Significance
It is well characterized that a unique combination of transcription factor binding sites within an enhancer gives rise to a well-defined boundary of target gene expression. However, it is yet to be understood how each binding site contributes to the overall transcriptional dynamics. With live imaging and quantitative analysis, we show that two transcription factors affect different aspects of transcription to regulate gene expression boundaries. Transcription factor Dorsal mostly changes the timing of transcriptional activation by affecting bursting frequency, while a pioneer factor Zelda influences both the timing of activation and amplitude, hence changing both bursting frequency and duration. Our study provides two distinct mechanisms with which transcription factors regulate target gene expression to ensure normal development.
Keywords: Drosophila, MS2, live imaging, t48, transcription factor
Abstract
The regulatory specificity of a gene is determined by the structure of its enhancers, which contain multiple transcription factor binding sites. A unique combination of transcription factor binding sites in an enhancer determines the boundary of target gene expression, and their disruption often leads to developmental defects. Despite extensive characterization of binding motifs in an enhancer, it is still unclear how each binding site contributes to overall transcriptional activity. Using live imaging, quantitative analysis, and mathematical modeling, we measured the contribution of individual binding sites in transcriptional regulation. We show that binding site arrangement within the Rho-GTPase component t48 enhancer mediates the expression boundary by mainly regulating the timing of transcriptional activation along the dorsoventral axis of Drosophila embryos. By tuning the binding affinity of the Dorsal (Dl) and Zelda (Zld) sites, we show that single site modulations are sufficient to induce significant changes in transcription. Yet, no one site seems to have a dominant role; rather, multiple sites synergistically drive increases in transcriptional activity. Interestingly, Dl and Zld demonstrate distinct roles in transcriptional regulation. Dl site modulations change spatial boundaries of t48, mostly by affecting the timing of activation and bursting frequency rather than transcriptional amplitude or bursting duration. However, modulating the binding site for the pioneer factor Zld affects both the timing of activation and amplitude, suggesting that Zld may potentiate higher Dl recruitment to target DNAs. We propose that such fine-tuning of dynamic gene control via enhancer structure may play an important role in ensuring normal development.
Highly coordinated movement of cells, such as mesoderm invagination during early Drosophila embryogenesis, is a fundamental process in morphogenesis, ensuring robust and reproducible development of the embryo (1, 2). Pulsatile myosin contraction generates mechanical forces, initiating apical constriction that leads to the invagination of hundreds of ventrally located cells in an embryo (3, 4). Myosin accumulation is the strongest in ventralmost cells, consistent with the most constricted apical surface (5, 6). Such geometrical and mechanical constraints mediated by the myosin gradient are the minimal requirements for proper cell shape changes (7, 8). Indeed, disruptions in the myosin gradient lead to defects in ventral furrow formation and the prevention of normal embryonic development (5).
In addition to studies exploring the cellular aspects of ventral furrow formation, precise transcriptional regulation of cytoskeletal genes is also crucial for robust morphogenesis (9–11). Our previous work demonstrated that the Rho-GTPase pathway component t48 and folded gastrulation (fog) exhibit graded gene expression patterns along the dorsoventral (DV) axis, contributing to the myosin gradient and subsequent gastrulation processes (12). We suggested that the kinetics of t48 and fog transcriptional activity preshadow morphogenesis, implying that transcriptional dynamics may play an important role in maintaining the robustness of this process. Yet, the transcriptional mechanisms through which this temporal gradient is regulated remain largely unknown.
The spatial expression pattern of a gene is determined by a unique set of transcription factor binding sites located in the enhancer (13, 14). The combination of these binding sites, their relative binding affinities for target transcription factors, and even their spacing along the enhancer have all been found to play roles in gene regulation. This complex encoding of information in regulatory DNA is referred to as enhancer grammar. The t48 enhancer contains binding sites for well-known transcription factors like Dorsal (Dl) and Zelda (Zld) (12). Dl is an NF-κB homolog in Drosophila whose nuclear protein expression is distributed in a graded manner along the DV axis, with a peak concentration in the ventralmost point (15, 16). Dl is known to regulate the spatial boundaries of its target genes in a concentration-dependent manner (17–19). Enhancers of ventrally expressed genes like twist (twi) and snail contain low-affinity Dl binding sites, such that these genes are expressed only in the ventral region where high Dl level is present. Conversely, short gastrulation and rhomboid enhancers have high-affinity Dl binding sites, and thus the genes can be expressed in the lateral region with intermediate Dl level. Moreover, the Dl level itself increases over nuclear cycles, contributing to the kinetics of its target genes (20, 21).
On the other hand, the zinc-finger protein Zld is known to regulate the kinetics of gene expression (22, 23). Uniformly expressed in embryos, Zld mediates global zygotic genome activation during early Drosophila embryogenesis (24). Many zygotic gene activations are delayed or accelerated upon removing or adding Zld binding sites to enhancers, suggesting Zld’s contribution to temporal regulation of the target genes (25). Zld is also known to operate as a pioneer factor to lower the nucleosome barrier and make chromatin more accessible for binding to other transcription factors (26). How does localized Dl and uniformly expressed Zld differentially contribute to regulation of the target genes? Moreover, an enhancer typically contains multiple low- and high-affinity binding sites of the same transcription factors (14). How do these multiple transcription factor binding sites coordinate with each other to determine the target gene’s spatial and temporal expression?
With a combination of live imaging and quantitative analysis, we show that the boundary of t48 expression is modulated mainly by changes in the timing of transcriptional activation. Initiation of transcription was significantly expedited or delayed by changing the binding affinity of a single Dl binding site within the enhancer, while the amplitude of transcription mostly remained the same, except when transcriptional activity was drastically changed (∼90%). Contrary to Dl binding-site modifications, changes in the pioneer factor Zld binding site led to changes in both the timing and the amplitude of transcriptional activity. To further characterize differential contribution of Dl and Zld, we used a two-state model to show that bursting frequencies, but not bursting durations, were affected upon Dl site modifications, whereas both bursting frequency and durations increased upon optimization of the Zld site. Our results suggest that Zld may regulate transcriptional kinetics by allowing more activators like Dl to bind to the target binding site and recruiting more Pol II to the promoter. This study presents a detailed analysis on how individual transcription factor binding sites within an enhancer contribute to transcriptional dynamics, especially in the context of the differential role of two transcription factors, Dl and Zld.
Results
Differential Transcriptional Activation of t48 Leads to Graded Expression along the DV Axis.
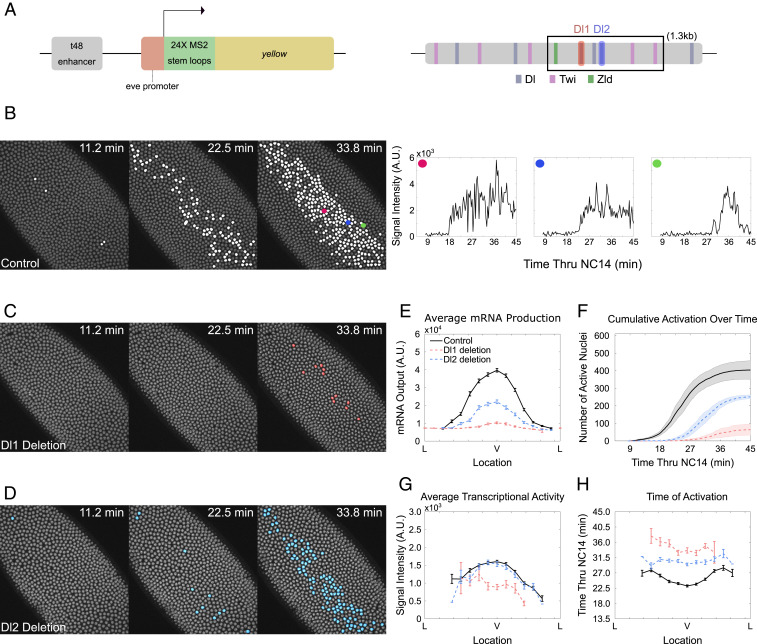
T48 is a transmembrane protein that recruits RhoGEF2 to activate the Rho-GTPase signaling pathway, which results in localized myosin activation and initiates ventral furrow formation in Drosophila embryos (27, 28). We first analyzed transcriptional dynamics of the MS2-yellow reporter gene driven by the wild-type 1.3-kb t48 intronic enhancer (12). Maternally provided MCP-GFP fusion proteins bind to the MS2 stem loops upon transcription, and nascent transcripts produced from the t48>MS2-yellow reporter construct can be visualized as fluorescent foci (Fig. 1A). Since t48 transcription is initiated in nuclear cycle (NC) 14, all analysis corresponds to NC14.
Fig. 1.
Graded t48 expression results from differential timing of transcriptional activation. (A, Left) Schematic of t48 enhancer>evePr-MS2-yellow construct. (A, Right) Dl, Twi, and Zld binding sites within the t48 enhancer. The boxed region highlights the minimal t48 enhancer. (B, Left) Snapshots of an embryo expressing t48-WT>MS2-yellow at different times during NC14. Actively transcribing nuclei are false-colored in white. (B, Right) Trajectories of MS2-yellow reporter gene for individual nuclei chosen from the middle to the edge of t48 expression domain (red, blue, and green, respectively). (C and D) Snapshots of an embryo expressing (C) t48-Dl1deletion>MS2-yellow and (D) t48-Dl2deletion>MS2-yellow at different times during NC14. Actively transcribing nuclei are false-colored in red and blue, respectively. (E) Average total mRNA production of t48-WT>MS2-yellow (black), t48-Dl1deletion>MS2-yellow (red), and t48-Dl2deletion>MS2-yellow (blue) of all nuclei across the t48 expression domain. (F) Cumulative number of active nuclei during NC14 for t48-WT>MS2-yellow (black), t48-Dl1deletion>MS2-yellow (red), and t48-Dl2deletion>MS2-yellow (blue). (G) Average transcriptional activity of t48-WT>MS2-yellow (black), t48-Dl1deletion>MS2-yellow (red), and t48-Dl2deletion>MS2-yellow (blue) from initial activation time to the end of NC14 for each nucleus across the t48 expression domain. (H) Average time point at which nuclei first became active within each bin. Error bars in E, G, and H are SEM, while error bars in F are SD; 3,622, 1,766, and 1,661 nuclei were examined from six, three, and three biological replicate embryos of t48-WT>MS2-yellow, t48-Dl1deletion>MS2-yellow, and t48-Dl2deletion>MS2-yellow, respectively.
t48>MS2-yellow exhibits a dynamic expression pattern, where nuclei along the ventral midline are the first to begin transcription, with more lateral nuclei becoming active as NC14 progresses (Fig. 1B and Movie S1). In order to plot the distribution of t48 messenger RNA (mRNA) along the DV axis, we divided the entire t48 expression domain of an embryo into 16 bins along the DV axis and measured the average mRNA production per bin by taking the sum of MS2-yellow fluorescent intensity during NC14. Consistent with previous results, t48>MS2-yellow transcription revealed a “bell curve” distribution of mRNA output, with mRNA production highest near the ventralmost point and decreasing away from the ventral midline (Fig. 1E, black) (5, 12). We proceeded to ask what dynamic processes drive graded expression of t48 by measuring the average amplitude of active transcription and the timing of transcription initiation. The amplitude exhibited graded distribution with peak amplitude in the ventralmost point and ∼70% decrease in intensity at the lateral border (Fig. 1G, black). In addition, transcription started ∼5 min earlier in the ventral nuclei compared to the lateral nuclei (Fig. 1H, black). Our results indicate that modulations in both the timing and the amplitude of transcriptional activation along the DV axis lead to the steep gradient of t48 within a narrow spatial boundary.
Deletion of Dl Binding Site Results in Narrower t48 Expression through Delayed Activation.
We asked whether this dynamic transcriptional regulation by the t48 enhancer is encoded in the transcription factor binding site arrangement of the enhancer itself. We hypothesized that tuning transcription factor binding site strength could serve as a way for enhancer grammar to reflect temporal information. We first characterized the 523-base pair (bp) minimal enhancer that successfully recapitulates endogenous t48 expression patterns (Fig. 1A). Using the published position weight matrix, we found three Dl, two Twi, and one Zld binding sites in the t48 minimal enhancer (Fig. 1A) (29–31). All Dl binding sites show relatively low binding affinity (TGGGATTTCTT, CCTAATTCCCA, and CGGGATTTCTC), providing an explanation for the narrower expression domain of t48 compared to other low-affinity Dl target genes like twi or snail. Modifying Twi binding sites, in contrast with Dl and Zld, resulted in little to no effect on the dynamics of t48 expression, and hence we focused our analysis on the latter sites only.
The minimal t48 enhancer contains three low-affinity Dl binding sites, requiring a high nuclear Dl level. We wondered whether these sites work linearly to recruit more transcription factors to the enhancer, or if they are redundant and independently sufficient. We chose two Dl binding sites to perturb, with one (CGGGATTTCTC, referred to as Dl1, red) showing stronger binding affinity than the other (TGGGATTTCTT, referred to as Dl2, blue) (Fig. 1A). We first examined the effect of deleting one Dl binding site at a time by replacing the binding site with nucleotides that show no affinity to Dl binding. We did not expect the removal of only one of three Dl binding sites within the t48 enhancer to induce large perturbations in transcriptional activity. Surprisingly, however, deletion of the stronger Dl1 site abolished most of the transcriptional activity from the embryo, suggesting that the remaining weaker Dl binding sites are not sufficient to maintain transcription (Fig. 1C and Movie S1). Deletion of the Dl1 site resulted in ∼90% reduction in mRNA production (Fig. 1E, red). Only approximately one-eighth of nuclei produced active transcripts in t48-Dl1deletion>MS2-yellow embryos and all active nuclei were located along the ventralmost domain, indicating that only a very high level of Dl can initiate transcription (Fig. 1 C and F, red). Transcriptional activity from those few active nuclei exhibited a significantly lower amplitude (Fig. 1G, red). In addition, transcriptional initiation was significantly delayed by about 13 min (Fig. 1H, red). Together, both the reduced amplitude and delayed transcriptional activation contribute to a significant reduction in mRNA production.
Multiple Dl Binding Sites Function Synergistically in Gene Regulation.
We wondered whether the stronger Dl1 site plays a dominant role in t48 regulation. If Dl1 and Dl2 sites work linearly with respect to their binding affinity, deletion of the weaker Dl2 site should lead to only about a 10% reduction in mRNA production. However, replacing the Dl2 site into nucleotides with no binding affinity to Dl resulted in ∼50% reduction in mRNA production (Fig. 1E, blue). The expression boundary was narrowed to ∼7-cell width, compared to the ∼14-cell width in wild type (Fig. 1D and Movie S1). This is consistent with findings that Dl regulates the expression boundary of target genes in a concentration-dependent manner (32, 33). Surprisingly, the average amplitude of transcriptional activity was not reduced upon Dl2 site removal (Fig. 1G, blue). Instead, changes in the spatial boundary and mRNA production seem to be mediated mainly through modulation in the kinetics of transcriptional activation. Transcription was delayed by ∼7 min in t48-Dl2deletion>MS2-yellow, uniformly across the DV axis (Fig. 1 F and H and SI Appendix, Fig. S1C, blue). Nuclei were no longer activated in a graded manner from ventral to lateral side. Rather, all of the nuclei within the narrowed t48 domain were activated ∼30 min into NC14 (Fig. 1H, blue).
Our results show that the removal of a single Dl site can lead to significant changes in transcriptional activity, manifested in a narrowed expression domain and reduced mRNA production. Importantly, the moderate change in gene expression level is attributed mostly to the delay in transcriptional activation, rather than the decrease in amplitude of transcription (Fig. 1 G and H, blue). Only when the mRNA output is drastically decreased (∼90% in Dl1 deletion) was transcriptional amplitude reduced (Fig. 1 G and H, red). Our results also suggest that the two Dl binding sites work synergistically with each other to induce even higher transcriptional activity. Indeed, mRNA production driven by the wild-type t48 enhancer is significantly higher than the sum of mRNA production driven by the t48 enhancer with a single Dl1 or Dl2 site (SI Appendix, Fig. S1B). Taken together, arranging two weak Dl binding sites in the t48 enhancer seems to allow high transcriptional activity in a narrow expression domain that requires a high Dl level.
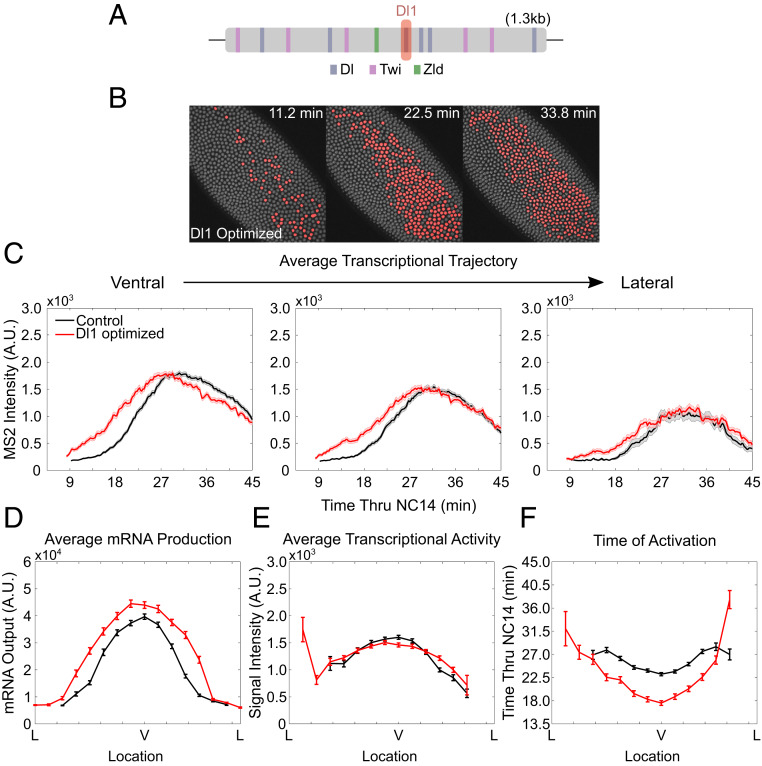
Higher Dl Affinity Binding Site Leads to Precocious Transcriptional Activation.
Next we hypothesized that converting a low-affinity Dl binding site within the t48 enhancer into a high-affinity consensus Dl binding site may lower the required Dl threshold to activate t48 and result in an expansion of the expression boundary. We first converted the Dl1 site into the consensus Dl binding site (GGGGAATTCCC) (Fig. 2A) (34). We observed wider expression of MS2-yellow, which was expected, since the high-affinity Dl site would allow even intermediate levels of Dl proteins to bind to the t48 enhancer (Fig. 2B and SI Appendix, Fig. S2G and Movie S2). What was surprising, however, was the precocious transcriptional activity of t48-optimizedDl1>MS2-yellow. Looking at the average trajectory of nuclei, we found that transcription began earlier across the t48 expression domain, leading to a wider and higher mRNA production profile (Fig. 2D). However, the amplitude was comparable to the wild-type t48>MS2-yellow (Fig. 2C).
Fig. 2.
Modulation of a single Dl binding site affects gene expression dynamics. (A) Schematic of t48 enhancer showing the location of Dl, Twi, and Zld binding sites. The Dl1 site is shown in red. (B) Snapshots of t48-optimizedDl1>MS2-yellow embryo at different time points in NC14. Actively transcribing nuclei in a given frame are false-colored. (C) Average trajectory of t48-WT>MS2-yellow (black) and t48-optimizedDl1>MS2-yellow (red) over NC14 of ventrally located nuclei (Left), off-ventral nuclei (Center), and lateral nuclei (Right). (D–F) (D) Average total mRNA production, (E) average transcriptional activity, and (F) average timing of transcriptional initiation of nuclei along the t48 expression domain for t48-WT>MS2-yellow (black) and t48-optimizedDl1>MS2-yellow (red). Only active nuclei were analyzed for C, E, and F; 2,028 nuclei were examined from three biological replicate embryos of t48-optimizedDl1>MS2-yellow embryos. All error bars indicate SEM.
Upon further analysis, we found that the kinetics of transcriptional activity was affected most significantly, resembling the case observed with the Dl2 site deletion. Transcription was initiated ∼10 min earlier than the wild type (Fig. 2F). The expression pattern was changed as well with activation beginning within the first few minutes as a thicker band of ∼10 cells wide, expanding to a final width of ∼18 cells (Fig. 2B). On the contrary, despite the increase in mRNA production and expansion of the gene expression boundary, the amplitude of transcription did not change significantly upon enhancing the binding affinity of Dl1 (Fig. 2E). Our result implies that the t48 enhancer with a single enhanced Dl binding site is sufficient to increase mRNA production and expand the gene expression boundary, mainly by quickening transcriptional activation, not by changing amplitude.
Upper Limit on Dl-Mediated Transcriptional Activation of t48.
Since the binding affinity of Dl to the Dl2 site is significantly weaker than that of Dl1, optimizing the Dl2 site into the consensus site should result in higher overall binding affinity of Dl to the t48 enhancer. To test this, we measured transcriptional activity driven by the t48 enhancer with the consensus Dl2 site. Interestingly, the overall mRNA production level of t48-optimizedDl2>MS2-yellow was similar to that from the embryos with the optimal Dl1 site (SI Appendix, Fig. S2C and Movie S2). All other parameters, including mRNA distribution, amplitude, and timing of activation were comparable between the t48-optimizedDl1>MS2-yellow and t48-optimizedDl2>MS2-yellow embryos, suggesting saturation of the t48 enhancer with Dl (SI Appendix, Fig. S2 A–E). As observed in t48-optimizedDl1>MS2-yellow embryos, transcription was initiated ∼10 min early compared to the wild type, while average amplitude stayed comparable (SI Appendix, Fig. S2F). However, unlike t48-WT and t48-optimizedDl1–mediated transcription, where the level decreases after ∼30 min into NC14, high transcriptional activity was maintained until the end of NC14 in t48-optimizedDl2>MS2-yellow embryos (SI Appendix, Fig. S2F). This indicates that the optimized Dl2 site allows Dl to be bound for a longer duration to maintain high transcriptional activity in late NC14 (35, 36). Taken together, our results suggest that the transcription factor Dl regulates the spatial boundary of its target gene t48 expression domain by mainly modulating the kinetics of transcriptional activation along the DV axis (except for the case in Dl1 site deletion, where 90% reduction in mRNA output was observed, due to lower amplitude and delayed activation). Optimization of each Dl binding site, however, did not result in further increase in transcriptional activity, possibly due to an upper limit of the amount of Dl that can bind to the t48 enhancer.
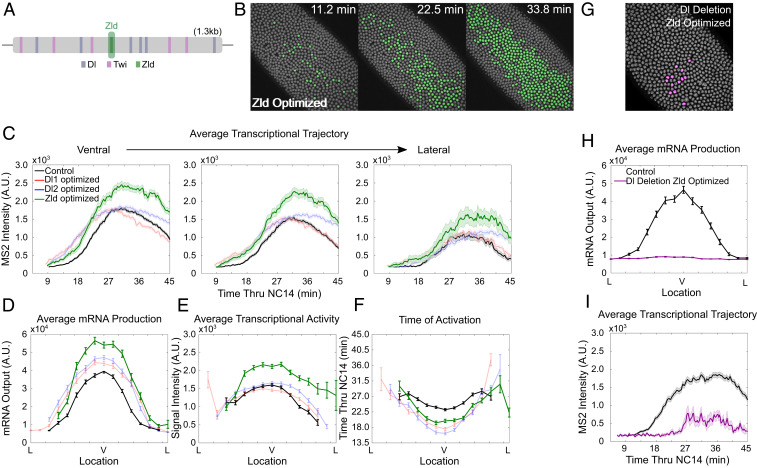
Optimization of Zld Binding Site Can Further Increase Transcription Potential.
The zinc-finger protein Zld is a well-known pioneer factor that mediates zygotic genome activation during early fly embryogenesis (24). We optimized the weak Zld site (GCAGGAAG) in the t48 enhancer into the consensus Zld binding site (CAGGTAG), to compare the effect of suboptimal to optimal Zld binding to DNA (Fig. 3A) (22). The expression boundary of the MS2-yellow reporter gene driven by the t48-optimizedZld enhancer was almost the same as the boundaries driven by the enhancers with optimized Dl1 or Dl2 sites, confirming Zld’s role in the transcriptional regulation (Fig. 3B and SI Appendix, Fig. S2G and Movie S3). t48-optimizedZld>MS2-yellow embryos showed expedited transcriptional activation as expected, but with slower kinetics compared to the embryos with optimized Dl sites (Fig. 3F). Rather, transcriptional amplitudes driven by the t48-optimizedZld enhancer were much higher, implying different contributions of Dl and Zld to gene regulation (Fig. 3 C and E).
Fig. 3.
Optimizing Zld site achieves greater gene expression through increase of transcriptional amplitude. (A) Schematic of t48 enhancer showing the location of Dl, Twi, and Zld binding sites. The Zld site is shown in green. (B) Snapshots of t48-optimizedZld>MS2-yellow embryo at different time points in NC14. Actively transcribing nuclei in a given frame are false-colored. (C) Average trajectory of t48-WT (black), t48-optimizedZld (green), t48-optimizedDl1 (red), and t48-optimizedDl2 (blue) >MS2-yellow embryos over NC14 of ventrally located nuclei (Left), off-ventral nuclei (Center), and lateral nuclei (Right). (D–F) (D) Average total mRNA production, (E) average transcriptional activity, and (F) average timing of transcriptional initiation of nuclei along the t48 expression domain for t48-WT (black), t48-optimizedZld (green), t48-optimizedDl1 (red), and t48-optimizedDl2 (blue) >MS2-yellow embryos. A Welch’s t test was conducted on the average transcriptional intensity data and resulted in a majority of the bins showing significant difference (P < 0.05) between Zld-optimized and the control, and no significant difference (P > 0.05) for a majority of the bins between both Dl1 and Dl2 optimized and the control. (G–I) (G) Snapshot of t48-Dldeletion-optimizedZld>MS2-yellow embryo showing false color of all actively transcribing nuclei. (H) Average total mRNA production, and (I) average trajectory of t48-WT (black) and t48-Dldeletion-optimizedZld (purple) >MS2-yellow embryos over NC14 of ventrally located nuclei. Only active nuclei were analyzed for E, F, and I; 1,976 nuclei were examined from three biological replicate embryos of t48-optimizedZld>MS2-yellow embryos and 1,713 and 1,605 nuclei were examined from three biological replicate embryos of t48-WT>MS2-yellow and t48-Dldeletion-optimizedZld>MS2-yellow, respectively. All error bars indicate SEM.
With precocious transcriptional activation and higher amplitude, significantly more mRNA was produced by t48-optimizedZld>MS2-yellow (Fig. 3D, green). We asked whether Zld works as a direct activator of t48, or as a pioneer factor that increases chromatin accessibility and allows more Dl to bind to the t48 enhancer. To distinguish between these two possibilities, we designed another construct where both the Dl1 and Dl2 sites were replaced with nonaffinity sites, and the Zld site was optimized to the consensus site. These embryos showed hardly any transcriptional activity (Fig. 3G and Movie S4). Between 3∼13 nuclei exhibited active transcription in each of the three biological replicates. For those few that showed active transcription, transcriptional initiation was significantly delayed, with very low amplitude (Fig. 3 H and I). Both parameters were decreased from those observed in t48_Dl1deletion>MS2-yellow embryos (Fig. 1, red). In addition, transcriptional activity was comparably low when only the Dl1 and Dl2 sites were removed from the t48 enhancer with original Zld site (SI Appendix, Fig. S2 H–J). Our results suggest that Zld alone does not directly activate t48. Rather, t48 activation seems to require synergistic activity among multiple transcription factors and a strong Zld site would allow more transcription factors like Dl to bind to the enhancer. Zld’s role of modulating the nuclear microenvironment of a Dl target enhancer has also been reported recently, supporting our observation (37). While there seems to be a limit at which enhanced Dl binding affinity can induce transcription, optimizing the Zld site is able to overcome that upper limit and enhance transcriptional activity even further.
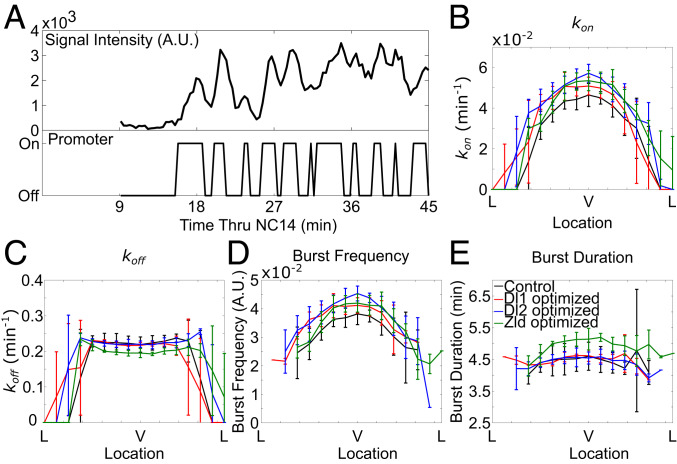
A Two-State Model Shows Modulation of Bursting Frequency upon Binding-Site Modifications.
As shown in many studies, transcriptional activity is not continuous, but rather stochastic, comprising multiple episodic bursts (38–40). Using a simple mathematical model of promoter activity, we asked whether the changes observed in transcription factor arrangement modulation in the t48 enhancer are due to changes in bursting characteristics. The “two-state” model of transcriptional bursting is one of the most commonly used models to describe the dynamics of gene expression in a single-cell resolution (41–43). We implemented a hidden Markov model (HMM) approach to infer the parameters of a two-state model to understand which transcriptional property is affected upon changes in transcription factor binding-site arrangement in the t48 enhancer. The modeling protocol as well as the calculation of kon and koff values is described in more detail in Materials and Methods. Briefly, we used fluorescence traces to infer the time-dependent switching of promoter state; an active promoter was expected to cause an increase in fluorescence and periods of inactivity would be expected to result in a decrease in fluorescence (Fig. 4A). The maximal likelihood transition matrix between the on and off states was used to recover the kon and koff values, corresponding to the rate per minute of switching between the two.
Fig. 4.
A simple mathematical modeling recapitulates experimental observations. (A) A representative trace of t48-WT>MS2-yellow over the course of NC 14 and its inferred promoter states based on a HMM. (B–E) Calculated values of (B) kon, (C) koff, (D) burst frequency, and (E) burst duration across the DV axis of MS2-yellow reporter gene trajectories driven by t48-WT (black), t48-optimizedDl1 (red), t48-optimizedDl2 (blue), and t48-optimizedZld (green) enhancers. All error bars are SD.
We plotted average values of kon and koff for all active nuclei along the DV axis for various t48 enhancer variants. We found that kon displayed a DV gradient similar to total mRNA production (Fig. 4B). koff, however, resulted in a flat expression along the DV axis (Fig. 4C). We found that optimization of Dl1, Dl2, or Zld sites led to an increase in kon across the expression domain while koff remained unchanged when optimizing Dl1 and Dl2 but decreased for Zld (Fig. 4 B and C). This result is supported by the current model of Zld as modulating proximal chromatin; a decrease in koff suggests slower unbinding which may be allowed by a more transcriptionally permissive chromatin state. Using the values of kon and koff, extracted using the HMM, we were able to calculate other kinetic parameters (42). Two kinetic parameters that stood out were burst frequency and burst duration. Burst frequency showed a slight increase for each optimized binding site while burst duration was longer only when optimizing the Zld site (Fig. 4 D and E). We also looked at the kinetics of the deletion of Dl1 and Dl2 and found a reduction of kon across the expression domain while koff remained the same (SI Appendix, Fig. S3 A and B). This corresponded to a lower burst frequency and no difference in burst duration compared to the control (SI Appendix, Fig. S3 C and D). In short, the two-state model indicates that modifications in Dl regulate transcription primarily by modulating burst frequency of transcriptional activation while Zld regulates transcription through both burst frequency and burst duration.
Discussion
The concept of enhancer grammar suggests that the highly tuned arrangement and strengths of binding sites on enhancers can contain both spatial and temporal information about gene control (18). Much work has been done to identify how a unique set of transcription factor binding sites in an enhancer asserts spatial control of gene expression (17, 44). However, much remains to be understood about how each binding site contributes to the overall transcription, and which aspect of transcription (e.g., timing of activation, amplitude, frequency of Pol II loading, etc.) is modulated. Here, we asked the question of whether enhancer grammar may encode temporal gene expression information, by modulating the strengths of binding sites for two key trascription factors that interact with the Drosophila t48 enhancer, Dl and Zld. We find that tuning the binding-site affinity of each transcription factor is responsible for both the spatial and temporal pattern of t48 expression. The initiation time of transcription serves as a “coarse-grained variable” that precedes all subsequent “fine-tuned” regulation, and our results suggest that this variable can be highly tuned by enhancer grammar.
It is notable that the expansion and reduction in t48 expression boundary upon Dl site modifications can be mostly attributed to the changes in activation kinetics, rather than the change in transcription amplitude (Fig. 2). Dl1 and Dl2 motifs contributed to overall t48 expression differently, with modifications in stronger Dl1 site resulting in bigger changes. While their relative affinity was different, we would also like to mention that the Dl1 site is located closer to the Zld site, and there could be a positioning effect between the two. Moreover, each Dl binding site works synergistically to contribute to the overall transcriptional dynamics of t48, such that an enhancer with multiple binding sites can produce more mRNA than the sum of mRNAs produced by enhancers with a single binding site (SI Appendix, Fig. S1B). We believe that such synergistic interactions between weak Dl binding sites allowed a high level of t48 mRNAs to be expressed in a narrow expression domain of t48 that requires a high Dl level.
Unlike Dl binding sites, changes in the Zld binding site led to significant changes in both the timing and the size of transcription (Fig. 3). This led the t48-optimizedZld enhancer to produce higher mRNA (Fig. 3D). In order to distinguish whether Zld regulates the amplitude of transcription by recruiting more Dl, or by working directly as a strong activator for t48, we used a construct where the t48 enhancer lacks both Dl1 and Dl2 sites but has an optimal Zld site and found that the transcriptional activity driven by Zld alone was minimal. While it is known that Zld works as a direct activator for a number of early genes, t48 seems to require multiple transcription factors to be activated, presumably due to a high nucleosome barrier (45). We believe that Zld works mainly to remodel the nucleosomes in the t48 enhancer, such that the t48 enhancer with an optimal Zld site has more accessible chromatin. This would allow more Dl to bind to the t48 enhancer, resulting in earlier transcriptional activation with a higher amplitude. Indeed, our results agree with other previous works that reported Zld’s role in promoting accumulation of other activators (37, 46). Finally, we successfully characterized the differential contribution of Dl and Zld using a simple mathematical model of transcription. Modifications in Dl binding site led to changes in bursting frequency only, while optimizing the Zld binding site resulted in both higher and longer bursting frequency and duration, respectively. It is a compelling result that two transcription factors Dl and Zld have a distinct role in regulation of gene expression dynamics.
Taken together, we suggest that changes in a single transcription factor binding site can significantly alter transcriptional kinetics, and the timing of transcriptional activation is a key parameter in gene control. The t48 gradient regulates myosin gradient, whose graded pattern is important for robust ventral furrow formation (5). It remains to be seen whether disruptions in t48 dynamics due to changes in transcription factor binding site arrangements in the enhancer lead to any phenotypic changes as well. Our results provide a paradigm in which an enhancer uses multiple transcription factor binding site interactions to drive robust spatiotemporal gene expression.
Materials and Methods
Motif Analysis of t48 Enhancer.
The previously described t48 enhancer is defined as the full-length one in this paper (12). Transcription binding sites in the full-length t48 enhancer were searched by FIMO (Find Individual Motif Occurrences) analysis with the motif matrices of Dl, Twi in ref. 30, and that of Zld (vfl_SANGER_5) at Fly Factor Survey (29, 47). The cut-off P value was set as P < 0.001.
Plasmids.
The transgenic lines were established using PhiC31-mediated site-specific integration (48). The minimal t48 enhancer was characterized by dividing the 1.3-kb full-length t48 enhancer into three fragments and finding the one that recapitulates the full pattern. Within the t48 minimal enhancer (523 bp), two Dl sites (Dl1 and Dl2) and one Zld site were mutated by PCR-mediated mutagenesis. The mutated enhancer was cloned into the MS2 reporter plasmid and sequenced for confirmation. The primers used for mutagenesis are listed in SI Appendix, Table S1.
Live Imaging.
yw;His2Av-mRFP;nanos>MCP-GFP virgins were crossed with males carrying the MS2 reporter genes (12). Embryos from the cross were then dechorionated and mounted in Halocarbon oil 27 (Sigma) between a semipermeable membrane (Sarstedt) and coverslip (18 mm × 18 mm). Embryos for Fig. 3 G–I and SI Appendix, Fig. S2 H–J were imaged using a Zeiss LSM 800 and Plan-Apochromat 40×/1.3 numerical aperture (N.A.) oil-immersion objective. Pixel size was set to 390 nm and a single image was 512 × 512 pixels. Images were created by taking the maximum projection of 16 z-stacked images separated by 0.75 µm. All other figures were imaged using a Zeiss LSM 880 and Plan-Apochromat 40×/1.3 N.A. oil-immersion objective. Pixel size was set to 461 nm and a single image was 512 × 512 pixels. The maximum projection of 30 z-stacked images separated by 0.5 µm was taken. The time resolution for each frame of the movie is 20 s. Images were captured in 16 bit. Three biological replicates were taken for each reporter line except for the control group where six replicates were taken.
Image Analysis.
Segmentation, nuclei tracking, and signal measurements were performed as described in ref. 12. Background subtraction was performed by subtracting the mean intensity of initial (nontranscribing) values within all nuclei of an individual embryo. All analyses were conducted using MATLAB (R2017b; MathWorks). Due to the variance in developmental time among replicates, the duration of NC14 was individually normalized for each embryo and we set the analysis window of NC14 to 45 min. Time of activation was set at the point where the MS2 signal went above 80% of the mean of the MS2 channel. To get spatial data, each embryo was divided into 16 bins across the DV axis. mRNA production was measured by taking the area under the curve of each MS2 trajectory over time and averaged over all nuclei within each bin. Averaging data from each bin for every replicate was carried out for all other spatial plots. The expression domain was calculated by interpolating nuclei output data at each time point, then taking the difference between the locations at half the maximum output.
Two-State Model Fitting.
Obtained nuclear traces were first subjected to the same background subtraction as described in the image analysis method. Because our subsequent analysis was to be a correlation between instantaneous fluorescence and instantaneous promoter state, we smoothened bursting trajectories with a moving window average with a width of four time points, corresponding to roughly 2 min, closer to the timescale of active transcription (a RNA Poll II molecule traversing the length of the gene).
An HMM approach was used to model promoter on/off states for fluorescent trances. We binned all trajectories into 16 bins along the DV axis in order to discretize states of the promoter. We then took the derivative of our binned trajectories to measure the change in fluorescence at every time point. We initialized a set of fully stochastic transition probabilities for the switching rates of the promoter and assumed that a positive change in fluorescence would indicate an active promoter state while a decrease or no change would correspond to an inactive promoter state. We did not assume the emission probabilities for different positive or negative fluorescence changes, and rather initialized them as equally probable. Using these initialization conditions, we fit state vectors to an HMM using the Baum–Welch algorithm (49). To increase the accuracy of spatial measurements, we binned the nuclei across the t48 expression domain and ran the Baum–Welch algorithm on the set of trajectories in each bin. The spatial bins were used to measure a “consensus” set of transition probabilities and emission probabilities for each spatial bin, and then these probabilities were reapplied to each individual burst trace in order to find the most likely sequence of (on/off) states, using the Viterbi dynamic programming algorithm (50). The learned transition matrix was used to calculate the kinetics of on/off transitions (kon/koff), by calculating the probability of transitioning per minute using experimental time steps. Using the kon and koff kinetics, we calculated burst frequency [] and burst duration ().
Data Availability.
Detailed methods are available in Materials and Methods and in SI Appendix. The scripts used in the paper are freely available on GitHub (https://github.com/limlab-upenn/t48).
Supplementary Material
Acknowledgments
We thank Evangelos Gatzogiannis for assistance with imaging and members of the B.L. laboratory for discussions. We also thank Mike Levine, Stas Shvartsman, and Arjun Raj for helpful comments on the manuscript. S.H.K. is funded from the Ashton Fellowship. S.G.J. is funded by NIH Grant F31AR075398. This work is funded by NIH Grant R35GM133425 to B.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The scripts used in the paper are freely available on GitHub (https://github.com/limlab-upenn/t48).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917040117/-/DCSupplemental.
References
- 1.Heisenberg C.-P., Bellaïche Y., Forces in tissue morphogenesis and patterning. Cell 153, 948–962 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Leptin M., Drosophila gastrulation: From pattern formation to morphogenesis. Annu. Rev. Cell Dev. Biol. 11, 189–212 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Martin A. C., Kaschube M., Wieschaus E. F., Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason F. M., Tworoger M., Martin A. C., Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat. Cell Biol. 15, 926–936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heer N. C. et al., Actomyosin-based tissue folding requires a multicellular myosin gradient. Development 144, 1876–1886 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spahn P., Reuter R., A vertex model of Drosophila ventral furrow formation. PLoS One 8, e75051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He B., Doubrovinski K., Polyakov O., Wieschaus E., Apical constriction drives tissue-scale hydrodynamic flow to mediate cell elongation. Nature 508, 392–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doubrovinski K., Swan M., Polyakov O., Wieschaus E. F., Measurement of cortical elasticity in Drosophila melanogaster embryos using ferrofluids. Proc. Natl. Acad. Sci. U.S.A. 114, 1051–1056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leptin M., Grunewald B., Cell shape changes during gastrulation in Drosophila. Development 110, 73–84 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Xie S., Martin A. C., Intracellular signalling and intercellular coupling coordinate heterogeneous contractile events to facilitate tissue folding. Nat. Commun. 6, 7161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa M., Wilson E. T., Wieschaus E., A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell 76, 1075–1089 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Lim B., Levine M., Yamazaki Y., Transcriptional pre-patterning of Drosophila gastrulation. Curr. Biol. 27, 286–290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J., Levine M., Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72, 741–752 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Ramos A. I., Barolo S., Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rushlow C. A., Han K., Manley J. L., Levine M., The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell 59, 1165–1177 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Steward R., Relocalization of the dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell 59, 1179–1188 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Hong J.-W., Hendrix D. A., Papatsenko D., Levine M. S., How the dorsal gradient works: Insights from postgenome technologies. Proc. Natl. Acad. Sci. U.S.A. 105, 20072–20076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markstein M., Markstein P., Markstein V., Levine M. S., Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 99, 763–768 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stathopoulos A., Levine M., Whole-genome analysis of Drosophila gastrulation. Curr. Opin. Genet. Dev. 14, 477–484 (2004). [DOI] [PubMed] [Google Scholar]
- 20.DeLotto R., DeLotto Y., Steward R., Lippincott-Schwartz J., Nucleocytoplasmic shuttling mediates the dynamic maintenance of nuclear Dorsal levels during Drosophila embryogenesis. Development 134, 4233–4241 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Kanodia J. S. et al., Dynamics of the Dorsal morphogen gradient. Proc. Natl. Acad. Sci. U.S.A. 106, 21707–21712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nien C.-Y. et al., Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dufourt J. et al., Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat. Commun. 9, 5194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang H.-L. et al., The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foo S. M. et al., Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr. Biol. 24, 1341–1346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y. et al., Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolsch V., Seher T., Fernandez-Ballester G. J., Serrano L., Leptin M., Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science 315, 384–386 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Brodland G. W. et al., Video force microscopy reveals the mechanics of ventral furrow invagination in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 22111–22116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yáñez-Cuna J. O., Dinh H. Q., Kvon E. Z., Shlyueva D., Stark A., Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 22, 2018–2030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A. et al., JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stathopoulos A., Levine M., Dorsal gradient networks in the Drosophila embryo. Dev. Biol. 246, 57–67 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Chopra V. S., Levine M., Combinatorial patterning mechanisms in the Drosophila embryo. Brief. Funct. Genomics Proteomics 8, 243–249 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Ip Y. T., Kraut R., Levine M., Rushlow C. A., The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell 64, 439–446 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Reeves G. T. et al., Dorsal-ventral gene expression in the Drosophila embryo reflects the dynamics and precision of the dorsal nuclear gradient. Dev. Cell 22, 544–557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim B. et al., Kinetics of gene derepression by ERK signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 10330–10335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada S. et al., The Drosophila pioneer factor Zelda modulates the nuclear microenvironment of a dorsal target enhancer to potentiate transcriptional output. Curr. Biol. 29, 1387–1393.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raj A., Peskin C. S., Tranchina D., Vargas D. Y., Tyagi S., Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chubb J. R., Trcek T., Shenoy S. M., Singer R. H., Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukaya T., Lim B., Levine M., Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peccoud J., Ycart B., Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 48, 222–234 (1995). [Google Scholar]
- 42.Zoller B., Little S. C., Gregor T., Diverse spatial expression patterns emerge from unified kinetics of transcriptional bursting. Cell 175, 835–847.e25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berrocal A., Lammers N., Garcia H. G., Eisen M. B., Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene. bioRxiv:10.1101/335901 (11 June 2018). [DOI] [PMC free article] [PubMed]
- 44.Farley E. K. et al., Suboptimization of developmental enhancers. Science 350, 325–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stampfel G. et al., Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature 528, 147–151 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Mir M. et al., Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 7, e40497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enuameh M. S. et al., Global analysis of Drosophila Cys2-His2 zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 23, 928–940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venken K. J. T., He Y., Hoskins R. A., Bellen H. J., P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Baum L. E., Petrie T., Statistical inference for probabilistic functions of finite state Markov chains. Ann. Math. Stat 37, 1554–1563 (1966). [Google Scholar]
- 50.Viterbi A., Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory 13, 260–269 (1967). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed methods are available in Materials and Methods and in SI Appendix. The scripts used in the paper are freely available on GitHub (https://github.com/limlab-upenn/t48).