Abstract
Plasma fibrinogen (F1) and fibronectin (pFN) polymerize to form a fibrin clot that is both a hemostatic and provisional matrix for wound healing. About 90% of plasma F1 has a homodimeric pair of γ chains (γγF1), and 10% has a heterodimeric pair of γ and more acidic γ′ chains (γγ′F1). We have synthesized a novel fibrin matrix exclusively from a 1:1 (molar ratio) complex of γγ′F1 and pFN in the presence of highly active thrombin and recombinant Factor XIII (rFXIIIa). In this matrix, the fibrin nanofibers were decorated with pFN nanoclusters (termed γγ′F1:pFN fibrin). In contrast, fibrin made from 1:1 mixture of γγF1 and pFN formed a sporadic distribution of “pFN droplets” (termed γγF1+pFN fibrin). The γγ′F1:pFN fibrin enhanced the adhesion of primary human umbilical vein endothelium cells (HUVECs) relative to the γγF1+FN fibrin. Three dimensional (3D) culturing showed that the γγ′F1:pFN complex fibrin matrix enhanced the proliferation of both HUVECs and primary human fibroblasts. HUVECs in the 3D γγ′F1:pFN fibrin exhibited a starkly enhanced vascular morphogenesis while an apoptotic growth profile was observed in the γγF1+pFN fibrin. Relative to γγF1+pFN fibrin, mouse dermal wounds that were sealed by γγ′F1:pFN fibrin exhibited accelerated and enhanced healing. This study suggests that a 3D pFN presentation on a fibrin matrix promotes wound healing.
Keywords: Fibrin, Fibronectin, Nanoclusters, Wound healing
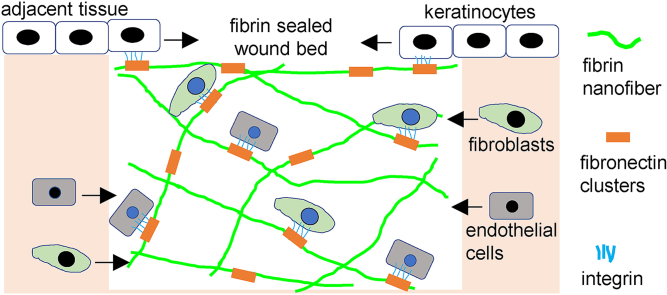
Graphical abstract
The novel fibrin sealant forms a 3D matrix at the wound bed. Nanoscale fibronectin clusters are formed on the fibrin nanofibers. These nanoclusters can stimulate the pro-healing cells such as endothelial cells, fibroblasts and keratinocytes and promote their migration, survival, proliferation and function, resulting in faster and better wound healing.
Highlights
-
•
A novel fibrin matrix exclusively from a 1:1 (molar ratio) complex of γγ’F1 and pFN was made.
-
•
In this new matrix, fibrin nanofibers were decorated with nano-scale pFN clusters (termed γγ’F1:pFN fibrin).
-
•
This matrix significantly enhances mouse skin wound healing.
-
•
This study suggests that a 3D pFN nanoclusters presented on a fibrin matrix can promote wound healing.
1. Introduction
Skin wound healing consists of four phases: hemostasis, inflammation, proliferation, and remodeling. After an injury in healthy people, plasma fibrinogen (F1) and plasma fibronectin (pFN) (10:1 at M ratio) are polymerized by thrombin and factor XIII (FXIII) to form a fibrin clot to stop bleeding (i.e., the hemostasis) [1]. In the clot, fibronectins are attached covalently to the fibrin nanofibers [2]. In a material sense, F1 and pFN are both diverse macromolecules that greatly contrast each other in structure. F1 is a linear, hexameric, 340 kDa glycoprotein having two alpha, two beta, and two gamma chains [[3], [4], [5]]. pFN is a 440 kDa globular protein having two chains with cellular binding domains. Both F1 and pFN have cellular binding domains that remain cryptic until crosslinking by activated Factor XIII (FXIIIa) into the fibrin matrix [6]. About 90% of F1 circulates as a homodimeric pairing of γ-chains (γγ F1) while the remaining 10% presents a heterodimeric pairing of γ with a more acidic γ′-chain (γγ′F1)[6]. The γ′-chain occurs from an alternative transcript having 20 additional amino acids at the carboxy-terminal that has been correlated with a pleiotropic impact on vascular health [[6], [7], [8]]. Importantly, pFN and γγ′F1 both occur at about the same concentration in plasma of about 300 μg/mL [[7], [8], [9], [10], [11]] and, therefore, their presence in fibrin clots.
The hemostasis begins with thrombin-mediated activation of F1 and FXIII. The activated F1 produces a semi-soluble, viscoelastic fibrin aggregate [3,12,13] containing pFN entrained from plasma. The FXIII is inherently present within the polymerizing fibrin aggregate as a complex with 1 out of every 100 γγ′F1 molecules [7]. It disassociates upon the formation of activated FXIII that enables efficient intra-fibrin cross-linking activity to produce an insoluble fibrin matrix [[14], [15], [16], [17], [18]], and to simultaneously anchor the matrix to the wound surface in about 3 min [7,19]. This clotting process causes protein conformational changes that induce interactions between pFN, fibrin, and also wound healing cells.
After hemostasis, the fibrin clot acts as a provisional matrix for different cell types to heal the wound [20]. Neutrophils and inflammatory (or M1) macrophages first enter the matrix to clear bacteria and tissue debris. Neutrophils secrete anti-microbial peptides, proteases and reactive oxygen species (ROS) to remove bacteria and damaged tissue [21]. These chemicals are toxic to surrounding healthy cells as well [21]. The M1 macrophages phagocytosis bacterial and damaged tissue [[22], [23], [24], [25], [26], [27], [28], [29], [30]]. Both cell types directly bind to fibrin nanofibers via their αMβ2 integrin receptors. Once the wound is debrided (takes ~3 days [31]), neutrophils undergo apoptosis, and M1 macrophages switch to the regenerative M2 phenotype that secretes cytokines and growth factors to recruit and stimulate healing cells such as endothelial cells, fibroblasts and keratinocytes to heal the wound (i.e., the proliferation phase) [[32], [33], [34]]. Endothelial cells form new blood vessels, and fibroblasts deposit new extracellular matrix proteins to form a granulation tissue (takes ~7 days) [[32], [33], [34]]. Keratinocytes migrate on the granulation tissue to form a new epidermis to close the wound (i.e., re-epithelization) [33]. Healing cells adhere to the fibrin matrix by binding to fibronectins with their integrin receptors [[32], [33], [34], [35]]. The binding also triggers downstream signalings that are required for these cells to migrate, proliferate, and repair the wound [[32], [33], [34], [35], [36]]. Fibronectins also bind, enrich, protect, orientate, and present growth factors locally to potentiate their functions [[37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48]]. In short, the fibrin-based provisional matrix is the central stage for cells to heal a wound, and fibronectins are essential for adhering and stimulating the healing cells. Consequently, insufficient provisional matrix or fibronectins or fibronectin-induced cell signalings lead to pathological wound healings [[32], [33], [34], [35],37]. For instance, small wounds can heal in about 14 days, while large wounds take much longer time to heal partially due to the insufficient fibrin-fibronectin provisional matrix.
We hypothesize that increasing the pFN content or FN-cell interaction in the fibrin matrix will promote wound healing via enhancing the migration, proliferation, and function of healing cells. The natural fibrin matrix only contains 10% (molar ratio) fibronectins [1]. Previously, we reported a novel method to increase the pFN content up to 50% (molar ratio) in the fibrin matrix and to present the pFN as nanoclusters on the fibrin nanofibers [8]. In this report, we showed that this novel pFN: γγ′F1 fibrin enhanced the adhesion, proliferation, and morphogenesis of healing cells in vitro, and accelerated re-epithelization and granulation tissue formation in mouse dermal wounds. This engineered fibrin sealant has attributes useful to both hemostasis and to promote faster wound healing, including improvement of granulation tissue quality.
Fibrin sealants using fibrinogen purified from human plasma are commercially available for various biomedical applications such as hemostat, tissue sealant, and tissue adhesive [49,50]. They are also widely used for tissue engineering, and drug and cell delivery. However, studies on using purified fibrin sealant alone to enhance wound healing are limited [[51], [52], [53], [54]]. The outcomes are positive but inconsistent and limited. Research showed cells did not infiltrate the topically applied fibrin sealant in rodent skin wounds [55]. Instead, cells migrated underneath the sealant. The limited success may because fibrin sealants typically use high fibrinogen concentrations (e.g.,>25 mg/mL). The resultant fibrin matrices have large and dense fibrin fibers that prevent cell infiltration and proliferation [56]. Additionally, the fibronectin content and presentation in the matrices are not defined. Some studies use fibronectin-deprived sealants. Others use sealants that contain small amounts of fibronectin (1 fibronectin per 10 fibrinogens). Our sealant is novel and different from these commercial fibrin sealant. First, it has low protein concentrations (<9 mg/mL), and the resultant matrix has similar micro- and nano-structures to the natural fibrin provisional matrix. Second, it contains high fibronectin content (e.g., 1 fibronectin per 10 fibrinogens). Third, fibronectins are presented as nanoclusters on the fibrin fibers.
2. Methods
2.1. Materials
All reagents were obtained from Sigma Chemical Company (St. Louis MO) unless otherwise noted. Human source plasma was provided by the U.S. Army Materials Command (Fort Detrick, MD.). Recombinant human thrombin was purchased from ZymoGenetics (Seattle, WA). DEAE Sepharose fast flow, Superose 6, and Gelatin Sepharose were purchased from GE Healthcare (Uppsala, Sweden). 4–12% NuPage Bis-Tris SDS polyacrylamide gels, Colloidal Blue stain, and See Blue molecular weight markers were from Invitrogen (Carlsbad, CA). Polyclonal antibody (GMA-034) for human fibrinogen was purchased from Green Mountain Antibodies (Burlington, VT). Antibody (ab2413) for human fibronectin was from Abcam.
2.2. Production of recombinant factor XIII
Recombinant active Factor XIII (rFXIIIa) was expressed in Pichia Pastoris following published methods [[56], [57], [58]]. The activity was determined using Pefakit (Pentapharm, Norwalk, CT).
2.3. Preparation of fibrinogen and fibronectin from human plasma [8]
Three units of human plasma that had been stored at −80 °C were thawed at 4 °C. The plasma was centrifuged at 4000 rpm for 20 min. The cryoprecipitate was re-suspended in 45 mM sodium citrate, 100 mM 6-aminocaproic acid, pH 7.0 at 37 °C. The solution was then centrifuged for 25 min at 4000 rpm. The supernatant was treated with a solvent detergent to inactivate the virus by addition of 0.15% TnBP, 0.5% Triton X-100 and stirred at room temperature for 60 min. The supernatant was adjusted to 1 M ammonium sulfate. The sample was stirred at room temperature for 30 min and then centrifuged at 2000 rpm for 15 min at room temperature. The pellet was re-suspended in 20 mM sodium citrate, 100 mM NaCl. The sample was dialyzed overnight at room temperature against the same buffer. The dialyzed sample was centrifuged at 2000 rpm for 15 min at room temperature.
The supernatant was fractionated to isolate γγF1, γγ′F1 and pFN using DEAE Sepharose Fast Flow chromatography at room temperature [59]. The sample was applied to a DEAE column then washed with 10 column volumes of 20 mM sodium citrate, 100 mM NaCl, pH 7.4. The unabsorbed fraction was γγF1. Bound protein was eluted with a linear gradient of 0.1 M–1.0 M NaCl. The pooled eluate produced an approximately equimolar mixture of γγ′F1 and pFN that was dialyzed against 20 mM citrate buffer, 20 mM NaCl at pH 6.8. The dialyzed mixture was concentrated using a 10 kDa centrifugal ultrafiltration device at 4000 g for 30 min.
Pure pFN and γγ′F1 were produced by application of the DEAE eluate pool to gelatin-Sepharose. Briefly, a 10 mL γγ′F1:pFN (“γγ’F1:pFN” that we will name from here as “complex”) was applied to a 3 mL analytical gelatin-Sepharose column. The column was washed with 10 vol of the dialysis buffer then eluted with the same buffer containing 6 M urea. pFN was eluted by urea while the γγ′F1 fell through the column.
2.4. 2D adhesion assay
Human foreskin fibroblasts were isolated and cultured in DMEM with 10% FBS. The use of primary human fibroblasts from anonymous donors was approved by the Institutional Review Board at the University of Nebraska Medical Center and by the Research and Development Committee at the Omaha VA Medical Center. Passage 4 to 6 cells were used. HUEVCs were purchased from Lonza and cultured in EGM2 medium. Passage 3 and 4 cells were used for the experiments. 100 μl fibrin matrix was made in one well of the 96-well plate. 1 × 104 cells were placed in each well with 200 μl medium. Three hours after plating, medium, and unattached cells were removed. Cells were washed with 200 μl PBS once. 100 μl fresh medium with 10 μl Alamar blue reagent was added and incubated for 3 h. 100 μl medium was collected for measuring the fluorescence that was calibrated to a standard curve to calculate the numbers of attached cells within each well. After the quantification, cells were fixed with 4% PFA and stained with DAPI. The nuclei were recorded with a fluorescence microscope.
2.5. 3D cell culture within the fibrin matrix
100 μl fibrin matrix with 2 × 104 cells were made in individual wells of a 96-well format plate. 200 μl medium was added to each well. At varied time points after plating, cell morphologies were recorded by phase-contrast microscopy. The medium was removed and 100 μl fresh medium with 10 μl Alamar blue reagent were added and incubated for 3 h. 100 μl medium was collected for measuring the fluorescence that was calibrated to a standard curve to calculate the numbers of cells in each matrix. The viability of cells within the gel was accessed with a Live/Dead cell assay kit (Invitrogen) according to the Manufacturer's instruction.
2.6. Fibrin formulation
To make the fibrin matrix for the above cell culture experiment, the following components were mixed to the final concentration of fibrinogen (γγF1, γγ′F1) at 2.5 mg/mL; thrombin at 1 U/mL; rFXIII at 15.4 U/mL; and pFN at 3.3 mg/mL. F1 and pFN were at a 1 to 1 M ratio. The mixtures were incubated at 37 °C for 15 min to form the fibrin matrix.
2.7. SEM and confocal microscopy
Fibrin matrices were made according to the formulation above. For SEM, samples were fixed with 2.5% glutaraldehyde in 100 mM phosphate buffer (PH 7.0) at room temperature for 1 h, then at 4 °C overnight. Samples were rinsed with phosphate buffer twice, 10 min each. Ethanol dehydration series were performed: 30%, 50%, 70%, 2 × 95%, 2 × 100%, 5 min for each procedure. Then samples were treated with hexamethyldisilazane (HMDS) as the following: 33% HMDS, 66% HMDS, 2 × 100% HMDS, 2 min for each procedure. Samples were left in 100% HMDS to air-dry at least overnight before sputter-coated with gold-palladium and imaged with the scanning electron microscope (Hitachi S4700 Field-Emission SEM, Hitachi, Tokyo, Japan) at 10 kV. For confocal microscopy, samples were fixed with 4% paraformaldehyde (PFA) at 4 °C overnight, washed with PBS for 3 times, and blocked with 5% goat serum for 1 h before incubating with primary antibodies at 4 °C overnight. After extensive washing, secondary antibodies were added and incubated for 2 h at room temperature. Samples were then washed with PBS before imaged with confocal microscopy (Nikon A1-R confocal system on a Nikon Eclipse 90i upright fluorescence microscopy).
2.8. Animal research
All animal protocols were approved by the Institutional Animal Care and Use Committee of the State University of Campinas (protocol number 4467–1/2017) and the University of Nebraska-Lincoln (protocol 1571). All experimental procedures were performed following the guidelines of the Institutional Animal Care and Use Committee of the University of Campinas and the University of Nebraska-Lincoln and. For the surgical procedure, mice were anesthetized using intraperitoneal of xylazine (10 mg/kg) and ketamine (100 mg/kg). Individual doses were calculated using Labinsane App (Labsincel, 2017). A deep anesthetic plane was verified by the loss of pedal and corneal reflexes. Two symmetrical 6-mm full-thickness excisional wounds were created on the back of each mouse as described previously [60]. Donut-like splints with an 8 mm diameter of the center hole and 15-mm diameter of the disc were created. The splint was carefully placed around the wound and secured to the skin with eight interrupted sutures. After surgery, animals were maintained in a warm bed for anesthetic recovery. We applied intraperitoneal injection of tramadol (25 mg/kg) at 0, 12, and 24 h after surgery as an analgesic treatment.
2.9. Mice fibrin sealant treatment
First, we cleaned the unwounded surrounding skin with a saline solution to remove any traces of biological residue. We sealed the wound with 50 μL fibrin. The fibrin sealant contained 2.5 mg/mL fibrinogen, 3.3 mg/mL fibronectin, 15.4U/mL rFXIIIa, 5 U/mL thrombin, 16 ng/mL VEGF, 5 mM CaCl2 in the Ringer solution. Control animals received the same amount of CaCl2 and VEGF in the Ringer solution. The wounds were covered with a sterile transparent Tegaderm dressing. On days 2, 4, 6, and 9, we cleaned and disinfected the surrounding skin as described above and reapplied 25 μL fibrin sealant.
2.10. Wound healing process documentation and analysis
Macroscopic evaluation of the healing process was evaluated by photographs taken at 0, 2, 4, 6, 9, and 11 days after wounding using a Nikon D610 (Nikon Systems, Inc., Tokyo, Japan) camera with an AF-S DX NIKKOR 18–55 mm f/3.5–5.6G VR objective. The same distance (wound to the objective lens), top white fluorescence illumination, and operator were used on each occasion. We calculate Wound Area as follows: Wound Area (%) = [(wound area of day 0 - wound area of day x)/wound area of day 0] × 100. In each group, we use 16 to 20 photographs per day of analysis. The percentage of wounds with complete closure was calculated by the number of mice that achieve 100% of reepithelization over the total of mice. Wound re-epithelization was examined by ImageJ Software (1.49v).
2.11. Histology
The skin samples containing wounded and unwounded areas were carefully harvested using an 8 mm biopsy punch. The samples were fixed in cold 4% (wt/vol) formaldehyde in PBS at 4 °C overnight. Formaldehyde was discarded, and the tissues were washed 3 times using PBS 1x. We divided the circular sample in the middle of the circle to standardize the diameter of the sectioned samples. Then, the samples were treated with increasing concentration of alcohol (70%, 80%, 95%, and 100%) followed by xylol and paraffin and finally fixed in paraffin blocks. We sectioned at 5.0 μm and set 4–6 slices on microscope slides pretreated with poly-l-lysine. Following this procedure, we stained the skin sections with hematoxylin and eosin (Sigma). We incubated the slices with hematoxylin for 30 s, rinsed them in distilled water, incubated them for 30 s with eosin, washed them over in distilled water, and dehydrated them. For Picrosirius red staining (Picro Sirius Red Stain Kit, Sigma), we soaked the section for 30 min in distilled water. Then we incubated the section with Picro-Sirius red solution for 60 min. Slides were washed with acetic acid solution, and then they were washed with absolute alcohol. For Masson's Trichrome Staining, we incubated slides in preheated Bouin's Fluid for 60 min and cool for 10 min. We rinsed it in distilled water, and then we incubated in Weigert's Iron Hematoxylin for 5 min, Biebrich Scarlet Acid Fuchsin solution for 15 min, differentiate in the phosphomolybdic acid solution for 15 min, Aniline Blue solution for 10 min and acetic acid solution for 5 min. Finally, we mounted the slices on Entellan®, digitally captured the images under bright field microscopy (Leica DM4500B), and processed with Leica Application Suite 3v for Full view reconstruction.
2.12. Collagen quantification
To analyze the birefringence patterns, we used microscopy (Nikon® Corp., Kanagawa, Japan) with two CPL polarized filters. We captured the images with a 10x or 40x objective and assessed them for collagen Type III (bright green color) and collagen Type I (red color), with a total magnification of 100X and 400X. The birefringence for Type I collagen was dense, strongly birefringent red fibers, whereas Type III collagen showed thin, green, and softly birefringents [61]. We used Image Pro Plus (version 4) to measure the stained collagen fibers as the percentage of total pixels in each wound pictures.
2.13. Full view reconstruction
To increase the precision of evaluation, we reconstructed the whole skin section using unwounded and wounded images. We used a Nikon Microscopy to take 10-15 adjacent microphotographs (with 30% of common areas, 5x objective) and then reconstructed each skin section. We applied the Photomerge tool with the perspective layout in Adobe Photoshop (19.1v).
2.14. Transcript expression determination by real-time polymerase chain reaction (PCR
We used sample aliquots with 5.0 ng RNA for reverse transcription, applying random hexamer primers and Superscript Moloney-MLV reverse transcriptase. PCR reactions were performed in real-time using the TaqMan™ system (Applied Biosystems). The GAPDH gene (TaqMan™—Applied Biosystems) was employed as the endogenous control for the reaction to which the expression levels of the gene of interest in different samples were normalized. After calculating the efficiencies of amplification, a scatter plot was constructed to define the series of concentrations for which the system was competent. Primers for the target genes were purchased from Applied Biosystems and IDT DNA Technology against the TGF-1β (Mm.PT.58.43.479940), TNF-α (Mm00443258_ m1), IL-6 (Mm00446190_m1), VEGF-α (Mm. PT.58.14200306), FGF-1 (Mm.PT.56a.41158563), IGF-1 (Mm.PT.58.5811533), IL-1β n(Mm00434228_m1), MCP-1 (Mm99999056_m1), IL-10 (Mm01288386_m1), SDF-1 (Mm00445553_m1), ITGA (NM_010577.3), Plaur (ENSG00000011422), MMP9 (ENSG00000100985), Procr (ENSG00000101000) FLRT2 (ENSG00000185070), F4/80 (Mm00802529_m1), Cd11b (Mm00434455_m1), iNos (Mm.PT.5843705194), Cd163 (Mm00474091_m1), Arg1 (Mm.PT.58.8651372) and GAPDH (4352339E). We Used the StepOnePlus Real-Time PCR System™ (Thermo Fisher Scientific). For transcript quantification, the PCR in real-time were carried out in duplicate in reactions containing 3 μl of TaqMan Universal PCR Master Mix 2X, 0.25 μl of primers and probe solution, 0.25 μl of water and 4.0 μl of cDNA. The negative control used 4.0 μl of water in place of cDNA. The cycling conditions were as follows: 45 cycles of 95 °C for 2 min, 95 °C for 5 s, and 60 °C for 30 s. The relative gene expression values were obtained by analyzing the results in the StepOne™ 2.2.2v (Applied Biosystems).
2.15. Statistical analysis
The data are presented as the mean ± S.D. We used an unpaired t-test to compare two groups and one-way ANOVA to compare more than two groups. P < 0.05 was considered statistically significant. We used GraphPad Prism 6 for Windows 6.01v to perform statistical analysis.
3. Results
3.1. Highly active factor XIII was critical to forming a stable fibrin matrix at low sealant concentration
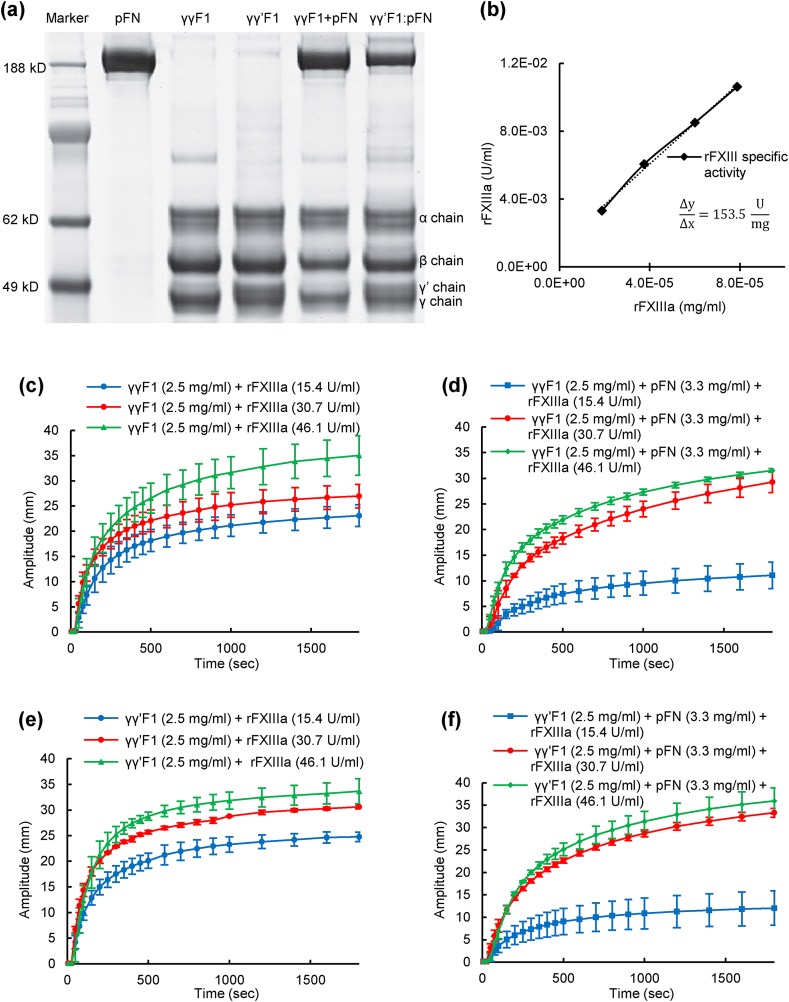
Recently, we developed a scalable process to separate γγF1 and γγ′F1 from human plasma at production scale [8]. We observed that γγ′F1 and pFN isolated as a noncovalent complex at about 1:1 M ratio [8]. This complex could be dissociated by passing through a gelatin affinity column where the pFN was absorbed, and γγ′F1 passed through the affinity column. We also found that the dissociated γγ′F1 and pFN could quickly re-associate at room temperature [62]. The high purity of the compositions used in this paper consisting of pFN, γγF1, γγ′F1, γγ′F1:pFN and a 1:1 mixture of γγF1 and pFN (γγF + pFN) were confirmed with reduced SDS-PAGE (Fig. 1a).
Fig. 1.
a) Reducing SDS-PAGE gel. b) rFXIIIa activity (153.5 ± 19 U/mg) was measured using the 5-(biotinamido) pentylamine incorporation assay. The effect of rFXIIIa on the viscoelastic and clotting kinetics of γγF1 (c), γγF1+pFN mixture (d), γγ′F1 (e), and γγ′F1:pFN complex (f) were evaluated via TEG assay.
The normal concentration of fibrinogen in blood plasma is 2–4 mg/mL [63]. The natural fibrin clot is strengthened by platelets. In literature, the fibrin matrix is typically prepared at high protein concentration, such as >25 mg/mL F1, to provide sufficient stability and strength [56]. However, our previous research found human cells typically did not grow well in fibrin matrix >9 mg/mL due to the dense fiber structures [56]. Thus, we recommended using <9 mg/mL fibrin for 3D in vitro cell culture, cell delivery, and wound healing application [56]. However, at such low concentration, it required FXIII-catalyzed covalent crosslinking to form a stable fibrin matrix [56]. To achieve this, we expressed a recombinant FXIII [56] with a specific cross-linking activity of 154 U/mg, which was about 6-fold higher than plasma-derived FXIII (Fig. 1b).
We then tested if stable fibrin matrix could be formed with low F1 concentration using this recombinant FXIIIa, and how the pFN influenced the gelation kinetics and resultant matrix mechanical properties. We used a commercially available, biotherapeutic grade recombinant thrombin and our rFXIIIa at different concentrations to make fibrin matrices with γγF1 alone, γγ′F1 alone, γγ′F1:pFN complex, or a 1:1 mixture of γγF1 and pFN. All sealants were formulated at an F1 concentration of 2.5 mg/mL with 3.3 mg/mL pFN for both γγ′F1:pFN and analogous γγF1+pFN mixtures. All groups formed stable matrices with higher concentrations of rFXIIIa observed to accelerate gelation and improve the mechanical properties of the fibrin (Fig. 1c–f). Thus, this highly active rFXIIIa allowed us to use a low concentration fibrin matrix for our following in vitro cell culture and in vivo studies.
3.2. 3D fibronectin presentation on the γγ′F1:pFN matrix
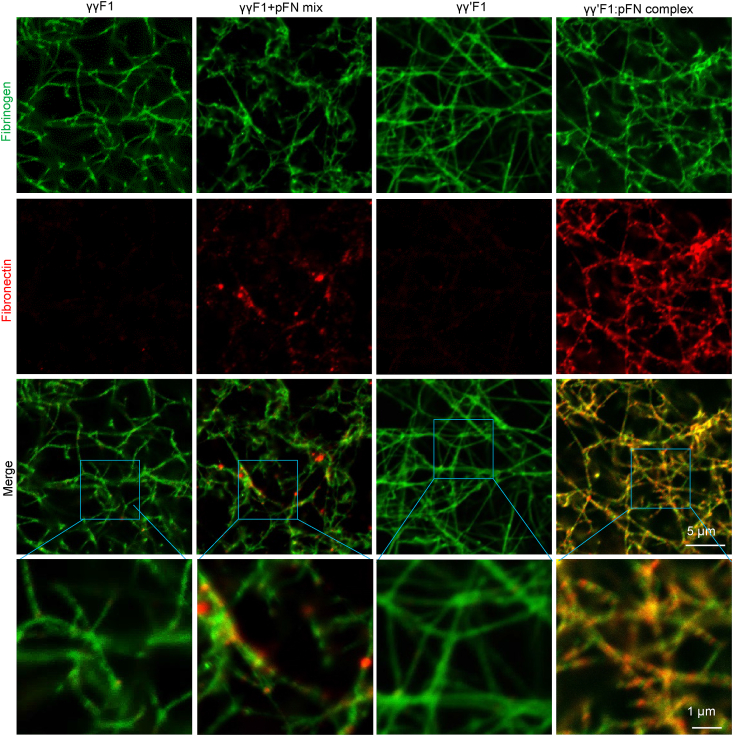
We used immunostaining and confocal microscopy to study the distribution of pFN and F1 in the fibrin matrices (Fig. 2). We found that all formulations produced similar fibrin fiber networks. Most fibers were between 100 and 400 nm in diameter while having a similar degree of branching. Large depots of pFN sporadically occurred within the fibrin matrix made from the γγF1+pFN mixture. The fibrin made from the γγ′F1:pFN complex presented pFN throughout the matrix as nanoclusters superimposed on the continuous γγ′F1 fiber network.
Fig. 2.
Confocal microscopy fibrin matrices made from γγF1, γγ′F1, 1:1 mixture of γγF1 and pFN, or γγ′F1:pFN complex.
3.3. The 3D fibronectin presentation enhanced the adhesion, migration, proliferation, and function of healing cells
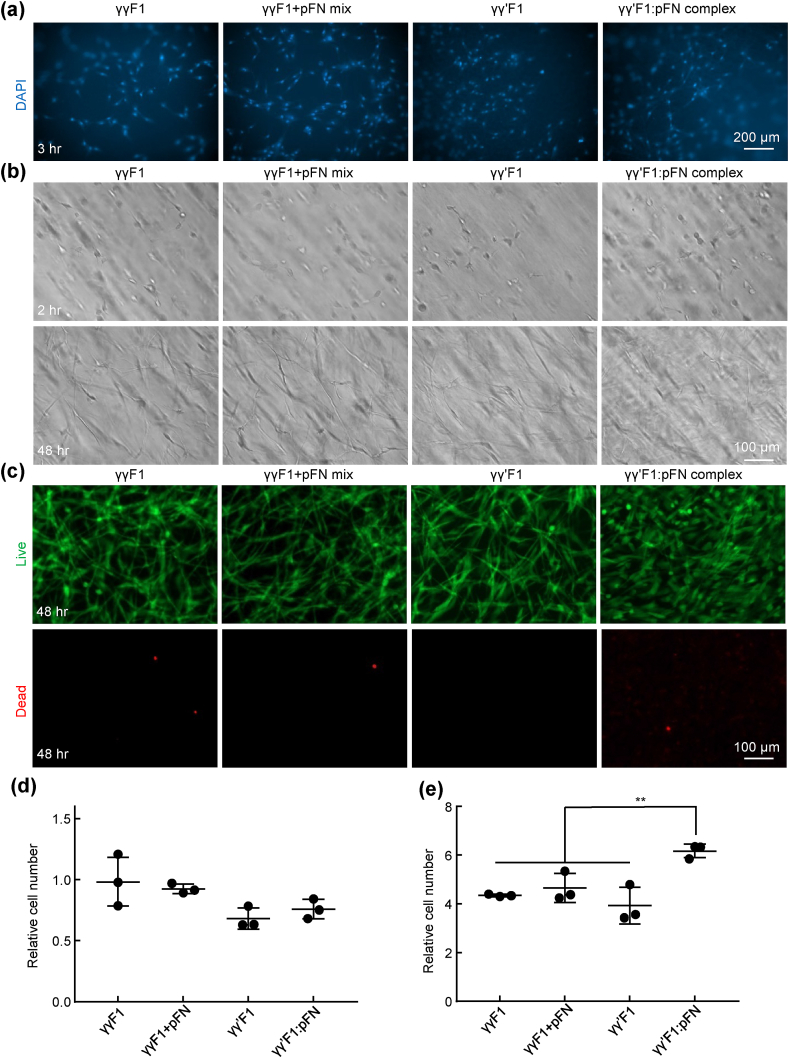
As previously documented, healing cells interact with the fibrin provisional matrix through binding fibronectins with their integrin receptors [[32], [33], [34], [35]]. We hypothesized that the 3D fibronectin nanoclusters could potentiate the cell-matrix interaction. We first tested the interactions of fibroblasts with these 4 different fibrin matrices. The adhesion of fibroblasts to fibrin made with γγF1, γγ′F1, 1:1 γγF1+pFN mixture, and γγ′F1:pFN complex were examined through culturing them on top of the matrix. Cells were allowed to attach to the matrix for 3 h, a time that is sufficient for cellular adhesion. The nuclei of the attached fibroblasts were stained by DAPI (Fig. 3a). Adhered cells were also quantified with Alamar blue assay (Fig. 3d). The results showed that fibroblasts had similar adhesion to these matrices.
Fig. 3.
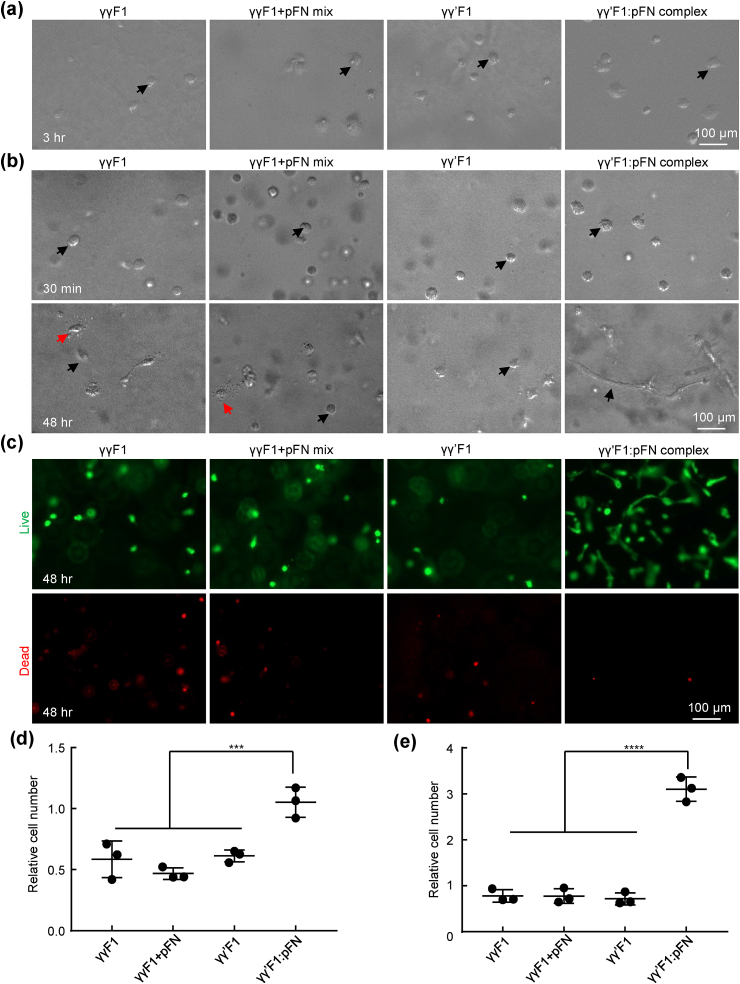
Culturing primary human fibroblasts on top of (a, d) and within 3D fibrin matrices (b, c, e). a) The nuclei of cells attached to fibrin matrices at 3 h. b) Fibroblasts in 3D fibrin matrices at 2 h and 48 h. c) Live and dead cells at 48 h. d) Cells attached to the fibrin matrices were quantified with alamar blue assay. e) Cells in 100 μl fibrin matrices at 48 h were quantified with alamar blue assay. Cell numbers are related to the initial seeded cells. **: p < 0.01.
For efficient wound healing, the provisional matrix also needs to support the survival, proliferation, and morphogenesis of both fibroblasts and endothelial cells (ECs) in addition to good adhesion. Since cells in vivo are in the 3D matrix, we also used 3D culture to study these phenomena in the 4 matrices. Fibroblasts were cultured within the fibrin matrix. After 48 h, fibroblasts exhibited extended morphology, indicating good adhesion in all matrices (Fig. 3b). Fibroblasts survived well in all matrices, as shown by the live/dead cell staining (Fig. 3c). However, the numbers of cells in the γγ′F1:pFN complex matrix was significantly higher (~1.5 fold) than in other matrices (Fig. 3e). These results showed the γγ′F1:pFN matrix or the 3D pFN presentation promoted the proliferation of fibroblasts.
The interactions of HUVECs with these 4 matrices were also investigated. HUVECs adhered to the surfaces of all fibrin matrices after 3 h. The numbers of HUVECs adhered to the γγ′F1:pFN matrix was 2 fold of these to the γγF1, γγ′F1, and 1:1 γγF1+pFN mixture matrix, indicating HUVECs had stronger adhesion to γγ′F1:pFN complex matrix (Fig. 4a and 4d). After 48 h of culturing in 3D fibrin matrices, HUVECs in γγF1, γγ′F1, and γγF1+pFN matrix showed spherical morphology, indicating poor adhesion to the matrices. Some cells showed blebs, a sign of apoptosis (Fig. 4b). The existence of apoptosis was confirmed by live/dead cell staining (Fig. 4c). HUVECs in γγ′F1:pFN complex matrix showed no apoptosis. Also, they showed a high level of morphological sprouting. Quantification found 400% more cells in the γγ′F1:pFN matrix than other formulations (Fig. 4e). These results showed the γγ′F1:pFN complex matrix or the 3D pFN presentation significantly promoted the adhesion, survival, proliferation, and morphogenesis of HUVECs.
Fig. 4.
Culturing primary human umbilical vein endothelial cells (HUVECs) on top of (a, d) and within 3D fibrin matrices (b, c, e). a) HUVECs attached to fibrin matrices at 3 h. b) HUVECs in 3D fibrin matrices at 30 min and 48 h. Red and black arrows point to apoptotic and live cells respectively. c) Live and dead cells at 48 h. d) Cells attached to the fibrin matrices were quantified with alamar blue assay. e) Cells in 100 μl fibrin matrices at 48 h were quantified with alamar blue assay. Cell numbers are related to the initial seeded cells. ***: p < 0.001 and ****: p < 0.0001.
3.4. The 3D fibronectin presentation accelerated skin wound healing in healthy mice
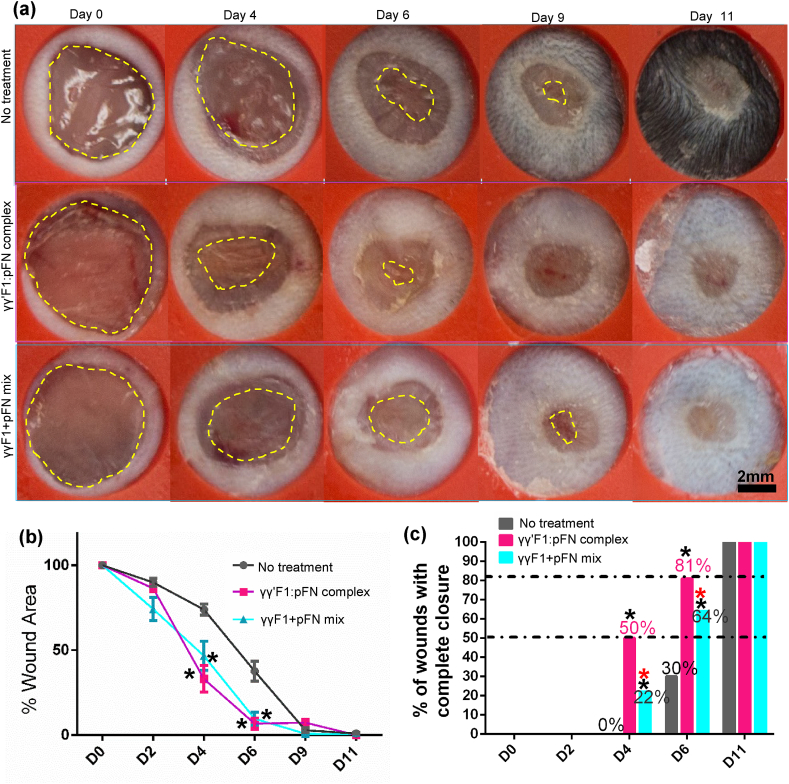
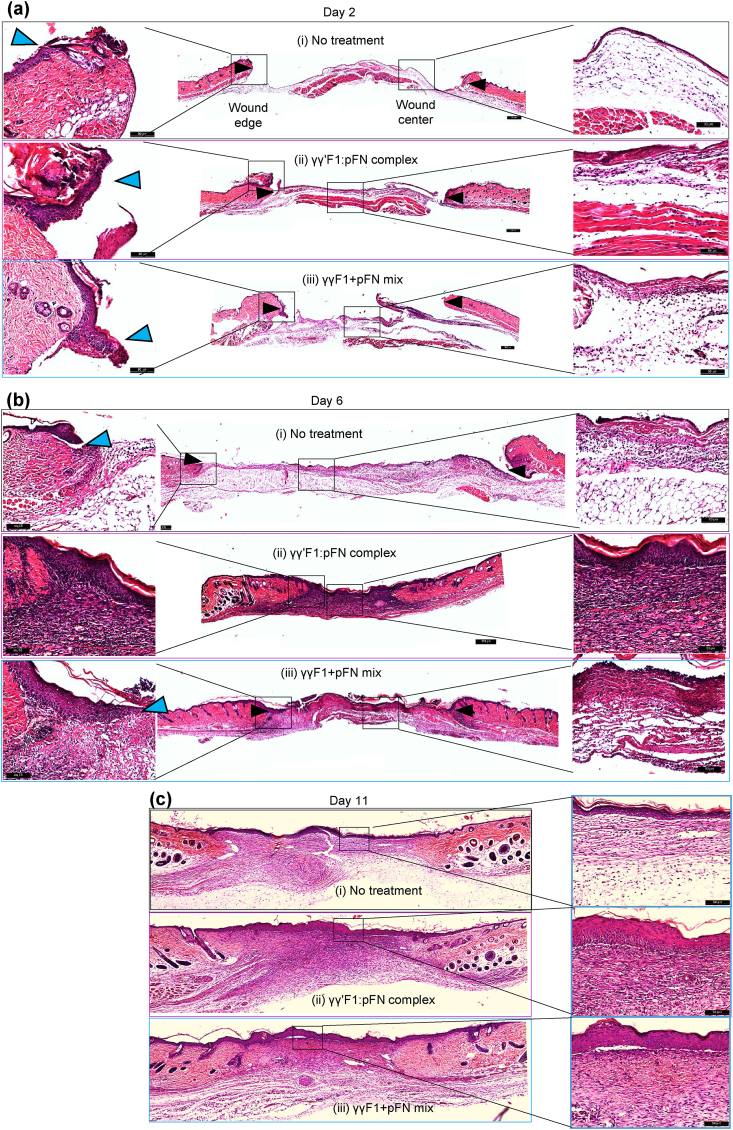
Based on the in vitro cell culture data, we hypothesized that the novel matrix could enhance the granulation tissue formation and wound closure in vivo. We tested this hypothesis in a full-thickness model in healthy mice. Wounds were treated with γγ′F1:pFN complex, or γγF1 and pFN mixture. The macroscopic evaluation showed the γγ′F1:pFN complex significantly speeded up wound closure (Fig. 5a). Indeed, 50% and 81% of wounds treated with the γγ′F1:pFN complex achieved full re-epithelization on days 4 and 6, respectively. On the other hand, 22% and 64% of wounds treated with the mixture achieved full re-epithelization on days 4 and 6, respectively, and 0% and 30% of the control wounds (without treatment) achieved full re-epithelization on day 4 and 6 respectively (Fig. 5c). The microscopic evaluation confirmed these observations. The migration of epidermal cells was significantly increased in the γγ′F1:pFN complex treated group just (i.e., the leading edge) 2 days post-treatment (Fig. 6a) and showed a complete epithelization over the wound bed on day 6 (Fig. 6b). Histology also showed dense granulation tissue at the wound center only in the γγ′F1:pFN complex treated group (Fig. 6b). These data supported that the novel fibrin sealant/matrix or the 3D pFN presentation promoted epidermal cell migration, resulting in faster re-epithelization in vivo.
Fig. 5.
(a) Macroscopic wound morphology on different days. Yellow dash line highlights the leading edge of the new epithelium. (b) The percentage of wound area, and (c) the percentage of mice achieved 100% re-epithelization on different days. 10 mice with 20 wounds were used. *: p < 0.05; *:compared to no treatment group; *: compared to complex.
Fig. 6.
H&E staining of days 2, 6 and 11 wounds. Reprehensive images from 5 animals or 10 wounds.
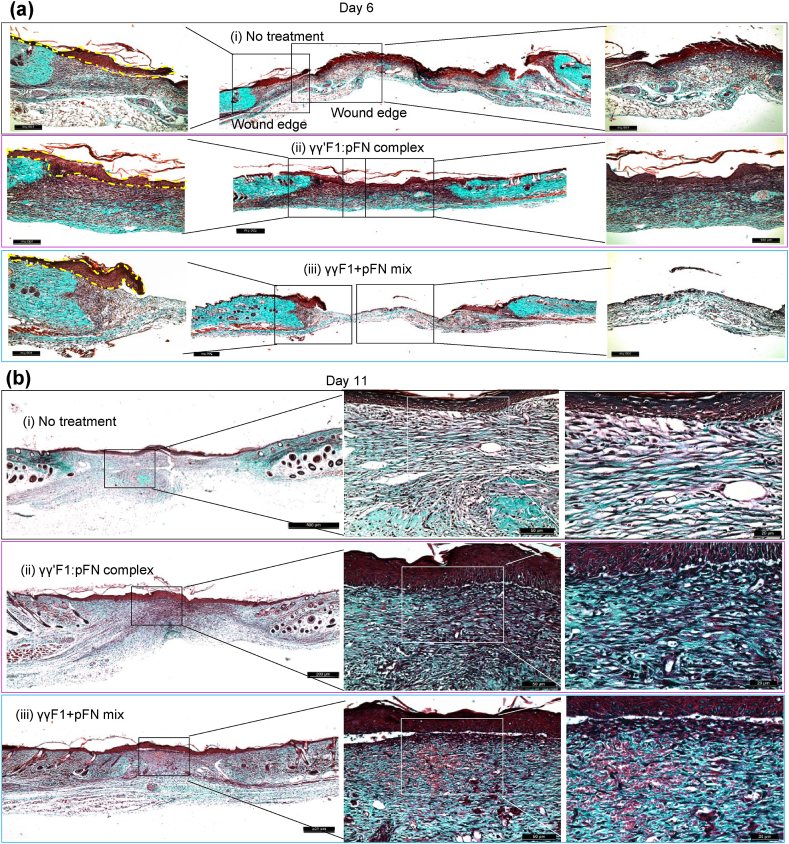
We also evaluated the extracellular matrix formation via histocolorimetric analysis of collagen content (Fig. 7). Total collagen deposition was identified by Masson's trichrome staining on days 6 and 11 post-wound, period when the highest production of collagen is reached [64]. Animals treated with γγ′F1:pFN complex or γγF1+pFN mixture treatments showed no differences in total collagen deposition. However, the quality of collagen was different. Staining showed dense and aligned reticular collagen fibers in the γγ′F1:pFN complex group on day 6, suggesting high-quality granulation tissue (Fig. 7a). The other treatment groups had a distinctly more sparse and randomly oriented collagen layer showing less mature granulation tissue.
Fig. 7.
Masson's trichrome staining was done to evaluate the total collagen deposition. Collagen (blue) on the wound bed was much less than the surrounding normal skin. Reprehensive images from 5 animals or 10 wounds.
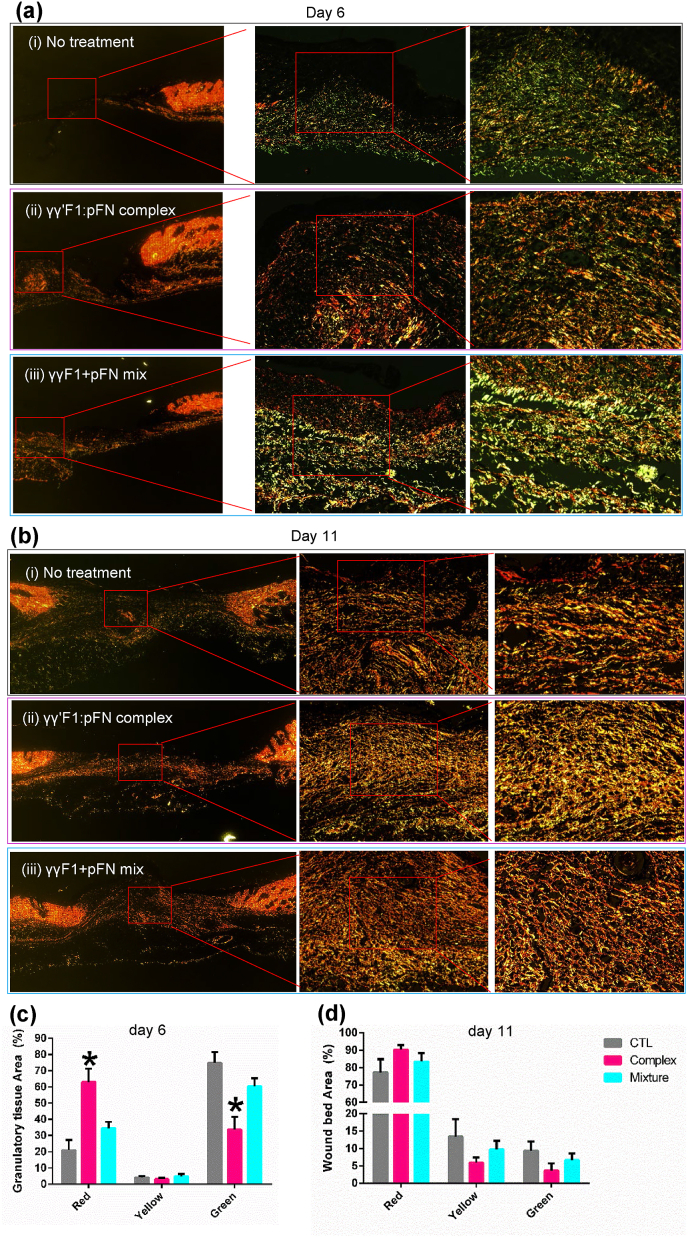
We evaluated the collagen subtype progression through Picrosirius Red Staining, which stains collagen III in green and collagen I in red (Fig. 8). In physiological wound healing, granulation tissue mainly consists of collagen III in the proliferative phase that gradually changes to mesh-like collagen I in the remodeling phase. Our data of control wounds agreed with this (Fig. 8). However, wounds treated with the γγ′F1:pFN complex on day 6 showed higher collagen I content. Even more, γγ′F1:pFN complex on day 6 was similar to day 11 of control group wounds (Fig. 8a–c). On day 11, the γγ′F1:pFN complex group had the mesh-like collagen organization (Fig. 8b). These data showed the γγ′F1:pFN complex shifted the granulation formation, collagen remodeling about 5 days earlier, and improved the quality of the new tissue.
Fig. 8.
Picrosirius red staining on collagen I (red) and III (green) on day 6 (a, c) and 11 (b, d). Reprehensive images from 5 animals or 10 wounds. Standard error is presented in (c, d). *:p < 0.05. CTL: control without treatment. Complex: γγ′F1:pFN complex. Mixture: 1:1 mixture of γγF1 and pFN.
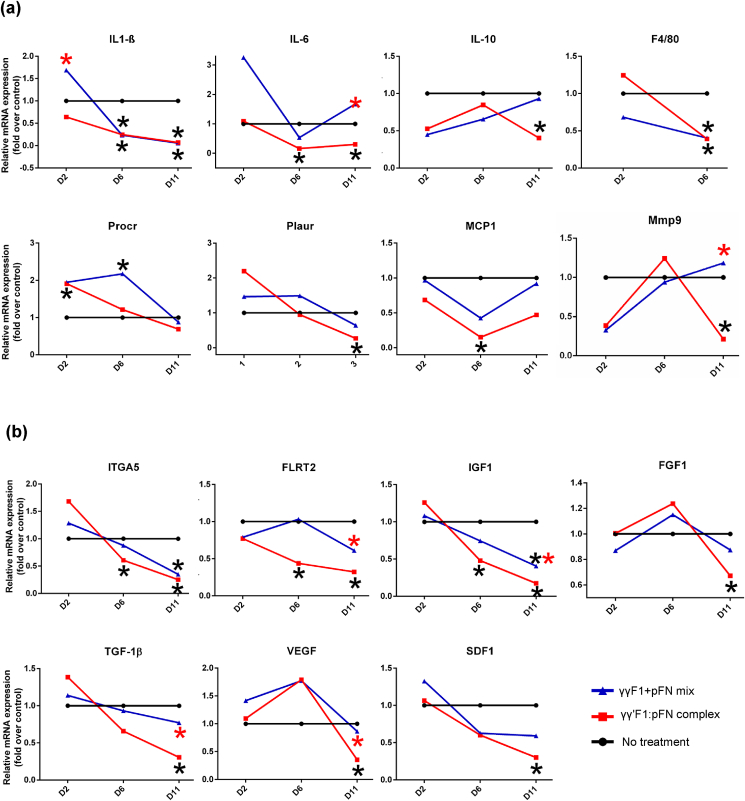
We assessed the expression of 20 signature genes associated with the transitioning between sequences of inflammation, cell migration, and proliferation [65] that could corroborate macroscopic changes in dermal tissue structure. Several statistically significant changes in gene expression related to the sequence of these processes were observed among the broad panel of assays presented here (Fig. 9a). For signals associated primarily with an active inflammatory phase, we observed by day 2 the γγ′F1:pFN fibrin treatment group had decreased expression of inflammatory cytokines IL-β1 and IL-10 relative to the other treatment groups(Fig. 9a). Furthermore, by day 6 both relative levels of IL-β1 and IL-6 had receded to very low relative levels as had the macrophage marker F4/80 and monocyte chemoattractant MCP-1. These already diminished signal profiles related to the inflammatory phase indicates that the inflammatory phase was resolved very early in the healing process.
Fig. 9.
The relative expression of genes related to inflammation (a), and cell migration and proliferation (b). RNA level was normalized to the untreated group. *:p < 0.05, *:compared to no treatment group; *: compared to complex.
The transition from inflammation to signaling needed for keratinocyte migration also occurred early. We found that PROCR, a gene highly expressed by migrating keratinocytes [66], was increased by the complex and Mixture treatments on day 6 (Fig. 9a). Moreover, the complex group showed an early expression of PROCR on day 2. The Fibronectin Leucine Rich Transmembrane Protein 2 (FLRT2), a cell-cell adhesion marker for keratinocytes, is highly expressed in the leading edge during wound healing [65]. FLRT2 is not expressed in healthy skin epidermis but is expressed during the granulation tissue formation. Our result showed that the FLRT2 expression was decreased on days 6 and 11 in the γγ′F1:pFN complex group, supporting that the cell migration was completed before day 6 in the complex group (Fig. 9b). Keratinocytes bind to the fibrin matrix using alpha 5 integrins (ITGA5), in order to migrate. We found ITGA5 expression increased on day 2 and significantly decreased on days 6 and 11 for the complex group, supporting that cell migration was completed before day 6 (Fig. 9b). These data agree with the acceleration of tissue structure observed, such as the complete re-epithelization and granulation tissue structure found on day 6 for the γγ′F1:pFN treatment group.
Growth factor signaling is critical for granulation formation. We found the γγ′F1:pFN treatment decreased expression of TGF-β1, FGF1, SDF1, VEFG-α and IGF1 genes on day 11, supporting that cell proliferation, granulation tissue formation, and re-epithelization were completed before day 11 in the γγ′F1:pFN complex group (Fig. 9a). Even more, IGF1 early declined on day 6, showing the end of the proliferative phase. Taking all together, animals treated with the γγ′F1:pFN complex showed a non-inflammatory, non-proliferative, and non-migrative profile on day 11, suggesting an early resolution of the wound healing process. In short, our data showed the γγ′F1:pFN complex or the 3D pFN nanoclusters significantly accelerated skin wound healing in healthy mice.
4. Discussion
The topical application of the fibrin matrix represents a promising approach to enhance the healing of skin wounds. The natural provisional matrix contains a small number of fibronectins [1,[67], [68], [69]]. Previous reports have shown that raising the fibronectin content in the matrix promotes wound healing. For instance, human fetus [[70], [71], [72]] and invertebrate animals newts [73] heal wounds much faster because their provision matrices contain much more fibronectins that allow cells to migrate and proliferate faster. The topical application of pure soluble fibronectins enhances normal and diabetic wound healing in animals [70,[74], [75], [76], [77], [78]]. These soluble fibronectins aggregate or attach to surrounding tissue, but do not form a 3D matrix in the wound bed. A recent study shows that if processed into a 3D nanofiber scaffold, fibronectins can heal normal skin wound much faster [70]. However, processing fibronectin nanofibers at a large scale is extremely challenging, limiting their clinical applications.
Both receptor occupancy and clustering are required for integrin signaling [[79], [80], [81]]. Most recent research shows that one way to boost integrin signaling is to present fibronectins as nanoclusters that can cluster the integrin receptors. For example, when cells are cultured on a uniformly fibronectin-coated substrate, high fibronectin density is required to trigger the integrin signaling. On the other hand, if fibronectins are patterned into nanoclusters, they trigger a much higher integrin clustering, signaling, and cell function [[81], [82], [83], [84], [85], [86], [87]]. However, nano-patterning fibronectin has only be achieved on a 2D substrate to date. How to pattern fibronectins as nanoclusters in a 3D matrix, especially on the fibrin nanofibers, and how the 3D nanoscale presentation influences the cell-matrix interaction and cell signaling are still unknown.
In our previous paper [8], we reported a bioprocess to separate γγF1 and γγ′F1 from plasma, and we found that γγ′F1 and pFN formed a tight complex. The γγ′F1 and pFN occur at nearly a 1:1 M ratio in normal human plasma [9,10,88,89], thus the γγ′F1:pFN complex we isolated may already exist in human plasma (Fig. 1). The γγ′F1:pFN complex formed a matrix with fibronectin nanoclusters on the fibrin nanofibers in the presence of highly active thrombin and rFXIIIa (Fig. 2). We showed that the fiber diameters and branch points are markedly similar for all of the fibrins studied. Thus, the fibronectin nanoclusters likely arose from interactions of pFN with the γ′ tails that are uniquely displayed by the γγ′F1 fibrils. Future research should make clear the dynamics of forming these nanoclusters, as well as how the sealant composition affects the matrix structure and properties. This report successfully reproduced all our previous findings (Fig. 1, Fig. 2), showing the robustness of the separation process, the γγ′F1:pFN complex, and the novel nanostructures.
We found that the γγ′F1:pFN matrix potentiated the adhesion of key cells that participate in the wound healing process, including fibroblasts (Fig. 3) and endothelial cells (Fig. 4) in vitro. This enhanced interaction led to boosted cell survival, proliferation, migration, and morphogenesis in 3D culture. This was particularly clear for endothelial cells. Because of the similarities in core fiber matrix structure among all fibrins studied here, the enhanced interaction of ECs with γγ′F1:pFN fibrin was likely due to the pFN nanoclusters. As said, angiogenesis is the rate-limiting step of granulation tissue formation, and granulation tissue formation is the rate-limiting step of wound healing. Thus the capability to stimulate endothelial cells has a significant impact on wound healing. The plasma-derived fibrin has a 10:1 ratio of F1 to pFN [7]. This fibrin has been previously shown to support the colonization of fibroblasts [90], but not EC. FGF-2 and or VEGF supplementation has been reported as the primary catalyst of EC colonization in plasma-derived fibrin [[91], [92], [93]]. In contrast, we observed an accelerated EC colonization in the γγ′F1:pFN fibrin without using these growth factors. Future research should systematically make clear the dynamics of cell-matrix interaction and integrin clustering, and how the interaction alters the global gene expression and cell phenotypes. It is also very valuable to systematically study how the sealant composition and matrix structure influence the matrix interaction with the major cell types of the wound healing process.
Our in vivo studies agreed well with the in vitro cell culture results. The novel nanostructure significantly accelerated the re-epithelization (Fig. 5, Fig. 6), granulation tissue formation, and modeling (Fig. 7, Fig. 8). The whole wound healing process was speeded up about 5 days. Additionally, the quality of the granulation tissue was significantly improved. This conclusion was supported by the macroscopic morphology (Fig. 5) and microscopic structure analysis (Fig. 6, Fig. 7, Fig. 8) as well as molecule profiles (Fig. 9). Here we have shown that animals treated with γγ′F1:pFN complex accelerated cell migration. The previous report showed that cells at the leading edge in direct contact with the fibrin matrix do not proliferate actively but migrate to the center of the wound [65,94]. Animal treated with the γγ′F1:pFN complex modulated two key membrane proteins that guide cell migration and cell adhesion. It will be valuable to systematically study the cellular profiles at different time points along the healing process in the future.
As said, large acute skin wounds after surgery, trauma, and burn heal slowly (e.g., take >1 month) and result in scarred skin that has an abnormal appearance, structure, and function. We believe our novel matrix has a high potential to promote the healing of these wounds. Since all materials used in our matrix are human proteins, their safety is expected. Thus our matrix can be quickly translated to bedside once the efficacy and underlying mechanisms are made clear. Since our matrix is fibrin-based, it can be used for both hemostasis and enhancing wound healing. In addition to the acute wounds, our matrix may be used to treat the chronic wound as well. Skin wounds in the aged population, people with diabetes, obesity, and vascular diseases present serious difficulties to heal [95,96]. Current treatments have limited success [[95], [96], [97], [98]]. It will be very valuable to study treating chronic wounds with our novel sealant in the future.
In conclusion, we confirmed that the 10% γγ′F1 in plasma could be separated from the 90% γγF1 using our previously reported method [8], as well as γγ′F1 formed a complex with pFN. We also confirmed that the γγ′F1:pFN complex formed a fibronectin-nanocluster-on-fibrin-nanofiber structure. We found that the 3D nano-scale presentation of the FN could significantly potentiate the cell-matrix interaction in vitro and accelerate the granulation formation, re-epithelization in a mice skin wound.
CRediT authorship contribution statement
Carlos Poblete Jara: Investigation, Methodology, Data curation, Formal analysis, Writing - original draft. Ou Wang: Investigation, Methodology, Data curation, Formal analysis, Writing - original draft. Thais Paulino do Prado: Investigation. Ayman Ismail: Investigation. Frank Marco Fabian: Investigation. Han Li: Investigation. Licio A. Velloso: Conceptualization, Writing - review & editing. Mark A. Carlson: Resources. William Burgess: Investigation. Yuguo Lei: Funding acquisition, Conceptualization, Supervision, Writing - original draft, Writing - review & editing, Project administration. William H. Velander: Funding acquisition, Conceptualization, Supervision, Writing - original draft, Writing - review & editing, Project administration. Eliana P. Araújo: Funding acquisition, Conceptualization, Supervision, Writing - original draft, Writing - review & editing, Project administration.
Declaration of competing interest
All authors declared no competing interests.
Acknowledgements
This research was supported by the University of Nebraska Research Initiative 2018–2019 (YL and WV); the University of Nebraska-Lincoln start-up (YL); the Nebraska DHHS Stem Cell Grant 2019 (YL and WV); the U.S. Army GRANT10824516 (WV); and the Department of Defense, USA, W81XWH-BAA-11-1 (WV). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 88882.434714/2019–01 (EPA and LAV). Confocal microscopy imaging was done in the Morrison Microscopy Core Research Facility at the University of Nebraska, Lincoln.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Yuguo Lei, Email: ylei14@unl.edu.
William H. Velander, Email: wvelander2@unl.edu.
Eliana P. Araújo, Email: earaujo@unicamp.br.
References
- 1.Clark R.a. Fibrin and wound healing. Ann. N. Y. Acad. Sci. 2001;936:355–367. doi: 10.1111/j.1749-6632.2001.tb03522.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y. Plasma fibronectin supports hemostasis and regulates thrombosis. J. Clin. Invest. 2014;124:4281–4293. doi: 10.1172/JCI74630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKee P.A., Mattock P., Hill R.L. Subunit structure of human fibrinogen, soluble fibrin, and cross-linked insoluble fibrin. Proc. Natl. Acad. Sci. U. S. A. 1970;66:738–744. doi: 10.1073/pnas.66.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spraggon G., Everse S.J., Doollttle R.F. Crystal structures of fragment D from human fibrinogen and its crosslinked counterpart from fibrin. Nature. 1997;389:455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 5.Weisel J.W., Stauffacher C.V., E Bullitt C.C. A model for fibrinogen: domains and sequence. Science. 1985;80:1388–1391. doi: 10.1126/science.4071058. [DOI] [PubMed] [Google Scholar]
- 6.Maurer L.M., Ma W., Mosher D.F. Dynamic structure of plasma fibronectin. Crit. Rev. Biochem. Mol. Biol. 2016;51:213–227. doi: 10.1080/10409238.2016.1184224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Willige S.U., Standeven K.F., Philippou H., Ariëns R.A.S. The pleiotropic role of the fibrinogen γ′ chain in hemostasis. Blood. 2009;114:3994–4001. doi: 10.1182/blood-2009-05-217968. [DOI] [PubMed] [Google Scholar]
- 8.Ismail A.E.A. The isolation of a plasma-derived γγ’ fibrinogen: fibronectin mixture that forms a novel polymeric matrix. Process Biochem. 2018;75:257–265. [Google Scholar]
- 9.Mosher D.F. Physiology of fibronectin. Annu. Rev. Med. 1984;35:561–575. doi: 10.1146/annurev.me.35.020184.003021. [DOI] [PubMed] [Google Scholar]
- 10.Hynes R.O., Yamada K.M. Fibronectins: multifunctional modular glycoproteins. J. Cell Biol. 1982;95:369–377. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariëns R.A.S., Lai T.S., Weisel J.W., Greenberg C.S., Grant P.J. Role of factor XIII in fibrin clot formation and effects of genetic polymorphisms. Blood. 2002;100:743–754. doi: 10.1182/blood.v100.3.743. [DOI] [PubMed] [Google Scholar]
- 12.Chen R., Doolittle R.F. Identification of the polypeptide chains involved in the cross-linking of fibrin. Proc. Natl. Acad. Sci. U. S. A. 1969;63:420–427. doi: 10.1073/pnas.63.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosesson M.W. Fibrin polymerization and its regulatory role in hemostasis. J. Lab. Clin. Med. 1990;116:8–17. [PubMed] [Google Scholar]
- 14.Mosher D.F., Johnson R.B. Specificity OF fibronectin‐fibrin cross‐linking. Ann. N. Y. Acad. Sci. 1983;408:583–594. doi: 10.1111/j.1749-6632.1983.tb23275.x. [DOI] [PubMed] [Google Scholar]
- 15.Corbett S.A., Wilson C.L., Schwarzbauer J.E. Changes in cell spreading and cytoskeletal organization are induced by adhesion to a fibronectin-fibrin matrix. Blood. 1996;88:158–166. [PubMed] [Google Scholar]
- 16.Mosher D.F. Cross-linking of cold-insoluble globulin by fibrin-stabilizing factor. J. Biol. Chem. 1975;250:6614–6621. [PubMed] [Google Scholar]
- 17.Takagi T., Doolittle R.F. Amino acid sequence studies on factor XIII and the peptide released during its activation by thrombin. Biochemistry. 1974;13:750–756. doi: 10.1021/bi00701a018. [DOI] [PubMed] [Google Scholar]
- 18.Makogonenko E., Tsurupa G., Ingham K., Medved L. Interaction of fibrin(ogen) with fibronectin: further characterization and localization of the fibronectin-binding site. Biochemistry. 2002;41:7907–7913. doi: 10.1021/bi025770x. [DOI] [PubMed] [Google Scholar]
- 19.Siebenlist K.R. Studies on the basis for the properties of fibrin produced from fibrinogen-containing γ′ chains. Blood. 2005;106:2730–2736. doi: 10.1182/blood-2005-01-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin P., Nunan R. Cellular and molecular mechanisms of repair in acute and chronic wound healing. Br. J. Dermatol. 2015;173:370–378. doi: 10.1111/bjd.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilgus T.A., Roy S., McDaniel J.C. Neutrophils and wound repair: positive actions and negative reactions. Adv. Wound Care. 2013;2:379–388. doi: 10.1089/wound.2012.0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzyszczyk P., Schloss R., Palmer A., Berthiaume F. The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front. Physiol. 2018;9:1–22. doi: 10.3389/fphys.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goren I. A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme m-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. Am. J. Pathol. 2009;175:132–147. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirza R., DiPietro L.A., Koh T.J. Selective and specific macrophage ablation is detrimental to wound healing in mice. Am. J. Pathol. 2009;175:2454–2462. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fadok V.A. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms. J. Clin. Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khanna S. Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PloS One. 2010;5 doi: 10.1371/journal.pone.0009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wicks K., Torbica T., Mace K.A. Myeloid cell dysfunction and the pathogenesis of the diabetic chronic wound. Semin. Immunol. 2014;26:341–353. doi: 10.1016/j.smim.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Shin-ichiro Okizaki. Suppressed recruitment of alternatively activated macrophages reduces TGF-β1 and impairs wound healing in streptozotocin-induced diabetic mice. Biomed. Pharmacother. 2015;70:317–325. doi: 10.1016/j.biopha.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Bannon P. Diabetes induces stable intrinsic changes to myeloid cells that contribute to chronic inflammation during wound healing in mice. Dis. Model. Mech. 2013;6:1434–1447. doi: 10.1242/dmm.012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirza R.E. Macrophage PPARγ and impaired wound healing in type 2 diabetes. J. Pathol. 2015;236:433–444. doi: 10.1002/path.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer A.J., Clark R.A.F. Cutaneous wound healing. N. Engl. J. Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 32.Greiling D., Clark R.a. Fibronectin provides a conduit for fibroblast transmigration from collagenous stroma into fibrin clot provisional matrix. J. Cell Sci. 1997;110:861–870. doi: 10.1242/jcs.110.7.861. [DOI] [PubMed] [Google Scholar]
- 33.Clark R. a F. Fibronectin and fibrin provide a provisional matrix for epidermal cell migration during wound reepithelialization. J. Invest. Dermatol. 1982;79:264–269. doi: 10.1111/1523-1747.ep12500075. [DOI] [PubMed] [Google Scholar]
- 34.Tonnesen M.G., Feng X., Clark R. a F. Angiogenesis in wound healing. J. Invest. Dermatol. Symp. Proc. 2000;5:40–46. doi: 10.1046/j.1087-0024.2000.00014.x. [DOI] [PubMed] [Google Scholar]
- 35.Lenselink E.a. Role of fibronectin in normal wound healing. Int. Wound J. 2015;12:313–316. doi: 10.1111/iwj.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knox P., Crooks S., Rimmer C.S. Role of fibronectin in the migration of fibroblasts into plasma clots. J. Cell Biol. 1986;102:2318–2323. doi: 10.1083/jcb.102.6.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawicka K.M., Seeliger M., Musaev T., Macri L.K., Clark a.F.R. Fibronectin interaction and enhancement of growth factors: importance for wound healing. Adv. Wound Care. 2015;4:469–478. doi: 10.1089/wound.2014.0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J., Clark R. a F. Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM-GF paradigm. J. Invest. Dermatol. 2014;134:895–901. doi: 10.1038/jid.2013.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balanis N., Carlin C.R. Mutual cross-talk between fibronectin integrins and the EGF receptor Molecular basis and biological significance. Cell. Logist. 2012;2:46–51. doi: 10.4161/cl.20112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita M. Cross-talk between integrin α6β4 and insulin-like growth factor-1 receptor (IGF1R) through direct α6β4 binding to IGF1 and subsequent α6β4-IGF1-IGF1R ternary complex formation in anchorage-independent conditions. J. Biol. Chem. 2012;287:12491–12500. doi: 10.1074/jbc.M111.304170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant M.B., Caballero S., Bush D.M., Spoerri P.E. Fibronectin fragments modulate human retinal capillary cell proliferation and migration. Diabetes. 1998;47:1335–1340. doi: 10.2337/diab.47.8.1335. [DOI] [PubMed] [Google Scholar]
- 42.Sieg D.J. FAK integrates growth-factor and integrin signals to promote cell migration. Nat. Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 43.Veevers-lowe J., Ball S.G., Shuttleworth A., Kielty C.M. Mesenchymal stem cell migration is regulated by fibronectin through a 5 b 1-integrin-mediated activation of PDGFR- b and potentiation of growth factor signals. J. Cell Sci. 2011;124:1288–1300. doi: 10.1242/jcs.076935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultz G.S., Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. Interact. 2009;17:153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 45.Rahman S. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol. 2005;6:8. doi: 10.1186/1471-2121-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin F. Fibronectin growth factor-binding domains are required for fibroblast survival. J. Invest. Dermatol. 2011;131:84–98. doi: 10.1038/jid.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuwada S.K., Li X. Epithelial cell proliferation through epidermal growth factor receptor activation. Mol. Biol. Cell. 2000;11:2485–2496. doi: 10.1091/mbc.11.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J., Asthagiri A.R. Matrix stiffening sensitizes epithelial cells to EGF and enables the loss of contact inhibition of proliferation. J. Cell Sci. 2011:1280–1287. doi: 10.1242/jcs.078394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spotnitz W.D. Fibrin sealant: past, present, and future: a brief review. World J. Surg. 2010;34:632–634. doi: 10.1007/s00268-009-0252-7. [DOI] [PubMed] [Google Scholar]
- 50.Spotnitz W.D. Fibrin sealant: the only approved hemostat, sealant, and adhesive-a laboratory and clinical perspective. ISRN Surg. 2014;2014:203943. doi: 10.1155/2014/203943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amrani D., Diorio J., Delmotte Y. Wound healing: role of commercial fibrin sealants. Ann. N. Y. Acad. Sci. 2001;936:566–579. [PubMed] [Google Scholar]
- 52.Gugerell A. Thrombin as important factor for cutaneous wound healing: comparison of fibrin biomatrices in vitro and in a rat excisional wound healing model. Wound Repair Regen. 2014;22:740–748. doi: 10.1111/wrr.12234. [DOI] [PubMed] [Google Scholar]
- 53.Eby J.B. The effect of fibrin sealant on the healing of laser-resurfaced skin. Aesthetic Surg. J. 2001;21:509–517. doi: 10.1067/maj.2001.120423. [DOI] [PubMed] [Google Scholar]
- 54.Yoon H.S., Na Y.C., Choi K.H., Huh W.H., Kim J.M. Wound healing effect of regenerated oxidized cellulose versus fibrin sealant patch: an in vivo study. Arch. Craniofacial Surg. 2019;20:289–296. doi: 10.7181/acfs.2019.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mogford J.E., Tawil B., Jia S., Mustoe T.A. Fibrin sealant combined with fibroblasts and platelet-derived growth factor enhance wound healing in excisional wounds. Wound Repair Regen. 2009;17:405–410. doi: 10.1111/j.1524-475X.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- 56.Wang O. A totally recombinant fi brin matrix for mesenchymal stem cell culture and delivery. J. Biomed. Mater. Res. 2018;6:3135–3142. doi: 10.1002/jbm.a.36508. [DOI] [PubMed] [Google Scholar]
- 57.Park D.S., Kim J.H., Lee S.W., Jeong J.M. Secretory expression of the alpha-subunit of human coagulation factor XIII in the yeast Pichia pastoris. Biotechnol. Lett. 2002;24:97–101. [Google Scholar]
- 58.Carlson M.A. A totally recombinant human fibrin sealant. J. Surg. Res. 2014;187:334–342. doi: 10.1016/j.jss.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 59.Siebenlist K.R., Meh D.A., Mosesson M.W. Plasma factor XIII binds specifically to fibrinogen molecules containing $γ$’ chains. Biochemistry. 1996;35:10448–10453. doi: 10.1021/bi9606206. [DOI] [PubMed] [Google Scholar]
- 60.Wang X., Ge J., Tredget E.E., Wu Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc. 2013;8:302–309. doi: 10.1038/nprot.2013.002. [DOI] [PubMed] [Google Scholar]
- 61.Junqueira L.C.U., Cossermelli W., Brentani R. Differential staining of collagens type I, II and III by Sirius red and polarization microscopy. Arch. Histol. Jpn. 1978;41:267–274. doi: 10.1679/aohc1950.41.267. [DOI] [PubMed] [Google Scholar]
- 62.Fabian F.M., Ismail A.E.A., Wang O., Lei Y., Velander W.H. Reversible associations between human plasma fibronectin and fibrinogen γγ’ heterodimer observed by high pressure size exclusion chromatography and dynamic light scattering. Anal. Biochem. 2020;598:113701. doi: 10.1016/j.ab.2020.113701. [DOI] [PubMed] [Google Scholar]
- 63.Ogston C.M., Ogston D. Plasma fibrinogen and plasminogen levels in health and in ischaemic heart disease. J. Clin. Pathol. 1966;19:352–356. doi: 10.1136/jcp.19.4.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinha M. Direct conversion of injury-site myeloid cells to fibroblast-like cells of granulation tissue. Nat. Commun. 2018;9:1–19. doi: 10.1038/s41467-018-03208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aragona M., Dekoninck S., Rulands S., Lenglez S., Mascré G., Simons B.D., Blanpain C. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 2017;8:14684. doi: 10.1038/ncomms14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKelvey K., Jackson C.J., Xue M. Activated protein C: a regulator of human skin epidermal keratinocyte function. World J. Biol. Chem. 2014;5:169–179. doi: 10.4331/wjbc.v5.i2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okada M., Blombäck B., Chang M.D., Horowitz B. Fibronectin and fibrin gel structure. J. Biol. Chem. 1985;260:1811–1820. [PubMed] [Google Scholar]
- 68.Corbett S.A., Lee L., Wilson C.L., Schwarzbauer J.E. Covalent cross-linking of fibronectin to fibrin is required for maximal cell adhesion to a fibronectin-fibrin matrix. J. Biol. Chem. 1997;272:24999–25005. doi: 10.1074/jbc.272.40.24999. [DOI] [PubMed] [Google Scholar]
- 69.Ramanathan A., Karuri N. Fibronectin alters the rate of formation and structure of the fibrin matrix. Biochem. Biophys. Res. Commun. 2014;443:395–399. doi: 10.1016/j.bbrc.2013.11.090. [DOI] [PubMed] [Google Scholar]
- 70.Chantre C.O. Production-scale fibronectin nano fibers promote wound closure and tissue repair in a dermal mouse model. Biomaterials. 2018;166:96–108. doi: 10.1016/j.biomaterials.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Longaker B.M.T. Studies in fetal wound Healing : III . Early deposition of fibronectin. J. Pediatr. Surg. 1989;24:799–805. doi: 10.1016/s0022-3468(89)80540-8. [DOI] [PubMed] [Google Scholar]
- 72.Moore A.L. Scarless wound healing : transitioning from fetal research to regenerative healing. Wiley Interdiscip Rev Dev Biol. 2018;7:2. doi: 10.1002/wdev.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calve S., Odelberg S.J., Simon H.G. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol. 2010;344:259–271. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiu Z., Ph D., Kwon A., Kamiyama Y. Effects of plasma fibronectin on the healing of full-thickness skin wounds in streptozotocin-induced diabetic rats. J. Surg. Res. 2007;138:64–70. doi: 10.1016/j.jss.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 75.Hamed S. Fibronectin potentiates topical erythropoietin-induced wound repair in diabetic mice. J. Invest. Dermatol. 2011;131:1365–1374. doi: 10.1038/jid.2011.15. [DOI] [PubMed] [Google Scholar]
- 76.McCulley J.P., Horowitz B., Husseini Z.M., Horowitz M. Topical fibronectin therapy of persistent corneal epithelial defects. Trans. Am. Ophthalmol. Soc. 1993;91:367–386. [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon A.H., Qiu Z., Hiraon Y. Topical application of plasma fibronectin in full-thickness skin wound healing in rats. Exp. Biol. Med. 2007;232:935–941. [PubMed] [Google Scholar]
- 78.Johnson M.B. Topical fibronectin improves wound healing of irradiated skin. Sci. Rep. 2017;7:3876. doi: 10.1038/s41598-017-03614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chester D., Brown A.C. The role of biophysical properties of provisional matrix proteins in wound repair. Matrix Biol. 2017;60–61:124–140. doi: 10.1016/j.matbio.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Miyamoto S. Integrin function: molecular hierarchies of cytoskeletal and signaling molecules. J. Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slater J.H., Frey W. Nanopatterning of fibronectin and the influence of integrin clustering on endothelial cell spreading and proliferation. J. Biomed. Mater. Res. 2008;87:176–195. doi: 10.1002/jbm.a.31725. [DOI] [PubMed] [Google Scholar]
- 82.Maheshwari G., Brown G., Lauffenburger D.A., Wells A., Griffith L.G. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 2000;1686:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 83.Arnold M., Cavalcanti-adam E.A., Glass R., Eck W. Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 84.Le Saux G. Spacing of integrin ligands influences signal transduction in endothelial cells. Biophys. J. 2011;101:764–773. doi: 10.1016/j.bpj.2011.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petrie T.A. Multivalent integrin-specific ligands enhance tissue healing and biomaterial integration. Sci. Transl. Med. 2010;2:45. doi: 10.1126/scitranslmed.3001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gautrot J.E. Cell sensing of the physical properties of the microenvironment at multiple scales: cells sense their mechanical microenvironment at the nanoscale level. Mechanobiol. Exploit. Med. Benefit chapter. 2017;19:297–329. [Google Scholar]
- 87.Di Cio S., Gautrot J.E. Cell sensing of physical properties at the nanoscale: mechanisms and control of cell adhesion and phenotype. Acta Biomater. 2016;30:26–48. doi: 10.1016/j.actbio.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 88.Chung D.W., Davie E.W. Gamma and gamma’ chains of human fibrinogen are produced by alternative mRNA processing. Biochemistry. 1984;23:4232–4236. doi: 10.1021/bi00313a033. [DOI] [PubMed] [Google Scholar]
- 89.Ariens R.A.S. Role of factor XIII in fibrin clot formation and effects of genetic polymorphisms. Blood. 2002;100:743–754. doi: 10.1182/blood.v100.3.743. [DOI] [PubMed] [Google Scholar]
- 90.Carlson M.A., Prall A.K., Gums J.J., Lesiak A., Shostrom V.K. Biologic variability of human foreskin fibroblasts in 2D and 3D culture: implications for a wound healing model. BMC Res. Notes. 2009;2:229. doi: 10.1186/1756-0500-2-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheresh D.A. Human endothelial cells synthesize and express an Arg-Gly-Asp-directed adhesion receptor involved in attachment to fibrinogen and von Willebrand factor. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6471–6475. doi: 10.1073/pnas.84.18.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sahni A., Francis C.W., Dc W. Vascular endothelial growth factor binds to fibrinogen and fibrin and stimulates endothelial cell proliferation Vascular endothelial growth factor binds to fibrinogen and fibrin and stimulates endothelial cell proliferation. Blood. 2014;96:3772–3778. [PubMed] [Google Scholar]
- 93.Sahni A., Sporn L.A., Francis C.W. Potentiation of endothelial cell proliferation by fibrin(ogen)-bound fibroblast growth factor-2. J. Biol. Chem. 1999;274:14936–14941. doi: 10.1074/jbc.274.21.14936. [DOI] [PubMed] [Google Scholar]
- 94.Park S. Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nat. Cell Biol. 2017;19:155–163. doi: 10.1038/ncb3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han G., Ceilley R. Chronic wound healing: a review of current management and treatments. Adv. Ther. 2017;34:599–610. doi: 10.1007/s12325-017-0478-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oo, N. I. J. S.-. Skin Substitutes for Treating Chronic Wounds. Technol. Assess. Program, Agency Healthc. Res. Qualtiy; 2019. https://www.ahrq.gov/sites/default/files/wysiwyg/r [Google Scholar]
- 97.Jeffcoate W.J., Vileikyte L., Boyko E.J., Armstrong D.G., Boulton A.J.M. Current challenges and opportunities in the prevention and management of diabetic foot ulcers. Diabetes Care. 2018;41:645–652. doi: 10.2337/dc17-1836. [DOI] [PubMed] [Google Scholar]
- 98.Armstrong D.G., Boulton A.J.M., Bus S.A. Diabetic foot ulcers and their recurrence. N. Engl. J. Med. 2017;376:2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]