Abstract
The CXCR5 (C-X-C motif chemokine receptor 5) is chemokine transmembrane receptor, acting via its ligand CXCL13 and plays a crucial role in controlling the trafficking of inflammatory cells into and from the sub-retinal space, which contributes to the pathogenesis of AMD. We have previously described the genetic ablation of CXCR5 deficiency causes RPE/choroid abnormalities and retinal degeneration (RD) in aged mice. Here we report the transcriptome data (RNA-Seq) of 24 months old CXCR5 knockout (KO) and age-matched C57BL/6 controls (WT). RNA sequencing was performed on the Illumina HiSeq 2500, providing up to 300 GB of sequence information per flow cell. The quality of RNA-seq libraries, RNA intensity were validated by Agilent Technologies Bioanalyzer-2100. The raw datasets contains on average 292,004,59 reads (after trimming 284,862,43 reads) in retina and 272,527,90 reads (after trimming 266,173,11 reads) in choroid samples. The mapped reads showed that a total of 1586 genes in retina and 1462 genes in choroid are differentially expressed in this experiment. The raw datasets were deposited into NCBI Sequence Read Archive (SRA) database and can be accessed via accession number PRJNA588421.
Keywords: CXCR5, aging, mice, retina, choroid, retinal degeneration, RNA-Seq, FastQC
Specifications Table
| Subject | Genetics, Genomics and Molecular Biology |
| Specific subject area | Mice transcriptomics (RNA-Seq) |
| Type of data | Transcriptome sequences Reads, Tables, and Figures. |
| How data were acquired | Sequencing RNA from the choroid and retinal tissues of CXCR5-deficient and WT control mice 24 months of age. |
| Data format | Raw (FASTQ) and analyzed (Tables) |
| Parameters for data collection | The [B6.129S2(Cg)-CXCR5tm1Lipp/J] (CXCR5 KO) and [C57BL/6J] (WT) mice strains were purchased from Jackson Laboratory. B6.129S2(Cg)-CXCR5tm1Lipp/J. All mice were fed standard chow diets and provided with water ad libitum. Mice were sacrificed at 24 months of age. RPE/choroid and retinal tissues were isolated from C57BL6 WT mice and CXCR5 KO mice and used for total RNA extraction, cDNA library preparation, and sequencing. |
| Description of data collection | The transcriptome dataset was collected from paired-end sequencing of mice cDNA libraries using Illumina HiSeq 2500 platform with 15 million paired 50 bp reads were obtained per sample. The raw reads were recorded in a FastQ file. Raw reads were filtered to remove reads containing adapter or reads of low quality, and clean reads were mapped to reference genome mouse (mm10). Total mapped reads and the number of transcripts were estimated from transcript assembly with a threshold of FPKM ≥ 0.2. |
| Data source location | University of Missouri School of Medicine, Mason Eye Institute, Ophthalmology, Columbia, Missouri-65201, USA. |
| Data accessibility | Repository name: NCBI Sequence Read Archive (SRA) Data identification number: PRJNA588421 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA588421 |
Value of the Data
-
•
These datasets provide the transcriptome profile of retina and choroid of aged CXCR5 KO and WT mice.
-
•
Aged CXCR5 KO mice retina and choroid tissue datasets can be useful to experimental mouse models of retinal degeneration.
-
•
The RNAseq datasets, together with human genomic data are important for the identification of functional gene markers such as diferentialy expressed genes (DEGs), single nucleotide polymorphisms (SNPs) related to age-related macular degeneration and retinal degeneration diseases research.
-
•
24-months old RNAseq WT mouse datasets can provide the baseline in the eye aging studies as producing such animal data is time and resource consuming.
1. Data Description
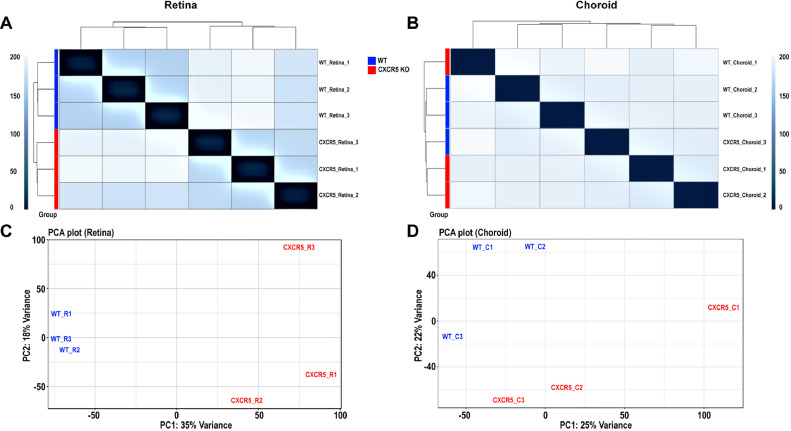
We have recently reported retinal degeneration phenotype in aged CXCR5-deficient (CXCR5 KO) mice associated with age-related macular degeneration (AMD) associated proteins accumulation. [1] Here, we present the whole transcriptome datasets from RPE/choroid and retinal tissues from 24 months old CXCR5 KO mice and age-matched WT controls. The schematic presentation of the experimental design was present in Fig. 1.RNA sequencing was performed on the Illumina HiSeq 2500, providing up to 300 GB of sequence information per flow cell. The quality of the RNA-seq libraries, RNA integrity, was evaluated with a Bioanalyzer-2100 (Agilent Technologies). The volume, concentration, and RNA Integrity Number (RIN) values of the RNA samples were presented in Table 1. RIN value ≥ 7.0 was set as the cut-off for sample inclusion for downstream processing for RNA sequencing analysis. Data were trimmed with Trimmomatic software to remove the adaptors and failed reads, the results of trimmed of datasets were shown in Table 2, with an average of 2.38% reads removed. The data trimming had minimal effect on evaluated parameters, indicating the high quality of the original raw datasets. The trimmed data were mapped to the mouse reference genome (mm10) by HISAT2 with default parameters. The key alignments were shown in Table 3, with an average of 95.1% reads mapped. The log2-transform count data were visualized by box plots (Fig. 2A, C) and density plots (Fig. 2B, D) based on the count distributions within the samples. We further validated the reproducibility across the samples using hierarchical clustering for retina and choroid samples (Fig. 3A, B). The principal component analysis (PCA), using the replicates log2-transformed counts after library-size normalization and variance stabilization (Fig. 3C, D). The hierarchical clustering plots showed control, and CXCR5 KO samples that were well separated and clustered together from each other by their expression difference in both retina and choroid tissues. The PCA plot showed control, and CXCR5 KO samples (retina and choroid) that were well separated from each other by their difference in the first PC, which explained 35% and 25% of the variance in the expression variables. The aligned reads were used to calculate the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) based on transcript abundance by HTSeq-count (Supplementary file1 and 2). The counts were applied to the package DESeq2 v1.26 (Bioconductor) to identify differentially expressed genes (DEGs). The raw reads were deposited into NCBI Sequence Read Archive (SRA) database and can be accessed via accession number PRJNA588421.
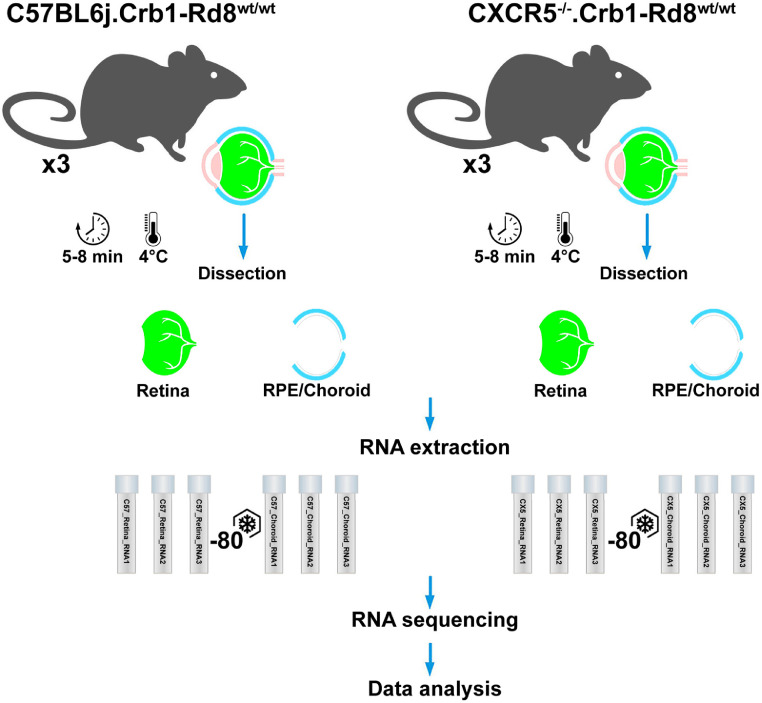
Fig. 1.
Schematic presentation of the experimental design. 24-months old WT, CXCR5 KO mice were sacrificed, and retina and choroid tissues were dissected on ice. Then, total RNA was purified from the tissues and used for RNA sequencing.
Table 1.
RNA quality control summary parameters and samples.
| S. No | File name | Novogene ID | Concentration (ng/ul) | Volume (uL) | Amount (ug) | RIN | Result | Sample | Tissue |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C57_R1 | USR18008997 | 137.49 | 45 | 6.187 | 7.0 | Pass | WT 1 | Retina |
| 2 | C57_R2 | USR18008998 | 138.91 | 46 | 6.390 | 7.2 | Pass | WT 2 | Retina |
| 3 | C57_R3 | USR18008999 | 136.83 | 45 | 6.123 | 7.0 | Pass | WT 3 | Retina |
| 4 | CXCR5_R1 | USR18009003 | 83.16 | 39 | 3.243 | 7.2 | Pass | CXCR5 KO1 | Retina |
| 5 | CXCR5_R2 | USR18009004 | 57.01 | 42 | 2.394 | 7.8 | Pass | CXCR5 KO2 | Retina |
| 6 | CXCR5_R3 | USR18009005 | 41.55 | 40 | 1.662 | 7.3 | Pass | CXCR5 KO3 | Retina |
| 7 | C57_C1 | USR18009000 | 77.50 | 17.1 | 1.325 | 8.6 | Pass | WT 1 | Choroid |
| 8 | C57_C2 | USR18009001 | 102.17 | 17.5 | 1.788 | 8.5 | Pass | WT 2 | Choroid |
| 9 | C57_C3 | USR18009002 | 83.24 | 18.9 | 1.780 | 8.6 | Pass | WT 3 | Choroid |
| 10 | CXCR5_C1 | USR18009006 | 40.87 | 30 | 1.226 | 7.6 | Pass | CXCR5 KO1 | Choroid |
| 11 | CXCR5_C2 | USR18009007 | 48.45 | 31 | 1.502 | 8.5 | Pass | CXCR5 KO2 | Choroid |
| 12 | CXCR5_C3 | USR18009008 | 47.81 | 32 | 1.530 | 8.2 | Pass | CXCR5 KO3 | Choroid |
Table 2.
Trimmomatic results summary, before and after trimmed datasets.
| S. No | File name | Before trimmed | After trimmed | Surviving reads (%) | Dropped reads (%) | Sample | Tissue |
|---|---|---|---|---|---|---|---|
| No of input reads | No of output reads | ||||||
| 1 | C57_R1 | 27712397 | 27085386 | 97.74 | 2.26 | WT 1 | Retina |
| 2 | C57_R2 | 27596737 | 26929856 | 97.59 | 2.41 | WT 2 | Retina |
| 3 | C57_R3 | 32760542 | 31915161 | 97.42 | 2.58 | WT 3 | Retina |
| 4 | CX5_R1 | 29072482 | 28380053 | 97.62 | 2.38 | CXCR5 KO1 | Retina |
| 5 | CX5_R2 | 30374628 | 29487454 | 97.07 | 2.93 | CXCR5 KO2 | Retina |
| 6 | CX5_R3 | 27685968 | 27119548 | 97.95 | 2.05 | CXCR5 KO3 | Retina |
| 7 | C57_C1 | 28405466 | 27846296 | 98.03 | 1.97 | WT 1 | Choroid |
| 8 | C57_C2 | 27248598 | 26552842 | 97.44 | 2.56 | WT 2 | Choroid |
| 9 | C57_C3 | 28759607 | 28149692 | 97.88 | 2.12 | WT 3 | Choroid |
| 10 | CX5_C1 | 28380944 | 27541862 | 97.04 | 2.96 | CXCR5 KO1 | Choroid |
| 11 | CX5_C2 | 24969365 | 24422598 | 97.81 | 2.19 | CXCR5 KO2 | Choroid |
| 12 | CX5_C3 | 25752760 | 25190580 | 97.81 | 2.19 | CXCR5 KO3 | Choroid |
Table 3.
HISTA2 alignment summary, the reads in these datasets have aligned to the mouse reference genome (mm10).
| S. No | Sample Name | No of input reads | Ave. input read length | Mapped reads (%) | Unmapped reads (%) | Sample | Tissue |
|---|---|---|---|---|---|---|---|
| 1 | C57_R1 | 27085386 | 136 | 97.02 | 2.98 | WT 1 | Retina |
| 2 | C57_R2 | 26929856 | 136 | 96.84 | 3.16 | WT 2 | Retina |
| 3 | C57_R3 | 31915161 | 136 | 97.06 | 2.94 | WT 3 | Retina |
| 4 | CXCR5_R1 | 28380053 | 136 | 96.64 | 3.36 | CXCR5 KO1 | Retina |
| 5 | CXCR5_R2 | 29487454 | 136 | 96.52 | 3.48 | CXCR5 KO2 | Retina |
| 6 | CXCR5_R3 | 27119548 | 136 | 96.25 | 3.75 | CXCR5 KO3 | Retina |
| 7 | C57_C1 | 27846296 | 136 | 87.52 | 12.48 | WT 1 | Choroid |
| 8 | C57_C2 | 26552842 | 136 | 87.21 | 12.79 | WT 2 | Choroid |
| 9 | C57_C3 | 28149692 | 136 | 96.81 | 3.19 | WT 3 | Choroid |
| 10 | CXCR5_C1 | 27541862 | 136 | 96.89 | 3.11 | CXCR5 KO1 | Choroid |
| 11 | CXCR5_C2 | 24422598 | 136 | 96.84 | 3.16 | CXCR5 KO2 | Choroid |
| 12 | CXCR5_C3 | 25190580 | 136 | 96.67 | 3.33 | CXCR5 KO3 | Choroid |
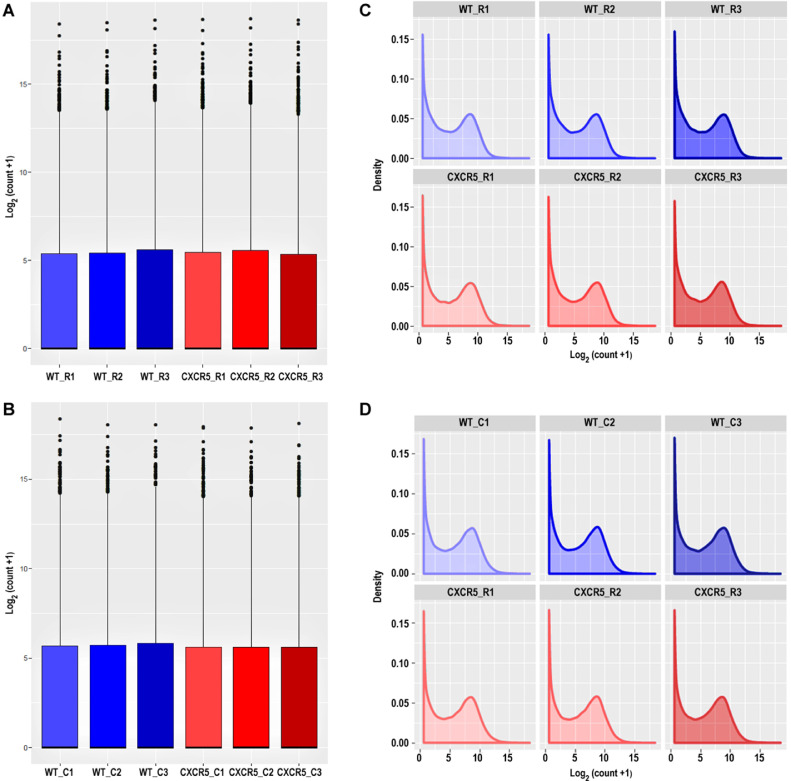
Fig. 2.
The log2-transform count data results were visualized by box plots based on the count distributions within the samples in the retina (A) and choroid (B) tissue. The log2-transform count data results were visualized by density plots based on the count distributions within the samples in the retina (C) and choroid tissue (D).
Fig. 3.
(A) Matrix correlation plot of three control samples (WT_Retina_1, WT_Retina_2, WT_Retina_3) and three CXCR5 KO samples (CXCR5_Retina_1, CXCR5_Retina_2, CXCR5_Retina_3) using the euclidean distance of the log2-transformed counts (after library size normalized and variance stabilized) in retina tissue. (B) Matrix correlation plot of three control samples (WT_Choroid_1, WT_Choroid_2, WT_Choroid_3) and three CXCR5 KO samples (CXCR5_Choroid_1, CXCR5_Choroid_2, CXCR5_Choroid_3) using the euclidean distance of the log2-transformed counts (after library size normalized and variance stabilization) in choroid tissue. A darker color means a smaller Euclidean distance, i.e., more correlated. (C) Principle component analysis in retina samples: X and Y axis show principal component 1 and principal component 2 that explain 35% and 18% of the variance. (D) Principle component analysis in choroid samples: X and Y axis show principal component 1 and principal component 2 that explain 25% and 22% of the variance.
2. Experimental Design, Materials and Methods
2.1. Animals
All experiments were approved in the University of Missouri Institutional Animal Care and Use Committee (protocol number: 9520) and were performed following the statement for the use of animals in ophthalmic and vision research of the association for research in vision and ophthalmology (ARVO). The [B6.129S2(Cg)-CXCR5tm1Lipp/J] (CXCR5 KO) (https://www.jax.org/strain/006659) and [C57BL/6J] (WT) mice strains were purchased from Jackson Laboratory. B6.129S2(Cg)-CXCR5tm1Lipp/J mice are on a C57/BL6J background with a small component of C57/BL6N genes (http://jaxmice.jax.org/strain/006659.html). The CXCR5 gene was replaced by the neomycin resistance gene in 129S2/SvPas-derived D3 embryonic stem cells. Resulting mutant mice were then backcrossed to C57BL/6 mice for eight generations [2]. Both CXCR5 KO and control mice were housed at the special pathogen-free animal facilities of the Bone Life Sciences Center at the University of Missouri and were fed normal chow diets and provided with water ad libitum.
2.2. Animal genotyping
Genotyping was performed with the assistance of Transnetyx: Outsourced PCR Genotyping Services (www.transnetyx.com) using custom-designed genotyping primers (TransnetYX) complementary to the sequence fragments of CXCR5 and neomycin resistance genes. RD8 genotyping was conducted using the custom Rd8 genotyping probe based on our previous Sanger sequencing data of the region 3600-3700 of the Crb1 gene (canonical transcript M_133239). [1] All animals were found to be Rd8 mutation free.
2.3. Sample collection and total RNA isolation
CXCR5 KO and WT control animals (age 24 months) were euthanized by intraperitoneal injection of ketamine hydrochloride (300 mg/kg body weight). Mouse choroid and retina tissues were isolated, as reported previously [1]. The time from the enucleation to the dissected retina and choroid tissue samples stabilization in RNAlater™ Stabilization Solution (AM7020; Thermo Fisher Scientific, Waltham, MA, USA) was 5-8 minutes. Total RNA from CXCR5 KO and controls tissues (retina and choroid) was extracted using the RNeasy Plus Mini Kit extraction kit (Qiagen, Gaithersburg, MD, USA) according to the extraction protocol. Tissue material from both eyes of each animal was pooled into a single sample. gDNA was removed by the gDNA eliminator column supplied with the RNeasy Plus Mini Kit.
2.4. Library preparation and sequencing
Sample preparation and Illumina sequencing were carried out at Novogene Leading Edge Genomic Services & Solutions, CA, USA. The samples were further DNase-treated to remove residual gDNA, and RNA quality was determined using the Agilent bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA integrity number (RIN) value greater than 7.0 was set as the cut-off for sample inclusion for downstream processing for RNA sequencing analysis. RIN is an RNA integrity and level of degradation and fragmentation established by Agilent Technologies. The values greater than 7 are considered sufficient for whole transcriptome analysis. [3] RNA sequencing was performed on the Illumina HiSeq 2500 using the latest versions of sequencing reagents and flow cells, providing up to 300 GB of sequence information per flow cell. TruSeq library generation kits were used according to the manufacturer's instructions (Illumina, San Diego, CA, USA).
2.5. Sequencing quality, and reference genome mapping
The paired-end fastq raw reads were trimmed for adapters, and low-quality reads using Trimmomatic version 0.36 with default parameter [4]. The trimming was performed with the minimal length set at 36, leading set 5, slindingwindow set 4:15, headcrop set 12, and maintaining a phred-score ≤ 30. After adapter removal, the quality of each paired-end sequence file was assessed using FastQC analysis (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [5]. The splice-mapping algorithm of HISAT2 (2.1.0) (http://ccb.jhu.edu/software/hisat2/index.shtml) was used to perform genome mapping of the pre-processed reads [6]. The clean data were then mapped to the mouse reference genome (mm10) with HISAT2 [7], and the default parameters were used.
2.6. Tissue purity validation using marker genes
We confirmed the lack of contamination of RNA samples obtained from the retina and choroid with different tissue material (i.e., optic nerve, muscle cells) during the eyecup dissection procedure. Gene markers that are typical for transcription in the retina (MATH5; BRNB3; PDE7A; PDE12; RHOT2) and RPE (RPE65; RLBP1; MITF and respectively were validated in all samples (Table 4).
Table 4.
RNA sample tissue origin validation based on retinal and choroidal tissue gene markers
| Gene name | Abbreviation | Ensembl | Retina | Choroid |
|---|---|---|---|---|
| Atonal BHLH Transcription Factor 7 | Math5 (Atoh7) | ENSG00000179774 | + | - |
| Brain-3B | POU4F2 (BRNB3) | ENSG00000151615 | + | - |
| Phosphodiesterase 7A | PDE7A | ENSG00000205268 | + | - |
| Phosphodiesterase 12 | PDE12 | ENSG00000174840 | + | - |
| Ras Homolog Family Member T2 | RHOT2 | ENSG00000140983 | + | - |
| Retinoid Isomerohydrolase RPE65 | RPE65 | ENSG00000116745 | - | + |
| RalA Binding Protein 1 | RLBP1 | ENSG00000017797 | - | + |
| Melanocyte Inducing Transcription Factor | MITF | ENSG00000187098 | - | + |
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Ethics Statement
All experiments were approved by the Institutional Animal Care and Use Committee of the University of Missouri School of Medicine (protocol number: 9520) and were in accordance with the guidelines of the Association for Research in Vision and Ophthalmology Statement for the use of animals in ophthalmic and vision research.
Acknowledgments
The authors wish to acknowledge the contribution of the Center for Biomedical Informatics (CBMI) University of Missouri (Columbia, MO, USA) for computer application facilities. Ms. Allen Raye (University of Missouri Department of Biomedical Sciences, Columbia, Missouri, USA) for assistance with animal resources; Ms. Lijuan Fan (Department of Ophthalmology, University of Missouri School of Medicine, Columbia, Missouri) for RNA extraction assistance. Mr. Dmitry Rumyancev (Belgorod, Russia) for the graphical abstract artwork design. Ms. Amy A. Folkerts from the University of Missouri, Department of Ophthalmology, Scientific Grants Submission Office (SGSO) for language corrections. The authors like to acknowledge the fincianl support from National Institute of Health (NIH) R01 grant (EY027824, H.H.) and Missouri University start-up fund.
Authors Contributions
This datasets deposition was conceived and designed by M.S.S., A.L., and H.H. A.L. performed ocular tissue dissection and RNA extraction. M.S.S. has performed quality checks, reference genome assembly, gene expression analysis. M.S.S has conducted DESeq2 and R program analysis. M.S.S and A.L have conducted figures design and statistical analysis. The manuscript was written by M.S.S, A.L., H.H., and critically revised by H.H. All Authors reviewed and accepted the final version of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105915.
Contributor Information
Anton Lennikov, Email: lennikov@gmail.com, lennikova@health.missouri.edu.
Hu Huang, Email: huangh1@missouri.edu.
Appendix. Supplementary materials
References
- 1.Lennikov A., Saddala M.S., Mukwaya A., Tang S., Huang H. Autoimmune-Mediated Retinopathy in CXCR5-Deficient Mice as the Result of Age-Related Macular Degeneration Associated Proteins Accumulation. Frontiers in immunology. 2019;10:1903. doi: 10.3389/fimmu.2019.01903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waehre A., Halvorsen B., Yndestad A., Husberg C., Sjaastad I., Nygard S., Dahl C.P., Ahmed M.S., Finsen A.V., Reims H., Louch W.E., Hilfiker-Kleiner D., Vinge L.E., Roald B., Attramadal H., Lipp M., Gullestad L., Aukrust P., Christensen G. Lack of chemokine signaling through CXCR5 causes increased mortality, ventricular dilatation and deranged matrix during cardiac pressure overload. PLoS One. 2011;6:e18668. doi: 10.1371/journal.pone.0018668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui C., Benard E.L., Kanwal Z., Stockhammer O.W., van der Vaart M., Zakrzewska A., Spaink H.P., Meijer A.H. Infectious disease modeling and innate immune function in zebrafish embryos. Methods Cell Biol. 2011;105:273–308. doi: 10.1016/B978-0-12-381320-6.00012-6. [DOI] [PubMed] [Google Scholar]
- 4.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H., Lennikov A., Saddala M.S., Gozal D., Grab D.J., Khalyfa A., Fan L. Placental growth factor negatively regulates retinal endothelial cell barrier function through suppression of glucose-6-phosphate dehydrogenase and antioxidant defense systems. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:13695–13709. doi: 10.1096/fj.201901353R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saddala M.S., Lennikov A., Bouras A., Huang H. RNA-Seq reveals differential expression profiles and functional annotation of genes involved in retinal degeneration in Pde6c mutant Danio rerio. BMC genomics. 2020;21:132. doi: 10.1186/s12864-020-6550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.