Significance
Five small and homologous domains—referred to as CC domains—in the cell surface protein RECK play essential roles in signaling via the WNT7A and Wnt7B ligands in the context of central nervous system vascular development. We have determined the three-dimensional structure of one of these domains and find that it consists of four short α-helices stabilized by three disulfide bonds, a structure that has not been seen previously in the extracellular domain of any vertebrate cell surface or secreted protein. Homology searches of genome sequences show that the CC domain arose early in the evolution of multicellular animals, but, unlike many other small folded domains, it is only detected in a single gene (RECK).
Keywords: Wnt signaling, blood–brain barrier, protein evolution, extracellular domain, four-helix bundle
Abstract
Five small protein domains, the CC-domains, at the N terminus of the RECK protein, play essential roles in signaling by WNT7A and WNT7B in the context of central nervous system angiogenesis and blood–brain barrier formation and maintenance. We have determined the structure of CC domain 4 (CC4) at 1.65-Å resolution and find that it folds into a compact four-helix bundle with three disulfide bonds. The CC4 structure, together with homology modeling of CC1, reveals the surface locations of critical residues that were shown in previous mutagenesis studies to mediate GPR124 binding and WNT7A/WNT7B recognition and signaling. Surprisingly, sequence and structural homology searches reveal no other cell-surface or secreted domains in vertebrates that resemble the CC domain, a pattern that is in striking contrast to other ancient and similarly sized domains, such as Epidermal Growth Factor, Fibronectin Type 3, Immunoglobulin, and Thrombospondin type 1 domains, which are collectively present in hundreds of proteins.
RECK (Reversion-inducing Cysteine-rich Protein with Kazal Motifs) is a multidomain glycosyl-phosphatidyl-inositol (GPI)-anchored protein that was originally identified based on its ability to induce morphological reversion in K-RAS–transformed NIH 3T3 cells (1). As judged by the phenotypes associated with loss-of-function mutations in mice, RECK plays important roles in multiple developmental processes, including angiogenesis, neurogenesis, and limb patterning (2–5). RECK possesses matrix metalloproteinase inhibitor activity (6), and one mechanism of RECK action is via its inhibition of metalloproteinase-dependent release of NOTCH ligands from the plasma membrane (2). Like the NOTCH ligands that it regulates, RECK’s local concentration can be altered by membrane release, with glycerophosphodiester phosphodiesterase 2 (GDE2)-mediated cleavage of RECK’s GPI-anchor leading to release of RECK from the plasma membrane, thereby decreasing RECK’s access to its membrane-associated metalloproteinase targets (7).
More recently, a second activity has been ascribed to RECK: stimulation of canonical WNT signaling (referred to hereafter as “BETA-CATENIN signaling”) in vascular endothelial cells (ECs) by neuron- and glia-derived WNT7A and WNT7B (8, 9). This signal plays an essential role in central nervous system (CNS) angiogenesis and blood–brain barrier (BBB) formation and maintenance. Current evidence has led to a model in which RECK acts in conjunction with G protein coupled receptor (GPR) 124, a seven-pass transmembrane protein, to enhance WNT7A and WNT7B signaling via FRIZZLED receptors and low-density lipoprotein receptor-related protein (LRP) 5/LRP6 coreceptors (10–15). The stimulatory activity exhibits striking ligand specificity: in transfected cells, stimulation is limited to WNT7A and WNT7B, with no measurable effect on signaling by any of the other 17 mammalian WNTs or by NORRIN (a cystine-knot growth factor and a BETA-CATENIN signaling activator). Interestingly, normal CNS angiogenesis requires Reck expression in both ECs and developing neural cells, as determined by cell type-specific gene deletion of a floxed Reck allele (12, 16).
Functional analyses of RECK in transfected cells has revealed an essential role for its most amino terminal domains in stimulating BETA-CATENIN signaling (12–14). This region is composed of five tandem and highly divergent repeats of a ∼70-aa sequence (Fig. 1A). The repeats can be recognized because each repeat unit contains six cysteines with characteristic spacing, including two that are adjacent (1). We refer to these units, which are presumed to comprise autonomously folding domains, as “CC domains,” with individual domains named CC1 to CC5. BETA-CATENIN signaling assays, together with binding assays to cell-surface signaling complexes, fragments of GPR124, full-length WNT7A/WNT7B, and WNT7A/WNT7B-derived peptides implicate RECK(CC1) in GPR124 recognition and RECK(CC4) in WNT7A/WNT7B recognition (12–15).
Fig. 1.
Domain architecture of the mRECK protein and sequence alignment of mRECK(CC4) and C. elegans HER-1. (A) Schematic of the domain architecture of mRECK. The other domains include a FRIZZLED-like cysteine-rich domain (Fz-like CRD), two Epidermal Growth Factor-like domains (EGF-like 1 and 2), and three Kazal-like motifs (Kazal-like 1 to 3). (B) Sequence alignment showing the predicted mRECK(CC4) secondary structure and disulfide bonding pattern based on the HHpred comparison with the nHER-1 crystal structure. Red boxes represent conserved cysteines (numbered 1 to 7). Disulfide bridges are shown above and below as connecting lines. α-Helical regions are indicated by magenta and blue rectangles. The two gaps in the sequence alignment are based on the structural superposition of the nHER-1 crystal structure and the mRECK(CC4) computational model shown in SI Appendix, Fig. S1.
To gain insight into the function and evolution of the RECK(CC) domains, we have determined the three-dimensional structure of mouse RECK (mRECK) CC4 by X-ray crystallography. The structure reveals a compact four-helix bundle with three disulfide bonds. This architecture is unlike any known vertebrate extracellular domain. Sequence and structure homology searches show that, despite its ancient origins and its retention in widely divergent Metazoa, CC-like domains are generally present only in the single RECK homolog in present-day species.
Results
Distant Homology between mRECK(CC4) and Caenorhabditis elegans HER-1.
We began our structural analysis of the CC domain by searching for homologous sequences to assist in computationally predicting domain boundaries and domain architecture. Distant homology searches with the Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) and FUGUE (17, 18) failed to reveal sequences other than RECK orthologs. However, searches against proteins of known structure using predictions of secondary structure similarity and pairwise comparisons of Hidden Markov models (HHpred; ref. 19) identified a single sequence, C. elegans HER-1 (nHER-1), in which the spacing of six cysteines closely resembles the cysteine spacing in RECK(CC) domains, as shown in Fig. 1B for mRECK(CC4). nHER-1 is a secreted protein that functions in C. elegans sex determination (20). We used the nHER-1 structure (21) to generate a three-dimensional model for mRECK(CC4) (SI Appendix, Fig. S1). This model, which generalizes to the other RECK(CC) domains, starts 10 residues before the first cysteine (the first cysteine in the pair of adjacent cysteines) and ends one residue beyond the sixth cysteine (Fig. 1B).
Protein Expression and Purification of mRECK(CC4).
Based on the predicted CC-domain boundaries, we used HEK293T cells to produce mRECK(CC4) constructs with an N-terminal signal peptide and C-terminal mVenus and 12×His tags (SI Appendix, Fig. S2). We developed a time- and cost-efficient method based on immobilized metal affinity chromatography (IMAC) followed by in-gel fluorescent imaging to quantify the level of secreted mVenus fusion proteins from conditioned medium (SI Appendix, Fig. S2). Small variations in the locations of the N and C termini have little effect on the yield of the mRECK(CC4)–mVenus fusion protein (SI Appendix, Fig. S3). As the mammalian secretory pathway has stringent quality-control machinery to ensure that secreted proteins are folded correctly (22), these data imply that all of the tested variants encompass the core folding unit. The construct with the highest level of expression (Construct 9; mRECK residues 206 to 270) was selected for large-scale protein production in HEK293T cells in the presence of the Class I α-mannosidase inhibitor kifunensine to minimize heterogeneity from any N-linked glycosylation events (23). This recombinant protein was produced with a yield of ∼2 mg/L and purified to apparent homogeneity, but it failed to crystallize.
To address the challenge of crystallization, mRECK(CC4) was fused to the C terminus of engineered Maltose Binding Protein (eMBP; SI Appendix, Fig. S4 A and B). eMBP is a derivative of Escherichia coli MBP with multiple genetically encoded alterations in surface residues that promote crystal lattice formation (24–28). Based on the known eMBP structure and the mRECK(CC4) model, the fusion protein was designed to minimize the length and flexibility of the connecting linker between the two domains. To eliminate any potential N-linked glycosylation events and increase protein production, eMBP-mRECK(CC4) was expressed in E. coli rather than in mammalian cells. For this purpose, we used E. coli SHuffle cells, a strain with a modified redox potential that permits disulfide bond formation in the cytoplasm (29). Soluble eMBP-mRECK(CC4) was produced at >20% of soluble cell protein, purified by IMAC, and recovered in a monodisperse state by size-exclusion chromatography (SEC; SI Appendix, Fig. S4 C and D).
eMBP-mRECK(CC4) Crystallization and Structure Solution.
Extensive screening of crystallization conditions using the vapor-diffusion method showed that eMBP-mRECK(CC4) requires maltose (the MBP ligand) and zinc ions for high-quality crystal growth. The zinc requirement is consistent with the presence of surface histidine mutations in eMBP that facilitate metal-induced interactions between neighboring monomers to enhance crystal lattice contacts (SI Appendix, Fig. S4B; ref. 25). X-ray diffraction data were collected to a resolution of 1.65 Å. To obtain initial phase information, we utilized a molecular replacement approach with MBP as a search model and identified additional electron density corresponding to mRECK(CC4) (SI Appendix, Fig. S5A). Iterative density modification was then used to generate an interpretable electron density map for model building and refinement of mRECK(CC4), revealing one eMBP–mRECK(CC4) molecule in the crystallographic asymmetric unit of the P21212 space group (SI Appendix, Fig. S5 B and C; SI Appendix, Table S1, provides data collection and refinement statistics). The maltose ligand, amino acid side chains, zinc ions, and disulfide bonds are clearly visible in the final electron density map (SI Appendix, Fig. S5 D–J). The two domains of the eMBP–mRECK(CC4) fusion protein are loosely packed against one another (SI Appendix, Fig. S6), and multiple contacts are present between eMBP domains in adjacent crystallographic symmetry units (SI Appendix, Fig. S7).
The 1.65-Å mRECK(CC4) structure reveals a compact antiparallel four-helix bundle with three short connecting loops (Fig. 2). The first helix (helix A) consists of three consecutive 310-helix turns. Six cysteine residues form three disulfide bonds that connect helices A+B, A+D, and B+C (Fig. 2A). Each of these cysteine residues is present in each RECK(CC) domain in all RECK orthologs identified thus far, as described more fully below. An analysis of the electrostatic surface potential of mRECK(CC4) reveals a negatively charged solvent-exposed groove formed by the N-terminal part of helix A and helices C and D (Fig. 2B). Most of the negatively charged residues are substituted by neutral residues in mRECK(CC1) (Fig. 3A). The potential significance of the CC4 groove for WNT7A/7B recognition is discussed in the Discussion.
Fig. 2.
Crystal structure of mRECK(CC4). (A) Ribbon diagram of mRECK(CC4) in rainbow coloring corresponding to the linear schematic (Upper Right). Circles denote the N and C termini. The six labeled cysteine residues form three disulfide bonds shown as sticks. (B) Electrostatic surface potential of mRECK(CC4). The color scale is calibrated from red (acidic; −5 KbT/ec) to blue (basic; 5 KbT/ec). The orientations of mRECK(CC4) in B are the same as in A. The locations of Pro256 and Trp261 are indicated. The negatively charged groove is denoted by dotted lines (Left) and a dotted circle (Right).
Fig. 3.
Sequence alignment of mRECK CC domains and modeling mRECK(CC1) based on the mRECK(CC4) crystal structure. (A) Sequence alignment of CC domains. α-Helical regions A to D correspond to the four helices in Fig. 2. The six conserved cysteine residues are highlighted. Blue and magenta indicate the key residues that were identified in previous studies (12, 13) for GPR124 binding (CC1) and stimulation of WNT7A signaling (CC4), respectively. (B) Percent sequence identity and similarity (shown in parentheses) among mRECK CC domains. (C) A sequence-based phylogenetic tree of mRECK CC domains. Horizontal branch lengths correspond to evolutionary distance (Materials and Methods). (D and E) Structural comparison of the mRECK(CC4) crystal structure with the modeled structure of mRECK(CC1). The key residues highlighted in A are rendered as sticks.
Implications of mRECK(CC4) for the Structure and Function of Other RECK(CC) Domains.
With the six conserved cysteines serving as anchors, an alignment of mRECK(CC1)-mRECK(CC5) revealed high variability in noncysteine residues, none of which are absolutely conserved among the five CC domains (Fig. 3A). Pairwise comparisons show that the percent identity—including the six cysteines, which account for ∼10% of the amino acids—ranges from 17 to 29% and the percent similarity+identity ranges from 30 to 49% (Fig. 3 B and C).
Previous work has shown that mRECK(CC1) mediates binding to the N-terminal extracellular leucine-rich repeat (LRR) and immunoglobulin (Ig) domains of GPR124, and that alanine substitution mutations at Arg69, Pro71, and Tyr73 in mRECK(CC1) greatly decrease binding (12). Additionally, when RECK(CC1) amino acids 68 to 73 were all converted to alanines by CRISPR-mediated targeting in the mouse germline, WNT7A/WNT7B signaling and CNS angiogenesis were impaired (12). By modeling mRECK(CC1) based on the RECK(CC4) structure, the solvent-accessible surface areas (SASAs) for mRECK(CC1) Arg69, Pro71, and Tyr73 were calculated to be 97.5, 52.4, and 106.8 Å2, respectively. The corresponding amino acids in RECK(CC4)—Gly245, Ile247, and Gly249—have SASA values of 15.3, 74.6, and 36.1 Å2, respectively (Fig. 3 D and E). The high solvent exposure of Arg69, Pro71, and Tyr73 are consistent with a model in which they interact directly with the GPR124 LRR and Ig domains (Fig. 3 D and E). Interestingly, Trp261, an amino acid in RECK(CC4) that is critical for WNT7A/WNT7B signaling, is conserved in mRECK(CC1) (Trp82). Other residues in RECK(CC1) and RECK(CC4) that were implicated in signaling by the earlier mutagenesis studies (12, 13) are not conserved between these two domains (Fig. 3 D and E).
Structural Similarities between mRECK(CC4) and Other Four-Helix Bundle Proteins.
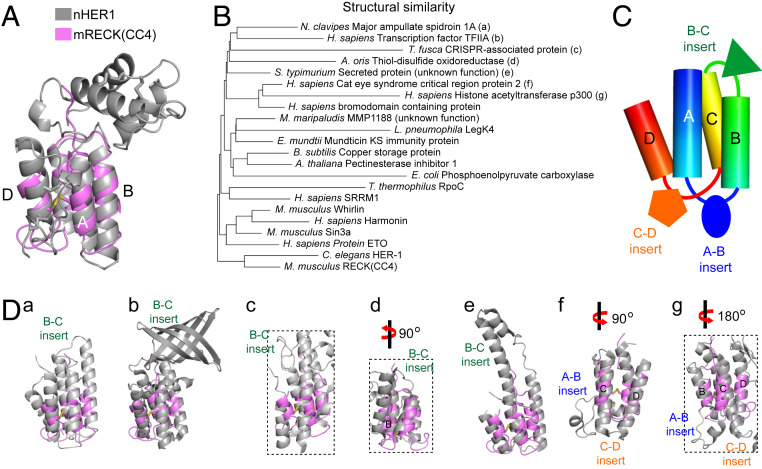
To assess the degree to which mRECK(CC4) resembles other proteins with four-helix bundles, we performed a 3D structure search of the Protein Data Bank (PDB) using the DALI server (30). Similar results were obtained with queries comprising full-length mRECK(CC4) or the helical core of mRECK(CC4) without the connecting loops. This approach has been used previously to assess divergent evolutionary relationships and deformations in the evolution of protein folds (30). As predicted by our initial modeling (SI Appendix, Fig. S1), mRECK(CC4) is a close structural match to the four-helix bundle domain of nHER-1, with 2.2 Å rmsd of Cα atoms (Fig. 4A and SI Appendix, Table S2). Among PDB proteins, nHER-1 is the closest structural match to mRECK(CC4), as reflected in the branching pattern of a structural similarity dendrogram (Fig. 4B). The nHER-1 protein does not have any orthologs in vertebrates. Interestingly, many four-helix bundle proteins are close structural matches to the mRECK(CC4) four-helix bundle despite extremely low similarity at the sequence level (5 to 15%). These include structural proteins, enzymes, transcription factors, and other diverse proteins from bacteria, plants, and animals (Fig. 4B and SI Appendix, Table S2).
Fig. 4.
Structure-based homologies between RECK(CC4) and diverse four-helix bundle proteins. (A) Superposition of mRECK(CC4) with nHER-1. (B) Structural similarity dendrogram. The distance matrices were calculated from pairwise superpositions of mRECK(CC4) with the indicated four-helix bundle proteins. (C) Cartoon representation of diverse sequence insertions in four-helix bundle proteins in the A-B, B-C, or C-D loops. (D) Superposition of mRECK(CC4) (magenta) with various four-helix bundle proteins (gray). The black dotted boxes represent the four-helix bundle region within the context of larger proteins. SI Appendix, Figs. S8 and S9, provide additional pairwise superpositions.
Detailed comparisons between mRECK(CC4) and its structural homologs reveal two trends: 1) loops A-B, B-C, and C-D can accommodate widely different insertions and 2), except for nHER1, the other four-helix bundle proteins lack the disulfide bonds present in mRECK(CC4), and nearly all have longer helices (Fig. 4 B–D and SI Appendix, Figs. S8 and S9). This second trend suggests that, with respect to structural stability, the relatively short helices in the mRECK(CC) domains are offset by the high density of disulfide bonds.
Strikingly, the DALI search with mRECK(CC4) did not identify homologous domains among secreted or cell surface vertebrate proteins (Fig. 4B and SI Appendix, Table S2). This negative finding is in contrast to other families of autonomously folding domains, many of which are widely dispersed throughout the extracellular proteomes of vertebrates. For example, the Epidermal Growth Factor (EGF), Ig, Fibronectin Type-3, and Thrombospondin Type-1 Repeat domains are found in 167, 551, 184, and 66 human proteins, respectively (31) (Fig. 5). Among 34 domains present in the human extracellular proteome that were studied by Vogel and Chothia (31), only one (class II MHC-associated invariant chain ectoplasmic trimerization domain) was represented by a single human protein.
Fig. 5.
A comparison of the number and diversity of vertebrate transmembrane and secreted proteins containing CC domains or four other extracellular domains. (Top) Ribbon diagram showing the domain structure [in rainbow coloring from N terminus (blue) to C terminus (red)] for RECK(CC4), epidermal growth factor (EGF; PDB ID code 1JL9), immunoglobulin (Ig; PDB ID code 5NST), fibronectin (PDB ID code 1TEN), and thrombospondin (TSP) type-I repeat (PDB ID code1LSL) domains. A partial list of mammalian proteins containing the latter four domains is below. Domain content was confirmed using the “Identify conserved domains” function in the NCBI protein database. The CC domain is found only in RECK.
Evolution of RECK, WNT, and GPR124.
We next sought to explore the evolutionary history of the CC domain. Wnt signaling is present broadly among Metazoa and it is absent in non-Metazoa, such as plants, fungi, and choanoflagellates (Wnt homepage; refs. 32 and 33). The repertoire of WNT proteins varies greatly among species, with multiple examples of lineage-specific WNT acquisition and loss (34, 35). To compare the evolutionary histories of WNT, RECK, and GPR124 proteins, BLASTP searches against GenBank proteins (inferred from DNA sequences) were performed with individual phyla or classes using mRECK, mRECK(CC1-5), and mGPR124 as queries.
Queries with mRECK and mRECK(CC1-5) identified homologs across diverse Metazoa—including sponges, corals, sea anemones, mussels, snails, insects, and vertebrates—that aligned convincingly along the entire length of the mRECK sequence (Fig. 6A and SI Appendix, Fig. S10). Weak homologies were observed between mRECK domains other than the CC domains and non-RECK proteins, but convincing homologies to mRECK(CC) domains were only observed with full-length RECK proteins. In those organisms possessing a RECK homolog, there was generally a single family member per genome, and, in the vast majority, all five CC domains were present; one exception is the sponge Amphimedon queenslandica, which appears to have a RECK homolog with only three CC domains (XP_019857591.1). Surprisingly, RECK homologs are absent from nematodes. RECK sequences are also absent from plants, fungi, or choanoflagellates.
Fig. 6.
RECK evolution and intron-exon structure. (A) Evolutionary trees for RECK, WNT, and GPR124. Representatives of major groups of organisms are indicated, with their associated phyla or clades shown in parentheses. Red or black branches indicate the presence or absence, respectively, of members of the indicated protein families (as determined by BLASTP searches). Branch lengths are not calibrated. The connectivity of the Placozoa branch is not certain. (B) Alignment of RECK(CC) amino acid sequences from mouse (Upper) and D. melanogaster (Lower). Intron locations for mouse (labeled A, B, C…) and Drosophila (labeled V, W, X…) are indicated by arrowheads. The location of each intron with respect to the RECK reading frame is shown in parentheses for each intron. Cysteines (“C”) are highlighted in red, and the five CC domains are delineated by red lines beneath the alignment.
To broadly assess RECK(CC1-5) sequence conservation/variation across the phylogenetically diverse species that harbor a RECK gene, we aligned RECK orthologs from mouse (NP_057887.2), zebrafish (XP_009295477.1), Drosophila (NP_001261853.1), termite (XP_021916359.1), spider (GBM32250.1), snail (XP_025096258.1), mussel (XP_013384099.1), sea anemone (XP_001635685.1), and coral (XP_027044662.1; SI Appendix, Fig. S10). This alignment revealed the presence of all 30 cysteines (5 domains with 6 cysteines per domain) across all sequences, but only 12 noncysteine positions were identical across all sequences. Among the additional 271 positions [using mRECK(CC1-5) as the standard for protein length], only 36 positions were highly conserved. Significantly, four of five residues in mRECK(CC1) that have been shown experimentally to be essential for binding to the N-terminal LRR and Ig domains of GPR124 are not conserved in more distant Metazoa (residues 69 to 73; ref. 12). Of the two positions in mRECK(CC4) that have been shown experimentally to be essential for WNT7A/WNT7B signaling, Trp261 is conserved but Pro256 is not conserved (13).
Despite the high degree of sequence diversity among RECK(CC) domains in different species, secondary structure predictions show that nearly all have high α-helical propensity. Using the Quick2D program, which compares the outputs of 5 secondary structure prediction algorithms (19), each of the 45 CC domains represented by the 9 RECK sequences in SI Appendix, Fig. S10, shows α-helix propensity in all 4 predicted helices, with only 1 exception: helix A in CC2 of termite RECK.
A similar evolutionary analysis with an mGPR124 query revealed the presence of proteins homologous to GPR124 and GPR125 (the ADGRA subfamily of orphan GPCRs) in diverse Metazoa, but, unlike RECK, a GPR124 homolog was not found in the existing sample of Porifera genomes (Fig. 6A). As with RECK(CC1-5), the N-terminal LRR and Ig domains of GPR124—the regions implicated thus far in WNT7A/WNT7B signaling—are highly divergent.
The presence of a RECK gene in widely divergent Metazoa implies that it arose >1 billion years ago. Thus, there has been ample time for exons encoding RECK(CC) domains to migrate to other locations in the genome and insert into genes coding for cell surface and secreted proteins. The likelihood that such an event would produce a functional protein depends in part on the intron-exon arrangement of the RECK(CC) gene. On first principles, it is reasonable to assume that the most facile spread of exonic sequences occurs when 1) the mobile exon(s) code for a protein segment that corresponds to one or more autonomously folding units (i.e., it corresponds precisely to one or more CC domains, all of which appear to fold autonomously based on their efficient production in mammalian cells and E. coli; refs. 12 and 13 and this work) and 2) the total length of the mobile exon(s) is a multiple of three nucleotides so that the inclusion of the new exon(s) in the mature mRNA does not create a frame-shift mutation in the target gene. Even with those most favorable arrangements, the reading frame of the inserted exons may not match the reading frame of the flanking exons in the target gene or, even if the reading frames match, the inserted protein segment may disrupt the folding or stability of the target protein.
Fig. 6B shows the locations of introns for mouse and D. melanogaster Reck, with the former serving as an example of the arrangement among mammalian Reck genes. In mRECK, introns A and D precisely delineate CC1 and are both in reading frame +1, introns D and G precisely delineate CC2 and are both in reading frame +1, and introns G and H precisely delineate CC3 and are both in reading frame +1. In Drosophila RECK, introns V and X precisely delineate CC1 and are both in reading frame +1, and introns X and Z precisely delineate CC2+CC3 and are both in reading frame +1. Thus, in these two examples, Reck intron-exon structure is optimal on both counts (as described in the preceding paragraph) for the migration of functional versions of mammalian CC1, CC2, and CC3 and Drosophila CC1 and CC2+CC3 to new genomic loci.
In sum, the evolutionary data show roughly similar times of appearance and radiation of WNT, RECK, and GPR124 families. With respect to RECK(CC) domain evolution, the most striking findings are 1) the presence of a Reck gene in widely divergent Metazoa; 2) the absence of recognizable RECK(CC) domain homologs in non-RECK proteins; 3) the high evolutionary diversity of most noncysteine positions, including those implicated in WNT7A/WNT7B signaling; and 4) an intron-exon arrangement favorable for RECK(CC) domain migration, but no evidence that such migration has occurred.
Discussion
The present study reveals the canonical RECK(CC) domain structure to a be a compact, triply disulfide-bonded four-helix bundle. Our strategy for its structure determination, which may be of general utility, consisted of 1) identifying and using a distant structural homolog (nHER1) to constrain a preliminary model, 2) defining a stable and biochemically well-behaved construct by small-scale expression in mammalian cells and in-gel fluorescence of mVenus fusion proteins, 3) fusion to eMBP to facilitate crystallization and permit structure solution by molecular replacement, and 4) large-scale expression of the eMBP fusion with a native disulfide bond arrangement in redox mutant E. coli cells.
Current evidence implicates RECK(CC4) in WNT7A/WNT7B recognition (13–15). Specifically, a protruding “finger” region in the amino terminal domain of WNT7A/WNT7B that is divergent among the 19 Wnt homologs (36, 37) has been implicated by site-directed mutagenesis and peptide binding experiments as a target of RECK(CC4) binding (13–15). As noted in the Results, the electrostatic surface potential of mRECK(CC4) shows a negatively charged solvent-exposed groove (Fig. 2B). Two amino acids in mRECK(CC4)—Pro256 and Trp261—that are essential for RECK-mediated WNT7A/WNT7B signaling reside at the base of this groove (13). Several positively charged residues in the WNT7A/WNT7B finger have been shown to be important for RECK-stimulated signaling, and alanine substitutions of Pro256 and Trp261 in mRECK(CC4) synergize with mutations in the WNT7A/WNT7B finger to reduce signaling in a cell culture system (13–15). It will be interesting to determine whether the negatively charged CC4 groove interacts directly with the positively charged finger in WNT7A/WNT7B, and, if so, whether that interaction is specific to CC4 and WNT7A/WNT7B relative to the other CC domains and Wnts.
An intriguing feature of the CC-domain is its ability to accommodate high sequence diversity at noncysteine residues. Like other small and heavily disulfide-bonded proteins, such as many snake venom proteins and invertebrate toxins, this feature likely reflects the contributions of the three disulfide bonds to structural stability. One consequence of this feature is that none of the noncysteine residues are absolutely conserved across all CC domains and fewer than 20% are highly conserved. Despite this lack of sequence conservation, secondary structure predictions showed that, in our sampling of 45 CC domains across widely divergent species, nearly all of the presumptive helical regions have a propensity to form α-helices. Since the CC domain exhibits an unusually high tolerance for insertion, deletions, and substitutions, it may be useful as a scaffold for protein engineering.
In light of 1) the enormous sequence diversity that can be accommodated by the CC domain, 2) the origin of the Reck gene at or close to the dawn of Metazoan evolution, and 3) the division of RECK(CC)-domain coding sequences into exons in mammals and insects in a manner that is optimal for productive exon migration, it is surprising that recognizable CC domains are detected in present-day Metazoa only in the context of the RECK protein. This limited repertoire stands in contrast to the expansion of several dozen other extracellular domain families into diverse targets that encompass thousands of distinct proteins. In addition to the examples shown in Fig. 5, these include the Cadherin, C-type lectin, CUB, C1q, Discoidin, EMI, Fasciclin-1, IL1-like, Kazal, Kringle, LRR, SEA, Sema, SCR, TGF-β, Von Willebrand factor A, and Zona pellucida domain families.
Finally, RECK’s ancient origin and high sequence diversity suggest that RECK initially evolved in primitive Metazoa to perform a function other than stimulating WNT7A/WNT7B signaling. This suggestion is supported by the presence of a RECK ortholog in diverse Metazoa, such as D. melanogaster, that do not have a recognizable WNT7 branch in their more limited repertoire of WNT family members. This line of reasoning implies that RECK was coopted, along with GPR124, early in vertebrate evolution to facilitate WNT7A/WNT7B signaling. At present, the more ancient function of the RECK(CC) domains remains to be determined.
Materials and Methods
Structure-Based Searches for Distant Homologs and Computational Modeling.
The sequence encompassing mRECK(CC1-CC5) (UniProtKB Q9Z0J1; residues 23 to 345) was used as the query to search for homologs in the protein structure database of the Protein Data Bank (PDB) using the HHpred server (19, 38). Computational models of mRECK(CC4) based on nHER-1 (PDB ID code 1SZH) and of mRECK(CC1) based on the crystal structure of mRECK(CC4) were generated with MODELLER (39).
Construct Design and Cloning.
For expression in mammalian cells, mRECK(CC4) coding segments were PCR-amplified from full-length mRECK (12) and cloned into the pHLsec-mVenus-12H vector, which contains a C-terminal human Rhinovirus (HRV)-3C protease cleavage site followed by a linker, monomeric (m)Venus, and a tandem pair of 6×His tags (23, 40). For expression in E. coli, a DNA segment coding for mRECK(CC4) residues 206 to 270 was cloned into a modified pET-11d vector (pET-11d-eMBP-8H). This vector has an N-terminal engineered (e)MBP fragment (PCR-amplified from pHLmMBP construct 3; ref. 24) and a C-terminal 8×His tag (SI Appendix, Fig. S4A). All constructs were confirmed by sequencing.
Mammalian Expression and Analysis of mVenus Fusion Proteins.
HEK293T (ATCC CRL-11268) cells were maintained in a humidified 37 °C incubator with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM; MilliporeSigma) supplemented with 2 mM L-glutamine (L-Glu; Gibco), 0.1 mM nonessential amino acids (NEAAs; Gibco), and 10% (vol/vol) fetal bovine serum (FBS; Gibco). The FBS concentration was lowered to 2% (vol/vol) after transfection. For small-scale transfection, HEK293T cells were grown in 12-well plates and transfected with mRECK(CC4) plasmids using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Conditioned media were collected 2 d after transfection. For immobilized metal affinity chromatography (IMAC) purification, each sample of 1 mL of conditioned medium was supplemented with 1 M Hepes, pH 7.5, to a final concentration of 20 mM, and then 10 μL Ni Sepharose Excel resin (GE Healthcare Life Sciences) was added. The samples were gently rotated for 60 min and then centrifuged at 2,000 rpm for 2 min. After discarding the medium, the IMAC resin was washed with 20 mM Hepes, pH 7.5, 0.5 M NaCl, 20 mM imidazole, and 10% glycerol, and the bound proteins were eluted in 20 mM Hepes, pH 7.5, 0.15 M NaCl, 0.5 M imidazole. For electrophoretic separation followed by in-gel fluorescent imaging, the eluted proteins were subjected to nonreducing sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) (NuPAGE 4 to 12% Bis-Tris Protein Gels; Thermo Fisher Scientific) with electrophoresis at 120 V at 4 °C. The mVenus fluorescent signal was detected using the Odyssey Fluorescent Imaging System (LiCor).
E. coli Expression and Protein Purification.
The pET-11d-eMBP-8H plasmid with the mRECK(CC4) insert was transformed into E. coli SHuffle T7 cells (New England Biolabs) and induced with 0.2 mM isopropyl β-thiogalactopyranoside in Luria broth containing 100 μg/mL ampicillin (MilliporeSigma) at room temperature (∼25 °C) overnight. For cell disruption, the cell pellets were harvested by centrifugation and resuspended in B-PER bacterial protein extract reagent (ThermoFisher) supplemented with 50 mM Hepes, pH 7.5, 0.3 M NaCl, 30 mM imidazole, 1 mM MgCl2, 500 U Benzonase (MilliporeSigma), 0.2 mg/mL lysozyme, and “cOmplete” Protease Inhibitor Cocktail (MilliporeSigma). The cell lysate was clarified by centrifugation, and the supernatant was filtered using a 0.45-μm Steritop filter (MilliporeSigma). Proteins were purified by IMAC using Ni Sepharose 6 Fast Flow resin (GE Healthcare Life Sciences). The resin was washed with 20 mM Hepes, pH 7.5, 0.5 M NaCl, 30 mM imidazole, and 10% glycerol, and eluted in 20 mM Hepes, pH 7.5, 0.15 M NaCl, 0.5 M imidazole. The eluted sample was subjected to size-exclusion chromatography (SEC) using HiLoad Superdex 200 pg (GE Healthcare Life Sciences) in 10 mM Hepes, pH 7.5, and 0.15 M NaCl.
Crystallization and Data Collection.
Purified eMBP-mRECK(CC4) protein was concentrated to 57.5 mg/mL in a buffer containing 10 mM Hepes, pH 7.5, and 50 mM NaCl. Prior to crystallization, recombinant eMBP-mRECK(CC4) protein was supplemented with 1.76 mM zinc acetate and 10 mM maltose. Using a Mosquito LCP crystallization robot (TTP Labtech), crystallization screens were set up using sitting-drop vapor diffusion in 96-well MRC 2 Well UVXPO plates (Hampton Research) and consisting of 150 nL protein solution and 150 nL reservoir. Crystals were grown in 0.2 M ammonium sulfate, 0.1 M sodium acetate, pH 4.6, 25% polyethylene glycol (PEG) 4K. Crystals were transferred into a reservoir solution supplemented with 15% PEG400 and then cryocooled in liquid nitrogen. X-ray diffraction data were collected at 100 °K using a Rigaku FR-E SuperBright source with a PILATUS 3R 200K-A detector (DECTRIS) at the Department of Biophysics and Biophysical Chemistry (Johns Hopkins University) X-ray facility and on the 17-ID-1 AMX beamline using an EIGER X 9M detector (DECTRIS) at the National Synchrotron Light Source II (NSLS II), Brookhaven National Laboratory (Upton, NY). Diffraction data were indexed, integrated, and scaled using the XIA2 expert system (41) coupled with DIALS (42), POINTLESS (43), and AIMLESS (44). A randomly selected subset of 5% of the diffraction data was used for calculating Rfree (45).
Structure Determination and Refinement.
The structure of eMBP-mRECK(CC4) was determined by molecular replacement in PHASER (46) using the MBP structure (PDB ID code 3SET) as a template to obtain the initial phases. The resulting map showed an additional electron density corresponding to mRECK(CC4). The electron density map of mRECK(CC4) was improved after density modification with PARROT (47) and subsequently fed into BUCCANEER in the CCP4 suite (48, 49) for initial model building. The mRECK(CC4) model was completed by manual building in COOT (50), and refinement was performed using REFMAC5 (51) and PHENIX Refine (52) with translation-libration-screw (TLS) parameterization. MOLPROBITY (53) was used to validate the models. The crystallographic statistics are listed in SI Appendix, Table S1.
Structure Analysis and Graphic Presentation.
Electrostatic potential calculations were generated using APBS tools (54). High-quality images of the molecular structures were generated with the PyMOL Molecular Graphic System (version 2.2; Schrödinger). Schematic figures and other illustrations were prepared using Corel Draw (Corel) and Illustrator (Adobe). The solvent-accessible surface area for individual residues was calculated by AreaIMol in the CCP4 software suite (49). Extracellular proteins with the domains listed in Fig. 5 were selected from the following databases: Evolutionary Classification of Protein Domains (prodata.swmed.edu/ecod/; ref. 55) and Structural Classification of Proteins (scop.mrc-lmb.cam.ac.uk; ref. 56). The secondary structure predictions for CC-domains were calculated using Quick2D, which compares the outputs of five secondary structure prediction algorithms (19).
Sequence-Based Phylogenetic Analysis.
The amino acid sequences of mRECK CC1-5 were aligned by ClustalW (57), and the phylogenetic tree was constructed with the neighbor-joining method using MEGA7 (58, 59). The optimal tree with the sum of branch lengths = 3.53685486 is shown. The phylogenetic tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the tree. The evolutionary distances were computed using the Poisson correction method (60) and are in the units of the number of amino acid substitutions per site. The analysis involved five amino acid sequences. All positions containing gaps and missing data were eliminated. There was a total of 44 positions in the final dataset.
Structure Search and Comparison of mRECK(CC4) with Four-Helix Bundle Proteins.
Searches for structure-based similarities to mRECK(CC4) were performed against the Protein Data Bank (PDB) using the DALI server (30). The queries consisted of the full-length mRECK(CC4) structure and a truncated derivative that includes only the four helices. The results with the two queries were highly correlated. To reduce the number of redundant PDB structures in the search, we used the PDB90 database (i.e., structures having less than 90% sequence identity to each other). Two key scoring functions were applied: 1) the rmsd of Cα atoms in the optimally superimposed structures and 2) the plastic deformation required for superimposition (61). A total of 511 structures were identified in the first-round search. Of these, 391 have >5% sequence identity with mRECK(CC4) in the aligned regions, and were selected for the comparison with mRECK(CC4) and classification by visual inspection. Twenty-one representative structures were further selected for the construction of a structural similarity dendrogram based on the criteria of one structure per ortholog family and possession of a distinct protein function, and were used for the construction of a structural similarity dendrogram. The distance matrices shown in Fig. 4B were derived by average linkage clustering of the structural similarity matrix. Detailed information is listed in SI Appendix, Table S2.
Data Availability.
The atomic coordinates and structure factors of the mRECK(CC4) domain have been deposited in the Protein Data Bank (PDB) under ID codes 6WBH and 6WBJ.
Supplementary Material
Acknowledgments
The authors thank Jean Jakoncic and Alexei Soares at the 17-ID-1 AMX beamline (Brookhaven National Laboratory) for assistance with data collection; Luca Jovine (Karolinska Institute) for the pHLmMBP vector; Ray Owens (Protein Production UK) and David Drew (Stockholm University) for helpful discussions about in-gel fluorescent imaging; and Chris Cho, Amir Rattner, and Jie Wang for advice and/or helpful comments on the manuscript. This work was supported by the Howard Hughes Medical Institute, the National Eye Institute (NIH; Grant R01EY018637), and the Arnold and Mabel Beckman Foundation. T.-H.C. was supported by a Human Frontier Science Program long-term fellowship (Grant LT000130/2017-L). S.B.G. was supported by Department of Defense Congressionally Directed Medical Research Programs Grant BC151831 and the National Cancer Institute (NIH) Grant CA062924SBG. Work at the Center for Biomolecular Structure beamline AMX (17ID-1) | FMX (17ID-2) | LIX (16ID) at NSLS-II was supported by the National Institute of General Medical Sciences (NIH; Grant 1P30GM133893), the Office of Biological and Environmental Research (Department of Energy [DOE]; Grant BER- BO 070), and the Office of Basic Energy Sciences Program (DOE; Grant BES-FWP-PS001).
Footnotes
The authors declare no competing interest.
Data deposition: The structures of the mRECK(CC4) domain have been deposited in the Protein Data Bank, https://www.rcsb.org/ (PDB ID 6WBH and 6WBJ).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006332117/-/DCSupplemental.
References
- 1.Takahashi C. et al., Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc. Natl. Acad. Sci. U.S.A. 95, 13221–13226 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muraguchi T. et al., RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat. Neurosci. 10, 838–845 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Chandana E. P. et al., Involvement of the Reck tumor suppressor protein in maternal and embryonic vascular remodeling in mice. BMC Dev. Biol. 10, 84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto M. et al., The transformation suppressor gene Reck is required for postaxial patterning in mouse forelimbs. Biol. Open 1, 458–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Almeida G. M. et al., Critical roles for murine Reck in the regulation of vascular patterning and stabilization. Sci. Rep. 5, 17860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J. et al., The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107, 789–800 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Park S. et al., GDE2 promotes neurogenesis by glycosylphosphatidylinositol-anchor cleavage of RECK. Science 339, 324–328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanhollebeke B. et al., Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. eLife 4, e06489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulrich F. et al., Reck enables cerebrovascular development by promoting canonical Wnt signaling. Development 143, 147–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Nathans J., Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev. Cell 31, 248–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Posokhova E. et al., GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Rep. 10, 123–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho C., Smallwood P. M., Nathans J., Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and blood-brain barrier regulation. Neuron 95, 1056–1073.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho C., Wang Y., Smallwood P. M., Williams J., Nathans J., Molecular determinants in Frizzled, Reck, and Wnt7a for ligand-specific signaling in neurovascular development. eLife 8, e47300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eubelen M. et al., A molecular mechanism for Wnt ligand-specific signaling. Science 361, eaat1178 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Vallon M. et al., A RECK-WNT7 receptor-ligand interaction enables isoform-specific regulation of Wnt bioavailability. Cell Rep. 25, 339–349.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H. et al., RECK in neural precursor cells plays a critical role in mouse forebrain angiogenesis. iScience 19, 559–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S. F. et al., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J., Blundell T. L., Mizuguchi K., FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann L. et al., A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Hodgkin J., More sex-determination mutants of Caenorhabditis elegans. Genetics 96, 649–664 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamaoka B. Y., Dann C. E. 3rd, Geisbrecht B. V., Leahy D. J., Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A. Proc. Natl. Acad. Sci. U.S.A. 101, 11673–11678 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trombetta E. S., Parodi A. J., Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19, 649–676 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Chang T. H. et al., Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. eLife 4, e06554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokhove M. et al., Easy mammalian expression and crystallography of maltose-binding protein-fused human proteins. J. Struct. Biol. 194, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laganowsky A. et al., An approach to crystallizing proteins by metal-mediated synthetic symmetrization. Protein Sci. 20, 1876–1890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon A. F., Mueller G. A., Zhong X., Pedersen L. C., A synergistic approach to protein crystallization: Combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19, 901–913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker I. H., Hsieh P. C., Riggs P. D., Mutations in maltose-binding protein that alter affinity and solubility properties. Appl. Microbiol. Biotechnol. 88, 187–197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waugh D. S., Crystal structures of MBP fusion proteins. Protein Sci. 25, 559–571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobstein J. et al., SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 11, 56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel C., Chothia C., Protein family expansions and biological complexity. PLOS Comput. Biol. 2, e48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan J. F., Baxevanis A. D., Hox, Wnt, and the evolution of the primary body axis: Insights from the early-divergent phyla. Biol. Direct 2, 37 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holstein T. W., Watanabe H., Ozbek S., Signaling pathways and axis formation in the lower metazoa. Curr. Top. Dev. Biol. 97, 137–177 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Croce J. C., McClay D. R., Evolution of the Wnt pathways. Methods Mol. Biol. 469, 3–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borisenko I., Adamski M., Ereskovsky A., Adamska M., Surprisingly rich repertoire of Wnt genes in the demosponge Halisarca dujardini. BMC Evol. Biol. 16, 123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janda C. Y., Waghray D., Levin A. M., Thomas C., Garcia K. C., Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirai H., Matoba K., Mihara E., Arimori T., Takagi J., Crystal structure of a mammalian Wnt-frizzled complex. Nat. Struct. Mol. Biol. 26, 372–379 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Söding J., Biegert A., Lupas A. N., The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eswar N. et al., “Comparative protein structure modeling using modeller” in Curr. Protoc. Bioinf., (2006), Vol. Chapter 5, p. Unit-5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aricescu A. R., Lu W., Jones E. Y., A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Winter G., xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010). [Google Scholar]
- 42.Winter G. et al., DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol. 74, 85–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brünger A. T., Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr. D Biol. Crystallogr. 49, 24–36 (1993). [DOI] [PubMed] [Google Scholar]
- 46.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowtan K., Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowtan K., The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Winn M. D. et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov G. N. et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen V. B. et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng H. et al., ECOD: An evolutionary classification of protein domains. PLOS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreeva A., Kulesha E., Gough J., Murzin A. G., The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 48, D376–D382 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson J. D., Higgins D. G., Gibson T. J., CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 60.Zuckerkandl E., Pauling L., “Evolutionary divergence and convergence in proteins” in Evolving Genes and Proteins, Bryson V., Vogel H. J., Eds. (Academic Press, New York, 1965), pp. 97–166. [Google Scholar]
- 61.Hasegawa H., Holm L., Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 19, 341–348 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of the mRECK(CC4) domain have been deposited in the Protein Data Bank (PDB) under ID codes 6WBH and 6WBJ.