Significance
CAR T cells have shown significant promise in treating hematopoietic cancers but challenges remain, including antigen loss and identification of targets in solid tumors. Aberrant O-linked glycosylation in solid cancers is common, often resulting in expression of cell-surface neoantigens. We used directed evolution to engineer the binding site of an antibody so variants reacted more broadly with tumor-specific glycoprotein epitopes containing Tn (GalNAc-O-S/T). As CARs, these variants showed improved broader activity against mouse and human cancer cells with dysregulated O-glycosylation. Even cancer cells without MUC1, the most studied cancer-associated glycoantigen, were recognized by the engineered CARs. Cancer-specific recognition was retained due to Tn-antigen requirement, but broader activity against the glycoprotein backbone provides opportunities for potent use with minimal antigen-loss escape.
Keywords: CAR T cells, antibody engineering, neoantigen epitopes
Abstract
The potency of adoptive T cell therapies targeting the cell surface antigen CD19 has been demonstrated in hematopoietic cancers. It has been difficult to identify appropriate targets in nonhematopoietic tumors, but one class of antigens that have shown promise is aberrant O-glycoprotein epitopes. It has long been known that dysregulated synthesis of O-linked (threonine or serine) sugars occurs in many cancers, and that this can lead to the expression of cell surface proteins containing O-glycans comprised of a single N-acetylgalactosamine (GalNAc, known as Tn antigen) rather than the normally extended carbohydrate. Previously, we used the scFv fragment of antibody 237 as a chimeric antigen receptor (CAR) to mediate recognition of mouse tumor cells that bear its cognate Tn-glycopeptide epitope in podoplanin, also called OTS8. Guided by the structure of the 237 Fab:Tn-OTS8-glycopeptide complex, here we conducted a deep mutational scan showing that residues flanking the Tn-glycan contributed significant binding energy to the interaction. Design of 237-scFv libraries in the yeast display system allowed us to isolate scFv variants with higher affinity for Tn-OTS8. Selection with a noncognate human antigen, Tn-MUC1, yielded scFv variants that were broadly reactive with multiple Tn-glycoproteins. When configured as CARs, engineered T cells expressing these scFv variants showed improved activity against mouse and human cancer cell lines defective in O-linked glycosylation. This strategy provides CARs with Tn-peptide specificities, all based on a single scFv scaffold, that allows the same CAR to be tested for toxicity in mice and efficacy against mouse and human tumors.
Tn antigens are a unique class of cancer-associated neoantigens that provide an opportunity to target cancer cells, without the recognition and destruction of normal tissue. These antigens arise from dysregulation of the cellular glycosylation machinery, leading to abnormal glycosylation of surface proteins on cancer cells and appearance of truncated immature glycans such as Tn (1–4). Tn-dependent antibodies have been configured as chimeric antigen receptors (CARs) and bispecific T cell engaging agents (BiTEs) (5, 6). The 237-monoclonal antibody is an example of such an antibody that recognizes a Tn antigen (GalNAcα1-O-Ser/Thr O-glycan) specifically on the protein called OTS8 or podoplanin (referred to as Tn-OTS8 here on) expressed on the surface of the Ag104A murine cancer line (7, 8). The Tn antigen can arise due to a number of glycosylation defects, including mutations in the protein encoded by the Cosmc gene. Cosmc functions as a chaperone for the β-3-galactosyltransferase (C1GALT1) that extends the initial GalNAc residue in O-glycosylation of proteins such as the OTS8 protein (8–10).
The binding site of the 237-antibody interacts with both the GalNAc moiety and the peptide region of the Tn-OTS8 glycopeptide epitope (11). While peptide specificity is presumably conferred by the 237:Tn-OTS8 interaction, the exact contribution of the side chains of flanking residues has not been studied. To define this specificity, as a prelude to engineering the 237-antibody using yeast display (12) and structure-guided design principles (13), here we conducted a deep mutational scan of the entire OTS8 peptide epitope. We show that many of the residues flanking the Tn-threonine epitope, also provide binding energy to the interaction and thus confer a significant degree of peptide specificity. This finding accounts for the observed 237 specificity for the aberrant Tn-glycoform of OTS8 (8) and suggests that it should be possible to evolve the 237-binding site to bind to alternative peptide side chains.
The effectiveness of 237 in targeting Tn-OTS8 and recognizing Ag104A as a 237-CAR and as a 237-BiTE (bispecific T cell engager) has been demonstrated (5). We also showed that the 237-CAR was stimulated by (5), and eradicated (14), the human T cell leukemia line Jurkat, which does not express murine OTS8. However, Jurkat contains a truncating mutation in Cosmc and expresses all O-glycoproteins with the Tn-glycoform (15), and thus we presumed that other Tn-glycopeptide epitopes could serve as the target for 237-CAR on Jurkat. Although our studies (5, 14) indicated that the 237-CAR could recognize Tn-glycoprotein antigens on human cancers, the ability of Jurkat to stimulate 237-CAR cytokine release was not as robust as with the cognate antigen on Ag104A, possibly due to the lower affinity of 237 for these alternative Tn-peptide backbones.
With this in mind, here we decided to take two approaches to further understand and optimize the activity mediated by the 237-CAR. First, given previous studies showing that affinity of the scFv in CARs impacts activity (e.g., refs. 16–18), we used yeast display of 237-scFv-CDR libraries to isolate a variant with 30-fold higher affinity. Using the same libraries, we used a Tn-MUC1 glycopeptide to isolate 237-scFv specificity variants that reacted with Tn-MUC1 as well as Tn-OTS8. Each of the affinity-matured and specificity variants were expressed as CARs and tested for IFN-γ stimulatory activity against a variety of cell lines. The affinity-matured CAR showed only modestly higher levels of activity against mouse Ag104A and ID8 cell lines, compared to the wild-type (WT) CAR. However, the specificity variants mediated dramatically higher activity against the human tumor lines tested (Jurkat and SKOV3-Cosmc−/−, and their MUC1 knockouts [KO]), when compared to wild-type 237-CAR. The engineered CARs also retained strong activity for mouse tumor lines. Thus, structure-guided engineering of the single 237-scFv scaffold allowed for the selection of CARs with broader cross-reactivity with human O-glycoproteins carrying aberrant Tn-glycans that mediated more efficient recognition of these cancer-associated antigens.
Results
Deep Mutational Scan of the 237-Epitope.
To assess the binding contribution of the peptide side chains in the OTS8 epitope, we conducted a deep mutational scan. Deep mutational scans are a relatively recent approach to understanding, at a very detailed level, the role of each amino acid residue in a protein:protein interface (19, 20). In the past, alanine scans were performed to assess the role of individual side chains in the binding site of a protein (21). However, this approach does not sample the other 18 possible side chains, and their chemistries, that are relevant to the issue of specificity. To examine the OTS8 epitope, we developed a system that would allow single codon libraries (SCLs) (19, 20, 22–26) of OTS8 to be expressed as cell-surface proteins in Jurkat (SI Appendix, Fig. S1).
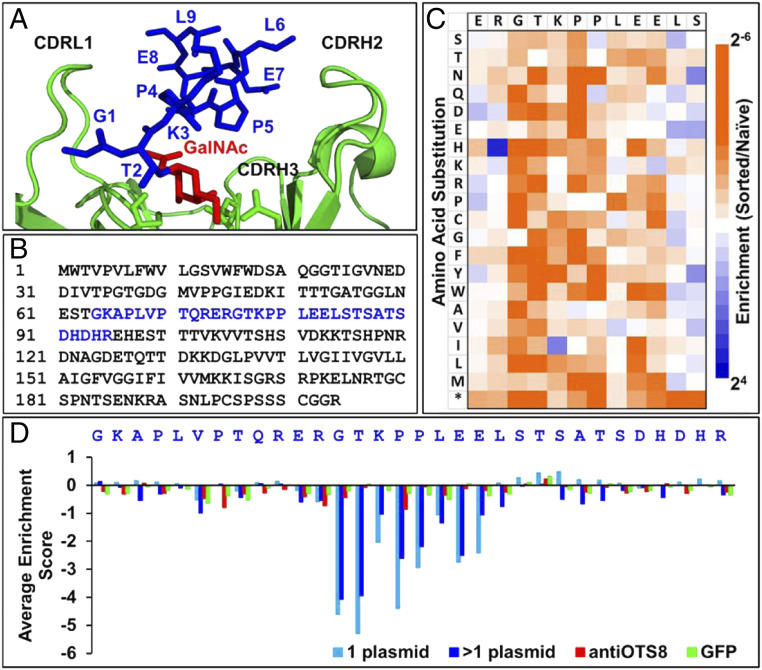
We generated libraries in each of the 10 codons for residues predicted to include the Tn-linked epitope, as observed in the 237-crystal structure (Fig. 1A). As controls, libraries in the 12 codons flanking the N terminus and, 10 codons flanking the C terminus of this region were also generated (Fig. 1B, full sequence scanned, in blue). After cloning into the pMFG retrovirus vector, PCR products of the SCL were transduced into Jurkat, yielding a library of 105 independent transduced cells (exceeding by about 100-fold the theoretical diversity of the combined SCL, 32 positions × 32 codons ∼103). Transduction efficiency was 5% and transduced cells were sorted for GFP to obtain a library of adequate purity and size. This population was sorted with three different probes: 1) 237-IgG, 2) an anti-OTS8 mAb, and 3) GFP. After selection of the top 1% fluorescent population, RNA was isolated, converted to cDNA, and subjected to deep sequencing. Bioinformatics software (22, 23) was used to analyze the numbers of sequences obtained for each mutant in the library, and these were compared to the frequencies in unselected library in order to generate heat maps of relative enrichment values (SI Appendix, Fig. S2). Blue indicates the mutant was found at higher frequencies than the wild-type OTS8, white indicates it was found at the same frequency as wild-type OTS8, and orange indicates it was found at lower frequencies than the wild-type OTS8. The scale is represented in log base2 units (e.g., a value of −4 indicates that the mutant was found at 16-times lower frequency than in the unselected library). Experimental variability, as discussed previously (19, 22), is revealed by the internal controls of this experiment (i.e., the 22 flanking codons of the epitope, and selections with the anti-OTS8, and with GFP).
Fig. 1.
Deep mutational scan of the 237-epitope. (A) Binding site of Tn-OTS8 peptide (blue, with GalNAc in red) in 237-monoclonal antibody (green) structure (Protein Data Bank [PDB]: 3IET). (B) Sequence of OTS8 protein, with residues that were subjected to deep mutational scan in blue. (C) Deep mutational scan of OTS8 peptide based on selection of SCLs with 237-IgG. Enrichment or depletion of substitutions was calculated relative to naive (unselected) libraries as a log2 ratio. Resultant enrichment scores were plotted on a color-coded scale ranging from ≤2−6 (orange) to ≥24 (blue). Stop codon is indicated by an *. (D) Average enrichment scores (log2 ratio) for all substitutions in OTS8 SCL when sorted with 237-IgG (blue), anti-OTS8 (red), and GFP (green).
The GFP-sorted heat map shows minimal enrichment or depletion of mutants, generally falling within the expected distribution around the value of 1. For the anti-OTS8 selected population, the primary mutations that yielded “orange” values were found in the stop codons for each of the 32 codon positions, as would be expected since selection required translation and surface expression of the full OTS8 protein (SI Appendix, Fig. S2). The selections that involved 237-IgG showed significant depletion of mutants with stop codons but also of mutations that were present within the epitope GTKPPLEE (Fig. 1C and SI Appendix, Fig. S2).
Analysis of the average depletion values for the library (Fig. 1D) showed that the order of residue impact was: T2 > G1 > P4 > P5 > E7 > E8 > K3 > L6. The results are consistent with the threonine being necessary for the O-linked GalNAc (8, 27), and the location of other residues within the binding site. Thus, the peptide region conferred a significant degree of specificity, and requirement for the threonine-linked GalNAc, which is found deep in the binding site (Fig. 1A). The binding contributions of peptide side chains supports the possibility that 237-scFv variants in complementarity determining region (CDR) residues that bind with improved affinity to alternative Tn-peptides could be selected.
Engineering Higher-Affinity 237-scFv Variants.
A scFv fragment used as a CAR requires optimal affinity for mediating activity, yet it also must remain cancer specific. Many scFv used as CARs have affinities that are higher than 237 (KD = 140 nM) (11). For example, the scFv used in the FDA-approved FMC63 CAR against CD19 for the treatment of acute lymphoblastic leukemia (ALL), and relapsed or refractory large B cell lymphoma exhibits an affinity of 5 nM as measured by titrations using flow cytometry, and 300 pM as measured for the scFv using surface plasmon resonance (SPR) (18, 28–30). Another high-affinity scFv against folate receptor beta (KD = 2.5 nM) was used as a CAR in a mouse model of human acute myeloid leukemia (AML), and against primary human AML (31). Density of the cancer antigen also has an impact on the affinity requirement—i.e., a low-density target may require a high-affinity CAR for efficient targeting; however, its affinity may need fine tuning to discriminate tumor from normal tissue expressing low levels of antigen (17, 32, 33).
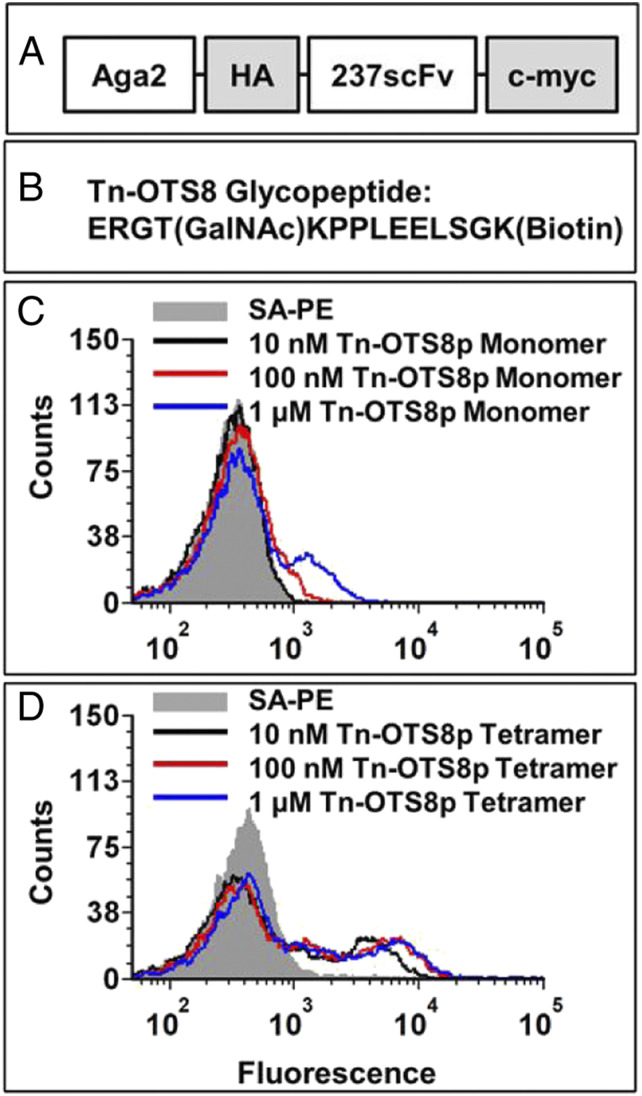
To determine if the 237-CAR could be improved by increasing its affinity against cognate antigen (Tn-OTS8), the 237-scFv was engineered using yeast display (Fig. 2A). A biotinylated synthetic peptide, Tn-OTS8p (ERGT[GalNAc]KPPLEELSGK-biotin) (Fig. 2B) was used as both a monomer and tetramer to stain yeast cells expressing the wild-type 237-scFv. Monomer staining showed detectable binding at 100 nM and above (Fig. 2C), consistent with the reported KD value (140 nM) of the 237-Fab fragment (11). Tetramers on the other hand showed significantly greater binding, even at 10 nM, due to the avidity of the interaction (Fig. 2D).
Fig. 2.
Yeast surface display and antigen binding of 237-scFv. (A) Schematic of 237-scFv cloned in yeast display vector. (B) Sequence of Tn-OTS8 peptide. (C) Yeast-displayed 237-scFv was stained with various concentrations of biotinylated Tn-OTS8 peptide, followed by streptavidin-phycoerythrin (SA-PE). Binding was measured by flow cytometry. The staining profile of yeast cells stained with streptavidin-PE only, is shown in gray. Similar results were obtained in more than three independent experiments. (D) Yeast-displayed 237-scFv was stained with various concentrations of tetramers of Tn-OTS8 peptide made with streptavidin-PE. Binding was measured by flow cytometry. The staining profile of yeast cells stained with streptavidin-PE only, is shown in gray. Similar results were obtained in more than three independent experiments.
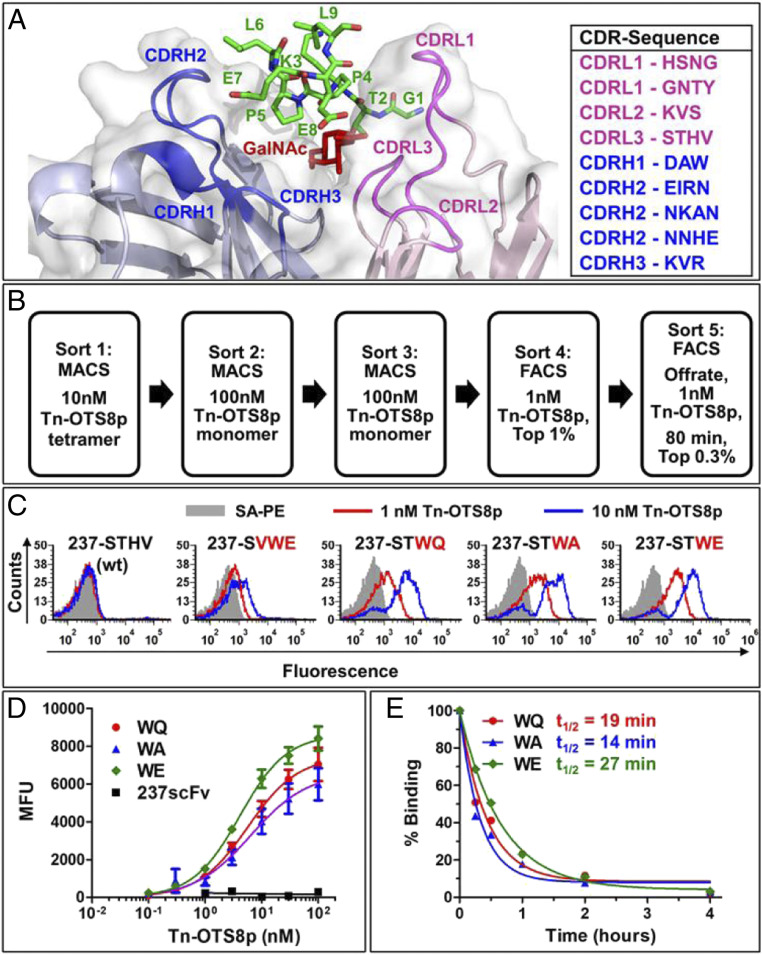
A structure-guided approach was used to design libraries of 237-scFv mutants in residues that are in proximity, or mediate binding, to the antigen (i.e., the sugar-binding, and peptide-binding residues of 237 that contact Tn-OTS8) (11). Nine libraries in CDRs of the heavy and light chains of the 237-scFv (Fig. 3A) were constructed. Each library showed diversity that exceeded the theoretical maximum (SI Appendix, Table S1). The pooled yeast libraries were induced and subjected to a combination of magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS) with tetrameric or monomeric Tn-OTS8 peptide (Fig. 3B). After sorts 4 and 5, 10 237-scFv mutants in each were isolated and analyzed for binding to low concentrations (1 or 10 nM) of Tn-OTS8 peptide, and compared with the parent 237-scFv. All mutants exhibited higher binding with Tn-OTS8 peptide compared to the parental 237-scFv (SI Appendix, Fig. S3). Sequencing indicated that there were seven different mutants, each derived from a library in CDR3 of the 237-light chain (CDR3L), which is in proximity to the GalNAc on OTS8 peptide (Fig. 3A) (11).
Fig. 3.
Engineering 237-scFv for higher affinity to Tn-OTS8 peptide. (A) Crystal structure of 237-monoclonal antibody with CDRs in proximity to the Tn-OTS8 peptide. Wild-type residues in each CDR where libraries were generated are shown on the Right. (B) Sorting scheme used to isolate 237-scFv mutants that bind with high affinity to Tn-OTS8 peptide. (C) Yeast-displayed 237-scFv as well as selected mutants in CDRL3 were stained with low concentrations (1 and 10 nM) of biotinylated, Tn-OTS8 monomeric peptide, followed by streptavidin-PE. Binding was measured by flow cytometry. The staining profile of yeast cells stained with streptavidin-PE only, is shown in gray. Similar results were obtained for the binding of 237 (WT), WQ, WA, and WE mutants to Tn-OTS8 peptide in three independent experiments. VWE mutant was analyzed in a single experiment. (D) Yeast-displayed 237-scFv and mutants (WQ, WA, and WE) were stained with various concentrations of biotinylated, Tn-OTS8 peptide, followed by streptavidin-PE. Mean fluorescence units (MFUs) were plotted versus peptide concentration to obtain apparent dissociation constants for each mutant. Similar results were obtained for the binding of WE mutant to Tn-OTS8 peptide in four independent experiments. The 237 (WT), and mutants WQ and WA were analyzed in two independent experiments. (E) In order to determine off rates, each yeast-displayed 237-scFv mutant was stained with 30 nM biotinylated Tn-OTS8 peptide. After washing, the mutants were incubated with 100 nM OTS8 peptide (not biotinylated). The decrease in binding to biotinylated Tn-OTS8 peptide by each mutant was measured over a period of 4 h, to calculate half-lives (t1/2) and off rates (koff) in a single experiment. Error bars in D represent SEM.
Three 237-scFv mutants (WQ, WA, and WE) showed among the highest levels of staining (Fig. 3C). These were further analyzed by flow cytometry-based Tn-OTS8 peptide binding titrations (Fig. 3D). Estimated KD values, based on the half-maximal binding concentrations, were shown to be 6 ± 1 nM, 6 ± 2 nM, and 4 ± 1 nM, respectively. Thus, each of these exhibit affinities that are about 30 times higher than the parental scFv. We also performed an off-rate assay using the yeast-displayed variants, showing that the WQ, WA, and WE variants exhibited t1/2 values of 19 min, 14 min, and 27 min, respectively (Fig. 3E). Although the off rate of the parental 237-scFv was too fast to measure in this assay (full dissociation even at the first time point), in comparison to the off rate measured previously by SPR (11), the WQ, WA, and WE variants all exhibited significantly longer off rates, consistent with the affinity measurements.
The wild-type residues (His93 and Val94) both lie within 5 Å of the GalNAc in the 237/Tn-OTS8 structure (SI Appendix, Fig. S4A). To gain some understanding of the mechanism of the higher-affinity mutations, the tryptophan and glutamic acid of the WE variant were modeled into the cocrystal structure of 237-IgG with Tn-OTS8 peptide (11). The position of these residues in the model (SI Appendix, Fig. S4B) suggested that one mechanism responsible for the increase in affinity may be additional polar contacts of the GalNAc moiety with the 237-WE mutant.
Engineering 237-scFv Variants with Broader Tn-Peptide Specificities.
As evidenced by the deep mutational scan, the 237-antibody has binding energy contributed by peptide side chains of the Tn-OTS8 protein. We reasoned that if we could broaden this reactivity and affinity for additional Tn-peptide epitopes, we could use such variants as CARs with more effective activity against tumors that express alternative Tn-linked targets. Since the original cloning and characterization of mucins (34–36), MUC1 has been among the Tn antigens that have been most studied (e.g., ref. 37). Thus, we used enzymatic synthesis to generate glycosylated MUC1 peptide for selections of the 237-scFv-yeast displayed libraries. Synthetic MUC1-derived peptide (biotin-KVTSAPDTRPAGSTAPPAHG) was incubated with N-acetylgalactosaminyltransferase-2, and the product was analyzed by ELISA with the Tn-MUC1 antibody 5E5 (37–39). Only the enzymatically treated peptide was found to be reactive with the 5E5 antibody (SI Appendix, Fig. S5).
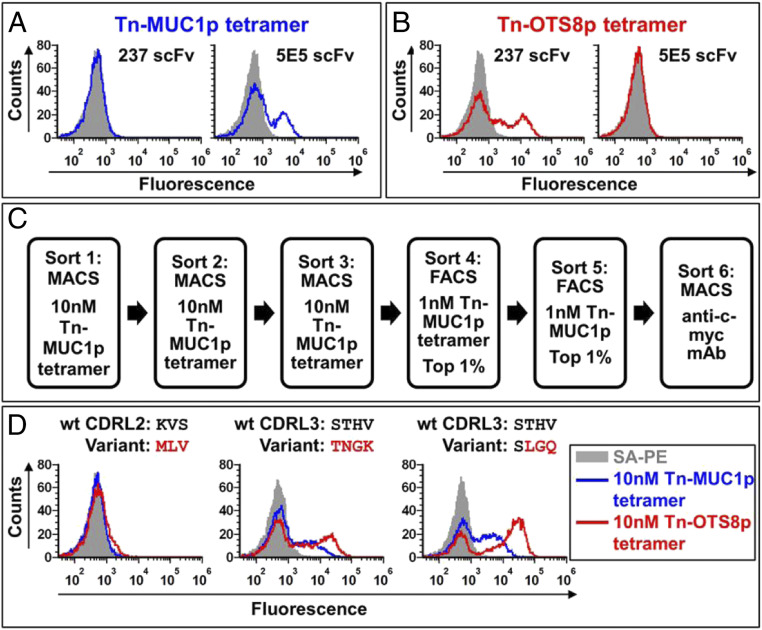
To show that the Tn-MUC1 reagent could be used for selections of the yeast display library, we cloned the 5E5 scFv into the yeast display vector and showed that it, but not the 237-scFv yeast, stained with a tetramer of the Tn-MUC1p (Fig. 4A). Conversely, the 237-scFv displayed on yeast, but not the 5E5, stained with the Tn-OTS8p tetramer (Fig. 4B).
Fig. 4.
Engineering 237-scFv for broader Tn-peptide specificity. (A) Flow cytometric measurement of binding of yeast-displayed 237-scFv or 5E5 scFv to 10 nM Tn-MUC1p tetramers prepared with streptavidin-PE (blue). Staining profile of cells stained with streptavidin-PE only, is shown in gray. Similar results were obtained in more than three independent experiments. (B) Flow cytometric measurement of binding of yeast-displayed 237-scFv or 5E5 scFv to 10 nM Tn-OTS8p tetramer prepared with streptavidin-PE (red). Staining profile of cells stained with streptavidin-PE only, is shown in gray. Similar results were obtained in more than three independent experiments. (C) Sorting scheme used to isolate 237-scFv mutants that have broader Tn-peptide specificity. (D) Yeast-displayed 237-scFv mutants (MLV, TNGK, and LGQ) were stained with 10 nM Tn-MUC1p or Tn-OTS8 peptide tetramers made with streptavidin-PE. Binding was measured by flow cytometry. The staining profile of yeast cells stained with streptavidin-PE only, is shown in gray. Similar results were obtained for the binding of TNGK and LGQ variants to the tetramers in two independent experiments. MLV variant was analyzed in a single experiment.
To identify 237-scFv variants that would react more effectively with other Tn-peptides, we sorted the pooled 237-CDR libraries through various rounds with the biotinylated Tn-MUC1p using both MACS and FACS (Fig. 4C). Because sequences of clones after the fourth and fifth sorts revealed some mutants with stop codons or frameshifts, we also included a final sort with anti-c-myc antibody against the C-terminal tag. Sequencing of 17 clones from these sorts yielded some contaminating clones with the 5E5 sequence, but also three unique, in-frame 237-scFv variants, one in CDRL2 and two in CDRL3. The two CDRL3 variants contained the sequences TNGK or SLGQ in place of the parental sequence STHV, and both of these variants were reactive with Tn-MUC1 and Tn-OTS8 (Fig. 4D). Thus, despite having libraries in all six CDRs, both affinity selections with Tn-OTS8 and specificity selections with Tn-MUC1 identified variants only in the single CDR, L3.
Activity of T Cells Expressing a 237-CAR with Higher Affinity.
To determine if 30-fold higher affinity resulted in enhanced activity against the cognate antigen, Tn-OTS8, the WE mutations were introduced into 237-CAR (237scFv-CD28-CD3zeta) to generate 237-WE-CAR. Activated T cells from C57BL/6 mice were transduced and cell surface expression of the parental 237 and the WE-CARs was confirmed with the biotinylated Tn-OTS8 tetramers (SI Appendix, Fig. S6A). The transduced T cells, compared to mock T cells, were incubated with Ag104A which expresses the cognate Tn-OTS8 antigen. As a control, the mouse cell line 58−/− was used at various target-to-effector ratios. Both 237- and WE-CARs were stimulated effectively to release IFN-γ by Ag104A, but WE-CAR was only slightly more effective than the wild-type 237-CAR. The antigen-negative line 58−/− was unable to stimulate either CAR (SI Appendix, Fig. S6B).
Activity of 237-scFv-Derived CAR T Cells with Immobilized Synthetic Glycopeptides.
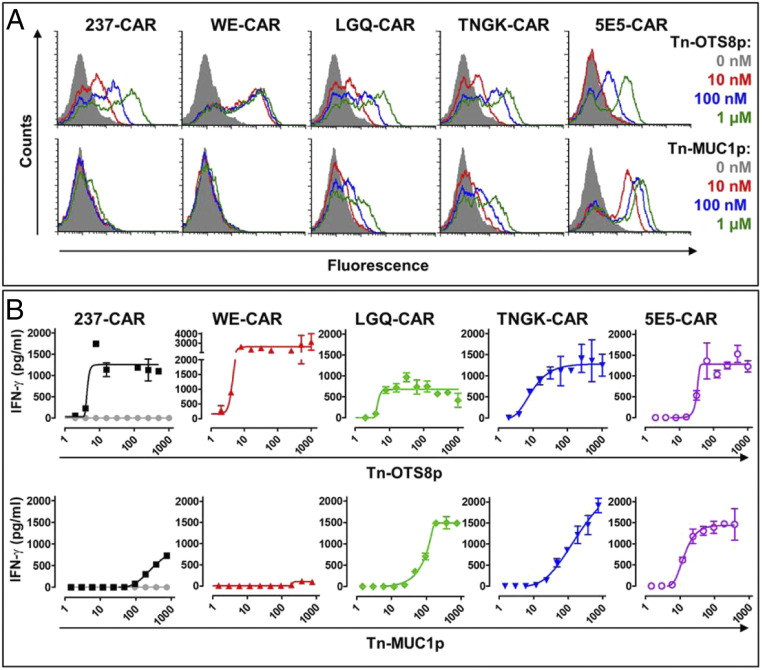
To examine if the Tn-MUC1 selected variants would demonstrate enhanced activity against noncognate Tn targets when used as CARs, we cloned the two variant sequences (TNGK and LGQ) into the parental 237-scFv CAR and transduced activated T cells with these, the 237-parental CAR, the higher-affinity variant WE-CAR, and the 5E5-CAR. To compare the affinities of the surface expressed CARs, we titrated the transduced T cells for each of the CARs with various concentrations of the Tn-OTS8 or Tn-MUC1 monomers (Fig. 5A). All five CARs bound to the Tn-OTS8 monomers, although the WE-CAR showed the highest affinity and the 5E5-CAR bound with the lowest affinity, as expected. Also as expected, the TNGK- and LGQ-CARs both bound well to the Tn-MUC1 monomer (estimated KD values of about 100 nM), but below the affinity of the 5E5-CAR. The parental 237-CAR revealed slight binding to the highest concentration (1,000 nM) of Tn-MUC1, whereas binding of the high-affinity WE variant appeared negligible even at the highest concentration.
Fig. 5.
Tn-peptide binding and activity of CAR-transduced T cells. (A) Splenic T cells from C57/BL6 mice were mock transduced or transduced with 237-CAR (the wild-type), WE-CAR, LGQ-CAR, TNGK-CAR, or 5E5-CAR. Transduced T cells were stained with various concentrations of Tn-OTS8 or Tn-MUC1 biotinylated monomeric peptides, followed by streptavidin-PE to assess binding by flow cytometry. Histograms of cells stained with streptavidin-PE only are shown in gray. Similar results were obtained in three independent experiments. (B) To assess activation, mock (shown in gray), 237-CAR, WE-CAR, LGQ-CAR, TNGK-CAR, or 5E5-CAR transduced T cells were added to plates containing various concentrations of immobilized Tn-OTS8 or Tn-MUC1 peptide and incubated at 37 °C, 5% CO2. After 24 h, IFN-γ released in the culture supernatants was measured by ELISA. Activation of transduced cells was measured in duplicate. The 237-, TNGK-, and 5E5-CARs were assessed in three independent experiments. WE- and LGQ-CARs were assessed in one experiment. Error bars indicate SEM.
The CAR-transduced T cells were examined for the ability of plate-immobilized Tn-glycopeptides Tn-OTS8p and Tn-MUC1p to stimulate IFN-γ release (Fig. 5B). All five CARs mediated significant activity when stimulated with immobilized Tn-OTS8p, although the highest-affinity variant WE exhibited a slight increase in sensitivity and maximal IFN-γ release. Among the 237-derived CARs, the TNGK-and LGQ-CARs exhibited the greatest activity with immobilized Tn-MUC1p, whereas the 237-parental CAR showed about 10-fold lower sensitivity. As expected, since it was generated against Tn-MUC1p (38), 5E5 had the greatest sensitivity against immobilized Tn-MUC1p. The WE affinity variant showed minimal activity, consistent with its negligible binding to Tn-MUC1p.
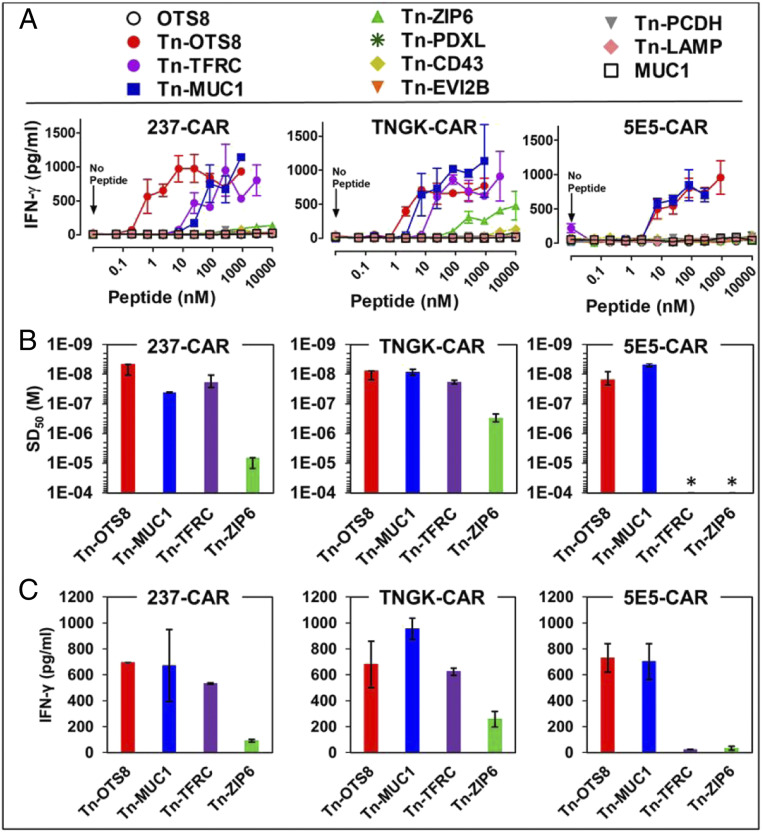
Recently, we showed that 237-CAR T cells are stimulated by several known human Tn-glycopeptides that were originally identified in Jurkat cell line (14, 40). It is possible that selection of 237-scFv variants for increased binding to the Tn-MUC1 glycopeptide could yield reductions in, or even loss of, binding to these other Tn-peptides. To examine this, we immobilized the Tn-peptides and examined cytokine release from 237-, TNGK-, and 5E5-CARs (Fig. 6A). The TNGK-CAR not only retained activity against the other peptides, but demonstrated increased sensitivity (lower SD50, Fig. 6B) against several of them (Tn-MUC1, Tn-ZIP6, and Tn-TFRC). Interestingly, these peptides exhibited some similarity to the residues in the OTS8 antigen, in that the residues around the Tn-linked threonine were conserved to some extent (i.e., GTKPP in OTS8, GSTAPP in MUC1, GTESP in TFRC, and STPPS in ZIP6) (Fig. 1C). In contrast, the 5E5-CAR exhibited detectable activity against only Tn-OTS8 and Tn-MUC1 (Fig. 6C). These results suggest that the TNGK-CAR is preferable, as it has broader reactivity against cancers that express different Tn O-glycoproteins.
Fig. 6.
Activity of 237-derived CAR T cells against additional human Tn-peptides. (A) To assess activation, mock, 237-CAR, TNGK-CAR, or 5E5-CAR transduced T cells were added to plates containing various concentrations of the indicated Tn-peptides and incubated at 37 °C, 5% CO2. After 24 h, IFN-γ released in the culture supernatants was measured by ELISA. Activation of transduced cells under each condition was measured in duplicate in two independent experiments. Error bars indicate SEM. (B) Sensitization doses that yielded half-maximal stimulation (SD50) of IFN-γ release were calculated for the active Tn-peptides using nonlinear regression analysis (GraphPad Prism 5). 5E5-CAR was not activated with TFRC and ZIP6; hence the SD50 values were assigned values of >10−4 M (indicated by an *). SD50 values from two independent experiments are plotted as replicates. (C) Maximum IFN-γ release for the active peptides was determined at the 1-μM concentration. Mean ± SD is plotted for B and C.
Activity of 237-scFv-Derived CAR T Cells with Tumor Cell Lines.
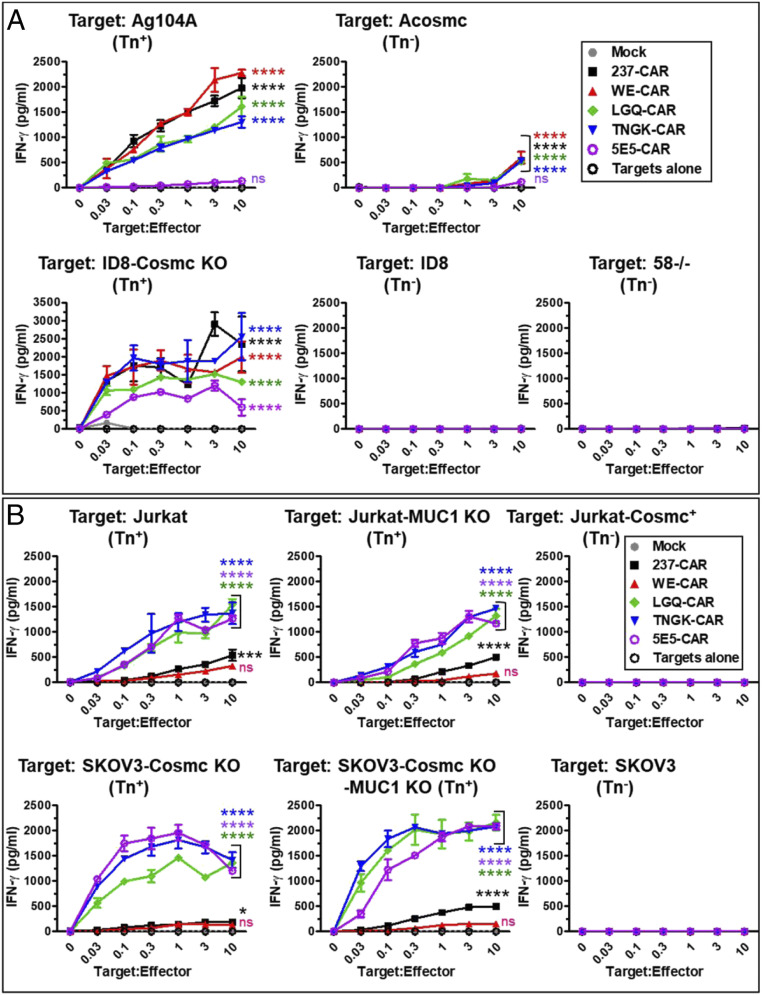
To determine the impact of the engineered CARs on tumor recognition, CAR-transduced T cells were incubated, at various target-to-effector ratios, with a panel of mouse tumor cell lines: Ag104A, ACosmc (Ag104A with WT Cosmc), ID8, ID8-Cosmc KO, and the T lymphoma 58−/− (Fig. 7A). As might have been predicted from the Tn-OTS8 binding titrations, the four 237-derived CARs were very sensitive to stimulation by the cognate antigen expressed on Ag104A target cells. The 5E5-CAR exhibited lower cytokine release induced by Ag104A and ID8-Cosmc KO, consistent with lack of detectable binding to 10 nM Tn-OTS8p (Fig. 5A) and reduced sensitivity with immobilized Tn-OTS8 (Fig. 5B). All CARs were negative for stimulation with the control lines ACosmc and 58−/− (we speculate that the IFN-γ released at very high target-to-effector ratios of ACosmc may be due to release of intracellular, partially O-linked OTS8 at these high cell densities). Although all CARs were stimulated with ID8-Cosmc KO, the 5E5-CAR exhibited the lowest sensitivity and IFN-γ release.
Fig. 7.
Activity of 237-derived CAR T cells against various mouse and human cancer cell lines. (A) Mock, 237-CAR, WE-CAR, TNGK-CAR, LGQ-CAR, or 5E5-CAR transduced T cells were cocultured with Tn antigen expressing murine target cells (Ag104A or ID8 Cosmc KO) or with control cell lines (ACosmc, ID8, or 58−/−), for 24 h at 37 °C, 5% CO2 at various target-to-effector cell ratios. IFN-γ released in the supernatants under each coculture condition was measured by ELISA. Activation of transduced cells was measured in duplicate in two independent experiments. (B) Mock, 237-CAR, WE-CAR, TNGK-CAR, LGQ-CAR, or 5E5-CAR transduced T cells were cocultured with Tn antigen expressing human target cells (Jurkat, Jurkat-MUC1 KO, SKOV3-Cosmc KO, or SKOV3-Cosmc KO-MUC1 KO) or with control cell lines (Jurkat-Cosmc+ or SKOV3), for 24 h at 37 °C, 5% CO2 at various target-to-effector ratios. IFN-γ released in the supernatants under each coculture condition was measured by ELISA. Activation of transduced cells was measured in duplicate in two independent experiments. Error bars indicate SEM. Significance of difference in activity of each CAR compared to control (mock) was determined by two-way ANOVA followed by Dunnett’s test using GraphPad Prism 8.0. ns refers to P ≥ 0.05; * refers to P = 0.01 to 0.05, *** refers to P = 0.0001 to 0.001, and **** refers to P < 0.0001.
CAR-transduced T cells were also examined at various target-to-effector ratios with a panel of human tumor cell lines: Jurkat, Jurkat-MUC1 KO, Jurkat-Cosmc+, SKOV3, SKOV3-Cosmc KO, and SKOV3-Cosmc KO-MUC1 KO (Fig. 7B). Most importantly, CARs containing the two Tn-MUC1-selected variants of 237, TNGK and LGQ, showed significantly enhanced activity with Jurkat and SKOV3-Cosmc KO, compared to either the parental 237 or the WE affinity variant, consistent with their broader specificity toward Tn-glycoprotein targets. There was no activity without the Cosmc deficiency (i.e., with Jurkat-Cosmc+ and SKOV3), further showing that aberrant O-glycosylation was required and that normal cells without these deficiencies are not capable of activating the CARs.
To determine if the CARs engineered for broadened specificity were likely to recognize other Tn-peptides expressed by human cancer cells, we examined the MUC1 KO lines of Jurkat and SKOV3-Cosmc KO (Fig. 7B). The absence of MUC1 on both lines was verified with an antibody against the MUC1 protein (SI Appendix, Fig. S7). Both engineered CARs, TNGK and LGQ, were potently stimulated by the MUC1-deficient cell lines, indicating that these lines express other Tn antigens at sufficient densities to induce activity. Low levels of IFN-γ release were observed with the parental 237-CAR. Stimulation by the SKOV3-Cosmc KO-MUC1 KO line was slightly greater for TNGK- and LGQ-CARs than the 5E5-CAR, supporting the well-known observation that cancer cell lines differ in their array of Tn-glycosylated cell surface proteins (e.g., ref. 41).
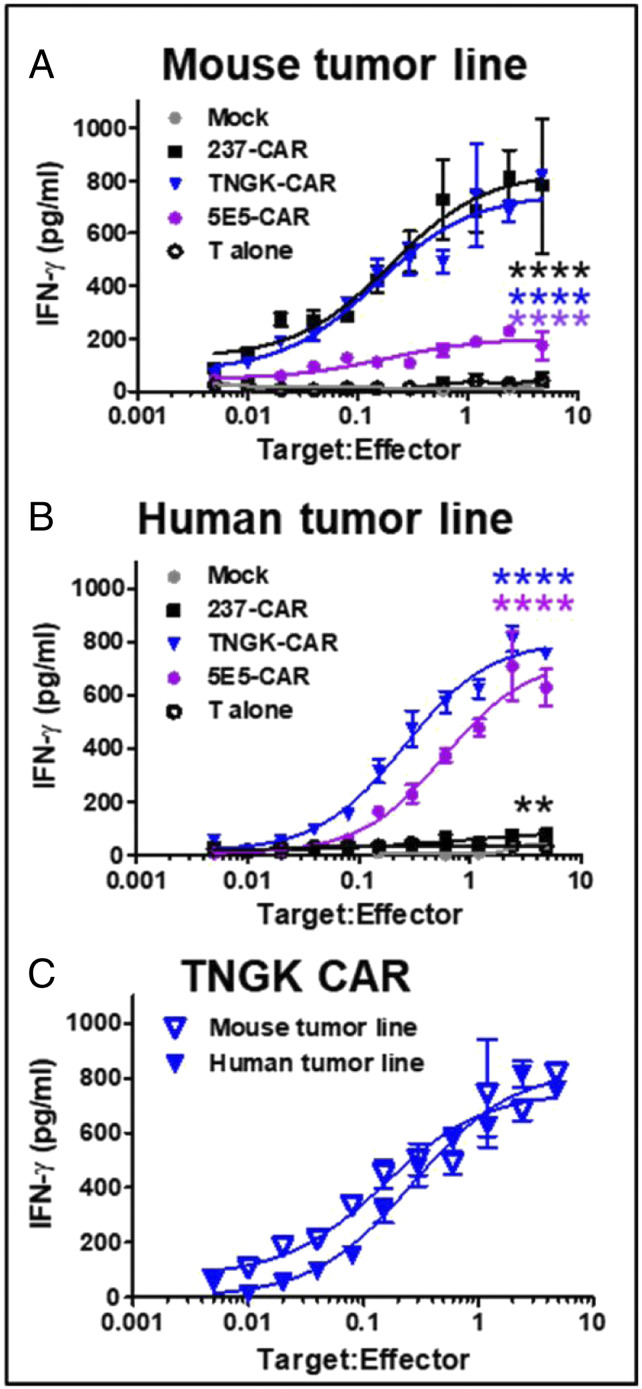
The engineered TNGK-CAR exhibited sensitive stimulation by both mouse and human tumor cell lines, whereas 237 was preferentially stimulated by mouse tumor lines, and 5E5-CAR was preferentially stimulated by human tumor lines. This finding suggests that TNGK-CAR would be an excellent candidate to use the same CAR for mouse toxicity testing, and efficacy studies in mouse and human models. To better assess this possibility, we conducted experiments in which the mouse and human tumor targets, mouse ID8 Cosmc KO (Fig. 8A) and human SKOV3 Cosmc KO MUC1 KO (Fig. 8B), were used in expanded, twofold titrations with preparations of 237-, TNGK-, and 5E5-CARs. These two cell lines (ID8 and SKOV3) were chosen because they represent the most well-studied ovarian tumor lines, one from mouse and one from human. This will allow studies in syngeneic immunocompetent mice with ID8 and xenotransplant studies in NSG mice (SKOV3). The MUC1-deficient cell line was used in order to determine what the activity might be for those cancers that express lower levels of MUC1 (transcriptional levels of MUC1 vary significantly not only among different types of cancers, but among patients with the same cancer, SI Appendix, Fig. S8). While 237-CAR was effective against the mouse cell line and 5E5-CAR was effective against the human cell line, the TNGK-CAR exhibited high sensitivity and activity against both cell lines with activity levels even below 0.1 tumor targets per TNGK-CAR T cell (Fig. 8C). In fact, TNGK-CAR was severalfold more sensitive than 5E5-CAR against the human MUC1-deficient line, suggesting that it will be useful against cancers that express low levels of MUC1.
Fig. 8.
Comparison of activity of 237-derived CAR T cells and 5E5-CAR T cells against mouse and human cancer cell lines. Mock, 237-CAR, TNGK-CAR, or 5E5-CAR transduced T cells were cocultured with Tn antigen expressing murine target cells, ID8 Cosmc KO (A), or Tn antigen expressing human target cells, SKOV3 Cosmc KO MUC1 KO (B), for 24 h at 37 °C, 5% CO2 at various target-to-effector ratios. Amount of IFN-γ released in the culture supernatants was measured by ELISA. Activation of transduced cells was measured in duplicate in two independent experiments. Activity of TNGK-CAR against the murine (A) and human (B) tumor line was compared in C. Error bars indicate SEM. Significance of difference in activity of each CAR compared to control (mock) was determined by two-way ANOVA followed by Dunnett's test using GraphPad Prism 8.0. ** refers to P = 0.001 to 0.01 and **** refers to P < 0.0001.
Discussion
We describe a strategy in which the exquisite cancer specificity of a CAR (237) could be retained, while engineering its potency against an expanded array of cancers. The cancer specificity of the 237-antibody stems from the nature of its binding to the GalNAc O-glycan, deep within the 237-binding site (8, 11). In the case of the cognate 237-epitope, the GalNAc residue is found on the mouse podoplanin protein called OTS8 (8, 27). The GalNAc Tn O-glycan represents the first step in protein O-glycosylation, and in normal cells the O-glycosylation proceeds with addition of further sugars to mask the immature Tn-glycans such that there are no detectable Tn O-glycans on surface proteins (42). Since the GalNAc moiety lies deep in the 237-antibody binding site (11), any extension of the sugar prevents binding. However, the peptide epitope flanking the threonine-linked GalNAc is located within the binding site, and as shown in our deep mutational scan, the side chains of the peptide contribute to the binding energy, and hence the specificity, of 237.
Our previous study showed that 237-CAR T cells could kill human Jurkat cells (5), and more recently we showed that Jurkat-derived tumors could be completely eliminated by 237-CAR T cells in a mouse xenotransplant model (14). Jurkat lacks the murine OTS8 protein (and hence the Tn-OTS8 epitope) and as shown in previous studies and here, Jurkat stimulates minimal IFN-γ release from 237-CAR T cells (5). This observation, together with the lower threshold required for target cytotoxicity compared to cytokine induction, is consistent with low affinity of the 237-scFv against Tn O-glycans on human O-glycoproteins expressed by Jurkat cells. The sensitivity of the CAR format allows it to be used for targeting antigens that bind with low affinity to the scFv recognition domains. We suggest that a similar low-affinity interaction explains the activity of the 5E5-CAR against the Tn-OTS8+ target Ag104A.
The relationship between CAR (scFv) affinity and T cell activity was further addressed in the present study. From the comparison of the activity of two CARs (237 and 237-WE) that differ in affinity by 30-fold, it is clear that there is only a minimal improvement in either sensitivity or activity of the higher-affinity version, against the cognate Tn-OTS8 antigen. Thus, the KD value of the 237-parental antibody appears to be sufficient when targeting the Tn-OTS8 antigen, and is already near a threshold for activity. These results are in accordance with other studies where it has been shown that an “affinity ceiling” or “affinity threshold” exists for CARs (17, 18, 33, 43), much like the case of T cell receptors for use in adoptive T cell therapy wherein increase in affinity does not translate to increase in potency. Both TCR and CAR affinities appear to be characterized by relatively sharp activity thresholds, and both are impacted by antigen density on target cells (44–46). The considerably lower affinity of the 237-CAR for alternative Tn O-glycoproteins found on Jurkat yields suboptimal ability to induce IFN- γ release. In fact, the modified 237-WE-CAR appears to have lost some activity against Tn-MUC1 and Jurkat, most likely because the CDR mutations resulted in a slightly modified binding site with less energy directed at the side chains of the MUC1 (or other) peptides compared to the OTS8 side chains.
Based on the structure of the 237-antibody (11) and the deep mutational analysis described here, we reasoned that the peptide fine specificity could be redirected toward a broader repertoire of Tn O-glycoproteins that are widely expressed in other types of cancers (2, 39). MUC1 represents one of these target proteins (39, 47), and was thus used here to select for 237-mutants that would bind with higher affinity to Tn-MUC1 but retain binding to Tn-OTS8. Two 237-scFv mutants isolated in these selections were converted to CARs and both showed significantly greater activity against human target cells (Jurkat and SKOV3) with aberrant Tn O-glycosylation. These mutant CARs also retained strong activity against the two mouse cancer lines Ag104A and ID8-Cosmc KO. The two engineered CARs (LGQ and TNGK) were stimulated potently by Jurkat and SKOV3-Cosmc KO with deleted MUC1. As these lines lack both Tn-OTS8 and Tn-MUC1, the LGQ- and TNGK-CARs must also bind well to other Tn-glycoproteins. In fact, a screen of several synthetic Tn O-glycopeptides identified originally in the human cell line, Jurkat (40), showed that the TNGK-CAR exhibited improved activities against Tn-TFRC and Tn-ZIP6 glycopeptides. In contrast, the 5E5-CAR did not show cross-reactivity with these targets. Unlike 237- and 5E5-, the TNGK-CAR has potent activity and exquisite sensitivity against both mouse and human tumors. This allows critical preclinical safety studies in mice with the exact same CAR to be used against human tumors. Most CARs against human targets lack the ability to fully examine toxicity due to the absence of the target in mice.
Broad Tn-peptide reactivity of the TNGK-CAR is important, especially in the context of cancers that do not express high levels of MUC1. In this regard, the TNGK-CAR showed activity that was equal to, or better than, the 5E5-CAR with the MUC1-deficient cell lines. Analysis of the RNAseq data available in the Expression Atlas database (EMBL-EBI website, The European Bioinformatics Institute) and The Cancer Genome Atlas (TCGA) database showed that MUC1 transcript levels vary by up to 1,000-fold among different cancer types and up to 100-fold within a cancer type (e.g., ovarian cancers) (SI Appendix, Fig. S8). Accordingly, in cancers where MUC1 is low, broadly reactive CARs like TNGK could be more applicable than CARs that were generated from antibodies against synthetic Tn-MUC1 immunogen, such as 5E5.
While the structure of the 5E5:Tn-MUC1 complex is unknown, the GalNAc moiety is presumed to provide significant binding energy as 5E5 does not bind to nonglycosylated or normally glycosylated MUC1 (37, 38). Another antibody AR20.5 that shows Tn selectivity, does bind to nonglycosylated protein at lower affinity, consistent with the location of GalNAc at the surface of the AR20.5 binding site (48). The structural basis of the scFv fragments LGQ and TNGK engineered here for higher affinity for Tn-MUC1 and the other Tn-glycoprotein targets remains to be determined, but it almost certainly retains the GalNAc moiety deep in the binding site. It is interesting that the one mutation shared between LGQ and TNGK, His93Gly, resides near the N terminus of the Tn-OTS8 epitope. This OTS8 region includes the sequence GTKPP, shown to be important in 237 binding (Fig. 1 C and D). A glycine substitution in CDR3L residue 93 could provide flexibility to accommodate the corresponding sequences of MUC1, TFRC, and ZIP6 (STAPP, GTESP, and STPPS, respectively).
There are numerous emerging options to configure antigen-recognition domains as CARs (49). The selection of the cancer-associated antigen as a therapeutic target is critically important for potent T cell engaging strategies (50, 51). Hematopoietic cancer cell antigens such as CD19 are tractable as targets because despite their expression on normal B cells, the B cell population can be replenished through hematopoiesis. Solid tumors of diverse origins do not have these capabilities. However, even antigens such as CD19 have limitations due to antigen loss variants that arise at a significant frequency, making such cancers resistant to CD19-targeted therapies (18, 51, 52). Recent approaches have attempted to address this challenge by using multiple antigen-recognition domains directed against multiple targets (e.g., ref. 53). The 237-CAR derivatives described here represent a single antigen-recognition module that could avoid such antigen loss variants as they are potently stimulated by many Tn-glycoprotein targets, and the aberrant glycosylation defects that yield Tn antigens are strongly associated with the malignant phenotype (2). Whereas the affinity for the parental 237-scFv is 140 nM for Tn-OTS8, and the parental 5E5 is 2 nM for Tn-MUC1, the affinities for cross-reacting Tn-peptides are unknown and likely quite variable. Nevertheless, the potency of the engineered 237-CARs relative to the wild-type no doubt stems from their increased affinity toward a broader repertoire of human Tn O-glycopeptide epitopes, including the selection target Tn-MUC1.
In conclusion, we show here that the 237-antibody provides an ideal platform for developing a panel of CARs that are cancer specific, yet exhibit broadened and improved activity against many different Tn O-glycoproteins found on diverse cancers. The 237-scFv libraries generated in these studies provide a discovery platform for virtually any Tn O-glycopeptide epitopes found in analysis of human cancers. The engineered TNGK-CAR that reacts potently with mouse and human targets allows critical preclinical safety studies in mice with the exact same CAR to be used against human tumors.
Materials and Methods
Peptide Synthesis, Glycosylation, and Flow Cytometry.
OTS8 peptide [ERGT(GalNAc)KPPLEELS-GK(biotin)] was synthesized and glycosylated by Sussex Research. MUC1 peptide (biotin-KVTSAPDTRPAPGSTAPPAHG) was synthesized by GenScript. For glycosylation, 50 μg MUC1 peptide was added to a 50 μL reaction mixture containing 10 mM MnCl2, 0.25% Triton X-100, 2 mM UDP-GalNAc, 25 mM Cacodylate buffer pH 7.4, in the presence of 0.5 mU of polypeptide GalNAc-T2 enzyme (SBH Sciences), and incubated for 4 h at 37 °C (54). Glycosylation of MUC1 peptide was confirmed by an ELISA with Tn-MUC1 5E5 antibody (38).
Additional Tn O-glycopeptides, originally identified in the Jurkat line (40), were synthesized by Synpeptide. Each peptide was biotinylated at N terminus and a GalNAc was added on the threonine residue indicated by (T*): TFRC [biotin-EPKTECERLAG(T*)ESPVRE], ZIP6 [biotin-GKLFPKDVSSS(T*)PPSVTS], PDXL [biotin-SSHSVTTDLTS(T*)KAEHL], CD43 [biotin-PPLTMATVSLE(T*)SKGTSG], EVI2B [biotin-SRKQITVHNPS(T*)QPTSTV], LAMP1 [biotin-TRCEQDRPSPT(T*)APPAPP] and PCDH [biotin-LNISHINAATG(T*)SASLVY] (14). Tetramers of Tn-glycosylated OTS8 or MUC1 peptides were prepared by mixing the biotinylated peptides with streptavidin-PE (BD Pharmingen) or streptavidin-Alexa Fluor 647 (Invitrogen) at 20:1 molar ratio. To assess binding by flow cytometry, yeast cells expressing 237-scFv (or its mutants) or 5E5 scFv, or mammalian cells expressing 237-CARs (or its mutants) or 5E5-CARs were stained either with various concentrations of fluorescent Tn-peptide tetramers or biotinylated, monomeric Tn-peptides. After washing, bound Tn-peptides were detected by staining cells with streptavidin-PE or streptavidin-647.
Cell Lines.
Ag104A, a spontaneous fibrosarcoma isolated from an aging C3H/HeN mouse (7) with a spontaneous mutational deletion in Cosmc (8); and its mutant cell line with wild-type Cosmc chaperone (ACosmc) (8) were maintained in complete Dulbecco's Modified Eagle Medium (DMEM) . ID8, a murine ovarian cancer cell line derived from C57BL/6 ovarian surface epithelial cells, and its mutant line with Cosmc deleted (ID8 Cosmc KO) were maintained in complete DMEM supplemented with insulin-transferrin-sodium selenite supplement. A murine T cell hybridoma, 58−/−, was maintained in complete Roswell Park Memorial Institute (RPMI) medium. Human T cell leukemia cell line (Jurkat, and its mutants MUC1 KO and Cosmc+) were maintained in complete RPMI. Human ovarian cancer cell line (SKOV3, and its mutants Cosmc KO and combination Cosmc and MUC1 KO) were maintained in complete DMEM.
As described (14), CRISPR/Cas9-mediated gene knockouts were generated using guiding sequences (sgRNA) using the sgRNA designer from the Broad Institute (55) and cloned over a BbsI site into the vector pSPCas9(BB)-2A-GFP (PX458, Addgene) as described (56). Cell lines were transfected by calcium phosphate with the respective PX458 vectors. Exon 1 of the human COSMC gene was targeted with the single guide RNA (sgRNA) 5′-TCACTATGCTAGGACACATT-3′. Likewise, exon 1 of murine Cosmc was targeted by the sgRNA 5′-CGAGATATCGTCTTTGTTAG-3′. COSMC-negative cell lines were confirmed by staining for Tn-positive cell populations using the Tn-specific antibody 5F4. Generation of human MUC1 KO cell lines was done by targeting exon 1 with the sgRNA 5′-TGAAGCTGGTTCCGTGGCCG-3′ and MUC1 negativity was confirmed by staining with the MUC1-specific antibody 5E10 or HMFG2. Sorting of GFP-positive populations and subsequent sorting of 5F4-positive or 5E10-negative cell populations was done by using the FACSAria II (BD Biosciences). Genomic knockout was verified by sequencing. Samples were incubated with primary antibodies followed by secondary stain using APC-labeled polyclonal antibody of goat anti-mouse IgG(H+L) (SouthernBiotech). Cytometry data were collected on LSR II (BD Biosciences) and analyzed by FlowJo (TreeStar).
Deep Mutational Scan of the 237-Epitope.
SCLs were constructed by PCR, in a stretch of 32 amino acids (GKAPLVPTQRERGTKPPLEELSTSATSDHDHR) in OTS8 protein, with 10 amino acids constituting the Tn-linked epitope (in bold) observed in the 237-crystal structure (11), and 12 and 10 amino acids flanking its N and C termini respectively. Primers containing degenerate codons (NNK) for each residue (synthesized by Integrated DNA Technologies) were used to introduce degeneracy at each codon, one at a time, by splice overlap extension PCR. Pooled PCR products representing each SCL were ligated into pMFG vector, and the resultant libraries were transformed into Escherichia coli Stellar chemically competent cells (Clontech). More than 10e5 colonies were picked to obtain library DNA that exceeded by two orders of magnitude the potential diversity of the library (32 positions × 32 codons = 1,024). Resultant plasmid DNA library was transduced into Jurkat cells using Phoenix Amphotropic packaging system with an efficiency of ∼5%.
The IRES-linked GFP in the pMFG vector was first used to sort 10e7 transduced Jurkat cells. Populations containing low GFP or high GFP were sorted separately to distinguish between populations containing one or more than one copies of the insert. Following sorts of the OTS8, SCLs were conducted by staining 10e7 Jurkat cells with 237-monoclonal antibody or anti-OTS8 antibody, or those expressing GFP. The top 1% fluorescent population was collected from each sorting condition. Jurkat expressing unsorted (naive) libraries, and each sorted library population was cultured for 24 to 48 h, following which cells were lysed and mRNA was isolated (GeneJET RNA purification kit, Thermo Scientific). cDNA was generated by RT-PCR (AccuScript PfuUltra II RT PCR kit, Agilent), and resultant PCR products were gel purified and subjected to deep sequencing on a Illumina HiSEq 2500 sequencer. Bioinformatic analysis was conducted on resultant sequences to obtain enrichment ratio for each substitution at each SCL location compared to the wild-type residue, and heat maps were generated as described (22).
Yeast Surface Display and Structure-Guided Engineering of 237-scFv.
The 237-scFv was cloned as an Aga-2 fusion into yeast display vector pCT302 with an N-terminal hemagglutinin (HA) tag, and a C-terminal c-myc tag. The expression and folding of 237-scFv on yeast cell surface was confirmed by flow cytometry, by assessing binding of Tn-OTS8 peptide as monomer or tetramer to the scFv. A structure-guided approach was taken to engineer 237-scFv for binding with higher affinity to Tn-OTS8 peptide, or to broaden its specificity to other Tn-linked peptides. For this purpose, nine libraries were constructed in residues of the six CDRs of 237-scFv, including those that were in proximity to, or contacted the sugar, or the peptide, or both, in the crystal structure (11). Either three or four residues in the CDRs were mutated at a time.
To obtain variants of 237-scFv that bound with high affinity to Tn-OTS8 peptide, pooled CDR libraries were subjected to a sorting scheme consisting of MACS and FACS, with tetrameric or monomeric Tn-OTS8 peptide. After fourth and fifth sorts, 10 mutants were isolated and analyzed for binding with 1 or 10 nM Tn-OTS8 peptide. DNA sequencing was performed to characterize each mutation. Selected mutants were titrated with varying concentrations of Tn-OTS8 peptide to determine approximate dissociation constant (KD) by flow cytometry. In addition, the mutants were subjected to an off-rate assay, where they were allowed to bind with saturating concentration of biotinylated Tn-OTS8 peptide, followed by competing the binding with unbiotinylated Tn-OTS8 peptide over a period of 4 h.
To obtain specificity variants of 237-scFv that could bind not only to Tn-OTS8 peptide but also to Tn-MUC1 peptide, pooled libraries were subjected to a sorting scheme consisting of MACS and FACS with tetrameric or monomeric Tn-MUC1 peptide. An additional sort with anti-c-myc antibody was conducted to get rid of frame-shift mutations in 237-scFv that resulted in truncated variants. Finally, three unique, in-frame mutants were characterized by DNA sequencing and flow cytometry.
T Cell Transduction and Activation Assays.
In order to assess the impact of various mutations in 237-scFv that were obtained by screening the libraries (either with OTS8 peptide or MUC1 peptide), the mutations were introduced into 237scFv-CD28-CD3zeta CAR construct (pMP71) (5) by Quikchange Lightning Kit (Agilent Technologies). The CARs were retrovirally transduced into total T cells from C57BL/6 mice. For transduction, Plat-E retroviral packaging cells were plated at a concentration of 10e6 cells per well on a poly-l-lysine-coated six-well tissue culture plate in DMEM supplemented with blasticidin and puromycin. After 24 h at 37 °C, 5% CO2, PlatE cells in each well were transfected with 10 μg plasmid DNA (237-CARs) or no DNA (mock). Retroviral supernatants were harvested 48 h after transfection. Total T cells were isolated from spleens of C57BL/6 mice using Dynabeads untouched mouse T cells kit (Invitrogen), and activated with anti-CD28 and anti-CD3 beads (Gibco), and IL-2 (Roche) in Iscove's Modified Dulbecco's Medium (IMDM) for 24 h at 37 °C, 5% CO2. After 24 h, activated T cells were transduced with filtered (0.45 μm) retroviral supernatants, in presence of Lipofectamine 2000 (Life Technologies) and IL-2. Transduced T cells were allowed to expand for a total of 72 h, with a 1:1 split in IMDM after 48 h. Transduction efficiencies (or cell surface CAR expression) were measured by flow cytometry by staining CAR-transduced T cells with fluorescent, Tn-OTS8 peptide or Tn-MUC1 peptide tetramers. For activation assays, various CAR-transduced T cells (effectors) were cocultured with immobilized Tn-OTS8 or Tn-MUC1 peptides, or with various target cell lines (Ag104A, ACosmc, ID8-Cosmc KO, ID8, 58−/−, Jurkat, Jurkat-MUC1 KO, Jurkat-Cosmc+, SKOV3-Cosmc KO, SKOV3-CosmcKO-MUC1KO, or SKOV3) in 96-well plates for 24 h at 37 °C, 5% CO2 at various target-to-effector ratios. The quantity of IFN-γ released in the supernatants under each coculture condition was measured by standard ELISA using mouse IFN gamma uncoated ELISA kit (Invitrogen).
Data Availability.
All data are available in the main text and SI Appendix. Protocols have been described in Materials and Methods. Any details regarding data, protocols, and materials will be made available to the readers by sending a request to the corresponding authors (P.S. and D.M.K).
Supplementary Material
Acknowledgments
We thank the staff of the Roy J. Carver Biotechnology Center at the University of Illinois for assistance in DNA sequencing, high-speed cell sorting, and flow cytometry; and the high performance computing group (HPCBio) for assistance in RNAseq data analysis. We also thank Carl June, Ed Roy, Erik Procko, and Dan Harris for helpful discussions. This work was supported by NIH grants R01CA178844 and R21CA238628 (to D.M.K.), R01CA022677 and R01CA37156 (to H.S.), and the Danish National Research Foundation (DNRF107) (to H.C.).
Footnotes
Competing interest statement: P.S., Y.H., K.S., H.S., and D.M.K. are listed as inventors on a US patent application involving this study. Q.C. is employed by Gilead Sciences, Inc. D.M.K. is a consultant for AbbVie. H.C. is a co-inventor on patents related to antibodies to Tn-glycopeptides, including Tn-MUC1 held by the University of Copenhagen. H.C. is co-founder of GlycoDisplay Aps and GO Therapeutics Inc., formerly GlycoZym Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920662117/-/DCSupplemental.
References
- 1.Springer G. F., T and Tn, general carcinoma autoantigens. Science 224, 1198–1206 (1984). [DOI] [PubMed] [Google Scholar]
- 2.Radhakrishnan P. et al., Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc. Natl. Acad. Sci. U.S.A. 111, E4066–E4075 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RodrÍguez E., Schetters S. T. T., van Kooyk Y., The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Tarp M. A., Clausen H., Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 1780, 546–563 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Stone J. D., Aggen D. H., Schietinger A., Schreiber H., Kranz D. M., A sensitivity scale for targeting T cells with chimeric antigen receptors (CARs) and bispecific T-cell Engagers (BiTEs). OncoImmunology 1, 863–873 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Posey A. D. Jr. et al., Engineered CAR T cells targeting the cancer-associated Tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity 44, 1444–1454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward P. L., Koeppen H., Hurteau T., Schreiber H., Tumor antigens defined by cloned immunological probes are highly polymorphic and are not detected on autologous normal cells. J. Exp. Med. 170, 217–232 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schietinger A. et al., A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science 314, 304–308 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Schietinger A., Philip M., Schreiber H., Specificity in cancer immunotherapy. Semin. Immunol. 20, 276–285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju T., Aryal R. P., Kudelka M. R., Wang Y., Cummings R. D., The Cosmc connection to the Tn antigen in cancer. Cancer Biomark. 14, 63–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks C. L. et al., Antibody recognition of a unique tumor-specific glycopeptide antigen. Proc. Natl. Acad. Sci. U.S.A. 107, 10056–10061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boder E. T., Wittrup K. D., Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Mendoza J. L. et al., Structure of the IFNγ receptor complex guides design of biased agonists. Nature 567, 56–60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y. et al., Multiple cancer-specific antigens are targeted by a chimeric antibody receptor on a single cancer cell. JCI Insight 4, e130416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju T., Cummings R. D., A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudecek M. et al., Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin. Cancer Res. 19, 3153–3164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caruso H. G. et al., Tuning sensitivity of CAR to EGFR density limits recognition of normal tissue while maintaining potent antitumor activity. Cancer Res. 75, 3505–3518 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghorashian S. et al., Enhanced CAR T cell expansion and prolonged persistence in pediatric patients with ALL treated with a low-affinity CD19 CAR. Nat. Med. 25, 1408–1414 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Fowler D. M. et al., High-resolution mapping of protein sequence-function relationships. Nat. Methods 7, 741–746 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead T. A. et al., Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 30, 543–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham B. C., Wells J. A., High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085 (1989). [DOI] [PubMed] [Google Scholar]
- 22.Procko E. et al., Computational design of a protein-based enzyme inhibitor. J. Mol. Biol. 425, 3563–3575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Procko E. et al., A computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein induces apoptosis in infected cells. Cell 157, 1644–1656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris D. T. et al., Deep mutational scans as a guide to engineering high affinity T cell receptor interactions with peptide-bound major histocompatibility complex. J. Biol. Chem. 291, 24566–24578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris D. T. et al., An engineered switch in T cell receptor specificity leads to an unusual but functional binding geometry. Structure 24, 1142–1154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma P., Kranz D. M., Subtle changes at the variable domain interface of the T-cell receptor can strongly increase affinity. J. Biol. Chem. 293, 1820–1834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steentoft C. et al., Characterization of an immunodominant cancer-specific O-glycopeptide epitope in murine podoplanin (OTS8). Glycoconj. J. 27, 571–582 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Sommermeyer D. et al., Fully human CD19-specific chimeric antigen receptors for T-cell therapy. Leukemia 31, 2191–2199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maude S. L. et al., Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maude S. L. et al., Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynn R. C. et al., High-affinity FRβ-specific CAR T cells eradicate AML and normal myeloid lineage without HSC toxicity. Leukemia 30, 1355–1364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma P., Kranz D. M., Recent advances in T-cell engineering for use in immunotherapy. F1000 Res. 5, 2344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X. et al., Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 75, 3596–3607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gendler S. J. et al., Cloning of partial cDNA encoding differentiation and tumor-associated mucin glycoproteins expressed by human mammary epithelium. Proc. Natl. Acad. Sci. U.S.A. 84, 6060–6064 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siddiqui J. et al., Isolation and sequencing of a cDNA coding for the human DF3 breast carcinoma-associated antigen. Proc. Natl. Acad. Sci. U.S.A. 85, 2320–2323 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kufe D. W., Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 9, 874–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarp M. A. et al., Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology 17, 197–209 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Sørensen A. L. et al., Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology 16, 96–107 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Lavrsen K. et al., Aberrantly glycosylated MUC1 is expressed on the surface of breast cancer cells and a target for antibody-dependent cell-mediated cytotoxicity. Glycoconj. J. 30, 227–236 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Steentoft C. et al., Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Steentoft C. et al., Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandall H. H. et al., Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 70, 1306–1313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chmielewski M., Hombach A., Heuser C., Adams G. P., Abken H., T cell activation by antibody-like immunoreceptors: Increase in affinity of the single-chain fragment domain above threshold does not increase T cell activation against antigen-positive target cells but decreases selectivity. J. Immunol. 173, 7647–7653 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Harris D. T. et al., Comparison of T cell activities mediated by human TCRs and CARs that use the same recognition domains. J. Immunol. 200, 1088–1100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone J. D. et al., A novel T cell receptor single-chain signaling complex mediates antigen-specific T cell activity and tumor control. Cancer Immunol. Immunother. 63, 1163–1176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chervin A. S. et al., The impact of TCR-binding properties and antigen presentation format on T cell responsiveness. J. Immunol. 183, 1166–1178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kufe D. W., MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene 32, 1073–1081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Movahedin M. et al., Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen. Glycobiology 27, 677–687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holzinger A., Abken H., CAR T cells: A snapshot on the growing options to design a CAR. HemaSphere 3, e172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.June C. H., Riddell S. R., Schumacher T. N., Adoptive cellular therapy: A race to the finish line. Sci. Transl. Med. 7, 280ps7 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Sadelain M., Tales of antigen evasion from CAR therapy. Cancer Immunol. Res. 4, 473 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Jackson H. J., Brentjens R. J., Overcoming antigen escape with CAR T-cell therapy. Cancer Discov. 5, 1238–1240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balakrishnan A. et al., Multispecific targeting with synthetic ankyrin repeat motif chimeric antigen receptors. Clin. Cancer Res. 25, 7506–7516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett E. P. et al., Cloning of a human UDP-N-acetyl-alpha-D-Galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J. Biol. Chem. 273, 30472–30481 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Doench J. G. et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ran F. A. et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and SI Appendix. Protocols have been described in Materials and Methods. Any details regarding data, protocols, and materials will be made available to the readers by sending a request to the corresponding authors (P.S. and D.M.K).