Significance
We present the third high-quality genome to be determined from a Neandertal. Patterns of variation in the genome suggest that her ancestors lived in relatively isolated populations of less than 60 individuals. When we analyze this genome together with two previously sequenced Neandertal genomes, we find that genes expressed in the striatum of the brain may have changed especially much, suggesting that the striatum may have evolved unique functions in Neandertals.
Keywords: genetics, Neandertals, human evolution, genome
Abstract
We sequenced the genome of a Neandertal from Chagyrskaya Cave in the Altai Mountains, Russia, to 27-fold genomic coverage. We show that this Neandertal was a female and that she was more related to Neandertals in western Eurasia [Prüfer et al., Science 358, 655–658 (2017); Hajdinjak et al., Nature 555, 652–656 (2018)] than to Neandertals who lived earlier in Denisova Cave [Prüfer et al., Nature 505, 43–49 (2014)], which is located about 100 km away. About 12.9% of the Chagyrskaya genome is spanned by homozygous regions that are between 2.5 and 10 centiMorgans (cM) long. This is consistent with the fact that Siberian Neandertals lived in relatively isolated populations of less than 60 individuals. In contrast, a Neandertal from Europe, a Denisovan from the Altai Mountains, and ancient modern humans seem to have lived in populations of larger sizes. The availability of three Neandertal genomes of high quality allows a view of genetic features that were unique to Neandertals and that are likely to have been at high frequency among them. We find that genes highly expressed in the striatum in the basal ganglia of the brain carry more amino-acid-changing substitutions than genes expressed elsewhere in the brain, suggesting that the striatum may have evolved unique functions in Neandertals.
Neandertals and Denisovans are the closest evolutionary relatives of present-day humans. Analyses of their genomes showed that they contributed genetically to present-day people outside sub-Saharan Africa (1, 2). However, to date, the genomes of only two Neandertals and one Denisovan have been sequenced to high quality. One of these Neandertal genomes (Vindija 33.19) comes from an individual found in Vindija Cave in Croatia (3), whereas the other Neandertal genome (Denisova 5 or the “Altai Neandertal”) (4) and the Denisovan genome (Denisova 3) (5) both come from specimens discovered in Denisova Cave, in the Altai mountains in Siberia.
A number of archaic genomes of moderate quality (one- to threefold genomic coverage) have yielded additional insights into Neandertal history. For example, genome sequences from five late Neandertals from Europe have shown that they carried little genetic variation (6, 7) and were more closely related to the Vindija 33.19 than to the Denisova 5 Neandertal. A genome sequence from a morphologically undiagnostic bone from Denisova Cave, Denisova 11, belonged to the direct offspring of a Neandertal mother and a Denisovan father (8), indicating that the two groups met in the Altai region. The Neandertal mother of Denisova 11 was more closely related to Vindija 33.19 than to Denisova 5, indicating that a replacement of Neandertal populations in the Altai Mountains occurred (8).
Here, we present the high-coverage genome sequence of a Neandertal from Chagyrskaya Cave, located 106 km to the west of Denisova Cave (9–12) (Fig. 1). This genome provides insights into Neandertal population structure and history and allows the identification of genomic features unique to Neandertals.
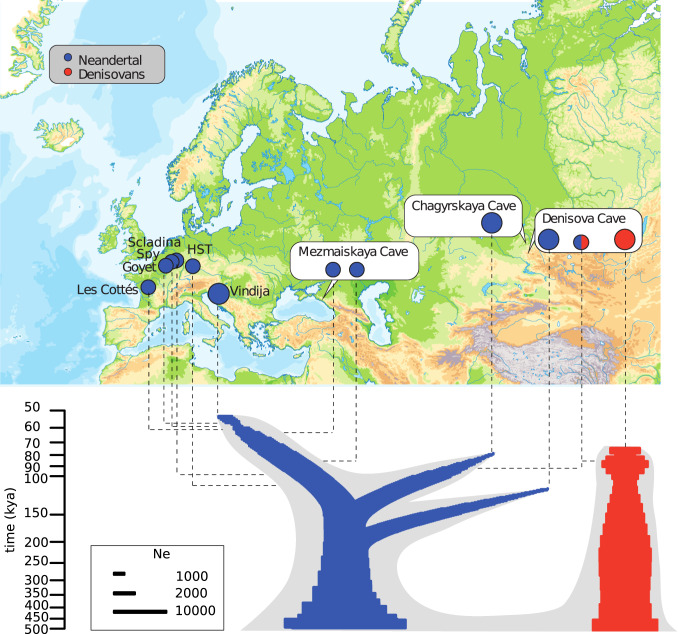
Fig. 1.
The Chagyrskaya 8 Neandertal and its relationship with other archaic individuals. (Top) Locations of Chagyrskaya Cave and other sites where archaic specimens analyzed here were found are indicated. (Bottom) Schematic illustration of the relationship among archaic genomes. The gray outline describes relationships between high-coverage genomes (SI Appendix 7). Population split times estimated using the F(A|B) statistics (3, 4, 8) (SI Appendix 7) between high-coverage and low-coverage genomes are indicated by horizontal dashed line. Within the gray outlines, colored silhouettes schematically indicate population sizes (Ne) over time as estimated in SI Appendix 6. Neandertals are indicated in blue, Denisovans in red. Genomes determined to high genomic coverage (>27-fold) are indicated by large circles. The genomes from Spy, Goyet, and Les Cottés are reported in ref. 6, Scladina and Holehnstein-Stadel in ref. 13, other genomes in refs. 2–6, 8.
Results
Genome Sequencing and Age Estimates.
We sampled 100 mg of bone powder from Chagyrskaya 8, a phalanx found in 2011 at Chagyrskaya Cave in layer 6b (SI Appendix 1). The DNA extracted (14) allowed the nuclear genome to be sequenced (SI Appendix 2) to an average coverage of 27.6-fold (SI Appendix 3). Less than 1% of the DNA fragments sequenced were estimated to originate from contamination by present-day modern human DNA (SI Appendixes 4 and 5).
We estimated the age of Chagyrskaya 8 using two different methods (SI Appendix 6). First, we counted the proportion of “missing” derived substitutions compared to present-day genomes (3–5, 15). We also used a method similar to Fu et al. (15) that takes advantage of the shared evolutionary history of the three high-coverage Neandertal genomes. Under the assumption that Neandertals had the same mutation rate (1.45 × 10−8 mutations per generation per base pair) (15) and generation time as present-day humans (29 y), both methods suggest that Chagyrskaya 8 lived ∼80 kya (1,000 y ago), i.e., ∼30 ky (1,000 y) after Denisova 5 and ∼30 ky before Vindija 33.19. This estimate is older than the optically stimulated luminescence dates of ∼60 kya (10, 12) for the archaeological layer in which Chagyrskaya 8 was found. Excluding redeposition from lower, older layers (10, 12), this might indicate that the genetic dates, based on the current mutation rate in humans, are incorrect. Possible explanations could be that Neandertals had a lower mutation rate than modern humans or that that the modern human mutation rate decreased recently (16). Additional high-quality genomes determined from well-dated Neandertal remains are needed to address these possibilities. Nevertheless, the Chagyrskaya 8 Neandertal and the Denisovan Denisova 3 display similar proportions of missing mutations relative to present-day humans, suggesting that they lived approximately at the same time (SI Appendix 6).
Relationship to Other Neandertals and Denisovans.
Chagyrskaya 8 shares more derived alleles with Vindija 33.19 and other later Neandertals in the Caucasus and in Europe than with the older Denisova 5 Neandertal from the Altai (Fig. 2). With the Denisovan Denisova 3, Chagyrskaya 8 shares fewer derived alleles than does Denisova 5.
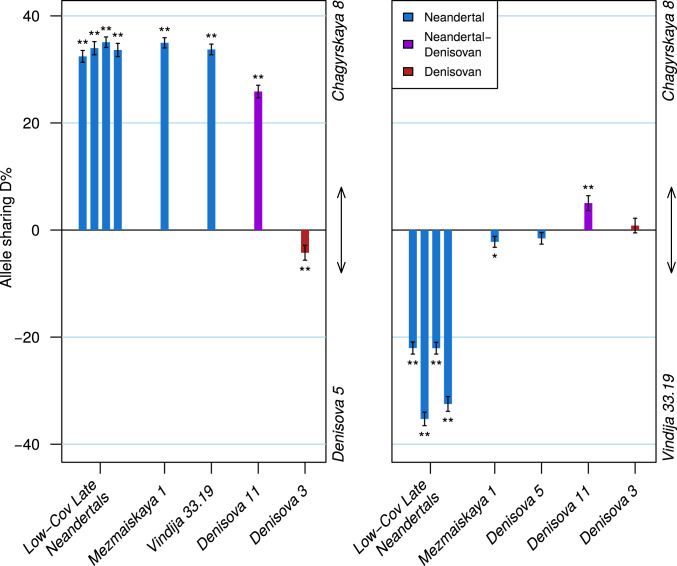
Fig. 2.
Relative sharing of derived allele among the Chagyrskaya 8 and other archaic genomes. Positive values in the D statistic indicate more allele sharing with Chagyrskaya 8 than with Denisova 5 (A) or than with Vindija 33.19 (B). Error bars indicate one SE. One asterisk indicates Z > 2; two asterisks indicate Z > 3. Late Neandertals of low genomic coverage refer to the genomes in ref. 6.
When compared to Vindija 33.19 (Fig. 2), Chagyrskaya 8 shares fewer derived alleles with other Neandertals that lived in Europe ∼50 kya, i.e., approximately at the same time as Vindija 33.19. However, Chagyrskaya 8 shares more derived alleles than Vindija 33.19 with Denisova 11, a first-generation Neandertal–Denisovan offspring (8). Since Vindija 33.19 and Chagyrskaya 8 do not differ in their sharing of derived alleles with the Denisovan Denisova 3, this indicates that Chagyrskaya 8 is most closely related among currently known Neandertal to the mother of Denisova 11 found at Denisova Cave (Fig. 1 and SI Appendix 7).
Relationship with Modern Humans.
Non-African present-day humans carry ∼2% (1, 3) Neandertal ancestry as a result of gene flow from Neandertals that occurred between 50 and 90 kya (1, 17). Genomewide, Chagyrskaya 8 shares more alleles with present-day human populations outside Africa than Denisova 5 does, and a similar proportion of alleles as Vindija 33.19 (SI Appendix 7). However, if the analysis is restricted to previously detected Neandertal haplotypes introgressed in present-day humans (18), or to derived alleles that occur at low frequencies in present-day non-African populations and that are therefore more likely to be introgressed from Neandertals, Vindija 33.19 shares more alleles with present-day populations than Chagyrskaya 8. This indicates that Vindija 33.19 is more closely related than Chagyrskaya 8 to Neandertal populations that contributed the majority of the DNA to present-day populations.
To test if any modern human population carries an additional genetic contribution from Neandertals more closely related to Chagyrskaya 8 than to Vindija 33.19, we made use of previously published haplotypes inferred to come from Neandertals that are found today exclusively in East Asia, Europe, India, or Oceania. Among 300 genomes from the Simons Genome Diversity Panel (19) and 89 Papuan genomes (20, 21), the proportions of alleles shared with Chagyrskaya 8 and with Vindija 33.19 are similar (SI Appendix 7), giving no indication that Neandertals closer related to Chagyrskaya 8 than to Vindija 33.19 contributed to the populations tested. Thus, within the limits of the resolution of these analyses, we conclude that if several Neandertal populations contributed to the genomes of different modern human populations in different proportions (22, 23), these Neandertal populations were similarly related to Chagyrskaya 8 and to Vindija 33.19.
Small Population Size and Inbreeding.
The Neandertal genome from Denisova Cave, Denisova 5, carries a high proportion of long tracts that are homozygous by descent (HBD tracts) (4). Whereas tracts that are over 10 cM long indicate that the parents of Denisova 5 were closely related, HBD tracts between 2.5 and 10 cM indicate that the population from which Denisova 5 comes was of small size over ∼100 generations before the individual lived (SI Appendix 8).
Compared to Denisova 5, the Chagyrskaya 8 genome carries fewer HBD tracts longer than 10 cM, but more HBD tracts of intermediate length (SI Appendix 8). In fact, all three high-coverage Neandertal genomes available carry more HBD tracts of intermediate size than almost all present-day and prehistoric modern human genomes, as well as the Denisovan genome (Denisova 3) (Fig. 3A). We show by coalescent simulations that this cannot be explained by an overall small but panmictic population. Rather, it suggests that Neandertal populations were subdivided (SI Appendix 8).
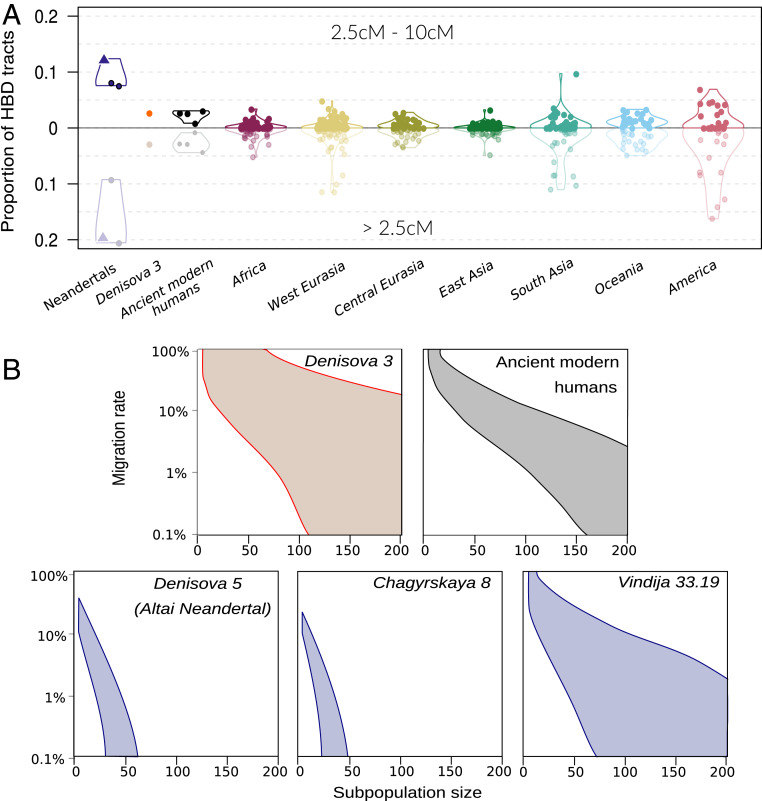
Fig. 3.
Homozygous tracts in archaic genomes. (A) Proportion of the genome spanned by HBD tracts of size 2.5–10 cM (Top, dark) and all HBD tracts (Bottom, light) for archaic genomes, ancient modern humans and present-day humans from the Simons Genome Diversity Project (18). Values for Chagyrskaya 8 are indicated by a triangle. The genetic size of the tracts was estimated assuming a uniform recombination rate of 1.3 × 10−8 recombinations/bp. (B) Estimated group size (x axis) and migration rate (y axis) for three Neandertals, one Denisovan, and ancient modern human genomes based on HBD tracts >2.5 cM long. The colored areas delimit 95% confidence intervals of likelihood ratios. Ancient modern humans give estimates for the genomes of four individuals dated to between ∼45 and ∼8 kya [“Ust’Ishim” (15), “Sunghir 2” (24), “Loschbour” (25), and “LBK-Stuttgart” (25)].
By coalescent modeling, we infer that Chagyrskaya 8 and Denisova 5 may have lived in subpopulations of 60 or fewer individuals. In contrast, current and past modern human populations as well as Denisovans (based on the Denisova 3 genome) lived in subpopulations of more than 100 individuals, assuming a migration rate between subpopulations of 1% or less (Fig. 3B and SI Appendix 8). Interestingly, the Vindija 33.19 Neandertal seems to have lived in a subpopulation of larger size than the two Siberian Neandertals, although this difference is only marginally statistically significant when the proportion of the genomes covered by all HBD tracts longer than 2.5 cM is considered (likelihood-ratio test, P = 0.05).
Derived Genomic Features in Neandertals.
We used the three high-coverage Neandertal genomes to identify biological pathways where protein-coding genes show more derived nonsynonymous substitutions fixed in the three Neandertals than expected from the silent and polymorphic changes. We identified 993 fixed nonsynonymous substitutions among 889 genes, and 2,952 polymorphic nonsynonymous substitutions in the three Neandertals.
No groups of genes associated with a specific known biological functions or phenotypes (26) show a higher ratio of nonsynonymous to synonymous fixed changes relative to the ratio of nonsynonymous to synonymous polymorphic changes (MacDonald–Kreitman ratio) (27), compared to other groups of genes (family-wise error rate > 0.1)(28 and SI Appendix 9).
However, when analyzing genes preferentially expressed in different brain regions according to the Allen Brain Atlas (29, 30), we find that genes expressed in the striatum in individuals 12–19 y of age show a higher MacDonald-Kreitman ratio (1.02, family-wise error rate = 0.029) than genes expressed in other brain regions and at other ages (0.53–0.83) (SI Appendix 9). This may indicate that negative selection in genes expressed in the striatum was relaxed in Neandertals. Alternatively, some of the proteins encoded by these genes might have been the targets of positive selection. In addition, genes expressed in the prenatal striatum carry more substitutions in their untranslated regions than genes expressed elsewhere (family-wise error rate = 0.049) and at other times. Among genes expressed in the striatum, those carrying fixed nonsynonymous changes in Neandertals are more often present in genomic regions that carry little or no DNA introgressed from Neandertals in present-day humans than striatal genes not carrying such changes (Fisher’s exact test P = 0.026). This pattern is not observed for all genes carrying fixed nonsynonymous substitutions in Neandertals (P > 0.1), suggesting that some substitutions in Neandertal striatal genes might have been negatively selected in modern humans. Besides the striatum, genes expressed prenatally in the posterior parietal cortex, in the ventrolateral prefrontal cortex, and in the primary somatosensory cortex carry more fixed substitutions in their regulatory regions in Neandertals than genes expressed in other brain regions and at other times (SI Appendix 9).
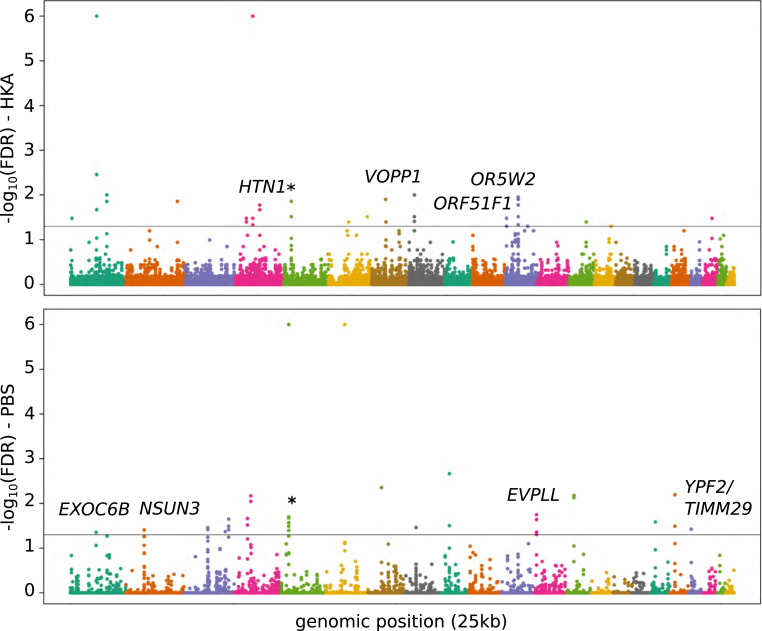
In a further attempt to detect positive selection along the Neandertal lineage, we performed the Hudson, Kreitman, Aguadé (HKA) test (31) and the population branch statistic (PBS) (32) in 25-kb sliding windows across the genome (SI Appendix 10). We estimated the probability of obtaining the observed values of the different statistics by coalescent simulations, and retained windows with a false discovery rate <5% (Fig. 4). On the autosomes, we identify a total of 35 separate candidate regions. One of these candidates, a 75-kb-long region identified by PBS on chromosome 5, overlaps two separate windows identified by HKA. This overlap is lower than if expected by change (P value < 4 × 10−4 and SI Appendix 10). The candidate regions identified overlap genes involved in neural development (EXOC6B), immunity, and wound healing (HTN1, EVPLL) and mitochondrial functions (NSUN3, TIMM29). For both statistics, we find an overlap with genomic regions previously identified as positively selected in modern humans (33) (enrichment test, P values = 0.010 and 0.056 for HKA and PBS, respectively).
Fig. 4.
Positive selection on the Neandertal lineage. Manhattan plots of HKA (Top) and PBS (Bottom) tests for positive selection applied to three Neandertal genomes. Different colors indicate different autosomes, from 1 (Left) to 22 (Right). The y axis indicates −log10 of false discovery rates (FDR). The gray line indicates FDR = 5%. Candidate windows that overlap exons are indicated by the name of the corresponding gene. The regions that are significant for both tests are indicated by an asterisk.
Discussion
Chagyrskaya 8 is more closely related to Vindija 33.19 and other late Neandertals in western Eurasia than to the Denisova 5 Neandertal who lived earlier in the Altai Mountains (Fig. 2). Chagyrskaya 8 is thus related to Neandertal populations that moved east sometime between 120 and 80 kya (13). Interestingly, the artifacts found in Chagyrskaya Cave show similarities to artifact assemblages in central and eastern Europe (10 and SI Appendix 1), suggesting that Neandertal populations coming from western Eurasia to Siberia may have brought their material culture with them (10, 34). Some of these incoming Neandertals encountered local Denisovan populations, as shown by Denisova 11, who had a Denisovan father and a Neandertal mother related to the population in which Chagyrskaya 8 lived.
In this regard, it is interesting that the Chagyrskaya 8 and Denisova 5 Neandertals lived in smaller populations than the Vindija 33.19 Neandertal in Croatia, the Denisovan Denisova 3, and modern humans (24) (Fig. 3B). Neandertals in the Altai region may have lived in smaller and more isolated populations than Neandertals elsewhere as that region represented the periphery of their geographical distribution and may have been an area where Denisovans were more continuously present. However, more detailed studies of the population history of Denisova Cave and other sites will be necessary to clarify this.
When analyzing genetic changes on the Neandertal lineage using the three Neandertal genomes, the number of changes in genes expressed in the striatum during adolescence stands out. One possibility is that these changes accumulated in Neandertals due to their small population size, perhaps in combination with a relaxation of selection on genes expressed in the striatum. Interestingly, genes expressed in the striatum overlap more frequently than expected with genomic regions where Neandertal introgressed fragments in modern human genomes are rare. We speculate that striatal genes may carry Neandertal-specific changes that were disadvantageous when introduced into modern humans. This, as well as positive selection for derived changes in the modern human lineage, may underlie so-called Neandertal deserts in present-day human genomes, i.e., regions that are depleted of Neandertal ancestry (20, 35).
As more high-quality Neandertal genomes become available it will be possible to more comprehensively explore genes and groups of genes that carried functionally relevant changes in Neandertals. Currently, there is suggestive evidence that such findings may be forthcoming. For example, in addition to genes expressed in the striatum, untranslated regions and promoters of genes expressed in the posteroventral (inferior) parietal cortex, a brain region that has been associated with speech and mathematical cognition (36), carry more changes in the three Neandertals than expected by chance. In addition, among the top phenotypes associated with changes in regulatory regions in Neandertals are abnormalities in parts of the skeleton where Neandertal morphology stand out, such as the nasal bridge and the rib cage (e.g., ref. 37).
Methods
DNA was extracted from ∼100 mg of bone power after sodium hypochlorite treatment (SI Appendix 2). Sequencing, filtering of the data, and genotyping was carried out as previously described (ref. 3 and SI Appendix 3). Present-day human DNA contamination was estimated using mitochondrial, autosomal data, and from the proportion of DNA fragment mapping on the Y chromosome (SI Appendixes 4 and 5). Demographic histories and split times were estimated using the Pairwise Sequentially Markovian Coalescent (38 and SI Appendix 6) and the F(A|B) method (refs. 35 and SI Appendix 7), respectively, and coestimated with momi2 (39 and SI Appendix 7). HBD tracts were computed as previously published (refs. 35 and SI Appendix 8). Selection analyses were performed using the R packages GOfuncR (https://github.com/sgrote/GOfuncR), ABAEnrichment (ref. 29 and SI Appendix 9), and by computing HKA and a modified PBS statistic in 25-kb windows (SI Appendix 10).
Data Availability Statement.
The genome sequence of Chagyrskaya 8 can be downloaded from http://ftp.eva.mpg.de/neandertal/Chagyrskaya/VCF/. Its mitochondrial DNA genome sequence has been deposited in GenBank (accession ID MK388903).
Supplementary Material
Acknowledgments
This study was funded by the Max Planck Society; the European Research Council (Grant Agreement 694707 to S.P.); and the Russian Science Foundation (Project 19-48-04107 to K.A.K.). We thank Sarah Nagel, Birgit Nickel, Barbara Schellbach, and Antje Weihmann for laboratory work; and Heiko Temming for computed tomography scans.
Footnotes
The authors declare no competing interest.
Data deposition: The genome sequence of Chagyrskaya 8 can be downloaded from http://ftp.eva.mpg.de/neandertal/Chagyrskaya/VCF/. Its mitochondrial DNA genome sequence has been deposited in GenBank (accession ID MK388903).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004944117/-/DCSupplemental.
References
- 1.Prüfer K. et al., A high-coverage Neandertal genome from Vindija cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajdinjak M. et al., Reconstructing the genetic history of late Neanderthals. Nature 555, 652–656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prüfer K. et al., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green R. E. et al., A draft sequence of the Neandertal Genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich D. et al., Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer M. et al., A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prüfer K., snpAD: An ancient DNA genotype caller. Bioinformatics 34, 4165–4171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slon V. et al., The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561, 113–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudaya N. et al., Palaeoenvironments during the period of the Neanderthals settlement in Chagyrskaya cave (Altai Mountains, Russia). Palaeogeogr. Palaeoclimatol. Palaeoecol. 467, 265–276 (2017). [Google Scholar]
- 10.Kolobova K. A. et al., Archaeological evidence for two separate dispersals of Neanderthals into southern Siberia. Proc. Natl. Acad. Sci. U.S.A. 117, 2879–2885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolobova K. A. et al., The significance of bifacial technologies in Altai Middle Paleolithic. Anthropologie 123, 276–288 (2019). [Google Scholar]
- 12.Roberts R. G., Jacobs Z., Li B., “Optical dating of sediment samples from Chagyrskaya Cave” in Multidisciplinary Studies of Chagyrskaya Cave–A Middle Paleolithic Site in Altai, Shunkov M. V., Ed. (Russian Academy of Sciences Publishing, Novosibirsk, 2018), pp. 353–369. [Google Scholar]
- 13.Peyrégne S. et al., Nuclear DNA from two early Neandertals reveals 80,000 years of genetic continuity in Europe. Sci. Adv. 5, eaaw5873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korlević P. et al., Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Fu Q. et al., Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besenbacher S., Hvilsom C., Marques-Bonet T., Mailund T., Schierup M. H., Direct estimation of mutations in great apes reconciles phylogenetic dating. Nat. Ecol. Evol. 3, 286–292 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Sankararaman S., Patterson N., Li H., Pääbo S., Reich D., The date of interbreeding between Neandertals and modern humans. PLoS Genet. 8, e1002947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skov L. et al., Detecting archaic introgression using an unadmixed outgroup. PLoS Genet. 14, e1007641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallick S. et al., The Simons genome diversity project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vernot B. et al., Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352, 235–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malaspinas A. S. et al., A genomic history of Aboriginal Australia. Nature 538, 207–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vernot B., Akey J. M., Complex history of admixture between modern humans and Neandertals. Am. J. Hum. Genet. 96, 448–453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villanea F. A., Schraiber J. G., Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol. 3, 39–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikora M. et al., Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science 358, 659–662 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Lazaridis I. et al., Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler S. et al., The human phenotype ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 42, D966–D974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald J. H., Kreitman M., Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Prüfer K. et al., FUNC: A package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics 8, 41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grote S., Prüfer K., Kelso J., Dannemann M., ABAEnrichment: An R package to test for gene set expression enrichment in the adult and developing human brain. Bioinformatics 32, 3201–3203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawrylycz M. J. et al., An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson R. R., Kreitman M., Aguadé M., A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi X. et al., Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peyrégne S., Boyle M. J., Dannemann M., Prüfer K., Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 27, 1563–1572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derevianko P., et al. , Multidisciplinary studies of Chagyrskaya Cave–A Middle Paleolithic site in Altai, Shunkov M. V., Ed. (Institute of Archaeology and Ethnography, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, 2018). [Google Scholar]
- 35.Sankararaman S. et al., The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S. S. et al., Functional heterogeneity of inferior parietal cortex during mathematical cognition assessed with cytoarchitectonic probability maps. Cereb. Cortex 19, 2930–2945 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver T. D., Out of Africa: Modern human origins special feature: The meaning of Neandertal skeletal morphology. Proc. Natl. Acad. Sci. U.S.A. 106, 16028–16033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamm J., Terhorst J., Durbin R., Song Y. S., Efficiently inferring the demographic history of many populations with allele count data. J. Am. Stat. Assoc. 20, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence of Chagyrskaya 8 can be downloaded from http://ftp.eva.mpg.de/neandertal/Chagyrskaya/VCF/. Its mitochondrial DNA genome sequence has been deposited in GenBank (accession ID MK388903).