Summary
Volume electron microscopy enables the ultrastructural analysis of biological tissue. Currently, the techniques involving ultramicrotomy (ATUM, ssTEM) allow large fields of view but afford only limited z-resolution, whereas ion beam-milling approaches (FIB-SEM) yield isotropic voxels but are restricted in volume size. Now we present a hybrid method, named ATUM-FIB, which combines the advantages of both approaches. ATUM-FIB is based on serial sectioning of tissue into “semithick” (2–10 μm) sections collected onto tape. Serial light and electron microscopy allows the identification of regions of interest that are then directly accessible for targeted FIB-SEM. The set of semithick sections thus represents a tissue “library” which provides three-dimensional context information that can be probed “on demand” by local high-resolution analysis. We demonstrate the potential of this technique to reveal the ultrastructure of rare but pathologically important events by identifying microglia contact sites with amyloid plaques in a mouse model of familial Alzheimer's disease.
Subject Areas: Imaging Anatomy, Biological Sciences, Cellular Neuroscience, Experimental Systems for Structural Biology
Graphical Abstract
Highlights
-
•
Fast nanometer-resolution relocation and 3D imaging of preselected structures
-
•
Transparent tape-based multiscale light and volume electron microscopy
-
•
Heated ultramicrotomy at 2–10 μm with precured epoxy resin
Imaging Anatomy; Biological Sciences; Cellular Neuroscience; Experimental Systems for Structural Biology
Introduction
Since the completion of the first connectomics dataset (White et al., 1986) volume electron microscopy (EM) techniques have been substantially refined and advanced. Although the interest in deciphering neuronal networks was the major driving force behind these technological developments, three-dimensional (3D) ultrastructural analysis has attracted considerable recent attention in a wide range of biological fields (Titze and Genoud, 2016). Applications range from classical cell biological questions to developmental biology, neuro- and cancer biology, botany, and microbiology (Karreman et al., 2014). Currently, 3D ultrastructure can be solved by destructive techniques including serial block-face electron microscopy (SB-SEM) (Mikula and Denk, 2015; Denk and Horstmann, 2004; Helmstaedter et al., 2013; Briggman et al., 2011) or focused ion beam-scanning electron microscopy (FIB-SEM) (Sonomura et al., 2013; Knott et al., 2008; Heymann et al., 2006). Although these methods benefit from high alignment accuracy, they lack the option of reacquisition and hierarchical imaging (Kornfeld and Denk, 2018). Alternatively, serial sectioning for transmission electron microscope camera array (TEMCA) (Lee et al., 2016; Bock et al., 2011; Zheng et al., 2018) or automated tape-collecting ultramicrotomy (ATUM) (Hayworth et al., 2014; Kasthuri et al., 2015; Mikula and Denk, 2015; Schalek et al., 2011; Tomassy et al., 2014; Hildebrand et al., 2017; Morgan et al., 2016; Terasaki et al., 2013) allow repetitive acquisition of regions of interest. However, as ultramicrotomy-based approaches are limited by their poor maximal z resolution of 30 nm, ion-milling techniques are required, if isotropic high-resolution voxels are needed. Still, the imaging volume in FIB-SEM is limited to a few tens of microns owing to the accumulation of milling artifacts caused by high-energy gallium ions (Xu et al., 2017). Despite recent advances in the application of alternative milling strategies (Kornfeld and Denk, 2018), targeted imaging is still required to restrict the acquisition volume. This is mainly achieved by correlated workflows involving targeted trimming guided by endogenous (Luckner et al., 2018) and artificial landmarks (Bishop et al., 2011; Karreman et al., 2014, 2016; Villani et al., 2019). X-ray micro computed tomography (microCT) (Villani et al., 2019; Karreman et al., 2014; Bushong et al., 2015; Sengle et al., 2012) has emerged as a tool for facilitated region of interest (ROI) relocation within the processed EM sample. This not only bridges multiple scales, from millimeter to micrometer dimensions, but also puts the site of interest into a broader morphological context (Maire and Withers, 2014). However, so far, microCT imaging options are not commonly accessible and the technique only provides a map for subsequent guided destructive sample preparation. An alternative prescreening of embedded tissue at a larger scale is implemented by rendering it accessible to light and electron microscopy (EM) modalities. Ultrathick sectioning at 10 to 20 μm thickness by the hot knife method produces samples that are accessible to large-scale FIB-SEM (Hayworth et al., 2015; Luckner and Wanner, 2018) enabling seamless reconstruction of large tissue blocks. Although this method was designed for a complete reconstruction of big volumes, it would be desirable to reduce time and data load for biological questions that require targeted ultrastructural analysis.
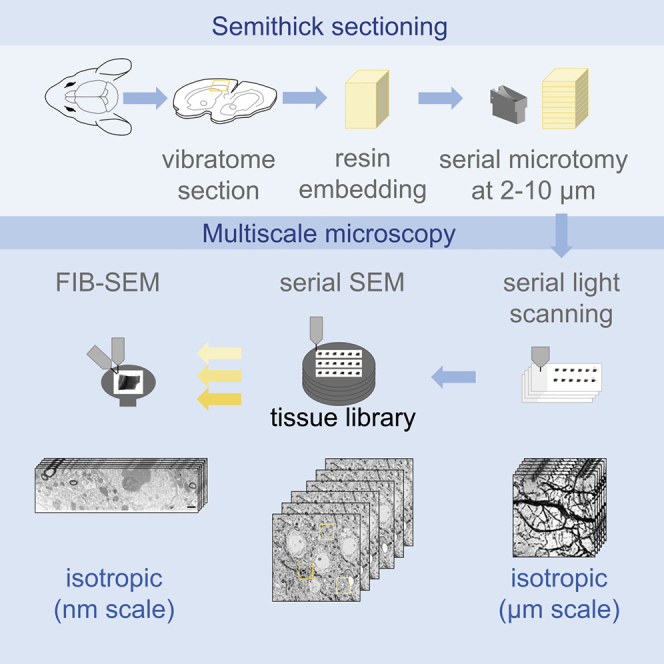
Here, we developed a multiscale method for targeted FIB-SEM on semithick (2–10 μm) sections named ATUM-FIB. Our method combines the advantages of both ultramicrotomy-based and serial block face imaging approaches. This approach is based on the ultramicrotomy of partially cured (Droz et al., 1975) resin-embedded samples facilitated by a custom-built diamond knife with temperature control. Serial thick sections are then collected onto carbon nanotube (CNT) tape (Kubota et al., 2018), compatible with both serial bright-field and scanning electron microscopy (SEM). While providing direct physical access to isotropic high-resolution imaging of multiple ROIs by FIB-SEM, this method simultaneously provides 3D tissue context information and allows archiving samples for future extended analysis.
Results
Semithick Sections Provide Suitable Information Content for Sparse Ultramicrotomy-Guided Targeting
Here, we developed a method that allows the search for an ultrastructural feature within a volume and the subsequent acquisition of a defined isotropic volume by FIB-SEM (Figure 1). Our goal was to expose surfaces at defined distances by serial sectioning. Although traditional ultrathin sectioning combined with sparse imaging saves imaging time, it is not compatible with the investigation of isotropic volumes. As the information content for FIB-SEM increases with section thickness, we explored whether “semithick” sections ranging from 2 to 10 μm would optimize ultramicrotomy time as well as screening efficiency (Figure 2A). Moreover, we aimed to apply water instead of an oil bath (as used in the ultrathick partitioning approach [Hayworth et al., 2015]) in order to avoid additional re-embedding and sectioning.
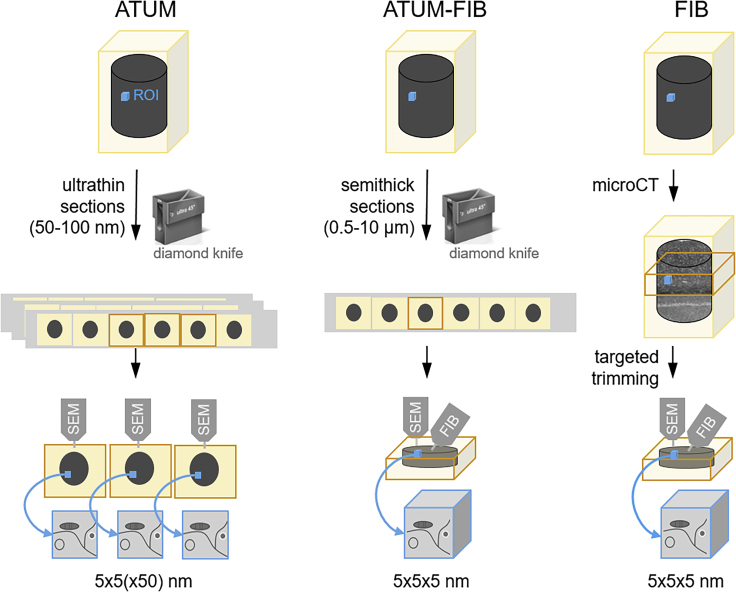
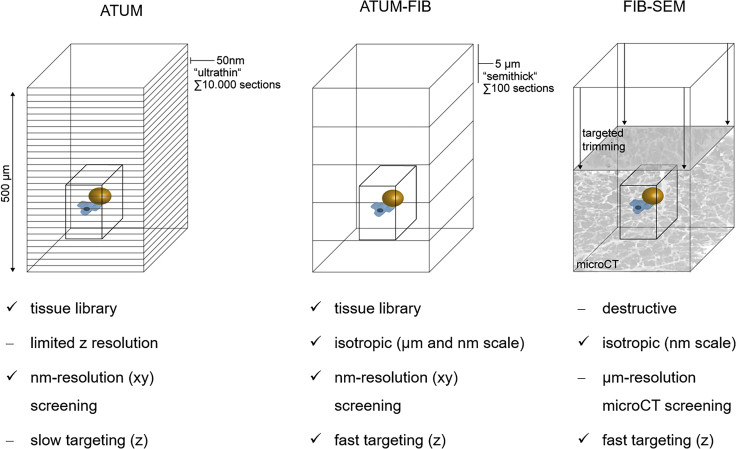
Figure 1.
Principle of ATUM-FIB as a Hybrid Volume EM Approach
Schematic of existing techniques for targeted volume SEM and comparison with the new ATUM-FIB approach (middle). Both ATUM and ATUM-FIB are microtomy-based, but ATUM-FIB generates semithick sections that can be selected by serial section light microscopy and SEM and subjected to further FIB-SEM investigation.
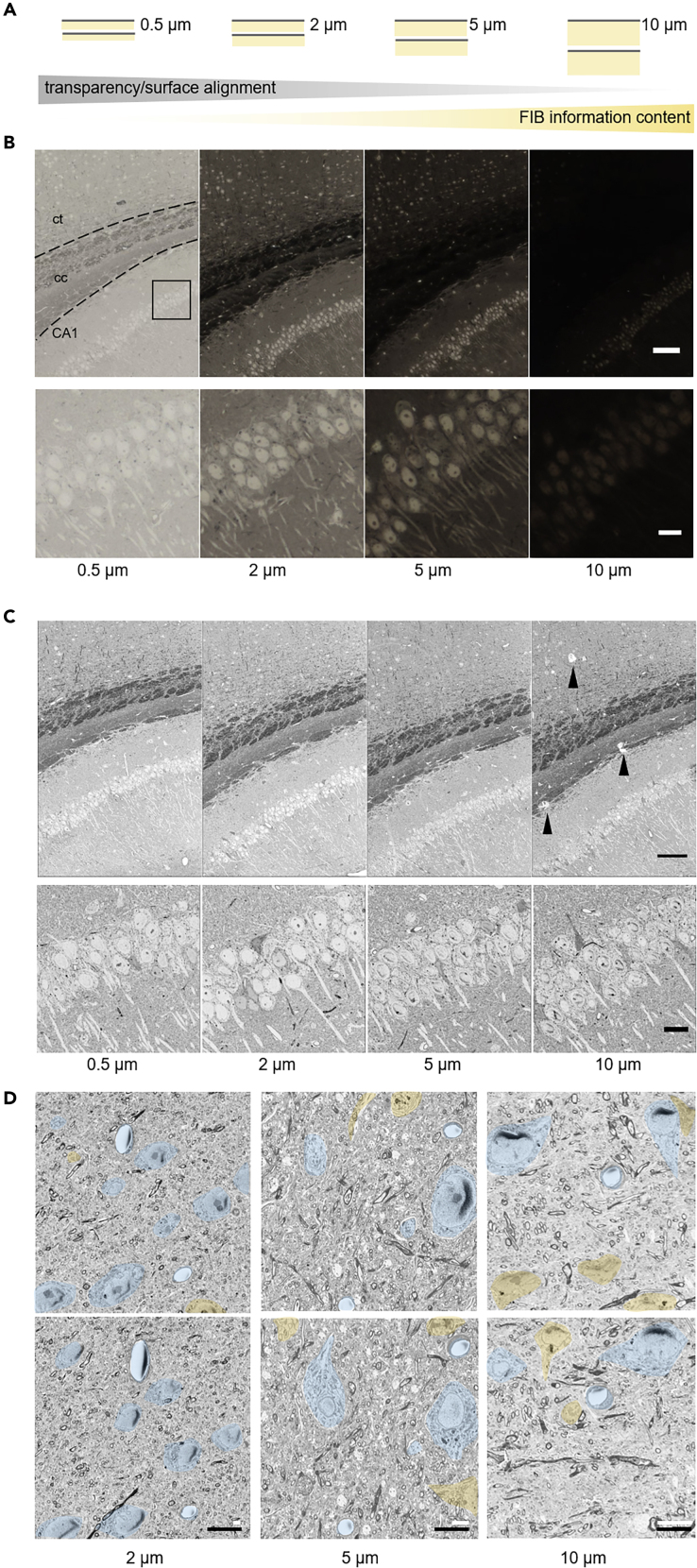
Figure 2.
Targeting Regions of Interest by Semithick Sectioning
(A) Cartoon of the information content of BSD surface imaging and the potential FIB-SEM analysis of sections of different thickness.
(B and C) (B) Transmitted light and (C) BSD images of semithin 0.5-μm and semithick 2-, 5-, and 10-μm sections of mouse brain tissue. cc, corpus callosum; ct, cortex; CA1, hippocampus CA1. Arrowheads indicate cracks, mainly in heavily stained tissue (corpus callosum). Scale bars 100 μm (top), 10 μm (bottom).
(D) Consecutive (upper, lower row) semithick (2, 5, 10 μm) sections allow tracing cell bodies and blood vessels across sections. Somata and blood vessels that can be followed from one to the next section are colored blue; others are shown in orange. Black scale bar 10 μm; white scale bars 2 (left), 5 (middle), and 10 μm (right). See Figure S1 for similarity of opposing surfaces of consecutive sections.
Semithick sections (0.5–10 μm) were generated using a diamond knife. Carbon nanotube (CNT) tape was chosen as a conductive and transparent section support for sequential light and electron microscopic assessment (Kubota et al., 2018). The tape strings with sections were placed onto a glass slide on a heating plate (60°C). A drop of water helped to adhere the strings flat on the glass slide and enable imaging with a slide scanner (Figure 2B). Transmitted light investigation was only possible for sections below 5 μm thickness, as in thicker sections reduced transparency masked morphological details. The sections were transferred onto a silicon wafer, and the surface was scanned by backscattered detection SEM (BSD), which enables the exploration of the entire semithick section range (Figure 2C). We generated two consecutive sections with 2, 5, and 10 μm thickness and looked at traceability of neuronal cell bodies and blood vessels. Although most of the structures could be recognized on the next 2-μm-thick section, alignment between two sections was difficult at thicknesses above 5 μm (Figure 2D). In summary, combined light microscopy and SEM with maximal FIB-SEM information content is optimal at 2 μm thickness, whereas electron microscopy is possible at 2–5 μm thickness depending on the needed alignment precision of the biological structure of interest.
Sequential Resin Curing and Heating Enables Semithick Sectioning and FIB Milling on the Same Sample
Diamond knives and resins have been successfully applied for ultrathin sectioning for decades. In order to routinely generate smooth surfaces by ultramicrotomy of serial sections with thickness of 2 μm or more, we speculated that the optimization of resin characteristics and ultramicrotomy parameters would be necessary. We therefore explored different contrasting methods and resins for semithick sectioning and subsequent SEM imaging. First, for contrasting, we settled on a standard rOTO (reduced osmium thiocarbohydrazide osmium) protocol without lead aspartate impregnation (Hua et al., 2015). Leaving out some heavy metal preembedding staining (e.g., in a rO protocol) would entail the need for post-contrasting strategies like the incubation in uranylacetate in ethanol at 60°C for 2–24 h (Locke and Krishnan, 1971). However, post-contrasting strategies carry the risk of uneven stain distribution within and between thick sections (Hayworth et al., 2015), so we avoided them (Table 1). Second, based on the rOTO stain, we optimized the resin choice as resin and contrasting requirements for thick sectioning and ion milling are conflicting: for semithick sectioning, softer resin and less heavy metal staining would be favored, whereas FIB-SEM requires hard resin and good contrasting to avoid charging artifacts. Various methacrylate (Norris et al., 2017) as well as epoxy resins including epon and durcupan (Hayworth et al., 2015) have been used for standard ultrathin serial sectioning. We investigated rOTO-processed mouse cortex samples embedded in different resins by semithick sectioning and subsequent backscattered detection SEM (Figure 3A). Regarding the sectioning characteristics, epon formulations as well as durcupan, which would be preferable for FIB-SEM (Xu et al., 2017), showed uneven surface topology at thicknesses above 1 μm (Figure 3A). Tissue embedded in epoxy resin LX112-embedded tissue could be sectioned up to 2-μm thickness without major surface defects (Figure 3A). LX112 resin (Ladd Research Industries) has been used for serial sectioning and is a replacement for Epon 812, which is not commercially available anymore (Ellis, 2014; Kizilyaprak et al., 2015). We reasoned that a two-step curing (Droz et al., 1975) would yield in resin characteristics that are optimal for both sectioning and FIB milling. Tissue embedded in LX112 was precured at 60°C for 10 and 48 h. Semithick sections of up to 10-μm thickness could be generated from the 10-h-cured blocks (Figure 3A). After shorter curing periods, resin blocks were too soft and sticky for sectioning (data not shown). Following ultramicrotomy, sections were post-cured for 2 days at 60°C ensuring the beam resistance required for both serial SEM and FIB-SEM imaging. Still, semithick sectioning proved difficult even in this softer pre-cured resin, therefore we explored a “hot knife” approach (Hayworth et al., 2015).
Table 1.
En bloc Embedding Protocol
| Reagent | Temperature | Microwave Protocol |
Bench Protocol |
||
|---|---|---|---|---|---|
| Time | Vacuum | Power (On-Off-On) | Time | ||
| 0.1 M cacodylate buffer | 23°C | 3 × 10 min | off | 250 W | 3 × 10 min |
| 2% osmium tetroxide, 2.5% potassium hexacyanoferrate in 0.1 M cacodylate buffer | 23°C | 6 × 1 min | on | 300 W | 2 h |
| 23°C | 1h (shaker) | – | – | ||
| 0.1 M cacodylate buffer | 23°C | 2 × 10 min | off | 250 W | 2 × 10 min |
| Water | 23°C | 1 min | on | 250 W | 10 min |
| 1% TCH | 40°C | 25 min | on | 200 W | 45 min |
| Water | 23°C | 3 × 10 min | on | – | 3 × 10min |
| 2% osmium tetroxide in water | 23°C | 6 × 1 min | on | 300 W | 2 h |
| 23°C | 1h (shaker) | – | – | ||
| Water | 23°C | 3 × 10 min | on | – | 3 × 10min |
| 1% uranylacetate in water | 4°C | over night | – | – | over night |
| 50°C | 2 h | – | – | 2 h | |
| Water | 23°C | 1 min | on | 250 W | 10 min |
| 15, 30, 40, 50, 60, 70, 80, 90, 100, 100, 100% ethanol | 23°C | 1 min each | off | 200 W | 10 min each |
| 25, 50, 75, 90% LX112 | 23°C | 10 min | on | 250 W | |
| 100% LX112 | 23°C | 10 min | on | 250 W | over night |
| 23°C | over night | – | – | ||
| 100% LX112 | 23°C | 10 min | on | 250 W | 3-6 h |
| 23°C | 3-6 h | – | – | ||
| Curing | 60°C | 10, 48 h | 10, 48 h | ||
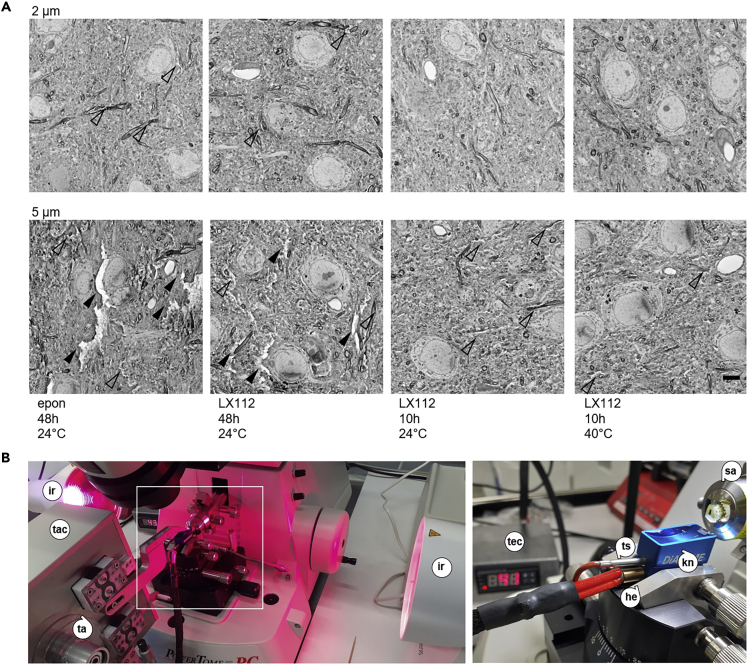
Figure 3.
Generation of Semithick Sections by Precuring and Elevated-Temperature Microtomy
(A) Comparison of resin performance for semithick sections of mouse cortex tissue. BSD images were acquired of 2 (top) and 5 μm (bottom) semithick sections of epon and LX112 blocks cured for 48 or 10 h and sectioned at room temperature or at 40°C. Left to right: epon cured for 48 h, sectioned at 24°C; LX112 cured for 48 h, sectioned at 24°C; LX112 cured for 10 h, sectioned at 24°C; LX112 cured for 48 h, sectioned at 40°C. Scale bar, 5 μm. Arrowheads indicate major (filled) and minor (hollow) tissue cracks.
(B) Photographs of infrared lights installed at the sides of the microtome sample arm to warm the sample during semithick sectioning (left). The heated diamond knife is depicted with a lower heater and the upper sensor sticks and the temperature control (right). he, heating element; ir, infrared light; sa, sample arm; ta, tape; tac, tape collector; tec, temperature control; ts, temperature sensor.
See Figure S2 for details on the heated diamond knife construction.
In order to facilitate semithick sectioning, we established a heated diamond knife (Figure 3B) approach. We drilled holes into the 35° and 45° knife boats to insert a temperature sensor (3 mm diameter) and a heater (6 mm diameter). For further temperature stability of the sample itself, we installed infrared lights at both sides and adjusted the distance to the ultramicrotome sample arm to yield a sample temperature of 40°C (Figure 3B). At temperatures above 40°C, water evaporated onto the sample block, and consequently, the resin partially melted (data not shown). We compared sectioning of rOTO processed mouse cortex precured for 10 h in LX112 at 0.5- to 10-μm thickness at room temperature, 30°C, and 40°C. Higher temperatures increased the smoothness of the section surface (Figure 3A). Surfaces in cortical areas with cell bodies and non-myelinated neurites were smooth, whereas myelinated axons and other heavily stained structures, e.g., the corpus callosum, showed some remaining cracks (Figures 2C and 3A). At higher temperatures, the water pumping system supplying the knife boat had to be adapted to increased flow rates to compensate increased evaporation. Therefore, a good compromise for serial sectioning were temperatures in the range of 35°C–40°C. Comparison of 35° ultra-with 45° ultra-knives did not show any major differences in semithick sectioning (data not shown), but higher long-term robustness is expected for the 45° knife (Matzelle et al., 2003). Consequently, a combination of two-step curing of LX112 and heated ultramicrotomy allows for semithick sectioning at 2–10 μm.
Light and Electron Microscopy of Serial Semithick Sections Reveals Ultrastructural Details for Selection for FIB-SEM Imaging
For volume analysis of cortical mouse tissue, we collected serial semithick sections on CNT tape using a tape collector (ATUM system mounted on a PowerTome, RMC, Figure 4A). Sections were cut at a slow speed (0.2–0.3 mm/s), both to limit compression of the sample and in order to keep the section on the heated water bath longer to smoothen. The tape speed was set to values above 0.3 mm/s within the cutting window to ensure efficient uptake of the section onto the tape. In order to maximize the number of sections per tape length and reduce inter-section gaps, the tape speed was reduced below 0.2 mm/s outside the cutting window. For test purposes, we cut 50 sections of mouse cortex tissue at 5 μm thickness. After sectioning and collection, the CNT tape was cut into 5-cm strips and reversibly adhered onto glass slides by application of a drop of water and incubation at 60° on a heating plate (Figure S3A). For 5-μm-thick sections, we acquired section overview images by transmitted light microscopy (Figure 4A). Samples were mounted onto carbon double adhesive tape attached to a silicon wafer as previously described (Kasthuri et al., 2015; Djannatian et al., 2019) and postcured (Figure S3B). BSD images were captured at 4 kV (Figure 4B). Serial section images were taken at 0.2 × 0.2 × 5 μm and regions of interest at 0.01 × 0.01 μm. The beam dwell time was reduced to values below 2 μs/pixel in order to avoid charging artifacts originating from increased sample thickness. Still, some charging at resin-rich regions like blood vessels or nuclei could not be prevented. Larger morphological features (such as cell bodies) were accessible by light microscopy, whereas SEM was needed to resolve objects in the range of large non-myelinated axons. Maximum projections revealed the tissue environment, such as vasculature patterns (Figure 4B).
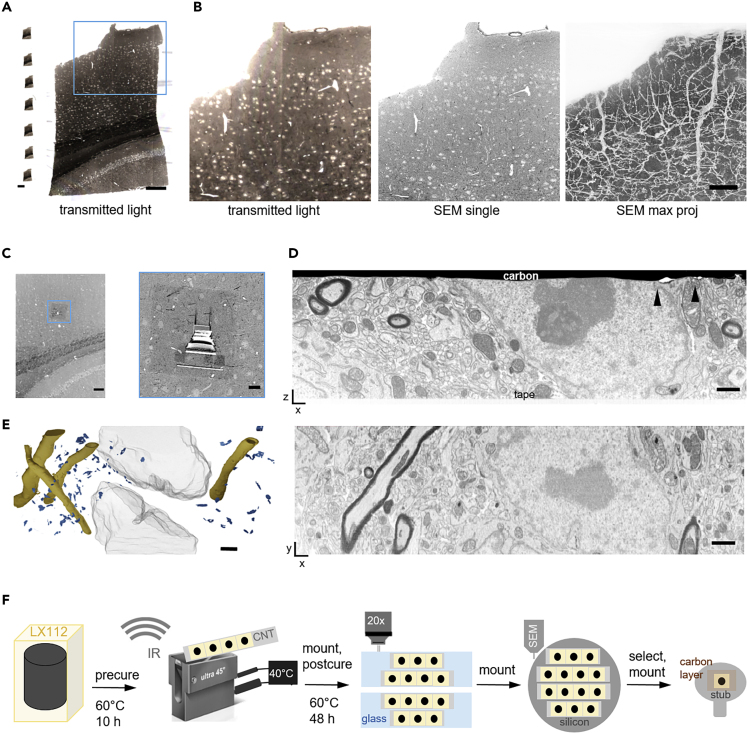
Figure 4.
Serial Semithick Sectioning for Targeted FIB-SEM.
Serial 5-μm-thick sections of mouse cortex were collected on CNT tape.
(A) Overview of several consecutive sections on tape (left) and a single transmitted light micrograph (20x objective, right). Scale bars, 2 mm and 100 μm, respectively.
(B) Transmitted light (left) and BSD images (10 × 10 × 5,000 nm resolution) of a selected region on a single section shown in (A) and maximum projection of all 93 sections revealing the vasculature morphology (right). Scale bar, 20 μm.
(C) A random cortical FIB target site was chosen (blue box) (left). The selected region was prepared for FIB-SEM by carbon deposition and trench milling (right). Scale bars, 100 and 10 μm, respectively.
(D) Cross section after FIB-SEM preparation showing the section—CNT tape adhesion site and the surface covered by a carbon layer (top). Defects in the surface layer topology are highlighted (black arrowheads). After the FIB-SEM run an xy section was reconstructed (bottom) from 2000 SE images (resolution 5 × 5 × 5 nm3, bottom). Scale bar, 1 μm.
(E) 3D model of the FIB-SEM stack revealing myelinated axons (yellow), post synaptic densities (blue), and neuronal nuclei (gray). Scale bar, 2 μm.
(F) Schematic of the ATUM-FIB strategy: sequential resin (LX112) curing and heated ultramicrotomy are required for semithick sectioning. Serial sections are attached onto glass slides for light microscopy and remounted onto silicon wafers for serial SEM imaging and target selection. A section of interest is remounted, adhered onto a standard sample holder, and carbon-coated for FIB-SEM examination.
See Figure S2 for photographs of the different processing steps.
Although FIB-SEM would be possible directly on the wafer, remounting onto FIB stubs is safer and gives much more freedom for the milling direction. It is possible to detach either the CNT tape directly by force or, preferably, to cut out the carbon adhesive carrying tape and 5-μm-thick section (Figure S3C). The latter option is preferred as it limits the risk of distorting the sections. The selected piece of section on tape(s) is then adhered onto a FIB-SEM stub by conducting carbon cement. A fine carbon layer was sputtered onto the sample providing surface accessibility for the electron beam while increasing conductivity. FIB-SEM was performed by milling an 8-μm-deep trench (Figure 4C). We acquired a FIB-SEM stack of a 30 × 30 × 5 μm cortical region data at 5 × 5 × 5 nm (Figure 4D). Ultrastructural features, such as post-synaptic densities and myelinated axons, were segmented and their three-dimensional orientation was depicted in a model (Figure 4E). Resin postcuring resulted in good milling characteristics without curtaining effects or other problems arising from samples embedded in soft epon. In summary, semithick sections can be imaged by light or electron microscopy to reveal (ultrastructural) details required for target site selection (Figure 4E). Selected regions on a particular section can then be subjected to targeted isotropic voxel acquisition by FIB-SEM.
Targeted FIB-SEM Enables Isotropic Ultrastructural Analysis of Amyloid Plaques in FAD
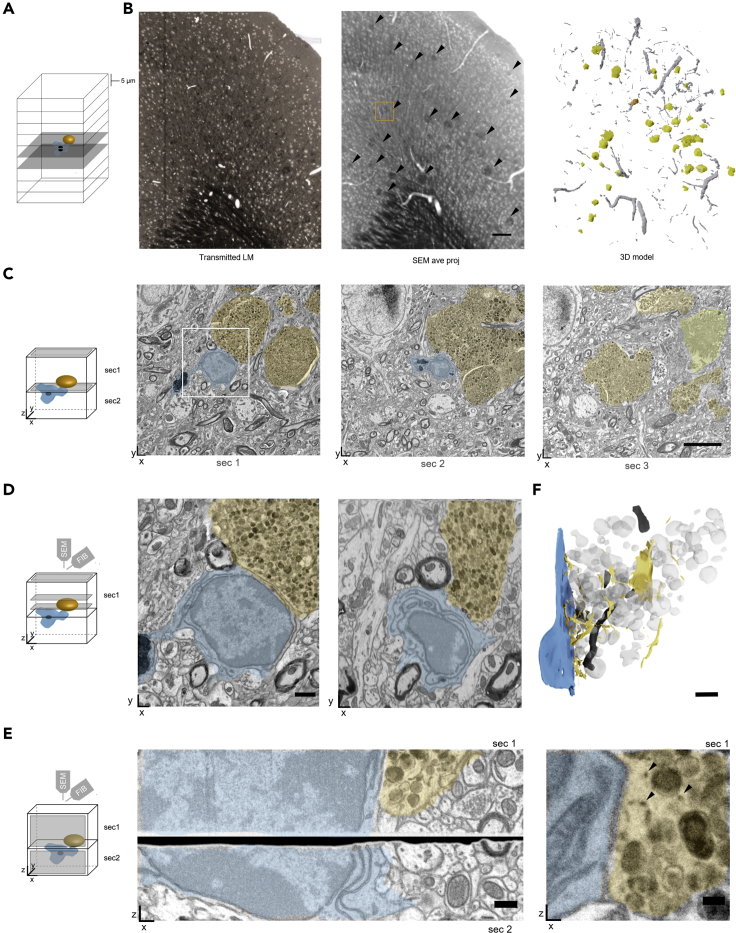
As a proof of concept, we combined serial semithick sectioning with targeted FIB-SEM to visualize microglia contacting dystrophic neurites in the cortex of a (familial) Alzheimer's disease (AD) mouse model. AD, the most common form of dementia in the elderly, causes gradual loss of memory, judgment, and the ability to function socially. It is characterized by the presence of extracellular plaques composed of amyloid-β (Aβ) peptides and intracellular tau aggregates (Kwak et al., 2020; Giacobini and Gold, 2013). The plaques are surrounded by microglia, phagocytic immune cells, which participate in the clearance of Aβ (Mattiace et al., 1990; Hemonnot et al., 2019). The ultrastructure of amyloid plaques has, so far, been studied by TEM (Gowrishankar et al., 2015; Terry et al., 1964) and also by correlated light microscopy and FIB-SEM (Blazquez-Llorca et al., 2017; El Hajj et al., 2019). Although the latter approach provides exact targeting of single events, ATUM-FIB allows unbiased sampling of cellular interactions around plaques. For ATUM-FIB, we collected 36 consecutive sections at 5 μm thickness onto CNT tape. Three-dimensional reconstruction of the light and electron micrographs revealed the amyloid plaque distribution in the resulting 180-μm-thick volume, as well as the vasculature pattern (Figures 5A and 5B).
Figure 5.
Targeted FIB-SEM on Selected Consecutive Sites of Semithick Sections in Familial Alzhiemer's Disease (FAD) Cortex Samples
(A) Schematic of ATUM-FIB on consecutive 5-μm-thick sections of mouse FAD cortex samples. Microglia (blue) contacts to plaques (orange) are targeted.
(B) Transmitted light image of one section (20x objective, left) and average projection SEM of 18 sections (middle) revealing plaques (arrowheads). The 3D model (right) was segmented from a set of 36 serial transmitted light images. Blood vessels (gray), plaques (yellow), and the target plaque (orange) are highlighted. Scale bar 100 μm.
(C–E) Schematics showing the imaging planes (gray) in the respective subfigures. (C) Three consecutive 5-μm-semithick sections of the FAD cortex sample on CNT tape. The microglial cell (blue), dystrophic neurites (orange), and amyloid core (yellow) are highlighted. The white box indicates the magnified image in (D). Scale bar, 5 μm. (D) Magnified surface image of the microglia to plaque contact site of section 1 (left). This region was targeted for FIB-SEM at 5 × 5 × 5 nm resolution. Reconstructions of two deeper levels within the semithick sections are shown (middle, right). Scale bar, 1 μm. (E) Stitched cross sections of corresponding areas in consecutive semithick sections 1 and 2 after FIB-SEM. Scale bar, 500 nm. (F) Region of interest of the selected section 1 of a FIB-SEM run. The 3D model shows the ER network (yellow) opposing the microglial plasma membrane (blue). Lysosomal structures are shown in light gray and mitochondria in dark gray (top). Scale bar, 500 nm. Representative SEM image revealing ER structures (black arrowheads) at sites of contact between microglia and dystrophic neurites (bottom). Scale bar, 200 nm.
We selected two consecutive sections containing a part of a plaque surrounded by a microglial cell for FIB-SEM examination (Figures 5A–5C). The ROI for the deposition of a protective carbon layer was relocated by overlaying the SEM image from the section series with a BSD image acquired at 8 kV. We imaged a 20 × 20 × 5 μm volume at 5 × 5 × 5 nm resolution by FIB-SEM. In order to reveal the strength of isotropic imaging we aligned the FIB-SEM volume and reconstructed another xz plane image of the microglial cell. The original SEM surface image was comparable with this virtual section (Figure 5D). Notably, we were able to stitch cross-sectional images of the two consecutive sections investigated (Figure 5E) (Xu et al., 2017), enabling extended z-volume analysis beyond 5 μm. Dystrophic neurites displayed ER structures at the contact sites opposing the microglial plasma membrane (Figure 5F). Although the target volume is resolved at high resolution and with isotropic voxels, light and electron microscopic images of the surrounding tissue provide a morphological context. Moreover, the non-destructive nature of serial sectioning on tape allows for reinspection of other ROIs at high resolution.
Discussion
Here, we introduce ATUM-FIB as a straightforward approach to combine overview imaging of tissue sections with targeted high-resolution three-dimensional reconstruction of subvolumes. In summary, ATUM-FIB has the following advantages over previously developed 3D EM volume workflows (Figure 6): (1) The ATUM-based generation of semithick sections allows screening of larger tissue volumes for rare events or specific sites of interest based on either light microscopic or low-resolution SEM exploration of the semithin sections. Such sites can then be targeted by FIB-SEM with ∼5 nm isotropic xyz-resolution. Thereby, the high-resolution volumes are put into a larger morphological context, similar to microCT investigations (Handschuh et al., 2013; Starborg et al., 2019), but with the advantage of providing direct access to target structures instead of coordinates. Moreover, this tissue context provides rich fiducial landmarks for correlated light/electron microscopy (Luckner et al., 2018). (2) In contrast to standard ultrathin ssTEM/SEM or ATUM techniques, our approach results in 100-fold fewer sections that need to be archived. This saves time, simplifies handling, and reduces cost. (3) Just as ATUM, ATUM-SEM is a non-destructive technique (except for the FIB-targeted areas) and thus preserves a library of semithick sections. These can be revisited to increase the number of detailed observations or explore new questions that emerge over time. The limitation in volume depth of ATUM-FIB due to the maximum thickness of semithick sections (≤5 μm) compared with investigations of whole blocks can be overcome be analyzing the same region in consecutive semithick sections. Stitching of regions across serial sections has been successfully applied before for ultrathick partitioning (Hayworth et al., 2015) as well as for (S)TEM tomography (Aoyama et al., 2008; He and He, 2014; Baumeister et al., 1999) and has been shown to result in minimal loss of information (Hayworth et al., 2015) (Figure S1). (4) The information in surface scans that are 2–5 μm apart are similar enough to be matched and aligned without the need for FIB-SEM information of the surrounding tissue section. Moreover, there is no need to section into oil instead of standard water baths compared with the ultrathick sectioning approach (Hayworth et al., 2015), which circumvents the need for re-embedding. (5) With the adaptation of standard diamond knives, our method can be applied in any EM laboratory with established FIB-SEM and ATUM workflows without the need of further equipment. This includes the standard rOTO staining method employed, which circumvents the need to establish novel en bloc contrasting protocols.
Figure 6.
Comparison of Volume SEM Techniques for Targeting Rare Events
Structures of interest like cell-cell interactions (microglia, magenta, dystrophic neurite, ocher) can be visualized by volume EM techniques. Current approaches for targeted imaging involve ATUM and FIB-SEM. ATUM-FIB combines the advantages of both approaches. High-precision targeting based on ultrastructural features is enabled, whereas increased section thickness reduces screening time and complexity compared with ATUM. Targeted FIB-SEM targeting is usually based on microCT data with limited xy resolution combined with trimming (arrows). A 500 μm z depth would be covered by 100 semithick sections (at 5 μm thickness, ATUM-FIB) instead of 10,000 ultrathin sections (at 50 nm thickness, ATUM). Comparable with FIB-SEM, ATUM-FIB allows for acquisition of the region of interest (black box) at isotropic voxels.
Plaques in AD and their complex cellular environment provide a good proof of concept of these advantages of ATUM-FIB: As Alzheimer plaques are sized roughly 10–100 μm diameter, these structures can be identified by light microscopy. The microglia-to-plaque contact sites can only be identified by serial SEM with a resolution of less than 10–100 nm. This would not be possible using microCT with a resolution limited to 500 nm at best. On top, with ATUM-FIB the identified regions of interest can be directly subjected to high-resolution FIB-SEM imaging, whereas microCT investigation requires targeted trimming. FIB-SEM on two consecutive semithick sections furthermore shows that not only several regions of interested can be analyzed but also information on semithick section images can be stitched.
As the implementation of ATUM-FIB is comparably simple and imaging modalities and section thickness can be flexibly adapted to scientific questions and biological tissues of interest, we envisage a broad range of applications. The information content of subsequent isotropic FIB-SEM investigations is especially suited for the analysis of organellar ultrastructure and cell-cell contacts in defined physiological or pathological circumstances, which are currently commonly screened on serial ultrathin sections. The fact that ATUM-FIB preserves the tissue library and provides histology-like context makes the method especially attractive in settings, where rare and precious samples are being archived for long-term reinvestigations. This includes material from complex treatment studies or correlated in vivo imaging/EM investigations (Karreman et al., 2014) in animals. Moreover, multi-scale investigations of human samples, e.g., brain biopsies, require cross-referencing with standard histopathology and will be increasingly needed to validate ultrastructural findings, e.g., from animal models of disease (Lewis et al., 2019; Jonkman et al., 2019; Shahmoradian et al., 2019).
Limitations of the Study
Similar to other serial sectioning techniques, ATUM-FIB has to be fine-tuned for the specific sample of interest, e.g., regarding block geometries (Baena et al., 2019). As standard volume EM embedding protocols differ, we recommend optimizing different contrasting and embedding conditions for each sample type. Adaptation of heavy metal load, resin composition, and curing time might be needed to achieve sections in the “semithickness” range. Transition regions from high to low contrast within a section like gray and white matter areas are especially prone to cause fractures. The approach presented here has been developed to target concrete structures but, in contrast to ultrathick partitioning (Xu et al., 2017; Hayworth et al., 2015), has not been optimized for a minimal loss of material between sections required for dense reconstruction and large-scale (connectomics) projects.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Martina Schifferer (martina.schifferer@dzne.de).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets supporting the current study have not been deposited in a public repository but are available from the corresponding author on request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy—ID 390857198) and the TRR 274/1 2020 (project Z01; ID 408885537). T.M.’s lab was also supported by DFG FOR2879, A03, and TRR274/1 2020, project B03, and the European Research Council (FP/2007-2013; ERC Grant Agreement n. 616791). M.K. and T.M. are jointly supported by the DFG through a common grant (Ke 774/5-1/Mi 694/7-1) and TRR274/1 2020, project C02. We thank Katalin Völgyi, Martin Dichgans, and Ozgun Gokce for providing fixed brain samples; Richard Schalek, Mark Terasaki, and Gerhard Wanner for valuable scientific and technical advice; Felix Beyer and Kerstin Karg for technical assistance; and Edgar Dawkins for reading the manuscript.
Author Contributions
Conceptualization, M. Schifferer and T.M.; Methodology, M. Schifferer, H.G., and G.K.; Investigation, G.K. and M. Schifferer; Resources H.G.; Visualization, G.K. and M. Schifferer; Supervision, Funding Acquisition, and Writing – Review & Editing, M.K., T.M., and M. Simons; Writing – Original Draft and Project Administration, M. Schifferer.
Declaration of Interests
The authors declare no competing interest.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101290.
Supplemental Information
References
- Aoyama K., Takagi T., Hirase A., Miyazawa A. STEM tomography for thick biological specimens. Ultramicroscopy. 2008;109:70–80. doi: 10.1016/j.ultramic.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Baena V., Schalek R.L., Lichtman J.W., Terasaki M. Serial-section electron microscopy using automated tape-collecting ultramicrotome (ATUM) Methods Cell Biol. 2019;152:41–67. doi: 10.1016/bs.mcb.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Grimm R., Walz J. Electron tomography of molecules and cells. Trends Cell Biol. 1999;9:81–85. doi: 10.1016/s0962-8924(98)01423-8. [DOI] [PubMed] [Google Scholar]
- Bishop D., Nikic I., Brinkoetter M., Knecht S., Potz S., Kerschensteiner M., Misgeld T. Near-infrared branding efficiently correlates light and electron microscopy. Nat. Methods. 2011;8:568–570. doi: 10.1038/nmeth.1622. [DOI] [PubMed] [Google Scholar]
- Blazquez-Llorca L., Valero-Freitag S., Rodrigues E.F., Merchán-Pérez Á., Rodríguez J.R., Dorostkar M.M., DeFelipe J., Herms J. High plasticity of axonal pathology in Alzheimer’s disease mouse models. Acta Neuropathologica Commun. 2017;5:14. doi: 10.1186/s40478-017-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock D.D., Lee W.C., Kerlin A.M., Andermann M.L., Hood G., Wetzel A.W., Yurgenson S., Soucy E.R., Kim H.S., Reid R.C. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman K.L., Helmstaedter M., Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- Bushong E.A., Johnson D.D., Kim K.-Y., Terada M., Hatori M., Peltier S.T., Panda S., Merkle A., Ellisman M.H. X-ray microscopy as an approach to increasing accuracy and efficiency of serial block-face imaging for correlated light and electron microscopy of biological specimens. Microsc. Microanal. 2015;21:231–238. doi: 10.1017/S1431927614013579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W., Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djannatian M., Timmler S., Arends M., Luckner M., Weil M.-T., Alexopoulos I., Snaidero N., Schmid B., Misgeld T., Möbius W. Two adhesive systems cooperatively regulate axon ensheathment and myelin growth in the CNS. Nat. Commun. 2019;10:4794. doi: 10.1038/s41467-019-12789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droz B., Rambourg A., Koenig H.L. The smooth endoplasmic reticulum: structure and role in the renewal of axonal membrane and synaptic vesicles by fast axonal transport. Brain Res. 1975;93:1–13. doi: 10.1016/0006-8993(75)90282-6. [DOI] [PubMed] [Google Scholar]
- El Hajj H., Savage J.C., Bisht K., Parent M., Vallières L., Rivest S., Tremblay M.-È. Ultrastructural evidence of microglial heterogeneity in Alzheimer’s disease amyloid pathology. J. Neuroinflammation. 2019;16:87. doi: 10.1186/s12974-019-1473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis E.A. No more epon 812: this product does not exist today. Microsc. Today. 2014;22:50–53. [Google Scholar]
- Giacobini E., Gold G. Alzheimer disease therapy—moving from amyloid-β to tau. Nat. Rev. Neurol. 2013;9:677–686. doi: 10.1038/nrneurol.2013.223. [DOI] [PubMed] [Google Scholar]
- Gowrishankar S., Yuan P., Wu Y., Schrag M., Paradise S., Grutzendler J., De Camilli P., Ferguson S.M. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl. Acad. Sci. U S A. 2015;112:E3699–E3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschuh S., Baeumler N., Schwaha T., Ruthensteiner B. A correlative approach for combining microCT, light and transmission electron microscopy in a single 3D scenario. Front. Zool. 2013;10:44. doi: 10.1186/1742-9994-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayworth K.J., Morgan J.L., Schalek R., Berger D.R., Hildebrand D.G., Lichtman J.W. Imaging ATUM ultrathin section libraries with WaferMapper: a multi-scale approach to EM reconstruction of neural circuits. Front. Neural Circuits. 2014;8:68. doi: 10.3389/fncir.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayworth K.J., Xu C.S., Lu Z., Knott G.W., Fetter R.D., Tapia J.C., Lichtman J.W., Hess H.F. Ultrastructurally smooth thick partitioning and volume stitching for large-scale connectomics. Nat. Methods. 2015;12:319. doi: 10.1038/nmeth.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., He Y. Electron tomography for organelles, cells, and tissues. In: Kuo J., editor. Electron Microscopy: Methods and Protocols. Humana Press; 2014. pp. 445–483. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M., Briggman K.L., Turaga S.C., Jain V., Seung H.S., Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- Hemonnot A.-L., Hua J., Ulmann L., Hirbec H. Microglia in Alzheimer disease: well-known targets and new opportunities. Front. Aging Neurosci. 2019;11:233. doi: 10.3389/fnagi.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann J.A., Hayles M., Gestmann I., Giannuzzi L.A., Lich B., Subramaniam S. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J. Struct. Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand D.G.C., Cicconet M., Torres R.M., Choi W., Quan T.M., Moon J., Wetzel A.W., Scott Champion A., Graham B.J., Randlett O. Whole-brain serial-section electron microscopy in larval zebrafish. Nature. 2017;545:345–349. doi: 10.1038/nature22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Laserstein P., Helmstaedter M. Large-volume en-bloc staining for electron microscopy-based connectomics. Nat. Commun. 2015;6:7923. doi: 10.1038/ncomms8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman L.E., Graaf Y.G.-d., Bulk M., Kaaij E., Pouwels P.J.W., Barkhof F., Rozemuller A.J.M., van der Weerd L., Geurts J.J.G., van de Berg W.D.J. Normal Aging Brain Collection Amsterdam (NABCA): a comprehensive collection of postmortem high-field imaging, neuropathological and morphometric datasets of non-neurological controls. Neuroimage Clin. 2019;22:101698. doi: 10.1016/j.nicl.2019.101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman M.A., Hyenne V., Schwab Y., Goetz J.G. Intravital correlative microscopy: imaging life at the nanoscale. Trends Cell Biol. 2016;26:848–863. doi: 10.1016/j.tcb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Karreman M.A., Mercier L., Schieber N.L., Shibue T., Schwab Y., Goetz J.G. Correlating intravital multi-photon microscopy to 3D electron microscopy of invading tumor cells using anatomical reference points. PLoS One. 2014;9:e114448. doi: 10.1371/journal.pone.0114448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N., Hayworth K.J., Berger D.R., Schalek R.L., Conchello J.A., Knowles-Barley S., Lee D., Vazquez-Reina A., Kaynig V., Jones T.R. Saturated reconstruction of a volume of neocortex. Cell. 2015;162:648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- Kizilyaprak C., Longo G., Daraspe J., Humbel B.M. Investigation of resins suitable for the preparation of biological sample for 3-D electron microscopy. J. Struct. Biol. 2015;189:135–146. doi: 10.1016/j.jsb.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Knott G., Marchman H., Wall D., Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld J., Denk W. Progress and remaining challenges in high-throughput volume electron microscopy. Curr. Opin. Neurobiol. 2018;50:261–267. doi: 10.1016/j.conb.2018.04.030. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Sohn J., Hatada S., Schurr M., Straehle J., Gour A., Neujahr R., Miki T., Mikula S., Kawaguchi Y. A carbon nanotube tape for serial-section electron microscopy of brain ultrastructure. Nat. Commun. 2018;9:437. doi: 10.1038/s41467-017-02768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S.S., Washicosky K.J., Brand E., von Maydell D., Aronson J., Kim S., Capen D.E., Cetinbas M., Sadreyev R., Ning S. Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer’s disease. Nat. Commun. 2020;11:1377. doi: 10.1038/s41467-020-15120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.C., Bonin V., Reed M., Graham B.J., Hood G., Glattfelder K., Reid R.C. Anatomy and function of an excitatory network in the visual cortex. Nature. 2016;532:370–374. doi: 10.1038/nature17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A.J., Genoud C., Pont M., van de Berg W.D., Frank S., Stahlberg H., Shahmoradian S.H., Al-Amoudi A. Imaging of post-mortem human brain tissue using electron and X-ray microscopy. Curr. Opin. Struct. Biol. 2019;58:138–148. doi: 10.1016/j.sbi.2019.06.003. [DOI] [PubMed] [Google Scholar]
- Locke M., Krishnan N. Hot alcoholic phosphotungstic acid and uranyl acetate as routine stains for thick and thin sections. J. Cell Biol. 1971;50:550–557. doi: 10.1083/jcb.50.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckner M., Burgold S., Filser S., Scheungrab M., Niyaz Y., Hummel E., Wanner G., Herms J. Label-free 3D-CLEM using endogenous tissue landmarks. iScience. 2018;6:92–101. doi: 10.1016/j.isci.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckner M., Wanner G. From light microscopy to analytical scanning electron microscopy (sem) and focused ion beam (fib)/sem in biology: fixed coordinates, flat embedding, absolute references. Microsc. Microanal. 2018;24:526–544. doi: 10.1017/S1431927618015015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire E., Withers P.J. Quantitative X-ray tomography. Int. Mater. Rev. 2014;59:1–43. [Google Scholar]
- Mattiace L.A., Davies P., Yen S.H., Dickson D.W. Microglia in cerebellar plaques in Alzheimer’s disease. Acta Neuropatholog. 1990;80:493–498. doi: 10.1007/BF00294609. [DOI] [PubMed] [Google Scholar]
- Matzelle T.R., Gnaegi H., Ricker A., Reichelt R. Characterization of the cutting edge of glass and diamond knives for ultramicrotomy by scanning force microscopy using cantilevers with a defined tip geometry. Part II. J. Microsc. 2003;209:113–117. doi: 10.1046/j.1365-2818.2003.01119.x. [DOI] [PubMed] [Google Scholar]
- Mikula S., Denk W. High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nat. Methods. 2015;12:541–546. doi: 10.1038/nmeth.3361. [DOI] [PubMed] [Google Scholar]
- Morgan J.L., Berger D.R., Wetzel A.W., Lichtman J.W. The fuzzy logic of network connectivity in mouse visual Thalamus. Cell. 2016;165:192–206. doi: 10.1016/j.cell.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris R.P., Baena V., Terasaki M. Localization of phosphorylated connexin 43 using serial section immunogold electron microscopy. J. Cell Sci. 2017;130:1333. doi: 10.1242/jcs.198408. [DOI] [PubMed] [Google Scholar]
- Schalek R., Kasthuri N., Hayworth K., Berger D., Tapia J., Morgan J., Turaga S., Fagerholm E., Seung H., Lichtman J. Development of high-throughput, high-resolution 3D reconstruction of large-volume biological tissue using automated tape collection ultramicrotomy and scanning electron microscopy. Microsc. Microanal. 2011;17:966–967. [Google Scholar]
- Sengle G., Tufa S.F., Sakai L.Y., Zulliger M.A., Keene D.R. A correlative method for imaging identical regions of samples by micro-CT, light microscopy, and electron microscopy: imaging adipose tissue in a model system. J. Histochem. Cytochem. 2012;61:263–271. doi: 10.1369/0022155412473757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmoradian S.H., Lewis A.J., Genoud C., Hench J., Moors T.E., Navarro P.P., Castaño-Díez D., Schweighauser G., Graff-Meyer A., Goldie K.N. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 2019;22:1099–1109. doi: 10.1038/s41593-019-0423-2. [DOI] [PubMed] [Google Scholar]
- Sonomura T., Furuta T., Nakatani I., Yamamoto Y., Unzai T., Matsuda W., Iwai H., Yamanaka A., Uemura M., Kaneko T. Correlative analysis of immunoreactivity in confocal laser-scanning microscopy and scanning electron microscopy with focused ion beam milling. Front. Neural Circuits. 2013;7:26. doi: 10.3389/fncir.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starborg T., OSullivan J.D.B., Carneiro C.M., Behnsen J., Else K.J., Grencis R.K., Withers P.J. Experimental steering of electron microscopy studies using prior X-ray computed tomography. Ultramicroscopy. 2019;201:58–67. doi: 10.1016/j.ultramic.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Shemesh T., Kasthuri N., Klemm R.W., Schalek R., Hayworth K.J., Hand A.R., Yankova M., Huber G., Lichtman J.W. Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell. 2013;154:285–296. doi: 10.1016/j.cell.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry R.D., Gonatas N.K., Weiss M. Ultrastructural studies in Alzheimer’s presenile dementia. Am. J. Pathol. 1964;44:269–297. [PMC free article] [PubMed] [Google Scholar]
- Titze B., Genoud C. Volume scanning electron microscopy for imaging biological ultrastructure. Biol. Cell. 2016;108:307–323. doi: 10.1111/boc.201600024. [DOI] [PubMed] [Google Scholar]
- Tomassy G.S., Berger D.R., Chen H.H., Kasthuri N., Hayworth K.J., Vercelli A., Seung H.S., Lichtman J.W., Arlotta P. Distinct profiles of myelin distribution along single axons of pyramidal neurons in the neocortex. Science. 2014;344:319–324. doi: 10.1126/science.1249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A., Benjaminsen J., Moritz C., Henke K., Hartmann J., Norlin N., Richter K., Schieber N.L., Franke T., Schwab Y., Peri F. Clearance by microglia depends on packaging of phagosomes into a unique cellular compartment. Dev. Cell. 2019;49:77–88.e7. doi: 10.1016/j.devcel.2019.02.014. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Xu C.S., Hayworth K.J., Lu Z., Grob P., Hassan A.M., Garcia-Cerdan J.G., Niyogi K.K., Nogales E., Weinberg R.J., Hess H.F. Enhanced FIB-SEM systems for large-volume 3D imaging. Elife. 2017;6:e25916. doi: 10.7554/eLife.25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Lauritzen J.S., Perlman E., Robinson C.G., Nichols M., Milkie D., Torrens O., Price J., Fisher C.B., Sharifi N. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. 2018;174:730–743.e22. doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository but are available from the corresponding author on request.