Abstract
The relentless debate on postoperative adjuvant radiotherapy in gastric adenocarcinoma (GA) has been lasting for decades. In this study, a new biomarker, named promoter methylation burden of DNA repair genes (RPMB), was established to identify the subgroup of patients who might benefit from adjuvant radiotherapy. Methylation profiles of 397 GA tumor samples were downloaded from The Cancer Genome Atlas (TCGA). RPMB for a patient was defined as the ratio of methylated DNA repair genes to the number of all DNA repair genes. Subgroup analyses in term of overall survival (OS) and disease-free survival (DFS) indicated that most of the subgroups favored the high-RMPB group. Kaplan-Meier analysis showed that overall the patients with high RPMB after R0 resection had a significantly better clinical outcome regarding DFS (hazard ratio [HR] = 0.013, p = 0.042). Additionally, high-RPMB patients, who underwent adjuvant radiotherapy with both ≥T2 tumor and positive lymph nodes, showed superior DFS in comparison with the low-RPMB group (HR = 5.35 × 10−10, n = 26, p = 0.010). RPMB might be considered as a promising biomarker for decision-making with regard to postoperative adjuvant radiotherapy for GA patients.
Keywords: gastric adenocarcinoma, promoter methylation, adjuvant radiotherapy, disease-free survival
Graphical Abstract

To identify the subgroup of gastric adenocarcinoma (GA) patients who can benefit from postoperative adjuvant radiotherapy, An et al. proposed a new biomarker, named promotor methylation burden of DNA repair genes (RPMB), closely associated with patient prognosis, hopefully to provide guidance for treating GA patients after radical resection.
Introduction
Gastric adenocarcinoma (GA) is the third most common cancer in males (513,600 cases, 9.5% of all cancers) and the fourth most common cancer in females (269,100 cases, 6.5% of the all cancers) worldwide in terms of mortality.1 The clinical approaches to treat GA vary drastically between oriental and occidental worlds. Almost all of the Western clinical trials of GA failed to prove the advantage of extending lymphadenectomy (D2) compared with perigastric lymphadenectomy (D1) with regard to overall survival (OS), whereas D2 lymphadenectomy is the standard clinical practice strongly recommended in Japan.2, 3, 4 However, the less-extensive D1 lymphadenectomy is still deemed as the most common surgical management conducted in Western countries.5
Eastern and Western communities cannot agree with each other in the clinical setting of adjuvant therapy, either. Postoperative adjuvant chemotherapy leads to disappointing clinical outcomes in Western patients, since many meta-analyses have demonstrated no significant benefit,6, 7, 8 whereas the Japanese ACTS-GC trial and the Korean CLASSIC trial showed a significant prognostic benefit with adjuvant chemotherapy after D2 resection.9,10 Additionally, opposite attitudes were held between Eastern and Western communities toward postoperative adjuvant radiotherapy. The Intergroup 0116 trial randomized GA patients with a tumor size of at least T3, positive lymph nodes, or both to receive radical surgery alone or with postoperative concomitant chemoradiotherapy. OS and relapse-free survival (RFS) data demonstrated continued strong efficacy in the postoperative chemoradiotherapy group, rendering postoperative chemoradiation therapy the standard clinical procedure after R0 resection of GA patients.11 The Korean ARTIST trial was also implemented to evaluate the efficacy of postoperative chemotherapy, with or without radiotherapy (more than 45 Gy), in GA patients after D2 lymph node dissection. The results indicated that the addition of radiotherapy did not significantly prolong overall disease-free survival (DFS) or OS. However, patients with pathologic lymph nodes seemed to have superior DFS in the radiotherapy group compared with those who received chemotherapy alone.12 Unfortunately, the advantage was demonstrated to be depressingly controversial in subsequent ARTIST 2 interim analysis, which showed that there was no therapeutic superiority in the SOX (S-1 and oxaliplatin) plus radiotherapy arm compared with the SOX alone arm after complete resection of node-positive GA (ClinicalTrials.gov: NCT01761461). Thus, the Intergroup 0116 trial seemed to open a two-door gate for postoperative adjuvant radiation therapy for GA, and the ARTIST trial closed one door, while the ARTIST 2 trial closed the other one. The seemingly grim prospect of adjuvant radiotherapy implies that a biomarker is greatly needed to efficiently identify the subgroup of GA patients for whom postoperative adjuvant radiotherapy is potentially effective after R0 resection. Unfortunately, no such biomarker has ever been established to solve this problem, leading to a worldwide relentless debate on adjuvant radiotherapy.
DNA methylation is essential in promoting embryo development,13 aging,14 and many types of cancer,15, 16, 17, 18 by interfering with DNA and chromatin molecular structures.19 Dysregulation of the promoter region has been widely reported to be a pivotal epigenetic event in the process of carcinogenesis, prognostic biomarker discovery, and clinical implementations.20, 21, 22, 23 O6-methylguanine-DNA methyltransferase (MGMT) is a DNA repair protein closely related to drug resistance to alkylating agents in cancer chemotherapy,24,25 and the methylation level of the MGMT promoter was reported as the strongest indicator for clinical outcome for temozolomide therapy in glioblastoma.26 Moreover, the promoter methylation of MGMT has also been extensively found in many other cancer types.27, 28, 29, 30 Therefore, the proposition is very tempting that the promoter methylation of DNA repair genes might also be a strong biomarker that could effectively identify GA patients who could most likely benefit from adjuvant radiotherapy after R0 resection, since DNA damaging is the major molecular mechanism both adopted by chemotherapy and radiotherapy. In this study, we aimed to establish the correlation between promoter methylation of DNA repair genes and the therapeutic efficacy of adjuvant radiation therapy after R0 resection of GA patients. Notably, a new biomarker, named promoter methylation burden of DNA repair genes (RPMB), is introduced to identify the subgroup of GA patients who might benefit from postoperative adjuvant radiotherapy based on The Cancer Genome Atlas (TCGA) database.
Results
Methylation Levels of DNA Repair Genes Were Significantly Lower Than in Other Genes
We first compared the methylation level of 433 DNA repair genes with that of 433 randomly selected genes 1,000 times. The median value of the methylation level of DNA repair genes was 0.206, which was significantly lower than any other set of randomly selected genes, with all p values <0.001 (boxplots of 10 random gene sets are presented in Figure 1A). Additionally, we also compared the methylation levels of DNA repair genes with those in 10 other Gene Ontology (GO) terms, of which the biological processes were critically important in the carcinogenesis process, including immune response, cell death, apoptotic process, angiogenesis, cell migration, cell development, morphogenesis, cell proliferation, secretion, and cell adhesion. The results also indicated that the promoter methylation level of DNA repair genes was significantly lower than in genes within any other biological process (all p values <0.001, Figure 1B).
Figure 1.
Comparisons of Methylation Level between DNA Repair Genes and Other Genes
(A) Comparison of methylation level between DNA repair genes and other 10 groups of randomly selected genes. (B) Comparison of methylation level between DNA repair genes and those within 10 GO terms.
Patient Characteristics
Table 1 shows the demographics and baseline disease characteristics of GA patients included in the analysis. The GA patients were first divided into two groups (low versus high) according to the median value of RPMB, and the interaction between RPMB and clinical characteristics was extensively explored, including age, sex, ethnicity, location, histology, pathological grade, pathological tumor size (pT), pathological lymph node (pN), distant metastasis, microsatellite instability (MSI) status, and Helicobacter pylori (HP) infection. The χ2 and Fisher’s exact tests showed that RPMB was significantly associated with sex, location, grade, and MSI status (Table 1). Patients with high RPMB tended to be female (Wald χ2 = 45.38, p = 1.621 × 10−11), with tumors located in the antrum or fundus region (Wald χ2 = 20.44, p = 1.379 × 10−4), pathological grade 3 (Wald χ2 = 14.53, p = 1.380 × 10−4), and MSI-H (Wald χ2 = 14.53, p = 1.638 × 10−7). Other characteristics, including age, ethnicity, histology, pT status, pN status, metastatic disease, and HP infection, were all well balanced between the two groups.
Table 1.
Patient Baseline Characteristics
| Characteristics | Low RPMB | High RPMB | p |
|---|---|---|---|
| Age, years (n = 386) | |||
| <70 | 122 | 116 | 0.483 |
| ≥70 | 79 | 69 | |
| Sex (n = 395) | |||
| Male | 168 | 91 | 1.621 × 10−11 |
| Female | 39 | 97 | |
| Ethnicity (n = 356) | |||
| Asian | 41 | 48 | 0.169 |
| White | 135 | 118 | |
| Black or others | 10 | 4 | |
| Location (n = 385) | |||
| Antrum | 66 | 78 | 1.379 × 10−14 |
| Fundus | 68 | 74 | |
| Cardia | 28 | 25 | |
| Gastroesophageal junction | 38 | 8 | |
| Histology (n = 393) | |||
| Intestinal | 104 | 74 | 0.065 |
| Diffuse | 35 | 44 | |
| Unknown | 66 | 70 | |
| Grade (n = 386) | |||
| G1–G2 | 97 | 54 | 1.380 × 10−4 |
| G3 | 103 | 132 | |
| pT status (n = 395) | |||
| T1 | 13 | 8 | 0.345 |
| T2–T3 | 142 | 122 | |
| T4 | 52 | 58 | |
| pN status (n = 388) | |||
| N− | 58 | 66 | 0.187 |
| N+ | 144 | 120 | |
| Metastatic disease (n = 376) | |||
| Yes | 8 | 15 | 0.171 |
| No | 183 | 170 | |
| MSI status (n = 397) | |||
| MSS | 155 | 115 | 1.638 × 10−7 |
| MSI-H | 16 | 55 | |
| MSI-L | 36 | 20 | |
| HP infection (n = 188) | |||
| No | 99 | 69 | 0.778 |
| Yes | 13 | 7 |
Subgroup Analyses of OS and DFS
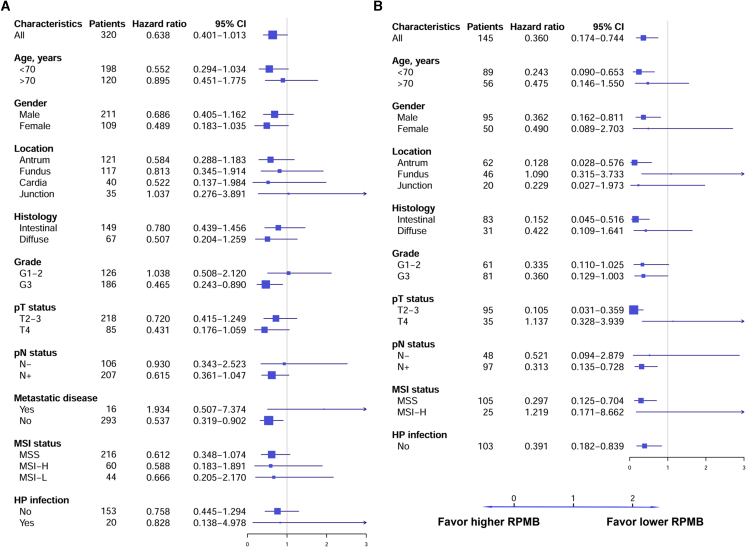
Forest plots for OS (Figure 2A) and DFS (Figure 2B) are illustrated as indicated. As for OS, 320 GA patients had OS information, and the overall hazard ratio (HR) was 0.638 (95% confidence interval [CI]: 0.401–1.013, p = 0.057), approximating significance. The subgroups were defined according to stratification factors and other baseline characteristics. The forest plots for OS indicated that almost all of the subgroups favored higher RPMB, except for tumors located at gastroesophageal junction (HR = 1.037, 95% CI: 0.276–3.891, p = 0.957), grade 1–2 (HR = 1.038, 95% CI: 0.508–2.120, p = 0.918), and distant metastatic disease (HR = 1.934, 95% CI: 0.507–7.373, p = 0.334). The subgroups significantly favoring higher RPMB included grade 3 tumors (HR = 0.465, 95% CI: 0.243–0.890, p = 0.021) and non-metastatic disease (HR = 0.537, 95% CI: 0.319–0.902, p = 0.019). As for DFS, data for 145 GA patients were collected with criteria that included the following: (1) receiving no kind of neo-adjuvant therapies, (2) non-metastatic disease, (3) undergoing R0 resection, and (4) containing detailed DFS information. The overall HR for DFS was 0.360 (95% CI: 0.174–0.744, p = 0.006). The HRs favored high RPMB levels across most of the subgroups in terms of DFS, except for tumor located at the fundus (HR = 1.090, 95% CI: 0.315–3.733, p = 0.892), pathological T4 tumors (HR = 1.137, 95% CI: 0.328–3.939, p = 0.840), and MSI-H (HR = 1.219, 95% CI: 0.171-8.662, p = 0.843). Although the sample size was comparatively limited, we were surprised to see that many subgroups were significantly associated with RPMB level in terms of DFS, including patients younger than 70 years of age (HR = 0.243, 95% CI: 0.090–0.653, p = 0.005), male (HR = 0.362, 95% CI: 0.162–0.811, p = 0.014), tumor located at antrum (HR = 0.128, 95% CI: 0.028–0.576, p = 0.007), intestinal histology (HR = 0.152, 95% CI: 0.045–0.516, p = 0.003), pathological T2–T3 (HR = 0.105, 95% CI: 0.031–0.359, p = 3.180 × 10−4), positive lymph nodes (HR = 0.313, 95% CI: 0.135–0.728, p = 0.007), microsatellite stable (MSS) (HR = 0.297, 95% CI: 0.125–0.704, p = 0.006), and no HP infection (HR = 0.391, 95% CI: 0.182–0.839, p = 0.016).
Figure 2.
Forest Plots of OS and DFS in Patient Subgroups
(A) Forest plots of OS. (B) Forest plots of DFS. HRs were estimated by an unstratified Cox model. Since the clinical data of TCGA database have many missing values, there were very limited numbers of patients in some subgroups. Subgroups containing fewer than 15 patients were excluded from subgroup analysis.
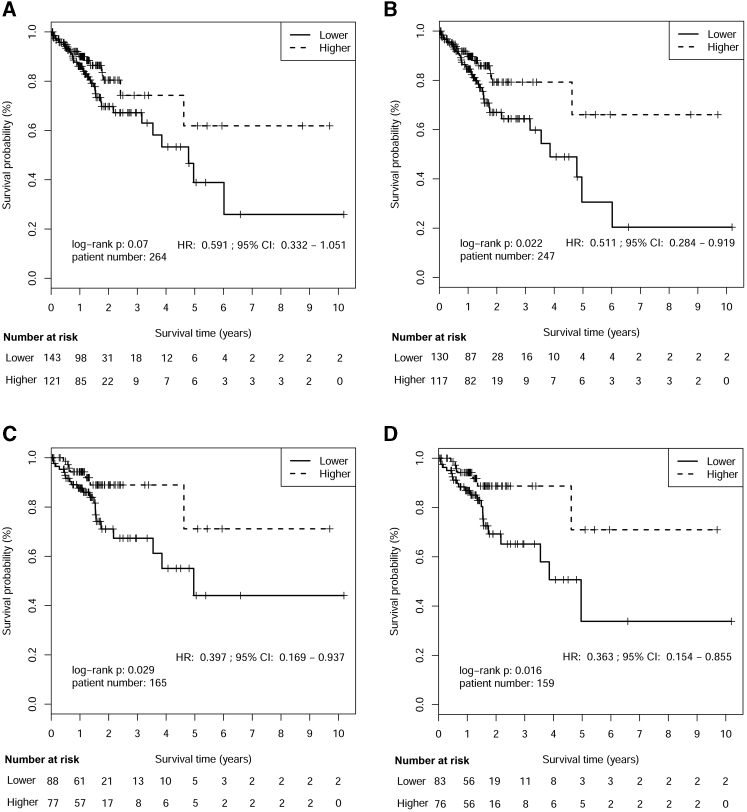
OS and DFS after R0 Resection
Median follow-up time for TCGA GA patients was 15 months. The OS difference between higher and lower RPMB patients was determined using Kaplan-Meier survival analysis. There were 264 patients with local GA after R0 resection containing OS information, and the overall HR was 0.591 (95% CI: 0.332–1.051), while the OS difference between the two groups was not significant (p = 0.07, Figure 3A). We further chose 247 out of 264 patients with pathological tumor ≥T2. The HR for aggressive disease was 0.511 (95% CI: 0.284–0.911), and the OS of these patients was significantly associated with RPMB level (p = 0.022, Figure 3B). Additionally, the OS of patients younger than 70 years of age (n = 165) was also shown to be significantly related to different RPMB levels (HR = 0.397, 95% CI: 0.169–0.937, p = 0.029, Figure 3C). Moreover, a smaller p value was obtained with fewer patients (n = 159) in the survival analysis of the younger patients (<70 years old) with ≥T2 tumor, suggesting the much stronger relationship between this subgroup of patients and RPMB level (HR = 0.363, 95% CI: 0.154–0.855, Figure 3D). The median OS for patients with lower RPMB was 4.96 years (95% CI: 3.55 to +∞ years) and median OS for those with higher RPMB had not been reached (95% CI: 4.62 to +∞ years).
Figure 3.
Kaplan-Meier Estimates of Overall Survival after R0 Resection by RPMB Level
(A) Kaplan-Meier analysis in the full analysis set. (B) Kaplan-Meier analysis in patients with pathological tumor ≥T2. (C) Kaplan-Meier analysis in patient with age <70 years. (D) Kaplan-Meier analysis in patient with both age <70 years and tumor ≥T2.
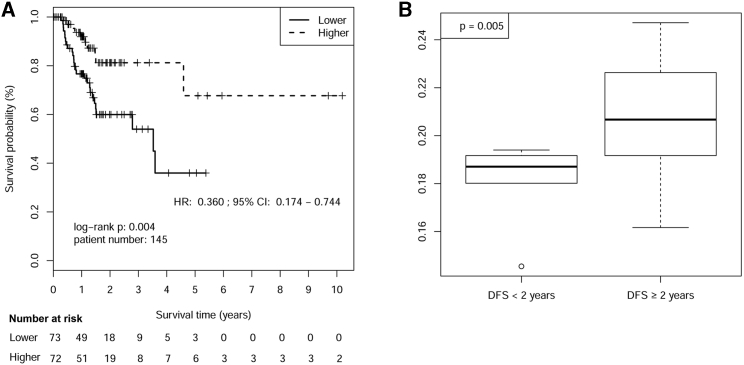
We further collected 145 patients with local disease after R0 resection containing DFS information. The results of Kaplan-Meier survival analysis indicated that patients with higher RPMB showed a significantly better benefit of DFS (HR = 0.360, 95% CI: 0.174–0.744, p = 0.004, Figure 4A). The unpaired t test demonstrated that the RPMB levels of patients who definitely recurred within 2 years (n = 33, median RPMB = 0.187) were significantly lower than those who recurred after 2 years (n = 112, median RPMB = 0.207, p = 0.005, Figure 4B).
Figure 4.
Disease-free Survival Analysis after R0 Resection by RPMB Level
(A) Kaplan-Meier analysis of disease-free survival after R0 resection. (B) RPMB level analysis between patients with DFS <2 years and DFS ≥2 years.
DFS and OS after Adjuvant Radiotherapy
The radiotherapy and DFS information contained many missing values, and thus only 34 patients with local disease were found to have undergone adjuvant radiotherapy (more than 40 Gy) with no neo-adjuvant treatments ever received. Kaplan-Meier analysis showed that patients with a higher RPMB had a significantly better DFS (HR = 0.013, p = 0.042, Figure 5A). Patients with ≥T2 tumors showed a more significant DFS difference between different RPMB groups (HR = 6.25 × 10−10, n = 32, p = 0.017, Figure 5B). Additionally, a significant DFS difference was also observed in patients with positive lymph nodes (HR = 6.21 × 10−10, n = 28, p = 0.016, Figure 5C). We eventually collected 26 patients with ≥T2 tumor and positive lymph nodes as well. The higher RPMB group showed far superior DFS compared to the lower RPMB group, and there was no recurrence found within the high PRMB group (HR = 5.35 × 10−10, n = 26, p = 0.010, Figure 5D). The limited number of patients rendered calculating the 95% CI of HR impossible (only HR values were present), and this also made the subgroup analysis of patients <70 years old impossible. Furthermore, survival analysis in terms of OS was further conducted in patients who received adjuvant radiotherapy after R0 resection. Although patients with higher RPMB showed a consistent trend of a benefit in aforementioned subgroups, the significant OS difference was not observed, probably due to the limited patient numbers (Figure S1).
Figure 5.
Kaplan-Meier Analysis of Disease-free Survival after Adjuvant Radiotherapy and R0 Resection by RPMB Level
(A) Kaplan-Meier analysis of disease-free survival for overall patients. (B) Kaplan-Meier analysis of disease-free survival with pathological tumor ≥T2. (C) Kaplan-Meier analysis of disease-free survival with positive lymph nodes. (D) Kaplan-Meier analysis of disease-free survival with both pathological tumor ≥T2 and positive lymph nodes.
Discussion
Adjuvant radiotherapy of GA has been under major debate in recent decades. Several milestone trials have been conducted in order to cope with this issue. Intergroup 0116 subset analyses implied robust treatment benefits in all subgroups observed except for patients with diffuse histology, although this trial was criticized primarily due to the suboptimum in surgical strategies (54% of patients underwent less than D1 lymphadenectomy).11 However, the ARTIST 2 trial from Korea firmly shut the door for adjuvant radiotherapy after D2 dissection in stage II/III GA patients, since no superiority was observed when compared with SOX chemotherapy alone. In general, most GA patients are actually at an advanced stage upon diagnosis, and the overall prognosis of advanced disease is suboptimal despite aggressive cancer treatment. Therefore, an effective biomarker is greatly needed to select proper patients who might obtain a prognostic benefit from adjuvant radiotherapy; unfortunately, however, there had been no such prognostic indicator ever found. In this study, RPMB was demonstrated to be a promising biomarker to fulfill this mission.
The intention to use RPMB as a GA biomarker was actually inspired by the remarkable prognostic value of one DNA repair gene, MGMT, in malignant glioma. More than two decades ago, a potential predictive value of MGMT protein levels was strikingly observed in malignant glioma determined through immunofluorescence microscopy.31,32 The hypermethylation of MGMT was proven responsible for the decreased levels of MGMT protein. MGMT promoter methylation was also reported to be a prognostic biomarker for benefits from alkylator-based chemotherapy, such as nitrosoureas33 or temozolomide,34 in glioma. Furthermore, MGMT promoter methylation was actually shown to prolong progression-free survival specifically in patients treated with temozolomide and radiotherapy in two clinical trials.35,36 The rationale of developing RPMB as predictor of adjuvant radiotherapy is as follows: (1) the theoretical foundation of radiation biology is to directly or indirectly compromise bioactive macromolecules, and DNA is the major target of radiotherapy. Inactivation of DNA repair genes through promoter hypermethylation might consolidate the DNA damaging biological effect of radiation, leading to a better prognosis of the patients. (2) The promoter methylation level of all of the DNA repair genes, rather than MGMT alone, might be a better measurement to predict the prognosis of radiotherapy. Therefore, we developed RPMB as a GA biomarker, with the primary intention to predict the prognosis of radiation therapy after R0 resection.
The promoter methylation level of DNA repair genes was significantly lower than the rest of genes, which could be referred to as a self-protecting maneuver to activate DNA repair genes, in order to “repair” the damages caused by a variety of cancer treatments, including radiotherapy and chemotherapy. This result increased our curiosity about the prognosis of the high-RPMB subgroup after radiotherapy. In this subgroup, the inactivation of DNA repair genes by virtue of a high RPMB level probably exposed tumor cells directly to the fierce attack of radiotherapy. In this manner, the tricky self-protecting maneuver was not in action in high-RPMB patients to keep cancer cells alive, probably leading to an encouraging clinical outcome after adjuvant radiotherapy.
Since endoscopy resection is also the standard treatment for patients with T1 tumor, we therefore conducted survival analysis in patients with ≥T2 tumors who underwent curative surgery. The comparatively shorter life expectancy and comorbidity of elderly patients might undermine the therapeutic response in high-RPMB patients and increase potential side effects of cancer treatments. The MOSAIC trial of colorectal cancer showed no statistically significant benefit (OS and DFS) for the addition of oxaliplatin to FL (fluorouracil with leucovorin) as adjuvant treatment for elderly patients ≥70 years of age, suggesting that the potent regimen of chemotherapy might not be proper for elderly patients.37 This trial, despite having been conducted on colorectal cancer, still provides a meaningful reference for the GA setting, since the regimen of oxaliplatin and fluorouracil is also the most commonly used combination in GA patients. Therefore, we carried out OS analysis in patients younger than 70 years old. The survival analysis indicated that patients with ≥T2 tumors and younger than 70 years old showed the most distinct prognoses between different RPMB groups, suggesting that patients with more aggressive disease and younger age could probably enhance the prognostic and predictive value of RPMB in the adjuvant clinical setting.
RPMB was not only associated with OS but it was also significantly associated with DFS after R0 resection, and DFS in patients with high RPMB was significantly longer than for those with low RPMB. We further explored the relationship between RPMB and the efficacy of adjuvant radiotherapy. The result indicated that the most distinct DFS curves were observed in patients with big tumor size and positive lymph nodes, and these two clinical characteristics were also proven to be the decision-making factors for subsequent radiotherapy after curative resection in the Intergroup 0116 trial, suggesting that the efficacy of adjuvant radiotherapy might be more notable in locally advanced disease, rather than in early stage disease. Although statistical significance was not achieved with respect to OS, the trend of the two curves separating from each other was quite obvious, implying that statistical significance might be reached after increasing the sample size. Based on the aforementioned analysis, the idea is very intriguing that RPMB might be a promising measurement to distinguish the patients who might be therapeutically responsive from the others after adjuvant radiotherapy.
The major limitation of this study is the limited patient numbers in subgroup analyses. TCGA clinical data contained many missing values, leading to limited patient numbers in some subgroups. For instance, only 34 patients were available for DFS analysis who underwent both R0 resection and adjuvant radiotherapy. Therefore, more patients are needed to increase the credibility of our conclusion. Second, to our knowledge, TCGA GA data were the only publically available dataset containing a global DNA methylation profile and survival information after adjuvant radiotherapy. Therefore, it is currently impossible to test the aforementioned results in independent cohorts, determine the exact β value threshold to categorize methylated or unmethylated promoters, or calculate the proper RPMB cutoff to identify ready-for-radiotherapy patients (only the median value was used as the cutoff). In the future, we will conduct a prospective study and enroll more patients to consolidate the validity of RPMB in predicting the prognosis after adjuvant radiotherapy, and determine the optimal two aforementioned cutoffs essential to quantify individual RPMB levels. Additionally, radiation oncologists should also consider the potential toxicities of adjuvant radiotherapy, which can certainly vary dramatically from patient to patient. The treatment volume for GA adjuvant therapy is relatively extensive, probably leading to serious radiation-related side effects. The best clinical management for GA patients should be individually tailored after balancing between both the potential risks and benefits.
Despite all of the shortcomings of this study due to limited available data, RPMB is the first genomic biomarker and indicator proven to be potentially effective in predicting clinical outcomes of postoperative adjuvant radiotherapy after curative radical surgery in GA patients. Hopefully, we can finally put an end to this intense debate lasting for decades after future thorough investigations on RPMB.
Materials and Methods
Data Retrieval
The methylation profile of GA patients and corresponding clinical information were obtained from the Bioconductor package RTCGA (https://rtcga.github.io/RTCGA). The methylation profile consisted of 397 GA tumor samples, generated from the platform of Illumina HumanMethylation450 chips. The flow diagram of inclusion and exclusion criteria is shown in Figure S2. The methylation level of each CpG site was already properly processed as β value, which was defined as the ratio of the signal of the methylated probes relative to the sum of all the probes, ranging from 0 (unmethylated) to 1 (fully methylated). The promoter region of a given gene was defined as the genomic region between 1,000 bp upstream of the transcription start site (TSS) and 300 bp downstream. If the probe of a CpG site was mapped to a given gene’s promoter region, its corresponding β value was adopted to quantify the promoter methylation of this gene. If multiple CpG sites were located within the same promoter region of a given gene, the mean value of their β values was defined as the methylation level of this gene. In this manner, the promoter methylation profile of 20,043 genes was obtained with DNA methylation data for GA.38
DNA Repair Genes Collection and Their Methylation Level Compared With the Others
DNA repair genes were downloaded from the GO (http://geneontology.org) term GO: 0006281. We obtained 450 DNA repair genes from the database, and 433 were found in our GA methylation data. We further compared the promoter methylation level of DNA repair genes with the others. First, 433 genes in the GA data, excluding 433 DNA repair genes, were randomly selected for 1,000 times, and then the promoter methylation level of 433 DNA repair genes was compared with each set of randomly selected genes using an unpaired t test. Second, we also compared the methylation level of DNA repair genes with those within other 10 GO terms, of which the biological processes play essential roles in carcinogenesis, including immune response (GO: 0006955), cell death (GO: 0008219), apoptotic process (GO: 0006915), angiogenesis (GO: 0001525), cell migration (GO: 0016477), cell development (GO: 0048468), morphogenesis (GO: 0000902), cell proliferation (GO: 0008283), secretion (GO: 0046903), and cell adhesion (GO: 0007155).
RPMB Calculation
The promoter methylation level of the 433 DNA repair genes was transformed into binary categories of “methylated” and “unmethylated,” with the threshold of the median β value (0.119) across the whole methylation profile of all the GA patients (hypermethylated when the value was >0.119, while hypomethylated when ≤0.0119). The RPMB for a patient was defined as the ratio of methylated DNA repair genes to the number of all of the DNA repair genes (n = 433).
Statistical Analysis
All statistical analyses in this research were conducted with R programming project software (version 3.6.1) and Bioconductor (version 3.9). Bioconductor annotation package org.Hs.eg.db (version 2.8.0) was used for gene retrieval within different GO terms.39 In the analysis of baseline characteristics, patients were divided into two equal-sized groups according to the median value of RPMB, and baseline characteristic of the two groups were compared using χ2 or Fisher’s exact tests for categorical variables (Table 1). Kaplan-Meier survival analysis was used to determine the OS difference between the two groups, which were also assigned based on the median value of RPMB, and a p value <0.05 was regarded as significant. We further estimated the treatment effects separately within levels of the factors and presented the results in two forest plots (OS and DFS, respectively), in order to explore potential relationships between different RPMB levels and variables. In the analysis of each factor, the patients were also divided based on the median value of RPMB, and Cox regression analysis was conducted to calculate the corresponding HR and 95% CI. The forest plots were carried out with R package rmeta (version 3.0).
Author Contributions
Conception and design: X.Y., N.A., Z.Y., H.-J.L., and Y.-C.Z. Collection and assembly of data: N.A., X.-J.H, Y.-Y.Z., and L.Y. Data analysis and interpretation: N.A., X.Y., X.-J.H., L.Y., and Y.-Y.Z. Manuscript writing: N.A., X.Y., Y.-C.Z., and Z.Y. Final approval of manuscript: all authors.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81802271 to N.A.), the National Natural Science Foundation of China (81801734 to X.Y.), and by the Natural Science Foundation of Shandong Province, China (ZR2019QH003 to X.Y.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.06.006.
Contributor Information
Yong-Chun Zhang, Email: zyc18661805058@163.com.
Hai-Jun Lu, Email: Lhj82920608@163.com.
Xue Yang, Email: yxue0409@outlook.com.
Supplemental Information
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Bonenkamp J.J., Hermans J., Sasako M., van de Velde C.J., Welvaart K., Songun I., Meyer S., Plukker J.T., Van Elk P., Obertop H., Dutch Gastric Cancer Group Extended lymph-node dissection for gastric cancer. N. Engl. J. Med. 1999;340:908–914. doi: 10.1056/NEJM199903253401202. [DOI] [PubMed] [Google Scholar]
- 3.Cuschieri A., Fayers P., Fielding J., Craven J., Bancewicz J., Joypaul V., Cook P., The Surgical Cooperative Group Postoperative morbidity and mortality after D1 and D2 resections for gastric cancer: preliminary results of the MRC randomised controlled surgical trial. Lancet. 1996;347:995–999. doi: 10.1016/s0140-6736(96)90144-0. [DOI] [PubMed] [Google Scholar]
- 4.Cuschieri A., Weeden S., Fielding J., Bancewicz J., Craven J., Joypaul V., Sydes M., Fayers P., Surgical Co-operative Group Patient survival after D1 and D2 resections for gastric cancer: long-term results of the MRC randomized surgical trial. Br. J. Cancer. 1999;79:1522–1530. doi: 10.1038/sj.bjc.6690243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ajani J.A., Lee J., Sano T., Janjigian Y.Y., Fan D., Song S. Gastric adenocarcinoma. Nat. Rev. Dis. Primers. 2017;3:17036. doi: 10.1038/nrdp.2017.36. [DOI] [PubMed] [Google Scholar]
- 6.Mari E., Floriani I., Tinazzi A., Buda A., Belfiglio M., Valentini M., Cascinu S., Barni S., Labianca R., Torri V. Efficacy of adjuvant chemotherapy after curative resection for gastric cancer: a meta-analysis of published randomised trials. A study of the GISCAD (Gruppo Italiano per lo Studio dei Carcinomi dell’Apparato Digerente) Ann. Oncol. 2000;11:837–843. doi: 10.1023/a:1008377101672. [DOI] [PubMed] [Google Scholar]
- 7.Earle C.C., Maroun J.A. Adjuvant chemotherapy after curative resection for gastric cancer in non-Asian patients: revisiting a meta-analysis of randomised trials. Eur. J. Cancer. 1999;35:1059–1064. doi: 10.1016/s0959-8049(99)00076-3. [DOI] [PubMed] [Google Scholar]
- 8.Hermans J., Bonenkamp J.J., Boon M.C., Bunt A.M., Ohyama S., Sasako M., Van de Velde C.J. Adjuvant therapy after curative resection for gastric cancer: meta-analysis of randomized trials. J. Clin. Oncol. 1993;11:1441–1447. doi: 10.1200/JCO.1993.11.8.1441. [DOI] [PubMed] [Google Scholar]
- 9.Sasako M., Sakuramoto S., Katai H., Kinoshita T., Furukawa H., Yamaguchi T., Nashimoto A., Fujii M., Nakajima T., Ohashi Y. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J. Clin. Oncol. 2011;29:4387–4393. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- 10.Bang Y.J., Kim Y.W., Yang H.K., Chung H.C., Park Y.K., Lee K.H., Lee K.W., Kim Y.H., Noh S.I., Cho J.Y., CLASSIC trial investigators Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379:315–321. doi: 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 11.Smalley S.R., Benedetti J.K., Haller D.G., Hundahl S.A., Estes N.C., Ajani J.A., Gunderson L.L., Goldman B., Martenson J.A., Jessup J.M. Updated analysis of SWOG-directed intergroup study 0116: a phase III trial of adjuvant radiochemotherapy versus observation after curative gastric cancer resection. J. Clin. Oncol. 2012;30:2327–2333. doi: 10.1200/JCO.2011.36.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J., Lim D.H., Kim S., Park S.H., Park J.O., Park Y.S., Lim H.Y., Choi M.G., Sohn T.S., Noh J.H. Phase III trial comparing capecitabine plus cisplatin versus capecitabine plus cisplatin with concurrent capecitabine radiotherapy in completely resected gastric cancer with D2 lymph node dissection: the ARTIST trial. J. Clin. Oncol. 2012;30:268–273. doi: 10.1200/JCO.2011.39.1953. [DOI] [PubMed] [Google Scholar]
- 13.Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan T., Jiao Y., de Jong S., Ophoff R.A., Beck S., Teschendorff A.E. An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet. 2015;11:e1004996. doi: 10.1371/journal.pgen.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Docherty S.J., Davis O.S.P., Haworth C.M.A., Plomin R., Mill J. DNA methylation profiling using bisulfite-based epityping of pooled genomic DNA. Methods. 2010;52:255–258. doi: 10.1016/j.ymeth.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Laird P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 17.Costello J.F., Plass C. Methylation matters. J. Med. Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baylin S.B. Tying it all together: epigenetics, genetics, cell cycle, and cancer. Science. 1997;277:1948–1949. doi: 10.1126/science.277.5334.1948. [DOI] [PubMed] [Google Scholar]
- 19.Akhavan-Niaki H., Samadani A.A. DNA methylation and cancer development: molecular mechanism. Cell Biochem. Biophys. 2013;67:501–513. doi: 10.1007/s12013-013-9555-2. [DOI] [PubMed] [Google Scholar]
- 20.De Carvalho D.D., Sharma S., You J.S., Su S.F., Taberlay P.C., Kelly T.K., Yang X., Liang G., Jones P.A. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deckers I.A., Schouten L.J., Van Neste L., van Vlodrop I.J., Soetekouw P.M., Baldewijns M.M., Jeschke J., Ahuja N., Herman J.G., van den Brandt P.A., van Engeland M. Promoter methylation of CDO1 identifies clear-cell renal cell cancer patients with poor survival outcome. Clin. Cancer Res. 2015;21:3492–3500. doi: 10.1158/1078-0432.CCR-14-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busche S., Ge B., Vidal R., Spinella J.F., Saillour V., Richer C., Healy J., Chen S.H., Droit A., Sinnett D., Pastinen T. Integration of high-resolution methylome and transcriptome analyses to dissect epigenomic changes in childhood acute lymphoblastic leukemia. Cancer Res. 2013;73:4323–4336. doi: 10.1158/0008-5472.CAN-12-4367. [DOI] [PubMed] [Google Scholar]
- 23.Choudhury J.H., Ghosh S.K. Promoter hypermethylation profiling identifies subtypes of head and neck cancer with distinct viral, environmental, genetic and survival characteristics. PLoS ONE. 2015;10:e0129808. doi: 10.1371/journal.pone.0129808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerson S.L. MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 25.Kaina B., Christmann M., Naumann S., Roos W.P. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst.) 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups. National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 27.Paz M.F., Avila S., Fraga M.F., Pollan M., Capella G., Peinado M.A., Sanchez-Cespedes M., Herman J.G., Esteller M. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in normal tissues and human primary tumors. Cancer Res. 2002;62:4519–4524. [PubMed] [Google Scholar]
- 28.Schneider B.G., Peng D.F., Camargo M.C., Piazuelo M.B., Sicinschi L.A., Mera R., Romero-Gallo J., Delgado A.G., Bravo L.E., Wilson K.T. Promoter DNA hypermethylation in gastric biopsies from subjects at high and low risk for gastric cancer. Int. J. Cancer. 2010;127:2588–2597. doi: 10.1002/ijc.25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.House M.G., Herman J.G., Guo M.Z., Hooker C.M., Schulick R.D., Lillemoe K.D., Cameron J.L., Hruban R.H., Maitra A., Yeo C.J. Aberrant hypermethylation of tumor suppressor genes in pancreatic endocrine neoplasms. Ann. Surg. 2003;238:423–431. doi: 10.1097/01.sla.0000086659.49569.9e. discussion 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schagdarsurengin U., Gimm O., Dralle H., Hoang-Vu C., Dammann R. CpG island methylation of tumor-related promoters occurs preferentially in undifferentiated carcinoma. Thyroid. 2006;16:633–642. doi: 10.1089/thy.2006.16.633. [DOI] [PubMed] [Google Scholar]
- 31.Belanich M., Pastor M., Randall T., Guerra D., Kibitel J., Alas L., Li B., Citron M., Wasserman P., White A. Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res. 1996;56:783–788. [PubMed] [Google Scholar]
- 32.Jaeckle K.A., Eyre H.J., Townsend J.J., Schulman S., Knudson H.M., Belanich M., Yarosh D.B., Bearman S.I., Giroux D.J., Schold S.C. Correlation of tumor O6 methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: a Southwest Oncology Group study. J. Clin. Oncol. 1998;16:3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- 33.Esteller M., Garcia-Foncillas J., Andion E., Goodman S.N., Hidalgo O.F., Vanaclocha V., Baylin S.B., Herman J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 34.Hegi M.E., Diserens A.C., Godard S., Dietrich P.Y., Regli L., Ostermann S., Otten P., Van Melle G., de Tribolet N., Stupp R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 35.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 36.Hegi M.E., Diserens A.C., Gorlia T., Hamou M.F., de Tribolet N., Weller M., Kros J.M., Hainfellner J.A., Mason W., Mariani L. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 37.Tournigand C., André T., Bonnetain F., Chibaudel B., Lledo G., Hickish T., Tabernero J., Boni C., Bachet J.B., Teixeira L., de Gramont A. Adjuvant therapy with fluorouracil and oxaliplatin in stage II and elderly patients (between ages 70 and 75 years) with colon cancer: subgroup analyses of the Multicenter International Study of Oxaliplatin, Fluorouracil, and Leucovorin in the Adjuvant Treatment of Colon Cancer trial. J. Clin. Oncol. 2012;30:3353–3360. doi: 10.1200/JCO.2012.42.5645. [DOI] [PubMed] [Google Scholar]
- 38.An N., Yang X., Cheng S., Wang G., Zhang K. Developmental genes significantly afflicted by aberrant promoter methylation and somatic mutation predict overall survival of late-stage colorectal cancer. Sci. Rep. 2015;5:18616. doi: 10.1038/srep18616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson M. 2013. org.Hs.eg.db: Genome wide annotation for human. R package version 2.8.0. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.