen, Valentin Ruano-Rubio, Andrea Saltzman, Kaitlin E. Samocha, Molly Schleicher, Cotton Seed, Matthew Solomonson, Jose Soto, Grace Tiao, Kathleen Tibbetts, Charlotte Tolonen, Christopher Vittal, Gordon Wade, Arcturus Wang, Qingbo Wang, James S. Ware, Nicholas A. Watts, Ben Weisburd, and Nicola Whiffin
Genome Aggregation Database Consortium:
Carlos A. Aguilar Salinas, Tariq Ahmad, Christine M. Albert, Diego Ardissino, Gil Atzmon, John Barnard, Laurent Beaugerie, Emelia J. Benjamin, Michael Boehnke, Lori L. Bonnycastle, Erwin P. Bottinger, Donald W. Bowden, Matthew J. Bown, John C. Chambers, Juliana C. Chan, Daniel Chasman, Judy Cho, Mina K. Chung, Bruce Cohen, Adolfo Correa, Dana Dabelea, Mark J. Daly, Dawood Darbar, Ravindranath Duggirala, Josée Dupuis, Patrick T. Ellinor, Roberto Elosua, Jeanette Erdmann, Tõnu Esko, Martti Färkkilä, Jose Florez, Andre Franke, Gad Getz, Benjamin Glaser, Stephen J. Glatt, David Goldstein, Clicerio Gonzalez, Leif Groop, Christopher Haiman, Craig Hanis, Matthew Harms, Mikko Hiltunen, Matti M. Holi, Christina M. Hultman, Mikko Kallela, Jaakko Kaprio, Sekar Kathiresan, Bong-Jo Kim, Young Jin Kim, George Kirov, Jaspal Kooner, Seppo Koskinen, Harlan M. Krumholz, Subra Kugathasan, Soo Heon Kwak, Markku Laakso, Terho Lehtimäki, Ruth J. F. Loos, Steven A. Lubitz, Ronald C. W. Ma, Daniel G. MacArthur, Jaume Marrugat, Kari M. Mattila, Steven McCarroll, Mark I. McCarthy, Dermot McGovern, Ruth McPherson, James B. Meigs, Olle Melander, Andres Metspalu, Benjamin M. Neale, Peter M. Nilsson, Michael C. O’Donovan, Dost Ongur, Lorena Orozco, Michael J. Owen, Colin N. A. Palmer, Aarno Palotie, Kyong Soo Park, Carlos Pato, Ann E. Pulver, Nazneen Rahman, Anne M. Remes, John D. Rioux, Samuli Ripatti, Dan M. Roden, Danish Saleheen, Veikko Salomaa, Nilesh J. Samani, Jeremiah Scharf, Heribert Schunkert, Moore B. Shoemaker, Pamela Sklar, Hilkka Soininen, Harry Sokol, Tim Spector, Patrick F. Sullivan, Jaana Suvisaari, E. Shyong Tai, Yik Ying Teo, Tuomi Tiinamaija, Ming Tsuang, Dan Turner, Teresa Tusie-Luna, Erkki Vartiainen, Marquis P. Vawter, James S. Ware, Hugh Watkins, Rinse K. Weersma, Maija Wessman, James G. Wilson, and Ramnik J. Xavier
Extended data
is available for this paper at 10.1038/s41586-020-2287-8.
Supplementary information
is available for this paper at 10.1038/s41586-020-2287-8.
References
- 1.Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry, G. H. et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 18, 1698–1710 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weischenfeldt, J., Symmons, O., Spitz, F. & Korbel, J. O. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 14, 125–138 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature10.1038/s41586-020-2308-7 (2020). [DOI] [PMC free article] [PubMed]
- 5.Walsh, R. et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 19, 192–203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green, R. C. et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 15, 565–574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karczewski, K. J. et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45 (D1), D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science316, 445–449 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talkowski, M. E. et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell149, 525–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurles, M. E., Dermitzakis, E. T. & Tyler-Smith, C. The functional impact of structural variation in humans. Trends Genet. 24, 238–245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, C. et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 44, 390–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloosterman, W. P. et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum. Mol. Genet. 20, 1916–1924 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Kosugi, S. et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 20, 117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hehir-Kwa, J. Y. et al. A high-quality human reference panel reveals the complexity and distribution of genomic structural variants. Nat. Commun. 7, 12989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang, C. et al. The impact of structural variation on human gene expression. Nat. Genet. 49, 692–699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandler, W. M. et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science360, 327–331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner, T. N. et al. Genomic patterns of de novo mutation in simplex autism. Cell171, 710–722.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werling, D. M. et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet. 50, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaisson, M. J. P. et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins, R. L. et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 18, 36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Audano, P. A. et al. Characterizing the major structural variant alleles of the human genome. Cell176, 663–675.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand, H. et al. Paired-duplication signatures mark cryptic inversions and other complex structural variation. Am. J. Hum. Genet. 97, 170–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature464, 704–712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watterson, G. A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975). [DOI] [PubMed] [Google Scholar]
- 27.Dennis, M. Y. & Eichler, E. E. Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 41, 44–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I. & Koonin, E. V. Selection in the evolution of gene duplications. Genome Biol. 3, research0008.1 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, N., Lee, I., Marcotte, E. M. & Hurles, M. E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6, e1001154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruderfer, D. M. et al. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 48, 1107–1111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature538, 265–269 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Redin, C. et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 49, 36–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47 (D1), D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neale, B. UK Biobank GWAS round 2; http://www.nealelab.is/uk-biobank/ (2018).
- 35.Xie, J. et al. Fine mapping implicates a deletion of CFHR1 and CFHR3 in protection from IgA nephropathy in Han Chinese. J. Am. Soc. Nephrol. 27, 3187–3194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Cid, R. et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 41, 211–215 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, D. T. et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 86, 749–764 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owen, D. et al. Effects of pathogenic CNVs on physical traits in participants of the UK Biobank. BMC Genomics19, 867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright, C. F. et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet385, 1305–1314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi, S. H. et al. Association between titin loss-of-function variants and early-onset atrial fibrillation. J. Am. Med. Assoc. 320, 2354–2364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natarajan, P. et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat. Commun. 9, 3391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehm, H. L. et al. ClinGen—the Clinical Genome Resource. N. Engl. J. Med. 372, 2235–2242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huddleston, J. et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pendleton, M. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods12, 780–786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, X., Weber, A. M. & Mills, R. E. A recurrence-based approach for validating structural variation using long-read sequencing technology. Gigascience6, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Battle, A., Brown, C. D., Engelhardt, B. E. & Montgomery, S. B. Genetic effects on gene expression across human tissues. Nature550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Figures 1-24, Supplementary Notes, Supplementary Methods, and additional references.
Reviewer reports and authors' response from the peer review of this Article at Nature.
This file contains Supplementary Tables 1-9.
Data Availability Statement
All gnomAD-SV site-frequency data for appropriately consented samples (n = 10,847) have been distributed in VCF and BED format via the gnomAD browser (https://gnomad.broadinstitute.org/downloads/), as well as from NCBI dbVar under accession nstd166. Furthermore, these SVs have been integrated directly into the gnomAD browser8. The architecture of the gnomAD browser is described in the main gnomAD study4, as well as instructions for how to access and query the data hosted therein.
The gnomAD-SV discovery pipeline is publicly available via a series of methods configured for the FireCloud/Terra platform (https://portal.firecloud.org/#methods) under the methods namespace ‘Talkowski-SV’. The svtk software package used extensively in the gnomAD-SV discovery pipeline is publicly available via GitHub (https://github.com/talkowski-lab/svtk). Most custom scripts used in the production and/or analysis of the gnomAD-SV dataset are publicly available via GitHub (https://github.com/talkowski-lab/gnomad-sv-pipeline). All code is made available under the MIT license, unless stated otherwise.