Significance
Temperature is a key control parameter of biological processes, but measuring and controlling temperatures on a cellular-length scale in living organisms remains an outstanding challenge. Applying nanoscale-thermometry techniques to early embryos, we study cell divisions in a highly controlled manner using local laser heating and real-time in vivo temperature readout. Nitrogen-vacancy centers in nanodiamonds, incorporated into the cells, allow us to map out the temperature distribution of a locally heated embryo with submicrometer spatial resolution and high sensitivity. The simultaneous cell-division imaging under controlled laser heating is used to achieve cell-cycle timing control and inversion, providing insights into timing-regulation mechanisms during early embryogenesis.
Keywords: quantum sensing, nanoscale thermometry, cell-division asymmetry, nitrogen-vacancy centers, cell-cycle control
Abstract
Understanding the coordination of cell-division timing is one of the outstanding questions in the field of developmental biology. One active control parameter of the cell-cycle duration is temperature, as it can accelerate or decelerate the rate of biochemical reactions. However, controlled experiments at the cellular scale are challenging, due to the limited availability of biocompatible temperature sensors, as well as the lack of practical methods to systematically control local temperatures and cellular dynamics. Here, we demonstrate a method to probe and control the cell-division timing in Caenorhabditis elegans embryos using a combination of local laser heating and nanoscale thermometry. Local infrared laser illumination produces a temperature gradient across the embryo, which is precisely measured by in vivo nanoscale thermometry using quantum defects in nanodiamonds. These techniques enable selective, controlled acceleration of the cell divisions, even enabling an inversion of division order at the two-cell stage. Our data suggest that the cell-cycle timing asynchrony of the early embryonic development in C. elegans is determined independently by individual cells rather than via cell-to-cell communication. Our method can be used to control the development of multicellular organisms and to provide insights into the regulation of cell-division timings as a consequence of local perturbations.
Cell-cycle asynchrony is a universal phenomenon and serves as a key to understanding microscopic cell-to-cell interactions that may govern the cell division (1–4). While correlations between division timing and cell temperatures can provide new insights into the microscopic mechanisms of cell division (5, 6), to date, controlled experiments at the cellular scale have been challenging (7–10). To study the role of cell-cycle asynchrony in embryogenesis, there have been attempts to synchronize the division timing by genetic modifications; however, such techniques have proven to be rather invasive, resulting in two identical blastomeres that do not possess an anteroposterior axis distinction and organisms that subsequently do not develop into regular adults (11–13).
Our approach makes use of a biocompatible temperature-control method to study and manipulate the early embryonic development (embryogenesis) of Caenorhabditis elegans, an ideal test bed for studying cell-cycle timings due to their stereotypical asynchronous cell cycles. We accurately control the cell-division timings with local laser heating, which imposes steep temperature gradients across the embryo, even in the absence of any heat-shock-response regulatory elements. The resulting temperature distributions are monitored in real time by using nanoscale diamond thermometers (14–22), in which nitrogen-vacancy (NV) centers in biocompatible nanodiamonds are used to measure the local temperature with high spatial resolution. The optical stability of NV centers circumvents the photo-bleaching drawbacks of existing fluorescence-based temperature-measurement techniques, thereby allowing the continuous monitoring of temperature gradients over extended periods of time. The combination of these techniques enables the study of the embryonic development of C. elegans in a quantitative manner, providing detailed information on how embryonic cells adjust their division timings under large temperature differences: We find, remarkably, that cell-selective heating leads to a pronounced inversion of division order in the two-cell stage.
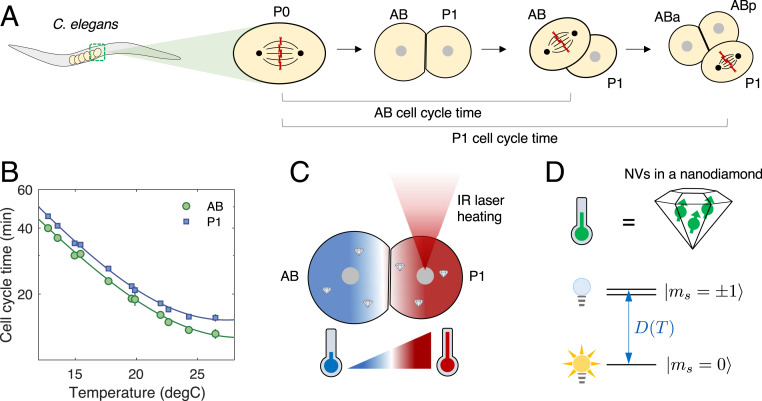
Fig. 1A illustrates the typical early embryonic development of C. elegans. Embryogenesis starts with a single P0 cell, which subsequently divides into smaller daughter cells, AB and P1. The AB cell, which is larger and develops into the anterior part of the embryo, always divides faster than the smaller, posterior P1 cell, regardless of the ambient global temperature, demonstrating robust cell-division order in the two-cell stage (Fig. 1B). Such highly ordered, asymmetric cell divisions can be found in many living organisms, raising intriguing questions regarding potential connections to biochemical signaling pathways between the cells (23, 24). We make use of the temperature dependence of the division timing, shown in Fig. 1B, to manipulate the cell-cycle duration. Specifically, we find that the cell-cycle times, , follow an exponential scaling with the inverse of absolute temperature, given by
| [1] |
where is the absolute temperature in Kelvin and are positive cell-dependent coefficients (SI Appendix, Table S1). The second term on the right-hand side is introduced to incorporate deviations from the simple Arrhenius law at high temperatures (25). This pronounced dependence opens the possibility of controlling cell-cycle durations by local laser heating, which introduces a steep temperature gradient over an embryo (Fig. 1C). In what follows, we investigate how the embryonic cells cope with such local thermal perturbations by monitoring cell-cycle-timing variations and correlating the changes with local temperatures measured by nanoscale in vivo thermometers.
Fig. 1.
Studying embryogenesis of C. elegans via nanoscale NV thermometry. (A) Early embryonic development of C. elegans. At the end of the one-cell stage, the chromosomes (red crosses) align at the center of the single cell P0 and are then split and pulled apart by spindle fibers (thin black arcs), resulting in two daughter cells, AB and P1. The two daughter cells exhibit asynchronous division, in which AB undergoes mitosis earlier than P1. The cell-cycle time of a given cell is defined as the separation between chromosome-splitting events of its parent cell and itself. (B) Cell-cycle times for AB and P1 as a function of global ambient temperature. Solid lines are fits to a modified Arrhenius equation (Eq. 1). (C) Local laser heating applied to a two-cell-stage embryo. A focused IR laser selectively illuminating P1 introduces a steep temperature gradient across the embryo (red, hot; blue, cold). Nanodiamond thermometers are incorporated inside the embryo to measure the resultant temperature distribution. (D) Principle of thermometry. The in vivo thermometer consists of an ensemble of NV centers (green arrows) in a nanodiamond. The NV center has three electronic spin states in its orbital ground state, , which are energetically separated by a temperature-dependent, zero-field splitting . The nearly degenerate states are optically dark, while the state is optically bright.
Experimental Setup
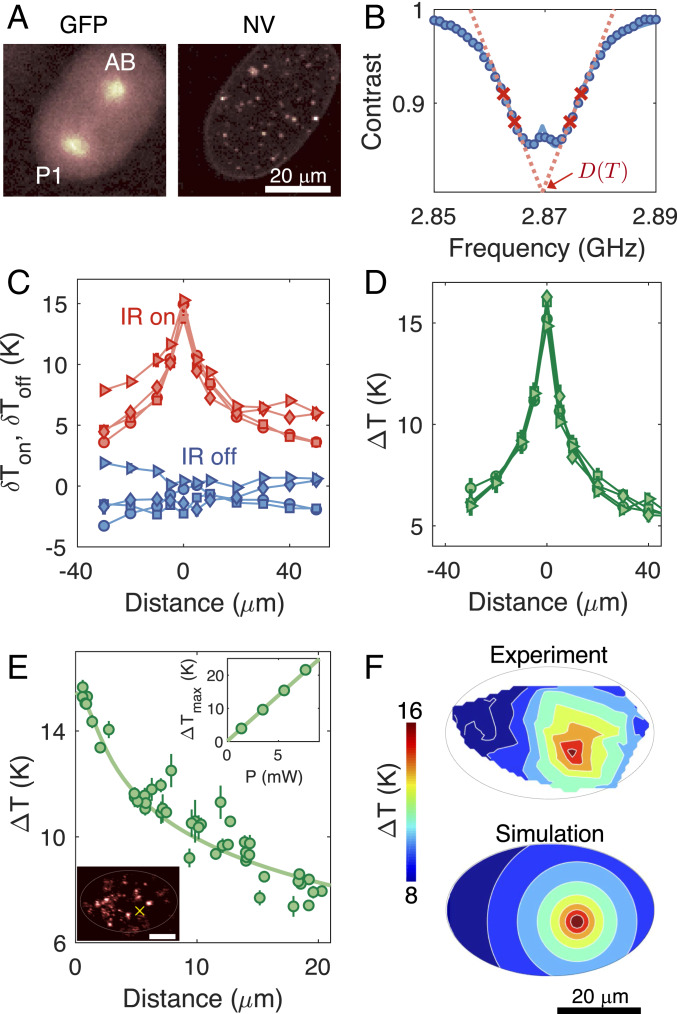
Our experimental apparatus consisted of the combination of a home-built confocal and wide-field microscope system for local laser heating, temperature monitoring, and embryo imaging (SI Appendix, Fig. S1). We used C. elegans (strain: TY3558) tagged with green fluorescent protein (GFP) in histones and tubulins to visualize cell division during early embryogenesis (Fig. 2 A, Left). For local heating, we employed an infrared (IR) laser at a wavelength of 1,480 nm (26, 27), selectively focused on targeted cells with a beam waist of 2 m. To accurately determine the temperature distribution under local laser illumination experimentally, it is crucial to monitor the temperature in vivo. To this end, we made use of NV-nanodiamond thermometers, which are optically bright and stable and have good biocompatibility after appropriate surface treatment (SI Appendix, Materials and Methods and Fig. S10). Measuring temperature using NV centers relies on optically detected electronic spin resonance of the ground-state triplet, (14). Under green excitation, the fluorescence-level difference between bright and dark states enabled readout of the NV center spin state, which, in turn, allowed the determination of the resonance frequency that depends on local temperature (Fig. 2B). Crucially, the resonance shift, , can be linearly translated into a change in local temperature as , with a temperature susceptibility of kHz/K (28). To identify , we employed a four-point measurement scheme (18), enabling faster temperature readout while maintaining robustness against fluorescence fluctuations and stray magnetic fields. By using a gonad-microinjection technique (29), the NV thermometers can be incorporated inside C. elegans embryos, allowing us to correlate cell-division dynamics with the local temperature distribution inside the embryo (Fig. 2 A, Right). In order to facilitate both cell-division imaging and temperature readout, we extracted the NV-injected embryos via dissection and placed them on top of a coplanar waveguide used for thermometer control and readout (SI Appendix, Fig. S2).
Fig. 2.
Local laser heating characterization. (A) Confocal images of the GFP-labeled embryo in the two-cell stage with GFP imaging (Left) and NV-nanodiamond fluorescence imaging (Right). (B) Optically detected magnetic resonance of an NV thermometer. Contrast is defined as the fluorescence ratio of the thermometer with and without the application of microwaves. In temperature measurements, the resonance curve is sampled at four optimized frequencies (red crosses) to extract the resonance position at a given local temperature (18). (C) In vivo temperature readout from an NV thermometer inside an embryo while sweeping the position of an IR laser relative to the thermometer. The IR laser at a fixed power of 5.7 mW is modulated on and off with a period of 30 s over the course of scanning. Each data point is averaged over 3 min, yielding a temperature uncertainty of 0.27 K. The different symbols correspond to different iterative runs. The temperature distribution curves fluctuate due to temporal variations in thermometer calibration parameters. (D) Differential temperature readout of , where are temperatures measured with the IR laser on and off. Such differential readouts reject common-mode noise, leading to different curves collapsing on top of each other and exhibiting robustness to experimental fluctuations. (E) In vivo temperature distribution measured by a collection of NV thermometers inside an embryo. Filled circles denote measured temperatures; the solid line denotes the simulated temperature profile (SI Appendix, Fig. S6). E, Lower Inset shows an image of the laser-illuminated embryo, and the yellow cross indicates the position of the IR heating laser. The bright spots in the image correspond to individual NV thermometers. (Scale bar: 20 m.) E, Upper Inset shows a linear scaling of the maximum temperature increase , as a function of the laser power . (F) The 2D temperature map of the laser-illuminated embryo, with comparisons between experiments (Upper) and simulations (Lower).
Measurement of Temperature Profile
We first characterized the temperature gradient in embryos subject to local IR laser illumination by scanning the laser across the NV nanodiamonds. In contrast to previous measurements, where temperature sensing was performed under static conditions (18), the free-floating NV thermometers inside embryos here are subject to strong positional drifts due to rapid fluid movement in cells. To address this issue, we combined a recently developed robust tracking algorithm for a Brownian particle (30) with the four-point microwave measurements, such that particle tracking was simultaneously performed with temperature readout (SI Appendix, Fig. S3). Under these reliable tracking conditions, we repeatedly read out the two temperature values with the IR laser on and off, , by periodically modulating the IR laser during the scan (Fig. 2C). While the repetitive scans showed different temperature profiles with fluctuating baselines associated with NV calibration drifts, the differential temperature, , produced consistent profiles that were insensitive to such systematic errors (Fig. 2D).
The measured profile is also consistent with a full heat map, where we fixed the IR heating position and instead used multiple NV thermometers in the embryo to map out local temperatures (Fig. 3A). The different sensors were individually calibrated to maximize their temperature sensitivities, with an optimal in vivo sensitivity of 2 K/ (SI Appendix, Fig. S5). We found that the obtained temperature distribution is in good agreement with a numerical simulation of the steady-state heat-conduction equation (Fig. 3B), taking into account the embryo geometry and sample substrate (SI Appendix, Fig. S6). Note that heat-convection effects were negligible due to the small size of the embryo, as also confirmed by independent simulations including such effects. More importantly, we achieved a steep temperature gradient in the vicinity of the IR heating spot, giving a full-width half-maximum of temperature increase of 20 m, much smaller than microfluidic approaches (31). At a laser power above 5 mW, the maximum local temperature difference between AB and P1 is estimated to be more than 10 K, sufficiently large to perturb the cell-division timing (Fig. 3B).
Fig. 3.
Selective acceleration of cell-cycle times and its correlation with local temperature changes. Cell-cycle times of a two-cell embryo subject to local heating as a function of laser power. (A) P1 nucleus heating (at least five measurements each). (B) AB nucleus heating (at least three measurements each). Solid and dashed lines are theoretical predictions based on average and nuclei temperatures of individual cells, respectively; see main text for details. The base temperature is maintained at 12.3 ○C. The error bars on the data markers denote the SD of the mean cell-cycle times. The bands on the theory predictions provide cell-cycle time uncertainties due to a 10% uncertainty in extracted cell volumes.
Local Cell Heating and Cell-Cycle Control
Having characterized the temperature distribution, we proceeded to examine the cell-cycle times as a function of laser-heating power and compared it to different hypothetical models based on the measured temperature profile. To maximize the temperature difference between AB and P1 in the two-cell stage while avoiding overheating, we operated at a low base temperature of 12.C, stabilized by air cooling. First, we heated the nucleus of the P1 cell with a focused IR laser beam, starting 200 s after chromosome separation so that the cytoplasm of AB and P1 were well separated. We heated at a range of IR laser powers to systematically control the temperature gradient between the two cells and monitored their cell-cycle changes. When the IR laser was focused on the nucleus of P1 (Fig. 3C), the cell-cycle times for both AB and P1 monotonically decreased with increasing IR power, with a greater decrease in P1, leading to a narrowing of the division-timing difference between the two cells. In contrast, AB nucleus heating produced an even larger division timing difference between the two cells (Fig. 3D).
The observed differences in cell-cycle acceleration rates can be attributed to a difference in local temperature under cell-selective laser heating. To understand the changes in the measured cell-cycle times, we introduced two competing hypotheses—does the early embryonic cell follow its local, nucleus temperature at the heating spot or follow the mean temperature averaged over its cell volume? The first one implies the importance of the nucleus in controlling cell-development rates, while the second one may suggest the importance of other replication and development processes occurring throughout the cytoplasm. To answer this question, we analyzed the temperature distribution measured by the NV thermometers to estimate expected cell-cycle times under the two hypotheses. We found that for both the AB and P1 heating cases, the cell-cycle times agreed well with expectations based on the average-temperature model (solid curves), in which we calculated the average temperature within each cell and determined the expected cell-cycle time based on the calibration curve in Fig. 1B. On the other hand, it significantly deviated from the nucleus temperature model (dashed curves), in which we used the nucleus temperature only to estimate the cell-cycle time. This suggests that the cell-cycle time is not solely determined by the DNA replication occurring in the nucleus of the cell, but is also dependent on the kinetics of cyclins and other proteins throughout the cytoplasm.
Observation of Cell-Cycle Inversion
One important consequence of the preceding cell-cycle time analysis is that under P1 heating, the P1 cell division can be substantially sped up relative to the AB cell, leading to an inversion from the normal cell-division order (Fig. 4 A and B). Examining the cell-cycle time differences in more detail in Fig. 4C, we indeed found that the P1 cell consistently divided faster than the AB cell at high laser powers. The inversion in cell-cycle ordering, combined with the good agreement of cell-cycle times with the average temperature model, is strong evidence that the fixed division order between AB and P1 cells under usual conditions is not mediated by cell-to-cell communication and that each cell follows its independent clock that sets cell-cycle timings. Following the inversion, we found that the cell-division orders of subsequent cell stages were preserved (SI Appendix, Fig. S9), as the division timing changes in the two-cell stage were not sufficient to modify the relative division order of later cell stages. In addition, we found that the relative cell-cycle durations at the four-cell stage were unmodified (Fig. 4D), with no noticeable differences between AB descendants (ABa and ABp) and P1 descendants (EMS and P2). This provided further evidence that the regulation of cell-division timings was performed independently by individual cells. Despite the inversion of cell-division times, the majority of heated embryos successfully hatched and grew into adult worms (8 of 12 under 4.5 mW) at rates that were only slightly less than, but still significantly different from, controls (10 of 11 without heating; P = 0.002 from a one-tailed test).
Fig. 4.
Observation of cell-cycle inversion. (A) Observation of inversion in cell-division order for a two-cell embryo. P1 is selectively heated with the IR laser at a power of 4.5 mW. The base temperature is held at 12.C. (B) GFP image of the cell-cycle inversion. (C) Cell-cycle time difference between the AB and P1 cells as a function of IR power. The top axis shows the difference in the local, average temperature increases between the two cells. Above 3-mW IR laser power, the cell-division order in the two-cell embryo is inverted as a result of local laser heating. (D) Cell-cycle durations monitored for the four-cell stage (ABa, ABp, EMS, and P2) after cell-cycle inversion between AB and P1 (see SI Appendix, Fig. S9 for later cell stages). The IR heating laser is turned off once the P1 cell division is completed. The bar plot shows fractional changes in their respective cell-cycle times comparing between with and without local heating. The error bars denote the SD of the mean values.
Discussions
Our experiments demonstrate a method to manipulate cell-cycle timings in early C. elegans embryos using local laser heating and in vivo nanodiamond temperature measurements. Our data imply that cell-cycle timings are controlled in a cell-autonomous manner, with no significant contribution from cell-to-cell communication. These results open multiple future directions, including the exploration of cell-cycle-timing control at later developmental stages and further investigations of the developmental consequence of perturbing cell-cycle times, as well as additional thermogenetic control (27, 32–34). In vivo nanoscale NV thermometry allows for the long-term readout of temperature at subcellular levels without photo-bleaching (10, 35) and may enable micro-Kelvin temperature resolution with future improvements utilizing magnetic-criticality-enhanced measurements (36). Moreover, various surface treatments of nanodiamonds also provide their attachment to specific cellular components, with minimal effects on cell viability (37, 38). Since nearly all biochemical processes are temperature-dependent, we expect that the methodology we developed here should be broadly applicable, including to the study of subcellular behaviors. For example, since locally heating the spindle impacts its structure and chromosome movement (39), our methodology could be used to dissect how the spindle segregates chromosomes and how spindle malfunctions lead to chromosome segregation errors in cancer cells (40).
Materials and Methods
Extended materials and methods are provided in SI Appendix.
Experimental Setup.
A schematic diagram of our home-built microscope system for embryo imaging and nanoscale thermometry is shown in SI Appendix, Fig. S1. Full details of the setup are described in SI Appendix, Materials and Methods.
Nanodiamond Injection Inside Embryos.
We used a microinjection microscope to supply a nanodiamond solution to the gonad of young, second-larval-stage hermaphrodites. The gonad-injected nanodiamonds were naturally incorporated inside embryos. To avoid aggregation of the particles in a cellular environment, the diamond surface was appropriately coated with polyethylene glycol polymer, which led to an enhanced injection success probability of 50%. Detailed procedures for the surface treatment and nanodiamond injection are described in SI Appendix, Materials and Methods.
Sample Preparation.
To monitor cell-cycle times under local heating, we placed the NV-incorporated C. elegans embryos on a glass cover slide and encapsulated them with M9 buffer solution, agar, and a coverslip to maintain humidity, as depicted in SI Appendix, Fig. S2. After monitoring from the single-cell stage up to the eight-cell stage, we removed the glass cover slide from the setup, added additional buffer solution, and either sealed the edges with wax or kept it in a humid environment to prevent drying of the sample. This allowed us to keep track of whether the locally heated embryos eventually hatched.
NV Thermometer Tracking.
For in-cell sensor tracking, we implemented a recently developed tracking algorithm (30) that can be applied to fluorescent objects subject to rapid drift and diffusion. Moreover, we eliminated tracking dead times by performing temperature measurements simultaneously with tracking, ensuring that we can continuously track a given NV thermometer over an extended period (SI Appendix, Fig. S3). A detailed description of our tracking algorithm is given in SI Appendix, Materials and Methods.
Temperature Measurement.
An NV thermometer can be used as a real-time relative thermometer if one keeps track of the temperature-dependent resonance shift, , over time. Here, is the linear temperature susceptibility of an NV center, and is the local temperature change. To extract , we employed the reported four-point method (18), which discretely samples a resonance curve at only four different microwave frequencies. We chose the four frequencies to be and , where denote the frequencies on the left and right sides with respect to the resonance frequency, respectively, and is a frequency modulation of 2 MHz . The resulting NV fluorescence under is given as
| [2] |
| [3] |
| [4] |
| [5] |
where corresponds to the fluorescence level under microwave illumination at frequency , and is the Zeeman shift due to an unknown static magnetic field from the local environment. For symmetrically chosen frequencies with and , the four measured data points can be used to estimate a change in local temperature as
| [6] |
As can be seen from Eq. 6, the four-point measurement is to leading order robust against global fluorescence changes and magnetic field fluctuations. In our experiments, both were independently optimized for different NV nanodiamonds to maximize their temperature sensitivities (SI Appendix, Fig. S5). The detailed measurement sequence for temperature readout is shown in SI Appendix, Fig. S4.
Simulation of Temperature Distribution.
We compared the measured temperature distributions to simulations based on the heat-conduction equation under local laser heating (SI Appendix, Fig. S6). The simulations were performed in COMSOL Multiphysics by taking into account the sample geometry and laser-heating condition. From the steady-state solution of the simulation, we obtained a two-dimensional (2D) temperature distribution in embryos. Independent simulations confirmed that heat-convection effects are negligible on these length scales, and we did not observe clearly distinguishable features caused by variations in the local thermal conductivity within the cell. This is presumably because the cell environment is predominantly water, while cell membranes are thin and have thermal conductivities on the same order of magnitude as water (41). By overlaying the temperature map with early embryo images, the average and nucleus temperatures of different cells can be calculated (SI Appendix, Fig. S7). See SI Appendix, Materials and Methods for details.
Cell Division of Later-Stage Embryos.
The embryonic cell divisions were characterized up to the eight-cell stage with and without the presence of local laser heating. The measurement results are summarized in SI Appendix, Figs. S8 and S9.
Data Availability.
Data included in this study are made publicly available at https://doi.org/10.7910/DVN/VYCUXB.
Supplementary Material
Acknowledgments
We thank A. Bentle, M. Fujiwara, C. Hunter, E. Kage-Nakadai, H. Knowles, V. Leon, J. Ni, A. Sawh, Y. Shikano, V. Susoy, A. Weisman, and Y. Zhang for insightful discussions and experimental help. This work was supported by the Center for Ultracold Atoms; NSF Grant PHY-1506284; the Army Research Office Multidisciplinary University Research Initiative; a Vannevar Bush Faculty Fellowship; the Moore Foundation; and a Samsung Fellowship.
Footnotes
The authors declare no competing interest.
Data deposition: All relevant data included in this study are available at Harvard’s Dataverse, https://dataverse.harvard.edu (https://doi.org/10.7910/DVN/VYCUXB).
See QnAs on page 14622.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922730117/-/DCSupplemental.
References
- 1.Gönczy P., Rose L. S., Asymmetric cell division and axis formation in the embryo, Wormbook 15, 1–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gönczy P., Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355–366 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Budirahardja Y., Gönczy P., PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development 135, 1303–1313 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Tavernier N., Labbé J., Pintard L., Cell cycle timing regulation during asynchronous divisions of the early C. elegans embryo. Exp. Cell Res. 337, 243–248 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Brauchle M., Baumer K., Gönczy P., Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13, 819–827 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Hubatsch L., et al. , A cell-size threshold limits cell polarity and asymmetric division potential. Nat. Phys., 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ermakova Y. G., et al. , Thermogenetic neurostimulation with single-cell resolution. Nat. Commun. 8, 15362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singhal A., Shaham S., Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat. Commun. 8, 14100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiyonaka S., et al. , Genetically encoded fluorescent thermosensors visualize subcellular thermoregulation in living cells. Nat. Methods 10, 1232–1238 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Arai S., et al. , Mitochondria-targeted fluorescent thermometer monitors intracellular temperature gradient. Chem. Commun. 51, 8044–8047 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kemphues K. J., Priess J. R., Morton D. G., Cheng N., Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311–320 (1988). [DOI] [PubMed] [Google Scholar]
- 12.Boyd L., Guo S., Levitan D., Stinchcomb D. T., Kemphues K. J., PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122, 3075–84 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Watts J. L., et al. , Par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122, 3133–40 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Mochalin V. N., Shenderova O., Ho D., Gogotsi Y., The properties and applications of nanodiamonds. Nat. Nanotechnol. 7, 11–23 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Wu Y., Jelezko F., Plenio M. B., Weil T., Diamond quantum devices in biology. Angew. Chem. Int. Ed. 55, 6586–6598 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Brites C. D. S., et al. , Thermometry at the nanoscale. Nanoscale 4, 4799–4829 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Vetrone F., et al. , Temperature sensing using fluorescent nanothermometers. ACS Nano 4, 3254–3258 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Kucsko G., et al. , Nanometre-scale thermometry in a living cell. Nature 500, 54–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann P., et al. , High-precision nanoscale temperature sensing using single defects in diamond. Nano Lett. 13, 2738–2742 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Toyli D. M., de las Casas C. F., Christle D. J., Dobrovitski V. V., Awschalom D. D., Fluorescence thermometry enhanced by the quantum coherence of single spins in diamond. Proc. Natl. Acad. Sci. U.S.A. 110, 8417–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laraoui A., et al. , Imaging thermal conductivity with nanoscale resolution using a scanning spin probe. Nat. Commun. 6, 8954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson D. A., et al. , Non-neurotoxic nanodiamond probes for intraneuronal temperature mapping. ACS Nano 11, 12077–12086 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Schierenberg E., Reversal of cellular polarity and early cell-cell interaction in the embryo of Caenorhabditis elegans. Dev. Biol. 122, 452–463 (1987). [DOI] [PubMed] [Google Scholar]
- 24.Guo S., Kemphues K. J., par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611–20 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Begasse M., Leaver M., Vazquez F., Grill S., Hyman A., Temperature dependence of cell division timing accounts for a shift in the thermal limits of C. elegans and C. briggsae. Cell Rep. 10, 647–653 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Kamei Y., et al. , Infrared laser-mediated gene induction in targeted single cells in vivo. Nat. Methods 6, 79–81 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Hirsch S. M., et al. , FLIRT: Fast local infrared thermogenetics for subcellular control of protein function. Nat. Methods 15, 921–923 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acosta V. M., et al. , Temperature dependence of the nitrogen-vacancy magnetic resonance in diamond. Phys. Rev. Lett. 104, 070801 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Mohan N., Chen C. S., Hsieh H. H., Wu Y. C., Chang H. C., In vivo imaging and toxicity assessments of fluorescent nanodiamonds in Caenorhabditis elegans. Nano Lett. 10, 3692–3699 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Fields A. P., Cohen A. E., Optimal tracking of a Brownian particle. Opt. Express 20, 22585 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Lucchetta E. M., Lee J. H., Fu L. A., Patel N. H., Ismagilov R. F., Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434, 1134–1138 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bath D. E., et al. , FlyMAD: Rapid thermogenetic control of neuronal activity in freely walking Drosophila. Nat. Methods 11, 756–762 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Churgin M. A., He L., Murray J. I., Fang-Yen C., Efficient single-cell transgene induction in Caenorhabditis elegans using a pulsed infrared laser. G3 Bethesda 3, 1827–1832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirsch L. R., et al. , Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc. Natl. Acad. Sci. U.S.A. 100, 13549–13554 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donner J. S., Thompson S. A., Kreuzer M. P., Baffou G., Quidant R., Mapping intracellular temperature using green fluorescent protein. Nano Lett. 12, 2107–2111 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Wang N., et al. , Magnetic criticality enhanced hybrid nanodiamond thermometer under ambient conditions. Phys. Rev. X 8, 011042 (2018). [Google Scholar]
- 37.Nagl A., Hemelaar S. R., Schirhagl R., Improving surface and defect center chemistry of fluorescent nanodiamonds for imaging purposes—a review. Anal. Bioanal. Chem. 407, 7521–7536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan M. S., et al. , Stepwise ligand-induced self-assembly for facile fabrication of nanodiamond–gold nanoparticle dimers via noncovalent biotin–streptavidin interactions. Nano Lett. 19, 2020–2026 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Nicklas R. B., Chromosome movement and spindle birefringence in locally heated cells: Interaction versus local control. Chromosoma 74, 1–37 (1979). [DOI] [PubMed] [Google Scholar]
- 40.Funk L. C., Zasadil L. M., Weaver B. A., Living in CIN: Mitotic infidelity and its consequences for tumor promotion and suppression. Dev. Cell 39, 638–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bastos A. R. N., et al. , Thermal properties of lipid bilayers determined using upconversion nanothermometry. Adv. Funct. Mater. 29, 1905474 (2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in this study are made publicly available at https://doi.org/10.7910/DVN/VYCUXB.