Significance
When a pathogen is engulfed by an immune cell, it is trapped in a transient organelle called the phagosome. The host cell recognizes the pathogen through receptors called Toll-like receptors (TLRs) that each recognize specific pathogen-associated molecular pattern (PAMP). When a TLR recognizes its cognate PAMP, it activates inducible nitric oxide synthase (NOS2), filling the phagosome with NO and destroying the pathogen. By developing a fluorescent reporter that directly images PAMP-triggered NOS2 activity in real time, we show that single-stranded RNA acts as a PAMP in zebrafish by activating its cognate TLR. Such a technology can help identify new PAMPs, pinpoint ligands for TLRs of unknown function, and suggest how PAMPs might synergize to help bugs evade the phagosomal deathtrap.
Keywords: nitric oxide synthase-2 (NOS2), live imaging, phagosome, nitric oxide, microglia
Abstract
Innate immune cells destroy pathogens within a transient organelle called the phagosome. When pathogen-associated molecular patterns (PAMPs) displayed on the pathogen are recognized by Toll-like receptors (TLRs) on the host cell, it activates inducible nitric oxide synthase (NOS2) which instantly fills the phagosome with nitric oxide (NO) to clear the pathogen. Selected pathogens avoid activating NOS2 by concealing key PAMPs from their cognate TLRs. Thus, the ability to map NOS2 activity triggered by PAMPs can reveal critical mechanisms underlying pathogen susceptibility. Here, we describe DNA-based probes that ratiometrically report phagosomal and endosomal NO, and can be molecularly programmed to display precise stoichiometries of any desired PAMP. By mapping phagosomal NO produced in microglia of live zebrafish brains, we found that single-stranded RNA of bacterial origin acts as a PAMP and activates NOS2 by engaging TLR-7. This technology can be applied to study PAMP−TLR interactions in diverse organisms.
Innate immune cells destroy pathogens by ingesting and trapping them within a transient, subcellular organelle called the phagosome (1, 2). The phagosome rapidly develops into a lethal hotspot of noxious chemicals such as reactive oxygen species (ROS) and nitric oxide (NO) that are produced by enzymes. In mammals, ROS is produced by reduced nicotinamide-adenine dinucleotide phosphate (NADPH) oxidase (NOX), while NO can be produced by three different isoforms of NO synthases (NOS): neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3) (3, 4). NOS1 and NOS3 are constitutively active. NOS1 is preferentially expressed in neurons and plays an important role in synaptic plasticity (5). NOS3 is expressed in endothelial cells where its activity is critical for vasodilation (6). NOS2 is majorly expressed in immune cells and is pivotal to neutralizing microbes within host phagosomes (7). Due to their differential susceptibilities to ROS and NO, different microbes are neutralized to different extents in the phagosome (7). We describe a DNA-based trigger-and-detect technology, that reports on NOS2 activity by mapping phagosomal NO in microglia of live zebrafish brains. By displaying various pathogen-associated molecular patterns (PAMPs) on these DNA-based reporters and mapping phagosomal NOS2 activity, we found that single-stranded RNAs (ssRNAs) of bacterial origin act as PAMPs in zebrafish. We identified in zebrafish, a putative Toll-like receptor (TLR), TLR-7, as the cognate receptor that activates NOS2.
Innate immune cells target pathogens for destruction by recognizing them through an array of endosome-resident or plasma membrane-resident receptors called pattern recognition receptors (PRRs) such as TLRs, RIG-like receptors, or NOD-like receptors (8). PRRs recognize conserved structural motifs of molecules present in the pathogen termed PAMPs (9). For example, TLR-4, TLR-3, and TLR-9 detect bacterial lipopolysaccharide (LPS), viral RNA (double-stranded RNA [dsRNA]), and bacterial DNA (CpG DNA), respectively (10). When a TLR receptor recognizes its cognate PAMP, it initiates the TLR signaling cascade to activate NOS2 at the phagosome. When macrophages are challenged with immunogens such as Mycobacterium smegmatis or CpG-containing sequences, there is an initial burst of NO within the first hour (11). This is followed by a second phase of NO production that lasts for hours over transcriptional time scales due to NOS2 expression induced by the transcription factor NF-κB as a result of TLR stimulation (11–13). The ability to directly map the initial burst of phagosomal NO in vivo could provide a new avenue to identify mechanisms that are operational at the early stages of pathogen infection.
NO produced due to PAMP−PRR recognition is generally quantified by the Griess assay that leverages the spontaneous oxidation of NO to nitrite (NO2−) which is then converted to nitrous acid (HONO), the detected species (14). Currently, the Griess assay detects NO in the extracellular milieu and does not provide single-cell−level information. Excellent small-molecule detection chemistries are available for NO, but the reacted probe molecules diffuse rapidly, obscuring spatial information (15, 16). Although such probes can be targeted to the lysosome using protonatable morpholine derivatives, it is difficult to address whether the probe molecules localize before or after reacting with NO (17). Additionally, both NOS2 activity and morpholine-based targeting are highly dependent on organellar pH. Thus it is difficult to distinguish between NO production efficiency and probe targeting efficiency (18). Current protein-based NO reporters lack the specificity and sensitivity of small-molecule probes, are pH sensitive, and need millimolar levels of Iron(II) supplementation for their function, thus precluding their utility in live-animal imaging (19).
We have recently described a DNA-based reporter technology called NOckout to quantify NO produced by NOS3 at the plasma membrane or in the trans-Golgi apparatus in cultured cells (20). This reporter combines the advantages of small-molecule probes with the stable spatial localization afforded by proteins (20). Here, we modify NOckout to display well-defined nucleic acid-based PAMPs in addition to displaying NO detection chemistry, and a different reference fluorophore better suited to in vivo imaging for quantitative ratiometry. All three functionalities can be integrated in a well-defined stoichiometry onto a single assembly by hybridizing complementary DNA strands, each bearing one of the functionalities. Thus, the resultant assembly is a DNA duplex that is recognized in the living brain as fragmented self-DNA, packaged into apoptotic bodies, which are then phagocytosed by microglia resident in the brain and thereby targeted to microglial phagosomes where they report on NOS2 activity.
We describe a series of NOckout probes, each displaying a different nucleic-acid based PAMP, but with identical NO detection characteristics. Only when specific PAMPs are displayed on NOckout probes do they trigger NOS2 activity. This allows one to assay NO levels due to NOS2 activation in situ in late endosomes (LEs) of cultured cells or in phagosomes of live zebrafish brains. We show that phagosomal NO is produced due to the engagement of a specific TLR by the PAMP on the NOckout probe (Fig. 1A). We applied this approach to show that ssRNAs derived from pathogenic sources activate NOS2 by engaging TLR-7.
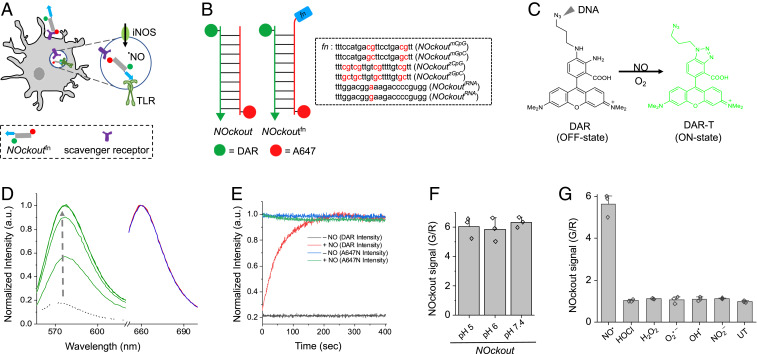
Fig. 1.
Design, sensitivity, and specificity of NOckout probes. (A) Schematic of TLR-mediated NOS2 activation by NOckoutFn. NOckoutFn, displaying a functional PAMP (Fn, blue), is phagocytosed and binds its cognate TLR-receptor (green hook) to activate NOS2 (green circle) and elevate phagosomal NO levels. (B) Schematic of the design of NOckout and NOckoutFn probes. NOckout is assembled from an ssDNA (24-mer) carrying an NO-sensing fluorophore (DAR, green circle) and a cDNA strand displaying a normalizing fluorophore (A647, red circle). NOckoutFn carries a functional (Fn) immunostimulatory CpG DNA or RNA motif as an overhang specific to mouse (NOckoutmCpG or NOckoutiRNA) or zebrafish TLR receptors (NOckoutzCpG or NOckoutiRNA). (C) Reaction with NO turns the nonfluorescent DAR probe (off-state) on NOckout into the highly fluorescent DAR-T (on-state). (D) Fluorescence spectra of NOckout (250 nM) in sodium phosphate buffer (pH 6.0, 50 mM) recorded in the DAR channel (green, λex = 550 nm, λem = 575 nm) and in the A647 channel (λex = 645 nm, λem = 660 nm) before (red) and after (blue) the addition of diethylamine NONOate (DEA NONOate, 50 μM, pH 6.0). Spectra in the DAR channel (G) were recorded in 30-s intervals (green traces). (E) Representative kinetic trace of NOckout in the DAR channel with (red) and without (black) DEA NONOate (50 μM, pH 6.0). Intensity in A647 channel before and after addition of DEA NONOate is shown in blue and green, respectively. (F) Fold change of NOckout represented as G/R values in pH 5, pH 6, and pH 7 buffers. (G) In vitro specificity of NOckout against various ROS and RNS. NOckout (250 nM, pH 6.0) was incubated with NO (50 μM of DEA NONOate), HOCl (5 μM), H2O2 (100 μM), O2.− (100 μM), OH· (100 μM), and NO2− (100 μM) for 6 min at room temperature, and the intensities were plotted as (G/R) values corresponding to DAR (G) to that of A647 (R). Error bars are SDs for n = 3 independent trials.
Design, Sensitivity, and Specificity of NOckout Probes
We designed a series of ratiometric, fluorescent NO-sensitive DNA-based probes denoted NOckoutfn, where fn denotes a functional DNA or RNA sequence that can potentially be a PAMP (Fig. 1B). The basic NOckout device comprises three modules, namely, the NO detection chemistry, a normalizing dye for ratiometric quantitation of NO, and a duplex DNA motif for internalization by specific cell types and stable compartmental localization thereafter (Fig. 1B). NOckout variants comprise a 24-base pair DNA duplex composed of two strands, S1 and S2, bearing four functionalities (Fig. 1B and SI Appendix, Table S1). The first is an NO-sensing dye based on diaminorhodamine (DAR) (21) which is attached via a polyethylene glycol linker to the 5′ end of S1 (SI Appendix, Scheme S1) (20). DAR is quenched due to intramolecular photoinduced electron transfer (PeT) from the aromatic diamino group (OFF state, ϕ = 0.0005, ɸ.ε ≈ 39 M−1⋅cm−1; Fig. 1C). Reaction with NO leads to a highly fluorescent triazole (DAR-T) due to relief of PeT (ON state, ϕ = 0.42, ɸ.ε ≈ 13,405 M−1⋅cm−1; Fig. 1C). DAR-T is highly photostable and bright, and its fluorescence is pH invariant (21).
The second functionality is a normalizing dye ATTO647N (A647) attached to the 5′-end of the S2 strand (Fig. 1B and SI Appendix, Scheme S1). A647 is the normalizing fluorophore, chosen for its pH insensitivity, molar brightness (ϕ = 0.65, ɸ.ε ≈ 97,500 M−1⋅cm−1), and chemical inertness toward ROS and reactive nitrogen species (RNS). All NOckout variants were assembled in near-quantitative yields by annealing equimolar ratios of S1 and S2 strands and characterized by gel electrophoresis (SI Appendix, Fig. S3).
The third functionality in every NOckout variant is a dsDNA module responsible for targeting the nanodevice into phagosomes of microglia in live zebrafish brains. This is essentially the 24-base pair DNA duplex formed by the hybridization of S1 and S2. This DNA duplex also facilitates endocytosis of all NOckout variants by macrophages in culture through the scavenger receptor-mediated endocytosis pathway (22, 23).
The fourth functionality in all NOckout variants is an immunogenic oligodeoxynucleotide (ODN) or oligoribonucleotide (ORN) phosphorothioate sequence denoted fn, and the resultant devices are denoted NOckoutfn. The ODN or ORN sequence in NOckoutfn devices is located at the 3′ end of S2 as single-stranded overhangs in the basic NOckout device (Fig. 1B and SI Appendix, Table S1). The ODN sequences 1826 and 2007 are well-characterized CpG sequences and agonists for specific TLRs in mouse and in zebrafish, respectively (13, 24). They trigger murine TLR-9 and zebrafish TLR-9/21 and, for clarity, are denoted mCpG and zCpG, respectively (Fig. 1B). The corresponding NOckoutfn devices are denoted NOckoutmCpG and NOckoutzCpG, respectively. Control ODN sequences were also used to create NOckoutmGpC and NOckoutzGpC devices. The sequences in these controls had the same base composition as mCpG and zCpG, but the CpG motifs were replaced with nonimmunostimulatory GpC motifs (Fig. 1B). Similarly, we created NOckoutiRNA displaying an immunostimulatory ribosomal RNA (iRNA) sequence of bacterial origin documented to activate the recently characterized mouse TLR-13 receptor (25, 26). A nonimmunostimulatory control, called NOckoutRNA, incorporated the same sequence with a single A to G point mutation (Fig. 1B). An important consideration in the design of NOckoutFn probes is that the lumen of every LE or phagosome of interest encounters the trigger and detector simultaneously. This means that the time between receptor engagement and NO detection is tightly controlled, which allows one to compare NO signals across endosomes or phagosomes at any given time.
To check the response characteristics of these NOckout variants to NO in vitro, we added the NO donor DEANONOate (50 μM) to 250 nM NOckout in phosphate buffer, pH 6.0, and monitored DAR-T and A647 fluorescence intensities as a function of time. DAR fluorescence (G) increased rapidly upon NO addition, and the time required for 50% reaction (t1/2) under these conditions was ∼30 s (Fig. 1D). The intensity of A647 (R) remained constant as a function of time (Fig. 1 D and E), and the ratio of DAR/A647 fluorescence intensities, that is, the G/R ratio, showed a ∼6.0-fold change upon complete reaction of the sensor (Fig. 1F). The NO response characteristics of all of the NOckout devices were similar within error and were insensitive to pH from pH 5 to 7.4 (Fig. 1F and SI Appendix, Figs. S4 and S5). They are thus suitable to map NO in acidic organelles.
All NOckout variants proved highly specific to NO over other reactive species both in vitro and in cells. The response of NOckout to 100 μM each of various ROS, that is, H2O2, O2•–, and OH•, was tested by measuring the in vitro fold change in G/R before and after addition of indicated reactive species. Just like the parent small-molecule NO probes DAR and DAF, NOckout, too, showed very high in vitro specificity for NO over other ROS (Fig. 1G).
To check the specificity of NOckout to NO in cells, endosomes of J774A.1 macrophages primed with LPS were labeled with NOckout and imaged in the DAR (G) and A647 (R) channels from where the G/R maps were obtained as described (Fig. 2A and Materials and Methods). Macrophages express scavenger receptors that bind and traffic dsDNA along the endolysosomal pathway (22). They also express the ROS- and NO-producing enzymes, NOX (27), myeloperoxidase (MPO), and NOS2 (28). Priming macrophages either with CpG oligonucleotides or endotoxins such as LPS induces the expression of NOS2 (SI Appendix, Figs. S8 and S9).
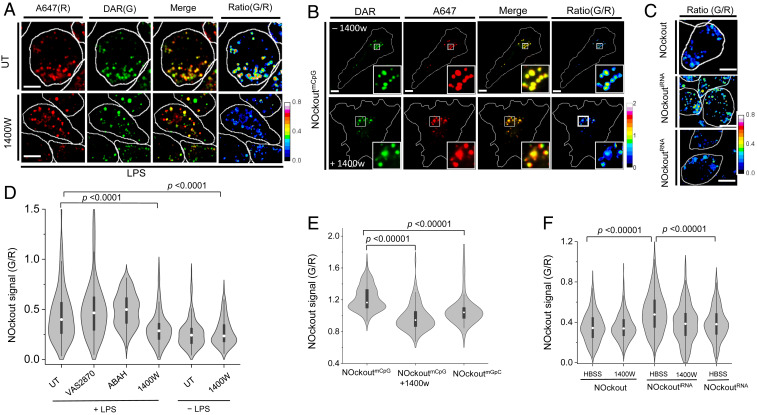
Fig. 2.
NOckoutFn detects NO in mouse primary microglia and in alveolar macrophages. (A) Representative confocal images of NOckout from LPS (1 μg/mL) primed J774A.1 macrophages in A647 (R) and DAR (G) channels in the absence (Upper) or presence (Lower) of NOS2 inhibitor 1400W. G/R intensities are represented as heat maps. (B) Representative confocal images of NOckoutmCpG from mouse primary microglia. Cells were incubated with NOckoutmCpG (500 nM) in the absence (Upper) or presence (Lower) of 1400W for 120 min in DMEM, imaged in DAR(G) and A647(R) channels, and converted into G/R heat maps. (C) Representative heat map (G/R) images of NOckout-, NOckoutiRNA-, and NOckoutRNA-treated J774A.1 cells. (D) Violin plot of the distribution of G/R values of ∼200 individual endosomes (n = 30 cells) of J774A.1 cells treated with VAS2870, ABAH, and 1400W in the presence and absence of LPS. (E) Violin plot of the distribution of G/R values of ∼100 individual endosomes (n = 20 cells) in primary microglia treated with NOckoutmCpG and NOckoutmGpC. (F) Violin plot of the distribution of G/R values of ∼200 individual endosomes (n = 30 cells) in J774A.1 macrophages treated with NOckoutRNA variants in the presence and absence of 1400W. All experiments were performed in triplicate. (Scale bar, 10 μm.) P values are obtained using Kruskal−Wallis statistical test across the dataset.
The in-cell specificity of NOckout was then evaluated by measuring the contribution of the reactive species produced by NOX, MPO, and NOS2 to the observed fold change in G/R ratio. We found that, if NOckout displayed no functional immunogenic sequence, it shows negligible NOS2 activation. Therefore, in order to trigger NOS2 activity for these experiments, the cells had to be explicitly activated by LPS treatment. We incubated J774A.1 cells primed with LPS, labeled with NOckout in the presence and absence of VAS2870 (29), 4-aminobenzoic acid hydrazide (ABAH) (30), or 1400W (31) that pharmacologically inhibit NOX, MPO, and NOS2, respectively (SI Appendix, Figs. S10 and S11). Fig. 2D shows the distribution of NOckout signal (G/R) from ∼200 endosomes for each experiment. The 1400W treatment significantly reduced G/R signal when compared to untreated cells (Fig. 2D). This was reconfirmed upon inhibiting NOS2 with L-NAME, a pan NOS blocker (SI Appendix, Fig. S13). Inhibiting NOX or MPO did not affect NOckout response, ruling out the contribution of ROS to the observed G/R signal (Fig. 2D and SI Appendix, Fig. S11).
NOckoutfn Devices Trigger NOS2 Activity in Microglia
Given that NOckout did not trigger NOS2 activity, we modified it to display CpG DNA and RNA sequences that function as PAMPs in order to engage the endosomal TLRs of the host cell and thereby activate NOS2 (Fig. 1 A and B). Nucleic acid-based PAMPs are recognized by the innate immune system through specific PRRs, namely TLRs (8). Specific TLRs recognize structurally distinct nucleic acid PAMPs present in “non−self-DNA” or “non−self-RNA” (32). Non−self-DNA is endocytosed by innate immune cells through scavenger receptors and is recognized by endosome-resident TLR-9, while non−self-RNA is recognized by TLR-7/8 or TLR-3, subsequently activating NOS2 and NOX (33–35).
We displayed mCpG, a ligand for murine TLR-9, on NOckout to give NOckoutmCpG. NOckoutmCpG is internalized by macrophages from the extracellular milieu by scavenger receptor-mediated endocytosis. Based on the mechanism of action of mCpG, endosomal NOckoutmCpG is expected to engage TLR-9, activate NOS2, and elevate NO in endosomes within 2 h of activation (Fig. 1A). We incubated NOckoutmCpG (500 nM) with primary mouse microglia for 90 min in the presence and absence of 1400W, and NOckout signals (G/R) of ∼100 individual endosomes from ∼50 cells were computed (Fig. 2 B and E). This revealed much higher levels of NO in endosomes and was comparable to the levels observed for LPS activation. The population of NO-rich endosomes was significantly enhanced in microglia treated with NOckoutmCpG (Fig. 2E). Next, the CpG motif in NOckoutCpG was substituted with a nonimmunostimulatory GpC motif that does not engage TLR-9, that is, a single base swap, to give NOckoutmGpC. This variant led to an overall reduction in NO-rich endosome population indicating that the mCpG moiety in NOckoutmCpG activates NOS2 (Fig. 2E). Next, colocalization studies were conducted to verify the identity of the compartments in J774A.1 cells containing NOckout. The time at which the steady-state NO measurements were performed was 90 min postpulsing. At 90 min, NOckout showed >60% colocalization with the late endosomal marker fluorescein isothiocyanate-labeled ovalbumin (SI Appendix, Fig. S15A). Immunofluorescence with anti−LAMP-1 showed that no lysosomes were labeled (SI Appendix, Fig. S15B). Thus, NOckout probes predominantly measure NO in LEs of J774A.1 cells.
Next, we quantified the immunogenicity of the various PAMPs alone and when they are displayed on NOckoutfn probes by detecting TNF-α levels arising from TLR stimulation. We treated J774A.1 cells with each PAMP motif, for example, mCpG, zCpG, RNA, or iRNA and their corresponding NOckout display variant and quantified TNF-α in the extracellular milieu by enzyme-linked immunosorbent assay (ELISA) (SI Appendix, Fig. S14 and Materials and Methods). Plain NOckout, devoid of any PAMP, did not generate TNF-α beyond basal levels in cell culture. This correlates well with our finding that NOckout alone does not activate NOS2. NOckoutmCpG and NOckoutiRNA treatment led to high TNF-α production. Importantly, these constructs were as immunogenic as the corresponding PAMP alone, indicating that PAMP display on the NOckout scaffold does not appreciably alter the immunogenicity of the PAMP. One base swap of CpG to GpC or single A to G mutation, that each converts the PAMPs to nonimmunogenic motifs in either NOckoutmGpC or NOckoutRNA, led to a dramatic reduction in TNF-α levels when the cells were treated with these probes. These supported our findings that the nonimmunogenic mGpC and RNA motifs alone were incompetent at TNF-α production (SI Appendix, Fig. S14). Importantly, NOckout probes do not stimulate their cognate TLR indefinitely. This is because, beyond 2 h in cell culture, the probe is degraded in lysosomes, likely by DNase II (36); NOckout probes therefore can reliably report on endosomal NO levels only up to 2 h postinternalization (SI Appendix, Fig. S12).
To study whether the NOckout scaffold could be more generally programmed to display specific PAMPs that could trigger their cognate PRRs, NOckout was designed to display an immunostimulatory RNA sequence that is a ligand for the murine TLR-13 receptor to give the variant NOckoutiRNA (25). TLR-13 was recently identified as the cognate receptor for a 13-base-long immunogenic RNA, denoted iRNA (Fig. 1B). The iRNA is derived from the domain V of ribosomal RNA, is conserved across several pathogenic Gram-negative bacteria, and effectively triggers NO production in innate immune cells (25, 26). NOckoutiRNA (500 nM) was incubated with J774A.1 cells for 90 min in the presence and absence of 1400W. NOckout signals (G/R value) of ∼200 endosomes from ∼50 cells were compared with the G/R value obtained for the plain NOckout probe (Fig. 2 C and F). NOckoutiRNA showed substantially higher endosomal NO levels that reduced to near-basal levels if the iRNA motif was dropped, or if NOS2 was inhibited (Fig. 2F). Further, a variant NOckoutRNA was made, where the PAMP displayed corresponds to a sequence where iRNA has a single A to G mutation that abolishes TLR-13 binding (Fig. 1B). This probe failed to elicit NOS2 activity in J774A.1 cells (Fig. 1F and SI Appendix, Fig. S18). We also obtained a similar result for these reagents in RAW264.7 macrophages that are a well-established model of TLR13 signaling (SI Appendix, Fig. S16) (26). This reveals that NOckoutiRNA can detect elevated NO in endosomes due to NOS2 activation. Thus, NOckout is an effective ratiometric NO detection platform to image NOS2 activity arising from PAMP−TLR engagement.
NOckoutUN Probes Localize in Phagosomes of Microglia in the Zebrafish Brain
Since NOckout probes can be rationally designed to trigger NOS2 activity through precise pathways in cultured cells, we applied it to study TLR signaling in vivo. Zebrafish is a powerful genetic model to dissect host−pathogen interactions, as one can image innate immune cells in live vertebrates with subcellular resolution (37, 38). Further, the innate immune signaling component can be specifically isolated in the larval stage due to the late onset of adaptive immunity in zebrafish (39). We therefore studied NOS2 activation in microglia of zebrafish by microinjecting PAMP-programmed NOckout probes into the optic tectum of larvae 3 d to 4 d postfertilization (dpf) (Fig. 3A and Movie S1).
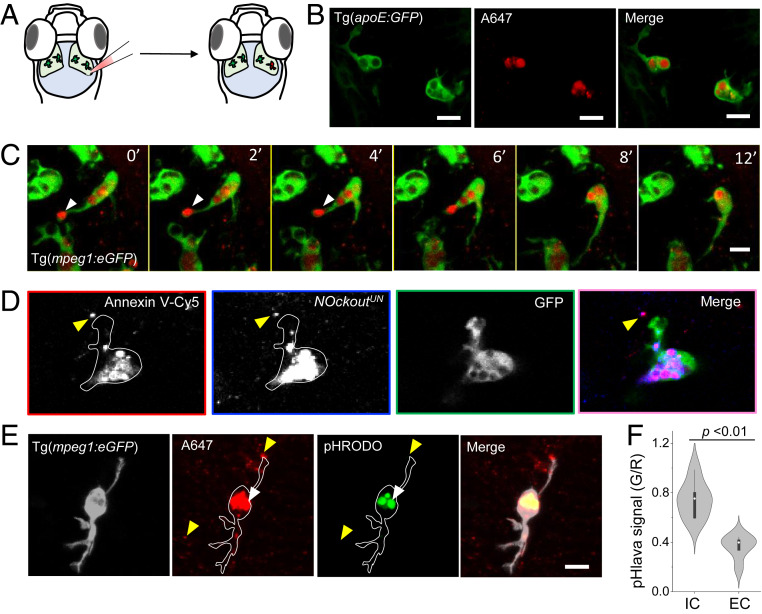
Fig. 3.
NOckoutUN localizes in phagosomes of zebrafish microglia in vivo. (A) Schematic of the larval transgenic zebrafish brain expressing GFP in microglia (Tg(apoE:GFP)). NOckoutUN (0.2 pmol in HBSS) was microinjected in the optic tectum of 3-dpf-old fish. (B) Images acquired 1 h postinjection of NOckoutUN (0.2 pmol in HBSS) shows probe localization in phagosomes of microglia in Tg(apoE:GFP) fish. (C) Time-lapse images (0 min to 12 min) shows NOckoutUN uptake by microglia. Fusion of newly formed phagosomes with existing phagosomes is also observed (white arrowhead). (D) Colocalization of NOckoutUN with apoptotic body marker annexin V−Cy5 prior to phagocytosis by microglia (yellow arrowhead). (E) Representative images of pHlava injected in Tg(mpeg1:eGFP) fish shows strong signal in phagosomes (white arrowhead) in the pHrodo channel (λem = 570 nm), while extracellular puncta show no signal (yellow arrowhead), indicating high and low acidities, respectively. (F) The pHlava signal from the phagosomes (IC) and the extracellular milieu (EC) are plotted as the ratio of pHrodo to A647 channel intensities (n = 16 phagosomes from n = 8 fish). All experiments were performed in triplicate. (Scale bar, 10 μm.) P values are obtained using Kruskal−Wallis statistical test.
First, we established the subcellular localization of extraneously introduced NOckout probes in the zebrafish brain. Since NOckout probes are essentially dsDNA bearing two fluorophores, we envisage their uptake in the mammalian brain would phenocopy that of fragmented self-DNA which would be internalized by scavenger receptors that are present on microglia (40, 41). We therefore injected NOckoutUN, a NOckout variant that has no DAR label nor PAMP, in the optic tectum of Tg(apoe:eGFP) larvae 3 dpf where microglia are marked with green fluorescent protein (GFP) (Fig. 3B and SI Appendix, Table S1). Within 30 min postinjection, NOckoutUN was internalized specifically by microglia. NOckoutUN was also internalized specifically by microglia when it was injected into the optic tectum of Tg(mpeg:eGFP) fish where all macrophages are marked with GFP. Live imaging revealed that several actively moving microglia rapidly phagocytosed particles of NOckoutUN, with many microglia performing multiple phagocytic events (Fig. 3C and Movies S2 and S3). The sizes of the intracellular compartments containing internalized NOckoutUN cargo were ∼2 μm to 4 μm, consistent with phagosomes rather than endosomes. Coinjection of tetramethylrhodamine (TMR)-labeled NOckoutUN with the apoptotic marker annexin V−Cy5 revealed substantial colocalization of NOckoutUN with apoptotic bodies prior to cellular uptake (Fig. 3D and SI Appendix, Fig. S19). Postuptake, NOckoutUN and annexin V−Cy5 extensively colocalized in phagosomes of microglia (Fig. 3D). This is consistent with other observations where extracellular DNA or extracellular RNA is integrated into apoptotic bodies to expedite their clearance by macrophages through phagocytosis (42, 43).
Since macrophage phagosomes have an acidic milieu, we first measured their luminal acidity using a DNA-based pH sensor denoted as pHlava (SI Appendix, Fig. S6). The pHlava comprises the pH-sensitive dye, pHrodo, attached to the 5′ end of the unlabeled strand in dsDNAA647. It ratiometrically reports pH, as protonation of pHrodo results in high fluorescence (G) while that of A647 is unchanged. The ratio of pHrodo to A647 (G/R) as a function of increasing acidity yielded a calibration curve (SI Appendix, Fig. S6). Injection of pHlava in Tg(mpeg:eGFP) larvae revealed a mean G/R ratio corresponding to a pH of 6.0 for the puncta inside microglia and near neutral for the extracellular puncta (Fig. 3 E and F). Thus, these large, acidic compartments in microglia labeled with pHlava are indeed phagosomes. Hence, extraneously introducing dsDNA probes in the optic tectum of zebrafish leads to their incorporation into apoptotic bodies followed by phagocytosis by microglia.
NOckout Detects NO in Microglial Phagosomes in Vivo
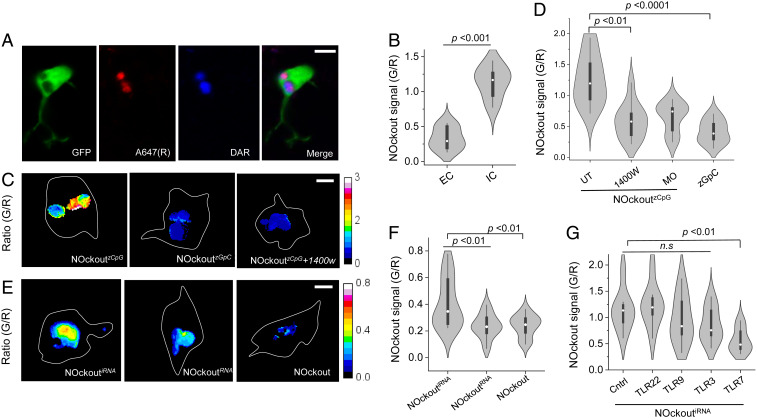
We then tested whether NOckout probes could detect NOS2 activity due to TLR receptor engagement in live brains by mapping the phagosomal NO generated in microglia. To achieve this, we made NOckoutzCpG which displayed the well-characterized CpG ODN, zCpG (Fig. 1B). The zCpG synergistically binds both TLR-9 and TLR-21 through a GTCGTT motif and triggers a strong immune response in zebrafish, producing IL-1, IFN-γ, and TNF-β (24). NOckoutzCpG was injected, internalized into phagosomes of microglia, and imaged as described earlier (Fig. 4A). Extracellular puncta of NOckoutzCpG showed a low NOckout signal (G/R) consistent with negligible NO, while NOckoutzCpG localized in phagosomes showed a high G/R ratio, consistent with high phagosomal NO (Fig. 4B). The G/R ratios of ∼20 such phagosomes were significantly higher than those labeled with plain NOckout, indicating effective NOS2 activation by NOckoutzCpG (Fig. 4 C and D). This was reduced by ∼50% when NOS2a, the NOS2 ortholog of zebrafish, was pharmacologically inhibited with 1400W or if the CpG motif in NOckoutzCpG was substituted with a nonimmunostimulatory GpC motif as seen in NOckoutzGpC (Fig. 4D). Morpholino knockdown of NOS2a also reduced the signal by ∼40% (Fig. 4D). Thus, the immunostimulatory sequences displayed on NOckout probes engage their cognate TLRs in the zebrafish brain and trigger NOS2 activity reflected in the elevated levels of phagosomal NO.
Fig. 4.
NOckoutzCpG and NOckoutiRNA elevate phagosomal NO in zebrafish microglia. (A) Representative three-color image showing phagosomal localization of NOckoutzCpG in microglia of 3-dpf Tg(mpeg1:eGFP) fish. A647 (R) is the reference fluorophore, and DAR signal (G) is shown in blue. (B) NOckoutzCpG signal from the EC and IC phagosomes are plotted as the ratio of DAR (G) to A647 channel intensities (n = 10 phagosomes from n = 5 fish). (C) Representative G/R heat maps of phagosomes in fish injected with NOckoutzCpG, NOckoutzGpC, and NOckoutzCpG + 1400W samples (0.2 pmol in HBSS). (D) Violin plot of G/R distribution in ∼10 individual phagosomes injected with NOckoutzCpG in different conditions (n = 5 fish). (E) Representative G/R heat maps of phagosomes labeled with NOckoutiRNA, NOckoutRNA, or NOckout (20 nL to 40 nL). (F) Violin plot of G/R distributions of ∼10 individual phagosomes (n = 5 fish). (G) G/R values of ∼10 phagosomes in morpholino knockdowns of the indicated TLR receptors (n = 5 fish per trial). All experiments were performed in triplicate. (Scale bar, 5 μm.) P values are obtained using Kruskal−Wallis statistical test across dataset.
Finally, in an illustrative example, we use NOckout technology to establish that pathogenic RNA of bacterial origin can act as PAMPs in zebrafish and thereafter identify its cognate receptor. Bacterial pathogens such as Mycobacterium marinum and Mycobacterim leprae in macrophages of zebrafish are effectively neutralized in the phagosome by NOS2 activation due to TLR engagement (44, 45). Ten out of the 20 putative TLR receptors in zebrafish have human orthologs, and the cognate ligands have been pinpointed for a few (SI Appendix, Table S2) (46). For example, TLR-3 and TLR-22 of zebrafish sense dsRNA and poly (I:C), while TLR-9 and TLR-21 sense CpG-ODNs (24, 47). However, it is not yet known whether ssRNA can trigger an immunogenic response in zebrafish.
Interestingly, the highly conserved iRNA sequence, derived from the ribosomal RNA of pathogens such as Staphylococcus aureus and Escherichia coli, engages murine TLR-13, elicits NO production, and is now a recognized PAMP in mice (25). This RNA sequence is also conserved in many natural aquatic pathogens that infect zebrafish, such as Edwardsiella tarda (48), Aeromonas hydrophila (49), and Francisella philomiragia (50) (SI Appendix, Fig. S21 and Table S3). We therefore sought to determine whether iRNA could act as a PAMP in zebrafish as well, and, if so, whether we could identify its cognate TLR using NOckout technology.
We displayed the iRNA sequence on NOckout to give NOckoutiRNA and tested its capacity to generate phagosomal NO. Postinjection of NOckoutiRNA in the brain, it was efficiently phagocytosed by microglia. The G/R ratios of ∼20 phagosomes revealed appreciable levels of NO compared to phagosomes labeled with plain NOckout (Fig. 4 E and F). Further, a single A to G mutation in the iRNA sequence that renders it nonimmunostimulatory in mice was used to create NOckoutRNA. Phagosomes labeled with NOckoutRNA in zebrafish brains revealed a significant drop in NO levels, revealing that the mutated RNA could not activate NOS2 (Fig. 4 E and F). This suggests that ssRNA can also act as a PAMP in zebrafish, and indicates a potentially evolutionarily conserved mechanism to detect ssRNAs (51). We therefore reasoned that the PRR responsible was also likely to be a TLR, given that the cognate PRR in mice is TLR-13. To identify the TLR responsible, we repeated the assay in zebrafish, where specific TLRs were knocked down. When we knocked down TLR-22, TLR-3, or TLR-9, we continued to observe high phagosomal NO corresponding to high NOS2 activity, suggesting that these are not the cognate PRRs (Fig. 4G). However, when we knocked down TLR-7 (SI Appendix, Fig. S22B), the phagosomes showed ∼40% signal reduction (Fig. 4G). This indicates that iRNA activates NOS2 by partly engaging zebrafish TLR-7.
In mammals, TLR-7 is an endosomal or phagosomal PRR that is known to engage ssRNA of viral or bacterial origin (52–54) and shows ∼56% sequence identity with zebrafish TLR-7. While the structure of mammalian TLR-7 is solved, that of zebrafish TLR-7 is not. A homology model of the predicted structure of zebrafish TLR-7 and macaque TLR-7 complexed with ssRNA reveals marked structural similarity (SI Appendix, Fig. S23) (55). Importantly, zebrafish TLR-7 showed conserved amino acid residues within its predicted ssRNA binding pocket (SI Appendix, Fig. S24). Further, zebrafish TLR-7 is highly expressed in 3-dpf larval zebrafish (SI Appendix, Fig. S22). We therefore suggest that selected ssRNAs act as PAMPs in zebrafish by engaging TLR-7 and activating NOS2a.
Conclusions
Small-molecule NO reporters are bright, photostable, specific, and pH insensitive; however, postreaction, the probe molecules diffuse rapidly, obscuring spatial information. Protein-based NO sensors offer spatial resolution, but have comparatively poor dynamic range, cannot be targeted to phagosomes, and are pH sensitive. NOckout technology combines the attractive reporter characteristics of small-molecule NO-sensitive dyes with the stable localization provided by biologics. NOckout probes leverage the 1:1 stoichiometry of DNA hybridization to display an NO-sensitive fluorophore, an internal reference dye, and a dsDNA domain to target the phagosome. Microinjecting NOckout nanodevices in live brains resulted in them being packaged into apoptotic bodies and being exclusively targeted to phagosomes in microglia. Since NOckout nanodevices are pH insensitive, they are well suited to map NO in the acidic phagosomal milieu.
Importantly, NOckout nanodevices can be programmed to display a specific nucleic acid PAMP with precise and uniform stoichiometries to give NOckoutfn devices that engage their cognate TLR receptor to thereby activate NOS2. This leads to phagosomal NO production that is detected by the NOckout probe. This modular design enables the creation of a suite of NOckout variants, each displaying a different PAMP, yet with identical reporting capabilities that can each engage a distinct TLR receptor. By displaying well-characterized immunogenic sequences such as mCpG or zCpG, we could activate murine TLR-9 and zebrafish TLR-9/TLR-21, respectively. We found that, while the basic NOckout scaffold was nonimmunogenic, NOckoutfn devices could activate NOS2 by engaging a specific TLR in primary microglia either in culture or in live zebrafish brains. NOS2 activation due to PAMP−TLR receptor recognition is well studied on longer time scales in cultured murine and human cells using biochemical assays. Now, using NOckout, one can map NO arising from NOS2 activity at early stages (<2 h) in single live cells in culture and in live brains, which was not previously possible.
NOckout can be used to map NO arising from NOS2 activation in the first few hours following PAMP−PRR association in vivo. It can be used to assay PAMP−TLR recognition, identify or validate ligands for PRRs with undetermined specificities, and potentially estimate their relative proclivities to activate NOS2. The DNA scaffold can be modified to display more than one PAMP in precise stoichiometries using three-way or four-way junctions, and the resultant combination NOckout devices can be applied to identify TLRs that could act synergistically or antagonistically. One can also envisage substituting NO detection chemistries for various ROS detection chemistries to map the in vivo activity of phagosomal ROS-producing enzymes arising from TLR activation. A technology that can map the dynamics of NO within phagosomes of innate immune cells in vivo could thereby offer new insights into the dynamics of host−pathogen interactions. Since resistant microbes use several mechanisms to bypass phagosomal degradation, NO mapping could help identify how resistant pathogens survive the phagosome.
NOckout reporters are applicable without further modification to transparent model organisms such as Caenorhabditis elegans and Drosophila melanogaster. One can also envisage NOckout being deployed in certain regions of mouse brain, for example, cerebral cortex. One caveat of NOckout technology is that, currently, it can be used to detect NO produced due to preexisting and actively translated NOS2 and is not suitable for sustained detection NO over transcriptional time scales. This limitation can be overcome by replacing dsDNA backbone by a peptide nucleic acid or L-DNA backbone to improve its cellular stability. This highly modular imaging platform can be applied to study the early stages of the innate immune response at very high chemical resolution.
Materials and Methods
Reagents.
All functionalized DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT) as a lyophilized powder. Oligonucleotides were dissolved in milli-Q water and quantified using an ultraviolet visible spectrophotometer (Shimadzu UV-3600, Japan) and subsequently stored in a refrigerator at −20 °C. Immunostimulatory ATTO647N-labeled DNA and RNA containing phosphorothioate backbone were custom synthesized by IDT. All of the chemicals used for the synthesis of DAR-N3 were purchased from Acros Organics and Sigma. Nitric oxide donor (DEANONOate), iNOS inhibitor (1400W), NADPH-oxidase inhibitor (VAS2870), MPO inhibitor (ABAH), and vacuolar H+-ATPase (V-ATPase) inhibitor (Bafilomycin A1) were purchased from Cayman Chemicals. Diamino polyethylene glycol linker (10 kDa) was purchased from Creative PEGWorks. Annexin V−Cy5 apoptosis kit was purchased from BioVision (catalog number K103), and pHrodo maleimide was purchased from Thermo Fisher Scientific (catalog number P35371). The iNOS antibody was obtained from Novus Biologicals (NB300-605). LAMP1 antibody was obtained from Abcam (ab62562). Morpholino sequences were purchased from Gene Tools. DAR-N3 was synthesized using previously reported procedure by Jani et al. (20).
NOckout Probe Synthesis.
NOckout probe was assembled by using a modified protocol reported by Jani et al. (20). The detailed description of the synthesis of NOckout and NOckoutFn probe is given in SI Appendix. The in vitro and in cellulo specificity and stability of NOckout against different ROS and RNS were investigated in detail, and the results are presented in SI Appendix.
Gel Electrophoresis.
Native polyacrylamide gels containing 15% acrylamide (30:1 acrylamide/bisacrylamide) were used for the gel electrophoresis assays. Gels were run in 1× Tris-borate-EDTA (TBE) buffer (90 mM Tris.HCl, 90 mM boric acid, and 2 mM ethylenediaminetetraacetic acid (EDTA), pH 8.3) at room temperature. Postrun, gels were first imaged with ChemiDoc MP imaging system (Bio-Rad) to visualize DAR- or pHrodo-conjugated (605/55 filter, green epi-illumination) and ATTO647N-conjugated (695/55 filter, red epi-illumination) DNA oligonucleotides. Ethidium bromide (1 μg/mL) was used to stain DNA duplex, and it was imaged with a GelDoc-It imaging system (UVP, λex = 302 nm).
In Vitro Fluorescence Measurements.
All fluorescence studies were carried out on a Fluoromax-4 (Horiba Scientific) spectrophotometer. Ten micromolar stock of NOckout or NOckoutFn sensors were diluted to a 100- to 250-nM final concentration in 50 mM sodium phosphate buffer, pH 5.0, 6.0, or 7.2. The emission spectra of DAR and A647N were acquired by exciting the sample at 550 nm and 650 nm, respectively. Fluorescence emission spectra were collected in the range of 560 nm to 620 nm (slit width = 2 nm) for DAR and 655 nm to 700 nm (slit width = 2 nm) for A647N. Diethylamine NONOate (DEA NONOate, Cayman Chemicals) was used as fast NO donor. The half-life of DEA NONOate is 2 min and 16 min at 37 °C and 25 °C, respectively, in 100 mM phosphate buffer (pH 7.4). DEA NONOate liberates 1.5 mol of NO per mol of parent compound (56). Fluorescence signal from NOckout (100 nM) was recorded sequentially in the DAR (G) and A647N (R) channels before and after the addition of DEANONOate (50 μM) at timed intervals (30 s). Emission intensity maximum of DAR was at 571 nm before NO˙ addition, and it shifted to 578 nm after NO addition. In vitro fold changes of all of the NOckout sensors were calculated and expressed as the ratio of DAR signal to that of the A647N signal, which is represented as a G/R value. In vitro pH sensitivity studies of NOckout were carried out on Fluoromax-4, and the detailed description is given in SI Appendix.
Synthesis of pHlava.
The pH sensor pHlava is a 24-mer dsDNA duplex carrying a pH-sensing fluorophore pHrodo (λex = 540 nm, λem = 580 nm) at the 5′ end of one of the ssDNA strands and ATTO647N fluorophore on the 5′ end of the complementary ssDNA (SI Appendix, Table S1). The synthesis, characterization, and pH-sensing properties of pHlava are described in SI Appendix.
Image Acquisition.
Wide-field microscopy was carried out on an IX83 inverted microscope (Olympus Corporation of the Americas) using either a 100× or 60×, 1.4 numerical aperture (NA), differential interference contrast (DIC) oil immersion objective (PLAPON, Olympus) equipped with an Evolve Delta 512 electron multiplying charge coupled device (EMCCD) camera (Photometrics). Filter wheel, shutter, and CCD camera were controlled using MetaMorph software (Molecular Devices). For NOckout stability assay, J774A.1 cells were excited simultaneously in the DAR (546 nm) and A647N (650 nm) channels. Confocal images from J774A.1 cells and primary microglia were captured with a Leica TCS SP5 II stimulated emission depletion (STED) laser scanning confocal microscope (Leica Microsystems, Inc.) equipped with a 63×, 1.4 NA, oil immersion objective. DAR was excited using a diode-pumped solid-state laser with 561-nm wavelength. A647N was excited using a He−Ne laser with 633-nm wavelength. Acousto-optical beam splitter (AOBS) with settings suitable for each fluorophore was employed and fluorescence emission was recorded using hybrid detectors.
Zebrafish brain was imaged in the optic tectum using an upright Zeiss LSM-710 confocal microscope equipped with water immersion objectives (20×, or 40×). Zebrafish larvae (3 dpf to 4 dpf) were mounted dorsally in a 1.5% low-melting-point agar bed on a confocal imaging dish (3.5-cm glass bottom). E3-containing 0.003% MS-222 was present in the imaging dish throughout the imaging section. For NOckout-injected samples, images were acquired simultaneously in GFP, DAR, and A647N channels using excitation wavelengths of 488, 560, and 645 nm from Argon-ion (488 nm), Helium-Neon (543 and 632 nm) laser sources, respectively. The pHrodo dye was imaged using the same settings as that of DAR.
Image Analysis.
We have used a custom-made MATLAB-based program called Findosome to calculate the (G/R) values of individual endosomes from the acquired images. Maximum Z-projected images created using Fiji (NIH) were fed to Findosome to pick endosomal compartments as regions of interest (ROIs) corresponding to individual endosomes based on the A647N channel intensity. For each ROI, the program calculates fluorescence intensities both in the normalizing channel (A647, R) and in the DAR (G) channel.
The results were exported as an Excel file. From the results, (G/R) value for individual endosomes can be calculated. Phagosomal (G/R) values from the zebrafish microglia are calculated using Fiji software. Images were imported to Fiji, and maximum Z-projected images were created. ROIs were manually drawn around clearly identifiable phagosomes (>200 μm from the injection site) using the normalizing channel A647N intensity. Fluorescence intensities in A647N and DAR channels were calculated. G/R values are either represented as heat map images using Fiji or as bar graphs using Origin software. Intracellular measurements of (G/R) values were performed in GFP colocalized microglia. For (G/R) ratio quantification experiments (e.g., Fig. 2A), we only consider those vesicles that are present in both green (G) and red (R) channels. We do this because the cell has ∼2 to 5% of vesicles that are highly autofluorescent in the green channel. We can estimate this percentage by labeling cells with a dsDNA carrying only an Atto647N label. Autofluorescent vesicles can be identified as puncta in the green channel (in such samples) that do not show up in the red channel and typically correspond to <5%. Therefore, we first have to identify and remove the autofluorescent vesicles during our analysis.
Cell Culture Protocol.
J774A.1 and RAW264.7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, 11960044) with 10% fetal bovine serum (Gibco, 26140079) containing 100 U/mL penicillin and 100 µg/mL streptomycin and maintained at 37 °C under 5% CO2. For all of the cell experiments, a passage number less than 30 was employed.
Neonatal Primary Microglia Isolation.
Mouse primary mixed cortical and hippocampal glial culture was isolated and cultured as described previously (57). The detailed description of the protocol and images showing the purity of the microglial culture obtained are given in SI Appendix.
Immunocytochemistry.
Primary microglia cells were treated with 1 μM NOckoutFn probe for 3 h. Cells were then washed three times with 1× phosphate-buffered saline (PBS) (pH 7.4) and fixed using 2.5% paraformaldehyde at room temperature for 15 min. The cells were subsequently washed three times with 1× PBS and followed by incubation with 3% bovine serum albumin (BSA) for blocking in PBS (1×) for 1 h at room temperature. Then the cells were incubated with anti-mouse iNOS antibody (Novus Biologicals, NB300-605, 1:30 dilution) in blocking solution (3% BSA in PBS) for 1 h at room temperature. To remove excess primary antibody, cells were washed three times with 1× PBS. Cells were then treated with Alexa Fluor 488 conjugated goat anti-rabbit secondary antibody (Invitrogen, a11008, 1:2,000 dilution) for 1 h followed by three washes with 1× PBS. Cells were incubated with Hoechst 33342 (5 μM) for 10 min to stain the nucleus. Cells were imaged on a confocal microscope as described elsewhere. LPS (1 μg/mL)-treated J774A.1 cells (for 12 h) were immunostained for iNOS and LAMP-1 using a similar protocol as described above for the microglial cells.
Pharmacological Inhibition.
Pharmacological inhibition of different enzymes was performed by employing known inhibitors. The iNOS inhibition was achieved using a specific and irreversible inhibitor, 1400W (10 μM) (Ki value against iNOS is 7 nM) (31). ROS produced in J774A.1 cells by NADPH-oxidase was inhibited by using a selective inhibitor VAS2870 (10 μM) (58). MPO activity to produce HOCl was blocked by using ABAH (50 μM) (59). V-ATPases were blocked by using a selective and reversible inhibitor Bafilomycin A1 (300 nM). Cells were bathed in inhibitors 1 h prior to the addition of NOckout probes during the specificity experiment performed in J774A.1 cells.
TNF-α Quantification.
To quantify the immunogenicity of NOckout probes and the corresponding immunogen alone, we performed ELISA for TNF-α in J774A.1 cells. Briefly, J774A.1 cells cultured in DMEM were incubated with 500 nM NOckout sensors, CpG nucleotides, RNA, or LPS at 37 °C for 2 h. Postincubation, 100 μL of extracellular medium from the culture was used to perform TNF-α quantification using ELISA kit (Cayman, catalog number 500850) according to manufacturer’s instructions. The calibration curve for TNF-α was generated using recombinant TNF-α as per the vendor’s instructions. The results are summarized in SI Appendix.
Zebrafish Breeding and Maintenance.
Zebrafish (Danio rerio) were maintained as described (60). Zebrafish were kept at 26 °C to 27 °C in a 14-h-light and 10-h-dark cycle. Embryos were collected by natural spawning and raised at 28 °C in E3 buffer. To avoid pigmentation, 0.003% 1-phenyl-2-thiourea was added at 1 dpf. All of the transgenic lines used in this study, Tg(mpeg:EGFP), Tg(apoe:EGFP), and Tg(HuC:Kaede), have been described previously (38, 61, 62). Tg(apoe:EGFP) fish was a kind gift from Prof. William Talbot’s laboratory, Stanford University, Stanford, CA. For microinjection of NOckout probes, embryos were obtained from wild-type AB, Tg(mpeg: EGFP), Tg(apoe:EGFP), and Tg(HuC:Kaede). Animals were housed in the fish facility according to the Institutional Animal Care and Use Committee regulation of University of Chicago (Protocol number 72468).
Microinjection in Larval Zebrafish.
About 20 nL to 30 nL of NOckout or NOckoutFn (20 μM stock solution in Hanks’ balanced salt solution [HBSS]) were injected to the optic tectum area of MS-222−treated 3-dpf zebrafish using a microinjector. Postinjection of the probes, fish were allowed to recover from anesthesia by placing them in fresh LPS-free E3 medium. Subsequently, fish were reanesthetized and embedded in agar for confocal imaging. Coinjection experiments of TMR-labeled NOckoutUN and Cy5−annexin V were performed by employing a similar protocol to that used for the NOckout injections. Briefly, dsDNATMR (20 μM in HBSS) and annexinV−Cy5 (10 μM) were mixed in a microfuge tube by vertexing. About 20 nL of the mixed solution was injected in the optic tectum of 3-dpf-old fish. Injected fish were allowed to recover from anesthesia by placing them in a fresh E3 medium, and the fish were further incubated for 1 h in E3 medium before imaging their brains using a confocal microscope (LSM-710) in the TMR and Cy5 channels. Morpholinos (Gene Tools) were dissolved in HBSS and injected at one cell stage. Morpholino sequences and primers used for RT-PCR are listed in SI Appendix.
RT-PCR.
Total RNAs were extracted from embryos and larvae using TRIzol reagent (Invitrogen) as per the manufacturer’s instructions. The cDNAs were then generated using SuperScript III reverse transcriptase. The cDNAs were used to generate gene-specific amplicon using primers listed in SI Appendix. Reverse transcription was performed at 50 °C for 1 h. PCR conditions were 2 min of initial denaturation at 94 °C, 45 cycles of denaturation at 94 °C for 20 s, annealing at 52 °C to 58 °C for 45 s, and extension at 72 °C for 1 min, followed by a final extension step at 72 °C for 10 min (63).
Data Availability.
All data relevant to this paper are available in the main text and SI Appendix. Full methods can be found in Movies S1–S3 and SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Victoria Prince and Noor Singh for help with zebrafish protocols. We thank Prof. William S. Talbot for the kind gift of Tg(ApoE-eGFP) fish. This work was supported by the Brain Research Foundation Scientific Innovation Award, University of Chicago Women’s Board, Mergel Funsky award, a pilot and feasibility award from National Institute of Diabetes and Digestive and Kidney Diseases Centre Grant P30DK42086 to the University of Chicago Digestive Diseases Research Core Centre, Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust Grant C-084 and University of Chicago start-up funds to Y.K., and Office of Naval Research, Department of Defense Grant N00014-18-126 to M.E.H. S.S.S. thanks Cure Alzheimer’s Fund for the support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003034117/-/DCSupplemental.
References
- 1.Stuart L. M., Ezekowitz R. A. B., Phagocytosis: Elegant complexity. Immunity 22, 539–550 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Underhill D. M., Ozinsky A., Phagocytosis of microbes: Complexity in action. Annu. Rev. Immunol. 20, 825–852 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Bedard K., Krause K.-H., The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 87, 245–313 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Knowles R. G., Moncada S., Nitric oxide synthases in mammals. Biochem. J. 298, 249–258 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuman E. M., Madison D. V., Nitric oxide and synaptic function. Annu. Rev. Neurosci. 17, 153–183 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Förstermann U., Sessa W. C., Nitric oxide synthases: Regulation and function. Eur. Heart J. 33, 829–837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wink D. A. et al., Nitric oxide and redox mechanisms in the immune response. J. Leukoc. Biol. 89, 873–891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi O., Akira S., Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Mogensen T. H., Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West A. P., Koblansky A. A., Ghosh S., Recognition and signaling by Toll-like receptors. Annu. Rev. Cell Dev. Biol. 22, 409–437 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Anes E. et al., Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell. Microbiol. 8, 939–960 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Gao J. J. et al., Cutting edge: Bacterial DNA and LPS act in synergy in inducing nitric oxide production in RAW 264.7 macrophages. J. Immunol. 163, 4095–4099 (1999). [PubMed] [Google Scholar]
- 13.Utaisincharoen P., Anuntagool N., Chaisuriya P., Pichyangkul S., Sirisinha S., CpG ODN activates NO and iNOS production in mouse macrophage cell line (RAW 264.7). Clin. Exp. Immunol. 128, 467–473 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giustarini D., Rossi R., Milzani A., Dalle‐Donne I., “Nitrite and nitrate measurement by Griess reagent in human plasma: Evaluation of interferences and standardization” Cadenas E., Packer L., Eds. (Methods in Enzymology, Elsevier, 2008), Vol. 440, pp. 361–380. [DOI] [PubMed] [Google Scholar]
- 15.Kojima H. et al., Detection and imaging of nitric oxide with novel fluorescent indicators: Diaminofluoresceins. Anal. Chem. 70, 2446–2453 (1998). [DOI] [PubMed] [Google Scholar]
- 16.McQuade L. E., Lippard S. J., Fluorescent probes to investigate nitric oxide and other reactive nitrogen species in biology (truncated form: Fluorescent probes of reactive nitrogen species). Curr. Opin. Chem. Biol. 14, 43–49 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Yu H., Xiao Y., Jin L., A lysosome-targetable and two-photon fluorescent probe for monitoring endogenous and exogenous nitric oxide in living cells. J. Am. Chem. Soc. 134, 17486–17489 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Pisano J. M., Firestone R. A., Lysosomotropic agents III. Synthesis of N-retinyl morpholine. Synth. Commun. 11, 375–378 (1981). [Google Scholar]
- 19.Eroglu E. et al., Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat. Commun. 7, 10623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jani M. S., Zou J., Veetil A. T., Krishnan Y., A DNA-based fluorescent probe maps NOS3 activity with subcellular spatial resolution. Nat. Chem. Biol. 16, 660–666 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Kojima H. et al., Bioimaging of nitric oxide with fluorescent indicators based on the rhodamine chromophore. Anal. Chem. 73, 1967–1973 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Veetil A. T. et al., Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol. 12, 1183–1189 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Chakraborty K., Veetil A. T., Jaffrey S. R., Krishnan Y., Nucleic acid-based nanodevices in biological imaging. Annu. Rev. Biochem. 85, 349–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh D.-W. et al., Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 110, 20711–20716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldenburg M. et al., TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337, 1111–1115 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Li X.-D., Chen Z. J., Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife 1, e00102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anrather J., Racchumi G., Iadecola C., NF-kappaB regulates phagocytic NADPH oxidase by inducing the expression of gp91phox. J. Biol. Chem. 281, 5657–5667 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Berlato C. et al., Involvement of suppressor of cytokine signaling-3 as a mediator of the inhibitory effects of IL-10 on lipopolysaccharide-induced macrophage activation. J. Immunol. 168, 6404–6411 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Wind S. et al., Comparative pharmacology of chemically distinct NADPH oxidase inhibitors. Br. J. Pharmacol. 161, 885–898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettle A. J., Gedye C. A., Winterbourn C. C., Mechanism of inactivation of myeloperoxidase by 4-aminobenzoic acid hydrazide. Biochem. J. 321, 503–508 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garvey E. P. et al., 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J. Biol. Chem. 272, 4959–4963 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Lee B. L., Barton G. M., Trafficking of endosomal Toll-like receptors. Trends Cell Biol. 24, 360–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalpke A. H. et al., Immunostimulatory CpG-DNA activates murine microglia. J. Immunol. 168, 4854–4863 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Iliev A. I., Stringaris A. K., Nau R., Neumann H., Neuronal injury mediated via stimulation of microglial toll-like receptor-9 (TLR9). FASEB J. 18, 412–414 (2004). [DOI] [PubMed] [Google Scholar]
- 35.To E. E. et al., Endosomal NOX2 oxidase exacerbates virus pathogenicity and is a target for antiviral therapy. Nat. Commun. 8, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash V. et al., Quantitative mapping of endosomal DNA processing by single molecule counting. Angew. Chem. Int. Ed. Engl. 58, 3073–3076 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Meijer A. H., Spaink H. P., Host-pathogen interactions made transparent with the zebrafish model. Curr. Drug Targets 12, 1000–1017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peri F., Nüsslein-Volhard C., Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133, 916–927 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Renshaw S. A., Trede N. S., A model 450 million years in the making: Zebrafish and vertebrate immunity. Dis. Model. Mech. 5, 38–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egensperger R., Maslim J., Bisti S., Holländer H., Stone J., Fate of DNA from retinal cells dying during development: Uptake by microglia and macroglia (Müller cells). Brain Res. Dev. Brain Res. 97, 1–8 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Li Y. et al., Microglial activation by uptake of fDNA via a scavenger receptor. J. Neuroimmunol. 147, 50–55 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sierra A. et al., Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazaheri F. et al., Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 5, 4046 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Madigan C. A. et al., A macrophage response to Mycobacterium leprae phenolic glycolipid initiates nerve damage in leprosy. Cell 170, 973–985.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cambier C. J. et al., Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Li Y., Cao X., Jin X., Jin T., Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell. Mol. Immunol. 14, 80–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuo A. et al., Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 181, 3474–3485 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Ishibe K. et al., Comparative analysis of the production of nitric oxide (NO) and tumor necrosis factor-alpha (TNF-alpha) from macrophages exposed to high virulent and low virulent strains of Edwardsiella tarda. Fish Shellfish Immunol. 27, 386–389 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez I., Novoa B., Figueras A., Immune response of zebrafish (Danio rerio) against a newly isolated bacterial pathogen Aeromonas hydrophila. Fish Shellfish Immunol. 25, 239–249 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Brudal E. et al., Establishment of three Francisella infections in zebrafish embryos at different temperatures. Infect. Immun. 82, 2180–2194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen S., Thomsen A. R., Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86, 2900–2910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C., Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Mancuso G. et al., Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat. Immunol. 10, 587–594 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Nishiya T., Kajita E., Miwa S., Defranco A. L., TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J. Biol. Chem. 280, 37107–37117 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z. et al., Structural analysis reveals that toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity 45, 737–748 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Maragos C. M. et al., Complexes of ⋅NO with nucleophiles as agents for the controlled biological release of nitric oxide. Vasorelaxant effects. J. Med. Chem. 34, 3242–3247 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Minter M. R. et al., Deletion of the type-1 interferon receptor in APPSWE/PS1ΔE9 mice preserves cognitive function and alters glial phenotype. Acta Neuropathol. Commun. 4, 72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.ten Freyhaus H. et al., Novel Nox inhibitor VAS2870 attenuates PDGF-dependent smooth muscle cell chemotaxis, but not proliferation. Cardiovasc. Res. 71, 331–341 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Kettle A. J., Gedye C. A., Hampton M. B., Winterbourn C. C., Inhibition of myeloperoxidase by benzoic acid hydrazides. Biochem. J. 308, 559–563 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westerfield M., The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed., (University of Oregon Press, Eugene, OR, 2000). [Google Scholar]
- 61.Ellett F., Pase L., Hayman J. W., Andrianopoulos A., Lieschke G. J., mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato T., Takahoko M., Okamoto H., HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis 44, 136–142 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Meijer A. H. et al., Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 40, 773–783 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to this paper are available in the main text and SI Appendix. Full methods can be found in Movies S1–S3 and SI Appendix, Materials and Methods.