Significance
Hematology analysis is an integral part of medicine but requires complex systems, expensive chemical reagents, time-consuming protocols, and highly trained personnel for operation. This paper presents a label-free optical assay based on molecular imaging with deep-ultraviolet microscopy that enables major advancements in hematological analysis. We show that our approach circumvents significant limitations of the current standard of care while delivering equal diagnostic power for peripheral blood and bone marrow analysis. Because our technology achieves fast, reliable, quantitative, and simple blood cell analysis in a low-cost configuration, it can simplify and improve current clinical hematological analysis. This has significant implications toward the development of a hematological analyzer with improved capabilities for clinical use, as a point-of-care device, and for low-resource settings.
Keywords: deep-UV microscopy, molecular imaging, hematology analysis, point-of-care diagnosis, label-free cell classification
Abstract
Hematological analysis, via a complete blood count (CBC) and microscopy, is critical for screening, diagnosing, and monitoring blood conditions and diseases but requires complex equipment, multiple chemical reagents, laborious system calibration and procedures, and highly trained personnel for operation. Here we introduce a hematological assay based on label-free molecular imaging with deep-ultraviolet microscopy that can provide fast quantitative information of key hematological parameters to facilitate and improve hematological analysis. We demonstrate that this label-free approach yields 1) a quantitative five-part white blood cell differential, 2) quantitative red blood cell and hemoglobin characterization, 3) clear identification of platelets, and 4) detailed subcellular morphology. Analysis of tens of thousands of live cells is achieved in minutes without any sample preparation. Finally, we introduce a pseudocolorization scheme that accurately recapitulates the appearance of cells under conventional staining protocols for microscopic analysis of blood smears and bone marrow aspirates. Diagnostic efficacy is evaluated by a panel of hematologists performing a blind analysis of blood smears from healthy donors and thrombocytopenic and sickle cell disease patients. This work has significant implications toward simplifying and improving CBC and blood smear analysis, which is currently performed manually via bright-field microscopy, and toward the development of a low-cost, easy-to-use, and fast hematological analyzer as a point-of-care device and for low-resource settings.
Hematological analysis relies on assessing alterations in blood cell morphology, blood cell enumeration, and molecular or cytogenetic properties to screen, diagnose, and monitor blood conditions and diseases, including allergic and inflammatory reactions, infections (1, 2), sepsis (3), immune system disorders (4), several types of cancers (5), and more (6–10). To this end, peripheral blood is collected and analyzed using a hematology analyzer to obtain a complete blood count (CBC). Modern hematology analyzers use a combination of techniques, including absorption spectroscopy, impedance measurement, and flow cytometry, to measure red blood cell (RBC) and platelet counts, hemoglobin (Hb) levels, and white blood cell (WBC) differentials (11) (i.e., neutrophil, eosinophil, basophil, lymphocyte, and monocyte counts). Although hematology analyzers are capable of rapid staining and automated analysis (12), they are costly and require multiple chemical reagents and regular calibration and maintenance. Thus, despite being one of the most common medical tests (13), CBC has to be performed by highly trained technicians at clinics or commercial laboratories that have the necessary equipment and infrastructure in place, which has the adverse effect of delaying critical (in some cases, life-saving) health information.
Moreover, for pathological samples or samples that produce atypical results, manual microscopic examination is often required to confirm CBC findings and to evaluate the morphology of blood cells (11). Microscopic analysis of peripheral blood is performed by first generating a blood smear and then fixing and staining the samples, typically using Romanowsky-type stains (including Giemsa), which are generally composed of a basic dye (e.g., methylene blue) to stain the nuclei and an acidic dye (e.g., eosin) used as a counterstain (11). Finally, accurate evaluation of peripheral blood smears requires well-stained samples, which are highly susceptible to variability, and microscopic analysis performed by trained personnel. Nearly identical procedures are used to evaluate bone marrow aspirates (14).
A number of methods have been explored to facilitate and improve hematological analysis, mostly aimed at developing point-of-care CBC devices and/or providing label-free quantitative analysis. Fluorescence-based methods, for example, have been applied for rapid and low-cost WBC detection/counting via image analysis (15–17) or flow cytometry (18, 19). While promising, fluorescence methods suffer from phototoxicity and photobleaching, which hinders quantification (20). Plus, the degree of cell differentiation comes at the expense of additional or more complex staining protocols and equipment. To address this issue, several label-free modalities have been applied, including Raman microscopy (21), hyperspectral imaging (22), fluorescence lifetime imaging microscopy (23), and quantitative phase imaging (20, 24–27). These methods present their own unique advantages and disadvantages; however, a trade-off still exists between the complexity, cost, or speed of the approach vs. the information content provided. Finally, the application of these technologies to other hematological practices, including bone marrow aspiration (14, 28), has not been explored.
Our approach to hematological analysis makes use of the deep-ultraviolet (UV) region of the spectrum (i.e., 200 to 400 nm), which has recently reemerged as a powerful tool for label-free, quantitative, molecular imaging (29–31). Deep-UV microscopy offers many advantages over traditional methods, such as higher spatial resolution due to the shorter wavelength of UV light and the ability to easily and quickly access quantitative information from many endogenous biomolecules that play an important role in cell structure and function (31, 32). Label-free imaging of live cells has been achieved using deep-UV microscopy over extended periods of time (∼6 h) without inducing apoptosis (29, 33). In addition, highly specific cell phenotyping can be realized using the information-rich UV images from which key intracellular architectural and biochemical features are readily available (29, 30, 34, 35).
In this work, we demonstrate a facile label-free analysis of live blood cells using a multispectral deep-UV microscope. Our approach provides quantitative endogenous molecular information from live cells and enables analysis and differentiation of blood cell types based on their molecular and structural signatures. We demonstrate the unique capabilities of our method by achieving a five-part differential WBC classification, as well as providing clear visualization of platelets, and normal, nucleated, and sickled RBCs. Quantitative Hb characterization is also achieved. In addition, we demonstrate a pseudocolorization scheme which accurately mimics the colors produced by standard Giemsa staining to enable fast, fixative-free, and label-free visual examination of blood smears and bone marrow aspirate slides. We further validate the clinical diagnostic utility of our wide-field pseudocolored UV images by performing a blind analysis, administered by a panel of hematologists, of samples collected from healthy donors and patients with thrombocytopenia and sickle cell disease (SCD). Ultimately, we show this UV-based assay enables fast, label-free, and stain-free hematological analysis without sacrificing diagnostic quality compared to the standard of care. The assay can be embodied in a low-cost, easy-to-use, and compact configuration. Thus, this approach has potential to simplify, improve, and advance CBC and blood smear analysis, and thus improve patient care.
Results
Deep-UV Microscopy of Live, Unlabeled Blood Cells from Whole Blood Samples.
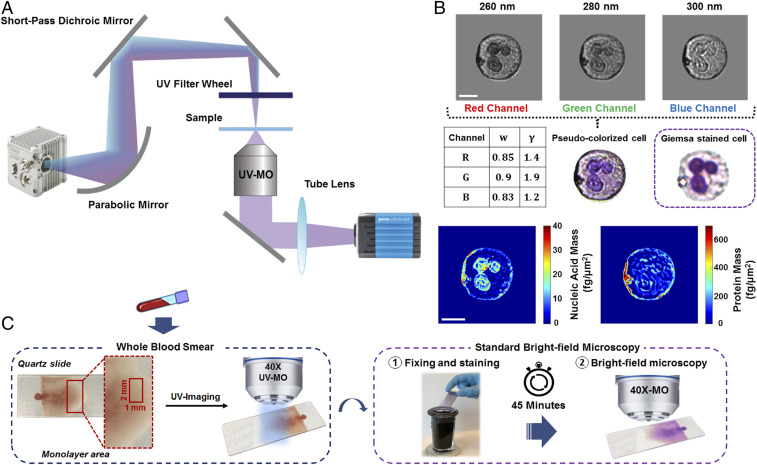
Our multispectral deep-UV microscope enables fast imaging of live, unstained cells at different discrete UV wavelengths. As depicted in Fig. 1A, the system uses a broad-band, laser-driven plasma source and a set of band-pass filters to tune the imaging wavelength to the absorption peaks of major biochemical components in human cells (more details in Materials and Methods). Here we use 260 nm and 280 nm, which correspond to absorption peaks from nucleic acids and proteins, respectively (29, 30, 34). We also acquire images with a center wavelength of 300 nm, specifically chosen because it does not correspond to an absorption peak of any endogenous molecule (31) and can thus be used as a virtual counterstain in our pseudocolorization scheme (see the pseudocolorization section in Materials and Methods). As discussed below, this wavelength is also critical for quantitative Hb-level analysis (Hb Mass Quantification in Individual RBCs). These three wavelengths were also strategically chosen because bright and inexpensive light-emitting diodes (LEDs) are commercially available at these spectral regions.
Fig. 1.
System setup and method for UV microscopy of blood samples. (A) Schematic of the deep-UV microscope consisting of an ultrabroadband plasma source, short-pass dichroic mirror, UV band-pass filters, UV microscope objective, and UV-sensitive camera. (B) Pseudocolorization scheme with optimized weights and gamma values for each channel along with an example colorized UV and its respective Giemsa-stained bright-field image. Nucleic acid and protein mass maps of the same cell are also shown. (Scale bar: 5 µm.) (C) Schematic of the blood smear sample preparation and imaging protocol. Blood smears were imaged immediately with the UV system, then the smears were fixed, stained, and imaged with a conventional bright-field microscope for comparison.
Fig. 1B shows example images of a neutrophil from the three chosen discrete wavelengths, as well as synthesized nucleic acid and protein mass maps of that cell (30). Clearly, the 260-nm image closely resembles the nucleic acid map, while the 280-nm image has some contributions from nucleic acid but more closely resembles the protein mass map. Finally, the 300-nm image has less intracellular structure, as expected, due to the less specific molecular absorption at this center wavelength. We do not discard potential contributions from other biomolecules (31), and it is worth emphasizing that our goal is not to quantify individual molecules but rather to enable cell identification and phenotyping.
Whole blood was collected from donors according to protocols approved by the Institutional Review Board of Georgia Institute of Technology and Emory University. As shown in Fig. 1C, blood smears were prepared on quartz microscope slides without any cell fixation, dilution, or staining. Images were obtained immediately after the smears dried. The acquired multispectral images were used individually to extract features of interest for hematological analysis or combined to produce a pseudocolor map that recapitulated Giemsa stains (Materials and Methods). After imaging with the UV system, the samples were fixed, stained, and imaged with a white-light bright-field microscope for comparison.
Pseudocolorization of Deep-UV Multispectral Images.
To enhance the utility of our deep-UV microscope in clinical settings, we developed a pseudocolorization scheme which transforms the grayscale multispectral UV images into a color image where blood cells appear with colors that accurately mimic those produced by conventional Giemsa staining. In this approach, we construct a color image in RGB color space by assigning the 260-, 280-, and 300-nm images to the red, green, and blue channels, respectively (as demonstrated in Fig. 1B). The weight factors and gamma values , listed in Fig. 1B for each channel, are optimized by comparing the color representation of each blood cell to their stained counterparts (see Materials and Methods for more details).
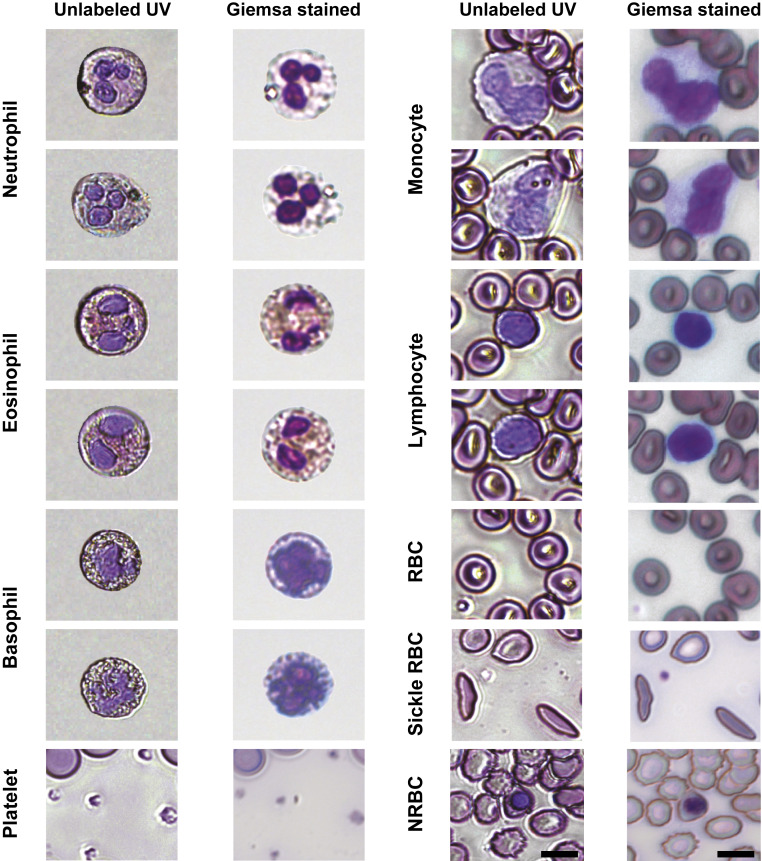
As shown in Fig. 2, our approach enables us to produce colorized images that faithfully recapitulate features of significant importance for blood cell phenotyping and differentiation using traditional staining protocols with bright-field microscopy. Again, the unique colors of each cell type produced with UV microscopy originate from spectral absorption differences of various cellular biochemical components. For example, absorption of nucleic acids in leukocyte nuclei gives rise to the well-known distinctive violet color observed in Giemsa-stained images. In addition to nuclear contrast, our UV images exhibit key cytoplasmic color differences which mainly stem from the different levels of protein (SI Appendix, Fig. S1). A notable example of this is the fidelity with which granulocytes (i.e., neutrophils, eosinophils, and basophils) appear under this pseudocolorization scheme compared to accepted staining protocols. For example, the granules in the cytoplasm of eosinophils maintain their unique characteristic orange hue due to higher amounts of cytoplasmic protein compared to the other granulocytes (SI Appendix, Fig. S1), which under stained samples depicts their eosinophilic morphology for which they are named. Moreover, the granularity of all granulocytes is recapitulated, in part, by the stronger scattering properties of the granules which affects all wavelengths, including 300 nm.
Fig. 2.
Mosaic collection of different blood cell types as depicted by the label-free pseudocolorized UV images and the Giemsa-stained images of the same cells. (Scale bars: 7 µm.)
Apart from color differences, the pseudocolorized UV microscopy images reveal detailed cellular structures and shapes that are important for diagnosing and monitoring blood diseases. SCD is a prominent example in which deformed crescent-shaped RBCs accumulate in blood. Such deformation is clearly identifiable in the UV images of sickled RBCs shown in Fig. 2. Platelets and nucleated RBCs are also clearly identified. We note that some edge effects are present in the UV images due to stronger diffraction and scattering, giving rise to a subtle halo-like structure; however, this does not strongly affect the cells’ appearance or qualitative and quantitative analyses discussed below.
Hb Mass Quantification in Individual RBCs.
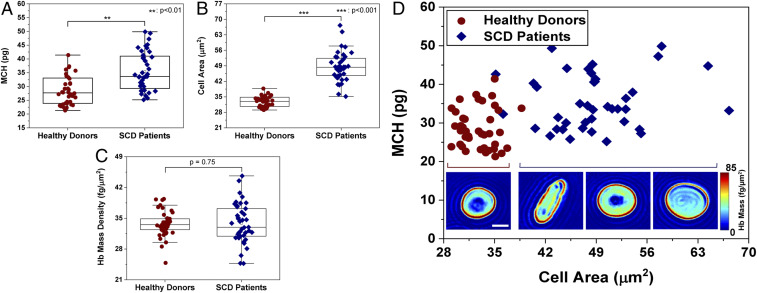
Quantification of RBC Hb mass is routinely performed in clinical hematology analysis by measuring the mean corpuscular Hb (MCH) using hematology analyzers. MCH refers to the total Hb mass per RBC and can be used to characterize many conditions, including anemias, such as SCD and thalassemia, where MCH values deviate from normal (typically, 29.5 ± 2.5 pg) (11).
Given the unique quantitative capabilities of our deep-UV microscope, we are able to quantify total Hb mass and average Hb mass per square area (related to Hb concentration) of individual RBCs. For this task, we leverage images acquired at 300 nm where the only significant absorber in RBCs is Hb. Here the absorbance images at this wavelength can be directly translated to Hb mass. Hb absorption is also high at 260 nm and 280 nm but proteins in the RBC membrane can interfere with Hb quantification. Further, 415 nm, which corresponds to the Soret peak, can also be used to gain higher sensitivity, but this would require an additional acquisition and, more importantly, absorption in this region is highly dependent on oxygenation state. On the other hand, 300 nm has a nearly identical absorbance for oxy and deoxy-Hb (36), thus further removing confounding factors in estimating Hb mass, concentration, and MCH.
As described in Materials and Methods, we assessed the Hb mass of RBCs from the blood smears of two healthy donors (n = 40 cells) and two SCD patients (n = 40 cells). Fig. 3A shows the retrieved MCH values, which were on average 28.6 ± 5.1 pg for RBCs from healthy donors and 35.4 ± 6.9 pg for SCD patients. These values indicate a statistically significantly higher MCH for SCD patients compared to RBCs from healthy donors (P < 0.01). According to the CBC results, MCH values were 29.7 and 34 pg for the healthy donors and SCD patients, respectively, in good agreement with our measurements. Fig. 3 B and D show the drastic increase in RBC size for SCD patients, while Fig. 3C shows that average Hb mass per square area (which again correlates with Hb concentration) is not statistically significant between the two groups (P = 0.75). This behavior is consistent with prior studies reporting on the production of larger RBCs with the same Hb concentration in SCD patients treated with hydroxyurea, which was indeed the case for our SCD population (37, 38).
Fig. 3.
Hb characterization in healthy and sickled RBCs. (A) MCH values obtained from two healthy (n = 40) and two SCD patients (n = 40). (B) Cell area from healthy and SCD donors. (C) Average Hb mass per square area for healthy and SCD RBCs. (D) Scatter plot of MCH vs. cell area for healthy and SCD samples, with insets showing example RBC Hb mass maps (in femtograms per square micrometer). (Inset scale bar: 5 µm.)
WBC Five-Part Differential.
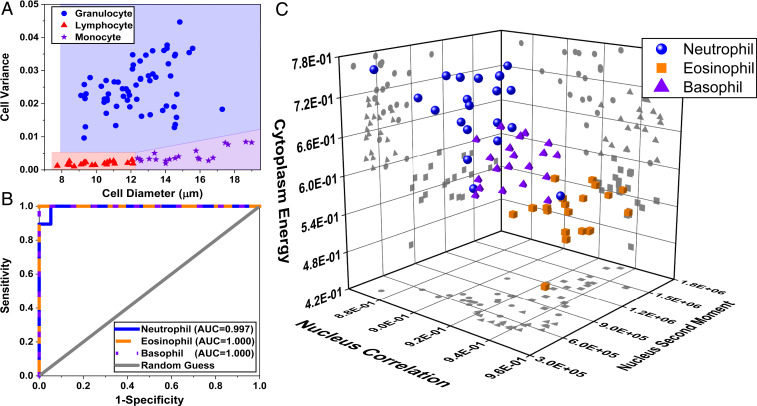
Differential cell counting is an essential part of laboratory hematological analysis for monitoring and diagnosing blood diseases. Here we employed a WBC classification process by extracting 58 features (listed in SI Appendix, Table S1) from all UV wavelengths (i.e., 260, 280, and 300 nm) obtained from 100 cells (20 lymphocytes, 20 monocytes, and 20 of each granulocyte subtype). Images of lymphocytes and monocytes were obtained from blood smears of four healthy donors. Each cell type was classified by a board-certified hematologist. Granulocytes (from up to five healthy donors), on the other hand, were first isolated using a magnetic antibody-based selection technique and then imaged (Materials and Methods). The latter procedure was used to produce a robust “ground truth” for granulocytes, since they can be difficult to differentiate in a whole blood smear. After imaging, individual cells were manually segmented. Then we calculated a series of morphological, statistical, and textural features (again, 58 in total) based on the gray-level cooccurrence matrix (GLCM) as well as biochemical masses from whole cell, cell nucleus, and cytoplasm to perform a five-part WBC differential (Materials and Methods and SI Appendix, Table S1).
First, we applied a three-part differential to classify lymphocytes, monocytes, and granulocytes, simply based on their size and granularity—similar to forward and side scattering with flow cytometry. As depicted in Fig. 4A, plotting the cell diameter against the variance of the intensity values throughout the cell gives a clear class separation between the three cell types (highlighted by three colors). Granulocytes with a higher granularity produce larger variations in the intensity, which leads to a higher pixel value variance compared to lymphocytes and monocytes. In addition, the monocytes are clearly larger than lymphocytes and have markedly low granularity, as expected. Again, this simple analysis resembles the forward versus side-scattering plots of WBCs in flow cytometry and is in agreement with the well-known morphological and textural properties of these cell types (39).
Fig. 4.
Five-part WBC differential from label-free UV images. (A) Cellular intensity variance plotted against cell diameter to perform a three-part WBC differential with blue color representing the granulocytes, red for lymphocytes, and purple for monocytes. (B) ROC curves associated with our machine learning-based granulocyte classification, thereby providing a five-part WBC differential. (C) Scatter plot of the granulocyte subtypes based on the three top-ranked features.
To complete the five-part cell differential analysis, we employed a machine learning algorithm, using support-vector machine (SVM) learning, trained using the extracted features from granulocytes. We evaluated the trained SVM model according to a fivefold cross-validation scheme which yields an accuracy of 98.3%, sensitivity of 95%, and specificity of 100% for classification of granulocyte subtypes (i.e., neutrophils, eosinophils, and basophils) using all features from the three UV wavelengths. The receiver operating characteristic (ROC) curves and confusion matrix associated with our classification, depicted in Fig. 4B and SI Appendix, Fig. S2, further illustrate the ability of our model to accurately identify granulocytes, misclassifying only one cell.
Finally, we applied a feature-ranking algorithm (Materials and Methods) to identify the three top-ranked features that uniquely identify each granulocyte subtype. Remarkably, we find that the most important features are cytoplasm energy, nucleus correlation, and nucleus second moment from a single wavelength: 260 nm (scatter plot in Fig. 4C). This finding is important as it indicates that an accurate five-part WBC differential can be acquired using a single wavelength, which has implications for the speed of the UV imaging assay (more on this below). Performance of the trained classifier with only these three features yields an accuracy of 96.7%, sensitivity of 95%, and specificity of 97.5% (refer to SI Appendix, Fig. S3 for ROC curves and the confusion matrix), which is only slightly lower than using 58 features from all three wavelengths.
For comparison, a similar classification algorithm using SVM was done on the bright-field images of the same cells after they were fixed and stained. Results show classification with an accuracy of 96.67%, sensitivity of 100%, and specificity of 95% (refer to SI Appendix, Fig. S4 for ROC curves and the confusion matrix), which is similar to those obtained using the single-UV-wavelength model.
Evaluation of Pseudocolorized UV-Based Diagnosis.
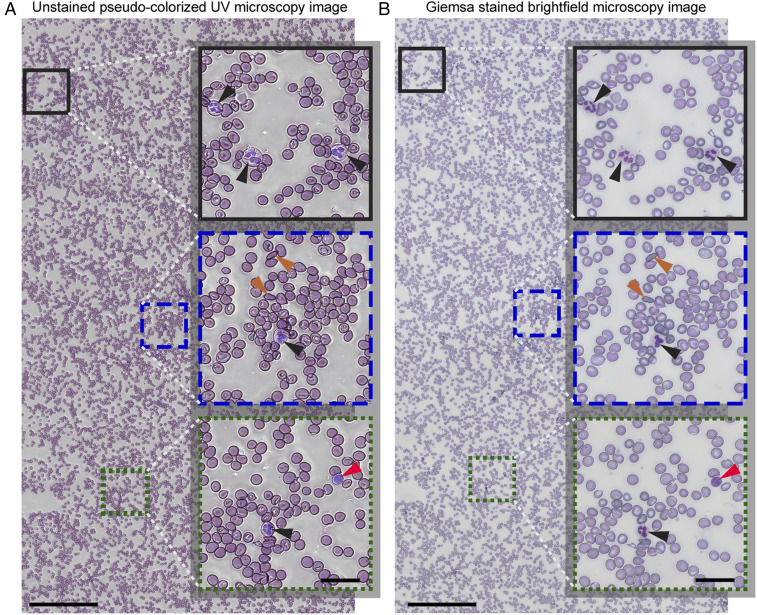
Given the ability of the pseudocolorized UV images to reveal unique morphological and molecular (and thus color) features for each blood cell type, we hypothesized that these images could serve as an alternative to the standard Giemsa-stained peripheral blood smear examination. The advantage of our approach is that it can provide immediate analysis/diagnosis without the need for any processing or chemical reagents. To test this hypothesis, we imaged blood smears from 13 individuals: 4 from healthy donors, 5 from patients with different degrees of thrombocytopenia, and 4 from patients with SCD. CBCs were collected and used as the ground truth to 1) confirm diagnosis and 2) determine the diagnostic accuracy of the visual blood smear examinations using the pseudocolorized, label-free UV images and conventional bright-field microscopy images of fixed and stained cells. As seen in Fig. 5, large areas (1 mm × 2 mm) were imaged to ensure enough cells (>20,000 cells) were included for a reliable diagnosis (see Materials and Methods for details). Due to the short exposure times, imaging of the entire area was performed in ∼3 min per wavelength (further improvement can be made by optimizing the translation stage routine). After imaging with the UV system, smears were fixed and stained and bright-field microscopy images from the same areas were acquired (Materials and Methods). Thus, we collected a total of 26 large-area blood smear images from 13 individuals (13 label-free UV images and the corresponding 13 bright-field stained images of the same region with the same cells) (40). An example large-area pseudocolorized UV image along with its corresponding Giemsa-stained, bright-field microscopy image is shown in Fig. 5. Careful inspection of magnified images reveal that the UV colorized images highlight key cellular features with outstanding clarity and contrast which are nearly equivalent to conventional stained images, demonstrating their potential to be used as an alternative for clinical diagnosis.
Fig. 5.
Large-area (1 × 2 mm2) fixative-free, unstained, pseudocolorized UV image of a whole blood sample collected from an SCD patient (A) along with the corresponding bright-field microscopy image after fixing and staining (B). (Scale bars: 200 µm.) The selected magnified insets highlight cellular features with black arrowheads pointing to neutrophils, orange arrowheads showing the sickled RBCs, and the red arrowheads point to lymphocytes. (Inset scale bars: 30 µm.)
A web-based survey was designed comprising two randomly distributed sets of large-area colorized UV images and conventional bright-field images from healthy and pathological samples, along with a questionnaire. Each set had one large-area blood smear image per donor (either UV or bright-field—again, randomly selected), for a total of 13 large-area images per set. Because the UV and bright-field images from an individual donor corresponded to the same area with same cells in the blood smear (Fig. 5), this study design avoids potential diagnostic bias arising from a hematologist reviewing images from the same donor twice. A panel of 10 board-certified and board-eligible hematologists, blinded to the patients’ condition and clinical history, reviewed the set of images (each hematologist was assigned one of the two sets). Hematologists also answered 10 questions pertaining to the cells’ appearance (i.e., morphology) and counts, as well as their overall diagnosis, diagnostic confidence, and diagnostic quality of each blood smear image. It is also worth noting that all hematologists in the panel had access to the same regions in the blood smears (with the same cells) from all 13 donors, just in a random assortment of different image types—colorized UV or conventional stained bright-field—depending on the assigned set. By the conclusion of the survey, each type of image (UV or bright-field) had 52 to 56 evaluations (13 smears, rated by four or five reviews), for a total of 109 evaluations. Questionnaire responses were recorded automatically and used for statistical analysis.
Results show an overall diagnostic accuracy from the hematologists’ visual inspection of 80.4% with the label-free UV images and a lower 76.5% with the stained bright-field images. In addition, the concordance between the colorized-UV and bright-field images is near-perfect, with a Cohen’s kappa value of κ = 0.84 for identifying healthy vs. diseased samples and combined κ = 0.83 for differentiating between all sample types (healthy vs. thrombocytopenia vs. SCD) (Table 1). Common smear artifacts in the transition region from the monolayer to the feathered region where RBCs lose their biconcave shape (SI Appendix, Fig. S5) are observed in both the label-free UV and stained images. This artifact was confounded with spherocytosis at a rate of 5.35% (3 out of 56 evaluations) for the UV images and 5.66% (3 out of 53 evaluations) for the bright-field images (mostly from the thrombocytopenia samples). Moreover, the presence of halo-like edge effects in UV images did not strongly affect visual inspection and was only noted by the hematologists at a rate of 1.83% (2 evaluations out of 109). Overall, hematologists expressed relatively high diagnostic confidence with an average rate of 76.45% (average 2.29 out of 3, corresponding to moderate to very confident) for the UV images and a nearly identical 75.74% (average 2.27 out of 3) for bright-field images. Moreover, the hematologist panel rated the suitability of UV and bright-field images for diagnosis at a rate of 94.48% and 90.88%, respectively. These results show that in our data the pseudocolorized UV images provide equal diagnostic power compared to Giemsa stains, which are the gold standard for manual diagnosis.
Table 1.
Results of panel review with Cohen’s kappa values and accuracy %
| Patient blood sample type | Imaging modality | No. of wide-field images | Accuracy % | Concordance (Cohen’s kappa, κ) |
| Healthy | UV | 4 | 85.0 | 0.84 |
| BF | 4 | 85.0 | ||
| Thrombocytopenia | UV | 5 | 76.2 | 0.75 |
| BF | 5 | 68.2 | ||
| SCD | UV | 4 | 80.0 | 0.90 |
| BF | 4 | 76.2 | ||
| Combined | UV | 13 | 80.4 | 0.83 |
| BF | 13 | 76.5 |
BF, bright-field.
Bone Marrow Aspirate Slides.
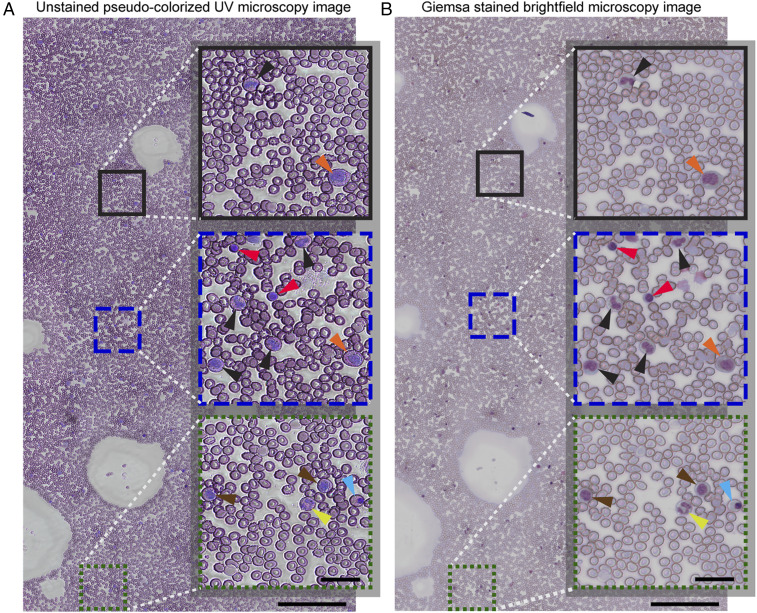
As a final demonstration of the capabilities and versatility of this UV-imaging assay, we imaged bone marrow aspirates slides. Slide preparation followed the same protocol as that used for the peripheral blood smears. Results, shown in Fig. 6, again reveal that UV-microscopy images closely resemble the Giemsa-stained slides. Importantly, even myelopoietic cells (e.g., promyelocytes, myelocytes, metamyelocytes, band neutrophils, and lymphocytes) and erythropoietic cells (e.g., normoblasts), which are not commonly found in peripheral blood, show the same color characteristics and morphology in the label-free UV images and in the stained images.
Fig. 6.
Wide-field unstained pseudocolorized UV image of a sample collected from the bone marrow of a healthy donor (A) along with the corresponding white-light bright-field microscopy image after staining (B). (Scale bars: 200 µm.) The selected magnified insets highlight cellular features of various myelopoietic and erythropoietic cells such as promyelocytes (orange arrowheads), myelocytes (brown arrowheads), metamyeloctes (black arrowheads), band neutrophils (yellow arrowheads), lymphocytes (red arrowheads), and normoblasts (blue arrowheads). (Inset scale bars: 30 µm.)
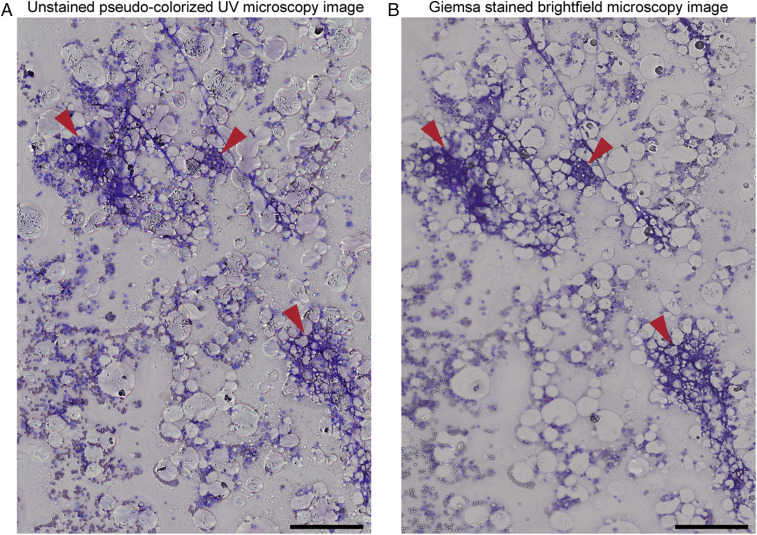
During aspiration, bone marrow samples are also inspected for the presence of bony spicules which are essential to ensure proper aspiration and avoid significant contamination from peripheral blood (41). Because fixing and staining bone marrow is time-consuming, current clinical practice involves taking a small sample during the aspiration procedure, making a smear without fixing or staining, and inspecting the unstained sample with the naked eye. This practice is difficult and requires highly trained technicians that can identify spicules in these relatively transparent samples. Of course, having technicians for this purpose is costly and may not be available at all hospitals; in addition, it is still subject to human error (42). Successful aspirations vary from fewer than 50% when no technician is available to more than 80% when technicians inspect the aspirates (42). This prevalent challenge can be mitigated by imaging unstained smears using the UV microscope, which can provide clear and immediate feedback. Fig. 7 illustrates that the spicules present in a bone marrow aspirate are easily identifiable by their characteristic deep blue hue in the unstained pseudocolorized UV image, which is nearly identical to the Giemsa-stained slide which take over 45 min to process. Our findings suggest that our UV imaging assay can aid clinicians and significantly improve the aspiration procedure’s quality and reliability by providing real-time feedback at the bedside to ensure spiculated bone marrow is obtained.
Fig. 7.
Wide-field unfixed and unstained pseudocolorized UV image of a spiculated bone marrow aspirate (A) and the corresponding white-light bright-field microscopy image after fixing and staining (B). The red arrowheads point to spicules present in the smear. (Scale bars: 200 µm.)
Discussion
We have introduced a quantitative assay based on multispectral deep-UV microscopy which provides fast and robust access to key hematological parameters to diagnose and monitor hematological conditions and diseases. The approach is label-free and fixative-free, thereby obviating the need for chemical reagents that are required in traditional hematological analyzers and blood smear analysis via standard microscopy. Additionally, no calibration is needed, and the system is simple and compact and can be constructed from inexpensive components (e.g., LEDs and compact UV-sensitive cameras) and supplies, including UV transparent slides using emerging UV transparent plastic materials. Currently, the cost of commercial hematology analyzers capable of performing CBC and Hb analysis is approximately $80,000 to $120,000 and require multiple reagents (∼7 to 14 reagents), which adds to the operation costs. In contrast, the cost of our system is approximately $25,000, which can be further reduced to approximately $5,500 with a few simple strategies (see Experimental Setup for more details). These unique attributes (i.e., quantitative, label-free, molecular sensitivity, high-resolution, wide-field, fast, simple-to-use, compact, and low-cost) make this UV-based assay ideal for hematological applications and provide a path for simplifying and improving the standard of care.
The devised pseudocolorization scheme, based on three strategic deep-UV wavelengths, produces color-accurate images that mimic those observed with the gold-standard Giemsa stains, enabling simple, fast, and reliable visual inspection of peripheral blood smears and bone marrow aspirates. This approach is highly translational to clinical and point-of-care settings. Indeed, other pseudocolorization schemes based on alternative label-free optical imaging technologies have been presented (mostly for tissue imaging to mimic hematoxylin and eosin stains) (43–47). This work presents a pseudocolorization scheme specifically designed to mimic the hematological gold standard (Giemsa stains). Further, these alternative label-free methods either lack the subcellular spatial resolution needed for hematological microscopic analysis and/or tend to be expensive and have a large footprint (43–47). Again, the ability of this UV assay to present images that are nearly identical to the gold standard without labels or fixative agents, in a compact and low-cost system, is not only important for translation but is also critical to enable detailed hematological analysis in previously inaccessible environments (e.g., point-of-care and low-resource settings).
We demonstrated a quantitative label-free, five-part differential analysis of WBCs which currently requires expensive and laborious methods. Accurate classification of WBC subtypes was achieved using a combination of morphological and biochemical properties. Notably, we found that the most important features that enable a five-part differential can be extracted from a single wavelength (260 nm). Thus, while all three wavelengths are needed to virtually stain the samples for visual inspection, this is not necessary if a user only seeks to quantify WBC differentials. In this case, the assay can be three times faster by only imaging at 260 nm. Similarly, quantitative Hb analysis only requires imaging at 300 nm, which yields MCH and, if the total blood volume is known, hematocrit. Therefore, the information content of the assay can be tuned depending on the specific test.
The diagnostic utility of deep-UV microscopy was assessed by a panel of hematologists performing a blind analysis on blood smears collected from healthy donors and patients diagnosed with thrombocytopenia and SCD. The near-perfect concordance between hematologists’ diagnoses made using our approach and the gold standard indicates the potential of our colorized-UV images to be applied as label-free alternative for diagnosis. Moreover, the versatile tool can also be used to monitor bone marrow aspirates, either for diagnostic purposes or to ensure a successful aspiration.
In conclusion, this deep-UV quantitative assay yields insight that currently requires CBC (for enumerating blood cells) and microscopic analysis (to evaluate blood cell morphology), and thus it has the potential to simplify and improve clinical hematological analysis overall. Moreover, visual inspection with deep-UV microscopy is appealing for situations where fast feedback is desired, in telemedicine applications, or in circumstances where the necessary reagents are not available, for example in low-resource settings. The brief exposure to UV light does not affect the cells, and thus samples can be preserved for other analyses, including Giemsa stains. Finally, the deep-UV imaging assay can be coupled with 1) flow-cytometry to provide complimentary label-free biomolecular and structural information or 2) microfluidic devices, which can pave the way for a low-cost point-of-care hematological analyzer.
Materials and Methods
Magnetic Granulocyte Isolation.
Live human granulocytes were isolated via negative magnetic antibody-based selection with the MACSxpress isolation kit (Miltenyi Biotec) and resuspended in RPMI media with l-glutamine and Hepes (Life Technologies). In order to induce and maintain a normal cell spreading (48) and adhesion to microscope slide surface, quartz slides were coated with a 1 nM solution of N-Formylmethionine-leucyl-phenylalanine (fMLP; Sigma-Aldrich) for 60 min then rinsed with distilled water and phosphate-buffered saline. The cell suspension was pipetted onto the coated slide and incubated for another 30 min. Slides were then washed with distilled water and dried before imaging.
The magnetic isolation protocol was used to ensure a robust ground truth for our granulocyte subtype classification. This isolation process provides a low yield, particularly for the rare subtypes such as basophils which typically have a very low concentration in whole blood. Thus, the number of cells per group was determined based on the most limiting blood cell subtype (i.e., basophils) to ensure a fair comparison and prevent bias. Nevertheless, the number of cells used in this analysis is on par with other studies using label-free techniques and cell isolation for unique blood cell identification (20, 27, 37).
Preparation of Whole Blood Smear Samples.
Whole blood was collected from healthy donors or patients and added to an anticoagulant solution (sodium citrate; Beckton Dickenson). All protocols were approved by the Institutional Review Boards of Georgia Institute of Technology and Emory University. Informed consent was obtained from donors. Blood smears were made on uncoated quartz slides by using 10 µL of whole blood. After drying the samples in air for 5 min, UV imaging was performed.
Preparation of Whole Bone Marrow Aspirate Smear Samples.
Whole human bone marrow aspirate was collected from healthy donors (AllCells) and added to Na heparin anticoagulant solution. Smears were made on uncoated quartz slides using 10 µL of whole bone marrow and immediately imaged using the UV microscope after drying in air for 5 min.
Fixing and Staining of Blood and Bone Marrow Samples.
Blood and bone marrow samples were first fixed using methanol (Thermo Fisher Scientific) for 7 and 15 min, respectively, and stained in May-Grünwald solution (MG500; Sigma-Aldrich) for 15 min. After a brief rinse, samples were put in a 1:10 diluted Giemsa stain solution (GS500; Sigma-Aldrich) for 20 min. Samples were then washed in a phosphate buffer solution (pH 6.6) and air-dried for bright-field microscopy.
Bright-Field Microscopy.
After imaging with the deep-UV microscopy setup, fixed and stained samples were imaged using an upright (Axioskop 2 Plus; Carl Zeiss) microscope equipped with a 40× objective (numerical aperture [NA] 0.6). By scanning the sample, a series of images from a 1 × 2-mm area on the sample was acquired. The tile scan images were stitched using the Zeiss Zen Black edition software (Carl Zeiss) to obtain a wide-field image.
Experimental Setup.
The developed deep-UV microscopy system consisted of an incoherent broadband laser-driven plasma light source (EQ-99X LDLS; Energetiq Technology). The output light from the broadband source was collected through an off-axis parabolic mirror (Newport Corporation) and relayed to the sample using a short-pass dichroic mirror (Thorlabs). Multispectral imaging was performed using UV band-pass filters (Chroma Technology) installed on a filter wheel, allowing acquisition of images at three wavelength regions centered at 260, 280, and 300 nm. The light intensity on the sample plane was measured to be 0.14, 4.5, and 0.22 mW at 260-, 280-, and 300-nm wavelengths, respectively. For imaging, we used a 40× UV microscope objective (NA 0.5) (LMU-40X; Thorlabs), by which we achieved an average spatial resolution of ∼280 nm. Images were then recorded using a UV-sensitive charge-coupled device (pco.UV; PCO AG) camera (integration time = 30 to 100 ms) while the sample was translated and adjusted for focusing via a three-axis high-precision motorized stage (MLS2031; Thorlabs). By raster-scanning the sample, a series of UV images from a 1 × 2-mm area were acquired at each wavelength. Imaging time is ∼3 min per wavelength (currently limited by the translation stage).
The deep-UV microscopy setup used in this study has been developed from research-grade components with an overall cost of approximately $25,000, with the broadband light source being the most expensive component. However, with recent advancements in the development of compact and inexpensive deep-UV LEDs and UV-sensitive cameras, the cost of the system can be further decreased to approximately $5,500. In this case, the UV objective (LMU-40X; Thorlabs) would constitute the most expensive component. Also, the use of single-color LEDs would obviate the need for the spectral filters, which, together with the more compact cameras, would result in a more compact system. Finally, the current need for quartz microscope slides adds to the overall cost of the procedure as the price of these slides is higher than conventional glass slides. However, this issue can be mitigated with the help of emerging UV-transparent plastics which are significantly less costly than quartz and can be easily manufactured into microscope slides.
Image Processing, Registration, and Pseudo-RGB Colorization.
After obtaining the tile image series at the three wavelengths, each image was normalized by a reference background image acquired from a blank area on the sample at each wavelength to remove any illumination artifacts. In the next step and in order to obtain accurate colorization of UV images, an intensity-based image registration algorithm (based on the imregister function), implemented in MATLAB (MathWorks) was used to coregister the corresponding images across all three wavelengths.
The registered intensity image stacks (260-, 280-, and 300-nm wavelength images) for each field of view were used to obtain pseudo-RGB colorized images. To form the pseudocolorized images, each color channel (i.e., R, G, and B channels) was formed according to the optimized weights (w) and gamma (γ) values (listed in Fig. 1B) based on: , , and . Finally, the colorized images were transformed to the HSV (hue, saturation, and value) color space, a constant hue offset of +0.05 was applied, and they were converted back to RGB color space.
Stitching of pseudocolorized images was performed using the Grid/Collection stitching plugin (49) of the Fiji (50) software, which calculates the overlap between each tile and linearly blends them into a single wide-field image. The obtained wide-field pseudocolorized images were then exported to the Zoomify (Zoomify Inc.) format, which enables viewing of large images using a standard web browser and uploaded to a custom-designed website for easy access and viewing by the clinical review panel.
Quantification of Biochemical Masses.
We quantified the nucleic acid and protein mass based on calculation of the optical density which is obtained by normalizing each UV image by a reference background image taken from an empty area on the sample at each wavelength. The OD maps obtained at 260-nm and 280-nm wavelengths are then used to calculate the mass maps assuming a linear contribution of species at each wavelength according to Eq. 1:
| [1] |
where ε is the extinction coefficient, is the optical path length, and c is the species concentration. Based on OD values for the two wavelengths, we can generate a set of equations and solve for the concentration–pathlength products ( terms) at each pixel. In our calculations, we use average extinction coefficients at 260 nm and 280 nm for nucleic acid and protein (29) and average OD values over the bandwidth of our UV filters. Further, we assume an average molar mass of 52,728 Da for protein and of 330 Da for nucleic acids to obtain mass values at each pixel.
For calculation of Hb mass in RBCs, we obtained the OD at 300-nm wavelength and used the molar extinction coefficient of oxygenated Hb (31, 36) to calculate the concentration–pathlength products ( terms) at each pixel. Then, by integrating these values over the entire cell area we obtained the total mass for each cell.
Feature Extraction and Ranking.
In order to train our unsupervised cell classifier, 58 different features from 100 WBCs (features are summarized in SI Appendix, Table S1) were extracted from whole cells, cell nuclei, and their cytoplasm. We extracted features based on morphology, statistical properties, and textural features based on the GLCM, from processing of intensity images obtained at 260-nm wavelength. Average biochemical masses from cells were also obtained using the 260-nm and 280-nm images according to the described method for quantification of biochemical masses.
To rank the extracted features, we used a ranking algorithm (rank features function, MATLAB) which calculates the area between the ROC curve and the random classifier slope and picked the three features that yielded the highest ROC areas for training of the single-wavelength classifier.
Cell Classification Model Construction and Performance Assessment.
A supervised cell classification model was constructed based on an SVM with a Gaussian kernel using the extracted features from WBCs. We assessed the trained SVM model according to a fivefold cross-validation scheme which randomly partitions the dataset into five equal-sized subsamples. A subsample was utilized for validation and testing the model while the remaining subsamples were used as training data. This process was repeated five times, with each of the five partitions used only once as the validation data. By doing this, we ensured that the whole dataset was used for both training and validation.
To assess the performance of the cell classification model, we obtained a number of performance metrics such as accuracy, which is the ratio of number of correct predictions to the total number of input dataset, sensitivity, which determines the percentage of true positive classification events, and specificity, which measures the percentage of true negatives determined by the model. The ROC curves were calculated for our multiclass classifier (with three classes, i.e., neutrophils, eosinophils, and basophils) using a one-versus-all scheme where the performance of the model is evaluated for each class against the two others. Sensitivity and specificity are calculated by treating each class as the true label consecutively. The ROC curves were then used for graphical illustration of model performance.
Survey Methodology.
A web-based survey consisting of 26 wide-field images (13 UV and 13 bright-field images) was developed and provided to a clinical review panel consisting of 10 board-certified and board-eligible hematologists who performed a blind analysis of the images. Each hematologist was presented with a random deidentified collection of pseudocolorized UV and bright-field images of healthy, thrombocytopenia, and SCD samples. In addition, each image was accompanied by an online 10-question questionnaire where the hematologists provided assessment of the population and morphology of each blood cell type and responded to questions about the diagnostic quality and their diagnostic confidence based on the images. Diagnostic quality was defined as whether the hematologists felt that the image quality enabled them to make a proper diagnosis on a scale from 1 to 3. Diagnostic confidence was defined as how certain the reviewers felt about their assessment. Questionnaire responses were recorded automatically and used for statistical analysis. This clinical panel review protocol (no. H19389) was Institutional Review Board-exempt and consent was waived.
Statistical Analysis.
For each hematologist, we calculated the concordance between their diagnosis (healthy vs. thrombocytopenia vs. SCD) based on UV and bright-field images using Cohen’s kappa (51), assuming that the data are categorical and that UV and bright-field images are taken on the same data, which they are. Cohen’s kappa values were also calculated for the diagnosis from UV and bright-field vs. ground truth (based on CBC performed at the collection site, Emory Hospital), which tells us how well each panel member’s diagnosis correlates with CBC. Finally, we calculated the concordance between every two hematologists to determine the reliability of their assessment.
Student’s t test was used for comparison of populations in our quantitative analysis of Hb mass in RBCs. JMP Pro software (version 14.0; SAS Institute Inc.) and MATLAB (MathWorks) was used for all statistical analyses.
Data Availability.
Files to support this study can be accessed through the associated Open Science Framework (DOI: 10.17605/OSF.IO/AYW4J)
Supplementary Material
Acknowledgments
We thank Dr. A. Aljudi of Children’s Healthcare of Atlanta (Department of Pathology) for providing the spiculated human bone marrow samples and M. E. Fay for assisting in cell isolation protocol. We also greatly acknowledge support for this work by the Massner Lane Family Foundation; Burroughs Wellcome Fund (CASI BWF 1014540); National Science Foundation (NSF CBET CAREER 1752011); and the Donaldson Charitable Trust Research Synergy Fund Award, a philanthropic award provided by the Winship Cancer Institute of Emory University, the Aflac Cancer & Blood Disorders Center at Children’s Healthcare of Atlanta, and the Wallace H. Coulter Biomedical Engineering Department at Emory University and the Georgia Institute of Technology.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Files to support this study can be accessed through the associated Open Science Framework (DOI: 10.17605/OSF.IO/AYW4J).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001404117/-/DCSupplemental.
References
- 1.Honda T., Uehara T., Matsumoto G., Arai S., Sugano M., Neutrophil left shift and white blood cell count as markers of bacterial infection. Clin. Chim. Acta 457, 46–53 (2016). [DOI] [PubMed] [Google Scholar]
- 2.van Wolfswinkel M. E. et al., Predictive value of lymphocytopenia and the neutrophil-lymphocyte count ratio for severe imported malaria. Malar. J. 12, 101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman T. B., Draper D., Puopolo K. M., Wi S., Escobar G. J., Combining immature and total neutrophil counts to predict early onset sepsis in term and late preterm newborns: Use of the I/T2. Pediatr. Infect. Dis. J. 33, 798–802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velo-García A., Castro S. G., Isenberg D. A., The diagnosis and management of the haematologic manifestations of lupus. J. Autoimmun. 74, 139–160 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Crawford J., Dale D. C., Lyman G. H., Chemotherapy-induced neutropenia: Risks, consequences, and new directions for its management. Cancer 100, 228–237 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Williams D. J. et al., Association of white blood cell count and C-reactive protein with outcomes in children hospitalized for community-acquired pneumonia. Pediatr. Infect. Dis. J. 34, 792–793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park M. S. et al., Leukopenia predicts remission in patients with inflammatory bowel disease and Behcet’s disease on thiopurine maintenance. Dig. Dis. Sci. 60, 195–204 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Langhorst J., Boone J., Lauche R., Rueffer A., Dobos G., Faecal lactoferrin, calprotectin, PMN-elastase, CRP, and white blood cell count as indicators for mucosal healing and clinical course of disease in patients with mild to moderate ulcerative colitis: Post hoc analysis of a prospective clinical trial. J. Crohn’s Colitis 10, 786–794 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Sitter T., Schmidt M., Schneider S., Schiffl H., Differential diagnosis of bacterial infection and inflammatory response in kidney diseases using procalcitonin. J. Nephrol. 15, 297–301 (2002). [PubMed] [Google Scholar]
- 10.Seebach J. D., Morant R., Rüegg R., Seifert B., Fehr J., The diagnostic value of the neutrophil left shift in predicting inflammatory and infectious disease. Am. J. Clin. Pathol. 107, 582–591 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Lewis S. M., Bain B. J., Bates I., Dacie and Lewis Practical Haematology, (Elsevier, 2006). [Google Scholar]
- 12.Winkelman J. W., Tanasijevic M. J., Zahniser D. J., A novel automated slide-based technology for visualization, counting, and characterization of the formed elements of blood: A proof of concept study. Arch. Pathol. Lab. Med. 141, 1107–1112 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Larsson A., Greig-Pylypczuk R., Huisman A., The state of point-of-care testing: A European perspective. Ups. J. Med. Sci. 120, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bain B. J., Bone marrow aspiration. J. Clin. Pathol. 54, 657–663 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majors C. E., Pawlowski M. E., Tkaczyk T., Richards-Kortum R. R., ““Low-cost disposable cartridge for performing a white blood cell count and partial differential at the point-of-care”” in 2014 IEEE Healthcare Innovation Conference, HIC 2014, (Institute of Electrical and Electronics Engineers Inc., 2014), pp. 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng S., Lin J., Kasdan H., Tai Y., Fluorescent labeling, sensing, and differentiation of leukocytes from undiluted whole blood samples. Sens. Actuators B Chem. 132, 558–567 (2008). [Google Scholar]
- 17.Osei-Bimpong A., Jury C., McLean R., Lewis S. M., Point-of-care method for total white cell count: An evaluation of the HemoCue WBC device. Int. J. Lab. Hematol. 31, 657–664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Geijn G. J. et al., Leukoflow: Multiparameter extended white blood cell differentiation for routine analysis by flow cytometry. Cytometry A 79, 694–706 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Chan L. L., Wilkinson A. R., Paradis B. D., Lai N., Rapid image-based cytometry for comparison of fluorescent viability staining methods. J. Fluoresc. 22, 1301–1311 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Yoon J. et al., Label-free characterization of white blood cells by measuring 3D refractive index maps. Biomed. Opt. Express 6, 3865–3875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramoji A. et al., Toward a spectroscopic hemogram: Raman spectroscopic differentiation of the two most abundant leukocytes from peripheral blood. Anal. Chem. 84, 5335–5342 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Verebes G. S. et al., Hyperspectral enhanced dark field microscopy for imaging blood cells. J. Biophotonics 6, 960–967 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Yakimov B. P. et al., Label-free characterization of white blood cells using fluorescence lifetime imaging and flow-cytometry: Molecular heterogeneity and erythrophagocytosis [Invited]. Biomed. Opt. Express 10, 4220–4236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalut K. J., Ekpenyong A. E., Clegg W. L., Melhuish I. C., Guck J., Quantifying cellular differentiation by physical phenotype using digital holographic microscopy. Integr. Biol. 4, 280–284 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Ekpenyong A. E. et al., Bacterial infection of macrophages induces decrease in refractive index. J. Biophotonics 6, 393–397 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Zangle T. A., Burnes D., Mathis C., Witte O. N., Teitell M. A., Quantifying biomass changes of single CD8+ T cells during antigen specific cytotoxicity. PLoS One 8, e68916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon J. et al., Identification of non-activated lymphocytes using three-dimensional refractive index tomography and machine learning. Sci. Rep. 7, 6654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley R. S. et al., A pathologist’s perspective on bone marrow aspiration and biopsy: I. Performing a bone marrow examination. J. Clin. Lab. Anal. 18, 70–90 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeskind B. J. et al., Nucleic acid and protein mass mapping by live-cell deep-ultraviolet microscopy. Nat. Methods 4, 567–569 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Ojaghi A., Fay M. E., Lam W. A., Robles F. E., Ultraviolet hyperspectral interferometric microscopy. Sci. Rep. 8, 9913 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soltani S., Ojaghi A., Robles F. E., Deep UV dispersion and absorption spectroscopy of biomolecules. Biomed. Opt. Express 10, 487–499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz R., “A, Alfan, “Optical biopsy-detecting cancer with light”” in Biomedical Optical Spectroscopy and Diagnostics, (OSA Trends in Optics and Photonics Series, Optical Society of America, 1996). [Google Scholar]
- 33.Zangle T. A., Teitell M. A., Live-cell mass profiling: An emerging approach in quantitative biophysics. Nat. Methods 11, 1221–1228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung M. C., Evans J. G., McKenna B., Ehrlich D. J., Deep ultraviolet mapping of intracellular protein and nucleic acid in femtograms per pixel. Cytometry A 79, 920–932 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung M. C. et al., Intracellular protein and nucleic acid measured in eight cell types using deep-ultraviolet mass mapping. Cytometry A 83, 540–551 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Prahl S., Optical absorption of hemoglobin. http://omlc.ogi.edu/spectra/hemoglobin. Accessed 10 October 2019.
- 37.Byun H. et al., Optical measurement of biomechanical properties of individual erythrocytes from a sickle cell patient. Acta Biomater. 8, 4130–4138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodgers G. P., Dover G. J., Noguchi C. T., Schechter A. N., Nienhuis A. W., Hematologic responses of patients with sickle cell disease to treatment with hydroxyurea. N. Engl. J. Med. 322, 1037–1045 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Adan A., Alizada G., Kiraz Y., Baran Y., Nalbant A., Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 37, 163–176 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Ojaghi A., Robles F. E., Label-free hematology analysis using deep-ultraviolet microscopy. Open Science Framework. https://osf.io/ayw4j/. Deposited 31 May 2020. [DOI] [PMC free article] [PubMed]
- 41.Braner D. A. V., Tegtmeyer K., Bone marrow aspiration and biopsy. Artic. New Engl. J. Med. 361, e28 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Bunting S., Atuan M., Castellino S., 119 improving the quality of bone marrow biopsy in a pediatric hospital. Am. J. Clin. Pathol. 149 (suppl. 1), S51 (2018). [Google Scholar]
- 43.Mayerich D. et al., Stain-less staining for computed histopathology. Technology (Singap World Sci) 3, 27–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivenson Y. et al., PhaseStain: The digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci. Appl. 8, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell M. et al., All-digital histopathology by infrared-optical hybrid microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 3388–3396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivenson Y. et al., Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 3, 466–477 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Orringer D. A. et al., Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 1, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettit E. J., Hallett M. B., Localised and global cytosolic Ca2+ changes in neutrophils during engagement of Cd11b/CD18 integrin visualised using confocal laser scanning reconstruction. J. Cell Sci. 109, 1689–1694 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Preibisch S., Saalfeld S., Tomancak P., Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J. et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen J., A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20, 37–46 (1960). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Files to support this study can be accessed through the associated Open Science Framework (DOI: 10.17605/OSF.IO/AYW4J)