Abstract
Recombinant adeno-associated virus (rAAV) vectors are a leading gene delivery platform, but vector manufacturing remains a challenge. New methods are needed to increase rAAV yields and reduce costs. Past efforts to improve rAAV production have focused on optimizing a single variable at a time, but this approach does not account for the interactions of multiple factors that contribute to vector generation. Here, we utilized a design-of-experiment (DOE) methodology to optimize rAAV production in a HEK293T suspension cell system. We simultaneously varied the transgene, packaging, and helper plasmid ratios, the total DNA concentration, and the cell density to systematically evaluate the impact of each variable across 52 conditions. The results revealed a unique set of parameters with a lower concentration of transgene plasmid, a higher concentration of packaging plasmid, and a higher cell density than previously described protocols. Using this DOE-optimized protocol, we achieved unpurified yields approaching 3 × 1014 viral genomes (VGs)/L of cell culture. Additionally, we incorporated polyethylene glycol (PEG)-based virus precipitation, pH-mediated protein removal, and affinity chromatography to our downstream processing, enabling average purified yields of >1 × 1014 VGs/L for rAAV-EGFPs across 13 serotypes and capsid variants.
Keywords: Adeno-associated virus, rAAV, suspension cells, design of experiment, affinity purification, production
Graphical Abstract
Zhao et al. utilized a design-of-experiment methodology to optimize rAAV production by simultaneously varying plasmid ratios, total DNA concentration, and cell density. They achieved unpurified yields approaching 3 × 1014 viral genomes (VGs)/L of cell culture and average purified yields >1 × 1014 VGs/L of rAAV-EGFP across multiple serotypes and capsid variants.
Introduction
Recombinant adeno-associated virus (rAAV) vectors are a leading gene delivery platform, and several rAAV-mediated therapies have recently been approved.1, 2, 3 Despite these advances in the clinic, rAAV vector manufacturing remains a challenge. As an example, a phase 1/2 trial for hemophilia B required over 400 ten-layer cell stacks to generate sufficient material for six patients.4,5 Although the trial was successful, this study highlights the need for new methods to improve vector generation. Increased production efficiency will reduce manufacturing costs, improve patient access, and make this emerging modality more feasible for large disease indications.6, 7, 8
Many factors impact rAAV vector production, including the production cell line, the cell density, the culture medium, the harvest time, the total amount of plasmid DNA, and the ratio of the transgene (pAAV), packaging (pRC, a plasmid encoding the AAV Rep and Cap genes), and pHelper plasmids. Historically, groups have optimized vector production by modifying one factor at a time (OFAT).9, 10, 11, 12, 13, 14 For example, Durocher et al.11 optimized rAAV2 production in HEK293 suspension cells by evaluating different plasmid ratios, cell densities, and harvest times. In their OFAT-optimized system, they achieved pre-purification yields approaching 3 × 1013 viral genomes (VGs)/L. Likewise, Grieger et al.14 achieved pre- and post-purification yields greater than 1 × 1014 and 1 × 1013 VGs/L, respectively, by individually varying the plasmid ratios, transfection reagent to plasmid DNA ratio, total plasmid DNA concentrations, and cell density in a HEK293 suspension cell system. Similar to these groups, we initially developed an internal rAAV production system in suspension cells using an OFAT-based approach. Using rAAV5 as a representative vector, we evaluated different cell lines, harvest times, cell densities, and plasmid concentrations to optimize virus production. Although we succeeded in improving rAAV5 vector generation, this protocol failed to produce other serotypes at similar yields. These results drove us to investigate new methods for optimizing rAAV production.
Design-of-experiment (DOE) methodology has been successfully used to optimize biotechnological processes, such as antibiotics production, antibody generation, and embryonic stem cell expansion.15 Unlike OFAT-based optimization, a DOE-driven approach allows one to evaluate the impact of multiple interdependent factors on a given output. In this study, we utilized a DOE methodology to optimize rAAV vector production in a HEK293T suspension cell system. To the best of our knowledge, this represents the first application of DOE methodology to optimize rAAV vector production.16 We simultaneously varied the concentrations of transgene, packaging, and helper plasmids, the total DNA concentration, and the cell density to systematically evaluate the impact of each variable across 52 different conditions. Data analysis revealed a unique set of parameters with a lower concentration of pAAV, a higher concentration of pRC, and a higher cell density compared with previously described methods using OFAT-based approaches. This DOE-optimized protocol allowed us to achieve unpurified yields approaching 3 × 1014/L cell culture. Additionally, we incorporated polyethylene glycol (PEG)-based virus precipitation, pH-mediated protein removal, and affinity chromatography to our downstream processing, enabling us to achieve average purified yields greater than 1 × 1014 VGs/L rAAV-EGFP vectors across 13 serotypes/capsid variants. These yields are significantly greater than the highest previously published rAAV yields of >1 × 1013 VGs/L by triple transfection in suspension cells.
Results
Creation of an OFAT-Optimized rAAV Suspension Cell Production Protocol
To create a suspension cell-based rAAV vector production system, we initially evaluated different production cell lines, cell densities, total plasmid DNA concentrations, and vector harvest times. For these studies, each parameter was assessed individually, and rAAV5-EGFP was used as a model serotype for optimizing production. We selected two cell lines for potential inclusion in our system: suspension HEK293T cells and suspension HEK293-6E cells. HEK293T cells express the SV40 large T antigen and they have previously been shown to produce more rAAVs than parental HEK293 cells.9,17 HEK293-6E cells are an improved variant of HEK293E cells and have increased transgene expression compared with HEK293E cells.18 We compared the transfection efficiency of HEK293-6E and HEK293T suspension cells using the plasmid pAAV-EF1α-EGFP (EF1α, elongation factor 1 alpha promoter) with Polyethylenimine MAX (PEIMAX) as a transfection reagent. The fluorescence intensity of transfected cells and the percentage of EGFP-positive cells were determined at 24 and 48 h post-transfection (HPT) by flow cytometry. HEK293T cells had an 18.2-fold and 5.1-fold higher mean fluorescence intensity than HEK293-6E cells at 24 and 48 HPT, respectively (Figure 1A). Whereas HEK293T and HEK293-6E cells had a similar percentage of EGFP-positive cells at 48 HPT, HEK293T cells had a 2-fold higher percentage of EGFP-positive cells at 24 HPT compared with HEK293-6E cells (Figure 1B). Given the robust transfection efficiency and higher levels of transgene expression, suspension-adapted HEK293T cells were selected for further optimization of rAAV vector production.
Figure 1.
Creation of an OFAT-Optimized rAAV5 Suspension Cell Production Protocol
(A and B) HEK293-6E and HEK293T suspension cells (20 mL) were seeded at 1 × 106/mL in a 125-mL flask shaking at 110 rpm and transfected in duplicate with pAAV-EF1α-EGFP (1 μg/mL) using PEIMAX at PEIMAX/plasmid DNA ratio of 3:1. The cells were collected at 24 and 48 HPT and fixed. The mean fluorescence intensity (A) and the percentage of EGFP-positive cells (B) were quantified by flow cytometry. (C) Optimization of the total amount of plasmid. HEK293T suspension cells (20 mL) were seeded in a 125-mL flask at 0.5 × 106/mL. Cells were transfected with PEIMAX at the DNA concentration indicated (PEIMAX/DNA = 3:1) with a plasmid ratio of 2:1:1 (pHelper/pRC/pAAV-EGFP weight ratio). About 18 HPT, sodium butyrate (5 mM) and TN1 (0.5%) were added to the suspension culture. At about 65 HPT, the cells were spun down and the resuspended pellets were subjected to three freeze-thaw cycles. After spinning down the cell debris, the supernatant was analyzed by immunoblot for capsid protein. The upper panel contains 15 μg protein per lane; the lower panel contains 10 μL cell lysate per lane. (D) rAAV was generated as in (C). Cells were seeded at different densities and transfected with 1.5 μg/mL total plasmid DNA. (E) rAAV was generated as in (D) using cell density of 0.5 × 106/mL. Cells were harvested at the time point indicated. (F) rAAV was generated as in (E) and harvested at 72 HPT. The impact of sodium butyrate and TN1 on rAAV capsid production was evaluated.
We next evaluated the impact of the total amount of plasmid on rAAV vector production. rAAV was generated by triple transfection using pHelper, pAAV, and pRC plasmids. The pRC plasmid was previously16 optimized to improve vector production based on a strategy similar to Li et al.,19 Xiao et al.,20 and Kozak21,22 (see Materials and Methods and Figures S1 and S2). In these experiments, the total DNA was varied, but the plasmid ratio was fixed at 2:1:1 (pHelper/pRC/pAAV weight ratios, ∼1:1:1 molar ratio). This ratio has been used previously for rAAV production in suspension cells and adherent cells.11,20 A cell density of 0.5 × 106/mL was utilized based on previously published work in HEK293T, HEK293, and HEK293F cells.9,11 rAAV5-EGFP production was carried out in 20-mL cultures, and immunoblots for rAAV capsid protein in cell lysate were used as an indirect measurement of rAAV vector yield. As shown in Figure 1C, concentrations at 1.5 and 2 μg/mL generated the most rAAV capsid protein. Thus, a concentration of 1.5 μg/mL was selected to reduce the amount of total DNA needed for vector production. Utilizing the optimized plasmid concentration of 1.5 μg/mL, we then assessed the role of cell density on capsid protein expression. Cell densities ranging from 0.5 to 2 × 106/mL were evaluated. Although we initially predicted higher cell densities would result in higher levels of viral capsid protein, we observed that cell concentrations greater than 1 × 106 cells/mL produced less capsid protein than lower cell densities (Figure 1D). Finally, we examined the optimal virus harvest time. The largest amount of AAV capsid proteins was produced between 72 and 96 HPT (Figure 1E). We observed reductions in cell viability at 96 HPT, so 72 HPT was selected as the final harvest time.
After optimizing the cell line, total DNA plasmid concentration, cell density, and harvest time, we explored the ability of different media additives to improve rAAV5 production. Histone deacetylase inhibitors (HDACis) can induce transcriptional activation and increase gene expression.23 The HDACi sodium butyrate has previously been shown to improve antibody production, and we wondered whether this compound could potentially increase rAAV vector generation.24 Similar to Grünberg et al.,24 we found a 5 mM concentration of sodium butyrate treatment improved rAAV5 capsid production (Figure 1F). Peptones have also been shown to increase recombinant protein production, and Hildinger et al.10 showed that soy peptone could increase rAAV vector production in HEK293 cells. Pham et al.25 previously screened 16 peptones, including soy peptone, for their ability to increase recombinant protein expression, and found Tryptone N1 (TN1; 0.5%) was the best peptone for recombinant protein expression in transient HEK293 expression assays. Given these studies, we evaluated whether 5 mM sodium butyrate and 0.5% TN1 could improve rAAV vector capsid protein expression. We found either TN1 or sodium butyrate increased capsid protein expression, and the addition of both TN1 and sodium butyrate augmented capsid protein expression more than either compound alone (Figure 1F). Given these findings, sodium butyrate and TN1 were added to our rAAV vector production protocol.
The OFAT-Optimized rAAV Production Protocol Works for rAAV5, But Not rAAV8
Using our OFAT-optimized parameters, we produced a 1-L batch of rAAV5-EGFP in suspension HEK293T cells and compared the yield with virus generated in adherent HEK293T cells using a 10-layer cell stack containing 1 L of medium. Virus generated in adherent cells was produced via a standard calcium phosphate transfection using 4 mg total DNA per cell stack and a plasmid ratio of 2:1:1 (pHelper/pRC/pAAV weight ratio) (see Materials and Methods for additional details). In these experiments, the production yield of the suspension culture system was comparable (∼80%) with rAAV5 produced in adherent cells (4.71 × 1013 VGs/cell stack for adherent cells and 3.77 × 1013 VGs/L for suspension cells). Given the successful generation of rAAV5 using the suspension cell protocol, we next evaluated whether this protocol could be used to effectively produce other rAAV serotypes. Unfortunately, a 1-L batch of rAAV8-EGFP produced using these parameters yielded only 18.5% of the total virus produced from one cell stack with 1 L of medium (2.41 × 1013 VGs/L suspension cells versus 1.38 × 1014 VGs/cell stack). These results suggested that the utility of OFAT-based optimization may be suboptimal and/or serotype specific. We therefore considered additional approaches for developing a suspension-cell-based rAAV production platform.
rAAV Production Can Be Optimized Utilizing DOE Methodology
DOE methodology is an efficient means of studying the interactions of multiple factors on a specified output. Unlike OFAT-based optimization, the DOE method allows users to manipulate several variables simultaneously to determine the true optimum for a given factor. In the next set of studies, we applied the DOE method to optimize rAAV8 production. We selected five input parameters: the cell density, the total DNA amount, the ratio of the pHelper/pRC plasmids, the ratio of the pHelper/pAAV plasmids, and the ratio of PEIMAX to total DNA. Multiple ranges for each parameter were identified based on the literature (Table 1, DOE I variation ranges), and 32 experimental conditions were generated via statistical software (Table S1). Transfections were conducted in 20 mL suspension cultures for each experimental condition. At approximately 18 HPT, sodium butyrate and TN1 were added to the cultures. Cells were harvested at about 65 HPT, and the amount of rAAV produced was quantified via dot blot analysis of cell lysates. This experiment was repeated with similar results. The percentage of virus produced in these cultures was compared with a 25-mL tissue culture flask containing adherent HEK293T cells (Table S1). Data analysis suggested low levels of total plasmid DNA and pAAV plasmid in combination with increased levels of pRC plasmid and high cell density were optimal for rAAV production (Table 1, DOE I optimized). These results differed significantly from the OFAT-optimized conditions for rAAV5 and the previously published ranges of these parameters discussed earlier. Therefore, we performed a second DOE study to confirm these results. The parameter ranges were set based on the results of the first study, and 20 different conditions (run in duplicate) were generated by statistical software (Table 1, DOE II variation ranges). To reduce the number of conditions tested, we fixed the PEIMAX/DNA ratio at 3:1.26 The results are shown in Table S2. Except for the total amount of plasmid DNA, the parameters identified in the DOE II study were strikingly similar to the DOE I experiments (Table 1, DOE II optimized). To ensure these findings would translate to larger-scale rAAV vector production, we tested the conditions identified in the DOE I and DOE II experiments for their ability to generate rAAV8-EGFP at a 1-L scale. For these studies, rAAVs were purified with AVB-Sepharose, and the titer was determined by the CyQuant method. The yields of rAAV for the DOE I and DOE II conditions were 1.11 × 1014 and 1.12 × 1014 VGs/L after affinity purification, respectively. These yields were an average of 4.6-fold higher than the yield of rAAV8-EGFP generated using the OFAT-optimized method discussed earlier (2.41 × 1013 VGs/L after affinity purification). In parallel to these studies, we also generated rAAV8-EGFP in adherent HEK293T cells grown in 10-layer cell stacks. Virus generated in adherent cells was produced via a standard calcium phosphate transfection using 4 mg total DNA per cell stack and a plasmid ratio of 2:1:1 (pHelper/pRC/pAAV weight ratio) (see Materials and Methods for additional details). The average yield from the two 10-layer cell stacks was comparable (1.38 × 1014 VGs/L) to the yields in suspension cells generated using the DOE-optimized parameters. Although the volumetric yields in suspension cells were roughly equivalent to the yield of the cell stack, it should be noted that more cells were used in the 1-L suspension culture than in the cell stack (∼2.5 × 109 cells/L versus ∼6.36 × 108 cells/10-layer cell stack). Although the DOE-optimized method produces similar rAAV yields to traditional production in adherent cells, this suspension-based protocol is more user-friendly and less labor-intensive due to the nature of working with a suspension cell culture. Because vector purification can result in the loss of vector, we also determined the genome-containing particles in crude lysate with the DOE II condition in suspension cells using a droplet digital polymerase chain reaction (ddPCR)-based titration method and achieved a yield of 2.93 × 1014 VGs/L of cell culture for rAAV8-EGFP.
Table 1.
Parameter Ranges Selected for Optimization and DOE-Optimized Values
| Parameters | DOE I Variation Range | DOE I Optimized | DOE II Variation Range | DOE II Optimized |
|---|---|---|---|---|
| Cell density (106/mL) | 0.25–3 | 2.67 | 1–4 | 2.45 |
| DNA amount (μg/mL) | 0.5–4 | 2.53 | 1.5–3.5 | 1.5 |
| pHelper/pTrans | 1:5 to 5:1 | 1:5 | 1:5 to 5:1 | 1:5 |
| pHelper/pCis | 1:5 to 5:1 | 1:0.23 | 1:1 to 6:1 | 1:0.31 |
| PEIMAX/DNA | 1:1 to 4:1 | 4:1 | 3:1 (fixed) | 3:1 (fixed) |
The DOE-Optimized Protocol Improves rAAV Production across Multiple AAV Serotypes
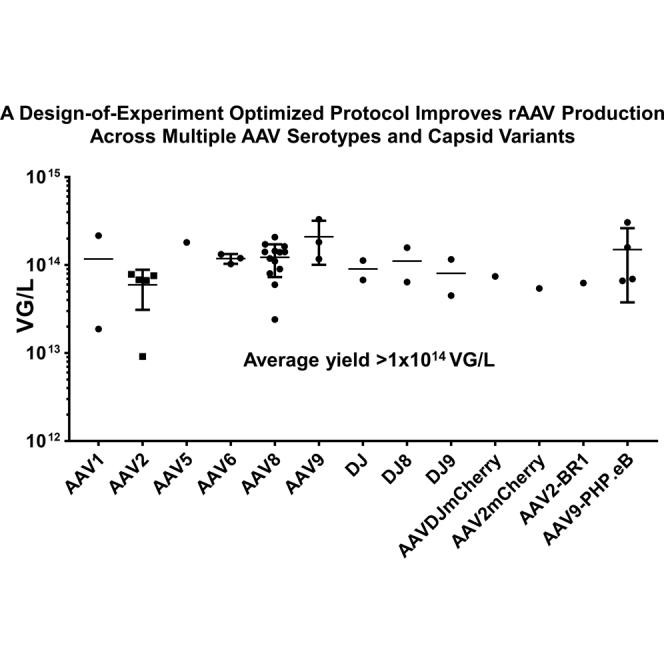
To determine whether the DOE-optimized method was specific to rAAV8, we compared virus production of rAAV1-, rAAV2-, rAAV5-, rAAV8-, and rAAV9-EGFP using the DOE- and OFAT-optimized protocols. For these studies we selected the conditions identified in the DOE II studies because this protocol required less total DNA. As shown in Figure 2A, the DOE-optimized method improved the average production yield by 6.1-fold compared with the rAAVs produced by the OFAT method. The yield of rAAV9 was not determined for this assay because the vector could not be purified by AVB-Sepharose. The cell lysates from these studies were also analyzed by immunoblot to quantify the amount of rAAV capsid produced. Based on band intensity, the DOE-optimized protocol generated 3.2-, 2.4-, 2.1-, 4.4-, and 2.9-fold more capsid proteins than the OFAT method in rAAV1, rAAV2, rAAV5, rAAV8, and rAAV9 vectors, respectively (Figure 2B). The DOE-optimized method was successfully used to produce large-scale rAAV-EGFPs for many serotypes, including rAAV1, rAAV2, rAAV5, rAAV6, rAAV8, and rAAV9, as well as the capsid engineered rAAVs DJ, DJ8, DJ9, DJ-mCherry, AAV2-mCherry, AAV2-BR1, and AAV9-PHP.eB (Figure 2C). The average yield for these affinity chromatography-purified viruses was 1.16 × 1014 VGs/L as determined by the CyQuant method (Figure 2C). We also examined whether DOE-derived plasmid ratios could be applied to adherent HEK293T cells using the calcium phosphate method for rAAV8 production. As shown in Figure 2D, we obtained comparable yields using the standard 2:1:1 ratio and the DOE-optimized ratio 1:5:0.31 (pHelper/pRC/pAAV) with less DNA (3 mg/L suspension cells for DOE-optimized method versus 4 mg/cell stack for adherent cells).
Figure 2.
The DOE-Optimized Protocol Improves rAAV Production across Multiple Serotypes
(A and B) Compare OFAT- and DOE-optimized methods for AAV1, AAV2, AAV5, AAV8, and AAV9 production. Twenty milliliters of suspension HEK293T cells were transfected using either the OFAT-optimized (optimized for rAAV5 vector production) or DOE-optimized (optimized for rAAV8 vector production) protocols in duplicate. At 65 HPT, the rAAVs were harvested and purified by AVB-Sepharose. The relative titers (A) and capsid proteins (B) in cell lysates were analyzed by bDNA assay and immunoblot (10 μL lysate per lane), respectively. The bands of immunoblot were quantified using Odyssey scanner at 700 nm, and relative intensities were shown (lower panel). (C) The yields of large-scale (1-L) rAAV-EGFP production of different serotypes and capsid variants using the DOE-optimized method. rAAVs were purified by one-step affinity chromatography, and the titers were determined by the CyQuant method. Note the mCherry in AAVDJmCherry and AAV2mCherry is on the surface of the capsid. (D) Adherent HEK293T cells were transfected by the CaPO4 method using 3 mg DNA per cell stack and a plasmid ratio of 1:5:0.31 (pHelper/pRC/pAAV weight ratios, left) or 4 mg DNA per cell stack and a plasmid ratio of 2:1:1 (right). rAAV8 vectors carrying different transgenes (n = 6 for each group) were purified, and the relative titers per cell stack are shown. Titers were determined by CyQuant method. Error bars show standard deviations.
Improvement of Downstream Processing for rAAV Production
In addition to optimizing the parameters involved in rAAV vector production, we also developed several methods to improve the downstream processing of rAAV. When harvesting rAAV, cells and media are usually separated and processed separately. Although rAAV2 and rAAV5 viral particles are mostly cell associated, significant amounts of viral particles can be found extracellularly in other rAAV serotypes.27 To save sample processing time, we added a PEG-NaCl solution directly to the whole-cell culture at the time of harvest to precipitate both virus-containing cells and extracellular virus in one step. Following PEG-NaCl treatment, the solution can be centrifuged and the pellet resuspended in a smaller volume. This protocol modification quickly reduces the volume of the culture to facilitate further downstream processing. Additionally, these smaller working volumes reduce the amount of costly reagents, such as DNase or Benzonase, needed for sample processing, and make downstream steps, such as centrifugation, freeze-thaw, and purification, more manageable.
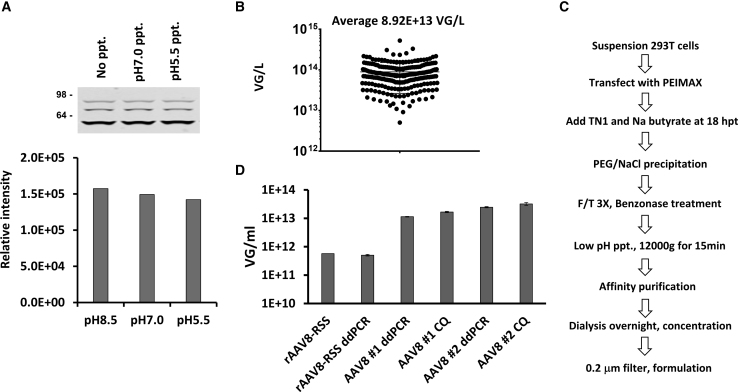
Following the PEG-NaCl precipitation and centrifugation, the resuspended pellets are subjected to three freeze-thaw cycles, Benzonase digestion, and centrifugation to remove cellular debris. The resulting supernatant contains rAAV, as well as large amounts of proteins and other impurities from the cells and medium. To remove these contaminants from the viral prep, we tested whether isoelectric-mediated protein precipitation could provide cleaner starting material for filtration, chromatography, or gradient purification. The isoelectric point (pI) of most cytoplasmic proteins is between pH 5 and 6, while the pI of rAAVs with a packaged genome (4.7 kb) is predicted to have a pI of 5.9.28,29 We evaluated whether reducing the pH could precipitate cellular proteins while a majority of the rAAV particles remain soluble. For these studies, we used rAAV1 encoding an empty vector. Following freeze-thaws, Benzonase digestion, and clarification of the lysate, we evaluated the amounts of protein precipitated and rAAV retained from supernatants at pH 5.5, 7.0, and 8.5. As shown in Table 2, adjusting the pH to 5.5 increased the subsequent precipitated pellet’s wet weight by 3.8-fold compared with pH 8.5. Importantly, an analysis of the corresponding viral supernatants showed that 90.5% of capsid proteins (based on band intensity) were recovered from pH 5.5 precipitation as compared with the control pH 8.5 (Figure 3A). We did not observe significant changes at pH 7.0. These results suggest a substantial amount of impurities can be removed prior to additional downstream processing, such as filtration, gradient centrifugation, or chromatography. We have successfully applied this pH-based precipitation method to multiple rAAV serotypes, except for rAAV2, which appears to precipitate at this lower pH (data not shown). These improvements combined with affinity purification (AVB-Sepharose, POROS CaptureSelect-AAV8, -AAV9, or -AAVX) enable us to purify rAAVs in less than 2 days. Since implementing this protocol in our lab, we have generated over 220 large-scale rAAVs (mostly 1-L scale) of different serotypes and genes of interest with an average production yield of 8.92 × 1013 VGs/L, demonstrating the reproducibility of this protocol (Figure 3B). A flow scheme providing an overview of the optimized rAAV production system is shown in Figure 3C.
Table 2.
pH-Mediated Precipitation of Cellular Proteins in rAAV1 Supernatants
| pH | Wet Weight (g) |
|---|---|
| 8.5 | 0.27 |
| 7.0 | 0.32 |
| 5.5 | 1.03 |
Figure 3.
Overview of the rAAV Production System
(A) Top: rAAV1-empty vector was produced in suspension HEK293T cells (1 L). Harvested cells and medium precipitation were resuspended, subjected to three freeze-thaw cycles and treated with Benzonase as described in the Materials and Methods. Parts of the total lysate were either not adjusted or adjusted to pH 7.0 or 5.5 and spun at 12,000 × g for 15 min. The pH of clarified supernatants was then adjusted to 8.0 and analyzed by immunoblot with anti-capsid protein antibody. Bottom: the bands of the immunoblot were quantified with Odyssey scanner. (B) The yields of 227 large-scale rAAV productions with different serotypes and different transgenes were produced using the DOE-optimized method. rAAVs were purified by affinity chromatography, and the titers were determined by the CyQuant (CQ) method. (C) Scheme of our rAAV production process (detailed in Materials and Methods). (D) Comparisons of in-house ddPCR titers with rAAV8 Reference Standard Stock (AAV8RSS) from ATCC. The ddPCR titers of internal rAAV8-EGFP preps (AAV8 #1 and AAV8 #2) were also compared with titers determined by the CQ method. Titers were performed in triplicate at three different dilutions. Error bars show standard deviations.
Our lab routinely titers rAAV by the CyQuant method and ddPCR using protocols adapted from Lock et al.30 with the primers and probe for BGHpA. Because titers can vary between researchers and labs, we compared our titers with the available rAAV8 reference standards generated via the AAV Reference Standard Working Group (AAVRSWG).31 As shown in Figure 3D, our ddPCR titers for the reference rAAV8 were well within the stated ranges and were equivalent to the titers defined by the AAVRSWG. In parallel, we also calculated the titers for two in-house AAV8-EGFP vectors. These titers were nearly equivalent (average 30.3% lower) to the titers calculated by the CyQuant method (Figure 3D). Because the CyQuant titration method is not PCR based, we were unable to titer the reference standards using the CyQuant method due to the low titer of the reference standard and the limited amounts of material available. These data validate our ability to achieve high rAAV yields using the DOE-optimized protocol in suspension cells.
Discussion
Despite the promising advances of rAAV-mediated therapies in the clinic, vector manufacturing remains a challenge. Previous efforts to improve vector production have focused on modifying single variables at a time, but these approaches do not account for the multiple interdependent factors that impact rAAV vector production.9, 10, 11, 12, 13, 14 To our knowledge, the DOE method has never been applied to rAAV vector production. In this study, we utilized DOE methodology to optimize rAAV production in a HEK293T suspension cell system by simultaneously varying the ratios of the transgene, packaging, and pHelper plasmids, the total DNA concentration, and the cell density. In total, we systematically evaluated over 52 different conditions and successfully identified a unique set of parameters for rAAV production that resulted in high yields of rAAV. Compared with previously published protocols that utilize the common 2:1:1 ratio (pHelper/pRC/pAAV weight ratio), we identified an optimal plasmid ratio of 1:5:0.31 (pHelper/pRC/pAAV weight ratio). Assuming the same amount of total input plasmid DNA, this results in a 5.1-fold decrease in the amount of pAAV DNA. Depending on the gene of interest, transgene expression from the pAAV plasmid can be toxic to production cells.32 Thus, a reduced amount of pAAV DNA may be beneficial for rAAV production with transgenes that impact cell viability and rAAV packaging. Another feature of the DOE-optimized protocol is the increased amount of pRC plasmid (1.19 mg/L), which comprises nearly 80% of the total plasmid DNA used in the transfection. Production of Cap proteins has been identified as a limiting factor for rAAV production, and increased expression of the cap gene by promoter modulation has previously been shown to increase rAAV production 10-fold.33 Thus, the increased amounts of pRC plasmid in the DOE protocol may contribute to the improved rAAV yields. Although the higher amounts of pRC may improve vector yields, the increased amounts of this plasmid may also result in higher numbers of empty particles. Additionally, although we obtained high-quality rAAV following affinity purification, this method does not specifically remove empty particles. We routinely administer these rAAVs to immune-competent mice and observe strong, sustained transgene expression in vivo, suggesting the levels of empty particles in these preparations do not have large, negative impacts on in vivo transduction. However, it is increasingly recognized that empty particles can impact rAAV-mediated gene delivery. Future studies will examine the changes in empty particle ratios between the OFAT and DOE protocols, and evaluate the possibility of implementing additional methods for empty particle removal. Lastly, in the DOE-optimized protocol, pHelper was reduced to one-third compared with plasmid ratios of 2:1:1 (pHelper/pRC/pAAV). Because the E2A and E4 genes may induce cytotoxicity, the reduction in the amount of the pHelper plasmid may be beneficial for rAAV production.34 In our initial studies using OFAT to optimize rAAV5 production, we observed viral capsid protein expression was decreased when the cell density was greater than 1 × 106 cells/mL (Figure 1D). Conversely, the data from the DOE-derived protocol suggested a cell density of 2.5 × 106/mL was optimal at the same total DNA concentration. This discrepancy could be due to the higher levels of the pRC plasmid in the DOE system, which may compensate for the higher cell density. These observations underscore the complex relationships between cell density and plasmid ratios, and highlight the advantages of the DOE methodology.
The DOE-optimized protocol allowed us to achieve yields approaching 3 × 1014 VGs/L of crude lysate as determined by ddPCR. Importantly, the pre-purification yields obtained via this DOE-optimized method were comparable (by volume) to those seen in baculovirus vector/sf9 cell systems (2.5–3.5 1014 VGs/L), which are currently viewed as one of the most promising platforms for rAAV vector production.35 Because these results were obtained utilizing an internally derived HEK293T suspension cell line and a modified pRC plasmid, this protocol may not be applicable to all HEK293T suspension cell lines or plasmids and will need to be evaluated on a case-by-case basis. In these studies, we also incorporated PEG-based virus precipitation, pH-mediated protein removal, and affinity purification to our downstream processing, enabling us to achieve average purified yields of 8.92 × 1013 VGs/L across many serotypes and transgenes. Future studies will focus on optimizing purification techniques and exploring continuous harvesting and perfusion technologies to further increase the production yield, improve the viral particle-to-VG ratio, and reduce the cost of rAAV vector production. In conclusion, we have developed a rAAV production system in HEK293T suspension cells and utilized a DOE-based approach to identify unique production parameters enabling us to achieve high yields of rAAV for multiple serotypes.
Materials and Methods
Cell Culture
HEK293T suspension cells were grown in FreeStyle 293 Expression Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 2% fetal bovine serum (FBS) and 50 μg/mL G418. These cells were derived from parental HEK293T cells as determined by short tandem repeat (STR) profiling (IDEXX, Westbrook, ME, USA) and adapted to suspension culture in-house. HEK293-6E cells were cultured in FreeStyle F17 Expression Medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 6 mM glutamine, 0.1% F68, and 25 mg/L G418. For small scale (20 mL), suspension cells were cultured in a 125-mL Erlenmeyer flask (Corning Life Science, Tewksbury, MA, USA) agitated at 110 rpm. For rAAV vector production in large scale, cells were cultured in 1-L medium in a 3-L baffled Erlenmeyer flask (Corning) agitating at 65 rpm. Adherent HEK293T cells were cultured in DMEM supplemented with 10% FBS and penicillin (100 U/mL), streptomycin (100 μg/mL), and glutamine (0.292 mg/mL) in 10-layer CellStack (Corning, Corning, NY, USA).
Plasmid Construction
pRC2/2 was generated by subcloning the AAV2 genome minus the inverted terminal repeats (ITRs) (bp 191–4,498) from pAV2 (ATCC 37216) into pBluescript II (Agilent Technologies).36 A modified AAV2 intron lacking the splice donor site was inserted upstream of the rep open reading frame (ORF) between the p5 promoter and the rep start codon to enhance AAV cap gene expression in a strategy similar to Li et al.,19 Xiao et al.,20 and Kozak.21,22 pACG2 and pXX2 were constructed according to Li et al.19 and Xiao et al.20 (see Figures S1 and S2). The pRC2/8 and other serotypes were constructed by replacing the cap ORF in pTrans2/2 with the AAV8 cap ORF or cap ORFs for other serotypes.16 To construct pAAV, we sub-cloned the ITRs of pAV2 into the pBluescript II backbone and then inserted the promoter/WPRE/BGHpA between the ITRs. The pHelper plasmids were constructed in-house using the similar strategy of Matsushita et al.37
rAAV Vector Production in Suspension Cells
Cells were transfected using PEIMAX (Polyscience) with three plasmids (pHelper, pRC, and pAAV). Plasmids were mixed in OptiMEM (Life Technology) in 1/20 vol of the cells to be transfected and incubated for 5 min. PEIMAX was then added to DNA diluted in OptiMEM, and after incubation for an additional 10 min, the DNA-PEIMAX complex was added to the cells. Eighteen hours post-transfection, sodium butyrate (5 mM; Sigma) and protein hydrolysate TN1 (0.5%; Organotechnie) were added to the culture, and rAAV vectors from cell lysate were harvested about 65 HPT. Alternatively, TN1 was replaced by Yeastolate (GIBCO Difco TC Yeastolate UF) at the same concentration. For large-scale production of rAAV vectors, rAAV vectors in both the cells and the medium were harvested (see below).
rAAV Vector Production in Adherent Cells
For large-scale rAAV vector production, adherent cells in a 10-layer CellSTACK (6,360 cm2; Corning) were transfected with 4 mg (plasmid ratio 2:1:1, pHelper/pRC/pAAV weight ratio) or 3 mg (plasmid ratio 1:5:0.31) of total DNA using a CaPO4 method in 2-L DMEM with 2% FBS, penicillin (100 U/mL), streptomycin (100 μg/mL), and glutamine (0.292 mg/mL). After 24 h, the transfection medium was replaced with 1 L fresh medium supplemented with TN1 and sodium butyrate, and rAAV vectors in cells and medium were harvested at about 65 HPT.
Purification of rAAV
For large-scale 1-L production, both cells and rAAV vectors in the medium were precipitated with 1/4 cell culture volume of 40% PEG 8000 (Sigma), 2.5 M NaCl. After incubation on ice for 2 h, the cells and rAAV in the medium were precipitated by centrifugation at 4,000 rpm for 30 min (J6-MI; Beckman Coulter). The precipitation was resuspended in 80 mL NT buffer (150 mM NaCl, 50 mM Tris [pH 8.50]) and subjected to three cycles of freeze and thaw. After Benzonase treatment (50 U/mL for 1 h at 37°C), the cell debris were spun down at 4,000 rpm for 30 min. The supernatant was then slowly adjusted to pH 5.5 with acetic acid (except for rAAV2) and centrifuged at 12,000 × g for 15 min. The pH was then adjusted to 8.0 with NaOH, and the starting material was filtered through a 0.45-mm filter. The filtrate was then loaded onto an affinity column (AVB-Sepharose or POROS CaptureSelect AAV8, AAV9, or AAVX) and eluted with 50 mM glycine-HCl (pH 3.0) after extensive wash with 10–20 times of column volume of phosphate-buffered saline (PBS) until the 280 nm absorbance was close to baseline. The eluted rAAV vectors were immediately neutralized with 1/10 vol of 1 M Tris-HCl (pH 8.0), dialyzed overnight against PBS-MK buffer (PBS with 1 mM MgCl2 and 2.5 mM KCl [pH 8.0]), and centrifuged at 3,000 rpm for 10 min to remove any precipitates. Purified rAAV vectors were then concentrated to a desired concentration in dialysis cassettes with Slide-A-Lyzer Concentrating Solution (Thermo Fisher) or spin filter (Amicon Ultra Centrifugal Filter Units [MilliporeSigma]; molecular cutoff, 100 kDa).
rAAV Titration
DNA Dot Blot Analysis
In brief, diluted cell lysates were treated with DNase I and then Proteinase K, loaded onto Nylon membrane, and hybridized overnight with a 32P-labeled EF1α promoter-specific probe. After washing with saline sodium citrate (SSC) buffer containing 0.1% SDS, the intensity of radioactivity was measured using a Storm A60 Scanner.
CyQuant Method
The titers of purified rAAV vectors from large-scale production were determined using the QuickTiter AAV Quantitation Kit (Cell Biolabs) according to the manufacturer’s instructions.
Titration of Purified rAAV and rAAV from Crude Cell Lysate and Medium Using ddPCR
Titration of purified rAAV was carried out as previously published.30 For crude cell lysate and medium, upon harvest, partial cell culture (including cells and medium) was removed immediately after mixing and subject to three times of freeze and thaw. After centrifugation, the supernatant was digested with DNase I and then proteinase K. The digested sample was serial 10× diluted, and the VG was determined by ddPCR with primers and probe for BGHpA as described by Lock et al.30
rAAV Titration Using Branched DNA (bDNA) Assay
For quantification of rAAV2/1, rAAV2, rAAV2/5, or rAAV2/8 produced in 20-mL cultures, rAAVs in cell lysates were first purified with AVB-Sepharose, and then the QuantiGene2.0 assay (Branched DNA Technology) was used to determine the VGs of rAAV using a rAAV prep with known viral VG as standard. The EF1α-specific probe sets for pAAV-EGFP were used. Serially diluted viral preps were incubated in lysis buffer with a specific probe set in bDNA capture plates at 55°C for overnight hybridization, and then chemiluminescent signals were read in a Perkin Elmer EnVision.
DOE Design and Statistical Methods
All experimental designs and statistical analyses were performed using JMP version 7.0.2 and version 9.0.0 under the Windows Vista System. For the purpose of characterizing the impact of cell density, total DNA amount, ratio of plasmid 1 (pHelper) to plasmid2 (pRC), and ratio of plasmid 1 to plasmid 3 (pAAV) on the production of rAAV8 vector in suspension HEK293T cells and maximizing such production with regard to these factors with TN1 and sodium butyrate fixed, two response surface design experiments were performed and statistically analyzed. Student’s t tests were performed where appropriate. p < 0.05 was considered statistically significant.
SDS-PAGE and Immunoblot
Proteins in cell lysates were separated on a 4%–20% reducing Tris-Glycine gel (Life Technologies). Following transfer, membranes were probed with anti-VP1, -VP2, and -VP3 monoclonal antibody (mAb) (Fitzgerald). The bands were visualized by either Pierce ECL Western Blotting Substrate (Pierce) or IRDye 800CW Secondary Antibodies and Odyssey Scanner. Protein bands were quantified on LI-COR Image Studio Software (Li-Cor). Protein concentration was determined by the BCA (Bicinchoninic Acid) method (Pierce).
Author Contributions
Conceptualization, H.Z., K.-J.L., S.W., and W.H.M.; Methodology, H.Z., M.D., K.-J.L., J.S. C.P., and W.H.M.; Resources, M.D., Y.L., T.W., and H.Z.; Investigation, H.Z., M.D., Y.L., T.W., J.S., C.P., K.-J.L., and W.H.M.; Writing – Original Draft, H.Z., K.-J.L., M.D., and W.H.M.
Conflicts of Interest
The authors are employees of Amgen Inc. (Thousand Oaks, CA, USA).
Acknowledgments
The studies were funded by Amgen Research, South San Francisco, CA, USA. The authors acknowledge Jessie Gu and Lei Zhou for DOE design and data analysis.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.06.004.
Supplemental Information
References
- 1.Ameri H. Prospect of retinal gene therapy following commercialization of voretigene neparvovec-rzyl for retinal dystrophy mediated by RPE65 mutation. J. Curr. Ophthalmol. 2018;30:1–2. doi: 10.1016/j.joco.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoy S.M. Onasemnogene Abeparvovec: First Global Approval. Drugs. 2019;79:1255–1262. doi: 10.1007/s40265-019-01162-5. [DOI] [PubMed] [Google Scholar]
- 3.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allay J.A., Sleep S., Long S., Tillman D.M., Clark R., Carney G., Fagone P., McIntosh J.H., Nienhuis A.W., Davidoff A.M. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 2011;22:595–604. doi: 10.1089/hum.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gene therapy’s next installment. Nat. Biotechnol. 2019;37:697. doi: 10.1038/s41587-019-0194-z. [DOI] [PubMed] [Google Scholar]
- 7.Colella P., Ronzitti G., Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2017;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J.Y., Lim B.P., Lee K., Kim Y.G., Jo E.C. Scalable production of adeno-associated virus type 2 vectors via suspension transfection. Biotechnol. Bioeng. 2006;94:416–430. doi: 10.1002/bit.20776. [DOI] [PubMed] [Google Scholar]
- 10.Hildinger M., Baldi L., Stettler M., Wurm F.M. High-titer, serum-free production of adeno-associated virus vectors by polyethyleneimine-mediated plasmid transfection in mammalian suspension cells. Biotechnol. Lett. 2007;29:1713–1721. doi: 10.1007/s10529-007-9441-3. [DOI] [PubMed] [Google Scholar]
- 11.Durocher Y., Pham P.L., St-Laurent G., Jacob D., Cass B., Chahal P., Lau C.J., Nalbantoglu J., Kamen A. Scalable serum-free production of recombinant adeno-associated virus type 2 by transfection of 293 suspension cells. J. Virol. Methods. 2007;144:32–40. doi: 10.1016/j.jviromet.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Feng L., Guo M., Zhang S., Chu J., Zhuang Y., Zhang S. Improvement in the suspension-culture production of recombinant adeno-associated virus-LacZ in HEK-293 cells using polyethyleneimine-DNA complexes in combination with hypothermic treatment. Biotechnol. Appl. Biochem. 2008;50:121–132. doi: 10.1042/BA20070081. [DOI] [PubMed] [Google Scholar]
- 13.Chahal P.S., Schulze E., Tran R., Montes J., Kamen A.A. Production of adeno-associated virus (AAV) serotypes by transient transfection of HEK293 cell suspension cultures for gene delivery. J. Virol. Methods. 2014;196:163–173. doi: 10.1016/j.jviromet.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grieger J.C., Soltys S.M., Samulski R.J. Production of Recombinant Adeno-associated Virus Vectors Using Suspension HEK293 Cells and Continuous Harvest of Vector From the Culture Media for GMP FIX and FLT1 Clinical Vector. Mol. Ther. 2016;24:287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandenius C.F., Brundin A. Bioprocess optimization using design-of-experiments methodology. Biotechnol. Prog. 2008;24:1191–1203. doi: 10.1002/btpr.67. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H., Lee K.J., Wolfe T., Plewa C., Sheng J.Z. 2014. High titer recombinant AAV vector production in adherent and suspension cells. US Patent US20160222356 A1, filed August 28, 2014, and published August 4, 2016. [Google Scholar]
- 17.Grimm D., Kern A., Pawlita M., Ferrari F., Samulski R., Kleinschmidt J. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 18.Durocher Y. 2006. Expression vectors containing a truncated Epstein Barr Nuclear Antigen 1 lacking the Gly-Gly-Ala domain for enhanced transient gene expression. US patent publication application WO2006096989, filed March 17, 2006, published September 13, 2007. [Google Scholar]
- 19.Li J., Samulski R.J., Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J. Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebbar P.B., Archer T.K. Chromatin remodeling by nuclear receptors. Chromosoma. 2003;111:495–504. doi: 10.1007/s00412-003-0232-x. [DOI] [PubMed] [Google Scholar]
- 24.Grünberg J., Knogler K., Waibel R., Novak-Hofer I. High-yield production of recombinant antibody fragments in HEK-293 cells using sodium butyrate. Biotechniques. 2003;34:968–972. doi: 10.2144/03345st02. [DOI] [PubMed] [Google Scholar]
- 25.Pham P.L., Perret S., Cass B., Carpentier E., St-Laurent G., Bisson L., Kamen A., Durocher Y. Transient gene expression in HEK293 cells: peptone addition posttransfection improves recombinant protein synthesis. Biotechnol. Bioeng. 2005;90:332–344. doi: 10.1002/bit.20428. [DOI] [PubMed] [Google Scholar]
- 26.Baldi L., Muller N., Picasso S., Jacquet R., Girard P., Thanh H.P., Derow E., Wurm F.M. Transient gene expression in suspension HEK-293 cells: application to large-scale protein production. Biotechnol. Prog. 2005;21:148–153. doi: 10.1021/bp049830x. [DOI] [PubMed] [Google Scholar]
- 27.Vandenberghe L.H., Xiao R., Lock M., Lin J., Korn M., Wilson J.M. Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing. Hum. Gene Ther. 2010;21:1251–1257. doi: 10.1089/hum.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz R., Ting C.S., King J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 29.Venkatakrishnan B., Yarbrough J., Domsic J., Bennett A., Bothner B., Kozyreva O.G., Samulski R.J., Muzyczka N., McKenna R., Agbandje-McKenna M. Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J. Virol. 2013;87:4974–4984. doi: 10.1128/JVI.02524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lock M., Alvira M.R., Chen S.J., Wilson J.M. Absolute determination of single-stranded and self-complementary adeno-associated viral vector genome titers by droplet digital PCR. Hum. Gene Ther. Methods. 2014;25:115–125. doi: 10.1089/hgtb.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moullier P., Snyder R.O. International efforts for recombinant adeno-associated viral vector reference standards. Mol. Ther. 2008;16:1185–1188. doi: 10.1038/mt.2008.125. [DOI] [PubMed] [Google Scholar]
- 32.Strobel B., Klauser B., Hartig J.S., Lamla T., Gantner F., Kreuz S. Riboswitch-mediated Attenuation of Transgene Cytotoxicity Increases Adeno-associated Virus Vector Yields in HEK-293 Cells. Mol. Ther. 2015;23:1582–1591. doi: 10.1038/mt.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent K.A., Piraino S.T., Wadsworth S.C. Analysis of recombinant adeno-associated virus packaging and requirements for rep and cap gene products. J. Virol. 1997;71:1897–1905. doi: 10.1128/jvi.71.3.1897-1905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao C., Li J., Skold A., Zhang X., Xiao X. Feasibility of generating adeno-associated virus packaging cell lines containing inducible adenovirus helper genes. J. Virol. 2002;76:1904–1913. doi: 10.1128/JVI.76.4.1904-1913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi P.R.H., Cervera L., Ahmed I., Kondratov O., Zolotukhin S., Schrag J., Chahal P.S., Kamen A.A. Achieving High-Yield Production of Functional AAV5 Gene Delivery Vectors via Fedbatch in an Insect Cell-One Baculovirus System. Mol. Ther. Methods Clin. Dev. 2019;13:279–289. doi: 10.1016/j.omtm.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laughlin C.A., Tratschin J.D., Coon H., Carter B.J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 37.Matsushita T., Elliger S., Elliger C., Podsakoff G., Villarreal L., Kurtzman G.J., Iwaki Y., Colosi P. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5:938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.