Abstract
Post-transcriptional RNA modification occurs on all types of RNA and plays a vital role in regulating every aspect of RNA function. Thanks to the development of high-throughput sequencing technologies, transcriptome-wide profiling of RNA modifications has been made possible. With the accumulation of a large number of high-throughput datasets, bioinformatics approaches have become increasing critical for unraveling the epitranscriptome. We review here the recent progress in bioinformatics approaches for deciphering the epitranscriptomes, including epitranscriptome data analysis techniques, RNA modification databases, disease-association inference, general functional annotation, and studies on RNA modification site prediction. We also discuss the limitations of existing approaches and offer some future perspectives.
Keywords: Epitranscriptome, RNA modification, Bioinformatics approaches, Recent progress, Future perspective
1. Background
Post transcriptional RNA modification occurs on all types of RNA and plays a vital role in regulating every stage of RNA life. More than 170 different types of RNA modifications have been identified, and the majority of them are methylation modification. Ribonucleoside modification contains many chemical components that are added to A, G, C or U. Some modifications come from non-enzymatic processes or oxidative damage. Others are RNA editing, which was originally described as a process of adding polyuridine residues to selected RNAs coding regions. Since then, editing has expanded to include the removal or addition of RNA bases, although further differences remain somewhat inconsistent. There are 111 modifications that can be found in tRNAs, 33 in rRNAs, 17 in mRNA, 11 in lncRNAs and other noncoding RNAs [1], [2].
However, due to the lack of effective means to detect RNA methylation, the research on RNA methylation had long been stagnant. For a long time, researchers had considered RNA modification as a mechanism of fine-tuning gene expression regulation, and mostly limited to noncoding tRNA and rRNA. The importance of RNA modifications was not fully aware of until the discovery of human fat mass gene FTO as a RNA m6A demethylase [3] and the invention of a transcriptome-wide m6A profiling approach MeRIP-seq (or m6A-seq) [4], [5]. These techniques give us a global view of mRNA modification by providing detailed maps of 12 modifications that can be incorporated into the transcriptome, including N6-methyladenosine (m6A), pseudouridine (ψ), N4-acetylcytidine (ac4C), N1-methyladenosine (m1A), N7-methylguanosine (m7G), 2′O-methylations (Cm, Am, Gm, Um), 5-methylcytidine (m5C), 5-hydroxymethylcytidine Cytidine (hm5c) and inosine (I). Please refer to two recent reviews in this perspective [1], [2].
N6-methyladenosine (m6A) is the most abundant and the most well studied chemical modification on eukaryotic mRNA [6], whose changes in level (and the consequent biological effects) are mediated by methyltransferase (writer), demethylases (eraser) and recognition proteins (reader) [7]. The methylation is catalyzed by the protein complexes of writer (including mainly METTL3, METTL14 and WTAP as well as KIAA1429, RBM15 and RBM15B) and is de-methylated by the demethylases such as ALKBH5 [8] and FTO [9]. Quite a few proteins involved in the formation of the reader complexes that specifically recognized m6A sites have been discovered. These include YTH family proteins (YTHDF1-3, YTHDC1) [10], [11], [12], [13], transcription initiation complex eIF3 [14], ribonucleoprotein HNRNPA2B1 [15] and HNRNPC [16]. Tens of thousands of m6A sites have been identified in the transcriptome, suggesting that this modification may have a wide-ranging effect on gene expression regulation [17]. For example, mediated gene silencing on the X chromosome by the long non-coding RNA X inactive specific transcripts (XIST) through YTHDC1 is due to the recognition of the m6A sites [18]. In addition, m6A can also affect translation extension by influencing the anticodon pairing rate and fidelity of both mRNA and tRNA [19]. Furthermore, m6A modification is also involved in the gene regulation of histone modifications [20] by recruiting CCR4-NOT complex which promoted degradation of targeted RNA after YTHDF2 binding to m6A site [11], [21]. The latter increases cap-independent translation under UV radiation or heat shock through the binding of the transcription initiation complex eIF3 binding to m6A sites in the 5′UTR region [22]. There are many reported functions of m6A modifications, including but not limited to, promoting the learning and memory capability of mice by m6A-mediated binding of the protein YTHDF1 to mRNA [23] and regulating the clearance of the mRNAs that affect embryonic development in zebrafish through YTHDC2 protein recognition of m6A sites on mRNAs [24]. In living organisms, enhancement of microRNA maturity can be achieved by m6A methylation of pri-miRNA [25]. Meanwhile, this kind of modification is also important for maintaining methyl-donor-S-adenosylmethionine (SAM) levels [26]. Inhibition of RNA methylation by reducing m6A demethylase can disrupt the circadian clock and extend the circadian clock cycle, while overexpression of demethylase shortens the cycle [27]. This is also supported by other studies that extensive mRNA stabilization occurs in cells, especially those mRNAs that encode proteins related to the circadian clock [28]. The m6A-mediated regulation of mRNA stability also plays important roles in stem cell differentiation [29], [30], [31]. Importantly, various changes in m6A methylation are presented in a large number of mRNAs encoded by genes associated with human diseases, especially cancers [32], suggesting that this modification could be targeted as the biomarkers for disease diagnosis or interference, for instance, in viral infection [33], cancer [34], [35], [36], acute myeloid leukemia [37], T Cell-related diseases [38] and certain brain disorders, such as autism, Alzheimer's disease and schizophrenia [39]. Inhibition of the activity of fat and obesity-related protein (FTO) by R-2-hydroxyglutaric acid (R-2HG) increases the overall m6A level in R-2HG sensitive leukemia cells which in turn decreases the stability of MYC/CEBPA transcripts, leading to suppression of MYC/CEBPA-related signaling pathways and thus inhibition of the proliferation of FTO-highly expressed cancer cells [40].
5-methylcytosine (m5C) is a wild-spread post-transcriptional RNA modification that has been detected in rRNA, tRNA, mRNA, lncRNA, viral RNA and etc. [41], [42], [43]. In mammals, m5C was primarily catalyzed by RNA methyltransferase DNMT2 and NSUN2 along with its homologs [44], [45], [46]. Likewise, the m5C marks on RNAs formed by NSUNs can contribute to promoting the overall protein synthetic level in cells in terms of protecting tRNA from shearing or promoting the formation of ribosomes [47], [48]. Another study indicated that the deficiency of NSUN2 in T cells may result in the loss of the m5C addition on the HIV-1 mRNA and perturb the translation of HIV-1 mRNA by inhibiting the recruitment of ribosomes and alternative splicing of viral RNA [49]. In human, YBX1 has been identified as a novel “reader” of m5C modification, preferentially recognizing methylation C by its cold shock domain (CSD) [50] and contributing to promote the maintenance, proliferation and differentiation of adult stem cells [51]. m5C deposition on RNAs will vary their stability among different RNA species. It may prevent mRNA decay though the binding of YBX1 with mRNA stabilizer PABPC1A [50] and another example proposed mRNA stability could be ensured by NSUN2 and YBX1, driving the pathogenesis of human bladder urothelial carcinoma [52]. While recently it has been reported that viral ncRNA level increased followed by the loss of m5C modification, which was mediated by the ablation of NSUN2 [53]. Latest studies in this year have related m5C modification with the regulation of the thermal adaptability in animal and plant cells by regulating the functions of tRNA and mRNA [54], [55]. A well-described hydroxymethylcytosine (hm5C) modification in DNAs was also observed in RNAs, which is oxidized form of m5C exhibiting important physiological roles in drosophila [56].
Adenosine-inosine (A-to-I) RNA editing is the main form of RNA editing in mammals [57], mediated by the members of the adenosine deaminase acting on RNA (ADAR) enzyme family, which will hydrolyze adenosine to inosine, first discovered in 1987 [58]. Guaranteed by the development of deep sequencing and advances in bioinformatics, 4.5 million A-to-I RNA editing sites have been confirmed by high throughput screening, either in the coding or non-coding region [59]. Due to the chemical properties of inosine are similar to guanosine, it will complement with cytidine to form base pairs [60]. This nucleotide conversion may affect gene expression, regulation and functions, in terms of the changes of amino-acid sequence, the process of miRNA expression and maturation, depending on the areas where the RNA editing events occur [61], [62]. Studies have showed that the dysregulation of A-to-I RNA editing in transcriptome or abnormal expression of ADAR is associated with various diseases, including neurological diseases, immune disease, cancer, viral infections and etc. For example, A-to-I RNA editing in a subunit of glutamate receptor 2 (GluR2) is essential for the survival of motor neurons, hence the inhibitors targeting non-A-to-I RNA editing receptors may serve as an additional tool for the treatment of amyotrophic lateral sclerosis (ALS) [63]. A-to-I RNA editing on some non-coding sites is believed to have some key roles for innate immunity [64], [65]. For instance, A-to-I RNA editing activities have a significant diversity in response to different subtypes of influenza A virus in human epithelial cells [66]. A latest example found the regulatory role of the A-to-I RNA editing in the post-transcriptional control of rheumatoid arthritis (RA), which could be used in clinical treatment as a therapeutic target [67]. During the occurrence and development of human tumors, the A-to-I RNA editing level on some oncogenes and tumor suppressor genes also been found in disorder, but either high or low is contingent on the specific cancer type [58], [68], [69]. A study indicated that inhibition of ADAR1 significantly inhibited proliferation, invasion and migration in a thyroid tumor cell model [70]. Another research proved that A-to-I RNA editing can contribute to the proteome diversity of breast cancer by the changes of amino acid sequence [71].
Our knowledge of RNA modification is continuing to expand rapidly. In addition to the widely studied m6A methylation, pseudouridine (ψ) is one of the most abundant [2] and extensively studied types of transcriptome modification of RNAs in living organisms. It is formed by isomerization of uracil nucleoside (U). The isomerization of uracil and pseudouracil is catalyzed by PUS enzymes alone or together with H/ACA ribonucleoproteins [72]. Ψ has a key function in guiding the process of protein translation in stem cells. It is of great potentials for the treatment of stem cell related diseases, such as human myelodysplastic syndrome [73]. Pseudouracil also reduces RNA conformational variability, enhances base pairing stability and polar interactions with proteins, and thereby regulating mRNA stability and gene expression [74]. It has been reported that pseudouracil nucleoside (ψ) is involved in heat shock response from yeast, but its molecular mechanisms remains unclear [75]. Several new types of RNA modifications have also been reported recently. For example, a well-described hydroxymethylcytosine (hm5C) modification in DNAs was also observed in RNAs, exhibiting important physiological roles in drosophila [56]. N4-acetylcytidine (ac4C) is revealed as a mRNA modification catalyzed by the acetyltransferase NAT10 [76]. N6,2′-O-dimethyladenosine (m6Am) in mRNA is another type of reversible methylation, and its modification status in the 5′ cap influences stability of cellular mRNAs [77], [78]. N1-methyladenosine (m1A) methylation is a newly discovered reversible epigenetic modification that can be de-methylated by RNA repair enzyme ALKBH1 [79], but there not have been identified yet about m1A methylation enzymes and its recognition proteins. Unlike m6A, m1A has even lower abundance, predominantly distributed in the 5′UTR region of mRNA which may be involved in regulating translation initiation process [80]. At last, 2′-O-methylation (Nm) of viral own genomes, such as HIV-1 and WNV RNA viruses, may help their escape from host innate immune responses [81], [82]. The above reviewing has that RNAs undergo a number of chemical modifications, which play various critical roles in physiological and pathological processes in nearly all organisms.
Thanks to the development of sequencing technologies, transcriptome-wide profiling of RNA modifications has been made possible by a number of different techniques such as MeRIP-seq and RNA BS-seq. With the accumulation of large amount of high-throughput datasets, bioinformatics approaches have been increasingly needed for unraveling the epitranscriptome as a cost effective avenue. We systematically reviewed in the following the emerging topics and recent progress in bioinformatics approaches for deciphering the epitranscriptomes, including epitranscriptome data analysis techniques, RNA modification databases, disease-association inference, general functional prediction, and RNA modification site prediction methods (especially deep learning approaches). The review is divided into the following sections. We firstly introduced the tools for epitranscriptome data analysis as well as some epitranscriptome profiling technologies. Then we summarized the algorithms for methylation sites prediction and the existing databases dedicated for RNA modifications. We in the next analyzed the disease marker and association prediction related to RNA modifications. In the end, we briefly discussed the limitations of existing epitranscriptiome bioinformatics approaches and offered some possible future perspectives (see Fig. 1).
Fig. 1.
The emerging topics in epitranscriptome bioinformatics.
2. Tools for epitranscriptome data analysis
With the advances in next-generation sequencing approaches, many experimental methods have been designed to profile various types of RNA modified nucleotides. Meanwhile, a number of computational programs were developed for the analysis of the massive high-throughput sequencing data generated. We briefly reviewed here a few software tools developed for epitranscriptome profiling data generated from MeRIP-Seq, RNA BS-Seq, etc.
2.1. MeRIP-Seq (or m6A-Seq)
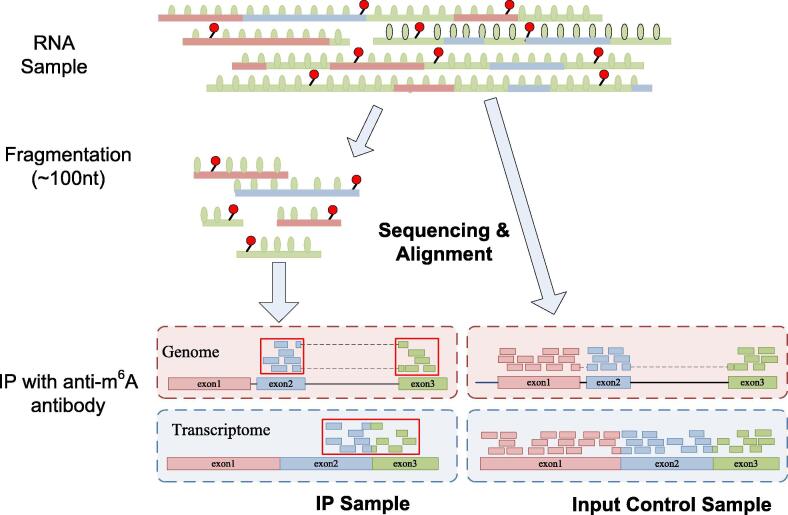
Methylated RNA Immunoprecipitation sequencing (MeRIP-Seq or m6A-Seq) [4], [5] is so far the most widely adopted experimental approach for profiling the transcriptome-wide distribution of RNA modification (see Fig. 2). Considered as a marriage of ChIP-Seq and RNA-Seq, it is possible to infer from the data generated from this antibody-based approach the location of RNA modification (peak calling) as well as the changes in methylation status between two different biological conditions (differential methylation analysis). Additionally, site clustering analysis of epitranscriptome data from multiple biological conditions may reveal co-methylation patterns, which may provide insights into the regulatory mechanisms of the epitranscriptome by relevant protein regulators (Writers, Erasers, and Readers). These perspectives are reviewed as follows:
Fig. 2.
Illustration of MeRIP-Seq Protocol. In MeRIP-Seq, two types of samples (IP and control samples) are generated. In the beginning of the protocol, RNA molecules are firstly sheared into fragments of around 100 nt. Through anti-m6A antibody, the IP sample provides unbiased measurement of the methylated RNA fragments; the control sample reflects the basal RNA abundance and is used as a negative control, by comparing to which, the peaks (or methylated sites) can be identified. The exomePeak approach seeks to identify enriched regions on pooled exons so that a single peak may not be split into multiple enriched regions on the genome.
2.1.1. Peak Calling (or Site Detection)
MeRIP-Seq immunoprecipitates the RNA fragments containing the modification with anti-m6A polyclonal (SYSY, RN131P) or monoclonal (17-3-4-1) antibodies in randomly interrupted RNA fragment library to construct the IP sample, so the segments carrying the methylation mark are over-represented in the IP samples compared with the input control sample, which can be viewed as the standard RNA-Seq library. Mapping the reads of IP sample to the reference genome will form a peak of reads coverage near the modification sites. Through the statistical analysis of IP over input enrichment on the genomic sliding windows, we can infer the locations of m6A modification sites across the genome or exome. Several computational tools have been developed for site detection (peak calling) from MeRIP-seq data (see Table 1), and among them, MACS [83] has been a popular peak calling tool originally developed for analyzing ChIP-Seq data, but it is also frequently used to process MeRIP-Seq data by many published studies [84]. Another widely applied software tool is exomePeak, which was specifically designed for epitranscriptome peak calling of MeRIP-Seq data [85], with a major update released recently (exomePeak2). The major advancement of exomePeak2 compared with MACS is its ability to account for the technical and biological variabilities that are predominant in RNA-Seq compared with DNA-Seq [86]. For example, exomePeak2 implements the functionality of sequence content bias correction. Consequently, it could dramatically reduce the systematic errors generated by PCR amplification during the library preparation of IP and input samples.
Table 1.
Site prediction tools from MeRIP-seq data.
| Tool | Input format | Description | URL/stand-alone package | Reference |
|---|---|---|---|---|
| MeRIP-PF | FASTQ | MeRIP-PF segments the reference genome by a fixed window, then compares reads count between control sample and IP sample to obtain p-value and adjusted p-value by Fisher’s exact test and Benjamini-Hochberg method. In order to form real peaks, the significant and adjacent windows are concatenated. | http://software.big.ac.cn/MeRIP-PF.html | [177] |
| BaySeqPeak | Reads count matrix | A Bayesian hierarchical model is proposed to detect methylation sites from MeRIP-Seq data in BaySeqPeak. By using zero expansion negative binomial model combined with hidden Markov model, the spatial dependence of enrichment of adjacent reading segments is explained, which has better stability under the condition of small samples. | https://github.com/liqiwei2000/BaySeqPeak | [178] |
| MACS2 | BAM | Use a Poisson model to detect peaks. It was originally developed for ChIP-seq data analysis, but has also been quite popular in MeRIP-seq data analysis for its reliability, speed and convenience. | https://github.com/taoliu/MACS/ | [83] |
| exomePeak | BAM | Exomepeak uses przyborowski and wilenski methods to compare the mean value (or C-test) of two Poisson distributions. It can detect the peak across the exon connection region of a specific gene exon set. The version 2 (exomePeak2) was released very recently. | https://rdrr.io/github/ZhenWei10/exomePeak2/ | [85] |
| m6AViewer | BAM | m6AViewer constrains the number of read segments and the width of the peak, and uses EM algorithm to find the most possible m6A methylation peak. | http://dna2.leeds.ac.uk/m6a | [179] |
| MeTPeak | BAM | MeTPeak models reads count, introduces the layer with beta variable to obtain the variance, and uses the hidden Markov model to describe the reading dependency near the site. | https://github.com/compgenomics/MeTPeak | [180] |
2.1.2. Differential methylation analysis
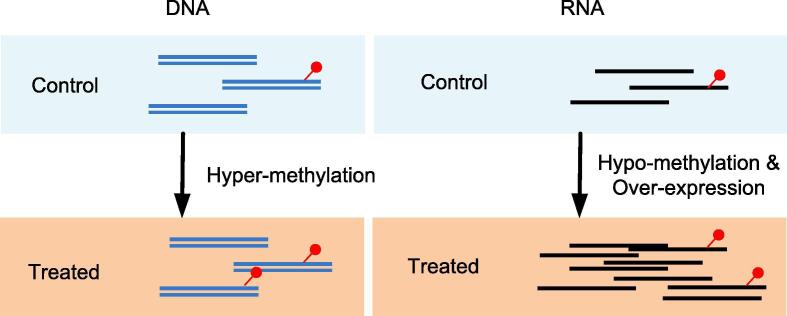
Differential methylation analysis focuses on the changes of modification status between two different biological conditions. Although changes in absolute abundance of methylation may be equally important, current formulation overwhelmingly focused on the relative abundance or methylation proportion, specifically, the ratio of methylated to total RNAs, as previously modeled by [87]. Different from DNA differential methylation, for differential RNA methylation analysis, it is important to account the changes in basal expression levels of the RNA transcripts, especially for the relative abundance comparison (See Fig. 3). We summarized the existing methods for RNA differential methylation in Table 2. Although classic approaches based on Fisher’s exact test (realized in the exomePeak R package) [87] has been quite popular in differential methylation analysis for its modeling robustness and implementation easiness, recent published methods such as RADAR [88] and QNB [89] have achieved higher accuracy through specifying refined statistical models.
Fig. 3.
RNA methylation and DNA methylation. Compared with the control group, both the absolute and relative amount of methylated DNA in the treatment group increased under the treated condition; however, for the two may not be consistent for RNA methylation. In the above example, the total amount of methylated RNA increased in the treatment group; however, due to the increased expression level (over-expression), the relative amount of methylated RNA decreased (hypo-methylation).
Table 2.
Differential methylation analysis tools from MeRIP-seq data.
| Tool | Input format | Description | URL/stand-alone package | Reference |
|---|---|---|---|---|
| exomePeak | BAM | The original exomePeak uses a rescaled version of Fisher’s exact test to detect differential methylation sites. The latest version of exomePeak2 uses a generalized linear model which considers the over-dispersion of reads count and GC content bias in sequencing data. | https://rdrr.io/github/ZhenWei10/exomePeak2/ | [87] |
| FET-HMM | BAM | FET-HMM divides the detected RNA methylation site into small bins and uses a hidden Markov model to detect differential methylation sites. | https://github.com/lzcyzm/RHHMM | [181] |
| MeTDiff | BAM | MeTDiff for differential methylation sites models reads variations with beta-binomial model. Then a likelihood ratio test based on the beta-binomial is developed to test the significance of differential methylation sites. | https://github.com/compgenomics/MeTDiff | [182] |
| RADAR | Reads count matrix | RADAR enables accurate identification of altered methylation sites by accommodating variability of pre-immunoprecipitation expression level and post-immunoprecipitation count using different strategies. | https://github.com/scottzijiezhang/RADAR | [88] |
| DRME | Reads count matrix | DRME aims at RNA differential methylation in small samples. It uses two independent negative binomial distributions to model the reads count in the methylation region, and uses two-dimensional local regression to estimate variance for solving the effect of transcription regulation, and carrying out RNA differential methylation analysis combined within-group biological variability difference of biological replication samples. | https://github.com/lzcyzm/DRME | [183] |
| QNB | Reads count matrix | QNB is based on four independent negative binomial distributions with the variances and means for reads count of the input control samples and IP samples, and linked by local regressions. QNB combined information from both input and IP samples to estimate gene expression, which could improve the testing performance for lowly expressed genes. | https://cran.rstudio.com/web/packages/QNB/ | [89] |
2.1.3. Clustering analysis of m6A sites (peaks)
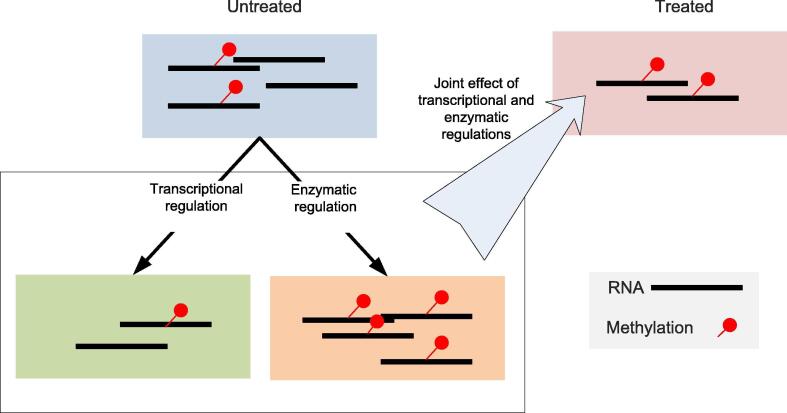
Epitranscriptome serve as an important layer of post-transcriptional regulation, and the count-based quantification in MeRIP-Seq data could potentially shed light on the mechanics of conditional specific regulation through RNA modification. Although the transcript specific regulatory mechanism of the epitranscriptome is still unclear, the clustering partition of the methylation sites on distance metric evaluated through the conditional specific methylation profiles may associate with the targeting of RNA modification regulators (Writers, Readers, Erasers) (See Fig. 4). Some relevant works in the field were summarized in Table 3. Although classical clustering algorithm such as hierarchical clustering and K-means clustering can be reasonably efficient under homogenous laboratory conditions [90], recent developments on technical independent quantification method could fundamentally improve the quality of the clustering partition after the stratification of the major technical variables from the methylation level estimates.
Fig. 4.
The regulation of RNA methylome. The dynamics in epitranscriptome are a result of a joint effect of both transcriptional and enzymatic regulations. On the one hand, transcriptional regulation directly changes the amount of RNA molecules and leads to coordinated changes in the absolute amount of methylated molecules, leaving the relative amount unchanged. On the other hand, enzymatic regulation of the RNA methylome by ‘methylation potential’ changes directly the percentage of methylated molecules. For the above illustration, under the joint effects of transcriptional down-regulation and enzymatic hypermethylation, the absolute amount of methylated RNA stays unchanged.
Table 3.
Summary of the studies for m6A methylation clustering.
| Method | Input format | Description | URL/stand-alone package | Reference |
|---|---|---|---|---|
| MeTCluster | BAM | MeTCluster, a novel algorithm and an open source R package, models the reads count variance and the underlying clusters of the methylation peaks by a hierarchical graphical model. It is evaluated on both simulated and real MeRIP-Seq datasets. | http://compgenomics.utsa.edu/metcluster | [87] |
| Binary Clustering | M-value | Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites | [184] | |
| Four clustering methods | M-value | Four different clustering approaches are used, including K-means, hierarchical clustering (HC), Bayesian factor regression model (BFRM) and nonnegative matrix factorization (NMF) to unveil the co-methylation patterns in epitranscriptome. | [185] | |
| Threshold-Based Measurement Weighting | Reads count matrix | A convenient measurement weighting strategy that can largely tolerate the artifacts of high-throughput sequencing data. | [186] | |
| DPBBM | Reads count matrix | DPBBM implements a beta-binomial model, which uses the original measurement value based on count instead of the estimation value to capture the clustering effect on the methylation level. In addition, the nonparametric Dirichlet process is used to automatically determine the optimal number of clusters, which avoids the common problem of model selection in cluster analysis. | https://cran.r-project.org/web/packages/DPBBM/ | [187] |
2.1.4. Quality assessment
The quality of MeRIP-seq data can be evaluated by the conventional 2nd generation sequencing quality control pipeline such as FASTQC. Recently, trumpet R package is developed for MeRIP-Seq specific quality examination [91]. The trumpet package takes the aligned BAM files as the inputs and returns an assessment report with a single line of R command concerning the quality statistics of sequencing reads distribution, the strength of the immunoprecipitation signal, and the comparison between different biological replicates. After the systematic examination of the published MeRIP-seq experiments, we observed substantial amount of technical biases in a significant proportion of the published MeRIP-seq samples. The technically effected samples should be handled carefully through the quantification methods independent of the source of errors while performing any downstream data analysis tasks.
2.2. Reverse transcription signature in sequencing
The combination of Reverse Transcription (RT) and high-throughput sequencing has emerged as an effective approach for identification of RNA modification through the analysis of the misincorporation during complementary DNA (cDNA) synthesis or abortive RT-products [92]. Before the advent of direct RNA sequencing technique nanopore [93], the modified RNA templates were reversed transcribed into cDNA to generate RNA-seq data. As the newly synthesized cDNA contains only four types of canonical deoxynucleotides, this process may lead to the partially or completely loss of information about RNA modifications stored on the original RNA templates [94]. To solve this problem, some chemical reagents were used to specifically react with a target modification, which in return alter the cDNA synthesis at modified RNA sites, i.e., the use of N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide-metho-p-toluenesulfonate (CMC) to leave a bulky group on pseudouridine modification and stop reverse transcription [74]. Besides, other RNA modifications with chemical groups added on their Watson-Crick face do not need chemical derivatization to alter cDNA synthesis. For example, the m1A RNA modification has a methyl group on the Watson-Crick face of adenosine, which resulted in cDNA synthesis differing from that of an unmodified adenosine on the RNA template. For the RNA templates containing m1A modification, the products of transcription arrest were presented in the synthesized cDNA, i.e., incorporation of mismatched dNTPs at modification position and abortive cDNA fragments, and this erroneous information generated during cDNA synthesis was termed as reverse transcription signatures. The signals of RT arrest were traditionally detected by either capillary electrophoresis or polyacrylamide gelelectrophoresis (PAGE) [95] before the development of deep sequencing methods. As the product of an A-to-I deamination, inosine is reversed transcribed for a cytidine rather than a thymidine during cDNA synthesis. And the first transcriptome-wide mapping of a RNA modification was benefited from this model misincorporation [96]. Also, combined analyses of both mismatch patterns plus defined RT arrest rate were performed to efficient identify 1-methyladenosine (m1A) modification [92]. The bioinformatics tools used for reverse transcription signature analysis were listed in Table 4.
Table 4.
Summary of the reviewed tools for RT signature analysis.
| Tool | Input format | Description | URL/stand-alone package | Reference |
|---|---|---|---|---|
| Coverage Analyzer (CAn) | SAM | CAn is a tool that offers the functions of inspection and visualization of deep sequencing data to identify RNA modification, which combines a pipeline to process data with flexible controls for differential or independent visualization and systemically screening for modification candidates using RT signatures. | https://zenodo.org/record/164811 (doi:https://doi.org//10.5281/zenodo.164811) or https://sourceforge.net/projects/coverageanalyzer/ | [188] |
| HAMR | BAM | HAMR is a tool that allows fast identification of RNA modification at single-nucleotide resolution using the nucleotide substitutions identified from RNA-seq datasets, which scans modification candidates either transcriptome-wide or specific locations by interested genomic coordinates. | http://wanglab.pcbi.upenn.edu/hamr | [189] |
| Galaxy modification calling pipeline | FASTQ | A modification calling pipeline based on Galaxy, which provides a versatile graphical workflow system for modification sites calling based on machine learning. The machine learning module in downstream analysis offers quality assessment parameters to help to improve the experimental parameters for both library preparation and sequencing. | https://github.com/HelmGroup | [94] |
2.3. RNA bisulfite sequencing
5-Methylcytosine (m5C) is a type of chemical modification on the carbon 5 atom of cytosine, which can be detected by high-throughput sequencing of RNA treated by bisulfite (RNA Bisulfite sequencing) that converts all unmodified cytosine into uracil leaving modified cytosine (m5C) unaffected [97]. Although with some intrinsic bias, it has been considered as the gold standard for profiling m5C epitranscriptome [98], [99]. One of the primary advantages of RNA Bisulfite sequencing in detection of modified cytosine (m5C) is that a transcriptome-wide view of m5C modification at single-based resolution can be provided. It is worth noting that, although both 5-methylcytosine and 5-hydroxymethylcytosine are resistant to deamination and thus can not be differentiated by bisulfite sequencing, the extremely low level of 5-hydroxymethylcytosine in human and mouse mRNAs [100] still makes RNA Bisulfite sequencing to become a robust and attractive technique for profiling m5C epitranscriptome. Several toolkits support the analysis of data generated from RNA Bisulfite sequencing (Table 5). It is important to note that a number of remedies have been taken to eliminate false positive sites reported from RNA Bisulfite sequencing technique, including statistical methods, excluding low quality reads, filtering of bisulfite conversion-resistant sites and SNPs, etc. [68], which are often necessary.
Table 5.
Summary of the reviewed tool kits to process data generated from RNA-BisSeq technique.
| Tool | Aligner | Description | Programlanguage | URL/stand-alone package | Reference |
|---|---|---|---|---|---|
| meRanTK | meRanT: Bowtie2 meRanG: STAR or TopHat2 |
meRanTk contains five multithreaded programs including meRanT, meRanG, meRanCall, meRanCompare, and meRanAnnotate, which is the first publicly available tool for high-throughput RNA cytosine methylation data analysis. | Perl | http://icbi.at/software/meRanTK | [104] |
| BS-RNA | HISAT2 | BS-RNA features in its ability to map ‘dovetailing’ reads using BEERS, compared with pervious published tool meRanTK. | Perl | http://bs-rna.big.ac.cn | [190] |
| BisRNA | BSMAP | BisRNA provides a computational tool that features in combining data-driven statistics modeling and tailored filtering together to reduce possible artifact introduced by bisulfite sequencing. | R | https://cran.r-project.org/web/packages/BisRNA | [191] |
| Episo | Bowtie | Episo is the only computational tool available to quantify the RNA m5C modification at the transcript isoform level, which distinguish m5C level between isoforms of the same gene. | GNU GPLv3 + license | https://github.com/liujunfengtop/Episo | [192] |
2.3.1. Quality control of raw RNA Bisulfite sequencing data
In the process of sodium bisulfite interaction and cDNA conversion, unmodified cytosine in mRNA will end up as thymine, and the GC contend is extremely low in mRNA Bisulfite sequencing data. Therefore, quality control of bisulfite sequencing reads should be implemented, low quality bases and adaptor sequences should be trimmed off from the raw data. Software tool such as Trimmomatic [101] can be used in this regard.
2.3.2. Alignment of RNA bisulfite sequencing reads
Reads generated from bisulfite sequencing can be mapped to either a reference transcriptome or an annotated genome, using aligners such as Bowtie2 [102] and HISAT2 [103]. For the alignment of bisulfite sequencing reads to a reference transcriptome, the longest transcript with the highest aligned score were considered, when it comes to issue that reads may be mapped to multiple transcripts of the same gene [104]. To increase the overall mapping rates of bisulfite sequencing data, sequencing reads may be aligned to an annotated genome first, and then a reference transcriptome can be used for further alignment against unmapped reads [105].
2.3.3. Methylation calling and elimination of false positive sites
To avoid the detection of false positive sites, strict filters and statistical methods were applied during methylation calling process [105], [106], [107], [108], along with the selection of bisulfite conversion-resistant bases using RNA secondary structure prediction tools [107]. Besides, the m5C methylation candidates were further filtered to remove those sites that overlapped with known genetic variants and RNA editing sites [109], using databases such as dbSNP [110] and REDIdb [111]. Several mRNA methylation studies have applied strict filtering pipelines to reduce false positive detection. Reads filters with strict criteria were set for the first step, this helps to remove PCR duplicates [107], [108] and reads with high level of unconverted cytosine rate [105], [106]. Sites filters were then applied for coverage depth, methylation level, base quality, and false discover rate (FDR). Furthermore, other filtering criteria were also implemented by different methylation studies to further evaluate candidate methylation sites, e.g., the candidate methylation sites should be detected in at least two biological replicates, or excluding 10 and 7 bases on the 5′ end of forward and reverse reads from methylation calling, respectively [107].
2.4. Other tools for RNA modification analysis
Besides the toolkits used for processing high-throughput sequencing data mentioned above, a number of downstream computational methods have been developed to effectively facilitate the analysis, annotation and exploration of RNA methylation data.
An open R/Bioconductor package Guitar [112] was developed to profile the transcriptomic view of RNA-related biological features represented by genome-based coordinates, which extracts the RNA coordinates relating to the landmarks of RNA transcripts and contributes to the efficiently analysis of massive amount of RNA-related biological features. RNAModR and MetaPlotR [113] were designed with similar purpose. RNAModR serves as an R-based package for the transcriptome-wide analysis and visualization of distribution of mRNA modifications, revealing the potential insights into the biological functions of these modified nucleotides. MetaPlotR was developed as a simple pipeline to help biologists with little bioinformatics knowledge to generate metagene plots of RNA modifications, protein binding sites, etc. A web application txCoords [114] can be used for peak re-mapping, therefore corrects the wrong labeled peaks and retrieves the true sequences.
RNAmod (https://bioinformatics.sc.cn/RNAmod) [115] is an interactive, one-stop, web-based platform for the automated analysis, annotation, and visualization of mRNA modifications in 21 species with 7 kinds of RNA modifications. RNAmod firstly extracts gene features (sequence length, GC content et.al) from the reference genome annotation, and then maps the submitted modification sites onto different RNA features. It then performs various coverage calculations, metagene analysis, and annotations focusing on mRNAs. The annotations include: (1) site distribution among different gene features and gene biotypes; (2) coverage analysis among RNA features; (3) site distribution around transcription start/end sites; (4) site distribution around translation start/end sites; (5) site distribution around splicing junction sites; (6) comparison of gene characteristics between modified genes and other genes; (7) modified site heatmap around translation start/end sites and transcription start/end sites; (8) mRNA metagene analysis; (9) motif enrichment analysis; (10) functional enrichments for modified genes. Three functional modules are separately provided by RNAmod, single case, group case and gene case to facilitate users who have different analysis requirements. The single case module allows users to annotate RNA modifications for a single sample. The group case modules allow users to annotate and compare the distribution of modifications between two samples or even more groups. The gene case analysis module can be used to analyze the modification distribution in the context of specific gene.
The nucleotide enrichment scores can be calculated by ToNER [116] (Transformation of Nucleotide Enrichment Ratio) through analyzing RNA-seq data generated from enriched and unenriched control libraries, in particular when obtaining data from experimental replicates, which may be used to analyze epitranscriptome data generated from enrichment-based approaches.
Furthermore, to explore the effect of genetic variants on RNA modifications, m6ASNP [117] was designed for the identification of m6A-associated variants that target m6A modification sites. A variant was defined as m6A-associated variants if it can cause the alteration of methylation status of a m6A site (gain or loss), and the potential impact of m6A modification on diseases was revealed by incorporating the information of disease-associated SNPs derived from different databases, including GWAS catalog [118], Johnson and O’Donnel [119] and ClinVar [120]. Lastly, an all-in-one toolkit RNA Framework [121] was recently developed, which is characterized by comprehensive analysis of most HGS-based RNA structure probing and post-transcriptional modification mapping experiments. To sum up, with the advances in high-throughput sequencing techniques and the increasing interest in RNA epitranscriptome, a variety types of upstream and downstream computational tools have been developed, to best share, annotate, analyze, and take advantage of the massive amount of NGS data generated.
3. RNA modification site prediction
Although is being reviewed lastly in this manuscript, computational prediction of RNA modification sites actually embodied the largest number of bioinformatics studies concerning epitranscriptome bioinformatics. At present, most of the computational prediction methods relied on gold standard datasets obtained from base-resolution epitranscriptome profiling approaches, extracted predictive features and utilized machine learning or deep learning classifiers to predict putative RNA modification sites. Among all the RNA modification types, m6A RNA methylation is the most widely studied, and it is also the predictive target of the earliest as well as the most sophisticated predictive approaches. We summarized in Table 6, Table 7 the prediction tools for m6A and other types of RNA modifications, respectively. These works together greatly improved our understanding of the distribution of multiple types of RNA modifications in different species (Please see a comprehensive review [122]). It may be worth noting that, according to a recent review [122], the WHISTLE approach, which was based on SVM algorithm and 35 additional genomic features as well conventional sequence features [123], achieved so far the best performance in m6A sites prediction, suggesting the value and importance of increased volume of high quality training data. When restricted to methods based on DNA/RNA sequences only, the deep learning-based method DeepPromise [122] achieved so far the best prediction performance.
Table 6.
Summary of m6A site prediction tools.
| Tool | Method | Encoding scheme | Species | URL/stand-alone package | Reference |
|---|---|---|---|---|---|
| iRNA-PseColl | SVM | NCP; ANF | Human | http://lin.uestc.edu.cn/server/iRNA-PseColl | [193] |
| WHISTLE | SVM | NCP; ANF; Genome features | Human | http://whistle-epitranscriptome.com | [123] |
| HMpre | XGBoost | Binary; CPD; k-mer; Site Location Related Features; Features Related to Entropy; SNP Features | Human | https://github.com/Zhixun-Zhao/HMpre | [194] |
| iRNA-Methyl | SVM | PseDNC | Yeast | http://lin.uestc.edu.cn/server/iRNA- Methyl | [195] |
| pRNAm-PC | SVM | PseDNC | Yeast | http://www.jcibioinfo.cn/pRNAm-PC | [196] |
| RAM-ESVM | SVM | PseDNC | Yeast | http://server.malab.cn/RAM-ESVM/ | [197] |
| m6Apred | SVM | NCP; ANF | Yeast | http://lin.uestc.edu.cn/server/m6Apred.php | [198] |
| RNAMethylPred | SVM | BPB; DNC; KNN score | Yeast | MATLAB package | [199] |
| TargetM6A | SVM | PSNP; PSDP; NC | Yeast | http://csbio.njust.edu.cn/bioinf/TargetM6A | [200] |
| iRNA(m6A)-PseDNC | SVM | PseDNC | Yeast | http://lin-group.cn/server/iRNA(m6A)-PseDNC.php | [201] |
| M6APred-EL | Ensemble | PS(k-mer)NP; PCPs; RFHC-GACs | Yeast | http://server.malab.cn/M6APred-EL/ | [202] |
| DeepM6APred | SVM | Deep features; NPPS | Yeast | http://server.malab.cn/DeepM6APred | [203] |
| iMethyl-STTNC | SVM | PseDNC; PseTNC; STNC; STTNC | Yeast | No | [204] |
| PXGB | XGBoost + PSO | PSNP; PSDP; NC | Yeast | No | [205] |
| M6APred-EL | Ensemble | PS(k-mer)NP; PCPs; RFHC-GACs | Yeast | http://server.malab.cn/M6APred-EL/ | [202] |
| Zhuang, Y., et al. | SVM + RF + LR | Compositional features; Position-specific features; Motif; Physiochemical features | Yeast | No | [206] |
| M6ATH | SVM | NCP; ANF | Arabidopsis | http://lin.uestc.edu.cn/server/M6ATH | [207] |
| AthMethPre | SVM | k-mer | Arabidopsis | http://bioinfo.tsinghua.edu.cn/AthMethPre/index.html | [208] |
| RFAthM6A | RF | PSNPF; PSDPF; KSNPF; KNF | Arabidopsis | https://github.com/nongdaxiaofeng/RFAthM6A | [209] |
| Zhang, J., et al. | SVM | NCP; ANF | E. coli | No | [210] |
| MethyRNA | SVM | NCP; ANF | Human; Mouse | http://lin.uestc.edu.cn/server/methyrna. | [211] |
| RNAMethPre | SVM | Binary; k-mer; MFE | Human; Mouse | http://bioinfo.tsinghua.edu.cn/RNAMethPre/index.html | [212] |
| SRAMP | RF | Binary; KNN; spectrum | Human; Mouse | http://www.cuilab.cn/sramp/ | [213] |
| Gene2vec | CNN | One-hot; Neighboring state; Word embedding; Gene2vec | Human; Mouse | http://server.malab.cn/Gene2vec/ | [124] |
| DeepM6ASeq | CNN + BLSTM | Binary | Human; Mouse | https://github.com/rreybeyb/DeepM6ASeq | [129] |
| iRNA-3typeA | SVM | NCP; ANF | Human; Mouse | http://lin-group.cn/server/iRNA-3typeA/ | [214] |
| Gene2vec | CNN | One-hot; Neighboring state; Word embedding; Gene2vec | Human; Mouse | http://server.malab.cn/Gene2vec/ | [124] |
| iRNA-3typeA | SVM | NCP; ANF | Human; Mouse | http://lin-group.cn/server/iRNA-3typeA/ | [214] |
| Dao, F.-Y., et al. | SVM | physical–chemical property matrix; Binary;NCP | Human; Mouse | http://lin-group.cn/server/iRNAm6A/service.html | [215] |
| iN6-Methyl | CNN | Word2vec | Human; Mouse; Yeast | https://home.jbnu.ac.kr/NSCL/iN6-Methyl.htm | [125] |
| RAM-NPPS | SVM | NPPS | Human; Yeast; Arabidopsis | http://server.malab.cn/RAM-NPPS/ | [216] |
| SICM6A | GRU | 3-mer | Mouse; Yeast; Arabidopsis | https://github.com/lwzyb/SICM6A | [217] |
| M6AMRFS | XGBoost | Dinucleotide Binary; Local Position-Specific Dinucleotide Frequency | Human; Mouse; Yeast; Arabidopsis | http://server.malab.cn/M6AMRFS/ | [218] |
| BERMP | RF + BGRU | ENAC; Word embedding | Human; Mouse; Yeast; Arabidopsis | http://www.bioinfogo.org/bermp | [127] |
Note: PseDNC (pseudo dinucleotide composition), ANF (accumulated nucleotide frequency), NCP (nucleotide chemical property), BPB (bi-profile bayes), DNC (dinucleotide composition), NC (nucleotide composition), PSNP (positionspecific nucleotide propensity), PSDP (position-specific dinucleotide propensity), NPPS (nucleotide pair position specificity), STTNC (split-tetra-nucleotide composition), PSNSP (position-specific nucleotide sequence profile), PSDSP (position-specific dinucleotide sequence profile), MFE (minimum free energy), PCPs (physical–chemical properties), KSNPF (K-spaced nucleotide pair frequencies), KNF(K-nucleotide frequencies), CPD (chemical property with density), ENAC (Enhanced nucleic acid composition), HPCR (heuristic nucleotide physicochemical property reduction), TNC (tri-nucleotide composition), TetraNC (tetra-nucleotide composition), mRMR (Minimum-redundancy and maximum-relevance).
Table 7.
Summary of prediction tools for non-m6A RNA modifications.
| Type | Tool | Method | Encoding scheme | Species | URL/stand-alone package | Ref |
|---|---|---|---|---|---|---|
| m5C | Feng, P., et al. | SVM | PseDNC | Human | No | [219] |
| iRNAm5C-PseDNC | RF | PseDNC | Human | http://www.jci-bioinfo.cn/iRNAm5C-PseDNC | [220] | |
| iRNA-PseColl | SVM | NCP; ANF | Human | http://lin.uestc.edu.cn/server/iRNA-PseColl | [193] | |
| M5C-HPCR | SVM | HPCR | Human | http://cslab.just.edu.cn:8080/M5C-HPCR/ | [221] | |
| pM 5 CS-Comp-mRMR | SVM | DNC; TNC; TetraNC; mRMR | Human | No | [222] | |
| RNAm5CPred | SVM | KNFs; KSNPFs; PseDNC | Human | http://zhulab.ahu.edu.cn/RNAm5CPred/ | [223] | |
| iRNA-PseTNC | SVM | PseDNC; seTNC; PseTetraNC | Human | No | [224] | |
| iRNA-m5C_NB | NB;RF;SVM;AdaBoost | BPB;k-mer;ENAC;XXKGAP;EIIP;PseEIIP | Human | No | [225] | |
| PEA-m5C | RF | Binary; k-mer; PseDNC | Arabidopsis | https://github.com/cma2015/PEA-m5C | [226] | |
| RNAm5Cfinder | RF | One-hot; NCP; ANF | Human; Mouse | http://www.rnanut.net/rnam5cfinder | [227] | |
| RNAm5Cfinder | RF | One-hot; NCP; ANF | Human; Mouse | http://www.rnanut.net/rnam5cfinder | [227] | |
| ψ | PPUS | SVM | binary | Human | http://lyh.pkmu.cn/ppus/ | [228] |
| PIANO | SVM | NCP; ANF; Genome features | Human | http://piano.rnamd.com | [229] | |
| iPseU-NCP | RF | NCP | Human; Yeast | https://github.com/ngphubinh/iPseU-NCP | [132] | |
| iRNA-PseU | SVM | NCP; ANF; PseKNC | Human; Mouse; Yeast | http://lin.uestc.edu.cn/server/iRNA-PseU | [230] | |
| PseUI | SVM | NC; DC; PseDNC; PSNP; PSDP | Human; Mouse; Yeast | http://zhulab.ahu.edu.cn/PseUI | [231] | |
| XG-PseU | XGBoost | NC; DNC; TNC; NCP; One-hot | Human; Mouse; Yeast | http://www.bioml.cn/ | [232] | |
| DeepMRMP | BGRU | One-hot | Human; Mouse; Yeast | No | [128] | |
| CNNPSP | CNN | DNC; NCP; ANF | Human; Mouse; Yeast | No | [130] | |
| iPseU-CNN | CNN | n-gram and multivariate mutual information (MMI) | Human; Mouse; Yeast | No | [233] | |
| EnsemPseU | SVM; XGBoost; NB; KNN; RF | k-mer; Binary; ENAC; NCP; ND | Human; Mouse; Yeast | https://github.com/biyue1026/EnsemPseU | [234] | |
| Nm | iRNA-2methyl | Ensemble; RF | Pse-in-One | Human | http://www.jci-bioinfo.cn/iRNA-2methyl | [235] |
| iRNA-PseKNC | CNN | One-hot | Human | No | [131] | |
| iRNA-2OM | SVM | NCP; ANF; Type 2 PseKNC | Human | http://lin-group.cn/server/iRNA-2OM | [236] | |
| Chen, W., et al. | SVM | NCP; ANF | Human; Mouse; Yeast | No | [237] | |
| m1A | iRNA-PseColl | SVM | NCP; ANF | Human | http://lin.uestc.edu.cn/server/iRNA-PseColl | [193] |
| ISGm1A | RF | NCP; ANF; Genome features | Human | https://github.com/lianliu09/m1a_prediction.git. | [238] | |
| iRNA-3typeA | SVM | NCP; ANF | Human; Mouse | http://lin-group.cn/server/iRNA-3typeA/ | [214] | |
| RAMPred | SVM | NCP; ANF | Human; Mouse; Yeast | http://lin.uestc.edu.cn/server/RAMPred | [239] | |
| A to I | iRNA-AI | SVM | NCP; ANF | Human | http://lin.uestc.edu.cn/server/iRNA-AI/ | [240] |
| EPAI-NC | LD-SVM | l-mers; n-gapped l-mers | Fly | http://epai-nc.info/ | [241] | |
| PAI | SVM | PseDNC | Fly | http://lin.uestc.edu.cn/server/PAI | [242] | |
| iRNA-3typeA | SVM | NCP; ANF | Human; Mouse | http://lin-group.cn/server/iRNA-3typeA/ | [214] | |
| m2G | iRNA-m2G | SVM | NCP; ANF | Human; Mouse; Yeast | No | [243] |
| m7G | iRNA-m7G | SVM | NCP; ANF; PseDNC; SSC | Human | http://lin-group.cn/server/iRNA-m7G/ | [244] |
| m7GFinder | SVM | NCP; ANF | Human | www.xjtlu.edu.cn/biologicalsciences/m7ghub | [245] | |
| D | iRNAD | SVM | NCP; ANF | Human; Mouse; Yeast | http://lin-group.cn/server/iRNAD | [246] |
| 5hmC | iRNA5hmC | SVM | k-mer; Binary | Fly | http://server.malab.cn/iRNA5hmC | [247] |
| ac4C | PACES | RF | One-hot;PSNSP;PSDSP;KNF;KSNPF;PseKNC | Human | http://www.rnanut.net/paces/ | [248] |
3.1. Deep learning in RNA modification sites prediction
One prominent trend is that, deep learning-based predictive approaches seem to be able to offer better overall prediction performance compared with classic machine learning methods. In contrast of traditional machine learning methods, deep learning (DL) model can automatically extract the non-linear features. Several deep learning-based methods have been developed. These methods firstly extracted the positive modification sites from existing studies and database like Met-DB and RMBase, and then selected negative samples to build a standard data set to train and test the proposed approach.
The most widely used deep learning models are convolutional neural network (CNN), which can effectively learn the motif related features from RNA sequence, and recurrent neural network (RNN), which can learn the non-linear sequential features from RNA sequence, including long short-term memory unit (LSTM) and gated recurrent unit (GRU). Gene2vec [124], DeepPromise [122], iN6-Methyl (5-step) [125] and Deep-m6A [126] built CNN models to predict m6A or m1A modifications; BERMP [127] employed a bidirectional Gated Recurrent Unit (BGRU) model to predict m6A; DeepMRMP [128] adopted bidirectional Gated Recurrent Unit (BGRU) and transfer learning to predict m6A, m1A, pseudouridine and m5C; the DL models of DeepM6ASeq [129] consists of two layers of CNN, one bidirectional long short-term memory (BLSTM) layer and one fully connected (FC) layer.
Although most of these approaches focused on m6A RNA methylation prediction, it is worth noting that deep learning has also been applied to other modifications as well. For example, CNNPSP [130] and iRNA-PseKNC [131] employed convolutional neural network (CNN) to predict pseudouridine and 2′-O-methylation, respectively. And DeepPromise [122] is also able to predict m1A sites. Please refer to Table 7 for a summary of prediction approaches for non-m6A modifications.
The input features of most DL-based methods are RNA or DNA sequence except Deep-m6A which embedded the MeRIP-Seq reads count with RNA sequence to predict condition-specific (e.g. disease or normal) m6A sites. There are diverse strategies to encode the input features. We summarize in the following 4 major kinds of encoding strategies, including:
-
(1)
one-hot embedding;
-
(2)
RNA/DNA nucleotides to vector embedding;
-
(3)
RNA sequence and MeRIP-Seq reads count embedding;
-
(4)
neighboring methylation state embedding.
One-hot encoding is widely used for sequence analysis [132], [133], which encodes the ‘A’, ‘U’ (or ‘T’), ‘C’ and ‘G’ to 4-dimensional binary vectors. DeepM6ASeq, Gene2vec, DeepPromise, DeepMRMP and Deep-m6A employed one-hot encoding as part of their feature encoding strategies. RNA/DNA nucleotides to vector embedding treated one or several nucleotides as words and the whole sequence as a sentence, then transfer nucleotides to numeric vectors based on semantic analysis. BERMP treated each nucleotide as a word and trained an embedding layer together with the BGRU model to convert the input nucleotide to vector; Gene2vec regarded 3 RNA nucleotides as an RNA word and developed a neural‐network-based model to generate a 100‐dimensional feature vector for each ‘word’ and also employed RNA word embedding which treated 3 RNA nucleotides as a word, then built a dictionary to embed the whole sentence and finally trained an embedding layer to transform each integral sequence into a data table; DeepPromise adopted the enhanced nucleic acid composition (ENAC) embedding, which can simultaneously depict the nucleotides’ composition and position information based on a length-fixed window slide on RNA sequence from 5′ to 3′ termini and also RNA-embedding which took 5 nucleotides as a word and adopted similar scheme with RNA word embedding adopted by Gene2vec to transform each integral sequence into a data table; iN6-Methyl (5-step) treated 3 nucleotides as a word and employed word2vec [134] to convert the ‘word’ to a 100-dimensional vector, similar with Gene2vec. Neighboring methylation state embedding is only adopted by Gene2vec, which embedded the 250 upstream and 250 downstream candidate m6A sites around the predicted m6A site to 501-dimensional binary vectors. RNA sequence and MeRIP-Seq reads count embedding is only adopted by Deep-m6A, which firstly encoded the RNA sequence using one-hot encoding and then embedded the normalized MeRIP-Seq IP sample reads count mapped to the corresponding nucleotide’s genome position to the binary vector of each nucleotide. The embedded MeRIP-Seq IP reads count can represent the m6A methylation level under specific condition like cancer, which provides the power to predict condition-specific m6A sites.
4. RNA modification databases
Knowledge bases with the comprehensive collection and curation of various information related to transcriptome-wide RNA modifications are often critical for elucidating their biological functions as well as for developing bioinformatics tools. A number of works have been accomplished addressing various aspects of RNA modifications including basic properties, pathway, distribution, disease association, visualization and GO functions. We review in the following a few databases related to RNA modifications.
4.1. RNAMDB
RNAMDB [135], [136] contains 109 RNA modifications with basic description of the RNA modification (chemical structure of the nucleoside, common chemical name, symbol, elemental composition and mass), type(s) of RNA in which the nucleoside occurs (tRNA, rRNA, mRNA, snRNA etc.), phylogenetic occurrence of the nucleoside (archaea, bacteria, eukarya and the corresponding literature citations for each, chemical abstracts registry numbers and chemical abstracts index name), literature citation to structure assignment of the nucleoside and literature citation to the first reported chemical synthesis of the nucleoside. Moreover, other informatics resources for RNA science are available in RNAMDB including different repositories of experimental protocols which comprise established procedures that are common practices in a typical RNA lab, related link database and sister database. The latest version was updated in 2011 which is freely accessible at https://mods.rna.albany.edu/.
4.2. MODOMICS
The MODOMICS database [137], [138] is currently the most comprehensive RNA modification pathway source. It displays the reactions linking a modified nucleoside to its precursor(s) and to hypermodificatons. Additionally, a typical entry of a modified ribonucleoside contains information about its fundamental chemical properties, chemical structure including the standard bases (A, U, C and G) they come from and the chemical groups they contain. MODOMICS also provides many other aspects to interpret and display the information above. From the aspect of RNA sequence, MODOMICS provides a collection of modified RNA sequences of different types. Sequences are visualized with all modifications highlighted and linked to the corresponding modification records. From the aspect of proteins, the MODOMICS database currently contains information above 340 functionally characterized proteins involved in RNA modification, both functional enzymes and protein co-factors necessary for multi-protein enzymes activities. MODOMICS also adds a catalogue of ‘building blocks’ for the chemical synthesis of naturally occurring modified nucleosides. The compilation is intended to facilitate solid phase synthesis of modified RNA, and thus to foster biophysical and biochemical studies. The database is freely accessible from: https://iimcb.genesilico.pl/modomics/.
4.3. MeT-DB
MeT-DB (MethyTranscriptome DataBase) [139] is the first comprehensive resource for m6A transcriptome methylation. The MeT-DB database includes three parts, Core DB, TREW DB and Functional DB. The Core database contains context-specific m6A peaks and single-base sites. For each predicted m6A peak, its chromosomal location, including start/end position, strand information, p-value, fold enrichment and q-value were reported. Moreover, m6A peaks, single-based m6A sites, motif, peak distribution plot, gene expression profiles were available and can be downloaded from the web page. The TREW database annotates target sites of m6A readers, writers and erasers, then the target site was further annotated with transcript regions (5′ UTR, CDS, 3′UTR, stop codon, transcription start sites and miRNA target site) and RNA type. In addition, some useful tools are provided in the MeT-DB web interface: the table view to facilitate researchers to explore and search the data in details, the genome browser to help the user visualize and compare m6A peaks and functions data, and the tool module that includes Guitar Plot and m6A-Driver for investigating the functions of m6A methyl transcriptome. It has undergone two versions [139], [140] and is currently available at http://compgenomics.utsa.edu/MeTDB/.
4.4. RMBase
RMBase [141], [142] currently has the most comprehensive collection of RNA modification sites, and is aimed to decode the map of RNA modifications from epitranscriptome sequencing data. RMBase v2.0 was expanded with 566 datasets and 1,397,244 modification sites from 47 studies among 13 species. To study the distribution of RNA modifications on the transcript products, RMBase mapped their sites onto the genomic coordinates of the genes with annotation including gene types and regions. RMBase also studied the relationships between RNA modification and post-transcriptome regulation, such as, miRNA binding sites, SNPs and RBPs. Besides, RMBase also annotated the RBPs as readers, writers and erasers. All SNPs and SNVs were intersected with the RNA modification regions to identify the SNPs and SNVs that might interact with the RNA modifications. Visualized logos of modification motifs and metagenes of RNA modification plotting along a transcript model are also available. The RMBase database is freely accessible at: http://rna.sysu.edu.cn/rmbase/.
4.5. m6AVar
m6AVar [143] is dedicated to the investigation of the functional association between genetic variants and m6A modification. Raw data resource of m6Avar can be mainly categorized into two parts, SNPs and m6A sites. In terms of SNPs, germline and somatic variants were obtained from dbSNP and TCGA database. A large number of disease-associated SNPs were obtained from GWAS, ClinVar etc. Furthermore, all of the SNPs were annotated by gene conservation scores and deleterious levels scoring from 0 to 5. In terms of m6A sites, they were acquired according to different confidence levels from high to low by using various strategies. The m6A sites with a high confidence level were derived from 7 miCLIP experiments and 2 PA-m6A-seq experiments. The m6A sites that have a medium confidence level were derived from 244 MeRIP-seq experiments and m6A sites that have a low confidence level were derived from a transcriptome-wide prediction based on Random Forest algorithm. Furthermore, the location of each m6A site was annotated by the transcript structure, including the CDS, 3′UTR, 5′UTR, start codon and stop codon. Combined with SNPs and m6A sites, m6AVar explored m6A-associated variants which were defined by evaluating whether it has the potential to alter the DRACH motif or other sequence features essential for m6A modification. Particularly, more than 2000 disease-related variants have been identified by linking the m6A-associated variants with GWAS and ClinVar data. m6AVar also provided an user-friendly web interface with multiple statistical diagrams and genome browser through which users can browse all of the m6A-associated variants and search data by various criteria. The m6AVar database is freely accessible at: http://m6avar.renlab.org/.
4.6. REPIC
REPIC [144] is a newly developed database dedicated to provide a new resource to investigate potential functions and mechanisms of m6A modifications across 11 species. To offer insights into the cell line- or tissue-specificity of m6A modification, REPIC supports query of m6A modifications by cell lines and tissue types. Peak annotation and sample annotation are available. Peak annotation includes genomic position, fold enrichment and genomic feature. Sample annotation includes the data source, read mapping statistics, metagene profiles and results from motif analysis. To better display multiple dimensional m6A modification information across the entire genome, REPIC provides a genome browser to visualize m6A peaks, fold enrichment and gene expression. REPIC is accessible at https://epicmod.uchicago.edu/repic/index.php.
4.7. RADAR
RADAR [145] is a rigorously annotated database of A-to-I RNA editing. It includes a comprehensive collection of A-to-I RNA editing sites identified in humans (Homo sapiens), mice (Mus musculus) and flies (Drosophila melanogaster), which contains 1 379 403 human, 8108 mouse and 2698 fly A-to-I RNA editing sites separately. Specifically, for each editing site, annotations are curated manually, which consist of the genome, strand, associated gene, functional region within the gene (coding sequence, untranslated region, intron), associated repetitive element (Alu, repetitive non-Alu, nonrepetitive), conservation of editing to other species and the reference study in which the site was first identified. In addition, for each editing site, RADAR also includes a catalog of tissue-specific editing levels from published RNA-seq datasets. RADAR allows the search for A-to-I RNA editing sites by using any combination of the abovementioned annotations. To facilitate more detailed searches, the UCSC genome browser is used to display the overlapping gene annotations, genomic nucleotide conservation, overlapping SNP database entries and overlapping repetitive elements. The RADAR is freely accessible at http://RNAedit.com.
5. Disease marker and association prediction
Recent studies demonstrated that aberrant m6A modifications is linked to a number of pathophysiological disorders, including obesity related traits [146], [147], [148], [149], diabetes [150], aberrant germ cell formation [151], circadian period elongation [27], developmental retardation [152]. Emerging evidence suggests that m6A modification is involved in multiple forms of human cancer and plays a crucial role in different cancer contexts, such as in breast cancer [153], [154], acute myeloid leukemia (AML) [155], [156], [157], [158], [159], glioblastoma [160], [161], [162], lung cancer [163] and liver cancer [164]. Precise identification of disease-associated m6A modification can be critical for understanding the disease pathogenesis. While wet lab experiments were often restricted by their costs in time and labor, computational approaches offered a viable avenue. We briefly summarized in the following some recent works related to in silico identification and prediction of disease association of RNA modifications including the relevant enzymes and the sites.
RNAMethyPro [165] used a biologically conserved signature of m6A regulators for prediction of survivals at pan-cancer level, which was based on 25 publically available datasets encompassing 13 cancer types. However, the construction of RNAMethyPro is based on silico analysis, which is controversial for obtaining the biological and clinical characteristics related to m6A, as well as for determining the specific functional modules of patients at high risk. Therefore, further mechanism and independent clinical verification are still appreciated to further validate that RNAMethyPro as a robust predictive signal in a variety of human cancers.
In a study led by Li et al. [166], the molecular alterations and clinical relevance of m6A regulators were analyzed across more than 10,000 subjects representing 33 cancer types, and revealed significant correlation between activities of cancer hallmark-related pathways and expression levels of m6A regulators. Besides, the authors revealed that m6A reader IGF2BP3 maybe a potential oncogene for Clear cell renal cell carcinoma (ccRCC), even though they cannot reliably predict the prognosis of ccRCC patients based on the risk score according to the mRNA expression of m6A regulatory genes.
The m6AVar database [167] established the association between individual m6A site and various diseases via disease-associated genetic mutations that may also lead to changes of RNA methylation status. To our knowledge, this is the first large-scale prediction study that linked individual RNA methylation sites to various diseases. In addition, it is a comprehensive database, which contains m6A related variables that may affect the m6A modification, which will help to interpret variables through the m6A function.
In the CVm6A database [168], 190,050 and 150,900 m6A sites were identified in cancer and non-cancer cells, which may demonstrate putative associations to cancer pathology. But due to the limitation of m6A sequence dataset, CVm6A, as well as most other databases, cannot fully determine the distribution of m6A on lncRNAs and other non-polyA RNAs.
Based on a random walk with restart approach, DRUM [169] successfully associated individual m6A sites to various diseases via a multi-layered heterogeneous network consisting of m6Asites, genes and diseases. The genes and sites were linked by association of expression levels and methylation levels, while genes and diseases are associated according to existing gene-disease association database.
By taking advantage of the guilt-by-association principle, m6Acomet [170] can infer putative GO functions of individual m6A sites from a RNA co-methylation network derived epitranscriptome profiling data using hub-based or module-based methods. This is the first study for large-scale prediction of GO functions for individual m6A sites. However, the two methods used in m6Acomet achieved only marginal improvement compared with random guesses. Furthermore, there are more data sources, which can be integrated with RNA comethylation network to obtain more accurate functional labeling.
Very recently, An et al developed a computational approach to systematically identify cell-specific trans regulators of m6A through integrating gene expressions, binding targets and binding motifs of large number of RNA binding proteins (RBPs) with a co-methylation network constructed using large-scale m6A methylomes across diverse cell states [171]. This study provides a new perspective for the regulation of m6A epitranscriptome.
6. Summary and outlook
With an increasing number of studies revealing the essence and importance of RNA modifications in general gene expression regulation and disease pathogenesis, RNA epigenetics [172] (or epitranscriptomics [173]) has captured growing attention. Bioinformatics capacity to analyze, digest, collect and share the rapidly growing epitranscriptome profiling data is sorely needed. We reviewed recent progress and emerging bioinformatics topics concerning RNA modifications, including epitranscriptome data analysis techniques, RNA modification databases, disease-association inference, functional annotation and RNA modification site prediction. Taken together, bioinformatics developments have greatly facilitated research in the area and have enhanced understanding of the biological meaning of RNA modifications.
Nevertheless, despite the rapid progress in epitranscriptome bioinformatics, there are still a number of limitations or open questions.
First, technological bias and limitations may not have received sufficient attention during development of bioinformatics tools. For example, most of the existing RNA bisulfite data interpretation tools failed to consider the abundant RNA secondary structures that may generate a large number of false positive errors [174]. Although it has been reported that there are major discrepancies between the results of different RNA modification profiling techniques (such as in m5C [98], [99]), few existing site prediction approaches have carefully considered it. Furthermore, most existing site prediction tools overlooked the bias induced by polyA selection during RNA-seq library preparation, which leads to under-representation of intronic and lncRNA sites.
Secondly, although existing studies suggested that RNA modification can affect the structure of RNA [175], it is not yet clear how it affects the 3D structure of RNA molecules in general [176]. It seems likely that many RNA modifications will exert at least some of their myriad functions through affecting structures so that methods considering and predicting this consequence of modification would be highly valuable.
Thirdly, some bioinformatics pipelines have not been extended to keep up with the emergence of novel modifications arising from the development of new technologies. For example, the site prediction and disease association frameworks developed for well-studied modifications (such as the WHISTLE [123] and m6AVar [167] frameworks for m6A modification site prediction and disease association) have not been extended to other relatively less studied RNA modifications (such as m1A and Nm), even though the extension should be fairly straightforward from a computational perspective. Such basic bioinformatics infrastructure is essential and should be established for all types of RNA modifications that can be profiled transcriptome-wide at base-resolution.
7. Author’s contribution
Jia Meng, Hui Liu and Xiujuan Lei initialized and coordinated the project. Yi Song, Rong Rong and Zhiliang Lu reviewed the biological background of RNA modifications; Lian Liu, Song-Yao Zhang, Kunqi Chen, Shao-Wu Zhang and Xiujuan Lei summarized sites prediction approaches; Lian Liu, Zhen Wei and Shao-Wu Zhang reviewed m6A-seq analysis approaches; Yujiao Tang, Xiangyu Wu, João Pedro de Magalhães and Daniel J. Rigden reviewed disease association and functional prediction; Bowen Song reviewed other bioinformatics tools for epitranscriptome data analysis; Jiani Ma, Hui Liu and Lin Zhang reviewed existing bioinformatics databases. All authors read, critically revised and approved the final manuscript.
Funding
This work has been supported by National Natural Science Foundation of China [61902230, 61972451, 31671373]; China Postdoctoral Science Foundation [2018 M640949]; Fundamental Research Funds for the Central Universities [GK201903083, GK201901010]; XJTLU Key Program Special Fund [KSF-T-01].
CRediT authorship contribution statement
Lian Liu: Writing - original draft, Methodology, Investigation. Bowen Song: Writing - original draft, Methodology, Investigation. Jiani Ma: Writing - original draft, Methodology, Investigation. Yi Song: Writing - original draft, Methodology, Investigation. Song-Yao Zhang: Writing - original draft, Methodology, Investigation. Yujiao Tang: Writing - original draft, Methodology, Investigation. Xiangyu Wu: Writing - original draft, Methodology, Investigation. Zhen Wei: Writing - review & editing. Kunqi Chen: Writing - review & editing. Jionglong Su: Writing - review & editing, Resources, Supervision. Rong Rong: Writing - review & editing, Resources, Supervision. Zhiliang Lu: Writing - review & editing, Resources, Supervision. João Pedro de Magalhães: Writing - review & editing, Resources, Supervision. Daniel J. Rigden: Writing - review & editing, Resources, Supervision. Lin Zhang: Writing - review & editing, Resources, Supervision. Shao-Wu Zhang: Writing - review & editing, Resources, Supervision. Yufei Huang: Writing - review & editing, Resources, Supervision. Xiujuan Lei: Writing - review & editing, Resources, Supervision, Funding acquisition, Project administration. Hui Liu: Writing - review & editing, Resources, Supervision, Funding acquisition, Project administration. Jia Meng: Writing - review & editing, Resources, Supervision, Funding acquisition, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.06.010.
Contributor Information
Xiujuan Lei, Email: xjlei@snnu.edu.cn.
Hui Liu, Email: hui.liu@cumt.edu.cn.
Jia Meng, Email: jia.meng@xjtlu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.McCown P.J. Naturally occurring modified ribonucleosides. WIREs RNA. 2020:e1595. doi: 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones J.D., Monroe J., Koutmou K.S. A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. WIREs RNA. 2020:e1586. doi: 10.1002/wrna.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia G. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominissini D. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 5.Meyer K.D. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155(6):1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20(15):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 8.Zheng G. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proceedings of the National Academy of Sciences. 2019;116(8):2919–2924. doi: 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roundtree I.A. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.X. Wang, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505 (7481) : p. 117-138. [DOI] [PMC free article] [PubMed]
- 12.Meyer K.D. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61(4):507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Wang X. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alarcón C.R. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature. 2015;518(7540):560. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominissini D. Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature. 2012;485(7397):201. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 18.Patil D P. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–394. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567(7748):414–441. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang Y. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543(7646):573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563(7730):249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B.S. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alarcon C.R. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pendleton K.E. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;165(9):824–835. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fustin J.M. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Vollmers C. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Met. 2012;16(6):833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geula S. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 31.Bertero A. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature. 2018;555(7695):256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteve-Puig R. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 2020;474:127–137. doi: 10.1016/j.canlet.2020.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y. N6-methyladenosine RNA modification–mediated cellular metabolism rewiring inhibits viral replication. Science. 2019;365(6458):eaax4468. doi: 10.1126/science.aax4468. [DOI] [PubMed] [Google Scholar]
- 34.Delaunay S., Frye M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019;21(5):552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 35.Choe J. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561(7724):556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han D. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270–291. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbieri I. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552(7683):126–156. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H.B. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon K.J. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell. 2017;171(4):877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su R. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCown P.J. Secondary structural model of human MALAT1 reveals multiple structure-function Relationships. INT. J. MOL. SCI. 2019;20(22):5610. doi: 10.3390/ijms20225610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motorin Y. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38(5):1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Squires J.E. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrenhofer-Murray Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules. 2017;7(1):14. doi: 10.3390/biom7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuorto F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct Mol. Biol. 2012;19(9):900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 46.Yang X. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanco S. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534(7607):335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heissenberger C. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019;47(22):11807–11825. doi: 10.1093/nar/gkz1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Courtney D.G. Epitranscriptomic Addition of m5C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host & Microbe. 2019;26(2):217–227.e6. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y.RNA. 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell. 2019;75(6):1188–1202. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 51.Zou F. Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. P. Natl. Acad. Sci. 2020;117(7):3603–3609. doi: 10.1073/pnas.1910862117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nature Cell Bio. 2019;21(8):978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 53.Henry B.A. 5-Methylcytosine Modification of an Epstein-Barr Virus Noncoding RNA Decreases its Stability. RNA. 2020 doi: 10.1261/rna.075275.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro I.C. Translational adaptation to heat stress is mediated by 5-methylcytosine RNA modification in Caenorhabditis elegans. boiRxiv. 2020 doi: 10.15252/embj.2020105496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Y. OsNSUN2-Mediated 5-Methylcytosine mRNA Modification Enhances Rice Adaptation to High Temperature. Dev Cell. 2020;53(3):272. doi: 10.1016/j.devcel.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 56.Delatte B. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351(6270):282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 57.Nishikura Functions and Regulation of RNA Editing by ADAR Deaminases. Annu Rev Biochem. 2010;79(1):321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan T.H.M. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014;63(5):832–843. doi: 10.1136/gutjnl-2012-304037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Picardi, E., et al., REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017. 45(D1): p. D750-D757. [DOI] [PMC free article] [PubMed]
- 60.Zipeto M.A. RNA rewriting, recoding, and rewiring in human disease. Trends. Mol. Med. 2015;21(9):549–559. doi: 10.1016/j.molmed.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Deffit S.N., Hundley H.A. To edit or not to edit: regulation of ADAR editing specificity and efficiency. Comput. Mol. Sci. 2016;7(1):113–127. doi: 10.1002/wrna.1319. [DOI] [PubMed] [Google Scholar]
- 62.Ota H. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153(3):575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamashita T. The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 2014;1584:28–38. doi: 10.1016/j.brainres.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 64.Han L. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cell. 2015;28(4):515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samuel C.E. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA–triggered innate immune responses. J. Biol. Chem. 2019;294(5):1710–1720. doi: 10.1074/jbc.TM118.004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao Y. A comprehensive study on cellular RNA editing activity in response to infections with different subtypes of influenza a viruses. BMC Genomics. 2018;19(1):925. doi: 10.1186/s12864-017-4330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlachogiannis N.I. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J Autoimmun. 2020;106:102329. doi: 10.1016/j.jaut.2019.102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shoshan E. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015;17(3):311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomaselli S. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015;16(1):5. doi: 10.1186/s13059-014-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramírez-Moya J. ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity. Oncogene. 2020:1–16. doi: 10.1038/s41388-020-1248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.X. Peng et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell 2018; 33(5): pp. 817–828. e7 [DOI] [PMC free article] [PubMed]
- 72.Schwartz S. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guzzi, N., et al., Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell, 2018. 173(5): p. 1204-1216 e26. [DOI] [PubMed]
- 74.Carlile T.M. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaffrey An expanding universe of mRNA modifications. Nat. Struct Mol. Biol. 2014;21(11):945. doi: 10.1038/nsmb.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dominissini D., Rechavi G. N4-acetylation of Cytidine in mRNA by NAT10 Regulates Stability and Translation. Cell. 2018;175(7):1725–1727. doi: 10.1016/j.cell.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 77.Mauer J. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2016;541(7637):371–394. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun H. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019;29(1):80. doi: 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu, F., et al., ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell, 2016. 167(3): p. 816-828 e16. [DOI] [PMC free article] [PubMed]
- 80.Dominissini D. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daffis, S., et al., 2Daffis, S., et of the viral mRNA cap evades host restriction by IFIT family members. Nature, 2010. 468(7322): p. 452. [DOI] [PMC free article] [PubMed]
- 82.Ringeard M. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019;565(7740):500–519. doi: 10.1038/s41586-018-0841-4. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dominissini D. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 85.Meng J. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics. 2013;29(12):1565–1567. doi: 10.1093/bioinformatics/btt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love M.I., Hogenesch J.B., Irizarry R.A. Modeling of RNA-seq fragment sequence bias reduces systematic errors in transcript abundance estimation. Nat. Biotechnol. 2016;34(12):1287. doi: 10.1038/nbt.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meng J. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69(3):274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Z. RADAR: differential analysis of MeRIP-seq data with a random effect model. Genome Biol. 2019;20(1):294. doi: 10.1186/s13059-019-1915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L. QNB: differential RNA methylation analysis for count-based small-sample sequencing data with a quad-negative binomial model. BMC Bioinf. 2017;18(1):387. doi: 10.1186/s12859-017-1808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schwartz S. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ Sites. Cell Rep. 2014;8(1):284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang T. trumpet: transcriptome-guided quality assessment of m(6)A-seq data. BMC Bioinf. 2018;19(1):260. doi: 10.1186/s12859-018-2266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hauenschild R. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 2015;43(20):9950–9964. doi: 10.1093/nar/gkv895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Byrne A. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt L. Graphical workflow system for modification calling by machine learning of reverse transcription signatures. Front Genet. 2019;10:876. doi: 10.3389/fgene.2019.00876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lempereur L. Conformation of yeast 18S rRNA. Direct chemical probing of the 5' domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible. Nucleic Acids Res. 1985;13(23):8339–8357. doi: 10.1093/nar/13.23.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Levanon E.Y. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22(8):1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 97.Schaefer M. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Motorin Y., Helm M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes. 2019;10(1):35. doi: 10.3390/genes10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hussain S. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14(11):215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huber S.M. Formation and abundance of 5-hydroxymethylcytosine in RNA. ChemBioChem. 2015;16(5):752–755. doi: 10.1002/cbic.201500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bolger A.M. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Langmead B. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim D. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rieder D. meRanTK: methylated RNA analysis ToolKit. Bioinformatics. 2016;32(5):782–785. doi: 10.1093/bioinformatics/btv647. [DOI] [PubMed] [Google Scholar]
- 105.Yang X. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang T. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019;26(5):380–388. doi: 10.1038/s41594-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 107.Amort T. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18(1):1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Edelheit S. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013;9(6) doi: 10.1371/journal.pgen.1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parker B.J. Statistical methods for transcriptome-wide analysis of RNA methylation by bisulfite sequencing. Methods Mol Biol. 2017;1562:155–167. doi: 10.1007/978-1-4939-6807-7_11. [DOI] [PubMed] [Google Scholar]
- 110.Sherry S.T. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Picardi, E., et al., REDIdb: the RNA editing database. Nucleic Acids Res, 2007. 35(Database issue): p. D173-7. [DOI] [PMC free article] [PubMed]
- 112.Cui X. Guitar: an R/bioconductor package for gene annotation guided transcriptomic analysis of RNA-Related genomic features. Biomed Res Int. 2016;2016:8367534. doi: 10.1155/2016/8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Olarerin-George A.O. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics. 2017;33(10):1563–1564. doi: 10.1093/bioinformatics/btx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan Z. txCoords: a novel web application for transcriptomic peak re-mapping. IEEE/ACM Trans Comput Biol Bioinform. 2017;14(3):746–748. doi: 10.1109/TCBB.2016.2568178. [DOI] [PubMed] [Google Scholar]
- 115.Liu Q., Gregory R.I. RNAmod: an integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 2019;47(W1):W548–W555. doi: 10.1093/nar/gkz479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Promworn Y. ToNER: A tool for identifying nucleotide enrichment signals in feature-enriched RNA-seq data. PLoS ONE. 2017;12(5) doi: 10.1371/journal.pone.0178483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang S. m6ASNP: a tool for annotating genetic variants by m6A function. Gigascience. 2018;7(5) doi: 10.1093/gigascience/giy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Buniello A. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics. Nucleic Acids Res. 2018;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johnson A.D. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Landrum M.J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2015;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Incarnato D. RNA Framework: an all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications. Nucleic Acids Res. 2018;46(16) doi: 10.1093/nar/gky486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen Z. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Briefings Bioinf. 2019:1–21. doi: 10.1093/bib/bbz112. [DOI] [PubMed] [Google Scholar]
- 123.Chen K. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res. 2019;47(7):e41. doi: 10.1093/nar/gkz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zou Q., Sr. Gene2vec: gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA. 2018;25(2):205–218. doi: 10.1261/rna.069112.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nazari I. iN6-Methyl (5-step): Identifying RNA N6-methyladenosine sites using deep learning mode via Chou’s 5-step rules and Chou’s general PseKNC. Chemomet Intell Lab. 2019:103811. [Google Scholar]
- 126.Zhang S.-Y. Global analysis of N6-methyladenosine functions and its disease association using deep learning and network-based methods. PLoS Comput. Biol. 2019;15(1) doi: 10.1371/journal.pcbi.1006663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang Y. BERMP: a cross-species classifier for predicting m6A sites by integrating a deep learning algorithm and a random forest approach. Int J Biol Sci. 2018;14(12):1669–1677. doi: 10.7150/ijbs.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun P. DeepMRMP: a new predictor for multiple types of RNA modification sites using deep learning. Mathemat Biosci Eng. 2019;16(6):6231–6241. doi: 10.3934/mbe.2019310. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y., Hamada M. DeepM6ASeq: prediction and characterization of m6A-containing sequences using deep learning. BMC Bioinf. 2018;19(19):524. doi: 10.1186/s12859-018-2516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fan Y., et al., CNNPSP. Pseudouridine Sites Prediction Based on Deep Learning. Intelligent Data Engineering and Automated Learning –. IDEAL, 2019. Cham: Springer International Publishing. p. 2019.
- 131.Tahir M., Tayara H., Chong K.T. iRNA-PseKNC(2methyl): Identify RNA 2'-O-methylation sites by convolution neural network and Chou's pseudo components. J. Theor. Biol. 2019;465:1–6. doi: 10.1016/j.jtbi.2018.12.034. [DOI] [PubMed] [Google Scholar]
- 132.Nguyen-Vo T.-H. iPseU-NCP: Identifying RNA pseudouridine sites using random forest and NCP-encoded features. BMC Genomics. 2019;20(10):971. doi: 10.1186/s12864-019-6357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meyer K.D. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bastian L. Single-nucleotide resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12(8):767. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rozenski J., Crain P.F., McCloskey J.A. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27(1):196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cantara W.A. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Machnicka M.A. MODOMICS: a database of RNA modification pathways-2013 update. Nucleic Acids Res. 2013;41(D1):D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boccaletto P. MODOMICS a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu H. MeT-DB: a database of transcriptome methylation in mammalian cells. Nucleic Acids Res. 2015;43(D1):D197–D203. doi: 10.1093/nar/gku1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu H. MeT-DB V2. 0: elucidating context-specific functions of N6-methyl-adenosine methyltranscriptome. Nucleic Acids Res. 2018;46(D1):D281–D287. doi: 10.1093/nar/gkx1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun W.-J. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016;44(D1):D259–D265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xuan J.-J. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46(D1):D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zheng Y. m6AVar: a database of functional variants involved in m(6)A modification. Nucleic Acids Res. 2018;46(D1):D139–D145. doi: 10.1093/nar/gkx895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu S., He C., Chen M.REPIC. A database for exploring N6-methyladenosine methylome. Genome Biol. 2020;21(1):100. doi: 10.1186/s13059-020-02012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ramaswami G. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42(D1):D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Scuteri A. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7) doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dina C. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39(6):724. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 148.Frayling T.M. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Davis W. The fat mass and obesity-associated FTO rs9939609 polymorphism is associated with elevated homocysteine levels in patients with multiple sclerosis screened for vascular risk factors. Metab. Brain Dis. 2014;29(2):409–419. doi: 10.1007/s11011-014-9486-7. [DOI] [PubMed] [Google Scholar]
- 150.Shen F. Decreased N6-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metabol. 2015;100(1):E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang Y. Increased N6-methyladenosine in human sperm RNA as a risk factor for asthenozoospermia. Sci. Rep. 2016;6:24345. doi: 10.1038/srep24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Daoud H. Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay. J. Med. Genet. 2016;53(3):200–207. doi: 10.1136/jmedgenet-2015-103399. [DOI] [PubMed] [Google Scholar]
- 153.Zhang C. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. 2016;113(14):E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang C. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217-and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7(40):64527. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bansal H. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28(5):1171. doi: 10.1038/leu.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kwok C.-T. Genetic alterations of m 6 A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10(1):39. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Barbieri I. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552(7683):126. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vu L.P. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23(11):1369. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li Z. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang S. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer cell. 2017;31(4):591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gong A.-H. FoxM1 drives a feed-forward STAT3-activation signaling loop that promotes the self-renewal and tumorigenicity of glioblastoma stem-like cells. Cancer Res. 2015;75(11):2337–2348. doi: 10.1158/0008-5472.CAN-14-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jin D.I. Expression and roles of W ilms' tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103(12):2102–2109. doi: 10.1111/cas.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lin S. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen M. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 165.Kandimalla R. RNAMethyPro: a biologically conserved signature of N6-methyladenosine regulators for predicting survival at pan-cancer level. NPJ Precis. Oncol. 2019;3 doi: 10.1038/s41698-019-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Y. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer. 2019;18(1):137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zheng Y. , et al. m6AVar: a database of functional variants involved in m6A modification.Nucleic Acids . Res. 2017 ; gkx895 - gkx895. [DOI] [PMC free article] [PubMed]
- 168.Han Y. A Visualization and Exploration Database for m(6) As in Cell Lines. Cells. 2019;8(2):168. doi: 10.3390/cells8020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tang Y. DRUM: inference of disease-associated m6A RNA methylation sites from a multi-layer heterogeneous network. Front. Genet. 2019;10:266. doi: 10.3389/fgene.2019.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu X. m6Acomet: large-scale functional prediction of individual m6A RNA methylation sites from an RNA co-methylation network. BMC Bioinf. 2019;20(1):223. doi: 10.1186/s12859-019-2840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.An S. Integrative network analysis identifies cell-specific trans regulators of m6A. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkz1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 2010;6(12):863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 173.Saletore Y. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13(10):175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xu X. Advances in methods and software for RNA cytosine methylation analysis. Genomics. 2019;112(2):1840–1846. doi: 10.1016/j.ygeno.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu N. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tanzer A. RNA modifications in structure prediction - status quo and future challenges. Methods. 2018;159:32–39. doi: 10.1016/j.ymeth.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 177.Li Y. MeRIP-PF: an easy-to-use pipeline for high-resolution peak-finding in MeRIP-Seq data. Genomics Proteom. Bioinform. 2013;11(1):72–75. doi: 10.1016/j.gpb.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang M. A Bayesian hierarchical model for analyzing methylated RNA immunoprecipitation sequencing data. Quant. Biol. 2018;6(3):275–286. doi: 10.1007/s40484-018-0149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Antanaviciute A. m6aViewer: software for the detection, analysis and visualization of N6-methyl-adenosine peaks from m6A-seq/ME-RIP sequencing data. RNA. 2017;23(10):1493–1501. doi: 10.1261/rna.058206.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cui X. A novel algorithm for calling mRNA m6A peaks by modeling biological variances in MeRIP-seq data. Bioinformatics. 2016;32(12):i378–i385. doi: 10.1093/bioinformatics/btw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang Y.-C. Spatially enhanced differential RNA methylation analysis from affinity-based sequencing data with hidden markov model. Biomed Res. Int. 2015;2015:12. doi: 10.1155/2015/852070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cui X. MeTDiff: a Novel Differential RNA Methylation Analysis for MeRIP-Seq Data. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2018;15(2):526–534. doi: 10.1109/TCBB.2015.2403355. [DOI] [PubMed] [Google Scholar]
- 183.Liu L. DRME: count-based differential RNA methylation analysis at small sample size scenario. Anal. Biochem. 2016;499:15–23. doi: 10.1016/j.ab.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 184.Schwartz S. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8(1):284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu L. Decomposition of RNA methylome reveals co-methylation patterns induced by latent enzymatic regulators of the epitranscriptome. Mol. BioSyst. 2015;11(1):262–274. doi: 10.1039/c4mb00604f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen K. Enhancing epitranscriptome module detection from m6A-seq data using threshold-based measurement weighting strategy. Biomed Res. Int. 2018 doi: 10.1155/2018/2075173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cui X. A hierarchical model for clustering m6A methylation peaks in MeRIP-seq data. BMC Genomics. 2016;17(7):520. doi: 10.1186/s12864-016-2913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hauenschild, R., et al., CoverageAnalyzer (CAn): A Tool for Inspection of Modification Signatures in RNA Sequencing Profiles. Biomolecules, 2016. 6(4). [DOI] [PMC free article] [PubMed]
- 189.Ryvkin P. HAMR: high-throughput annotation of modified ribonucleotides. RNA. 2013;19(12):1684–1692. doi: 10.1261/rna.036806.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liang F. BS-RNA: an efficient mapping and annotation tool for RNA bisulfite sequencing data. Comput Biol Chem. 2016;65:173–177. doi: 10.1016/j.compbiolchem.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 191.Legrand C. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27(9):1589–1596. doi: 10.1101/gr.210666.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu J. Episo: quantitative estimation of RNA 5-methylcytosine at isoform level by high-throughput sequencing of RNA treated with bisulfite. Bioinformatics. 2019;36(7):2033–2039. doi: 10.1093/bioinformatics/btz900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Feng P. iRNA-PseColl: identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Mol. Ther. Nucleic Acids. 2017;7:155–163. doi: 10.1016/j.omtn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao Z. Imbalance learning for the prediction of N 6-Methylation sites in mRNAs. BMC Genomics. 2018;19(1):574. doi: 10.1186/s12864-018-4928-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chen W. iRNA-Methyl: Identifying N6-methyladenosine sites using pseudo nucleotide composition. Anal. Biochem. 2015;490:26–33. doi: 10.1016/j.ab.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 196.Liu Z. pRNAm-PC: predicting N6-methyladenosine sites in RNA sequences via physical-chemical properties. Anal. Biochem. 2015 doi: 10.1016/j.ab.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 197.Chen W., Xing P., Zou Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci. Rep. 2017;7:40242. doi: 10.1038/srep40242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen W. Identification and analysis of the N6-methyladenosine in the Saccharomyces cerevisiae transcriptome. Sci. Rep. 2015;5:13859. doi: 10.1038/srep13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jia C.Z., Zhang J.J., Gu W.Z. RNA-MethylPred: a high-accuracy predictor to identify N6-methyladenosine in RNA. Anal Biochem. 2016;510:72–75. doi: 10.1016/j.ab.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 200.Li, G.Q., et al., TargetM6A: Identifying N6-methyladenosine Sites from RNA Sequences via Position-Specific Nucleotide Propensities and a Support Vector Machine. IEEE Transactions on NanoBioscience, 2016.15(7): 674-682. [DOI] [PubMed]
- 201.Chen W. iRNA(m6A)-PseDNC: identifying N6-methyladenosine sites using pseudo dinucleotide composition. Anal Biochem. 2018;561:59–65. doi: 10.1016/j.ab.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 202.Wei L. M6APred-EL: A sequence-based predictor for identifying N6-methyladenosine sites using ensemble learning. Mol Ther Nucleic Acids. 2018;12:635–644. doi: 10.1016/j.omtn.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wei L. Integration of deep feature representations and handcrafted features to improve the prediction of N 6 -methyladenosine sites. Neurocomputing. 2018;324:3–9. [Google Scholar]
- 204.Akbar S. iMethyl-STTNC: identification of N6-methyladenosine sites by extending the Idea of SAAC into Chou’s PseAAC to formulate RNA sequences. J. Theor. Biol. 2018;455:205–211. doi: 10.1016/j.jtbi.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 205.Zhao X. Identifying N6-methyladenosine sites using extreme gradient boosting system optimized by particle swarm optimizer. J. Theor. Biol. 2019;467:39–47. doi: 10.1016/j.jtbi.2019.01.035. [DOI] [PubMed] [Google Scholar]
- 206.Zhuang Y. A linear regression predictor for identifying N6-methyleadenosine sites using frequent gapped K-mer pattern. Mol. Ther. Nucleic Acids. 2019 doi: 10.1016/j.omtn.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen W. Identifying N6-methyladenosine sites in the Arabidopsis thaliana transcriptome. Mol. Genet. Genomics. 2016;291(6):2225–2229. doi: 10.1007/s00438-016-1243-7. [DOI] [PubMed] [Google Scholar]
- 208.Xiang S. AthMethPre: a web server for the prediction and query of mRNA m6A sites in Arabidopsis thaliana. Mol. BioSyst. 2016;12(11):3333–3337. doi: 10.1039/c6mb00536e. [DOI] [PubMed] [Google Scholar]
- 209.Wang X. RFAthM6A: a new tool for predicting m(6)A sites in Arabidopsis thaliana. Plant Mol Biol. 2018;96(3):327–337. doi: 10.1007/s11103-018-0698-9. [DOI] [PubMed] [Google Scholar]
- 210.Zhang J. Identifying RNA N6-methyladenosine sites in Escherichia coli genome. Front. Microbiol. 2018;9:955. doi: 10.3389/fmicb.2018.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chen W. MethyRNA: A web-server for identification of N-methyladenosine sites. J Biomol Struct Dyn. 2016;35(3):683–687. doi: 10.1080/07391102.2016.1157761. [DOI] [PubMed] [Google Scholar]
- 212.Xiang S. RNAMethPre: a web server for the prediction and query of mRNA m6A Sites. PLoS ONE. 2016;11(10) doi: 10.1371/journal.pone.0162707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhou Y. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44(10):e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen W. iRNA-3typeA: identifying three types of modification at RNA’s adenosine sites. Mol Ther Nucleic Acids. 2018;11:468–474. doi: 10.1016/j.omtn.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Dao F.-Y. Computational identification of N6-Methyladenosine sites in multiple tissues of mammals. Comput Struct Biotechnol. J. 2020;18:1084–1091. doi: 10.1016/j.csbj.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xing P. Identifying N6-methyladenosine sites using multi-interval nucleotide pair position specificity and support vector machine. Sci Rep. 2017;7:46757. doi: 10.1038/srep46757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wenzhong, L., SICM6A: Identifying m6A Site across Species by Transposed GRU Network. bioRxiv, 2019: p. 694158.
- 218.Qiang X. M6AMRFS: robust prediction of N6-methyladenosine sites with sequence-based features in multiple species. Front. Genet. 2018;9:495. doi: 10.3389/fgene.2018.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Feng P. Identifying RNA 5-methylcytosine sites via pseudo nucleotide compositions. Mol. BioSyst. 2016;12(11):3307–3311. doi: 10.1039/c6mb00471g. [DOI] [PubMed] [Google Scholar]
- 220.Qiu W.-R. iRNAm 5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget. 2017;8(25):41178–41188. doi: 10.18632/oncotarget.17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang M. Accurate RNA 5-methylcytosine site prediction based on heuristic physical-chemical properties reduction and classifier ensemble. Anal Biochem. 2018;550:41–48. doi: 10.1016/j.ab.2018.03.027. [DOI] [PubMed] [Google Scholar]
- 222.Sabooh M.F. Identifying 5-methylcytosine sites in RNA sequence using composite encoding feature into Chou's PseKNC. J Theor Biol. 2018;452:1–9. doi: 10.1016/j.jtbi.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 223.Fang T. RNAm 5CPred: Prediction of RNA 5-methylcytosine sites based on three different kinds of nucleotide composition. Mol. Ther. Nucleic Acids. 2019:739–748. doi: 10.1016/j.omtn.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Akbar S. iRNA-PseTNC: identification of RNA 5-methylcytosine sites using hybrid vector space of pseudo nucleotide composition. Front. Comput. Sci. 2019;14(2):451–460. [Google Scholar]
- 225.Dou, L., et al., iRNA-m5C_NB: a novel predictor to identify RNA 5-Methylcytosine sites based on the Naive Bayes classifier. . IEEE Access, 2020; 8: 84906 - 84917.
- 226.Song J. Transcriptome-wide annotation of m5C RNA modifications using machine learning. Frontiers Plant Sci. 2018:9(519). doi: 10.3389/fpls.2018.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Li J. RNAm 5Cfinder: a web-server for predicting RNA 5-methylcytosine (m5C) Sites based on random forest. Sci. Rep. 2018;8(1):17299. doi: 10.1038/s41598-018-35502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li Y.-H. PPUS: a web server to predict PUS-specific pseudouridine sites. Bioinformatics. 2015;31(20):3362–3364. doi: 10.1093/bioinformatics/btv366. [DOI] [PubMed] [Google Scholar]
- 229.Song B. PIANO: a web server for pseudouridine site (Ψ) identification and functional annotation. Front Genet. 2020;11(88) doi: 10.3389/fgene.2020.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chen W. iRNA-PseU: identifying RNA pseudouridine sites. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.He J. PseUI: pseudouridine sites identification based on RNA sequence information. BMC Bioinf. 2018;19(1):306. doi: 10.1186/s12859-018-2321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Liu K. XG-PseU: an eXtreme Gradient Boosting based method for identifying pseudouridine sites. Mol. Genet. Genomics. 2019;295(1):13–21. doi: 10.1007/s00438-019-01600-9. [DOI] [PubMed] [Google Scholar]
- 233.Tahir M., Tayara H., Chong K.T. iPseU-CNN: identifying RNA pseudouridine sites using convolutional neural networks. Mol Therapy Nucleic Acids. 2019;16:463–470. doi: 10.1016/j.omtn.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bi Y. EnsemPseU: Identifying pseudouridine sites with an ensemble approach. IEEE Access. 2020;8:79376–79382. [Google Scholar]
- 235.Qiu W.-R. iRNA-2methyl: identify RNA 2'-O-methylation sites by incorporating sequence-coupled effects into general PseKNC and ensemble classifier. Med. Chem. 2017;13(8):734–743. doi: 10.2174/1573406413666170623082245. [DOI] [PubMed] [Google Scholar]
- 236.Yang H. iRNA-2OM: a sequence-based predictor for identifying 2'-O-methylation sites in homo sapiens. J Comput Biol. 2018;25(11):1266–1277. doi: 10.1089/cmb.2018.0004. [DOI] [PubMed] [Google Scholar]
- 237.Chen, W., et al., Identifying 2 al., l., sed Predictor for Identifying 2'-O-leotide chemical properties and nucleotide compositions. Genomics, 2016. 107(6): p. 255-258. [DOI] [PubMed]
- 238.Lian, L., et al., ISGm1A: Integration of sequence features and genomic features to improve the prediction of human m1A RNA methylation sites. IEEE Access, 2020. 8(1): 81971 - 81977.
- 239.Chen W. RAMPred: identifying the N1-methyladenosine sites in eukaryotic transcriptomes. Sci. Rep. 2016;6:31080. doi: 10.1038/srep31080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Chen W. iRNA-AI: identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget. 2017;8(3):4208. doi: 10.18632/oncotarget.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ahmad A., Shatabda S. EPAI-NC: Enhanced prediction of adenosine to inosine RNA editing sites using nucleotide compositions. Anal. Biochem. 2019;569:16–21. doi: 10.1016/j.ab.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 242.Chen W. PAI: Predicting adenosine to inosine editing sites by using pseudo nucleotide compositions. Sci. Rep. 2016;6:35123. doi: 10.1038/srep35123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Chen W. iRNA-m2G: identifying N2-methylguanosine sites based on sequence derived information. Mol. Ther. Nucleic Acids. 2019;18:253–258. doi: 10.1016/j.omtn.2019.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chen W. iRNA-m7G: identifying N7-methylguanosine sites by fusing multiple features. Mol. Ther. Nucleic Acids. 2019;18:269–274. doi: 10.1016/j.omtn.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Song B. m7GHub: deciphering the location, regulation and pathogenesis of internal mRNA N7-methylguanosine (m7G) sites in human. Bioinformatics. 2020;36(11):3528–3536. doi: 10.1093/bioinformatics/btaa178. [DOI] [PubMed] [Google Scholar]
- 246.Xu Z.-C. iRNAD: a computational tool for identifying D modification sites in RNA sequence. Bioinformatics. 2019;35(23):4922–4929. doi: 10.1093/bioinformatics/btz358. [DOI] [PubMed] [Google Scholar]
- 247.Liu, Y., et al., iRNA5hmC: The First Predictor to Identify RNA 5-Hydroxymethylcytosine Modifications Using Machine Learning. Frontiers in Bioengineering and Biotechnology, 2020. 8(227). [DOI] [PMC free article] [PubMed]
- 248.Zhao W. PACES: prediction of N4-acetylcytidine (ac4C) modification sites in mRNA. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-47594-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.