Significance
RNA-mediated chromatin regulation is central to gene expression in many organisms. However, the mechanisms by which RNA influences the local chromatin environment are still poorly understood. Here, we show how RNA 3′ processing factors, which promote proximal polyadenylation of an Arabidopsis antisense transcript, physically associate with the chromatin modifiers FLD/LD/SDG26. The chromatin modifiers exist in a protein complex that inhibits H3K4me1 and H3K36me3 accumulation. By antagonizing transcription, the FLD/LD/SDG26-containing complex promotes H3K27me3 accumulation, reducing transcriptional initiation and elongation rates. This cotranscriptionally mediated chromatin silencing mechanism may be widely relevant for gene regulation in many organisms.
Keywords: non-coding RNA, chromatin, polycomb, FLC, Arabidopsis
Abstract
Noncoding RNA plays essential roles in transcriptional control and chromatin silencing. At Arabidopsis thaliana FLC, antisense transcription quantitatively influences transcriptional output, but the mechanism by which this occurs is still unclear. Proximal polyadenylation of the antisense transcripts by FCA, an RNA-binding protein that physically interacts with RNA 3′ processing factors, reduces FLC transcription. This process genetically requires FLD, a homolog of the H3K4 demethylase LSD1. However, the mechanism linking RNA processing to FLD function had not been established. Here, we show that FLD tightly associates with LUMINIDEPENDENS (LD) and SET DOMAIN GROUP 26 (SDG26) in vivo, and, together, they prevent accumulation of monomethylated H3K4 (H3K4me1) over the FLC gene body. SDG26 interacts with the RNA 3′ processing factor FY (WDR33), thus linking activities for proximal polyadenylation of the antisense transcripts to FLD/LD/SDG26-associated H3K4 demethylation. We propose this demethylation antagonizes an active transcription module, thus reducing H3K36me3 accumulation and increasing H3K27me3. Consistent with this view, we show that Polycomb Repressive Complex 2 (PRC2) silencing is genetically required by FCA to repress FLC. Overall, our work provides insights into RNA-mediated chromatin silencing.
Both long and short noncoding chromatin-associated RNA transcripts have emerged as key regulators of the chromatin environment (1). Detailed mechanisms of how 21- to 24-nt RNAs initiate and maintain heterochromatin have been elucidated (2). However, less is understood about the mechanisms linking long noncoding RNA, chromatin regulation, and transcription. The most well-studied example is the role of X inactive specific transcript (Xist) in X chromosome inactivation (3). Different repeats on Xist recruit an array of protein factors that silence and conformationally alter the X chromosome (4). The RNA-binding protein SPEN binds the Xist A repeat and has recently been shown to transcriptionally down-regulate X-linked genes and trigger Polycomb silencing in a process requiring nucleosome remodelers and histone deacetylases (5). Similar RNA-mediated chromatin mechanisms act at the single locus Arabidopsis FLOWERING LOCUS C (FLC), which encodes a MADS-box transcription factor that acts as a floral repressor in Arabidopsis thaliana. A well-understood process involving FLC is vernalization, the cold-induced epigenetic silencing that occurs during winter, enabling plants to flower in spring. Cold induces a set of antisense long noncoding transcripts at the FLC locus, called COOLAIR, which mediate transcriptional down-regulation of FLC, as a prelude to a Polycomb-induced epigenetic switch (6). However, in a second less well understood mechanism at FLC, transcription is quantitatively regulated by COOLAIR antisense transcript processing linked to chromatin regulation. This is mediated by a set of genes grouped into the autonomous floral pathway (some of which are putative equivalents of SPEN), which have widespread transcriptional functions in the Arabidopsis genome through RNA-mediated chromatin pathways (7).
The autonomous pathway component FCA is an RNA-binding protein that mediates alternative 3′ end processing of COOLAIR transcripts (8). FCA associates with a coiled-coil protein, FLL2, which promotes formation of liquid-like nuclear condensates that appear to concentrate 3′ processing factors and change their dynamics at specific poly(A) sites (9). The proximal processing of COOLAIR results in an FLC chromatin environment that reduces FLC transcriptional initiation and elongation rates (10). This process requires FLOWERING LOCUS D (FLD), which is a homolog of the H3K4 demethylase LSD1 (11). Nevertheless, how FCA-mediated RNA processing links to FLD remained to be elucidated.
We have investigated this mechanism further, and here we identify two proteins, LUMINIDEPENDENS (LD) and SET DOMAIN GROUP 26 (SDG26), that tightly associate with FLD. Like FLD, LD and SDG26 function genetically in the FLC-repression pathway with FCA. We find that SDG26 transiently interacts with FY, one of the RNA 3′ processing factors that associates with FCA, physically linking FCA to FLD. Through genetic and chromatin immunoprecipitation analysis, we determine that loss of the FLD/LD/SDG26, or FCA, leads to overaccumulation of histone modifications, including H3K4me1/me2 and H3K36me3. Thus, we can now physically link RNA 3′ processing of the COOLAIR transcripts with a chromatin modification complex that influences H3K4me1-H3K36me3 and transcriptional activity at the locus. By antagonizing transcription, FLD/LD/SDG26-containing complex promotes H3K27me3 accumulation, consistent with a requirement for Polycomb Repressive Complex 2 in the FCA-mediated repression of FLC. We propose that FLD/LD/SDG26 influences an active transcription module that antagonizes PRC2 function.
Results
FLD Associates with LD and SDG26.
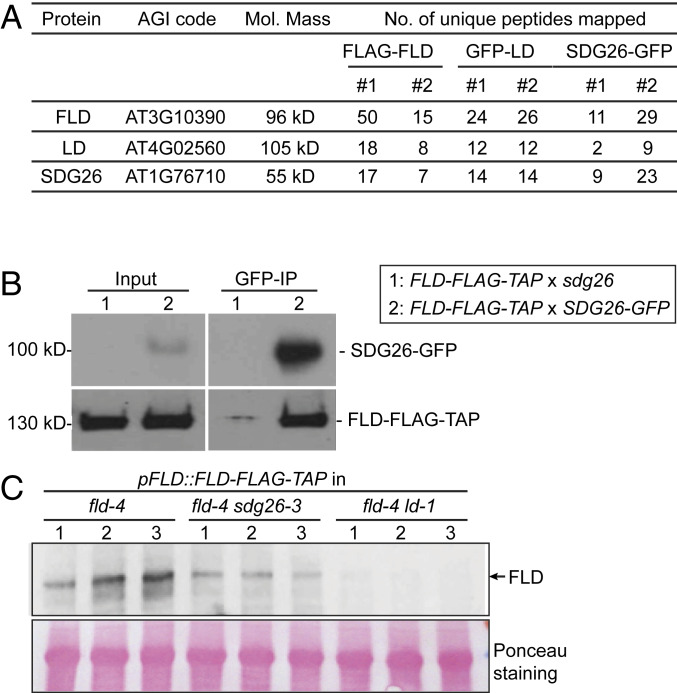
We previously performed a suppressor mutagenesis screen and identified FLD as one of the components required for FCA-mediated FLC regulation (11). To gain insights into how FLD represses FLC transcription, we used a proteomic approach to search for FLD interactors. We immunopurified FLD from a transgenic line expressing FLD tagged at the carboxyl terminus with FLAG-TAP epitopes (FLD-FLAG-TAP) (10). Mass spectrometric analyses of the FLD immunoprecipitation revealed that FLD tightly associates with LUMINIDEPENDENS (LD) and a SET domain protein, SDG26, in vivo (Fig. 1A and Dataset S1). Purifications from transgenic plants expressing GFP-tagged versions of each protein but not GFP only or Col-0 enriched the other two proteins of the complex (Fig. 1A and Datasets S2 and S3). The interaction between FLD and SDG26 was confirmed by coimmunoprecipitation (co-IP) in stable transgenic lines (Fig. 1B). Loss of LD or SDG26 caused a reduction in FLD protein levels (Fig. 1C and SI Appendix, Fig. S1). One possible explanation for this is that the interaction between FLD and LD/SDG26 may be required for FLD stability.
Fig. 1.
FLD forms a complex with LD and SDG26. (A) Table listing the number of unique peptides identified for FLD, LD, and SDG26 in FLAG-FLD, GFP-LD, and SDG26-GFP affinity purifications. Nontransgenic Col-0 was included in all purifications, and the transgenic line expressing GFP alone was included in GFP purifications as a negative control. The read counts from the negative controls were all zero for the listed proteins. (B) Co-IP in stable transgenic plants to detect the association of SDG26-GFP with FLD-FLAG-TAP. The FLD-FLAG-TAP transgenic line was crossed either with sdg26 mutant or SDG26-GFP transgenic line. F1-generation plants were used for co-IP. (C) The protein level of FLD-FLAG-TAP in the indicated genetic backgrounds as determined by Western blot. The numbers indicate three biological replicates. Ponceau staining served as a loading control.
LD was one of the first flowering regulators to be cloned based on a late-flowering phenotype of a T-DNA insertion (12), but how its function connected to other autonomous pathway components was unclear. LD encodes a protein carrying an N-terminal homeodomain (SI Appendix, Fig. S2A) and has been reported to bind DNA without sequence specificity (13). SDG26 is a close homolog of SDG8 (SI Appendix, Fig. S2A), the major histone H3K36 methyltransferase in the Arabidopsis genome; however, in vitro and in vivo analysis so far has provided no evidence that SDG26 is an H3K36 methyltransferase. In fact, sdg26 mutants show an opposite (late-flowering) phenotype compared to sdg8 (early flowering) through opposite effects on FLC expression, suggesting different functions or indirect effects (14, 15). We tested the subcellular localization of FLD, LD, and SDG26 in stable transgenic lines and found that they are all nuclear-localized (SI Appendix, Fig. S2B).
LD and SDG26 Function Genetically in the Same Pathway as FLD and FCA.
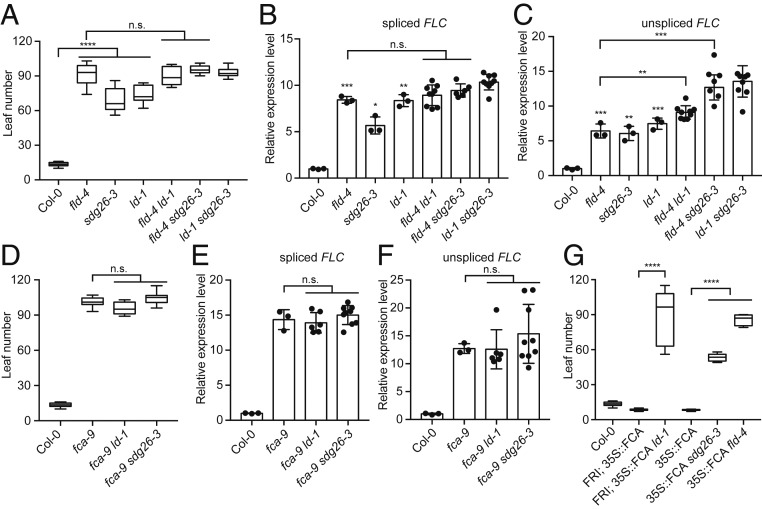
Similar to fld mutant, loss-of-function mutations of LD and SDG26 showed a late-flowering phenotype and increased FLC expression (Fig. 2 A–C). In order to dissect the genetic relationships between FLD, LD, and SDG26, we combined the mutations to create double mutants. The results showed that fld ld, fld sdg26, and ld sdg26 did not give any additional lateness (Fig. 2A) or increase in spliced FLC RNA levels (Fig. 2B), but did lead to higher unspliced FLC RNA levels (Fig. 2C), compared to the single mutants. The inconsistency between spliced and unspliced FLC suggests that, similar to Paf1C (16), FLD, LD, and SDG26 might have a concerted role in regulating the release of nascent FLC transcripts.
Fig. 2.
The FLD/LD/SDG26 complex functions genetically downstream of FCA to repress FLC. (A) Flowering time of indicated plants (assayed as total leaf number, produced by the apical meristem before it switched to producing flowers) grown in a long-day photoperiod. Data are presented as the mean ± SD (n ≥ 10). Asterisks indicate significant differences between the indicated plants (****P ≤ 2.42353E-09, two-tailed t test). n.s., not significant. (B and C) Expression of spliced FLC (B) and unspliced FLC (C) relative to wild-type Col-0 in the indicated mutants. Data are presented as the mean ± SD (n ≥ 3). Asterisks indicate significant differences between the indicated plants (*P ≤ 0.0121, **P ≤ 0.0028, ***P ≤ 0.0010, two-tailed t test). n.s., not significant. Each dot represents one biological replicate. (D) Flowering time of indicated plants grown in a long-day photoperiod. Data are presented as the mean ± SD (n ≥ 10). n.s., not significant. (E and F) Expression of spliced FLC (E) and unspliced FLC (F) relative to wild-type Col-0 in the indicated mutants. Data are presented as the mean ± SD (n ≥ 3). n.s., not significant. Each dot represents one biological replicate. (G) Flowering time of indicated plants grown in a long-day photoperiod. Data are presented as the mean ± SD (n ≥ 10). Asterisks indicate significant differences between the indicated plants (****P ≤ 4.68856E-14, two-tailed t test).
FLD has been shown to function in the same genetic pathway and downstream of FCA in that fld is not additive to fca with respect to flowering time, and fld suppressed the ability of FCA to down-regulate FLC (11). To test whether LD and SDG26 behave in the same way as FLD, we first combined ld and sdg26 with fca and found no additivity compared to fca with respect to flowering time (Fig. 2D) or FLC expression (Fig. 2 E and F). Combination of a 35S-FCA transgene, with and without the FLC activator FRIGIDA, with ld and sdg26 mutations then showed that both mutations compromised the effect of overexpressed FCA on FLC (Fig. 2G). Taken together, these data support that FLD, LD, and SDG26 exist in a complex that functions downstream of FCA to repress FLC expression.
SDG26 Transiently Interacts with the 3′ Processing Factor FY (WDR33).
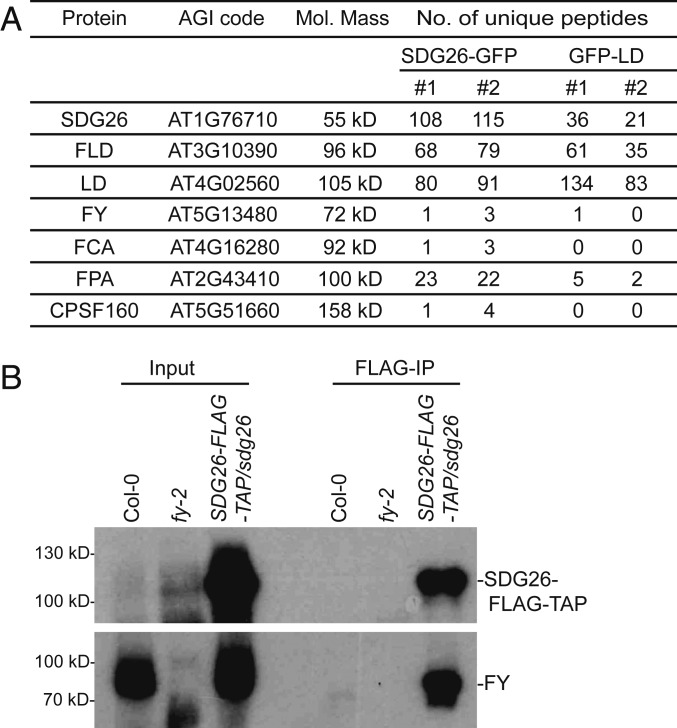
The strong genetic interactions between FLD/LD/SDG26 and FCA raised the question of how FCA function is molecularly linked to FLD. No in vivo physical interactions of FCA with 3′ processing factors or chromatin regulators had been found until our recent analysis using a technique termed cross-linked nuclear immunoprecipitation and mass spectrometry (CLNIP–MS) (9). We found FCA interacted with both RNA and a range of proteins and, in vivo, localizes to nuclear condensates that are highly dynamic (9). Those condensates are likely to concentrate 3′ processing factors and contribute to 3′-end processing of RNAs including COOLAIR (9). We reasoned that the interaction between the FLD/LD/SDG26-containing complex and FCA, if any, would also be transient and dynamic. To this end, we applied CLNIP–MS to SDG26. Surprisingly, we found that, in addition to finding FLD and LD with high peptide counts, some 3′ RNA processing factors were also detected (Fig. 3A and Dataset S4) in the SDG26 immunoprecipitation after cross-linking. These include FCA, as well as the RRM-containing protein FPA (8, 17), FY (18, 19), and Cleavage/Polyadenylation Specificity Factor 160 (CPSF160), all of which have been shown to associate with FCA and colocalize with FCA in the nuclear condensates (9). Purifications from Col-0 or a transgenic plant expressing a 35S-GFP fusion did not retrieve any of those proteins (Dataset S4). We then set out to confirm the interaction between SDG26 and FY, using an FY antibody raised in rabbits against the native recombinant protein (20). Using an SDG26-FLAG-TAP transgenic line, we performed cross-linked nuclear immunoprecipitation of SDG26 and probed against FY. The result showed that FY was readily detected (Fig. 3B). Without cross-linking, neither FY nor any of the 3′ processing factors were found in the SDG26 immunoprecipitation (Dataset S3). CLNIP-MS of LD also identified FY and FPA (Fig. 3A and Dataset S5). These data suggest that the interactions between the FLD/LD/SDG26-containing complex and 3′ processing factors provide a physical link, so that, when 3′ RNA processing of proximal COOLAIR occurs, the FLD/LD/SDG26-containing complex is brought in to repress FLC transcription.
Fig. 3.
SDG26 transiently associates with 3′ processing factors. (A) A partial list of proteins identified by SDG26 and LD affinity purifications after cross-linking. Nontransgenic Col-0 and transgenic line expressing GFP alone were included as negative controls in both purifications, and the read counts were all zero for the listed proteins. (B) Co-IP in stable transgenic plants after cross-linking to detect the association of SDG26 with FY.
Loss of FLD/LD/SDG26 Results in Overaccumulation of H3K4me1 at FLC.
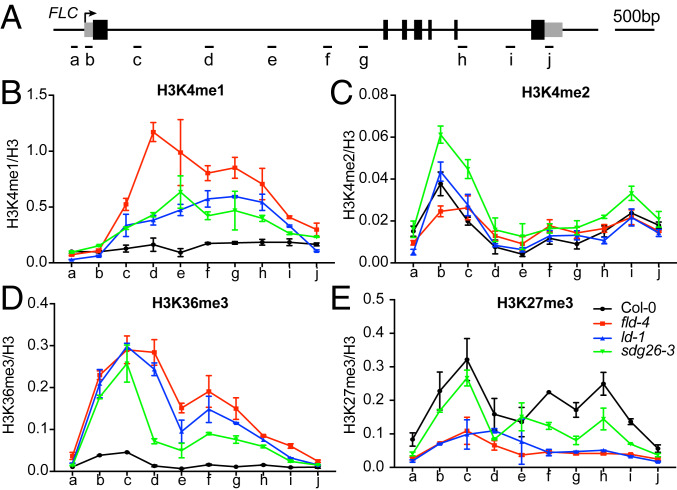
Our mathematical modeling and experimental evidence have shown that FLD-mediated repression of FLC is achieved in a manner consistent with a coordinated reduction of transcription initiation and Pol II elongation rates (10). Whether and how this is connected to histone modifications is not fully understood. Arabidopsis has four homologs of human LSD1, including FLD, LDL1, LDL2, and LDL3 (21). The fld mutation led to a limited 1.5- to 2-fold increase of H3K4me2 on FLC (10, 11). More recently, the ldl2 mutation was shown to increase gene body H3K4me1, which correlated positively with gene expression (22). We therefore decided to analyze the effect of FLD, LD, and SDG26 mutations on H3K4me1 and H3K4me2 levels at FLC. Chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR) showed a small increase of H3K4me2 at 1 to 4 kb beyond the TSS of FLC in fld (Fig. 4 A and C), consistent with previous reports (10, 11). Surprisingly, we observed a much more dramatic increase of H3K4me1 over the FLC gene body in fld (Fig. 4 A and B). ld and sdg26 also significantly overaccumulated H3K4me1 (Fig. 4B), indicating a major role of the FLD/LD/SDG26-containing complex in inhibiting H3K4me1 accumulation through the demethylase activity of FLD. It is also noteworthy that sdg26 accumulated more H3K4me2 than fld (Fig. 4C), suggesting a role for the FLD/LD/SDG26-containing complex in a stepwise removal of H3K4me2 and H3K4me1, with each component contributing differently to this activity. fca-9 showed a large increase in H3K4me1 and a similar increase in H3K4me2 as sdg26, in agreement with FLD/LD/SDG26 functioning genetically downstream of FCA (SI Appendix, Fig. S3 A–C). Given that SDG26 features a SET domain, a hallmark of histone methyltransferases, we sought to determine whether the FLD/LD/SDG26-containing complex, in addition to FLD-mediated demethylation, could also directly alter chromatin states through SDG26-mediated histone methylation. However, we failed to detect activity of SDG26 toward recombinant Arabidopsis nucleosomes in vitro for both heterologously expressed SDG26 or FLD/LD/SDG26 complex purified from Sf9 cells, nor for the endogenous FLD/LD/SDG26-containing complex purified via FLD-FLAG-TAP purification (SI Appendix, Fig. S4). Overall, these findings suggest demethylation of H3K4 is a major activity of the complex.
Fig. 4.
Measurements of histone modification levels upon the loss of the FLD/LD/SDG26 complex. (A) Schematic diagram showing FLC gene structure. Gray boxes represent untranslated regions, and black boxes represent exons. The other regions are represented by the black line. The arrow indicates the transcription start site (TSS). Short black lines indicate positions of primers used for qPCR amplification. (B–E) ChIP analysis of H3K4me1 (B), H3K4me2 (C), H3K36me3 (D), and H3K27me3 (E) levels at FLC in various genetic backgrounds. The letters on the x axis correspond to the positions indicated in A. Data are shown as relative to H3. Values are means ± SEM from three independent biological replicates.
SDG8 Is Epistatic to FLD/LD/SDG26 to Activate FLC.
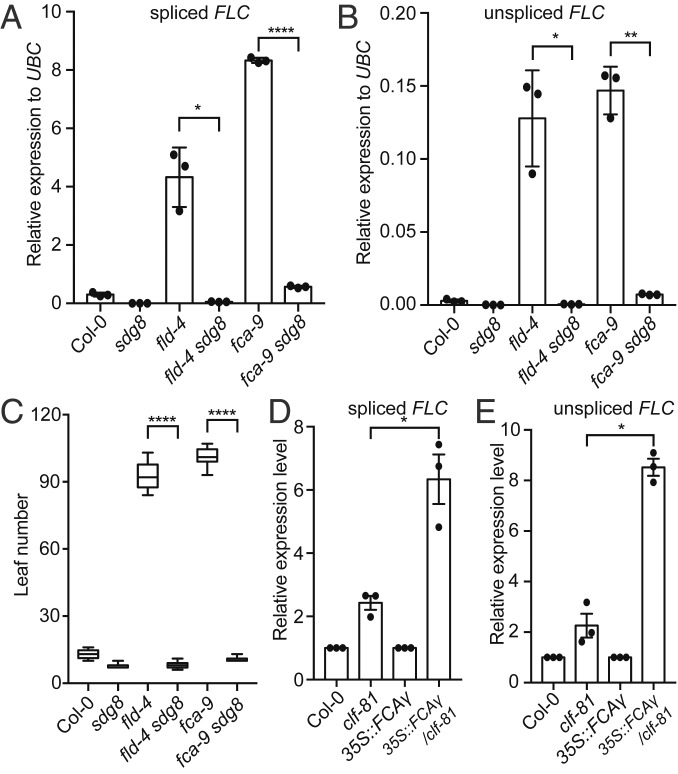
H3K4me1 is enriched at enhancers as well as gene bodies in mammalian cells (23). Recent studies suggested that H3K4me1 might fine-tune, rather than tightly control, enhancer activity and function (24–26). In plants, H3K4me1 is mainly found in gene bodies, removal of which mediates transcriptional silencing (22). Interestingly, the CW domain of Arabidopsis SDG8, an H3K36me3 methyltransferase, preferentially binds H3K4me1 (27, 28), providing a mechanism to link H3K4me1 to delivery of the active histone modification H3K36me3. Consistent with this, we found loss of the FLD/LD/SDG26-containing complex, as well as FCA, led to a large overaccumulation of H3K36me3 in the FLC gene body (Fig. 4D and SI Appendix, Fig. S3D), which mirrored the change of H3K4me1 (Fig. 4B and SI Appendix, Fig. S3B). In addition, H3K27me3, the mutually exclusive histone modification of H3K36me3, was greatly reduced in the fld-4, ld, and fca mutants (Fig. 4E and SI Appendix, Fig. S3E). Consistent with this, SDG8 ChIP did not show signal on FLC in the Col-0 background (29), where H3K4me1 was kept at a very low level (Fig. 4B). The connection between H3K4me1 and H3K36me3 raised the possibility that FLD/LD/SDG26 repressed FLC via removal of H3K4 methylation, thereby inhibiting SDG8-mediated H3K36me3 and indirectly promoting the accumulation of H3K27me3. To test this possibility, we generated the fld sdg8 double mutant and found that the sdg8 mutation completely suppressed both the fld-induced higher expression of FLC (Fig. 5 A and B) and the resulting delayed flowering time (Fig. 5C). This would suggest that the FLD/LD/SDG26 repression of FLC transcription involves inhibition of SDG8 function. In comparison, the sdg8 mutation largely, but not completely, reversed the expression of FLC (Fig. 5 A and B) and flowering time (Fig. 5C) caused by fca-9, suggesting that FCA can, to a limited extent, also repress FLC via a pathway that is independent of FLD and SDG8.
Fig. 5.
The genetic relationships of FCA and FLD with SDG8 and PRC2. (A and B) Expression of spliced FLC (A) and unspliced FLC (B) relative to UBC in the indicated genotypes. Data are presented as the mean ± SD (n = 3). Asterisks indicate significant differences between the indicated plants (*P ≤ 0.0217, **P = 0.0043, ****P = 6.27105E-05, two-tailed t test). Each dot represents one biological replicate. (C) Flowering time of indicated plants (assayed as total leaf number, produced by the apical meristem before it switched to producing flowers) grown in a long-day photoperiod. Data are presented as the mean ± SD (n ≥ 10). Asterisks indicate significant differences between the indicated plants (****P ≤ 2.26769E-09, two-tailed t test). (D and E) Expression of spliced FLC (D) and unspliced FLC (E) relative to UBC in the indicated genotypes. Note that expression level in the mutant background was separately normalized to its corresponding wild-type background. Data are presented as the mean ± SD (n = 3). Asterisks indicate significant differences between the indicated plants (*P ≤ 0.0458, two-tailed t test). Each dot represents one biological replicate.
FCA Requires PRC2 to Silence FLC.
The above data support a model where the alternative 3′ processing of COOLAIR by FCA mediates the silencing of FLC by Polycomb Repressive Complex 2 (PRC2) via inhibiting an active transcription module consisting of H3K4me1, H3K36me3, and transcription, which antagonizes the deposition of H3K27me3 (30). We tested this model by asking whether PRC2 is required by FCA to silence FLC. We took advantage of an Arabidopsis progenitor line carrying a single insertion of a 35S::FCAγ transgene in combination with an active FRIGIDA allele, in an otherwise wild-type background, which we had used to identify mutations suppressing the ability of FCA to down-regulate FLC (11). This sensitized background enhances FLC derepression and so is an efficient way to screen for factors required for FCA function. A weak allele of clf, reduced in PRC2 H3K27me3 methyltransferase activity (31), was introduced into this 35S::FCAγ genotype. clf-81 strongly released FLC expression, much more than in the Col background (Fig. 5 D and E), supporting that FCA requires PRC2 to silence FLC. In line with our findings, Tian et al. showed that CLF enrichment at the FLC locus requires FCA function (32).
Discussion
Studying the quantitative transcriptional regulation of the A. thaliana floral repressor FLC has led us into dissection of how alternative processing of antisense transcripts regulates local chromatin environment and thus transcriptional output (7). We find that dynamic interactions between RNA-binding proteins, 3′ processing factors, and the chromatin modifiers FLD/LD/SDG26 result in a chromatin state associated with low transcriptional initiation and slow elongation, marked by low H3K4me1, low H3K36me3, and high H3K27me3. Loss of any of the factors switches the locus to the opposing high transcriptional state, overaccumulation of H3K4me1 and H3K36me3 and reduction of H3K27me3. We propose that the FLD/LD/SDG26 exist in a complex that inhibits an active transcription module, so promoting the deposition of H3K27me3 (SI Appendix, Fig. S5). This process parallels with the cleavage and polyadenylation factor (CPF)-mediated facultative heterochromatin assembly in yeast (33), the exact mechanism of which is still unknown.
FCA associates dynamically with 3′ processing factors in FCA nuclear bodies (9). The fact that the interactions between SDG26 and 3′ processing factors were only detected after cross-linking suggested that the interactions are also dynamic, and raised the possibility that FLD/LD/SDG26 might colocalize in FCA nuclear bodies. LD, like FCA and FY, has been found to contain a prion-like domain (34) (SI Appendix, Fig. S2A), which was identified as a driver for ribonucleoprotein granule assembly (35), and LD formed distinct foci when expressed in yeast cells (34). However, under normal confocal microscopy and expressed at endogenous levels, neither FLD, LD, nor SDG26 formed obvious nuclear bodies (SI Appendix, Fig. S2B). One possible explanation is that FLD/LD/SDG26 form nuclear bodies in vivo that are too dynamic/small to be detected by normal confocal microscopy. Superresolution microscopy analysis of FLD, LD, and SDG26 subcellular localization will help to address this question. On the contrary, not all genes in the genome targeted by FCA for RNA processing also need the FLD/LD/SDG26-containing complex for silencing (36). This agrees with our finding that FCA immunoprecipitation after cross-linking did not recover FLD, LD, or SDG26 (9). In addition, genetic data suggested that, even at the FLC locus, FCA could function in FLD-independent pathways to achieve some measure of silencing (Fig. 5 A and B) (11). A recent study showed that FCA interacts with CLF in vitro and in vivo, suggesting an FLD-independent role of FCA in regulating H3K27me3 directly (32). However, we have not detected this interaction in FCA on in vivo immunoprecipitation-mass spectrometry (IP-MS) (9), and it was not detected in CLF on in vivo IP-MS (37).
An important question raised by this work is what is the active transcription module that FLD/LD/SDG26-containing complex inhibits. We were unable to find any histone methyltransferase activity in vitro for the FLD/LD/SDG26 complex (SI Appendix, Fig. S4), suggesting that additional components are required for the complex to exert its function. One tantalizing hypothesis is that the histone-modifying activity is tightly linked to the RNA polymerase II (Pol II) complex during transcription. Indeed, we detected Pol II subunits (e.g., NRPB1, NRPB2, and NRPB3) and factors involved in the regulation of transcription initiation and elongation (e.g., SPT5, SPT6, and SPT16) in the SDG26 CLNIP-MS list (Dataset S4). In addition, LD contains a PP1-AP–like domain shared with the transcription elongation factor TFIIS, suggesting a role for LD in transcriptional elongation (38). Further analysis of these possibilities will expand our understanding of how the RNA-binding protein FCA connects COOLAIR to antagonizing an active transcription module, thereby eventually leading to Polycomb silencing. Full dissection of this mechanism will reveal any further parallels between COOLAIR and Xist function, thus elaborating our evolutionary understanding of RNA-mediated chromatin silencing.
Materials and Methods
More detailed descriptions of the materials and methods used in this study are provided in the SI Appendix. A brief summary is provided here.
Plant Materials.
The progenitor lines C2 and 35S::FCA/Col (11) and the mutants fld-4 and fca-9 (11), sdg8 (39), and clf-81 (40) were described previously. The transfer-DNA (T-DNA) insertion line ld-1 (CS876430) and sdg26-3 (GK-087B12) were obtained from the Nottingham Arabidopsis Stock Centre.
Flowering Time Analysis.
The flowering time was determined essentially as described (9). Briefly, plants were grown in long-day conditions, and the total leaf number (TLN) produced before the initiation of flowering was counted to measure variation in flowering time.
RNA Analysis.
RNA analysis was performed as described previously (9). Briefly, total RNA was extracted, treated with DNase, and reverse-transcribed by SuperScript IV Reverse Transcriptase (Invitrogen) using gene-specific reverse primers. Quantitative reverse transcription and PCR (qPCR) analysis was performed on a LightCycler480 II (Roche), and qPCR data were normalized to UBC. Primer pairs for amplifying unspliced FLC, spliced FLC, and UBC are listed in SI Appendix, Table S1.
Immunoprecipitation and Immunoblot.
Extracts were prepared and immunoprecipitated with either anti-FLAG M2 Magnetic Beads (Sigma-Aldrich; M8823) or GFP-Trap Magnetic Agarose (ChromoTek; gtma-10).
For immunoblot analysis, protein extracts or immunoprecipitates were separated by SDS/PAGE, transferred to PVDF membranes, and detected by GFP (Roche; no. 11814460001), FLAG (Sigma-Aldrich; F3165), or FY (20) antibodies.
Materials and Data Availability.
Full lists of mass spectrometry are provided as Datasets S1–S5. All of the other raw data and materials that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank all members of the C.D. and M.H. laboratories for discussions and Gerhard Saalbach for assistance with proteomics. We are particularly grateful to Prof. Tetsuji Kakutani and Dr. Soichi Inagaki of the National Institute of Genetics, Japan, for information regarding FLD activity on H3K4me1 prior to publication. The work was supported by the Biotechnology and Biological Sciences Research Council Institute Strategic Programme Genes in the Environment (BB/P013511/1), the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Grant (800318), a Wellcome Senior Investigator grant (210654/Z/18/Z), and ERC Advanced Grant EPISWITCH-833254 to C.D. Work in the Voigt lab is supported by the Wellcome Trust (104175/Z/14/Z, Sir Henry Dale Fellowship to P.V.) and through funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-STG Grant Agreement no. 639253 to P.V.). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007268117/-/DCSupplemental.
References
- 1.Holoch D., Moazed D., RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martienssen R., Moazed D., RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 7, a019323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loda A., Heard E., Xist RNA in action: Past, present, and future. PLoS Genet. 15, e1008333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moindrot B., Brockdorff N., RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin. Cell Dev. Biol. 56, 58–70 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Dossin F. et al., SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578, 455–460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry S., Dean C., Environmental perception and epigenetic memory: Mechanistic insight through FLC. Plant J. 83, 133–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Z., Fang X., Zhu D., Dean C., Autonomous pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism. Plant Physiol. 182, 27–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F., Marquardt S., Lister C., Swiezewski S., Dean C., Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Fang X. et al., Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569, 265–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z. et al., Quantitative regulation of FLC via coordinated transcriptional initiation and elongation. Proc. Natl. Acad. Sci. U.S.A. 113, 218–223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F. et al., The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lee I. et al., Isolation of LUMINIDEPENDENS: A gene involved in the control of flowering time in arabidopsis. Plant Cell 6, 75–83 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aukerman M. J., Lee I., Weigel D., Amasino R. M., The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J. 18, 195–203 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Liu B. et al., Interplay of the histone methyltransferases SDG8 and SDG26 in the regulation of transcription and plant flowering and development. Biochim. Biophys. Acta 1859, 581–590 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Xu L. et al., Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28, 1348–1360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalik K. M. et al., The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature 520, 248–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornyik C., Terzi L. C., Simpson G. G., The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 18, 203–213 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Manzano D. et al., Altered interactions within FY/AtCPSF complexes required for Arabidopsis FCA-mediated chromatin silencing. Proc. Natl. Acad. Sci. U.S.A. 106, 8772–8777 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson G. G., Dijkwel P. P., Quesada V., Henderson I., Dean C., FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Henderson I. R., Liu F., Drea S., Simpson G. G., Dean C., An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132, 3597–3607 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Jiang D., Yang W., He Y., Amasino R. M., Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19, 2975–2987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inagaki S. et al., Gene-body chromatin modification dynamics mediate epigenome differentiation in Arabidopsis. EMBO J. 36, 970–980 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzman N. D. et al., Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Local A. et al., Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 50, 73–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorighi K. M. et al., MLL3 and MLL4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576 e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rickels R. et al., Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoppmann V. et al., The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30, 1939–1952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Huang Y., Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain of Arabidopsis histone methyltransferase SDG8. J. Biol. Chem. 293, 6470–6481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y. et al., The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol. 16, 79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H., Howard M., Dean C., Physical coupling of activation and derepression activities to maintain an active transcriptional state at FLC. Proc. Natl. Acad. Sci. U.S.A. 113, 9369–9374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodrich J. et al., A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Tian Y. et al., PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 5, eaau7246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vo T. V. et al., CPF recruitment to non-canonical transcription termination sites triggers heterochromatin assembly and gene silencing. Cell Rep. 28, 267–281 e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakrabortee S. et al., Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc. Natl. Acad. Sci. U.S.A. 113, 6065–6070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hennig S. et al., Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 210, 529–539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonmez C. et al., RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc. Natl. Acad. Sci. U.S.A. 108, 8508–8513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang S. C., et al. , Kicking against the PRCs–A domesticated transposase antagonises silencing mediated by Polycomb group proteins and is an accessory component of Polycomb repressive complex 2. PLoS Genet. 11, e1005660 (2015). Correction in: PLoS Genet.12, e1005812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grasser M. et al., Transcript elongation factor TFIIS is involved in arabidopsis seed dormancy. J. Mol. Biol. 386, 598–611 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Yang H., Howard M., Dean C., Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 24, 1793–1797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim G. T., Tsukaya H., Uchimiya H., The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206, 175–183 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.