Graphical abstract
Keywords: Bacteriophage-host, Viral diversity, Viral metagenomics, Microbial ecology, Bioinformatics
Highlights
-
•
Computational methodologies for studying double-stranded DNA viruses.
-
•
Tools for viral sequence identification from metagenomic data.
-
•
Computational approaches to assess viral diversity and the current challenges.
-
•
In-silico prediction of virus-host pairs.
-
•
Understanding viral dynamics within the context of the microbial community.
Abstract
Dynamic virus-host interactions play a critical role in regulating microbial community structure and function. Yet for decades prior to the genomics era, viruses were largely overlooked in microbial ecology research, as only low-throughput culture-based methods of discovering viruses were available. With the advent of metagenomics, culture-independent techniques have provided exciting opportunities to discover and study new viruses. Here, we review recently developed computational methods for identifying viral sequences, exploring viral diversity in environmental samples, and predicting hosts from metagenomic sequence data. Methods to analyze viruses in silico utilize unconventional approaches to tackle challenges unique to viruses, such as vast diversity, mosaic viral genomes, and the lack of universal marker genes. As the field of viral ecology expands exponentially, computational advances have become increasingly important to gain insight into the role viruses in diverse habitats.
1. Introduction
Viruses are abundant and dynamic members of all microbial communities. As obligate parasites, viruses play an important role in determining community structure and affect their environment, for example by modifying the metabolism of their hosts. The top-down control that viruses exert on microbial populations contribute to changes in community function, as viral infections often destroy large numbers of the host populations. For example, biogeochemical cycling of carbon is in part facilitated by lytic viral infections of carbon-fixing cyanobacteria, which release carbon stored as biomass as dissolved organic carbon [1]. Furthermore, viruses and their hosts are entangled in a co-evolutionary arms race to develop new infection and defense strategies, respectively [2], [3], which over time affects the microbial community structure and the fitness of the hosts. Virus-host interactions are a key part of comprehensively studying microbial ecology as they have been demonstrated to influence their hosts and environments in a variety of natural [4], host-associated [5] and engineered [6] environments.
Beyond cell death, viruses also influence microbial community structure and function by facilitating gene transfer between and across species via transduction [7]. Sometimes viruses also encode auxiliary metabolic genes (AMGs) that augment the host metabolic pathways to suit the production of viral particles [8]. Further, some proviruses (viruses that integrate their genome into the host genome) exhibit a mutualistic relationship by preventing other viruses from successfully infecting the cell – a phenomenon called “superinfection exclusion” [9].
Viruses are incredibly diverse in their structure, genetic material (ssRNA, dsDNA, etc.), host ranges and environments, making them challenging to study with traditional molecular methods as well as advancing computational techniques. Moreover, the absence of universal marker genes across viral lineages, such as those used to assess bacterial and archaeal phylogeny, compounds efforts to assess viral diversity.
Culture-based methods of virus discovery are biased towards lytic viruses whose hosts can be grown in the lab [10], often excluding proviruses and uncultivable hosts from the analysis. Culture-independent sequencing of microbial communities, namely shotgun metagenomics, coupled with downstream bioinformatics tools, has greatly enhanced the discovery of new viruses and their impacts in many diverse environments such as the ocean [11], [12], soil-permafrost interfaces [13], downhole hydraulic fracturing wells [14] and activated sludge treatment plants [6]. While standardized computational tools for studying viruses are not necessarily available, the Minimum Information about an Uncultivated Virus Genome (MIUVIG) standards provide guidelines for what to report on uncultivated viruses [15]. Here we review commonly used computational approaches in viral ecology, and their advantages and limitations.
2. Virus discovery before metagenomics
Classical experimental methods for studying viruses require pure cultures of either the viruses and/or the potential hosts for spot and plaque assays and viral tagging (fluorescent labeling and sorting of viruses) [16]. Cultured viruses are examined by electron microscopy and assays for information on their morphology, host range and replication cycles. The International Committee for the Taxonomy of Viruses (ICTV) used this information to classify viral lineages. However, these time-intensive classical techniques to isolate individual viruses are low-throughput. Additionally, the requirement of pure cultures renders the procedures impractical for viral analysis of environmental samples, where isolation of a bacterium or virus presents numerous challenges.
Universal prokaryotic marker genes such as the small subunit rRNA gene (or 16S) [17], as well as domain-specific marker genes used in the Genome Taxonomy Database (GTDB) [18] have been used to detect and classify microbes found in environmental samples. Studies targeting specific groups of viruses have used marker genes from these groups to detect viruses and assess their diversity in environmental samples through PCR-based fingerprinting [19], [20]. Examples of viral marker genes include major capsid proteins (for T4-like myoviruses), auxiliary metabolic genes (e.g. photosynthesis proteins in cyanophages), and DNA/RNA polymerases [19]. Marker genes have been defined primarily for tailed viruses of the order Caudovirales. However, there are several challenges with using marker genes to detect and classify new viruses. First, primer sets designed for marker genes are highly degenerate and require low annealing temperatures, suggesting that even conserved group-specific genes are diverse and not ideal for quantitative PCR [19], [21]. Second, primer sets are only available to specific viral groups and exclude a large part of the virome. Finally, PCR-based fingerprinting is inadequate for identifying novel viruses that do not possess any known marker genes. Therefore, the use of metagenomic analyses is imperative to the exploration of viruses and their community dynamics.
Viral metagenomics seeks to understand virus-host interactions and the impact they have on community structure and function in environmental samples. With a transition into the metagenomic era a vast number of viruses have been discovered through metagenomic studies, the largest study adding 125,000 new partial viral genomes [22] and creating a viromics pipeline and database called IMG/VR [23]. Improved computational approaches and the exploration of unique environments will continue to spark discovery of new viruses at an accelerated pace. Only a small portion of these will likely be isolated for individual study, making it impossible for low-throughput classical methods of host prediction and taxonomic classification to keep pace. Most approaches to metagenomic sequencing of microbial communities will only capture double-stranded DNA (dsDNA) viruses. While alternative protocols for RNA extraction [24] or for amplifying ssDNA [25] are also available to capture RNA and ssDNA viruses respectively, they have been used much less frequently.
Comprehensive in silico analysis of viruses requires identifying viral genomes, classifying them taxonomically and predicting virus-host pairs from metagenomic sequence data and metagenome-assembled genomes (MAGS). These are nontrivial tasks that require the use of multiple approaches at each step.
3. Identifying Viruses in Metagenomes
Here we discuss commonly used tools for virus identification in metagenomic data in depth and address additional relevant tools. These tools are summarized in Table 1, with approaches, advantages, and limitations. The first step in the viral analysis of a metagenomic dataset is to identify as many viral sequences as possible. As no standard protocol for comprehensively detecting viruses in a metagenome exists, the best practice may be to utilize several tools in parallel, as each approach will yield unique insights.
Table 1.
Summary table of various tools available for predicting viral sequences from genomic sequence data.
| Approach | Tools | Method | Advantages | Limitations | Reference |
|---|---|---|---|---|---|
| Homology-Based | VirusSeeker | BLAST-based data analysis pipeline | Identifies both eukaryotic and prokaryotic viruses | [27] | |
| VirMine | Removes non-viral reads/contigs by sequence similarity to known non-viral sequences | Uses large bacterial and archaeal databases to select for viruses | May falsely acquire unknown bacterial genes or plasmids as viral genes | [28] | |
| Phaster, Prophinder, Phage_finder | Uses a sliding window to look for viral genes flanked by bacterial genes | Specifically targets proviruses in bacterial genomes | Requires complete or near complete bacterial genomes | [32], [33], [34] | |
| VirSorter | Hidden Markov Models of viral proteins from NCBI Viral RefSeq and curated viral metagenomic datasets | Detects viral sequences similar to viruses from reference databases | Requires gene prediction; Cannot be used on unassembled reads | [26] | |
| PhiSpy | Homology to viral proteins + random forest classification using five homology-independent charactertistics | Targets novel proviruses in bacterial genomes by including homology-independent characteristics | Cannot be used on short contigs (<5kbp) or unassembled reads | [35] | |
| Machine Learning | VirFinder | Supervised machine learning based on k-mer frequency profiles | Does not require gene prediction; can be used on unassembled metagenomic reads | False positive detection of host sequences; Requires a training dataset from a similar environment | [36] |
| MARVEL | Random forest machine learning based on three virus-specific gene characteristics | Specifically targets viruses from Caudovirales order; Works best on long contigs (consisting of several genes) | Only uses information from coding regions on the genome; Requires information on gene placement on the genome | [37] | |
| VirMiner | Random forest model built on mapping viral genes to protein databases | Web-based viromics pipeline also includes functional annotation and host-prediction using CRISPRs | Virus-host predictions based only on CRISPR spacers remain incomplete; Virus taxonomic predictions are not benchmarked against other predictions or ICTV classifications | [43] | |
| Deep Learning | DeepVirFinder | A convolutional neural network trained on “genomic motifs” from viral sequences for predictions | Improved version of VirFinder by using convolutional neural networks | Training datasets must be customized to specific environments | [41] |
| VirNet | Uses 'deep attention' to predict viral sequences | Can identify novel viruses | Further testing on metagenomic data and third-party benchmarking required | [42] | |
| VIBRANT | Neural networks trained on protein family annotations from KEGG, Pfam and VOGs | Does not rely on sequence homology or genomic signatures of reference viruses; identifies proviruses, viral sequences, AMGs; Manually curated training dataset | New tool, requires third-party benchmarking | [44] | |
Currently, the most widely used tool for virus detection in metagenomic data is VirSorter [26], which detects both lytic viruses and proviruses. VirSorter uses Hidden Markov Models (HMMs), constructed from known viral hallmark proteins (e.g. major capsid proteins) to identify protein coding sequences in metagenomic sequences. Other markers used by VirSorter to identify viral sequences include “viral-like” genes, protein coding sequences that are short or have unknown functions and genes that are not associated with Caudovirales viruses. Based on these criteria, identified viral sequences are categorized and reported with confidence levels. VirSorter and other similarity-based tools [27], [28] are best at predicting known viruses, which represent only a small portion of the viruses that exist [29], as they depend heavily on the completeness of reference viral databases such as NCBI Viral RefSeq [30]. As 1200 new prokaryotic viral genomes have been added to Viral RefSeq v65 since VirSorter was built, viral detection can be improved by appending these new viral genomes as a custom database to VirSorter for a more contemporary viral analysis. Another protocol which utilizes HMMs of a broad range of viral protein families has been described by Paez-Espino [31] and was used to discover thousands of viral sequences from diverse environmental metagenomes [23]. The uniqueness of this method is the use of viral protein families from many diverse habitats, while VirSorter largely targets freshwater, marine and human microbiomes.
Programs that detect proviruses include Phaster [32], Prophinder [33], Phage_finder [34], PhiSpy [35]. Using a sliding window, these programs find viral genes sandwiched between bacterial genes and rely on sequence homology to known viruses. PhiSpy optimizes its predictions by using homology-independent viral characteristics such as protein sequence length, transcription strand direction, GC skew, and the abundance of viral-specific k-mers (see below) to classify viral sequences.
Newer tools for viral sequence detection, such as VirFinder [36] and MARVEL [37], use additional genomic features and machine learning approaches to distinguish between prokaryotic and viral sequences. VirFinder uses supervised learning of oligonucleotide frequency signatures. Oligonucleotides, or k-mers, are short sub-sequences of the genome of length k (generally ranging between 4 and 20). A k-mer frequency profile is a vector of the frequencies of all the k-mers in a genome and these profiles are unique to a given species or population of closely related bacteria, archaea or viruses. VirFinder does not rely on gene prediction or sequence similarity to known viruses and can be used on assembled or unassembled metagenomic reads. While VirFinder has better prediction accuracy and recall (true positives) for the identification of unknown viruses than VirSorter, its recall relies heavily on the composition of the training dataset, making it biased towards the most-represented viruses in the database. As viral communities typically differ between environments as a function of their hosts [38], this bias can be leveraged to identify viruses by using an environment-specific dataset to train VirFinder [38]. Sequences from prokaryotic and eukaryotic viruses often mimic the k-mer frequencies of their hosts, in an effort to avoid defense mechanisms or to integrate into the host genome as proviruses [39]. Consequently, VirFinder may also recruit prokaryote or eukaryote sequences as false positives [38].
MARVEL uses genomic features of known viruses to predict viruses on assembled contigs, which contain several genes. These features include how closely genes occur together (gene density), how frequently genes switch to the opposite strand (strand shift frequency) and number of significant hits to the pVOGs (prokaryotic virus orthologous groups) database [40]. MARVEL shows better recall and accuracy of viral sequence detection than both VirSorter and VirFinder, however specifically targets viruses from the Caudovirales order only, as it relies on a Caudovirales-specific database. Other tools utilizing machine learning, and in particular deep learning, to detect viral sequences include DeepVirFinder [41], VirNet [42], the VirMiner pipeline [43], and VIBRANT [44]. DeepVirFinder is an improvement upon VirFinder using convolutional neural networks, while VirNet uses ‘deep attention’, a technique commonly used for natural language processing. VIBRANT [44], a recently developed tool, utilizes protein family annotations from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [45], Pfam [46] and Virus Orthologous Group (VOG) [40] databases to train a neural network for the prediction of viral sequences in metagenomic data. Other virus detection tools to note that focus largely de novo detection of human pathogens (including viruses) from metagenomic data are SURPI [47], SUNBEAM [48], VIP [49] and VirusDetect [50].
4. Viral Phylogenomics and Taxonomic Classification
Phylogenomics utilizes whole genomes or large portions of the genome to reconstruct evolutionary histories of organisms. Viral phylogeny is fundamentally different in structure to phylogenies of cellular organisms as viral genomes are often a result of rapid mutation and lateral gene transfer [51]. Methods to describe viral phylogeny and assess relatedness between viruses are continually evolving as new viruses are discovered. Recently, a new framework for viral lineages was proposed by Koonin et al. that splits viruses into four realms and their subsequent hierarchies [52]. This reorganization is based on recent phylogenomic studies [56], [57] showing that hallmark genes are shared between groups of viruses with different replication-expression strategies and suggesting that the current Baltimore Classes [56] of viruses does not accurately describe viral evolutionary relationships. This new framework supports the classification of uncultured viruses based solely on their genomic content – which is a necessity in the metagenomic era.
4.1. Phylograms
Phylogenetic trees define hypothetical evolutionary relationships between multiple lineages and are based on sequence similarities between common genes. As viruses lack a universal marker gene, analysis of viral relatedness requires a phylogenomic approach, comparing whole viral genomes to generate a phylogram. The first exploration of a phylogenomic method to assess viral diversity was the Phage Proteomic Tree (2002) [57], where distances in the tree were calculated using the number of shared proteins between 105 reference viral genomes. The resulting viral groups in the Phage Proteomic Tree matched the ICTV classification of manually curated reference viruses [57]. A web-based interactive version, ViPTree [58], exists as a server that places user viral sequences among the updated reference viruses in the Phage Proteomic Tree.
Newer methods in viral phylogenomics have expanded upon the idea of a proteomic tree with modified methods to calculate distances between viral genomes. Genomic Lineages of Uncultured Viruses of Archaea and Bacteria (GL-UVAB) a pipeline for the automatic taxonomic classification of viral sequences from metagenomes uses the Dice coefficient, assigning taxonomy in strong agreement with current ICTV classifications [59]. The Dice coefficient has also been utilized to explore marine viral dynamics in several recent studies [4], [60]. Another example of a distance metric is the Genome BLAST Distance Phylogeny (GBDP) [61], as used in the program VICTOR [62] to construct phylograms and classify prokaryotic viruses.
Other approaches to construct phylograms and classify viruses include the use of single copy marker genes or HMMs for a specific group of viruses, and average nucleotide identity. In a recent study, 77 single copy marker genes were systematically identified from reference viruses from the Caudovirales order. These were used to construct a phylogram, which can be used to taxonomically classify new viruses in agreement with the ICTV classifications at the sub-family and genus levels, within the Caudovirales order [55]. ClassiPhage, a recently developed tool for classifying viruses from the families Myoviridae, Podoviridae, Siphoviridae, and Inoviridae, uses profile HMMs of proteins from reference viruses within these families known to infect Vibrionaceae [63]. The specificity of the HMMs in Classiphage may limit the usefulness of this tool for classifying viruses that do not infect Vibrionaceae. For classification at the species rank, the MIUVIG consensus suggests the use of whole-genome average nucleotide identity (95% identity cut-off over 85% of alignment fraction [15]) to viral sequences from NCBI Viral RefSeq [30] and IMG/VR [23].
4.2. Networks
While viral phylogenomics is a useful tool to understand viral relatedness and taxonomy, the inherently hierarchical tree structures are unable to represent the mosaicism present in viral genomes, and thus struggle to portray actual evolutionary trajectories. Further, different viral lineages may not fit on the same phylogram if they have no genes in common with the other taxa [55], [64], [65].
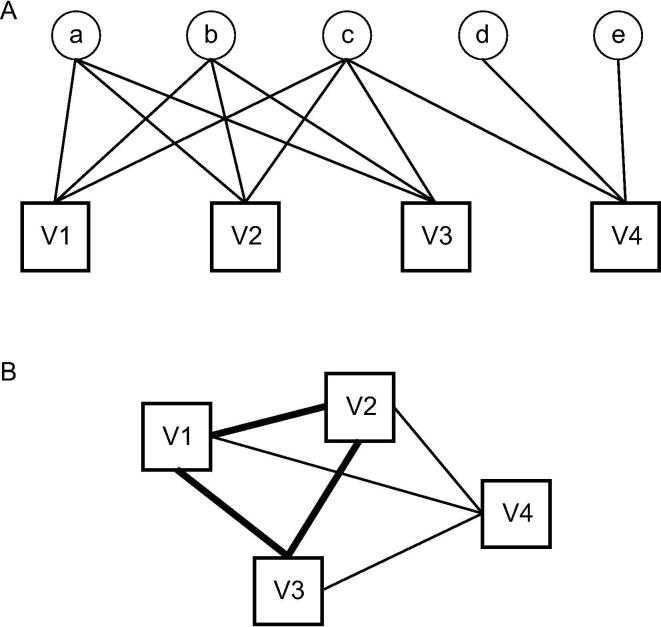
Trends in exploring viral lineages have moved towards using networks to consider the mosaic and highly diverse nature of viral genomes [51]. Mono- and bipartite networks have been explored for their use in viral classification. While both networks use shared proteins to link viral genomes, monopartite networks have weighted edges based on the total protein sequence similarity between two genomes as shown in Fig. 1(B).
Fig. 1.
Hypothetical viral genomes in squares represented by two types of networks. A) Bipartite network shows which gene families (in circles) are shared by the viral genomes (V1–V4). B) The network in (A) is simplified into a monopartite network between viral genomes with the thickness of the edges representing how many gene families are shared between genomes. This figure was recreated from an article by Iranzo et al. [64].
VConTACT2 [66] is a tool that uses a monopartite network of reference viral genomes to classify taxonomy of user viral sequences. Sequences that share proteins with reference viruses will cluster together within the network, while novel viral sequences can be identified as outliers to the network. VConTACT2 uses NCBI Viral Refseq as its database and will best work for dsDNA viruses of prokaryotes, as NCBI Viral Refseq is currently biased towards these. The same procedure for building and visualizing monopartite networks can be applied to other viruses, if an appropriate reference database is available. The biggest limitation of monopartite networks is the lack of information about which genes connect the viral clusters.
Bipartite networks show relatedness between viral genomes and also indicate which proteins are shared between groups of viruses [66] as shown by the circles in Fig. 1(A). Although there are currently no open-source tools for bipartite network analysis, it has enabled the identification of 14 hallmark genes most commonly shared by dsDNA viruses [53], [54]. This approach is an important piece in studying the evolutionary history of viruses and was used as evidence for the recently proposed reorganization of the of viral lineages [52].
5. Virus-Host Predictions
Virus-host interactions are a central part of viral research as viruses are obligate parasites and are assumed to affect their environment only through their hosts. Indicators of virus-host interactions manifest in the viral and host genomes and can be exploited to predict virus-host linkages in silico from metagenomic data. These indicators point towards which viruses infect which member(s) of the microbial community and depending on the type of indicator, what potential impact an infection might have on the host.
The Virus-Host Database [67] currently includes host information for all viral sequences from NCBI Viral RefSeq (release 99). This database has been populated from information collected from RefSeq [30], Genbank [68], UniProt [69], ViralZone [70], and manually curated literature surveys. Computational approaches to predict hosts of viruses often use sequence homology to uncover virus-host linkages. This homology is based on CRISPR spacers (snippets of viral DNA) found in prokaryotic genomes, bacterial tRNAs or auxiliary metabolic genes (AMGs) found in viral genomes, and parts of genes shared at recombination sites of temperate viruses (attP and attB in viruses and bacteria respectively).
Many bacteria and archaea carry CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) arrays as part of a microbial adaptive immune system to recognize invading viruses. The CRISPR-Cas mechanism integrates short snippets of viral DNA (25–50 bp) into the microbial genome referred to as ‘spacers’ [71]. CRISPR arrays can be found pre- or post-assembly using the tools Crass [72] and MinCED [73]. The sequences of the spacers from CRISPR arrays can be aligned to viruses found in the same metagenome using BLASTn’s short task command [74], which is optimized for alignments under 50 bp. To minimize the number of false positives for hosts, the Blastn-short task should be used with a mismatch cut-off of 1, as this is the most sensitive parameter for the alignment [16]. A cell often incorporates multiple spacers from the same viral invader [75]. Therefore, a more specific and reliable virus-host match can be established if a virus matches multiple spacers from the same CRISPR array. The spacers approach works best if spacers are matched to viruses from the same metagenome, as spacers are rapidly replaced and may only indicate recent viral invasions. As not all prokaryotes carry a CRISPR-Cas defense system, other host prediction approaches should be used in parallel to matching spacers.
Recombination sites for some lysogenic viruses (viruses that integrate into the host genome) have recognition sequences (called attP) that are similar to recombination sites on the microbial genome (attB). The attP sequence is typically situated close to DNA or RNA integrases on the viral genome. Sequences of attP and attB have a common core of 2–15 bp, which can be used to determine the host via an exact alignment match [16]. However, sequences < 15 bp also have a higher frequency of random occurrence in the genome and may lead to false positives, therefore the longest sequences for alignment should be used. Manual curation of the matches may also be required. AMGs can also be identified in viral sequences by sequence similarity to host MAGs but must be manually curated to not include host genes. Annotation of AMGs using protein families databases such as Pfam [46] or local alignments must use stringent criteria such as E-values < 10−3 and bitscores > 60 [11]. Additionally, these genes can be more confidently classified as AMGs if they are preceded and succeeded by common viral genes such as integrases, terminases or structural genes.
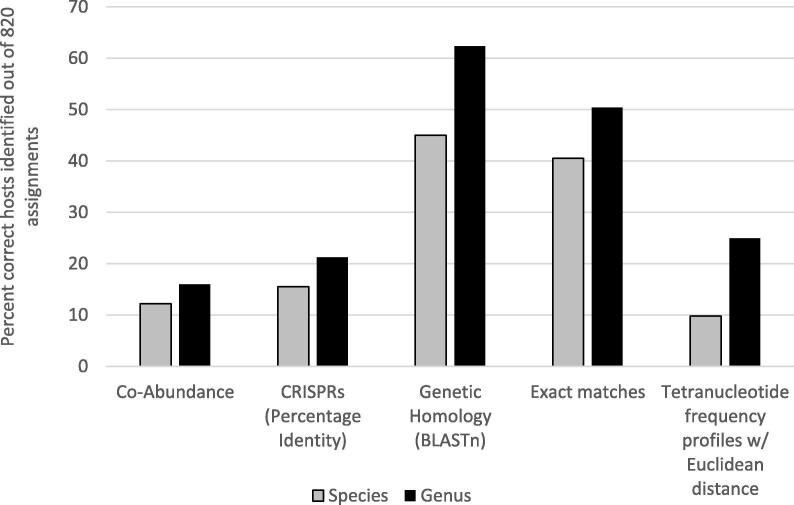
Host prediction approaches that do not use sequence homology include abundance profiles and oligonucleotide (k-mer) frequencies. Abundance profiles, the sequencing coverage of viral or host sequences across multiple samples, can be used for host prediction as viruses approximately mimic the same abundance patterns as their hosts. These patterns vary based on the type of infection (lytic versus lysogenic), predator–prey dynamics [76] or the number of integrated proviruses inside the host genome. For example, a single integrated provirus will have the exact same abundance profile as its host because it is replicated with the host genome. However, abundance profiles predict a relatively low number of correct hosts (Fig. 2) compared to other methods, as variations in host range and temperate viruses skew the abundances. As such, abundance profiles are best used with time-series metagenomic data where viral and microbial dynamics can be made obvious [77], [78]. K-mer frequency profiles, like those used by VirFinder to identify viral sequences, can also be used to predict viral hosts. This exploits the theory that prokaryotic viruses often have similar k-mer frequency profiles to their hosts to avoid recognition by cell defense mechanisms. Distances between tetranucleotide (4-mer) frequency profiles of viruses and their hosts are commonly used for predicting virus-host pairs, where the closest (smallest) Euclidean distance is indicative of the most likely host [4], [13], [16], [29], [11]. Tetranucleotide frequency profiles of viral and potential hosts sequences can be generated using Jellyfish [79].
Fig. 2.
Percentage of correct hosts identified out of 820 assignments by four methods at the genus and species level. This figure was recreated using supplementary data provided by Edwards et al. [16].
Tools that utilize k-mer frequency distributions include VirHostMatcher (k = 6) [80], HostPhinder (k = 16) [81] and Host Taxon Predictor (k = 1,2,3) [82] which also uses machine learning to differentiate between eukaryotic and prokaryotic viruses. K-mer frequency distributions are more robust when built from longer contigs (>1000 bp). For contigs shorter than 3000 bp, WIsH [83] can be used for host prediction. WIsH uses a homogenous Markov models, trained on bacterial and archaeal genomes and computes a likelihood that a viral sequence matches closely with the model. It is benchmarked to have comparable results to VirHostMatcher.
While tetranucleotide frequency profiles provide an alignment-free approach for host prediction, they are still limited by the variability of virus host ranges. Some groups of viruses have closer profiles to their hosts, while others are largely dissimilar, depending on whether they have narrow or broad host ranges respectively [80]. Tetranucleotide frequency profiles are also best at predicting hosts above the genus rank, as k-mer frequencies may not have enough differentiation at the species level. For example, Fig. 1 shows that the number of correct host predictions at the species level decrease considerably. This may result in the false prediction of hosts that are closely related to the true host, but may provide useful information, nevertheless.
Some predictive host-indicators, such as sequence-homology exact matches > 15 bp long, result in more specific matches and have higher prediction accuracy compared to other methods (Fig. 1). However, indicators with a lower specificity, such as tetranucleotide profiles, typically predict a larger number of potential virus-host pairs. A large portion of the host genome is necessary for comparison against viral sequences, for all methods of host-prediction. Viruses with broader host ranges infect multiple strains or species, and multiple viruses will target hosts of higher relative abundance. As a result, virus-host pairs are likely to resemble a many-to-many pattern, rather than one-to-one relationship. In silico virus-host pairs are best predicted from a metagenome where both the viruses and hosts come from the same environmental sample and were sequenced together. Using the consensus result of multiple approaches, including homology- and non-homology-based methods, will result in the most comprehensive, robust and accurate host prediction of viruses.
6. Conclusion
Advances in computational approaches provide an excellent avenue to explore the vast viral diversity present in our world. Yet, there remain many computational challenges to overcome in the discovery of new viruses and the dynamics of virus-host interactions. First, many of the described methods rely on reference databases, which are still biased towards well-studied viruses of cultivable hosts. These databases will continue to improve with every new virus discovery or complete assembly of a viral genome, and in-turn also improve analytical tools.
Second, the incredible diversity of viruses (structural, ssDNA, ssRNA etc.) makes the analysis of all possible virus types in a single metagenomic dataset an unfeasible task and the standardization of analytical tools impractical. Therefore, we recommend the use of multiple approaches/tools at every step in the virus discovery pipeline, to compensate for the limitations of individual approaches. Tailoring your analytical approach to answer specific research questions will also generally yield the best results.
Finally, methods to discover and describe viruses must be used in conjunction with studying the ecology of the community as a whole to understand the biological significance these viruses have in their environments. For example, CRISPR array analyses have shown that some globally-dispersed populations of bacteria are adapted to local phages by their viral defence mechanisms [84], and viruses encoding AMGs point to their role in complex carbon degradation in terrestrial environments [13].
As viral ecology becomes a tractable field through metagenomics and computation approaches, we can begin to understand the complex roles of viruses in microbial communities. Providing insights into the ever-evolving world of viruses is key to our understanding of biogeochemical cycling of nutrients in the natural environment, human health via the microbiome [85] and medicine [86] as well as engineered microbial processes [6].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wilhelm SW, Suttle C a. Viruses and Nutrient Cycles in the Sea. Bioscience 1999;49:781–8.
- 2.Koskella B., Brockhurst M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Houte S., Buckling A., Westra E.R. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol Mol Biol Rev. 2016;80:745–763. doi: 10.1128/MMBR.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutinho F.H., Gregoracci G.B., Walter J.M., Thompson C.C., Thompson F.L. Metagenomics sheds light on the ecology of marine microbes and their viruses. Trends Microbiol. 2018;26:955–965. doi: 10.1016/j.tim.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Ogilvie L.A., Jones B.V. The human gut virome: a multifaceted majority. Front Microbiol. 2015;6:1–12. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davenport R.J., Allen B.D., Sloan W.T., Brown M.R., Baptista J.C. Coupled virus – bacteria interactions and ecosystem function in an engineered microbial system. Water Res. 2019;152:264–273. doi: 10.1016/j.watres.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Jiang S.C., Paul J.H. Gene transfer by transduction in the marine environment. Appl Environ Microbiol. 1998;64:2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warwick-Dugdale J., Buchholz H.H., Allen M.J., Temperton B. Host-hijacking and planktonic piracy: how phages command the microbial high seas. Virol J. 2019;16:1–13. doi: 10.1186/s12985-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rostøl J.T., Marraffini L. (Ph)ighting phages: how bacteria resist their parasites. Cell Host Microbe. 2019;25:184–194. doi: 10.1016/j.chom.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012 doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roux S., Brum J.R., Dutilh B.E., Sunagawa S., Duhaime M.B. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho F.H., Silveira C.B., Gregoracci G.B., Thompson C.C., Edwards R.A. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson J.B., Roux S., Brum J.R., Bolduc B., Woodcroft B.J. Host-linked soil viral ecology along a permafrost thaw gradient. Nat Microbiol. 2018;3 doi: 10.1038/s41564-018-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly R., Roux S., Borton M., Morgan D., Johnston M. Viruses control dominant bacteria colonizing the terrestrial deep biosphere after hydraulic fracturing. Nat Microbiol. 2018 doi: 10.1038/s41564-018-0312-6. In review. [DOI] [PubMed] [Google Scholar]
- 15.Roux S., Adriaenssens E.M., Dutilh B.E., Koonin E.V., Kropinski A.M. Minimum information about an uncultivated virus genome (MIUVIG) Nat Biotechnol. 2019;37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards R.A., McNair K., Faust K., Raes J., Dutilh B.E. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev. 2016;40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woese C., Kandler O., Wheelis M. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:99–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 19.Adriaenssens E.M., Cowan D.A. Using signature genes as tools to assess environmental viral ecology and diversity. Appl Environ Microbiol. 2014;80:4470–4480. doi: 10.1128/AEM.00878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drosten C., Günther S., Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 21.Duhaime M.B., Sullivan M.B. Ocean viruses: rigorously evaluating the metagenomic sample-to-sequence pipeline. Virology. 2012 doi: 10.1016/j.virol.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 22.Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M. Uncovering Earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 23.Paez-Espino D., Chen I.A., Palaniappan K., Ratner A., Chu K. IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 2017;45:D457–D465. doi: 10.1093/nar/gkw1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L.Greninger A. A decade of RNA virus metagenomics is (not) enough. Virus Res 2018;244:218–29. [DOI] [PMC free article] [PubMed]
- 25.Roux S., Solonenko N.E., Dang V.T., Poulos B.T., Schwenck S.M. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ. 2016;2016:1–17. doi: 10.7717/peerj.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter : mining viral signal from microbial genomic data 2015:1–20. [DOI] [PMC free article] [PubMed]
- 27.Zhao G., Wu G., Lim E.S., Droit L., Krishnamurthy S. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology. 2017 doi: 10.1016/j.virol.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garretto A., Hatzopoulos T., Putonti C. VirMine: Automated detection of viral sequences from complex metagenomic samples. PeerJ. 2019 doi: 10.7717/peerj.6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux S., Hallam S.J., Woyke T., Sullivan M.B. Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. Elife. 2015;4:1–20. doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brister J.R., Ako-Adjei D., Bao Y., Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paez-Espino D., Pavlopoulos G.A., Ivanova N.N., Kyrpides N.C. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat Protoc. 2017;12:1673–1682. doi: 10.1038/nprot.2017.063. [DOI] [PubMed] [Google Scholar]
- 32.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lima-Mendez G., Van Helde J, Toussaint A., Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- 34.Fouts D. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 6AD;34:5839–51. [DOI] [PMC free article] [PubMed]
- 35.Akhter S., Aziz R.K., Edwards R.A. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity-and composition-based strategies. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren J., Ahlgren N.A., Lu Y.Y., Fuhrman J.A., Sun F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome. 2017;5:69. doi: 10.1186/s40168-017-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amgarten D., Braga L.P.P., Silva A.M., Setubal J.C. MARVEL, a Tool for prediction of bacteriophage sequences in metagenomic bins. Front Genet. 2018;9:1–8. doi: 10.3389/fgene.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponsero A.J., Hurwitz B.L. The promises and pitfalls of machine learning for detecting viruses in aquatic metagenomes. Front Microbiol. 2019;10:1–6. doi: 10.3389/fmicb.2019.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pride D.T., Wassenaar T.M., Ghose C., Blaser M.J. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grazziotin A.L., Koonin E.V., Kristensen D.M. Prokaryotic virus orthologous groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2017;45:491–498. doi: 10.1093/nar/gkw975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren J, Song K, Deng C, Ahlgren NA, Fuhrman JA, et al. Identifying viruses from metagenomic data by deep learning 2018. [DOI] [PMC free article] [PubMed]
- 42.Aly O. A, Mahmoud I. K, Elaraby M, Abbas H, Ali H. A. E. VirNet: Deep attention model for viral reads identification. 2018 13th Int Conf Comput Eng Syst 2018:623–6.
- 43.Zheng T., Li J., Ni Y., Kang K., Misiakou M.A. Mining, analyzing, and integrating viral signals from metagenomic data. Microbiome. 2019 doi: 10.1186/s40168-019-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kieft K, Zhou Z, Anantharaman K. VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of virome function from genomic sequences. BioRxiv n.d. [DOI] [PMC free article] [PubMed]
- 45.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999 doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naccache S.N., Federman S., Veeraraghavan N., Zaharia M., Lee D. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014 doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke E.L., Taylor L.J., Zhao C., Connell A., Lee J.J. Sunbeam: An extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome. 2019 doi: 10.1186/s40168-019-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Wang H, Nie K, Zhang C, Zhang Y, et al. VIP: An integrated pipeline for metagenomics of virus identification and discovery. Sci Rep 2016. [DOI] [PMC free article] [PubMed]
- 50.Zheng Y., Gao S., Padmanabhan C., Li R., Galvez M. VirusDetect: An automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology. 2017 doi: 10.1016/j.virol.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 51.Lima-Mendez G., Van Helden J., Toussaint A., Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 52.Koonin E.V., Dolja V.V., Krupovic M., Varsani A., Wolf Y.I. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev. 2020;84:1–33. doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iranzo J., Krupovic M., Koonin E.V. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio. 2016;7:1–21. doi: 10.1128/mBio.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iranzo J., Koonin Eugene V., Prangishvili V., Krupovic M. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsidless mobile elements. Virology. 2016;90:11043–11055. doi: 10.1128/JVI.01622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Low S.J., Džunková M., Chaumeil P.A., Parks D.H., Hugenholtz P. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat Microbiol. 2019;4:1306–1315. doi: 10.1038/s41564-019-0448-z. [DOI] [PubMed] [Google Scholar]
- 56.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohwer F., Edwards R. The phage proteomic tree: a genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishimura Y., Yoshida T., Kuronishi M., Uehara H., Ogata H. ViPTree: the viral proteomic tree server. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx157. [DOI] [PubMed] [Google Scholar]
- 59.Coutinho F.H., Edwards R.A., Rodríguez-Valera F. Charting the diversity of uncultured viruses of Archaea and Bacteria. BMC Biol. 2019;17:1–16. doi: 10.1186/s12915-019-0723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizuno C.M., Rodriguez-Valera F., Kimes N.E., Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henz S.R., Huson D.H., Auch A.F., Nieselt-Struwe K., Schuster S.C. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
- 62.Meier-Kolthoff J.P., Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chibani CM, Farr A, Klama S, Dietrich S, Liesegang H. Classifying the unclassified: A phage classification method. Viruses 2019;11. [DOI] [PMC free article] [PubMed]
- 64.Iranzo J., Krupovic M., Koonin E.V. A network perspective on the virus world. Commun Integr Biol. 2017;10:1–4. doi: 10.1080/19420889.2017.1296614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corel E., Lopez P., Méheust R., Bapteste E. Network-thinking: graphs to analyze microbial complexity and evolution. Trends Microbiol. 2016 doi: 10.1016/j.tim.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolduc B., Bin Jang H, Doulcier G., You Z.Q., Roux S. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ. 2017:1–26. doi: 10.7717/peerj.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mihara T., Nishimura Y., Shimizu Y., Nishiyama H., Yoshikawa G. Linking virus genomes with host taxonomy. Viruses. 2016 doi: 10.3390/v8030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Consortium TU UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;2019(47):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hulo C., de Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I.L.M.P. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39:D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horvath Philippe, Barrangou Rodolphe. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science:1179555. [DOI] [PubMed] [Google Scholar]
- 72.Skennerton C, Imelfort M. Crass: The CRISPR assembler (v0.3.11) 2015:1–11.
- 73.Bland C., Ramsey T.L., Sabree F., Lowe M., Brown K. CRISPR recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinf. 2007;8:1–8. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camacho C., Coulouris G., Avagyan V., Papadopoulos M.N.J., Bealer K. BLAST+: architecture and applications. BMC Bioinf. 2009;10 doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fineran P.C., Gerritzen M.J.H., Suárez-Diez M., Künne T., Boekhors J. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci USA. 2014;111:1629–1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thingstad T.F. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Am Soc Limnography Oceanogr. 2000;45:1320–1328. [Google Scholar]
- 77.Van Goethem M.W., Swenson T.L., Trubl G., Roux S., Northen T.R. Characteristics of wetting-induced bacteriophage blooms in biological soil crust. MBio. 2019;10:1–15. doi: 10.1128/mBio.02287-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arkhipova K., Skvortsov T., Quinn J.P., McGrath J.W., Allen C.C.R. Temporal dynamics of uncultured viruses: a new dimension in viral diversity. ISME J. 2018;12:199–211. doi: 10.1038/ismej.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marcais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahlgren N.A., Ren J., Lu Y.Y., Fuhrman J.A., Sun F. Alignment-free d2∗ oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 2017;45:39–53. doi: 10.1093/nar/gkw1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Villarroel J., Kleinheinz K.A., Jurtz V.I., Zschach H., Lund O. HostPhinder: a phage host prediction tool. Viruses. 2016 doi: 10.3390/v8050116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gałan W., Bąk M., Host Jakubowska M. Taxon predictor – A tool for predicting taxon of the host of a newly discovered virus. Sci Rep. 2019 doi: 10.1038/s41598-019-39847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galiez C., Siebert M., Enault F., Vincent J., Söding J. WIsH: who is the host? Predicting prokaryotic hosts from metagenomic phage contigs. Bioinformatics. 2017;33:3113–3114. doi: 10.1093/bioinformatics/btx383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koonin V., He S., Warnecke F., Peterson S.B., Garcia Martin H. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sausset R., Petit M.A., Gaboriau-Routhiau V., De Paepe M. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Nat Mucosal Immunol. 2020 [Google Scholar]
- 86.Lin D.M., Koskella B. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8:162–173. doi: 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]