Significance
Bacteria are known to add amino acids (aa) to membrane lipids to resist antimicrobials and escape immune responses. This surface lipid aminoacylation process requires diverting aminoacyl-tRNAs from protein synthesis. While widespread in bacteria, no analogous lipid remodeling system had thus far been evidenced in eukaryotes. We uncovered that most fungi tRNA-dependently add aspartate onto ergosterol (ergosteryl-3β-O-l-aspartate [Erg-Asp]), the major sterol found in fungal membranes. Asp addition is catalyzed by an ergosteryl-3β-O-l-aspartate synthase (ErdS) and its removal by a dedicated hydrolase (ErdH). This pathway is conserved across “higher” fungi, including pathogens. Given the central roles of sterols and derivatives in fungi, we propose that the Erg-Asp homeostasis system might impact membrane remodeling, trafficking, antimicrobial resistance, or pathogenicity.
Keywords: aminoacyl-tRNA, ergosterol, fungi, DUF2156, lipid aminoacylation
Abstract
Diverting aminoacyl-transfer RNAs (tRNAs) from protein synthesis is a well-known process used by a wide range of bacteria to aminoacylate membrane constituents. By tRNA-dependently adding amino acids to glycerolipids, bacteria change their cell surface properties, which intensifies antimicrobial drug resistance, pathogenicity, and virulence. No equivalent aminoacylated lipids have been uncovered in any eukaryotic species thus far, suggesting that tRNA-dependent lipid remodeling is a process restricted to prokaryotes. We report here the discovery of ergosteryl-3β-O-l-aspartate (Erg-Asp), a conjugated sterol that is produced by the tRNA-dependent addition of aspartate to the 3β-OH group of ergosterol, the major sterol found in fungal membranes. In fact, Erg-Asp exists in the majority of “higher” fungi, including species of biotechnological interest, and, more importantly, in human pathogens like Aspergillus fumigatus. We show that a bifunctional enzyme, ergosteryl-3β-O-l-aspartate synthase (ErdS), is responsible for Erg-Asp synthesis. ErdS corresponds to a unique fusion of an aspartyl-tRNA synthetase—that produces aspartyl-tRNAAsp (Asp-tRNAAsp)—and of a Domain of Unknown Function 2156, which actually transfers aspartate from Asp-tRNAAsp onto ergosterol. We also uncovered that removal of the Asp modifier from Erg-Asp is catalyzed by a second enzyme, ErdH, that is a genuine Erg-Asp hydrolase participating in the turnover of the conjugated sterol in vivo. Phylogenomics highlights that the entire Erg-Asp synthesis/degradation pathway is conserved across “higher” fungi. Given the central roles of sterols and conjugated sterols in fungi, we propose that this tRNA-dependent ergosterol modification and homeostasis system might have broader implications in membrane remodeling, trafficking, antimicrobial resistance, or pathogenicity.
Remodeling of membranes and lipid modifications are processes used by living cells to interact with and adapt to their environment. They are notably critical for host/pathogens interactions, antimicrobial resistance, and virulence in both bacteria (1, 2) and fungi (3). MprFs are bacterial virulence factors that transfer amino acids (aa) onto membrane glycerolipids in a so-called aminoacylation reaction (4). This process requires aminoacyl-transfer RNAs (aa-tRNA) that are first synthesized by aminoacyl-tRNA synthetases (aaRS) (5), prior transfer of the aa moiety onto a lipid acceptor substrate, that is, phosphatidylglycerol, cardiolipin, or diacylglycerol (4). The aminoacyl-tRNA transferase (AAT) module, that catalyzes the transfer, belongs to the DUF2156 family and recognizes both the aa-tRNA and the lipid substrates (6). Glycerolipid aminoacylation modifies the overall charge, fluidity, and permeability of bacterial membranes, which enhances antimicrobial resistance, explaining why these enzymes have been termed multiple peptides resistance factors (MprF) (4). Glycerolipid aminoacylations also affect host/pathogen interactions and have been shown to potentiate immune escape and to increase virulence of pathogens (7). In Pseudomonas aeruginosa (8), Enterococcus faecium (9), and Agrobacterium tumefaciens (10), extracytoplasmic esterases of the VirJ or α/β-hydrolase family participate in the homeostasis of aminoacylated lipids and hydrolyze the modifying aa from lipids.
So far, only glycerolipids have been shown to be aminoacylated and only by MprFs in prokaryotes. In the present study, we show that eukaryotes aminoacylate sterols in a tRNA-dependent modification pathway that mimics those described in bacteria. In higher fungi, we functionally characterized a novel enzyme, the ergosteryl-3β-O-l-aspartate synthase (ErdS), composed of a DUF2156/AAT domain fused to an aspartyl-tRNA synthetase (AspRS) paralog. We show that ErdS synthesizes ergosteryl-3β-O-l-aspartate (Erg-Asp), a previously undetected conjugated sterol. Moreover, most of higher fungi (Dikarya), including opportunistic human pathogens such as Aspergillus fumigatus (Afm), encode this ErdS and produce Erg-Asp, suggesting that the system is widespread in “higher” fungi. We also evidenced that these species express a dedicated Erg-Asp hydrolase (ErdH) that deacylates Erg-Asp and whose gene is almost always located in a two-gene cluster next to the erdS gene. In conclusion, our data unravel an evolutionary conserved sterol−aa conjugation pathway. We discuss the potential implications of this pathway in the context of lipid homeostasis and antimicrobial resistance.
Results
DUF2156 Proteins Are Present in Fungi.
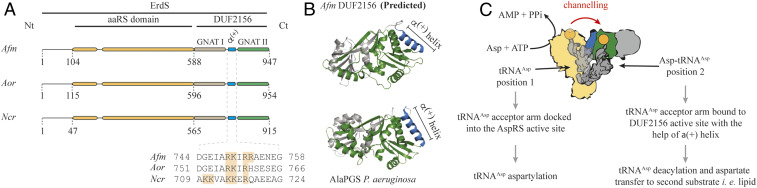
Although DUF2156 domains were thought to be absent or rare outside prokaryotes (11), we identified, in most higher fungi, DUF2156-containing proteins fused to the C terminus of an aaRS-like domain (Fig. 1A and SI Appendix, Fig. S1A), that we named ErdS for reasons that will be presented below. This protein architecture, previously detected in two fungi (12), is actually widespread across fungi, including the well-characterized Afm, Aspergillus oryzae (Aor), or Neurospora crassa (Ncr) (Fig. 1A). Firstly, sequence analyses and structure predictions of the detected DUF2156 domains suggested that they all adopt the characteristic double Gcn5-like N-acetyltransferase (GNAT) fold observed in bacterial MprFs (6). In fungal DUF2156 domains, the GNAT I (recognition of the acceptor substrate) and II (transferase activity) subdomains of the double GNAT fold are separated, like in bacteria, by a positively charged α(+) helix involved (Fig. 1 A and B) in the binding of aa-tRNAs (6), suggesting that 1) they could indeed be aa-tRNA−utilizing modules and 2) they might transfer the aa moiety of aa-tRNAs onto lipids. Second, the aaRS domains contain bona fide AspRS signature sequences including the known residues (13) involved in tRNAAsp, Asp, and adenosine 5′-triphosphate (ATP) binding ubiquitously found in functional AspRSs (SI Appendix, Fig. S1B). These considerations prompted us to propose the following working hypothesis: The AspRS domain of ErdS would generate Asp-tRNAAsp from ATP, l-Asp, and tRNAAsp, which would then be transferred to the DUF2156 module, thought to carry the AAT activity, to transfer the Asp residue onto a yet-to-be identified lipid (Fig. 1C).
Fig. 1.
Filamentous fungi AspRS-DUF2156, proteins termed ErdSs, are potential lipid aminoacylation factors. (A) In silico analyses predicted Afm, Aor, or Ncr erdS genes to encode proteins composed of an AspRS domain N-terminally fused to a DUF2156 domain. Protein domains were delimited by confronting data obtained from Protein Families database (PFAM) and from multiple alignments as described in SI Appendix, Supplementary Materials and Methods. Critical positively charged residues in the alpha helix (α(+)) separating both GNAT folds are indicated (orange). (B) Comparison of the DUF2156 structure of AlaPGS (alanyl-phosphatidylglycerol synthase, MprF) from P. aeruginosa to the Phyre2 prediction of Afm DUF2156. GNAT I and II subdomains are highlighted in gray and green, respectively, with the positively charged α(+) helix in blue. (C) Schematic representation of the hypothetical ErdS reaction mechanism during which tRNAAsp shift from the AspRS catalytic site (position 1: tRNAAsp aspartylation) to the DUF2156 active site (position 2: Asp transfer from Asp-tRNAAsp onto a lipid substrate). The α(+) helix is indicated in blue, and the active site is indicated in green. Aspartate is represented in orange.
ErdS Is Essential for the Synthesis of a Lipid in Filamentous Fungi.
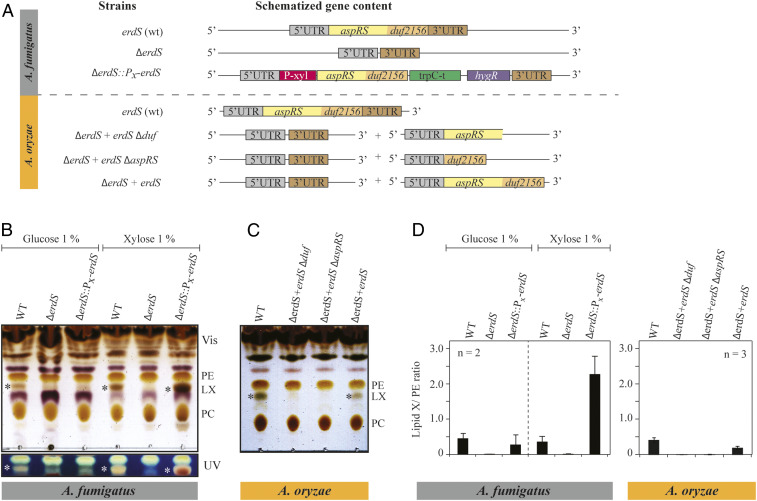
In order to test our hypothesis on the function of ErdSs, we deleted the corresponding erdS genes in two fungi (Fig. 2A and SI Appendix, Fig. S2), namely, the human opportunistic pathogen Afm and the species of biotechnological and industrial interest Aor. Both ΔerdS mutants grew normally on solid media (SI Appendix, Fig. S3), demonstrating that the genes are not essential under standard growth conditions. To visualize whether ErdSs are involved in lipid aminoacylation, we analyzed total lipids from mycelia of the wild-type (WT) and ΔerdS strains by thin-layer chromatography (TLC), and results revealed, in both fungi, the presence of an additional ErdS-dependent lipid (hereafter, lipid X or LX) with a distinctive brownish staining that migrates between phosphatidylethanolamine (PE) and phosphatidylcholine (PC) (Fig. 2 B and C).
Fig. 2.
Identification of an Aspergillus lipid species, whose synthesis requires ErdS. (A) Deletions of erdS from Afm and from Aor were performed by homologous recombination (see SI Appendix for details). The genotypes of the strains are indicated. For Afm, the ΔerdS strain shown corresponds to the strain after excision of the deletion cassette, whereas, for the complemented ΔerdS::PX-erdS strain, the selection marker is still present. For Aor, the ORF of erdS was replaced by a pyrG-containing module and subsequently excised by selection on 5-FOA medium. Complementation was operated by ectopic expression of erdS-ΔDUF, erdS-ΔAspRS, or erdS. (B and C) Total lipids extracted from the different strains described in A were analyzed by TLC and stained with a sulfuric acid/MnCl2 solution. TLC plates were observed either under white light or under UV light. Cultures were done in glucose or xylose containing media; * indicates the LX. (D) Quantification of the TLCs shown in B and C. LX signal (number of pixels) was normalized to that of PE (phosphatidylethanolamine) (LX/PE ratio). All TLCs are representative of at least two independent experiments (n = 2).
To verify the ErdS dependency of LX synthesis, we constructed an Afm ΔerdS::Pxyl-erdS strain, in which we reintroduced the WT erdS gene under the control of a xylose (Xyl)-inducible promoter (Pxyl-erdS) at the ΔerdS locus (Fig. 2A and SI Appendix, Fig. S2). Under noninduced conditions (in the presence of glucose), LX was barely undetectable in the ΔerdS::Pxyl-erdS strain compared to the WT, while, under induction conditions (in the presence of Xyl), strong accumulation of LX occurred (Fig. 2B). In Aor, complementation of the ΔerdS mutation with the WT erdS copy (Fig. 2A) at an ectopic locus under the control of its own promoter again restored LX synthesis (Fig. 2C), which proved that LX synthesis is dependent on ErdS expression. Interestingly, only the full-length ErdS could sustain efficient LX synthesis in Aor, while an ErdS mutant in which the AspRS domain was deleted (Fig. 2C, ΔerdS+erdS-∆aspRS) could not, at least not at detectable levels.
Fungal ErdS Catalyzes tRNA-Dependent Aspartylation of a Lipid.
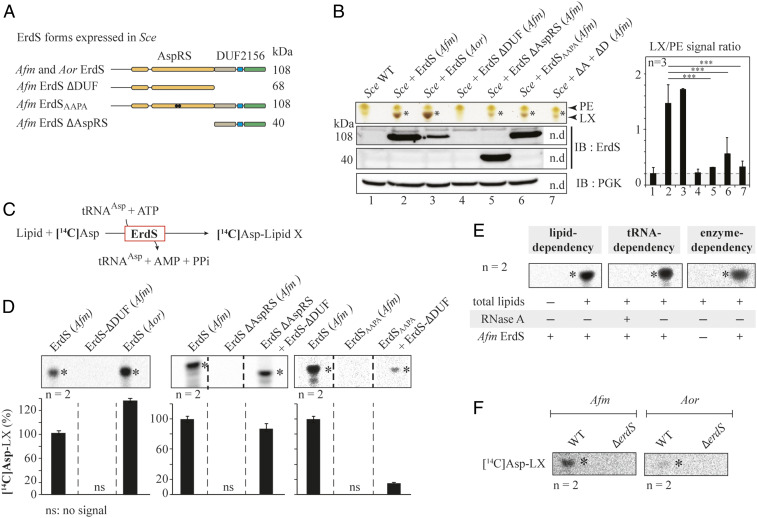
To dissect the mechanism of LX synthesis, we transferred the synthesis of LX, hypothesized to rely on the sole ErdS enzyme, to the budding yeast Saccharomyces cerevisiae (Sce), in which we overexpressed Afm ErdS. Switching to a heterologous yeast system was dictated for three main reasons: 1) Genetic engineering is far more time consuming in Afm than in Sce; 2) no DUF2156-containing proteins were detected in Sce (SI Appendix, Fig. S1A); and 3) the lipid synthesis pathways are conserved across ascomycetes (amigo.geneontology.org/amigo/term/GO:0006629), and thus the lipid contents of these species belonging to the same phylum are likely to be very similar (14). To start with and test whether the AspRS moiety was functional, Afm ErdS or ErdS-∆DUF, that is an ErdS form that comprises only the AspRS domain, was expressed in the Sce Δdps1 strain (15) (DPS1 encodes the essential AspRS), and complementation of the Δdps1 mutation lethality was tested by plasmid shuffling (Fig. 3A and SI Appendix, Fig. S4A). Results show that both ErdS and ErdS-∆DUF functionally replaced Sce AspRS in vivo. This was not observed with a catalytic null mutant (ErdSAAPA) of the AspRS domain (Fig. 3A and SI Appendix, Fig. S4A), in which the QSPQ signature motif (16) was mutated to AAPA. In addition, in vitro aminoacylation assays with purified recombinant Afm ErdS and Sce tRNAAsp confirmed ErdS's capacity to generate Asp-tRNAAsp (SI Appendix, Fig. S4B). Interestingly, full-length ErdS displays a lower complementation efficiency of the Sce Δdps1 strain compared to ErdS-∆DUF (SI Appendix, Fig. S4A). We interpreted that, in the full-length ErdS, the DUF2156 domain likely transfers Asp from Asp-tRNAAsp onto an accepting lipid, thereby decreasing the amount of Asp-tRNAAsp available for protein synthesis and impacting growth.
Fig. 3.
Dissecting ErdS lipid modification mechanism. (A) Schematized modular organization of full-length ErdS (108 kDa, Afm and Aor) and of its variants (Afm) that were expressed in the Sce heterologous model. ErdS-ΔDUF: AspRS standalone domain; ErdS-ΔAspRS: DUF2156 standalone domain (40 kDa); ErdSAAPA: catalytic null of the AspRS moiety (108 kDa). (B) TLC-based analysis of total lipids from Sce expressing ErdS variants described in A and Sce + (ΔD + ΔA) corresponds to the double expression of ErdS-ΔDUF + ErdS-ΔAspRS in Sce. The TLC plate stained with sulfuric acid/MnCl2 was cropped to the area of interest. LX signal (number of pixels) was normalized to that of PE in each case, and the LX/PE ratio is represented in a graph (Right). In the absence of LX, PG (phosphatidylglycerol) becomes visible; thus the PG/LX ratio obtained was considered “background signal” (gray background). However, the brown staining of LX (yellow for PG) made it possible to visually assess the presence of LX even for low LX/PE values. The Student's t test was used to assess the significance of the means of the data; ***P < 0.005. ErdS variants expression was analyzed by Western blot with an anti-Afm DUF2156 polyclonal antibodies (IB:ErdS), and loading control was performed with anti-PGK antibodies (IB:PGK). (C) Schematized reaction of the LA assay, described in Materials and Methods. (D) LX synthesis by the purified recombinant full-length and mutant ErdSs was measured using the LA assay. When mixed, proteins were in equimolar ratios. The [14C]-Asp lipid levels (percent) are provided below each TLC. (E) Verification of tRNA, lipid, and ErdS dependency of LX synthesis using LA assay; +: presence; −: absence. (F) Measurements of lipid X synthesis (ErdS activity) by WT and ΔerdS Aspergillus spp. crude extracts using LA assay; *: LX. The [14C]Asp lipids were revealed using phosphorimaging (D–F). TLCs and immunoblots are representative of two to three independent experiments, and the number of replicates (n) is indicated.
To determine whether the DUF2156 module of ErdS transfers Asp from Asp-tRNAAsp onto lipids in vivo, as suggested from the results obtained in Aspergillus spp. (Fig. 2 B and C), we analyzed the total lipid composition of a WT Sce strain bearing plasmids that express Afm or Aor full-length or truncated ErdSs (Fig. 3A). Expression of proteins was confirmed by Western blot with anti-ErdS antibodies that recognize both Afm and Aor ErdS (Fig. 3B), and analyses of total lipids by TLC revealed the presence of an additional brownish lipid, absent in WT Sce strains, when Afm and Aor ErdS were expressed. This lipid presents the same migration profile as the LX detected in Afm and Aor (SI Appendix, Fig. S4C), strongly suggesting that heterologous expression of Afm or Aor ErdS in Sce allows complete and correct synthesis of LX. Positive staining of LX with ninhydrin (detection of primary amine) and bromocresol green (detection of carboxyl) further suggested that it contained an aa moiety, likely l-Asp ester- or amide-linked through its α-carboxyl the lipid moiety, with free α-NH3+ and β-COO− groups (SI Appendix, Fig. S4D).
To investigate whether ErdS transfers Asp from Asp-tRNAAsp onto a lipid in a DUF2156-dependent manner, we analyzed the total lipids produced by ErdS-ΔDUF, ErdS-ΔAspRS, and ErdSAAPA expressed in Sce (Fig. 3B). Expression of the different constructs was confirmed by anti-ErdS Western blot (Fig. 3B). The ErdS-∆DUF could not be detected, because our antibodies were raised against the DUF2156 domain; however, complementation of the lethal ∆dps1 mutation in Sce by ErdS-∆DUF confirmed that the AspRS moiety was expressed and functional (SI Appendix, Fig. S4A). LX spots were quantified and normalized to those of PE (LX/PE ratio, presented in Fig. 3B). Of note is that, in the absence of LX, PG becomes visible at the same position in Sce, a reason for which the PG/PE ratio was used as a “background” signal (Fig. 3B, gray background). LX was not observed in Sce upon removal of the DUF2156 domain from ErdS (LX/PE ratio similar to the PG/PX ratio of the WT strain), and expression of the DUF2156 domain in trans in the Sce+ErdS-∆DUF strain restored LX synthesis (Fig. 3B, Sce+∆A+∆D), although at lower levels, with an LX/PE ratio close to that of the PG/PE ratio of the WT strain. Only the specific brown staining of LX confirmed its low-level production. These data demonstrate that synthesis of LX is DUF2156 dependent. The reduced level of LX in the Sce+ErdS-∆AspRS strain, despite proper expression of the enzyme, shows that the lipid synthesis activity of the standalone DUF2156 domain is weaker than when fused to its AspRS moiety (low LX/PE ratio), which recalls results obtained in Aor (Fig. 2C). Similarly, in the Sce strain that expresses ErdS with a catalytically null AspRS moiety (Sce+ErdSAAPA), LX levels mirrored those of the Sce+ErdS-∆AspRS and Sce+∆A+∆D strains (Fig. 3B). These experiments show, in addition, that the DUF2156 domain of ErdS can efficiently utilize Sce Asp-tRNAAsp, regardless of whether it was generated by the Afm AspRS moiety of ErdS or by the endogenous Sce AspRS.
To better characterize the ErdS catalytic mechanism, we developed an in vitro lipid aminoacylation assay (LA assay), using purified Afm or Aor recombinant ErdS incubated with purified Sce tRNAAsp, ATP, and [14C]-Asp in the presence of Sce total lipids (Fig. 3C). Reaction products from the LA assay were analyzed by TLC (Fig. 3 D and E). Results confirmed that Afm or Aor recombinant ErdSs catalyzed the formation of a [14C]-labeled lipid (Fig. 3D). Adding RNase or removing lipids from the LA assay abolished synthesis of [14C]-lipid, thereby proving the tRNA and lipid dependency of LX synthesis by Afm ErdS (Fig. 3E). When the recombinant Afm ErdS-ΔDUF protein was used, synthesis of the [14C]-lipid was abolished, proving that the reaction is, indeed, DUF2156 dependent (Fig. 3D). Likewise, when ErdS-∆AspRS or ErdSAAPA were used in the LA assay, synthesis of the [14C]-lipid was abolished (Fig. 3D), but fully restored upon addition of the AspRS domain of ErdS in trans. These data confirm results obtained in Sce but also that the DUF2156 is capable of capturing Asp-tRNAAsp formed by an AspRS expressed in trans (Fig. 3D).
Finally, crude extracts from WT Afm or Aor mycelia also catalyzed a [14C]-Asp-tRNAAsp−dependent [14C]-lipid synthesis in vitro, whereas crude extracts from ΔerdS mutant strains did not (Fig. 3F). However, for reasons that will be discussed below, the ErdS activity was hardly detectable in crude extracts, especially for Aor, when compared to recombinant ErdS.
Altogether, these results show that ErdS produces Asp-tRNAAsp from ATP, l-Asp, and tRNAAsp in its AspRS domain, and that the Asp-tRNAAsp product likely shifts from the AspRS active site to the DUF2156 module, that transfers the Asp acylating tRNAAsp onto a lipid, which mirrors the activity of bacterial DUF2156 proteins (4).
Liquid chromatography-MS/MS Reveals that ErdS Aspartylates Ergosterol.
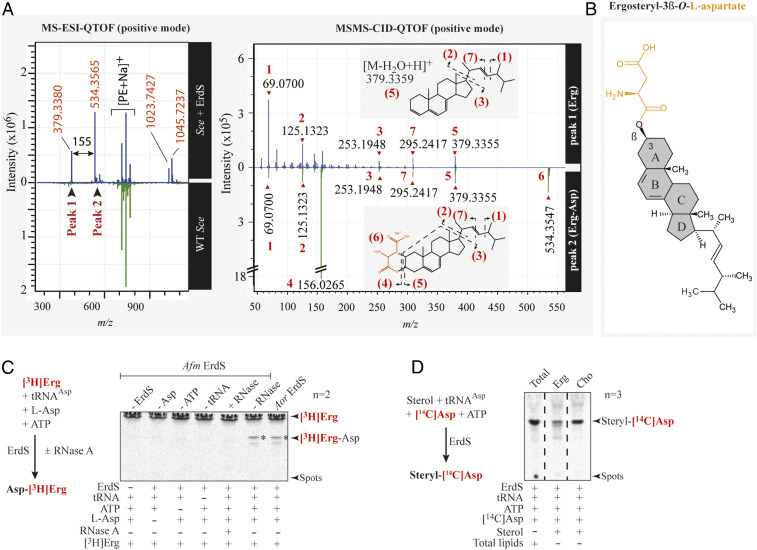
To identify the substrate lipid of ErdS, we fractionated total lipids from an ErdS overexpressing Sce strain by column chromatography and isolated LX at ∼80% purity (SI Appendix, Fig. S5A). Fractions were submitted to mass spectrometry (MS) analyses and compared to equivalent fractions obtained from total lipids of a WT yeast strain. Strikingly, MS electrospray ionization quadrupole time-of-flight (MS-ESI-QTOF) spectra revealed two peaks, absent in WT fractions, with m/z of 379.3380 and 534.3565 that were submitted to a second round of MS/MS collision-induced dissociation QTOF (MS/MS-CID-QTOF) (Fig. 4A). Contrary to bacterial MprF-aminoacylated lipids, results were not compatible with LX being a glycerolipid derivative. Fragmentation pattern of the first peak was rather consistent with a dehydrated ergosterol (Erg) (m/z 379.3355), which was supported by the presence of characteristic additional Erg fragmentation products (Fig. 4A) (17). The second peak gave similar fragmentation products together with that of Asp (m/z of 156.0265, [C4H7NO4Na]+) and of an aspartylated form of Erg (534.3547, [M∼aspartyl+Na]+). Results were consistent with an Erg moiety esterified with Asp on the β-OH group in position 3 of the A ring, making this species an Erg-Asp (Fig. 4B). Finally, liquid chromatography (LC)-ESI-MS/MS analyses of total lipids extracted directly from Afm confirmed that Erg-Asp was indeed present (SI Appendix, Fig. S5B). In parallel, we chemically synthesized Erg-Asp that we compared, on TLC, to Erg-Asp present in total lipids of Afm, Aor, or Sce expressing Afm ErdS and confirmed that, as expected, it shared the exact same migration properties (SI Appendix, Fig. S6).
Fig. 4.
Identification of the ergosteryl-3β-O-l-aspartate produced by ErdS in vivo and in vitro. (A) MS-ESI-QTOF spectrum (positive mode) of a lipid fraction containing LX extracted and purified from an Sce WT strain expressing Afm ErdS (upper spectrum, blue) or not (bottom spectrum, green). Peaks 1 and 2 have been analyzed by MS/MS collision-induced dissociation (CID) QTOF analysis in the positive mode. (B) Chemical structure of ergosteryl-3β-O-l-aspartate (Erg-Ap) corresponding to LX deduced from MS spectra shown in A. (C) The [3H]Erg-Asp synthesis was measured by LA assay in the presence of purified Afm ErdS, pure Sce tRNAAsp, radiolabeled [3H]Erg, and cold Asp in the presence (+) or absence (−) of the enzyme or of the indicated substrates. Tests included addition (+) or not (−) of RNase A. [3H]Erg-Asp is indicated with an asterisk. (D) Erg-[14C]Asp synthesis measured by LA assay using purified Afm ErdS, pure Sce tRNAAsp, radiolabeled [14C]Asp, and indicated sterols. LA assay using total lipids from WT Sce was used as a migration control. The adapted LA assay reactions are displayed (simplified) next to the corresponding TLCs. Radiolabeled compounds are highlighted in red, and the number of independent experiments (n) is indicated.
To confirm that Erg is the substrate of ErdS, we performed LA assays using either [3H]-Erg and cold Asp or cold Erg and [14C]-Asp. Analyses of reaction products by TLC revealed that ErdS indeed produced [3H]-Erg-Asp (Fig. 4C) or [14C]-Erg-Asp (Fig. 4D) in a tRNA-dependent manner, and that it requires l-Asp, ATP, and tRNAAsp for activity. Interestingly, in the presence of cholesterol (Cho), the animal equivalent of Erg and [14C]-Asp, ErdS also produced cholesteryl-aspartate (Cho-Asp) (Fig. 4D), indicating that it likely has a relaxed specificity toward its sterol substrate. Overall, our results demonstrate that ErdSs represent a novel type of bifunctional AspRS/ergosteryl-aspartate synthases that we named ergosteryl-3β-O-l-aspartate synthases or ErdS, with “Er” standing for “ergosteryl,” “d” for Asp, and “S” for synthase.
Fungi Having ErdS also Encode a Dedicated Erg-Asp Hydrolase Involved in Erg-Asp Turnover.
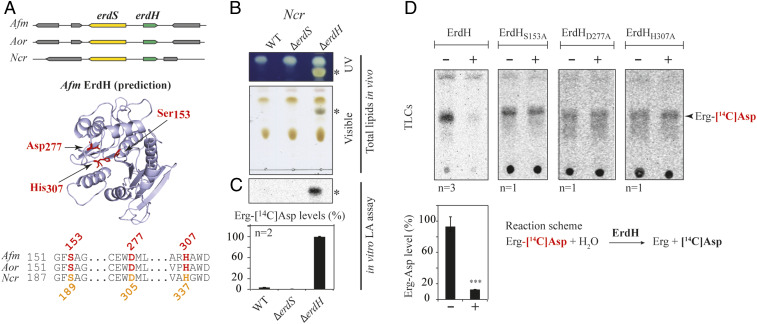
Bacteria that possess MprFs are also equipped with dedicated hydrolases that remove aa modifiers from lipids, and whose gene usually lays next to mprF genes (8, 9). In Afm, Aor, and Ncr, we observed that the erdS gene (AFUA_1g02570 in Afm Af293, AO090005000838 in Aor RIB40, and NCU007082 in Ncr OR74A) is found close to a gene encoding a protein belonging to the α/β-hydrolase family (PFAM: PF07859) (AFUA_1g02580, AO090005000837, and NCU007081, respectively) (Fig. 5A). These α/β-hydrolases all contain three conserved residues, Ser153, Asp277, and His307 (numbering of Afm), that are typical of the catalytic triad of esterases/lipases, a finding supported by structure predictions (Fig. 5A).
Fig. 5.
Detection and characterization of ErdH in fungi carrying ErdS. (A) Schematic representation of the genomic context of the erdS (yellow) and erdH (for Erg-Asp hydrolase, in green) genes locus from Afm, Aor, and Ncr that highlights that erdS and erdH are found in a divergent orientation. Phyre2-based α/β-hydrolase−like predicted structure of Afm ErdH and active site alignments of Afm, Aor, and Ncr ErdHs. Ser-Asp-His catalytic triads of α/β-hydrolases/lipases (S153, D277, and H307 in Afm ErdH) are displayed and highlighted on the structure prediction and the alignment. (B) Total lipids from WT, ∆erdS, and ∆erdH Ncr strains were separated by TLC and stained with sulfuric acid/MnCl2 and observed under UV or visible light (n = 3); * indicates Erg-Asp. (C) In vitro measurements of Erg-[14C]Asp synthesis by LA assay using protein extracts from the WT, ∆erdS, and ∆erdH Ncr strain protein extracts, using pure Sce tRNAAsp as a substrate (n = 2). (D) In vitro measurement of the Erg-[14C]Asp hydrolase activities of purified recombinant WT (n = 3) or catalytic mutant Ncr ErdHs (n = 1). (C and D) The Student's t test was used to determine significance of the means of the data; ***P < 0.005.
To determine whether these esterases were involved in the Erg-Asp metabolic pathway, we used Ncr because, in this fungus, despite the presence of an erdS gene (Fig. 5A and SI Appendix, Fig. S1A), we could not observe Erg-Asp in total lipids, although an ErdS activity could be detected using the LA assay in crude protein extracts (SI Appendix, Fig. S7). The WT, ΔerdS, and Δesterase deletion mutants of Ncr were obtained from the Fungal Genetic Stock Center (FGSC) (18, 19). We compared total lipids profiles of Ncr WT, ΔerdS, and Δesterase mutants by TLC (Fig. 5B). As stated, Erg-Asp was undetectable in the WT or ΔerdS strains of Ncr but accumulated at high levels in the Δesterase strain (Fig. 5B), suggesting that the enzyme was most likely responsible for the absence of detectable Erg-Asp in WT Ncr. This esterase, seemingly involved in Erg-Asp degradation, was therefore named ErdH, for Erg-Asp hydrolase, and the Δesterase strain was renamed ΔerdH (Fig. 5 A and B). Additionally, we monitored the Erg-[14C]-Asp synthesis activity by LA assay in protein extracts from WT Ncr, ΔerdS, and ΔerdH strains. Our data show that, while almost undetectable under the conditions tested in the WT and ΔerdS strains, Erg-[14C]-Asp levels were much higher (>23-fold) in the ΔerdH strain (Fig. 5C). This is again in agreement with ErdH being involved in Erg-Asp deacylation, which most likely masked the Erg-Asp synthesis activity of ErdS in the WT Ncr strain. Similarly, Erg-Asp synthesis was hardly monitored in vitro using total protein extracts of Afm and Aor (Fig. 3F), suggesting either low expression levels of ErdS or that ErdH hydrolyzed the Erg-Asp product.
In order to confirm that ErdH was indeed an Erg-Asp hydrolase, we used an in vitro Erg-Asp deacylation assay. Erg-[14C]-Asp incubated for 30 min with purified recombinant Afm ErdH was almost entirely hydrolyzed (88 ± 2%), while, in the same reaction performed without ErdH, Erg-[14C]-Asp levels remained stable (Fig. 5D). In addition, purified ErdH mutants of the catalytic triad (S153A, D277A, or H307A) did not hydrolyze Erg-[14C]-Asp in vitro (Fig. 5D). All these results demonstrate the presence of two enzymes in fungi that regulate the synthesis and degradation of Erg-Asp, namely ErdS and ErdH.
An Evolutionary Conserved Synteny between erdS and erdH.
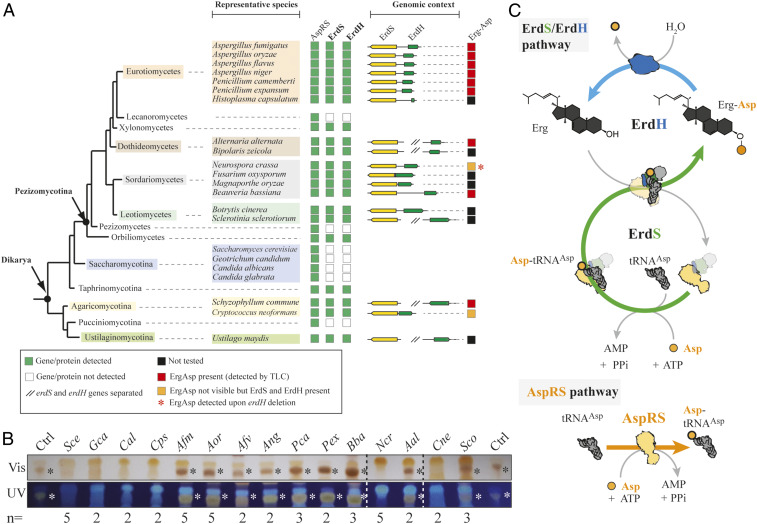
ErdS and ErdH are present in Afm, Aor, and Ncr, with their respective genes found at the same locus and encoded as divergent expression units. In order to analyze whether ErdS and ErdH are more generally found in fungi and/or other eukaryotes, we performed bioinformatics searches (SI Appendix, Supplemental Materials and Methods). We first searched ErdS sequences among eukaryotes using the basic local alignment search tool (BLAST) with the Afm ErdS sequence as a probe and found 7,584 protein sequences (only those with lengths 200 to 2,000 residues), that corresponded to ErdS, standalone DUF2156 domains, canonical AspRSs, and homologous aaRSs such as asparaginyl- and lysyl-tRNA synthetases. Among them, 1,006 (13.3%) were bona fide DUF2156-containing proteins, with 780 (77%) being ErdS with a minimal length of ∼800 aa (no N- or C-terminal extensions) and a maximal length of 1,417 aa (with long N- and/or C-terminal appendages). “Standalone” DUF2156 domains (23%) were also detected and often accounted for ErdS forms with either the AspRS or the DUF2156 domain truncated. A closer look at the phylogenomic distribution of complete ErdS (AspRS-DUF2156 fusions) highlighted that they are present only in “higher” fungi (Dikarya, both Ascomycota and Basidiomycota), and specifically among 9 out of the 13 fungal lineages analyzed (Fig. 6A), with the notable exception of Saccharomycotina (including Sce), Lecanoromycetes, and Pezizomycetes. ErdS is present in prominent human pathogens such as Afm (20), Histoplasma capsulatum, or Cryptococcus neoformans (Cne), plant pathogens such as Fusarium oxysporum, Magnaporthe oryzae, or Ustilago maydis, insect pathogens like Beauveria bassiana or Metarhizium spp., and also fungi of biotechnological interest like Aor or Ncr. We then visualized the synteny of the erdS gene in various fungi and observed that, in most of the cases, the erdH gene was almost always present next to erdS with the same divergent orientation in numerous species, showing that the linkage between the two genes is conserved. In fungi lacking erdS, the erdH genes were also absent, as judged from the absence of proteins with significant sequence homology to ErdH (Fig. 6A). Overall, the whole ErdS/ErdH enzymatic pathway seemed conserved across “higher” fungi.
Fig. 6.
The Erg-Asp lipid metabolic enzymes ErdS and ErdH are conserved across “higher” fungi. (A) The presence (green square) or absence (white squares) of a regular AspRS, ErdS (yellow), or ErdH (green) genes across fungal species is indicated, and the organization of the locus is shown; // indicates that erdS and erdH genes are interspaced. (B) TLC-based analysis of total lipids extracted from 15 species among the representative ones displayed in A. The TLC plates (number of replicates indicated for each strain) stained with sulfuric acid/MnCl2 solution and observed under white (Vis) or UV light were cropped to the area of interest; * indicates Erg-Asp synthesis (red squares in A) as revealed by the detection of a dark brown band with mobility equivalent to control Erg-Asp. Gca: Geotrichum candidum; Cal: Candida albicans; Cps: Candida parapsilosis; Afv: Aspergillus flavus; Ang: Aspergillus niger; Pca: Penicillium camemberti; Pex: Penicillium expansum; Bba: Beauveria bassiana; Aal: Alternaria alternata; Sco: Schizophyllum commune. (C) A schematic model representing the turnover between Erg-Asp synthesis, and hydrolysis. The green arrow shows ErdS activities (Asp-tRNAAsp production and Asp modification of Erg), and the blue arrow indicates the ErdH-dependent hydrolysis of Erg-Asp. All ErdS-containing fungi always possess a canonical AspRS to ensure Asp-tRNAAsp production for protein synthesis (AspRS pathway).
Finally, in order to confirm that Erg-Asp is synthesized in several representative ErdS/ErdH-containing fungi, we extracted and analyzed, by TLC, total lipids from a collection of 15 fungal species (13 Ascomycota and 2 Basidiomycota). As expected from the absence of an erdS gene, Erg-Asp was absent in the four tested Saccharomycotina. However, it was detected in eight Ascomycota and one Basidiomycota species tested (Fig. 6B). The only exceptions were Ncr (Ascomycota), as already explained above, and Cne (Basidiomycota), for which the erdS gene regulation and/or the activity of ErdH could also account for the absence of Erg-Asp. Taken together, our results confirm that Erg-Asp is likely widely distributed and conserved across fungi.
Discussion
A wide range of bacteria possess membrane proteins with an AAT activity, namely MprFs, that reroute aa-tRNAs from protein synthesis to cell surface remodeling, thereby intensifying antimicrobial resistance, pathogenicity, and/or virulence (4, 7, 11). Our work reveals that most of the fungal species found within the Dikarya subkingdom possess bifunctional enzymes, ErdS, that use ATP, l-Asp, and tRNAAsp to produce Asp-tRNAAsp in the AspRS domain, this latter product being used in a second step by the appended DUF2156/Asp-tRNA transferase module to transfer Asp from its tRNAAsp onto the 3β-OH group of Erg, yielding a novel form of conjugated sterol, Erg-Asp (Fig. 6C). Erg-Asp synthesis constitutes a change of paradigm for three main reasons: 1) this is a tRNA-dependent lipid modification process in eukaryotes; 2) the use of Asp as a lipid modifier, that has no equivalent in bacteria, and 3) the modification of Erg (instead of glycerolipids) with an aa that had, to our knowledge, never been described, despite the widespread distribution of Erg-Asp synthesis in fungi (Fig. 6B). Moreover, we found no evidence in the literature or databases of any 3β-O-aminoacylated sterols. Erg-Asp likely escaped previous detection because, as an ester, most of alkaline extraction procedures used for sterols extraction promote its hydrolysis. Erg-Asp is therefore a novel type of sterol conjugate, specific to fungi, that is performed by ErdSs enzymes, through a tRNA-dependent process. It is worth noting that the fusion of the AspRS and the transferase (DUF2156) domain is required for full activity, since the standalone DUF2156 domain was poorly active in the Sce heterologous model (Fig. 3B) or inactive in Aor (Fig. 2C). Channeling of Asp-tRNAAsp from the AspRS to the appended DUF2156 is the most plausible explanation. It would explain the better capacity of the AspRS domain alone, that releases Asp-tRNAAsp, to complement the Sce Δdps1 strain over the full-length ErdS (SI Appendix, Fig. S4A), that transfers Asp-tRNAAsp directly to the transferase domain for sterol aspartylation, likely without releasing it frequently. Of note is that all fungal species that encode ErdS also possess a regular cytoplasmic AspRS (Fig. 6A and SI Appendix, Fig. S1A), suggesting that the Asp-tRNAAsp generated by the AspRS moiety of ErdS is probably entirely dedicated to lipid modification, thereby necessitating a second AspRS to produce the Asp-tRNAAsp used for translation, as observed in all inspected fungi (Fig. 6 A and C and SI Appendix, Fig. S1A).
Characterization of ErdH, that hydrolyzes Erg-Asp in fungi, revealed that, like in bacteria, ErdS-containing fungi possess a homeostasis system that enables the tRNA-dependent aminoacylation of a lipid (ErdS) and its deacylation through hydrolysis (ErdH), suggesting that Erg-Asp levels might be tightly controlled. This provides an explanation for the apparent absence of Erg-Asp in total lipids from Ncr or Cne (Figs. 5B and 6B) under the growth conditions used. Of note is that this turnover cycle recalls that of the acyl-CoA transferase-dependent fatty acylation and lipase-dependent deacylation systems involved in sterols storage, turnover, and trafficking in eukaryotes, including fungi (21). The erdS and erdH genes tend to cluster as divergently expressed units within the same genomic locus, which resembles the cluster organization of genes with metabolically related functions found in many fungi (22). This suggests that erdS and erdH may be coregulated at the transcriptional level, likely to control the balance between Erg and Erg-Asp, a tempting hypothesis that remains to be analyzed. Erg-Asp levels could also depend on the turnover (expression and degradation) and regulation (posttranslational modifications, etc.) of ErdS and ErdH and/or on the relative subcellular localization of both enzymes. Such potential species-specific spatiotemporal regulations of Erg-Asp homeostasis could account for the different steady-state levels of Erg-Asp that we observed between the various fungal species, especially between Aspergillus spp. and Ncr. We have not been able, so far, to identify the subcellular localization of either ErdS or ErdH. Bioinformatics predictions (Wolf PSort, MitoProt II software), however, suggest that ErdH might be mitochondrial in Aspergillus spp., while ErdS is predicted to be cytoplasmic and/or nucleocytoplasmic, a fact that is not unusual for aaRSs (23). Also, no plausible membrane-spanning or membrane-anchoring helices could be predicted in ErdS or ErdH, which differs from bacterial homologs: MprFs have N-terminal membrane-spanning domains, and aminoacyl-glycerolipid hydrolases have N-terminal secretion signals to ensure correct periplasmic or extracellular localization (4). Very preliminary subcellular fractionation experiments suggest that, in Afm or in the heterologous model Sce, ErdS is not soluble but is associated with membranes, but the nature of those membranes remains unknown. Of note, in any case, is that membrane association of ErdS—to recognize Erg—requires that the enzyme be exposed to the cytoplasmic space, where its tRNAAsp, Asp, and ATP substrates are present (SI Appendix, Fig. S8).
Erg is the major sterol found in fungal plasma membranes (24). Its function is analogous to that of Cho in mammals, which includes modulating permeability and fluidity of the lipid bilayer (25, 26) and regulating the activity of membrane proteins, such as G protein-coupled receptors (27) or V-ATPases (28). Erg, together with sphingolipids, regulates membrane trafficking, which is illustrated in filamentous species, such as Aspergillus spp., by a constant supply of Erg at the tip of mycelia to form lipid microdomains that are crucial to polarized cell growth (29). Erg also plays multiple roles in pathogenicity and in antifungal resistance (24, 30, 31). Incidentally, because Erg biosynthesis pathway differs from that found in humans for Cho, it is one of the main targets of current antifungal drugs (32).
Although we deciphered the function of ErdS and ErdH, the role of Erg-Asp in the biology of fungi now remains to be discovered, and this will be the focus of our future work. The viability of the ΔerdS strains of Afm, Aor, and Ncr shows that neither ErdS nor ErdH are essential under standard growth conditions. However, as discussed below, this fact does not exclude functions that could become essential or contribute to the overall fitness of fungi under challenging conditions, such as in the presence of membrane-targeting antifungals or antimicrobial peptides, which may explain the evolutionary conservation of the enzymatic system across Dikarya. As a first hypothesis, aspartylation of Erg by ErdS might play a role similar to aminoacylated glycerolipids in bacteria, regarding antimicrobial resistance (4, 7, 11, 32). Indeed, addition of l-Asp on the neutral and highly hydrophobic Erg leads to a novel zwitterionic sterol, which may significantly change Erg physicochemical properties within membranes, by influencing its phase behavior, its lateral diffusion, and/or its interaction with other lipids. Local Erg-Asp synthesis might, in turn, influence membranes’ overall properties such as interface hydration, membrane proteins composition, permeability, and/or fluidity, or the activity and/or localization of membrane proteins. Therefore, Erg-Asp could participate in membrane remodeling processes that might have an impact on antifungal resistance, notably, in the case of polyenes, known to directly interact with Erg to form pores, since aspartylation of the 3β-OH could change their interactions with Erg (33), a hypothesis that we will explore. Erg-Asp levels (7 to 20% of free Erg in Aor; SI Appendix, Fig. S9) upon growth in rich medium seem incompatible with an overall surface remodeling; however, nothing is currently known on the regulation of Erg-Asp synthesis and the amount of modification that can occur under challenging conditions. As an illustration, in Sce, an Erg acetylation/deacylation cycle, that functionally resembles our ErdS/ErdH enzymatic system, involves the Atf2 acetyltransferase and the Say1 esterase. This nonessential enzymatic system protects cells from the accumulation of toxic Erg intermediates within membranes, and from the effect of membrane-disrupting agents such as the antifungal agent eugenol (34), suggesting that the ErdS/ErdH system and Erg aspartylation might be of importance in resistance when Erg biosynthesis inhibitors, such as azole derivatives, that trigger accumulation of toxic Erg intermediates, are used against fungi. This could be tested by determining the susceptibility of WT (Erg-Asp-containing), ΔerdS (Erg-Asp-deficient), ΔerdH (Erg-Asp-accumulating), or overexpressing (Erg-Asp-overproducing) strains in the presence of azole derivatives.
While lipid modifications are usually involved in cells’ surface remodeling and antimicrobial resistance in bacteria (7, 35), they also often participate in membrane trafficking and lipid-derived signaling in eukaryotes (3), like, for example, in the case of phosphoinositides or sphingolipids (36). Erg-Asp is produced at levels (7 to 20% of free Erg in Aor; SI Appendix, Fig. S9) that could be compatible with such cellular functions. Ergosteryl-3β-O-glucoside (Erg-Glc), another sterol conjugate, produced by Erg glycosylation, is nonessential, but known to actively participate in the regulation of autophagy in several yeasts and in Aor (37, 38), likely through the recruitment of protein partners on membrane structures (39). It is therefore possible that Erg aspartylation could intervene in the recruitment of protein factors on membranes (Erg-containing microdomains, etc.) and influence regulatory or trafficking pathways. Finally, bacterial esterases that remove aa modifications from glycerolipids also influence virulence in A. tumefaciens (10). Those observations in various bacterial lipid aminoacylation systems or fungal sterol modification pathways raise the question of the contribution of Erg aspartylation and deacylation to those processes in fungi. Given the conservation of the ErdS/ErdH enzymatic system across Dikarya, these aspects will now have to be addressed experimentally to decipher the function of Erg-Asp.
Materials and Methods
Materials, Strains, and Growth Condition.
The erdS codon-optimized synthetic gene was purchased from the Genscript Corporation. The anti-DUF2156 polyclonal antibody production was performed by Covalab (R&D in Biotechnology). Some of the fungal strains were ordered from the Fungal Genetics Stock Center. The CEA17 ΔakuBKU80 strain of Afm (40) and the RIB40 strain of Aor (41) have been used as parental strain for this study. The routine growth and maintenance of Afm and Aor are described in SI Appendix, Supplementary Materials and Methods. The strains (bacterial and fungi) used in this study are listed in SI Appendix, Table S1.
Total Lipid Extraction from Sce and from Filamentous Fungi.
Lipid extraction protocols for Sce and filamentous fungi were adapted from the Bligh and Dyer procedure (42). Briefly, Sce strains were grown in synthetic complete (SC) medium without leucine (SC-LEU) (MP Biomedicals) until optical density 600 nm (OD600 nm) reached 1.0. An equivalent of 150 OD600 nm of Sce cells were centrifuged at 4,000 × g for 15 min at 4 °C. The cell pellet was resuspended in 1 mL of Na-acetate 120 mM pH 4.5. Mechanical cell disruption was performed in a FastPrep-24 apparatus (MP Biomedicals, serial no. 10020698) in the presence of 400 µL of glass beads (ø 0.25 mm to 0.5 mm, Roth) at 6 m/s, for 1 min repeated 6 times, with cooling on ice between each step; 3.75 vol of MeOH:CHCl3 2:1 (v:v) were added, and the mixture was incubated on a rotating wheel at 4 °C for 3 h. Then, 1.25 vol of CHCl3 and Na-acetate 120 mM pH 4.5 were successively added and mixed by vortexing. The mixture was then centrifuged at 4,000 × g, 4 °C for 10 min to obtain two phases. The lower organic phase, containing the lipids, was transferred into a new tube, dried under vacuum, and stored at −20 °C until use or resuspended in 50 μL to 200 µL of MeOH:CHCl3 1:1 (vol/vol) for analysis by TLC.
For filamentous fungi, the same protocol was applied on 2 g of mycelia that were ground in a mortar with a pestle in liquid nitrogen to obtain a fine powder. Furthermore, the incubation on the rotating wheel was performed overnight at 4 °C.
Lipid X Purification.
LX was purified from adaptation of the flash chromatography procedure used for bacterial aminoacylated lipids (11). Total lipids extracted from 500 OD600 nm of Sce cells were solubilized in 200 µL of chloroform and loaded on a 1.5-mL silica gel (Sigma-Aldrich, pore size 60) glass column preequilibrated with CHCl3. After absorption of lipids on the column, nonpolar lipids and glycolipids were eluted with 10 mL of chloroform followed by 10 mL of acetone, respectively. To elute polar lipids, various ratios of CHCl3:MeOH from 9:1 to 6:4 (vol/vol) (20 mL each) were used, and elution fractions were collected in 2-mL samples in glass tubes. Fractions were dried under vacuum, resolubilized in 35 µL of MeOH:CHCl3 (1:1, vol/vol), and visualized by TLC.
TLC Analyses.
Dried lipid samples were resuspended in MeOH:CHCl3 (1:1, vol/vol) and spotted onto 10 × 20 cm silica gel on TLC Al foils (Sigma-Aldrich). TLC was developed with a CHCl3:MeOH:H2O (130:50:8 vol/vol/vol) mobile phase (10 min at room temperature [RT]), and plates were stained with either MnCl2/sulphuric acid or ninhydrin or bromocresol green. MnCl2/sulphuric acid, a universal staining (43), was prepared with 0.8 g of MnCl2 tetrahydrate dissolved in 240 mL of 50% (vol/vol) MeOH supplemented with 9 mL of concentrated sulphuric acid. To detect primary amines (pink color), ninhydrin 0.4% (wt/vol) (Sigma-Aldrich) was used. To reveal carboxylate-containing compounds (blue color) with a pKa below 5.0 (44), bromocresol green (0.04 g) (Sigma-Aldrich) dissolved in absolute ethanol and NaOH 0.1 M was used. In the case of MnCl2/sulphuric acid and ninhydrin stains, plates were heated at 100 °C until coloration developed, while direct ultraviolet (UV) (254 or 315 nm) visualization was performed with bromocresol green. Quantification of LX (Erg-Asp), PE, or PG spots was performed using the ImageJ software. Lipid spots signals (number of pixels, N) were normalized to that of PE (NLX/NPE). In the absence of LX (Erg-Asp), PG becomes visible at the same position; the PG/PE signal thus represents the background signal in the absence of LX.
MS Analyses.
Analyses of total purified lipid extracts were performed on a liquid chromatograph Surveyor Plus with an autosampler and coupled with an LTQ-XL ion trapanalyzer (Thermo Finnigan). Ten microliters of filtered lipid extracts were injected on a Cogent Diamond Hydride (250 × 4.6 mm, 4 µm, Microsolv) at a temperature of 40 °C as described in ref. 45 with minor modifications. Briefly, elution was performed at a flow rate of 1 mL/min in hydrophilic interaction liquid chromatography (HILIC) conditions with a binary gradient running from 99.7:0.3 (A:B) to 75:25 in 40 min, where A was acetonitrile and B was 40 mM aqueous ammonium formate. Both solutions contained 5.5 mM formic acid. The instrument was tuned by direct infusion of cholesterol (Avanti) in positive mode. Drying gas flow rate was 20 units, and temperature of the ESI was 380 °C. Full-scan spectra were collected in the 110 to 2,000 m/z range, and data-dependent Zoom and MS2 spectra were acquired on the 15 most intense peaks. Data were analyzed with XcaliburQual Browser.
In Vitro LA Assay.
Prior to lipid aminoacylation, the in vitro tRNAAsp aspartylation was performed as described in SI Appendix, Supplementary Materials and Methods. Lipid or sterols aspartylation assays were adapted from lipid aminoacylation protocols used for bacterial MprFs (42). Briefly, reactions were performed in the following aminoacylation mix: Na-Hepes 100 mM pH 7.2 buffer containing KCl 30 mM, MgCl2 12 mM, ATP 10 mM, bovine serum albumin 0.1 mg/mL, pure yeast tRNAAsp (10 µM) (46), [U-14C]-Asp (280 cpm/pmol, Perkin-Elmer, NEC268E050UC) in a final volume of 50 µL. To test the transfer of [14C]-Asp onto lipids, total lipids were added to a final concentration of 2 mg/mL, and commercial pure sterols (Erg and Cho) were added to a final concentration of 0.5 mg/mL. When [3H]-Erg (10 Ci/mmol, Hartmann Analytic) was used, 20,000 cpm were added to the reaction mix. The resulting suspension was sonicated for 30 s in a sonicator bath at RT, and the enzyme (0.1 to 0.5 µg) or crude extracts of Afm, Aor, or Ncr (20 µg total proteins) were added to initiate the reaction before a 40-min-long incubation at 30 °C (1 h for crude extracts). To test the tRNA dependency of the reaction, 1 µg of RNase A from bovine pancreas was added 10 min before initiation of the reaction.
After incubation, reactions were stopped with addition of 500 µL of CHCl3:MeOH:Na-acetate 120 mM pH 4.5 (130:50:8, vol/vol/v) and vortexing. Then, 130 µL of CHCl3 and 130 µL of Na-acetate 120 mM pH 4.5 were successively added, and the mixture was vortexed and centrifuged for 1 min at 5,000 × g (RT). The lower organic phase was recovered and dried under vacuum. Reaction products were dissolved in a CHCl3:MeOH (1:1, v:v) mixture, spotted on TLC plates, and separated with the CHCl3:MeOH:H2O mobile phase. TLC plates were exposed onto an imaging plate (Fuji Imaging plate) for at least 2 h. Radioactivity was detected using a Typhoon TRIO variable Mode Imager (GE Healthcare). Quantification of LX (Erg-Asp) radioactive spots was performed using the ImageJ software (number of pixels).
In Vitro Lipid Deacylation Assay.
To generate Erg-[14C]-Asp, we used the protocol described above for LA assay. RNase A was added and incubated for 20 min at 30 °C to hydrolyze tRNAAsp and stop the tRNA-dependent aspartylation catalyzed by ErdS. Then, 0.1 μM of purified recombinant ErdH or ErdH variants were added, and the reaction mixture was incubated for 30 min at 30 °C. Lipids were extracted with the Bligh and Dyer procedure as described above, dried, resuspended in CHCl3:MeOH (1:1, vol/vol), and analyzed on TLC with the CHCl3:MeOH:H2O (130:50:8, v:v:v) solvent, and results were analyzed by phosphorimaging as described above.
Statistical Analyses.
The Student's t test was used to determine significance of the means of the data.
Materials and Data Availability.
All available data are already included in the manuscript. All materials and strains are freely available (contact: h.becker@unistra.fr; frfischer@unistra.fr).
Supplementary Material
Acknowledgments
This work was supported by the Fondation pour la Recherche Médicale (FRM), to H.D.B. (Grant DBF20160635713), by ‘‘MitoCross’’ Laboratory of Excellence (Grant ANR-10-IDEX-0002-02 to H.D.B.), by the University of Strasbourg (to H.D.B.), by an IDEX from the University of Strasbourg (W17RAT81, to F.F.), the National Center for Scientific Research (H.D.B, B.S., F.F., and N.Y.), and the Meiji University (T.K., Y.S., S.T., D.Y., and H.N.). N.Y. was supported by a fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche, and N.M. was supported by a postdoctoral fellowship from the FRM (Grant DBF20160635713). H.R. and C.D.G. were supported by NIH Grant 1R21AI144481-01. We thank J.-P.L., Dr. I. Mouyna, Dr. A. Beauvais, and Dr. T. Fontaine (Institut Pasteur, Paris, France) for providing A. fumigatus strains and deletion cassette templates, for having trained F.F. on the manipulation of the strains, and for thoughtful discussions. We thank H.N. for A. oryzae strains and related plasmids and Maryline Brock (University of Strasbourg) for other fungal species. In addition, we thank Dr. Sylvie Friant for critical review, experimental suggestions, and careful reading of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003266117/-/DCSupplemental.
References
- 1.Bohuszewicz O., Liu J., Low H. H., Membrane remodelling in bacteria. J. Struct. Biol. 196, 3–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fozo E. M., Rucks E. A., The making and taking of lipids: The role of bacterial lipid synthesis and the harnessing of host lipids in bacterial pathogenesis. Adv. Microb. Physiol. 69, 51–155 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Singh A., Del Poeta M., Lipid signalling in pathogenic fungi. Cell. Microbiol. 13, 177–185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields R. N., Roy H., Deciphering the tRNA-dependent lipid aminoacylation systems in bacteria: Novel components and structural advances. RNA Biol. 15, 480–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibba M., Soll D., Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Hebecker S. et al., Structures of two bacterial resistance factors mediating tRNA-dependent aminoacylation of phosphatidylglycerol with lysine or alanine. Proc. Natl. Acad. Sci. U.S.A. 112, 10691–10696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slavetinsky C., Kuhn S., Peschel A., Bacterial aminoacyl phospholipids–Biosynthesis and role in basic cellular processes and pathogenicity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1310–1318 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Arendt W., Groenewold M. K., Hebecker S., Dickschat J. S., Moser J., Identification and characterization of a periplasmic aminoacyl-phosphatidylglycerol hydrolase responsible for Pseudomonas aeruginosa lipid homeostasis. J. Biol. Chem. 288, 24717–24730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith A. M., Harrison J. S., Sprague K. M., Roy H., A conserved hydrolase responsible for the cleavage of aminoacylphosphatidylglycerol in the membrane of Enterococcus faecium. J. Biol. Chem. 288, 22768–22776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenewold M. K. et al., Virulence of Agrobacterium tumefaciens requires lipid homeostasis mediated by the lysyl-phosphatidylglycerol hydrolase AcvB. Mol. Microbiol. 111, 269–286 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Smith A. M. et al., tRNA-dependent alanylation of diacylglycerol and phosphatidylglycerol in Corynebacterium glutamicum. Mol. Microbiol. 98, 681–693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datt M., Sharma A., Novel and unique domains in aminoacyl-tRNA synthetases from human fungal pathogens Aspergillus niger, Candida albicans and Cryptococcus neoformans. BMC Genomics 15, 1069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruff M. et al., Class II aminoacyl transfer RNA synthetases: Crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science 252, 1682–1689 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Klug L., Daum G., Yeast lipid metabolism at a glance. FEMS Yeast Res. 14, 369–388 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Ador L. et al., Active site mapping of yeast aspartyl-tRNA synthetase by in vivo selection of enzyme mutations lethal for cell growth. J. Mol. Biol. 288, 231–242 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Cavarelli J. et al., The active site of yeast aspartyl-tRNA synthetase: Structural and functional aspects of the aminoacylation reaction. EMBO J. 13, 327–337 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Headley J. V., Peru K. M., Verma B., Robarts R. D., Mass spectrometric determination of ergosterol in a prairie natural wetland. J. Chromatogr. A 958, 149–156 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Colot H. V. et al., A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCluskey K., Wiest A., Plamann M., The fungal genetics Stock center: A repository for 50 years of fungal genetics research. J. Biosci. 35, 119–126 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Abad A. et al., What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 27, 155–182 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Jacquier N., Schneiter R., Mechanisms of sterol uptake and transport in yeast. J. Steroid Biochem. Mol. Biol. 129, 70–78 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Nützmann H. W., Scazzocchio C., Osbourn A., Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52, 159–183 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Debard S. et al., Nonconventional localizations of cytosolic aminoacyl-tRNA synthetases in yeast and human cells. Methods 113, 91–104 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Rodrigues M. L., The multifunctional fungal ergosterol. MBio 9, e01755-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas L. M., Konopka J. B., Fungal membrane organization: The eisosome concept. Annu. Rev. Microbiol. 68, 377–393 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Kodedová M., Sychrová H., Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10, e0139306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morioka S. et al., Effect of sterol composition on the activity of the yeast G-protein-coupled receptor Ste2. Appl. Microbiol. Biotechnol. 97, 4013–4020 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y. Q. et al., Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6, e1000939 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer R., Zekert N., Takeshita N., Polarized growth in fungi—Interplay between the cytoskeleton, positional markers and membrane domains. Mol. Microbiol. 68, 813–826 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Alcazar-Fuoli L., Mellado E., Ergosterol biosynthesis in Aspergillus fumigatus: Its relevance as an antifungal target and role in antifungal drug resistance. Front. Microbiol. 3, 439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koselny K. et al., A genome-wide screen of deletion mutants in the filamentous Saccharomyces cerevisiae background identifies ergosterol as a direct trigger of macrophage pyroptosis. MBio 9, e01204-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazu T. K., Bricker B. A., Flores-Rozas H., Ablordeppey S. Y., The mechanistic targets of antifungal agents: An overview. Mini Rev. Med. Chem. 16, 555–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamiński D. M., Recent progress in the study of the interactions of amphotericin B with cholesterol and ergosterol in lipid environments. Eur. Biophys. J. 43, 453–467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiwari R., Köffel R., Schneiter R., An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae. EMBO J. 26, 5109–5119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuri R., Shprung T., Shai Y., Defensive remodeling: How bacterial surface properties and biofilm formation promote resistance to antimicrobial peptides. Biochim. Biophys. Acta 1848, 3089–3100 (2015). [DOI] [PubMed] [Google Scholar]
- 36.De Craene J. O., Bertazzi D. L., Bär S., Friant S., Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int. J. Mol. Sci. 18, 634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grille S., Zaslawski A., Thiele S., Plat J., Warnecke D., The functions of steryl glycosides come to those who wait: Recent advances in plants, fungi, bacteria and animals. Prog. Lipid Res. 49, 262–288 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Kikuma T., Tadokoro T., Maruyama J. I., Kitamoto K., AoAtg26, a putative sterol glucosyltransferase, is required for autophagic degradation of peroxisomes, mitochondria, and nuclei in the filamentous fungus Aspergillus oryzae. Biosci. Biotechnol. Biochem. 81, 384–395 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Yamashita S., Oku M., Sakai Y., Functions of PI4P and sterol glucoside are necessary for the synthesis of a nascent membrane structure during pexophagy. Autophagy 3, 35–37 (2007). [DOI] [PubMed] [Google Scholar]
- 40.da Silva Ferreira M. E. et al., The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5, 207–211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machida M. et al., Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Roy H., Ibba M., Monitoring Lys-tRNA(Lys) phosphatidylglycerol transferase activity. Methods 44, 164–169 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami S. K., Frey C. F., Manganous chloride spray reagent for cholesterol and bile acids on thin-layer chromatograms. J. Chromatogr. A 53, 389–390 (1970). [DOI] [PubMed] [Google Scholar]
- 44.Shokrollahi A., Firoozbakht F., Determination of the acidity constants of neutral red and bromocresol green by solution scanometric method and comparison with spectrophotometric results. Beni. Suef Univ. J. Basic Appl. Sci. 5, 13–20 (2016). [Google Scholar]
- 45.Cífková E., Hájek R., Lísa M., HolĿapek M., Hydrophilic interaction liquid chromatography-mass spectrometry of (lyso)phosphatidic acids, (lyso)phosphatidylserines and other lipid classes. J. Chromatogr. A 1439, 65–73 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Keith G., Gangloff J., Dirheimer G., Isolement des tRNATyr et tRNAAsp de levure de bière hautement purifiés. Biochimie 53, 123–125 (1971). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.