Abstract
Little is known about the correlations between the genetic susceptibility/resistance to Mycobacterium avium subsp. paratuberculosis (Map) infection and the estimated breeding values for type, production and functional traits. Previously, we identified 70 combinations of five single nucleotide polymorphisms (SNPs) in four bovine innate immune genes (SLC11A1, SP110, TLR2, CD209) that are associated with the genetic risk of paratuberculosis (PTB) or Johne's disease progression, which can be graded as low (LOWIN), latent (LATIN), or patent (PATIN) risk. Other possible combinations of these 5 SNPs were grouped in the average group (AVERIN). In the current study, differences in estimated breeding values (EBVs) for several traits were analyzed using linear models in a large cohort of Holstein cows (N = 15656) genotyped across Spain in 2016 or 2017. After the assignment of each genotyped cow to a risk group, cows within the PATIN risk group (N = 1448) had a superior combined genetic index (2797.57), type genetic index (524.62), milk yield (653.92 kg), protein yield (21.77 kg), fat yield (24.82 kg) and economic merit index (125 Euros) compared with the other three risk groups. Statistically significant differences in the longevity scores between the cows that were included in the PATIN risk group (108.85) and the LOWIN (107.82) and AVERIN (107.92) groups were also observed. The associations between the genetic risk groups and PTB diagnostic results were validated in a population of 99 cows from a Spanish farm with a high prevalence of PTB. Significant differences in ELISA readings between the PATIN (65.49 %) and the AVERIN (15.97 %), LATIN (2.11 %), and LOWIN (3.27 %) groups were observed. In addition, significant differences in Map DNA copies/gram of feces were observed between the PATIN and the other three risk groups. These results together with the substantial economic impact of PTB in dairy cattle support the selection of the animals with less susceptibility to PTB in the Spanish breeding program.
Keywords: Microbiology, Animal breeding, Animal science, Cattle, Gene mutation, Veterinary medicine, Paratuberculosis, Genetic susceptibility, Single nucleotide polymorphisms, Estimated breeding values
Microbiology; Animal breeding; Animal Science; Cattle; Gene Mutation; Veterinary Medicine; paratuberculosis; genetic susceptibility; single nucleotide polymorphisms; estimated breeding values
1. Introduction
Paratuberculosis (PTB), or Johne's disease (JD), is a chronic enteritis of domestic and wild ruminants caused by Mycobacterium avium susbp. paratuberculosis (Map). PTB is responsible for significant economic losses in dairy herds worldwide mainly due to reduced milk yield, reduced reproductive efficiency, premature culling of JD positive animals and reduced cull cow values (Ott et al., 1999; Hasonova and Pavlik, 2006; Gonda et al., 2007). Consequently, PTB infection affecting domestic and wild ruminants is a major issue for animal health, and the World Organization for Animal Health (OIE) requires member countries to maintain epidemiological surveillance with notification of disease cases. Map spreads through the fecal-oral route, and calves are considered more susceptible to the disease, likely due to their low level of immune-competence (Chase et al., 2008). The disease has a long latent period that depends on individual genetic resistance to infection, farm hygiene, seasonal changes and the level of exposure to Map (Coussens, 2004; Vázquez et al., 2014). Progressive weight loss, diarrhea and decreased milk yield are the main clinical signs, which are apparent only in advanced stages of disease (Sweeney, 2011). PTB is a worldwide problem, as almost all countries testing for the disease have found Map-infected cows. According to the framework of the Animal Health Law Regulation (EU, No, 2016/429), PTB can be considered eligible for the application of disease prevention and control rules. PTB is an OIE-listed disease and must be reported to the OIE as indicated in its Terrestrial Animal Health Code. Map might also be of public health concern since an association has been found between Map and Crohn's disease in humans (Juste et al., 2008).
The control of major mycobacterial diseases affecting livestock such as PTB is a major challenging issue. As the infection with Map is not treatable, it is important to control the infection by identifying and culling infected animals and by preventing Map transmission to susceptible animals using appropriate hygienic-sanitary strategies (Groenendaal et al., 2002; Garry, 2011). Since test and cull programs for JD are time-consuming, expensive, have poor sensitivity and only detect disease in later stages of infection, alternative control strategies are needed. Current inactivated vaccines against PTB are very successful in reducing the presence of Map in feces and tissues and in increasing both milk production and cattle productive life, but do not completely clear the infection (Juste et al., 2009; Bastida and Juste, 2011; Alonso-Hearn et al., 2012). Additional obstacles are interference with tests to identify animals infected with Mycobacterium bovis, the causal agent of bovine tuberculosis (Garrido et al., 2013). Because current prophylactic, diagnostic and therapeutic strategies for PTB have some limitations or are expensive to implement, improving host genetics through selective breeding could enhance natural resistance to Map infection and complement existing control strategies (Tsairidou et al., 2018).
Over the last decade, specific candidate genes have provided evidence for the existence of genomic variations associated with differences in Map susceptibility in cattle, sheep and goats (Purdie et al., 2011). This has been informed in part by studies on genetic susceptibility to other mycobacterial diseases, including tuberculosis in humans and cattle, with many correlates found. Most candidate gene discovery studies for Map susceptibility use a case-control design, where samples are selected from a set of cases (infected animals) and controls (animals exposed but not infected) in a population reference. Candidate genes have single nucleotide polymorphisms (SNPs) for which the frequency in cases and controls differs. Previously, we performed a case-control study in 636 Spanish Holstein cows that were slaughtered in Spanish abattoirs during a two year period. Animals were phenotyped for Map infection using a histopathological analysis of gut tissues and classified into three epidemiologically pathogenic forms of disease according to the severity of the lesions: i) apparently free of infection, cows with no detectable lesions; ii) latent PTB, cows with focal lesions; and iii) patent PTB; cows with more advanced lesions such as multifocal or diffuse lesions (Vázquez et al., 2013). Our case-control study identified 24 SNPs that were associated with susceptibility/resistance to Map infection in bovine genes that encode pattern recognition receptors in macrophages and dendritic cells such as the nucleotide-binding oligomerization domain 2 (NOD2), dendritic cell-specific ICAM-3-grabbing non-integrin dectin-1 (DC-SIGN or CD209), and the Toll-like receptors 2 and 4 (TLR2 and TLR4) (Ruiz-Larrañaga et al., 2010a, 2011, 2012). Other SNPs were identified in candidate genes encoding regulators of the innate immune response, such as the solute carrier family 11 member A1 gene (SLC11A1) and the nuclear body protein SP110 (Ruiz-Larrañaga et al., 2010b, 2010c). The 24 SNPs fit three quality parameters: sample call rate (>80 %), SNP call rate (>80 %) and Hardy-Weinberg equilibrium.
A previous genetic analysis suggested that genotypic combinations should provide more information than individual SNPs for association studies (Pinedo et al., 2009). Using combinations of the 24 previously identified SNPs, we found a set of five SNPs that best explained the variation in disease progression in our case-control population. This is important because the effect of host genotype on disease progression, shedding and immune response is not completely understood. More specifically, we proposed that 70 genetic combinations in 5 SNPs could explain differences in the infection progression, which was graded as low (LOWIN), latent (LATIN), or patent (PATIN) risk (Juste et al., 2018). Other possible combinations of these 5 SNPs were grouped in the average risk category (AVERIN). The main objective of the current study was to evaluate the relationships between these risk groups and the estimated breeding values (EBVs) for several type, production and functional traits. For this purpose, we genotyped and classified a large cohort of Spanish Holstein cows (N = 15656) in the genetic risk groups, and then we analyzed the EBVs for several traits included in the Spanish combined genetic index. In addition, the associations between the risk groups and PTB diagnostic results were validated in 99 lactating cows from a Spanish farm with a high PTB prevalence. This approach allowed us to validate the association between a high risk for clinical PTB and positive ELISA and fecal PCR results.
2. Materials and methods
2.1. Ethics statement
Experimental procedures were approved by the Animal Ethics Committee of the Servicio Regional de Investigation y Desarrollo Agroalimentario (SERIDA) and authorized by the Regional Consejeria de Agroganadería y Recursos Autoctonos of the Principality of Asturias (authorization code PROAE 29/2015). Experimental procedures were conducted according to the European Community Council Animal Welfare Guidelines (2010/63/EU). Blood samples were collected under the supervision of official veterinarians and in accordance with good veterinary practice. Otherwise, the study was restricted to routine on-farm observations and measurements that did not inconvenience or stress the animals.
2.2. Animals
Genotypes and EBVs corresponding to 15656 cows that were genotyped with the Illumina LD EURO G10K BeadChip in 2016 or 2017 were provided by the Spanish Federation of Holstein Cattle (CONAFE). For the second part of the study, blood and fecal samples were collected from 99 cows from a single commercial dairy farm in Asturias (Spain). This farm had a past history of PTB clinical cases, with some cows confirmed by necropsy examination. The mean prevalence of the disease in this farm as estimated by ELISA was 6.29 %. The mean age of the herd was 3.95 years (range, 1.05–10 years). Our study assesses associations between functional and genetic characteristics, and it adheres to the reporting of the genetic association studies (STREGA) initiative (Little et al., 2009).
2.3. Blood sampling, DNA extraction, genotyping and predicted risk of infection progression
Blood samples were collected from the coccygeal (tail) vein into EDTA Vacutainer tubes (BD Vacutainer System, Becton, Dickinson and Company, Sparks, MD, USA). Blood samples were centrifuged at 2500 x g for 20 min at 4 °C and buffy layers containing white blood cells were collected. Genomic DNA was extracted from the blood buffy coat using the QIAmp DNA Blood Mini Kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). Purified genomic DNA was quantified and subsequently genotyped using the Illumina LD EURO G10K BeadChip, which includes the following five SNPs in the bovine genes: SLC11A1 (SNP1: rs110090506), SP110 (SNP2: rs136859213, SNP3: rs1100480812), TLR2 (SNP4: rs41830058) and CD209 (SNP5: rs210748127) (Table 1). The EURO G10K BeadChip was developed by EuroGenomics and its collaborators. SNP1 and SNP2 are intronic. SNP3 is located in the coding region of exon 5 and causes an amino acid change from asparagine to serine at codon 196 (p.Asn196Ser). SNP4 in exon 2 and SNP5 in exon 8 do not result in any amino acid changes. DNA samples were genotyped at the molecular genetic laboratory service of the Spanish Federation of Holstein Cattle using the InfiniumTM iScan software (Illumina, San Diego, CA) for allele assignation. Animals were then assigned to a low (LOWIN), latent (LATIN), patent (PATIN) or AVERIN risk group according to 70 genetic combinations (Supplementary Table S1) (Juste et al., 2018). These 70 combinations contain 24 genotypes that belonged to the LOWIN group, 12 to the LATIN, 8 to the PATIN and 26 to the AVERIN risk group.
Table 1.
SNPs in the SLC11A1, SP110, TLR2, and CD209 genes used in the current study.
| Gene | SNP 1 | Chromosomal position (bp) | Major and minor alleles | Funtional Consequence | Reference |
|---|---|---|---|---|---|
| SLC11A1 | rs110090506 | Cr:2:107122737 | c.1157-91A > T | intron variant | Ruiz-Larrañaga et al, 2010c |
| SP110 | rs136859213 | Cr:2:119123998 | c.-1974C > T | intron variant | Ruiz-Larrañaga et al, 2010b |
| rs110480812 | Cr:2:119109955 | c.587A > G | missense | ||
| TLR 2 | rs41830058 | Cr:17:3951480 | c.1707T > C | synonymous codon | Ruiz-Larrañaga et al., 2011 |
| CD209 | rs210748127 | Cr:7:17811240 | c.762T > C | synonymous codon | Ruiz-Larrañaga et al., 2012 |
Genbank accession number.
2.4. Estimated breeding values (EBVs)
Genomic EBVs reflect the genetic potential or merit of an animal for a specific trait. The genomic EBVs are calculated by CONAFE for individual animals using the SNP BLUP method. This method is a BLUP using a genomic relationship matrix as a matrix of relationships between genotypes. BLUP assumes that all SNPs have the same variance, and it follows a normal distribution. The genomic matrix, called M, is a matrix of the codified SNPs in each animal. The genomic evaluation is a single trait assessment, where the linear model that is used to estimate the direct genomic values (VGDs) of each character is written as follows: y = 1μ+Mg + e, where y: De-regressed proofs (PDR) of the Reference Population animals for a given trait, calculated from the current MACE proofs published by Interbull (Jairath et al., 1998). μ: Population average. g: Random SNP effects vector. M: Design matrix relating the genetic effects of SNPs with phenotypic data (in this case, PDR). Normal distributions for genetic (g) and environmental (e) effects are assumed. Three times a year (March, June and November) the de-regressed proofs from the last MACE evaluation were calculated, and the Reference Population was updated in order to calculate the genomic EBVs of all genotyped animals recorded in the CONAFE database. More information about the genomic evaluation of the Holstein breed in Spain can be found at CONAFE (http://213.0.29.171/IndicesGen/Docs/evaluaciones_Metodologia_Genomica_EN.pdf). For large-scale genetic and genomic evaluations of Spanish dairy cattle, CONAFE uses the MiX99 software (MiX99 Development Team, 2017). Only records from cows in official milk recording, in the herd book and with known sire and dam were considered. Close lactations and records of culled cows were included and projected to 305 days, if days in milk ≥215. The records in progress were included, and projected to 305 days if days in milk ≥65 (Pena et al., 2001). Management variables such as diet were taken into account when the EBVs for milk yield were calculated. To eliminate the influence of other environmental factors affecting cattle productive life, all of the lactations for each individual cow were considered.
The EBVs for milk yield (kg), milk fat (kg), milk protein (kg), longevity score, days open (DO), functional merit index (ICOT), economic merit index (ECM), and the combined genetic index (ICO) were obtained from the 2019 CONAFE evaluation. ICO is the official index for total genetic merit. It combines the different traits according to their economic importance and their genetic correlations. It is used to rank the animals in the official lists of the best cows genotyped, with the breeding objective to improve future profitability of farms. The somatic cell score (SCS) is the arithmetic mean of monthly test of somatic cells that is transformed using a base-2 logarithmic function (Ali and Shook., 1980). The longevity score is measured as the number of days between the first calving and last milk record. DO is the time from when a cow calves until it conceives. The 2019-ICO consists of the following components: milk yield (11 %), fat yield (17 %), protein yield (21 %), feet and legs score (8 %), combined udder index (12 %), claw health index (4 %), SCS (8 %), longevity score (11 %), and DO (8 %). The functional merit index (ICOT) consists of the same traits as the ICO, excluding the production traits. The economic merit index (ECM) is expressed as a financial value in euros and reflects the expected increase in revenue per lactation relative to an animal with an ECM of zero (Charfeddine and Pérez-Cabal, 2019).
2.5. Enzyme-linked immunosorbent assay (ELISA) for the detection of Map-specific antibodies
Blood was collected from the coccygeal (tail) vein of each animal into 4.5 ml serum clot activator Vacutainer® tubes (Vacuette, Kremsmunster, Austria). After clotting, serum was separated by centrifugation (2500 x g for 20 min) and stored at -20 °C until use. The serum samples were tested in duplicate using the Mycobacterium paratuberculosis antibody test kit (IDEXX laboratories, Oofddorp, The Netherlands) according to the manufacturer's instructions. The optical density (OD) at 450 nm was measured in each well by an ELISA plate reader (model 680, Sigma, St. Lois, MO). The measured ODs were normalized, and the results were expressed as a percentage of the positive control OD according to the following formula: % relative OD sample/OD positive control = 100 X [(OD sample + Ag - OD sample - Ag)/(OD positive control + Ag – OD positive control - Ag). Samples with a ≥55 % relative OD/OD positive control were considered positive.
2.6. Fecal real-time polymerase chain reaction (PCR) and quantitative PCR (qPCR)
Isolation of genomic DNA from feces was performed using the MagMax Total Nucleic Acid Isolation kit according to the manufacturer's instructions (ThermoFisher Scientific, Lissieu, France). The kit employs mechanical disruption of the samples with zirconia beads in a guanidinium thiocyanate-based solution that rapidly releases nucleic acids while it simultaneously inactivates nucleases in the sample. Samples were then diluted with isopropanol, and paramagnetic beads with nucleic acid binding surface were added to the sample. For the detection of Map DNA, the LSI VetMax Triplex real-time PCR was used according to the manufacturer's instructions (ThermoFisher Scientific, Lissieu, France). Real-time PCR amplifications were performed using the MX3000P Real-Time PCR detection system (Stratagene, San Diego, USA) with the following conditions: 1 cycle at 50 °C for 2 min, 1 cycle of denaturation at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 60 s. Samples with Ct values lower than 45 for the IS900 and F57 targets were considered positive. Fecal samples were also tested with the ParaTB Kuanti-VK kit following the manufacturer's instructions (Vacunek, Bizkaia, Spain). The kit uses a F57 TaqMan probe that is labeled with the fluorescent reporter dye 5-carboxyfluorescein (FAM) at the 5′ end and primers that specifically amplify the single-copy F57 insertion sequence of Map. Inhibition of the amplification reaction is ruled out by including an internal hybridization probe labeled with 6-carboxy-4′,5′-dichloro-2′,7′-dimethosyfluorescein succinimidyl ester (JOE) at the 5′-end and specific primers. This internal amplification control molecule is co-amplified alongside the F57 diagnostic target in a duplex format. Quantification of Map titer (F57 copy numbers per gram of feces) was accomplished by preparing a standard curve using serial dilutions of a standard sample containing a known number of MAP DNA copies. Real-time qPCR amplifications were performed using the Step One Plus detection system (Applied Biosystems, Carlsbad, CA) with the following conditions: 1 cycle of denaturation at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 15 s, and annealing/extensions at 60 °C for 60 s. The results were analyzed with ABI Prism software version 1.4. This F57 real-time qPCR assay was able to quantify 10 to 107 copies/reaction.
2.7. Statistical analysis
We used a generalized linear model (GLM) or multivariate regression model to model the relationships between multiple independent (predictive or explanatory) variables and a single dependent (predicted) variable, which was the risk of PTB progression with the following four degrees: AVERIN, LOWIN, LATIN and AVERIN. The independent variables were the EBVs, ELISA readings and Map DNA copies/gram of feces. Differences between the four risk groups in the least squares means of the EBVs for several traits were estimated by analysis of variance using the GLM procedure of the SAS 9.1. statistical package for Windows (SAS Institute Inc, Cary, NC, USA). Pearson correlations between the risk groups and EBVs for several traits were calculated with the CORR procedure of the SAS 9.1. statistical package.
In addition, the associations between the genetic risk groups and diagnostic results (mean ELISA readings and Map DNA copies/gram of feces) were estimated in a population of 99 cows from a Spanish farm with a high prevalence of PTB by analysis of variance using the GLM procedure of the SAS 9.1 statistical package. The independent variables were the ELISA readings and Map DNA copies/gram of feces In all of the analyses, the least squares means of the independent variables were estimated and compared with the Tukey-Kramer correction for multiple comparisons. Statistical analyses resulting in P values lower than 0.05 were considered to be significant.
3. Results
3.1. Genotyping, risk group assignment and associations with estimated breeding values for several traits
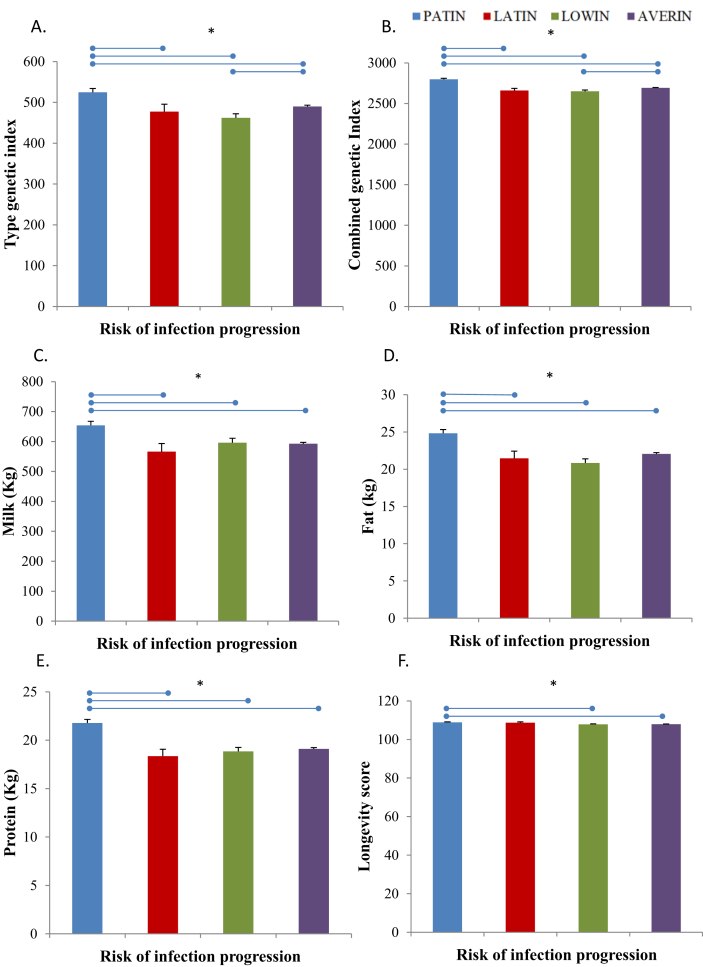
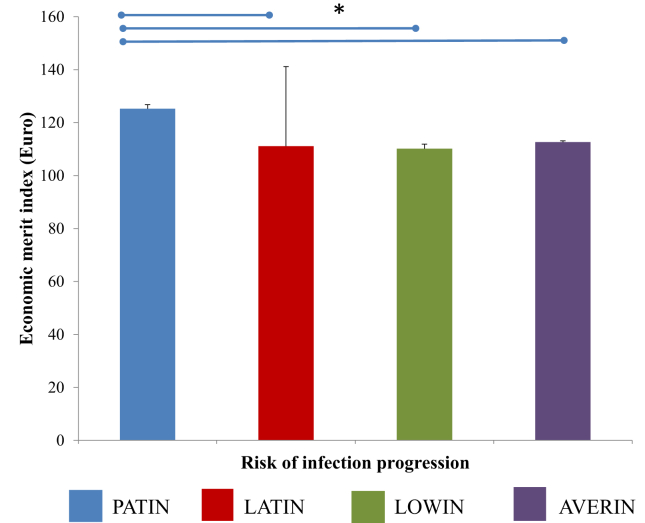
A large cohort of Spanish cattle (N = 15656) was genotyped and classified in four genetic risk groups as previously described (Juste et al., 2018). After the assignment of each individual cow to a risk group, we observed that 7.83 % of the cows were classified in the LOWIN risk group, 2.44 % in the LATIN, 9.24 % in the PATIN and 80 % in the AVERIN risk group (Table 2). Next, differences in the EBVs for several productions, types, and functional traits currently included in the Spanish breeding goal were analyzed. As seen in Figure 1, cows within the PATIN risk group (N = 1448) had a superior combined genetic index (2797.57), functional merit index (524.62), milk yield (653.92 kg), fat yield (24.82 kg), and protein yield (21.77 kg) compared with the other three risk groups. As seen in Figure 1F, statistically significant differences in longevity scores between the cows included in the PATIN risk group (108.85) and the LOWIN (107.82) and AVERIN (107.92) cows were also observed; P = 0.0132 and P = 0.0008, respectively. However, no significant differences in longevity scores were found between the PATIN (108.85) and the LATIN (108.66) cows. Differences between the risk groups in mean economic profit index were also estimated. As seen in Figure 2, cows within the PATIN risk group had superior economic merit index per year of productive life (125.25 €) compared to the cows included in the LATIN (111.11 €), LOWIN (110.17 €), and AVERIN (112.64 €) risk groups. Finally, the Pearson correlations between the cows classified in the risk groups and EBVs for several types, productions, and functional traits were calculated. As seen in Table 3, a trend is consistent for the different EBVs across the risk of infection. A positive correlation between the PATIN risk group and all of the variables that were included in the combined genetic index was observed (0.06), which suggests that animals that are prone to develop PTB clinical signs are also capable of producing higher yields. A high milk yield is a predisposing factor in the development of clinical PTB. In contrast, negative correlations between the LOWIN and AVERIN groups and the combined genetic indexes were estimated (-0.02). No statistically significant correlations were observed between the LATIN risk group and the EBVs of the analyzed traits.
Table 2.
Estimated risk of infection progression of 15656 genotyped cows.
| Risk group | Genotypes codes (Nº of cows with a specific genotype) | Animals (N) | Animals (%) |
|---|---|---|---|
| PATIN | 6 (13), 15 (38), 21 (119), 29 (179), 42 (84), 45 (267), 46 (690), 61 (58) | 1448 | 9.24 |
| LATIN | 8 (23), 25 (31), 26 (95), 32 (132), 35 (10), 37 (9), 38 (5), 39 (47), 60 (8), 65 (8), 70 (12) | 380 | 2.44 |
| LOWIN | 1 (4), 4 (57), 5 (125), 7 (17), 9 (48), 10 (21), 11 (46), 12 (13), 13 (3), 14 (2), 31 (53), 34 (78), 36 (41), 48 (255), 49 (6), 50 (66), 52 (10), 58 (80), 62 (153), 63 (30), 66 (17), 67 (30), 68 (68), 69 (3) | 1226 | 7.83 |
| AVERIN | 2 (35), 3 (117), 16 (283), 17 (897), 18 (42), 19 (442), 20 (1110), 22 (365), 23 (38), 24 (125), 27 (149), 28 (334), 30 (370), 33 (25), 40 (646), 41 (1604), 43 (804), 44 (2172), 47 (91), 51 (196), 54 (259), 55 (617), 56 (307), 57 (731), 59 (228), 64 (107), other genotypes (508) | 12602 | 80.49 |
LOWIN- Low risk of infection progression, LATIN- risk of progression to a latent form of disease, PATIN- risk of progression to a patent form of disease, AVERIN- animals with SNPs combinations that did not fit into any of the mentioned groups. SNPs combinations defining each risk group are included in Supplemental Table 1. Genotypes not previously identified were classified as new genotypes.
Figure 1.
Differences between the risk groups in mean estimated breeding values for several traits. The 15656 Holstein cows that were genotyped for the 5 SNPs in bovine CD209, SLC11A1, SP110, and TLR2 genes in 2016 or 2017 were assigned to a LOWIN, LATIN, PATIN or AVERIN risk of PTB progression. A linear model was used to test if the 2019 EBVs of the four risk groups differed. The EBV of the following traits were included in the analysis; type genetic index (A), combined genetic index (B), milk yield (C), fat yield (D), protein yield (E), and the longevity score(F). The blue bar represents the PATIN risk group, the red bar corresponds to the LATIN risk group, and the LOWIN and AVERIN groups are represented with green and purple bars, respectively. The variability of the mean values is represented by the standard error. Statistically significant differences are represented with an asterisk.
Figure 2.
Differences in the economic merit index between the four risk groups. The differences in the mean economic merit index of the four risk groups were estimated in a population of 15656 Holstein cows genotyped across Spain in 2016 or 2017. The blue bar represents the PATIN risk group, the red bar corresponds to the LATIN risk group, and the LOWIN and AVERIN groups are represented with green and purple bars, respectively. Statistically significant differences are represented with an asterisk.
Table 3.
Pearson correlation coefficients between the risk groups and EBVs for several traits.
| Functional genetic index | Combined genetic index | Milk (Kg) | Fat (Kg) | Protein (kg) | Longevity score | ||
|---|---|---|---|---|---|---|---|
| PATIN | Correlation | 0.0304 | 0.0626 | 0.0342 | 0.0439 | 0.0553 | 0.0302 |
| P | 0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0002 | |
| LATIN | Correlation | -0.0057 | -0.1189 | -0.0095 | -0.0061 | -0.0107 | 0.0116 |
| P | 0.4687 | 0.1368 | 0.2329 | 0.4403 | 0.1788 | 0.1400 | |
| LOWIN | Correlation | -0.0233 | -0.0269 | -0.0011 | -0.0207 | -0.0094 | -0.0066 |
| P | 0.0035 | 0.0070 | 0.8829 | 0.0093 | 0.2370 | 0.4056 | |
| AVERIN | Correlation | -0.0041 | -0.0229 | -0.0205 | -0.0156 | -0.0298 | -0.0220 |
| P | 0.6025 | 0.0042 | 0.0101 | 0.0505 | 0.0020 | 0.0057 | |
Significant correlations (P ≤ 0.05) are shown in bold.
3.2. Genotyping, risk group assignment and associations with diagnostic tests
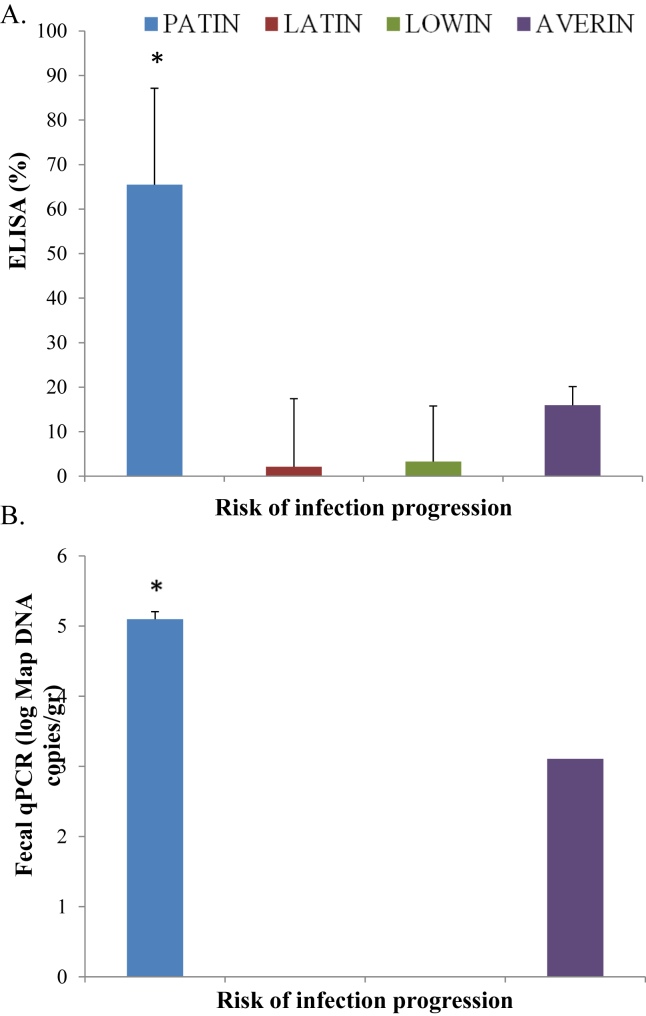
A total of 99 cows from a Spanish farm with a high prevalence of clinical PTB were genotyped for the 5 selected SNPs (Table 1). Animals were then assigned to the LOWIN, LATIN, PATIN or AVERIN risk groups as previously described (Juste et al., 2018) Three of the 99 cows belonged to the PATIN risk group (3.03 %), 6 were LATIN (6.06 %) and 9 LOWIN (9.09 %) (Table 4). The remaining 81.81 % of cows were classified within the AVERIN risk group. As shown in Figure 3A, significant differences in mean ELISA readings between the animals included in the PATIN risk group (65.49) and the AVERIN (15.97), LATIN (2.11), and LOWIN (3.27) groups were observed, at P = 0.027, P = 0.018 and P = 0.014, respectively. This result shows a positive association between the ELISA readings and the genetic combinations of the animals within the PATIN risk group. In addition, significant differences in mean log Map DNA copies/gr of feces were observed between the PATIN group (5.09) and the AVERIN (3.106), LATIN (0) or LOWIN (0) groups, at P = 0.013, P = 0.014, and P = 0.009, respectively (Figure 3B).
Table 4.
Classification of the cows from the on-farm study (N = 99) in risk groups.
| Risk Group | Genotypes codes (Nº of cows with a specific genotype) | Animals (N) | Animals (%) |
|---|---|---|---|
| PATIN | 15 (1), 29 (1), 45 (1) | 3 | 3.03 |
| LATIN | 25 (1), 26 (2), 32 (1), 39 (2) | 6 | 6.06 |
| LOWIN | 5 (1), 31 (1), 34 (2), 48 (2), 62 (2), 67 (1) | 9 | 9.09 |
| AVERIN | 16 (1), 17 (8), 19 (3), 20 (12), 22 (1), 24 (1), 28 (1), 30 (2), 40 (6), 41 (9), 43 (8), 44 (10), 47 (2), 54 (2), 55 (3), 56 (1), 57 (3), 59 (2). New genotypes (6). | 81 | 81.81 |
LOWIN- Low risk of infection progression, LATIN-risk of progression to a latent form of disease, PATIN- risk of progression to a patent form of disease, AVERIN- animals with SNPs combinations that did not fit into any of the mentioned groups. SNPs combinations corresponding to each risk group are included in Supplemental Table 1. Genotypes not previously identified were classified as new genotypes.
Figure 3.
Associations between the risk groups and PTB diagnostic tests in Holstein cattle from a commercial Spanish farm. (A) Mean ELISA readings for the detection of Map-specific antibodies. The measured ODs were normalized, and the results were expressed as a percentage of the positive control. (B) Mean log Map DNA copies/gr of feces quantified by qPCR. The blue bar represents the PATIN risk group, the red bar corresponds to the LATIN risk group, and the LOWIN and AVERIN groups are represented with green and purple bars, respectively. The variability of the results is represented by the standard error. Statistically significant differences are represented with an asterisk.
4. Discussion
It is generally accepted that intensive selection for high milk production, with limited emphasis on health and longevity traits, increases the incidence of health problems such as mastitis, milk fever, retained fetal membranes and gastrointestinal conditions during the periparturient period (Aleri et al., 2016). Pritchard et al. (2013) showed that higher yielders tend to be more commonly affected by mastitis accompanied by higher SCS, higher lameness incidence, increased calving interval and days to first service, increased number of inseminations and increased returns to service. However, there is no consensus in the literature regarding correlations between genetic susceptibility/resistance to Map infection and EBVs for several type, production and functional traits. Recently, Brito et al. (2018) estimated the genetic parameters for the Map-specific antibody response using milk ELISA scores in Canadian Holstein cattle as an indicator of resistance to PTB. Correlations between resistance to PTB and higher EBVs for several traits in Canada were considered favorable, with the exception of SCS. In the current study, the correlations between genetic risk groups of PTB progression and the 2019-EBVs for several production, type and functional traits were analyzed in 15656 Holstein cattle that were genotyped across Spain in 2016 and 2017. Our results showed a positive association between a high risk of clinical PTB and high EBVs for most of the traits included in the Spanish combined genetic index. Differences among both studies may be due to many factors such as variable diagnostic tests, variable disease incidence in the studied populations, different management practices, different trait definitions and others. It should be considered that our results are based on the genetic risk groups that are able to predict future PTB outcome early in life. Map infection occurs in calves at a young age, but clinical signs such as reduced milk yield appear later in life. Therefore, most of the animals in the PATIN group might be at a high risk of developing clinical infection without actually showing reduced milk yield yet. In fact, early work by Tiwari et al. found no association between Map and lower milk yield until lactation 4 or greater (Tiwari et al., 2007). Other studies showed that cows that ultimately tested positive had a higher milk production before their first positive test than cows that never tested positive (Nielsen et al., 2009; Smith et al., 2009).
According to Bo (2009) a breeding goal should include increased income (higher production of milk), reduced cost (better fertility, fewer diseases, and reduced culling rates), ease of management (temperament and milking speed), and advantages regarding the sale of products (animal welfare, ethics, and consumer concerns). Since only healthy cows can perform at high levels of production, breeding programs are focusing on reducing diseases and improving functional traits (Rauw et al., 1998; Zwald et al., 2010). The main challenge is to balance high levels of production and profit with fertility, longevity and health through genetic selection (Egger-Danner et al., 2014). Trait definitions, combined genetic indexes and models of analysis vary between countries. The Spanish combined genetic index (ICO) includes the lactation somatic cell score (SCS) and the combined udder score (ICU) as indicators of mastitis resistance (Pérez-Cabal and Charfeddine, 2013). More recently, the claw health index has been included in the ICO (Pérez-Cabal and Charfeddine, 2015). In the United Kingdom (UK), genetic selection for increased resistance to bovine tuberculosis has shown a favorable effect on cow longevity and on the profit lifetime index (PLI), which effectively combines production, durability, health and fertility in one single value. This is consistent with the UK PLI placing over 20 % of its emphasis on health and fertility traits (Banos et al., 2017).
The current study links the genetic risk for clinical PTB with production using a large cohort of Spanish cattle. In addition, the associations between the risk groups and diagnostic results were evaluated in 99 lactating cows from a PTB-infected dairy farm. Positive associations between the cows that were classified within the PATIN risk group and positive ELISA and fecal PCR results were obtained. By contrast, we could not detect any association between the positive diagnosis results and the genotypic combinations of the cows that were included in the LATIN risk group, probably due to the relatively late onset of fecal shedding and delayed production of antibodies during the natural course of Map infection, which only allows a definitive detection of animals in the advanced stages of PTB. Until a very sensitive diagnostic tool is developed that identifies cattle with early or chronic disease, the struggle of identifying and validating the genomic regions that are associated with PTB susceptibility/resistance will continue. Recent transcriptomic studies using RNA-Seq technology suggest that some host genes, which are biomarkers, are differentially expressed in the gut tissues and blood of PTB-infected animals depending on disease status (Alonso-Hearn et al., 2019). Using novel biomarker-based diagnostic assays, we could potentially detect infected cattle at an earlier age than is possible with current ELISA and PCR-based diagnostic assays.
Although we recognize that the population size used in the second part of the study is small (N = 99); the association between the genotypic combinations of the cows included in the PATIN risk group and positive ELISA and fecal PCR results was demonstrated. Using this independent population, we were able to validate the genotypes that were previously identified to be associated with the highest risk of disease progression by testing their association with ELISA and fecal PCR results. Overall, our results support the hypothesis that genetic variants in the bovine CD209, SLC11A1, SP110, and TLR2 genes are associated with the outcome of Map infection in cattle. However, we cannot exclude the possibility that polymorphisms in other genes involved in the host innate immune response might also be associated with disease progression.
The prospects of including PTB into the Spanish selection program are convincing despite the moderate hereditability of the trait. It should be acknowledged that while hereditability of clinical mastitis in dairy cattle is low, the SCS is included in selective breeding programs in many countries including Spain. The selection for health traits such as mastitis and PTB should reduce the need for involuntary culling due to disease. Reducing the occurrence of cattle at a high risk of progression to a clinical state through selective breeding would reduce Map infection and transmission between cattle over time and would particularly increase cattle productive life if used in combination with other control measures such as vaccination and biosafety practices. Similarly, the potential of a herd to pass infection to the environment, wild ruminants and reservoirs would also be reduced. Selection for PTB resistance would particularly benefit high risk geographic areas where the disease is highly prevalent, vaccination is not allowed and highly resistant sires are required. In addition, the genetic improvement of PTB resistance should in turn lead to simultaneous improvement of longevity, thus capturing a greater potential milk yield from older cows. Increasing the survival of milking cows is expected to have not only economic benefits but also social and environmental benefits by means of reducing farm costs.
In conclusion, our results showed a positive association between a high risk of clinical PTB and high EBVs for most of the traits included in the Spanish combined genetic index. This unfavorable correlation emphasizes the need for the inclusion of PTB resistance as a novel trait into the Spanish selection program. However, further research is required to determine optimal selection methods in order to exclude those cows with a high risk of clinical PTB.
Declarations
Author contribution statement
Maria Canive: Analyzed and interpreted the data; Wrote the paper.
Rosa Casais: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Jose A. Jimenez, Ramon A. Juste: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Cristina Blanco-Vazquez: Performed the experiments.
Javier Amado: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Joseba M. Garrido: Contributed reagents, materials, analysis tools or data.
Marta Alonso-Hearn: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by grants from the National Institute for Agricultural Research, European Funds for Regional Development (FEDER) and the Spanish Ministry of Science, Innovation and Universities (RTA2014-0009-CO2-01 and RTI2018-094192-R-C21).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank the Spanish Federation of Holstein Cattle (CONAFE) for including the 24 SNPs that are associated with susceptibility/resistance to Map infection in the LD EURO G10K Beadchip and for providing the genotype and trait data. We are also grateful to Medelin Ocejo and Jose Luis Lavin from NEIKER for assistance with data analysis. The authors want to express their gratitude to Andone Estonba, Mikel Aguirre and Begoña Jugo from the Department of Genetics, Physical Anthropology and Animal Physiology, University of the Basque Country (UPV/EHU) for their useful comments and suggestions. We are grateful to Kyle Hearn for the careful editing of the manuscript.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Suplemmentary Table 1.xlsx
References
- Aleri J.W., Hine B.C., Pyman M.F., Mansell P.D., Wales W.J., Mallard B., Fisher A.D. Periparturient immunosuppression and strategies to improve dairy cow health during the periparturient period. Res. Vet. Sci. 2016;108:8–17. doi: 10.1016/j.rvsc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Ali A.K.A., Shook G.E. An optimum transformation for somatic cell concentration in milk. J. Dairy Sci. 1980;63(3):487–490. [Google Scholar]
- Alonso-Hearn M., Canive M., Blanco-Vazquez C., Torremocha R., Balseiro A., Amado J., Varela-Martinez E., Ramos R., Jugo B.M., Casais R. RNA-Seq analysis of ileocecal valve and peripheral blood from Holstein cattle infected with Mycobacterium avium subsp. paratuberculosis revealed dysregulation of the CXCL8/IL8 signaling pathway. Sci. Rep. 2019;9:14845. doi: 10.1038/s41598-019-51328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Hearn M., Molina E., Geijo M., Vázquez P., Sevilla I.A., Garrido J.M., Juste R.A. Immunization of adult dairy cattle with a new heat-killed vaccine is associated with longer productive life prior to cows being sent to slaughter with suspected paratuberculosis. J. Dairy Sci. 2012;95(2):618–629. doi: 10.3168/jds.2009-2860. [DOI] [PubMed] [Google Scholar]
- Banos G., Winters M., Mrode R., Mitchell A.P., Bishop S.C., Woolliams J.A., Coffey M.P. Genetic evaluation for bovine tuberculosis resistance in dairy cattle. J. Dairy Sci. 2017;100(2):1272–1281. doi: 10.3168/jds.2016-11897. [DOI] [PubMed] [Google Scholar]
- Bastida F., Juste R.A. Paratuberculosis control: a review with a focus on vaccination. J. Immune Base Ther. Vaccine. 2011;9(1):8. doi: 10.1186/1476-8518-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo N. 2009. Practical Cattle Breeding in the Future _ Commercialized or Co-operative, across Borderlines between Countries and Organisations.https://journal.interbull.or/index.php/ib/article/viewFile/1122/1113 Retrieved 28 march 2014, from. [Google Scholar]
- Brito L., Mallikarjunappa S., Sargotzaei M., Koeck A., Chesnais J., Schenkel F.S., Meade K.G., Miglior F., Karrow N.A. The genetic arquitecture of milk ELISA scores as an indicator of Johne´s disease (paratuberculosis) in dairy cattle. J. Dairy Sci. 2018;101:1–14. doi: 10.3168/jds.2017-14250. [DOI] [PubMed] [Google Scholar]
- Charfeddine N., Pérez-Cabal M.A. Estudio económico para la actualización del ICO y definición de nuevos índices de mérito económico total. Frisona Española. 2019;2094:48–49. http://www.revistafrisona.com/Noticia/estudio-economico-para-la -actualizacion-del-ico-y-definicion-de-nuevos-indices-de-merito-economico-total [Google Scholar]
- Chase C.C.L., Hurley D.J., Reber A.J. Neonatal immune development in the calf and its impact on vaccine response. Vet. Clin. North Am. - Food Anim. Pract. 2008;24(1):87–104. doi: 10.1016/j.cvfa.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens P.M. Model for immune responses to Mycobacterium avium subsp. paratuberculosis in cattle. Infect. Immun. 2004;72(6):3089–3096. doi: 10.1128/IAI.72.6.3089-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger-Danner C., Cole J.B., Pryce J.E., Gengler N., Heringstad B., Bradley A., Stock K.F. Invited review: overview of new traits and phenotyping strategies in dairy cattle with a focus on functional traits. Animal. 2014;9(2):191–207. doi: 10.1017/S1751731114002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido J.M., Vázquez P., Molina E., Plazaola J.M., Sevilla I.A., Geijo M.V., Alonso-Hearn M., Juste R.A. Paratuberculosis vaccination causes only limited cross-reactivity in the skin test for diagnosis of bovine tuberculosis. PloS One. 2013;8(11):2–8. doi: 10.1371/journal.pone.0080985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry F. Control of paratuberculosis in dairy herds. Vet. Clin. North Am. - Food Anim. Pract. 2011;27(3):599–607. doi: 10.1016/j.cvfa.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Gonda M.G., Chang Y.M., Shook G.E., Collins M.T., Kirkpatrick B.W. Effect of Mycobacterium paratuberculosis infection on production, reproduction, and health traits in US Holsteins. Prev. Vet. Med. 2007;80(2–3):103–119. doi: 10.1016/j.prevetmed.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Groenendaal H., Nielen M., Jalvingh A.W., Horst S.H., Galligan D.T., Hesselink J.W. A simulation of Johne’s disease control. Prev. Vet. Med. 2002;54(3):225–245. doi: 10.1016/s0167-5877(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Hasonova L., Pavlik I. Economic impact of paratuberculosis in dairy cattle herds: a review. Vet. Med. 2006;51(5):193–211. [Google Scholar]
- Jairath L., Dekkers J.C.M., Schaeffer L.R.L., Liu Z., Burnside E.B., Kolstad B. Genetic evaluation for herd life in Canada. J. Dairy Sci. 1998;81(2):550–562. doi: 10.3168/jds.S0022-0302(98)75607-3. [DOI] [PubMed] [Google Scholar]
- Juste R.A., Elguezabal N., Garrido J.M., Pavon A., Geijo M.V., Sevilla I., Cabriada J.L., Tejada A., García-Campos F., Casado R., Ochotorena I., Izeta A., Greenstein R.J. On the prevalence of M. avium subsp. paratuberculosis DNA in the blood of healthy individuals and patients with inflammatory bowel disease. PloS One. 2008;3(7):3–8. doi: 10.1371/journal.pone.0002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juste R.A., Alonso-Hearn M., Molina E., Geijo M., Vázquez P., Sevilla I.A., Garrido J.M. Significant reduction in bacterial shedding and improvement in milk production in dairy farms after the use of a new inactivated paratuberculosis vaccine in a field trial. BMC Res. Notes. 2009;22(2):233. doi: 10.1186/1756-0500-2-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juste R.A., Vázquez P., Ruiz-Larrañaga O., Iriondo M., Manzano C., Agirre M., Estonba A., Geijo M.V., Molina E., Sevilla I.A., Alonso-Hearn M., Gómez N., Pérez V., Cortes A., Garrido J.M. Association between combinations of genetic polymorphisms and epidemiopathogenic forms of bovine paratuberculosis. Heliyon. 2018;4(2) doi: 10.1016/j.heliyon.2018.e00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J., Higgins J.P.T., Ioannidis J.P.A., Moher D., Gagnon F., Von Elm E., Khoury M.J., Cohen B., Davey-Smith G., Grimshaw J., Scheet P., Gwinn M., Williamson R.E., Zou G.Y., Hutchings K., Johnson C.Y., Tait V., Wiens M., Golding J., Van Duijn C., McLaughlin J., Paterson A., Wells G., Fortier I., Freedman M., Zecevic M., King R., Infante-Rivard C., Stewart A., Birkett N. Strengthening the reporting of genetic association studies STREGA: an extension of the STROBE statement. Genet. Epidemiol. 2009;337:581–598. doi: 10.1002/gepi.20410. [DOI] [PubMed] [Google Scholar]
- Nielsen S.S., Krogh M.A., Enevoldsen C. Time to the occurrence of a decline in milk production in cows with various paratuberculosis antibody profiles. J. Dairy Sci. 2009;92(1):149–155. doi: 10.3168/jds.2008-1488. [DOI] [PubMed] [Google Scholar]
- Ott S.L., Wells S.J., Wagner B.A. Herd-level economic losses associated with Johne’s disease on US dairy operations. Prev. Vet. Med. 1999;40(3–4):179–192. doi: 10.1016/s0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- Pena J., Ibañez M.A., Carabaño M.J., Janss L.L.G. Interbull Technical Workshop in Verden; Germany: 2001. New Genetic Parameters for National Evaluations of Production Traits in Spanish Holsteins Excluding Selected Base Animals from the Estimation of Genetic Variance. 2001. October 22-23 2000. [Google Scholar]
- Pérez-Cabal M.A., Charfeddine N. Genetic relationship between clinical mastitis and several traits of interest in Spanish Holstein dairy cattle. Interbull Bull. 2013;47:77–81. [Google Scholar]
- Pérez-Cabal M.A., Charfeddine N. Models for genetic evaluations of claw health traits in Spanish dairy cattle. J. Dairy Sci. 2015;98(11):8186–8194. doi: 10.3168/jds.2015-9562. [DOI] [PubMed] [Google Scholar]
- Pinedo P.J., Wang C., Li Y., Rae D.O., Wu R. Risk haplotype analysis for bovine paratuberculosis. Mamm. Genome. 2009;20(2):124–129. doi: 10.1007/s00335-008-9167-0. [DOI] [PubMed] [Google Scholar]
- Pritchard T., Coffey M., Mrode R., Wall E. Understanding the genetics of survival in dairy cows. J. Dairy Sci. 2013;96(5):3296–3309. doi: 10.3168/jds.2012-6219. [DOI] [PubMed] [Google Scholar]
- Purdie A.C., Plain K.M., Begg D.J., de Silva K., Whittington R.J. Candidate gene and genome-wide association studies of Mycobacterium avium subsp. paratuberculosis infection in cattle and sheep: a review. Comp. Immunol. Microbiol. Infect. Dis. 2011;34(3):197–208. doi: 10.1016/j.cimid.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Rauw W.M., Kanis E., Noordhuizen-Stassen E.N., Grommers F.J. Undesirable side effects of selection for high production efficiency in farm animals: a review. Livest. Prod. Sci. 1998;56(1):15–33. [Google Scholar]
- Ruiz-Larrañaga O., Garrido J.M., Iriondo M., Manzano C., Molina E., Koets A.P., Rutten V.P., Juste R.A., Estonba A. Genetic association between bovine NOD2 polymorphisms and infection by Mycobacterium avium subsp. paratuberculosis in Holstein-Friesian cattle. Anim. Genet. 2010;41(6):652–655. doi: 10.1111/j.1365-2052.2010.02055.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-Larrañaga O., Garrido J.M., Iriondo M., Manzano C., Molina E., Montes I., Vázquez P., Koets A.P., Rutten V.P., Juste R.A., Estonba A. SP110 as a novel susceptibility gene for Mycobacterium avium subspecies paratuberculosis infection in cattle. J. Dairy Sci. 2010;93(12):5950–5958. doi: 10.3168/jds.2010-3340. [DOI] [PubMed] [Google Scholar]
- Ruiz-Larrañaga O., Garrido J.M., Manzano C., Iriondo M., Molina E., Gil A., Koets A.P., Rutten V.P., Juste R.A., Estonba A. Identification of single nucleotide polymorphisms in the bovine solute carrier family 11 member 1 (SLC11A1) gene and their association with infection by Mycobacterium avium subspecies paratuberculosis. J. Dairy Sci. 2010;93(4):1713–1721. doi: 10.3168/jds.2009-2438. [DOI] [PubMed] [Google Scholar]
- Ruiz-Larrañaga O., Manzano C., Iriondo M., Garrido J.M., Molina E., Vázquez P., Juste R.A., Estonba A. Genetic variation of toll-like receptor genes and infection by Mycobacterium avium ssp. paratuberculosis in Holstein-Friesian cattle. J. Dairy Sci. 2011;94(7):3635–3641. doi: 10.3168/jds.2010-3788. [DOI] [PubMed] [Google Scholar]
- Ruiz-Larrañaga O., Iriondo M., Manzano C., Agirre M., Garrido J.M., Juste R.A., Estonba A. Single-nucleotide polymorphisms in the bovine CD209 candidate gene for susceptibility to infection by Mycobacterium avium subsp. paratuberculosis. Anim. Genet. 2012;43(5):646–647. doi: 10.1111/j.1365-2052.2011.02316.x. [DOI] [PubMed] [Google Scholar]
- Smith R.L., Grohn Y.T., Pradhan A.K., Whitlock R.H., Van Kessel J.S., Smith J.M., Wolfgang D.R., Schukken Y.H. A longitudinal study on the impact of Johne’s disease status on milk production in individual cows. J. Dairy Sci. 2009;92(6):2653–2661. doi: 10.3168/jds.2008-1832. [DOI] [PubMed] [Google Scholar]
- Sweeney R.W. Pathogenesis of paratuberculosis. Vet. Clin. North Am. - Food Anim. Pract. 2011;27(3):537–546. doi: 10.1016/j.cvfa.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Tiwari A., VanLeeuween J.A., Dohoo I.R., Keefe G.P., Haddad J.P., Tremblay R., Scott H.M., Whitting T. Production effects of pathogens causing bovine leukosis, bovine viral diarrhea, paratuberculosis and neosporosis. J. Dairy Sci. 2007;90(2):659–669. doi: 10.3168/jds.S0022-0302(07)71548-5. [DOI] [PubMed] [Google Scholar]
- Tsairidou S., Allen A., Banos G., Coffey M., Anacleto O., Byrne A.W., Skuze R.A., Glass E.J., Woolliams J.A., Doeschl-Wilson A.B. Can we breed cattle for lower bovine TB infectivity? Front. Vet. Sci. 2018;5:1–8. doi: 10.3389/fvets.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez P., Garrido J.M., Juste R.A. Specific antibody and interferon-gamma responses associated with immunopathological forms of bovine paratuberculosis in slaughtered friesian cattle. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez P., Ruiz-Larrañaga O., Garrido J.M., Iriondo M., Manzano C., Agirre M., Estonba A., Juste R.A. Genetic association analysis of paratuberculosis forms in Holstein-Friesian cattle. Vet. Med. Int. 2014 doi: 10.1155/2014/321327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwald N.R., Weigel K.A., Chang Y.M., Welper R.D., Clay J.S. Genetic analysis of clinical mastitis data from on-farm management software using threshold models. J. Dairy Sci. 2010;89(1):330–336. doi: 10.3168/jds.S0022-0302(06)72098-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suplemmentary Table 1.xlsx