Abstract
In the current investigation, bacterial strain Bacillus thuringiensis MB497 was examined for production of intracellular and extracellular organophosphorus phosphatase (OPP) enzymes. This strain produced significant amount of extracellular acidic and alkaline phosphatases. Production of neutral phosphatase was negligible. Production of OPP was generally highest at pH 11 and at 45–50 °C. However, activity and stability of OPP was highest at 37 °C and reduced at higher temperatures. OPP production was decreased after 48 h of incubation. Largely, OPP activity was inhibited by SDS and EDTA and significantly enhanced by metals (Zn++, Cu++ and Cd++). Both acidic and alkaline OPPs were capable of bio-precipitation of selected metals (Ni, Mn, Cr and Cd) up to 86–100%. When used against 50 mg/l of three OP pesticides (Chlorpyrifos, Triazophos, and Dimethoate), 81–94.6% degradation of pesticides was observed by alkaline OPP, while acidic OPP showed less degradation (61–70.5%) within 30 min of incubation.
Keywords: Microbiology, Phosphatase, Degradation, Organophosphate, Pesticide
Microbiology; Phosphatase; Degradation; Organophosphate; Pesticide.
1. Introduction
Enzymatic detoxification of organophosphate pesticides and other pollutants is a modern, cost effective and environmental friendly technique for the removal of these chemical pollutants from contaminated sites [1]. Recently, research is focused on the isolation of new enzymes/biocatalysts that exhibit stability against a wide range of environmental conditions (temperature, pH, salts etc.) in order to determine the potential use of enzymes in future applications.
Phosphatases belong to hydrolase enzymes and have been frequently applied against various compounds like OP pesticides, proteins, alkaloids and nucleotides in order to eradicate phosphate groups [2]. Phosphotriesterase (PTE), also known as organophosphorus hydrolase (OPH) is the earliest isolated enzyme that has potential to hydrolyze a variety of OP compounds by breaking P–S and P–O linkages [3]. These enzymes like OPH, OP acid anhydrolases (OPAA) and methyl parathion hydrolase (MPH), have been mostly derived from microorganisms including bacteria and have been proved very effective for elimination of OP pesticides [4]. Phosphotriesterase isolated from Flavobacterium sp. was tested against a variety of OP pesticides and was found specific in its catalytic activity [5]. Similarly, another much effective OP degrading enzyme related to OP hydrolases is OpdA originally extracted from Agrobacterium radiobacter [6]. Though both OPDA and OPH have secondary structure resemblance, but they have different substrate specificities because of their difference in active site structure. OpdA usually selects substrates with smaller alkyl substituents. There is a theory that due to mutation in active site, OPH evolved to OpdA naturally [7]. It was revealed by Gao et al. [8] that immobilized OpdA was very efficient in removing methyl parathion from solution. The basic and most important step during breakdown of OP compounds is hydrolysis, which makes them more vulnerable for further microbial degradation [9]. OP hydrolase and OPAA have been focus of most studies of organophosphorus degrading enzymes and can be used for the remediation of OP-polluted sites [10]. In recent times, new much effective OP degrading enzymes known as Phosphotriesterase-Like-Lactonase (PLL) have been reported with extraordinary thermal stability. These enzymes were isolated from extremophilic/hyperthermophilic bacteria and archaea [1]. Both phosphate monoesterases and diesterases belong to superfamily of alkaline phosphatase [11]. Phosphatases are classified as acidic, neutral and alkaline, on the basis of optimum pH required for their catalytic action [12]. These enzymes are capable of transforming insoluble organic phosphorous into soluble and available phosphorous during their active cellular metabolism, while their production has been reported by prokaryotes as well as eukaryotes [13, 14].
Therefore, an enzyme assay was designed to explore the potential of Bacillus thuringiensis MB497 to produce acidic, neutral and alkaline organophosphorus phosphatases (OPPs). Crude extracts of OPP enzymes were then optimized against different factors (pH, temperature, substrate concentrations, incubation time and metals) for their production, activity and stability. Bio-precipitation potential of OPP was also studied for different metals. Acidic and alkaline OPP were cross examined for their substrate specificity against Chlorpyrifos [O, O-diethyl O-(3, 5, 6-trichloro-2-pyridinyl] (CPF), Triazophos [diethoxy-[(1-phenyl-1, 2, 4-triazol-3-yl) oxy]-sulfanylidene-λ5-phosphane] (TAP) and Dimethoate [2-dimethoxyphosphinothioylsulfanyl-N-methylacetamide] (DM).
2. Materials and methods
Three OP pesticides (CPF, TAP and DM) with 40% EC were supplied by “Four Brothers Agri services, Pakistan. Other chemicals used during this investigation were purchased from Sigma-Aldrich/Merck. National Botanical Research Institute's phosphate growth broth (NBRIP) was used for screening of bacteria for phosphate solubilization potential and adopted from Nautiyal [15]. All experiments were conducted in triplicate to verify results.
2.1. Selection of microorganism
Bacterial strain Bacillus thuringiensis MB497 (accession number KP886829) was selected on the basis of its previous tested capability to degrade OP pesticides (CPF, TAP and DM). Therefore, this strain was strong candidate for the presence of OP degrading phosphatases responsible for the cleavage of P–O–C bond of these pesticides.
2.1.1. Screening for phosphate solubilization potential of bacterial isolate
Bacterial strain MB497 was cultured in National Botanical Research Institute's phosphate (NBRIP) medium [15] supplemented with calcium phosphate (as a substrate) and Agar-agar (1.5%) for an agar assay of phosphate solubilizing soil bacteria. Sterile toothpicks were used to inoculate bacterial strain on petri plates in triplicate and incubated at 37 °C for 10 days. At the end of given incubation period, bacterial potential to solubilize insoluble phosphate was measured by the solubilization index as given below [16]:
| Phosphate solubilization index (PSI) = (Colony diameter + Halo zone) / colony diameter |
2.1.2. Screening of extracellular organophosphorus phosphatase (OPP) and enzyme assay
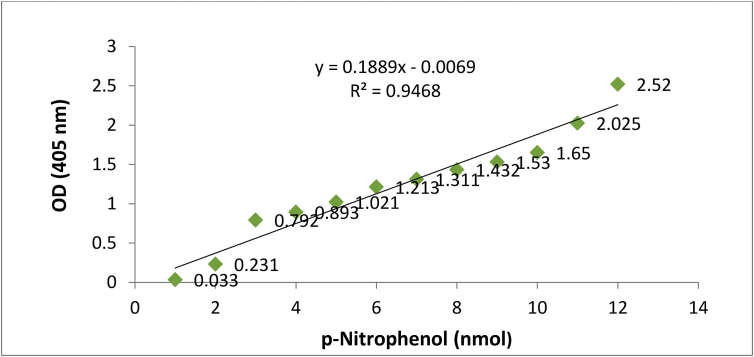
Fresh bacterial culture (500 μl) of MB497 was inoculated in 30 ml NBRIP broth and grown to the late logarithmic phase for three days incubation followed by centrifugation at 3,000 rpm for 10 min. Then, supernatant was used to check the production of extracellular Organophosphorus Phosphatase (OPP) (acidic, neutral and alkaline) with the help of p-nitrophenyl phosphate (p-NPP) as a colorless substrate, which is converted into yellow end product (p-nitrophenol) after removal of phosphate group due to hydrolysis by OPP [17]. Next, acidic, neutral and alkaline OPP activities were measured using buffer substrate mixtures which were prepared by dissolving 0.6 mg of p-nitrophenol phosphate/ml of respective buffer. For acidic, neutral and alkaline OPP, 0.1 M sodium acetate buffer (pH 5.2), 0.1 M sodium phosphate buffer (pH 7) and 0.1 M Tris HCl buffer (pH 9.5) were used respectively to dissolve substrate [18, 19]. Further, 3 ml of crude enzyme extract was mixed with 1 ml of respective buffer substrate (p-nitrophenyl phosphate) to prepare the reaction mixture followed by incubation at 37 °C for 20 min. The reaction was stopped by the addition of 1 ml of 1 N NaOH so as to increase pH of the reaction mixture. Both p-NPP and p-Np are colorless at acidic and neutral pH but at alkaline pH, p-nitrophenol so formed, becomes yellow in color. Enzyme activity was measured using a UV-VIS Spectrophotometer (BMS UV-160) at 405 nm [20]. Amount of enzyme that liberates 1 nmol of p-nitrophenol per minute at 37 °C is equivalent to one unit (U) of OPP activity [21]. The concentration of OPP produced was measured using standard curve of p-Nitrophenol (Figure 1) with the help of serial dilutions of p-NP (1–10 μg/l).
Figure 1.
Standard curve for p-Nitrophenol.
2.1.3. Screening of intracellular organophosphorus phosphatase (OPP)
Bacterial cell pellets were resuspended in 1 ml of phosphate buffer with pH 7 and cells were disrupted using silica/glass beads. The lysate was centrifuged at 30,000 g at 4 °C for 30 min to remove all cell debris and the supernatant was used as enzyme source for intracellular OPP activity [22]. The 3 ml of this supernatant enzyme extract was mixed with substrate buffer mixture (1 ml) after various time intervals and incubated for 30 min. The reaction was stopped by adding 1 ml of 1 N NaOH. All the experiments were performed in triplicates.
2.2. Factors effecting the production of organophosphorus phosphatase (OPP) enzyme
2.2.1. Effect of pH on OPP production
In order to study, the effect of pH on enzyme production, 24 h fresh bacterial culture (50 μl) was inoculated in 5 ml of NBRIP broth at different pH (6, 7, 8, 9, 10 and 11), supplemented with substrate p-NPP followed by incubation for 24 h at 37 °C. Earlier, before being autoclaved, pH of the medium was adjusted by 1 N HCl or 1 N NaOH. After incubation period, bacterial cultures were centrifuged at 3000 rpm for 10 min and supernatant was used for enzyme assay.
2.2.2. Effect of temperature on OPP production
Effect of temperature on OPP enzyme production was investigated by inoculating 50 μl of 24 h fresh bacterial cultures in 5 ml of NBRIP broth in test tubes followed by incubation at different temperatures (37, 45, 50 and 60 °C) at pH 7 in a rotary shaker for 24 h. Then, enzyme was extracted by centrifugation at 3000 rpm for 10 min. The supernatant was analyzed by using UV-VIS spectrophotometer at 405 nm for enzyme production.
2.2.3. Effect of incubation time on OPP enzyme production
Same procedure was repeated to study the effect of incubation time on the production of acidic, neutral and alkaline organophosphate phosphatase enzyme and reaction mixture was incubated at 37 °C in a rotary shaker at 120 rpm for different periods (24, 48 and 72 h) followed by steps as described above.
2.3. Factors affecting OPP activity
2.3.1. Effect of temperature on OPP activity
OPP activity was assayed at 37, 45, 50, 60 and 70 °C, while pH being maintained according to the acidic (pH 5.2), neutral (pH 7.0) or alkaline (pH 9.5) nature of enzyme. The enzyme activity was measured at 405 nm using UV-VIS Spectrophotometer as before immediately.
2.3.2. Effect of chemicals on OPP activity
In order to study the effect of different concentrations of SDS and EDTA on OPP activity, supernatant enzyme extract was mixed with substrate buffer and then augmented with SDS and EDTA (2.5, 5 and 7.5%) separately followed by incubation at 37 °C for 30 min. Enzyme activity was assayed using UV-VIS Spectrophotometer at 405 nm. Control was blank (without these chemicals).
2.3.3. Effect of metals on OPP activity
Similarly, the effect of different metals on the activity of acidic, neutral and alkaline OPP was checked by adding 2.5% of stock solutions of different metals (using their salts) like ZnSO4, CuSO4 and CdCl2 separately to enzyme substrate mixture followed by incubation at 37 °C for 30 min along with control (without metals). Enzyme activity was assessed using UV-VIS Spectrophotometer at 405 nm.
2.3.4. Effect of substrate concentration on OPP activity
The effect of substrate concentration on the activity of OPP was studied by using different substrate concentrations (0.06, 0.6, 0.8 and 1.1%) in the supernatant enzyme extract of phosphatases (acidic, neutral and alkaline) and then incubated for 30 min. Enzyme activity was measured with the help of UV-VIS Spectrophotometer at 405 nm.
2.3.5. Effect of incubation period on OPP activity
Likewise, enzyme activity at different incubation times of 30, 50, 70 and 90 min was investigated using p-NPP as substrate in the supernatant enzyme extract and then enzyme activity was monitored with the help of UV-VIS Spectrophotometer at 405 nm.
2.4. Metal bio-precipitation by OPP enzyme
Bio-precipitation capability of acidic, neutral and alkaline phosphatases was checked for different heavy metals i.e Ni+2, Cr+6, Mn+2 and Cd+2. For this purpose, reaction mixture was prepared by mixing buffer substrate mixture and extracellular phosphatase extract (supernatant) supplemented with respective metal ion stock solutions (1000 ppm) of Ni+2 (NiCl2), Cr+6 (K2Cr2O7), Mn+2 (MnCl2) and Cd+2 (CdCl2) at different incubation times (60, 120 and 180 min). Amount of bio-precipitated metal was measured by calculating the decline in the concentration of respective metal content in the supernatant before and after incubation using Atomic Absorption Spectrometer (AAS Shimadzu AA 7000), with the help of equation given by Chaudhury et al. [22]:
| X = (A-B)/A × 100 | (1) |
Where,
X = % of metal precipitated
A = Initial metal concentration in the aliquot
B = Final metal concentration in the aliquot
2.5. Substrate specificity of OPP enzyme against OP pesticides (CPF, TAP and DM)
The substrate specificity of OPP was cross checked against different OP insecticides (Chlorpyrifos, Dimethoate and Triazophos) by measuring the OPP activity against 50 mg/l of respective pesticides after 30 min of incubation at 37 °C followed by HPLC analysis [23, 24]. For this purpose, High-performance liquid chromatography (HPLC Shimadzu) LC-20AT equipped with a UV-Visible detector (SPD-20A) and a C18 column (0.46 × 15 cm) was used. Samples were filtered by syringe filters (0.45 μm) and directly analyzed by HPLC according to conditions given below in Table 1 for each pesticide respectively.
Table 1.
HPLC conditions used for the analyses of Chlorpyrifos, Triazophos and Dimethoate degradation by OPP.
| HPLC parameters | TAP | CPF | DM |
|---|---|---|---|
| Isocratic mobile phase | Acetonitrile and water (80:20) | Acetonitrile and water (70:30) | Acetonitrile and water (60:40) |
| Detection wavelength | 270 nm | 280 nm | 221 nm |
| Injection volume | 10 μl | 10 μl | 15 μl |
| Flow rate | 1.0 ml/min | 1.0 ml/min | 1.0 ml/min |
| Oven temperature | 40 °C | 40 °C | 30 °C |
| Protocol followed with modification | [25] | [26] | [27] |
As neutral OPP production was negligible by the isolates, only acidic and alkaline OPP were tested for substrate specificity. During HPLC investigation, the concentration of OP pesticide (mg/l) was measured by comparing peak areas in the chromatogram of enzyme reaction sample with that of the peak area of standard pesticide chromatogram as given below in Eq. (2) [28]:
| Concentration of OP in sample (mg/l) = Peak area of chromatogram of sample ÷ Peak area of chromatogram of standard OP compound × concentration of standard OP compound (mg/l) | (2) |
The OP pesticide degradation (%) was then calculated using the following equation [29]:
| B% = (Ca – Cb) ÷ Ca ×100 | (3) |
Where,
B = Pesticide degradation. Cb = the concentration of OP pesticide (mg/l) residues left in the medium containing OPP enzyme. Ca = the initial concentration of OP pesticide (mg/l) supplemented to the enzyme reaction.
3. Results
3.1. Screening for phosphate solubilization potential of Bacillus thuringiensis MB497
Isolate MB497 formed clear zones of phosphate solubilization and its phosphate solubilization potential was measured using phosphate solubilization index (Table 2).
Table 2.
Phosphate solubilization Index (PSI) of bacterial isolate MB497.
| Incubation period | PSI |
|---|---|
| Day 3 | 2.5 |
| Day 6 | 3.0 |
| Day 9 | 3.8 |
3.2. Screening for extracellular organophosphorus phosphatase (OPP) production by Bacillus thuringiensis MB497
There was less production (4.21 u/ml) of neutral phosphatase in contrast to acidic (18.68 u/ml) and alkaline phosphatases and maximum production (22.32 u/ml) of alkaline phosphatase (ALP).
3.3. Screening of intracellular organophosphorus phosphatase (OPP) production in Bacillus thuringiensis MB497
Among the intracellular phosphatases, more and equal production (3.44 u/ml) of alkaline and acidic OPP followed by neutral (3.16 u/ml) was observed. However, the intracellular OPP production and activity was much reduced as compared to extracellular phosphatases, therefore, enzyme assay was further carried out using only extracellular phosphatases.
3.4. Factors affecting the production of extracellular OPP by Bacillus thuringiensis MB497
3.4.1. Effect of pH on extracellular OPP production
PP production was highest at pH 11 by MB497. There was gradual increase in OPP enzyme production with increase in pH from 6 to 11 (Figure 2 a).
Figure 2.
Organophosphorus phosphatase production by bacterial strain MB497 at different pH (a) and at different temperatures (b). Error bars represent standard errors for values of three sample replicates.
3.4.2. Effect of temperature on extracellular OPP production
Strain MB497 exhibited maximum OPP enzyme production (65.16 u/ml) at 50 °C. Minimum enzyme production (3.10 u/ml) was observed at 37 °C (Figure 2b).
3.4.3. Effect of incubation time on OPP production
Alkaline and acidic OPP production by strain MB497 was maximum after 72 h, while neutral OPP was produced maximum after 24 h (Figure 3).
Figure 3.
Organophosphorus phosphatase production by bacterial strain MB497 at different incubation periods. Error bars represent standard errors for values of three sample replicates.
3.5. Factors affecting organophosphorus phosphatase (OPP) activity
3.5.1. Effect of temperature on OPP activity and stability
Significant enzyme activity and stability was noticed over a broad range of temperatures (37, 45, 50, 60 and 70 °C). There was less enzyme activity at higher temperatures but still active even at 70 °C. In strain MB497 maximum acidic and alkaline OPP activity was observed at 37 °C while neutral OPP was most active at 50 and 60 °C respectively.
3.5.2. Effect of substrate (p-NPP) concentration on OPP activity
In MB497, maximum acidic OPP activity was noticed at 0.6%, whereas neutral and alkaline OPP were most active at 0.8 and 0.06% of substrate (p-NPP) respectively.
3.5.3. Effect of incubation time on OPP activity
In strain MB497, maximum acidic, neutral and alkaline OPP activity was observed after 70 min of incubation time (Figure 4).
Figure 4.
Effect of incubation period on activity of Organophosphorus phosphatase produced by bacterial strain MB497. Error bars represent standard errors for values of three replicates.
3.6. Study of stimulatory or inhibitory effect of chemicals (SDS, EDTA and metals) on OPP activity
3.6.1. Effect of SDS on OPP activity
Acidic OPP activity increased at all given concentrations of SDS after 30 min of incubation period, whereas, neutral OPP activity was observed to enhance in SDS concentrations up to 2.5%, followed by inhibition at higher concentrations. On the other hand, alkaline OPP activity was not affected by SDS at all concentration.
3.6.2. Effect of EDTA on OPP activity
Acidic, neutral and alkaline OPP activities were inhibited at all given concentrations of EDTA (2.5, 5 and 7.5%) after 30 min of incubation period.
3.6.3. Effect of metals on OPP activity
Activities of acidic, neutral and alkaline OPP were greatly enhanced by all the metal ions used. In case of alkaline OPP activity, there was more stimulation by Cu++. Similarly, activity of neutral OPP was largely enhanced by Cd++, while there was only little promotion of acidic OPP activity by all the metals.
3.7. Metal bio-precipitation by OPP
In case of acidic, neutral and alkaline OPP extracted from MB497, significant bio-precipitation of different metals including Ni++ (94–98%), Mn++ (93–99%), Cr++ (86–90%) and Cd++ (99–100%) was observed within 180 min of incubation times.
3.8. Substrate specificity of OPP for CPF, TAP and DM
Acidic OPP was less active as there was less degradation of CPF, TAP and DM (70.5, 65.5 and 61% respectively) as compared to alkaline OPP which showed 94.6, 93 and 81% degradation of CPF, TAP and DM respectively (Figure 5).
Figure 5.
Degradation of OP pesticides by acidic and alkaline OPP extracted from strain MB497 as analyzed by HPLC after 30 min of incubation. Error bars represent standard errors for values of three replicates.
HPLC chromatograms for degradation of TAP (RT = 2.4 min), CPF (RT = 5.4 min) and DM (RT = 2.9 min) by alkaline OPP extracted from isolate MB497 have been demonstrated in Figures 6, 7, and 8.
Figure 6.
HPLC chromatograms showing degradation of 50 mg/l of TAP (RT = 2.4 min) by alkaline OPP after 30 min of incubation. a). Control, b). MB497.
Figure 7.
HPLC chromatograms showing degradation of 50 mg/l of CPF (RT = 5.4 min) by alkaline OPP after 30 min of incubation. a). Control, b). MB497.
Figure 8.
HPLC chromatograms showing degradation of 50 mg/l of DM (RT = 2.9 min) by alkaline OPP after 30 min of incubation. a). Control, b). MB497.
4. Discussion
Phosphate solubilizing activity by 6 bacterial isolates was reported by Islam et al. [30]. In general, if more is the intensity of dissolved phosphate, then, greater is the phosphatase production as there is positive correspondence between the two activities [31]. Bacterial requirements for inorganic phosphorus are fulfilled by the removal of phosphorus from organophosphate and phosphonate sources. Alkaline phosphatase has been considered as the most active dephosphorylating enzymes in the periplasmic space thus coping with the cell demand of inorganic phosphorus [32]. Alkaline phosphatases usually exhibit maximum catalytic activity at pH 8 or above [33]. Alkaline phosphatase (isolated from Rhizobium) was reported to have a broad range of pH optimum (6.8–11.8) with maximum activity at pH 9.8 [34]. Similarly, ALP isolated from Pyrococcus abyssi was reported to work best at pH 11 [35]. Earlier, optimum production of extracellular alkaline OPP (derived from Proteus mirabilis) within 28 h incubation time was reported [36].
Earlier, alkaline phosphatases from two bacterial strains (Escherichia coli and Alcaligenes faecalis) was reported to show activity at 80 and 45 °C respectively [37, 38]. Similarly, Shah et al. [39] reported maximum activity of alkaline phosphatase at 37 °C followed by decrease at higher temperatures. At higher temperatures, kinetic energy of the protein molecules in enzyme structure is amplified, thus breaking the bonds between the active amino acids, leading to loss of enzyme activity [40]. According to Danial and Alkhalf [41], the optimum temperature and pH for the acid phosphatase activity was 60 °C and 5.0 respectively. Moreover, the enzyme activity was enhanced by metals like Fe3+, Cu2+, Ca2+, Mg2+ and K+ but was strongly inhibited by Zn2+. It was revealed that alkaline phosphatase exhibited greater substrate specificity for p-nitro phenyl phosphate and showed more activity with the increased substrate concentration [37, 42]. According to Mori et al. [43], alkaline phosphatases are considered as metalo-dependent enzymes, especially associated with Zn and Mg for their improved activity and stability. It was revealed that Hg2+, Cu2+, and Cd2+ inhibited E. coli ALP, while Co2+ had little promoter effect, whereas divalent alkaline earth metals (like Ca2+ and Mg2+), stimulated the enzyme activity [44]. Current advanced industrialization has led to serious environmental metal pollution. Major challenge is to extract preferred/selected metals from such waste metal residues. Secondly, effective immobilization of metal pollutants is required to stop groundwater contamination [45]. At present, biotechnology has been successfully used for bio-precipitation/recovery of even very low concentrations of heavy metals (as metal phosphates) [46]. Effective bio-precipitation of metals (including uranium and cadmium) have been revealed, using acid phosphatase extracted from bacterial strains in an acidic-to-neutral pH range [47, 48, 49]. Likewise, Appukuttan et al. [50] demonstrated the bio-recovery of uranium from acidic or neutral wastes by a nonspecific acid phosphatase, which was extracted from genetically engineered radio-resistant bacterium (Deinococcus radiodurans R1). Recently, CIAP mediated bioremediation (white biotechnology) has been used for the bioremediation of soils and industrial effluents polluted with heavy metals [22]. Extracellular alkaline phosphatase (derived from recombinant Escherichia coli) was able to precipitate more than 90% of initial uranium within less than 2 h under alkaline condition [51]. Similar to current study results, Chaudhuri et al. [22] reported highest (80.99%) removal of Cd2+ followed by Ni 2+ (64.78%) > Cr 3+ (46.15%) > Co 2+ (36.47%) > Cr 6+ (32.33%) by calf intestinal alkaline phosphatase (CIAP) enzyme. Further, they revealed greater metal precipitation at pH 11 and at lower initial metal concentration (250 ppm).
In recent times, enzyme technology has been effectively used for the degradation of pesticides as a tool of bioremediation [52]. Being a class of hydrolases, phosphatase enzyme is able to hydrolyze its substrate, thus cleaves a phosphoric acid monoester into a phosphate ion and an alcohol [53].
It was reported previously that there was 50 and 85% degradation of 10 ppm CPF by intracellular alkaline phosphatase (extracted from Spirulina sp.) after 1 and 2 h of incubation respectively [54]. Tang and You [55] demonstrated degradation of TAP (84.4%) by intracellular extracts of Triazophos hydrolase derived from Bacillus sp. TAP-1 respectively within 1 h of incubation and suggested that the expression was constitutive. Similarly, Gothwal et al. [56] reported organophosphorus hydrolase (OPH) extracted from Brevundimonas diminuta capable of degrading methyl parathion up to 500 μM. There was 44 and 78% degradation of 100 ppm CPF by 25 and 50 μl of ALP respectively after 1 h of incubation [57]. Similarly, phosphatase production by the bacterial strains was positively correlated with increased degradation of five OP pesticides in skimmed milk by using ten lactic acid bacteria [58]. The growth of phosphate solubilizing microorganisms along with acid and alkaline phosphatase activities of the soil were significantly improved by the incorporation of the OP pesticides in soil, displaying more availability of water-soluble phosphorus in soil [59].
5. Conclusion
Present study proved that bacterial strain MB497 was phosphate solubilizing as well as capable of producing extracellular acidic, neutral and alkaline phosphatases. Three types (acidic, neutral and alkaline) of OPP were recorded to be active and stable over a wide range of pH, temperature, substrate (p-NPP) concentrations, metal ions and different chemicals like SDS and EDTA. Moreover, these enzymes unveiled great potential for bio-precipitation of different metals like Ni, Mn, Cd and Cr. Lastly, acidic and alkaline OPP exhibited a comprehensive substrate specificity against three OP pesticides (CPF, TAP and DM) with significant degradation as analyzed by HPLC. Therefore, current study results strongly recommend application of this strain and/or its phosphatase enzymes for bioremediation of soil and water resources contaminated with OP pesticides and heavy metals.
Declarations
Author contribution statement
Samina Ambreen: Analyzed and interpreted the data; Wrote the paper.
Azra Yasmin: Conceived and designed the experiments.
Satara Aziz: Performed experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Manco G., Porzio E., Suzumoto Y. Enzymatic detoxification: a sustainable means of degrading toxic organophosphate pesticides and chemical warfare nerve agents. J. Chem. Technol. Biotechnol. 2018;93(8):2064–2082. [Google Scholar]
- 2.Chu Y.H., Yu X.X., Jin X., Wang Y.T., Zhao D.J., Zhang P., Zhang Y.H. Purification and characterization of alkaline phosphatase from lactic acid bacteria. RSC Adv. 2019;9(1):354–360. doi: 10.1039/c8ra08921c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh B.K., Walker A., Wright D.J. Bioremedial potential of fenamiphos and chlorpyrifos degrading isolates: influence of different environmental conditions. Soil Biol. Biochem. 2006;38(9):2682–2693. [Google Scholar]
- 4.Schenk G., Mateen I., Ng T.K., Pedroso M.M., Mitić N., Jafelicci M., Jr., Ollis D.L. Organophosphate-degrading metallohydrolases: structure and function of potent catalysts for applications in bioremediation. Coord. Chem. Rev. 2016;317:122–131. [Google Scholar]
- 5.Ortiz-Hernandez M.L., Quintero-Ramirez R., Nava-Ocampo A.A., Bello-Ramírez A.M. Study of the mechanism of Flavobacterium sp. for hydrolyzing organophosphate pesticides. Fundam. Clin. Pharmacol. 2003;17(6):717–723. doi: 10.1046/j.1472-8206.2003.00202.x. [DOI] [PubMed] [Google Scholar]
- 6.Selleck C., Guddat L.W., Ollis D.L., Schenk G., Pedroso M.M. High resolution crystal structure of a fluoride-inhibited organophosphate-degrading metallohydrolase. J. Inorg. Biochem. 2017;177:287–290. doi: 10.1016/j.jinorgbio.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Yang H., Carr P.D., McLoughlin S.Y., Liu J.W., Horne I., Qiu X., Ollis D.L. Evolution of an organophosphate-degrading enzyme: a comparison of natural and directed evolution. Protein Eng. 2003;16(2):135–145. doi: 10.1093/proeng/gzg013. [DOI] [PubMed] [Google Scholar]
- 8.Gao Y., Truong Y.B., Cacioli P., Butler P., Kyratzis I.L. Bioremediation of pesticide contaminated water using an organophosphate degrading enzyme immobilized on nonwoven polyester textiles. Enzyme. Microb. Technol. 2014;54:38–44. doi: 10.1016/j.enzmictec.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Pinto G., Mazzone G., Russo N., Toscano M. Trimethylphosphate and dimethylphosphate hydrolysis by Binuclear CdII, MnII, and ZnII–FeII promiscuous organophosphate-degrading enzyme: reaction mechanisms. Chem. Eur J. 2017;23(55):13742–13753. doi: 10.1002/chem.201702379. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz-Hernandez M., Sanchez-Salinas E., Olvera-Velona A., Folch-Mallol J.L. Pesticides in the environment: impacts and its biodegradation as a strategy for residues treatment. In: Stoytcheva M., editor. Pesticides-Formulations, Effects, Fate. In Tech. 2011. p. 808. [Google Scholar]
- 11.Sunden F., AlSadhan I., Lyubimov A.Y., Ressl S., Wiersma-Koch H., Borland J., Herschlag D. Mechanistic and evolutionary insights from comparative enzymology of phosphomonoesterases and phosphodiesterases across the alkaline phosphatase superfamily. J. Am. Chem. Soc. 2016;138(43):14273–14287. doi: 10.1021/jacs.6b06186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shahbazkia H.R., Sharifi S., Shareghi B. Purification and kinetic study of bone and liver alkaline phosphatase isoenzymes in the dog. Comp. Clin. Pathol. 2010;19(1):81–84. [Google Scholar]
- 13.Eshanpour A., Amini F. Effect of salt and drought stress on acid phosphatase activities in alfalfa (Medigo sativa L.) explants under in vitro culture. Afr. J. Biotechnol. 2003;2(5):133–135. [Google Scholar]
- 14.Amlabu E., Nok A.J., Sallau A.B. Purification and biochemical characterization of lysosomal acid phosphatases (E.C. 3.1.3.2) from blood stream forms, Trypanosome brucei. Parasitol. Int. 2009;58:238–242. doi: 10.1016/j.parint.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Nautiyal C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999;170(1):265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 16.Premono M.E., Moawad A.M., Vlek P.L.G. Effect of phosphate-solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones. J. Crop Sci. 1996;11(1):13–23. [Google Scholar]
- 17.Hernandez I., Fernandez J.A., Niell F.X. A comparative study of alkaline phosphatase activity in two species of Gelidium (Gelidiales, Rhodophyta) Eur. J. Phycol. 1995;30(1):69–77. [Google Scholar]
- 18.Weinberg R.A., Zusman D.R. Alkaline, acid, and neutral phosphatase activities are induced during development in Myxococcus xanthus. J. Bacteriol. 1990;172(5):2294–2302. doi: 10.1128/jb.172.5.2294-2302.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plisova E.Y., Balabanova L.A., Ivanova E.P., Kozhemyako V.B., Mikhailov V.V., Agafonova E.V., Rasskazov V.A. A highly active alkaline phosphatase from the marine bacterium. Cobetia Mar. Biotechnol. 2005;7(3):173–178. doi: 10.1007/s10126-004-3022-4. [DOI] [PubMed] [Google Scholar]
- 20.Harishankar M.K., Sasikala C., Ramya M. Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech. 2013;3(2):137–142. doi: 10.1007/s13205-012-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhry G.R., Ali A.N., Wheeler W.B. Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the Opd gene from a Flavobacterium sp. Appl. Environ. Microbiol. 1988;54:288–293. doi: 10.1128/aem.54.2.288-293.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhuri G., Shah G.A., Dey P.S.G., Venu-Babu P., Thilagaraj W.R. Enzymatically mediated bio-precipitation of heavy metals from industrial wastes and single ion solutions by mammalian alkaline phosphatase. J. Environ. Sci. Health A. 2013;48(1):79–85. doi: 10.1080/10934529.2012.707851. [DOI] [PubMed] [Google Scholar]
- 23.Liang W.Q., Wang Z.Y., Li H., Wu P.C., Hu J.M., Luo N., Liu Y.H. Purification and characterization of a novel pyrethroid hydrolase from Aspergillus niger ZD11. J. Agric. Food Chem. 2005;53(19):7415–7420. doi: 10.1021/jf051460k. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y., Chen S., Hu M., Hu Q., Luo J., Li Y. Purification and characterization of a novel chlorpyrifos hydrolase from Cladosporium cladosporioides Hu-01. PloS One. 2012;7(6) doi: 10.1371/journal.pone.0038137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rani S., Dhiraj S. Development and validation of HPLC method for determination of triazophos pesticide in water. Int. J. Res. Chem. Environ. 2015;5(4):65–69. [Google Scholar]
- 26.Alvarenga N., Birolli W.G., Nitschke M., de O Rezende M.O., Seleghim M.H., Porto A.L. Biodegradation of chlorpyrifos by whole cells of marine-derived fungi Aspergillus sydowii and trichoderma sp. J. Microb. Biochem. Technol. 2015;7:133–139. [Google Scholar]
- 27.Bagyalakshmi J., Kavitha G., Ravi T.K. Residue determination of dimethoate in leafy vegetables (spinach) using RP-HPLC. Int. J. Pharma Sci. Res. 2011;2(2):62–64. [Google Scholar]
- 28.Bishnoi K., Sain U., Kumar R., Singh R., Bishnoi N.R. Distribution and biodegradation of polycyclic aromatic hydrocarbons in contaminated sites of Hisar (India) Indian J. Exp. Biol. 2009;47(3):210–217. [PubMed] [Google Scholar]
- 29.Eissa F.I., Mahmoud H.A., Massoud O.N., Ghanem K.M., Gomaa I.M. Biodegradation of chlorpyrifos by microbial strains isolated from agricultural wastewater. J. Am. Sci. 2014;10(3):98–108. http://www.jofamericanscience.org [Google Scholar]
- 30.Islam T.M., Deora A., Hashidoko Y., Atiqur Rahman A., Ito T., Tahara S. Isolation and identification of potential phosphate solubilizing bacteria from the rhizoplane of Oryza sativa L. cv. BR29 of Bangladesh. Z. Naturforsch. C. 2007;62(1-2):103–110. doi: 10.1515/znc-2007-1-218. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai M., Wasaki J., Tomizawa Y., Shinano T., Osaki M. Analysis of bacterial communities on alkaline phosphatase gene in soil supplied with organic matter. Soil Sci. Plant Nutr. 2008;54(1):62–71. [Google Scholar]
- 32.Kriakov J., Lee S.H., Jacobs W.R. Identification of a regulated alkaline phosphatase, a cell surface-associated lipoprotein, in mycobacterium smegmatis. J. Bacteriol. 2003;185(16):4983–4991. doi: 10.1128/JB.185.16.4983-4991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rina M., Pozidis C., Mavromatis K., Tzanodaskalaki M., Kokkinidis M., Bouriotis V. Alkaline phosphatase from the Antarctic strain TAB5. Eur. J. Biochem. 2000;267(4):1230–1238. doi: 10.1046/j.1432-1327.2000.01127.x. [DOI] [PubMed] [Google Scholar]
- 34.Kumar M., Kaur P.P., Ganjewala D. Isolation of periplasmic alkaline phosphatase from Rhizobium bacteria. Res. J. Microbiol. 2008;3(3):157–162. [Google Scholar]
- 35.Zappa S., Rolland J.L., Flament D., Gueguen Y., Boudrant J., Dietrich J. Characterization of a highly thermostable alkaline phosphatase from the euryarchaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 2001;67(10):4504–4511. doi: 10.1128/AEM.67.10.4504-4511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahesh M., Somashekhar R., Bagchi P., Puttaiah E.T. Optimization for the production of extracellular Alkaline phosphatase from Proteus mirabilis. J. Bioprocess. Biotech. 2015;5(3):1–5. [Google Scholar]
- 37.Behera B.C., Yadav H., Singh S.K., Sethi B.K., Mishra R.R., Kumari S., Thatoi H. Alkaline phosphatase activity of a phosphate solubilizing Alcaligenes faecalis, isolated from Mangrove soil. Biotechnol. Res. Innov. 2017;1(1):101–113. [Google Scholar]
- 38.Janeway C.M., Xu X., Murphy J.E., Chaidaroglou A., Kantrowitz E.R. Magnesium in the active site of Escherichia coli alkaline phosphatase is important for both structural stabilization and catalysis. Biochemistry. 1993;32(6):1601–1609. doi: 10.1021/bi00057a026. [DOI] [PubMed] [Google Scholar]
- 39.Shah A.Q., Iqbal S., Niazi Z. Partial purification and characterization of intracellular alkaline phosphatase from newly isolated strain of Bacillus subtilis KIBGE-HAS. Internet J. Microbiol. 2008;7(1):1–6. [Google Scholar]
- 40.Bryan L.W., Keith W. General principles of biochemical investigation. In: Wilson K., Walker J., editors. Principles and Techniques of Practical Biochemistry. second ed. Cambridge University Press; Cambridge: 1994. p. 586. [Google Scholar]
- 41.Danial E.N., Alkhalf M.I. Effect of physical and chemical parameters on the activity of purified phosphatase enzyme produced by Bacillus cereus. Int. J. Biotechnol. Wellness Ind. 2018;6(3):64–71. [Google Scholar]
- 42.Boulanger R.R., Kantrowitz E.R. Characterization of a monomeric Escherichia coli alkaline phosphatase formed upon a single amino acid substitution. J. Biol. Chem. 2003;278(26):23497–23501. doi: 10.1074/jbc.M301105200. [DOI] [PubMed] [Google Scholar]
- 43.Mori S., Okamoto M., Nishibori M., Ichimura M., Sakiyama J., Endo H. Purification and characterization of alkaline phosphatase from Bacillus stearothermophilus. Biotechnol. Appl. Biochem. 1999;29(3):235–239. [PubMed] [Google Scholar]
- 44.Alnuaimi M.M., Saeed I.A., Ashraf S.S. Effect of various heavy metals on the enzymatic activity of E. coli alkaline phosphatase. Int. J. Biotechnol. Biochem. 2012;8:47–59. [Google Scholar]
- 45.Lieser K.H. Radionuclides in the geosphere: sources, mobility, reactions in natural waters and interactions with solids. Radiochim. Acta. 1995;70–71:355–375. [Google Scholar]
- 46.Lokhande M., Achuthan P.V., Jambunathan U., Ramanujam A., Rao A.S., Mahajan S.K. Proceedings of BRNS-DAE National Symposium on Nuclear and Radiochemistry. Department of Chemistry, University of Pune; Pune, India: 2001. Studies on the recovery of uranium using enzyme mediated and chemical methods; pp. 224–225. [Google Scholar]
- 47.Renninger N., Knopp R., Nitsche H., Clark D.S., Keasling J.D. Uranyl precipitation by Pseudomonas aeruginosa via controlled polyphosphate metabolism. Appl. Environ. Microbiol. 2004;70:7404–7412. doi: 10.1128/AEM.70.12.7404-7412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelobolina E.S., Sullivan S.A., O'Neill K.R., Nevin K.P., Lovley D.R. Isolation, characterization, and U (VI)-reducing potential of a facultatively anaerobic, acid-resistant bacterium from Low-pH, nitrate-and U (VI)-contaminated subsurface sediment and description of Salmonella subterranea sp. nov. Appl. Environ. Microbiol. 2004;70(5):2959–2965. doi: 10.1128/AEM.70.5.2959-2965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez R.J., Beazley M.J., Taillefert M., Arakaki A.K., Skolnick J., Sobecky P.A. Aerobic uranium (VI) bio-precipitation by metal-resistant bacteria isolated from radionuclide-and metal-contaminated subsurface soils. Environ. Microbiol. 2007;9(12):3122–3133. doi: 10.1111/j.1462-2920.2007.01422.x. [DOI] [PubMed] [Google Scholar]
- 50.Appukuttan D., Rao A.S., Apte S.K. Engineering of Deinococcus radiodurans R1 for bio-precipitation of uranium from dilute nuclear waste. Appl. Environ. Microbiol. 2006;72:7873–7878. doi: 10.1128/AEM.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilgiriwala K.S., Alahari A., Rao A.S., Apte S.K. Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bio-precipitation of uranium from alkaline solutions. Appl. Environ. Microbiol. 2008;74(17):5516–5523. doi: 10.1128/AEM.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cycon M., Zmijowska A., Wojcik M., Piotrowska-Seget Z. Biodegradation and bioremediation potential of diazinon-degrading Serratia marcescens to remove other organophosphorus pesticides from soils. J. Environ. Manag. 2013;117:7–16. doi: 10.1016/j.jenvman.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 53.Liberti S., Sacco F., Calderone A., Perfetto L., Iannuccelli M., Panni S., Cesareni G. HuPho: the human phosphatase portal. FEBS J. 2012;280(2):379–387. doi: 10.1111/j.1742-4658.2012.08712.x. [DOI] [PubMed] [Google Scholar]
- 54.Thengodkar R.R.M., Sivakami S. Degradation of chlorpyrifos by an alkaline phosphatase from the cyanobacterium Spirulina platensis. Biodegradation. 2010;21(4):637–644. doi: 10.1007/s10532-010-9331-6. [DOI] [PubMed] [Google Scholar]
- 55.Tang M., You M. Isolation, identification and characterization of a novel triazophos-degrading Bacillus sp. (TAP-1) Microbiol. Res. 2012;167(5):299–305. doi: 10.1016/j.micres.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Gothwal A., Dahiya M., Beniwa P., Hooda V. Purification and kinetic studies of organophosphorus hydrolase from B. diminuta. Int. J. Pharm. Pharmaceut. Sci. 2014;6(10):341–344. [Google Scholar]
- 57.Khalid S., Hashmi I., Khan S.J., Qazi I.A., Nasir H. Effect of metal ions and petrochemicals on bioremediation of chlorpyrifos in aerobic sequencing batch bioreactor (ASBR) Environ. Sci. Pollut. Res. 2016;23(20):20646–20660. doi: 10.1007/s11356-016-7153-8. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y.H., Xu D., Liu J.Q., Zhao X.H. Enhanced degradation of five organophosphorus pesticides in skimmed milk by lactic acid bacteria and its potential relationship with phosphatase production. Food Chem. 2014;164:173–178. doi: 10.1016/j.foodchem.2014.05.059. [DOI] [PubMed] [Google Scholar]
- 59.Majumder S.P., Das A.C. Phosphate-solubility and phosphatase activity in Gangetic alluvial soil as influenced by organophosphate insecticide residues. Ecotoxicol. Environ. Saf. 2016;126:56–61. doi: 10.1016/j.ecoenv.2015.12.018. [DOI] [PubMed] [Google Scholar]