Significance
Membrane-binding toxins demonstrate both a public health challenge and a bioengineering opportunity due to their efficient internalization into cells. These toxins multivalently bind to naturally occurring lipid receptors at the plasma membrane and initiate endocytosis. This manuscript reports the importance of structured lipid receptor clustering for the induction of membrane bending. We also observed that the magnitude of membrane curvature was correlated to the stoichiometry of toxin-bound receptors. By identifying how these bacterial proteins initiate membrane curvature, these findings provide mechanistic insights into the early steps of pathogenic endocytosis.
Keywords: gangliosides, toxins, binding stoichiometry, curvature generation, clathrin-independent endocytosis
Abstract
AB5 bacterial toxins and polyomaviruses induce membrane curvature as a mechanism to facilitate their entry into host cells. How membrane bending is accomplished is not yet fully understood but has been linked to the simultaneous binding of the pentameric B subunit to multiple copies of glycosphingolipid receptors. Here, we probe the toxin membrane binding and internalization mechanisms by using a combination of superresolution and polarized localization microscopy. We show that cholera toxin subunit B (CTxB) can induce membrane curvature only when bound to multiple copies of its glycosphingolipid receptor, GM1, and the ceramide structure of GM1 is likely not a determinant of this activity as assessed in model membranes. A mutant CTxB capable of binding only a single GM1 fails to generate curvature either in model membranes or in cells, and clustering the mutant CTxB−single-GM1 complexes by antibody cross-linking does not rescue the membrane curvature phenotype. We conclude that both the multiplicity and specific geometry of GM1 binding sites are necessary for the induction of membrane curvature. We expect this to be a general rule of membrane behavior for all AB5 toxins and polyomaviruses that bind glycosphingolipids to invade host cells.
Cholera toxin (CTx) causes the massive and often deadly secretory diarrhea following enteric infection with the bacteria Vibrio cholerae; 2.9 million infections and 95,000 deaths are attributed to cholera annually (1). CTx typifies the AB5 family of bacterial toxins that includes the heat-labile Escherichia coli, Shiga toxin (STx), and pertussis toxins. The B subunits of these toxins multivalently bind and cross-link naturally occurring plasma membrane glycosphingolipids to induce cellular internalization and guide intracellular trafficking (2). CTx, STx, and simian virus 40 (SV40) are known to induce membrane bending, likely through binding to multiple copies of their glycolipid receptors with acyl tail dependencies (3–9). In the case of CTx, the B subunit (CTxB) binds the glycosphingolipid GM1 to engage endogenous mechanisms of endocytic uptake and retrograde trafficking from the plasma membrane into the endoplasmic reticulum (ER). Once in the ER, the enzymatically active A subunit coopts the machinery for ER-associated degradation to retrotranslocate into the cytosol and induce disease (10). Except for pertussis toxin, the B subunits of these toxins assemble into stable and perfectly symmetrical homopentamers that can bind five or more glycosphingolipids at once and localize the toxin within plasma membrane nanodomains enriched in cholesterol (11, 12). But, how these evolutionarily conserved and highly potent B subunits work to enable invasion of the host cell remains incompletely understood.
Here, we test the idea that CTxB requires defined stoichiometry of binding to induce de novo curvature in membranes (13–17) and to sort the toxin into such highly curved membrane structures (17–19). Because CTxB internalization in vivo does not exclusively rely on either clathrin- or caveolin-dependent processes (20–27), it is feasible that the inherent membrane bending capability of CTxB may drive cellular uptake (3, 4). We examine the mechanisms by which CTxB induces membrane curvature, using polarized localization microscopy (PLM). PLM provides superresolution information on membrane orientation and detects membrane orientation with order-of-magnitude higher sensitivity than comparable optical methods (28). Curvature induction was measured in model membranes to test the influence of the multivalent binding of CTxB and the molecular shape and lipid phase preference of GM1. We found that membrane bending in model membranes caused by CTxB can be explained by the stoichiometry of binding at least two GM1 molecules per CTxB, and no contribution was observed from the ceramide tail structure of the GM1 or membrane cholesterol content. Finally, the need for multivalent binding of CTxB to GM1 to induce membrane curvature in the plasma membrane of live cells was confirmed by superresolution imaging of cells exposed to pentavalent wild-type (wt) or a monovalent mutant CTxB (mCTxB) capable of binding only a single GM1.
Results
Stoichiometry of GM1 Binding to CTxB Dictates Membrane Curvature Induction.
We tested the hypothesis that AB5 toxins employ a structured cross-linking of their ganglioside receptor to induce membrane curvature. We tested this with CTxB that contains five potential binding sites for its receptor GM1 as a model system. To detect membrane curvature, we used a nanoscale membrane budding assay based on PLM (28). Using this assay, we previously showed the addition of fluorescently labeled CTxB to supported bilayers composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoch (POPC), 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI), and GM1 induced the growth of membrane buds with a radius of <50 nm that subsequently grow to form tubules with a radius of >200 nm. These membrane bending events were driven by the accumulation of CTxB at the base of the tubule, reflecting the preference of the toxin for negative Gaussian membrane curvature vs. planar or positive curvature regions.
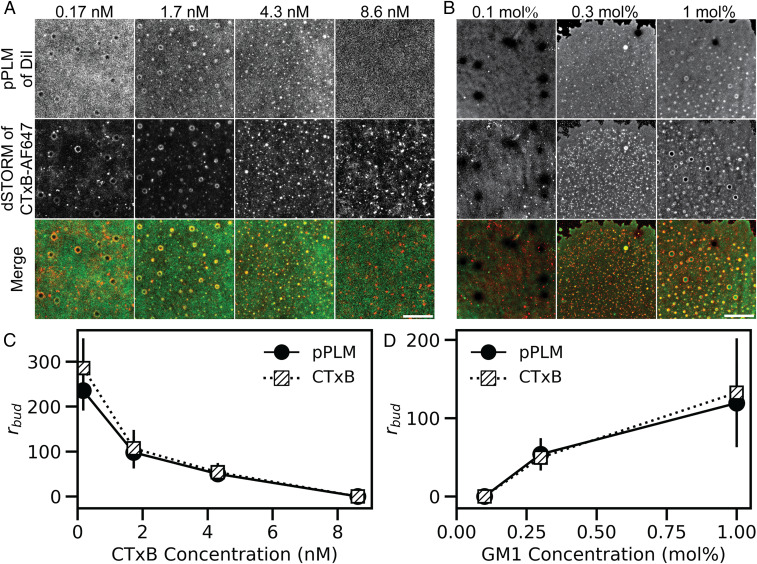
We first tested the effect of receptor clustering by altering the average number of GM1s bound per toxin. This was accomplished by either varying the concentration of CTxB while holding the levels of GM1 in the membrane constant at 0.3 mol % or varying the concentration of GM1 while holding the CTxB concentration constant at 4.3 nM (Fig. 1). Membrane curvature and CTxB localization were then determined independently via p-polarized localization microscopy (pPLM) and direct stochastic optical reconstruction microscopy (dSTORM), respectively (28, 29).
Fig. 1.
CTxB binding to supported bilayers containing GM1 induces buds that vary in size as a function of the concentration of both CTxB and GM1. (A) Planar SLBs containing 0.3 mol % GM1 were incubated with CTxB concentrations ranging from 0.17 nM to 8.6 nM and subsequently imaged by pPLM and dSTORM. (B) Planar lipid bilayers containing varying GM1 concentrations were incubated with 4.3 nM CTxB-AF647 prior to imaging using pPLM and dSTORM. The pPLM results quantify the membrane curvature, and dSTORM images show the CTxB distribution. Merged images of the DiI (green) and CTxB-AF647 (red) show strong colocalization of clustered CTxB at sites of membrane bud formation. (Scale bars, 2 μm.) (C) Measurements of membrane bud sizes from the superresolution images of bilayers labeled with varying concentrations of CTxB reveal bud size decreases with increasing CTxB concentrations. (D) Bud size increases with increasing GM1 concentrations. Error bars represent the SD of individual bud sizes from three or more repeats per condition.
Reconstructed images in the two channels revealed that regions of membrane budding were enriched in CTxB. Puncta of DiI localizations via pPLM indicated membrane buds and regions of the sample in which the membrane was perpendicular to the microscope coverslip (28). Interestingly, altering the GM1:CTxB ratio altered the radii of the membrane buds. Decreasing the GM1:CTxB ratio by increasing the concentration of CTxB from 0.17 nM to 4.3 nM also decreased the average bud size from a maximum value of 285 ± 66 nm to 53 ± 20 nm as detected by dSTORM and 116 ± 68 nm to 49 ± 14 nm as reported by pPLM (Fig. 1). A similar result was observed when the GM1:CTxB ratio was varied by changing GM1 concentrations. In this case, the bud size increased with increasing concentrations of GM1. Measured values at 0.3 and 1 mol % were 50 ± 14 nm and 132 ± 70 nm via dSTORM and 53 ± 20 nm and 119 ± 50 nm via pPLM, respectively (Fig. 1). No membrane bending was detected at the lowest GM1-to-CTxB ratios examined, including either a high concentration of CTxB (8.6 nM) or a low concentration of GM1 (0.1 mol %). These findings reveal the GM1:CTxB ratio is an essential factor underlying the induction of membrane curvature, with high ratios giving rise to the formation of large buds and low ratios entirely failing to initiate membrane bending. Based on our results, we hypothesized that, at low GM1:CTxB ratio, the predominant stoichiometry of membrane-bound CTxB is that of a single GM1 receptor bound to each CTxB, which does not induce membrane buds.
Monovalent CTxB Does Not Induce Membrane Budding.
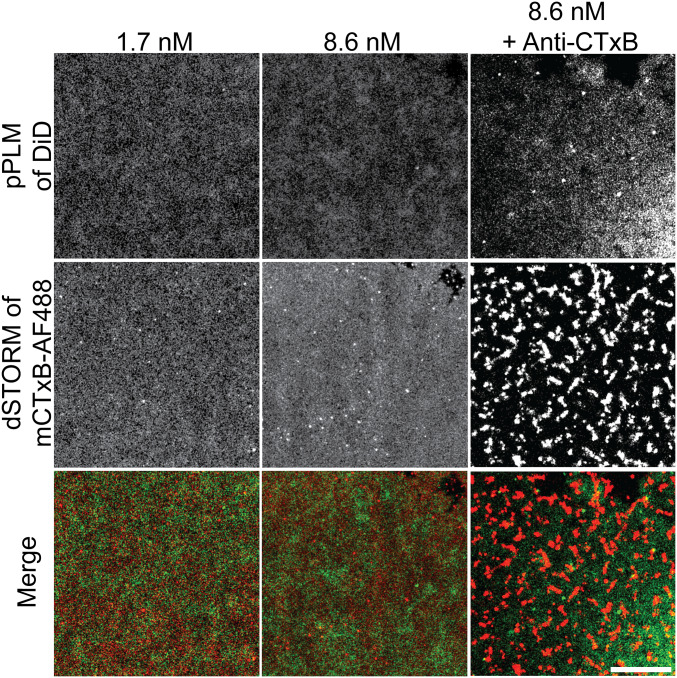
To test this hypothesis alternately, we determined the capacity of a monovalent variant of CTxB (mCTxB) to induce membrane curvature. Each mCTxB contains one native GM1-binding subunit and four subunits with G33D mutations that disrupt GM1 binding; each mCTxB can bind precisely one GM1 (8). We examined the ability of 1.7 nM or 8.6 nM mCTxB to induce budding in model membranes containing 0.3 mol % GM1. These concentrations of mCTxB were chosen to match the protein concentration and active GM1 binding site concentration, respectively, of the 1.7 nM wtCTxB condition that consistently induced membrane bending (Fig. 1). In contrast to the behavior of wtCTxB that contained five GM1 binding sites, no change in membrane topography was detected upon the addition of mCTxB to membranes at either concentration in model membranes (Fig. 2). To confirm the effect of valency on CTxB-induced curvature, we determined whether mCTxB was capable of inducing curvature in intact giant unilamellar vesicles (GUVs). CTxB induced membrane invaginations on GUVs as expected (3), whereas mCTxB caused no apparent change to the membrane shape (SI Appendix, Fig. S1). Thus, the binding of CTxB to a single GM1 is not sufficient to bend membranes.
Fig. 2.
No membrane curvature is induced by the addition of mCTxB to GM1-containing membranes, even when it is cross-linked. Planar SLBs containing 0.3 mol % GM1 were incubated with 1.7 nM mCTxB or 8.6 nM mCTxB. Cross-linking of the mCTxB was performed with a 1:100 dilution of anti-CTxB antibody. Membranes were subsequently imaged by pPLM and dSTORM. Cross-linking led to the formation of large-scale clusters in dSTORM images but nevertheless failed to generate significant levels of membrane budding. Lower concentrations of anti-CTxB demonstrate less mCTxB clustering and no membrane budding (SI Appendix, Figs. S2 and S3). Merged images of the DiD (green) and mCTxB-AF488 (red) show no coincident puncta, indicating that mCTxB fails to form clusters or induce membrane curvature. (Scale bar, 2 μm.)
Spatial Organization of GM1 Binding Is Necessary for Inducing Membrane Curvature.
We next tested the hypothesis that the spatial organization of binding sites on CTxB critically underlies the generation of membrane curvature. To test this idea, we asked whether the clustering of monovalent CTxB by antibody cross-linking could override the requirement for multivalent binding by CTxB. For these experiments, membrane-bound mCTxB was cross-linked with varying amounts of anti-CTxB antibodies. The highest antibody concentrations used (1:100 dilution) induced clustering of mCTxB into punctate structures (Fig. 2 and SI Appendix, Fig. S2) and significantly slowed the diffusion of mCTxB (SI Appendix, Fig. S3). However, no membrane bending was observed by pPLM at any anti-CTxB concentration examined.
We also tested whether a general protein-induced clustering of lipids is sufficient to induce curvature. We labeled membranes containing varying concentrations of l-α-dipalmitoyl phosphatidylethanolamine (DPPE)-biotin and cross-linked the lipids using streptavidin (SI Appendix, Fig. S4). The tetravalent binding of streptavidin to DPPE-biotin yielded no apparent variations in the membrane topography as assessed with pPLM at any of the streptavidin or DPPE-biotin concentrations tested, consistent with previous reports (30). Thus, not all protein-induced lipid clustering is sufficient to bend membranes, and the structural details of the GM1 binding sites in CTxB are essential for generating curvature.
Single-Particle Tracking Reveals Complexes with High GM1-to-CTxB Ratios Diffuse Extremely Slowly.
We used the diffusional behavior of CTxB−GM1 complexes to gain further insights into the stoichiometry of GM1 bound to CTxB in the membrane buds. To probe the dynamics of CTxB−GM1 complexes, the single-molecule blinking data used in the reconstruction of superresolution images for wtCTxB and mCTxB were analyzed using high-throughput single-particle tracking. To eliminate the possibility of topographical effects slowing diffusion, we excluded any single-particle trajectories that were coincident with punctate localizations in pPLM. As a control, an identical analysis was performed for DiI. Minimal effects of CTxB binding were observed on DiI diffusion. Under all experimental conditions tested, DiI exhibited diffusion consistent with a single population diffusion coefficient (D) of 0.46 ± 0.2 μm2/s.
The mCTxB, like DiI, demonstrated a single population of Brownian diffusers with a characteristic D of 0.74 ± 0.54 μm2/s. In contrast, wtCTxB displayed heterogeneous, multipopulation distributions of diffusion coefficients. Greater GM1 availability increased the abundance, but not the speed, of the slower moving populations of wtCTxB (Fig. 3 and SI Appendix, Fig. S5).
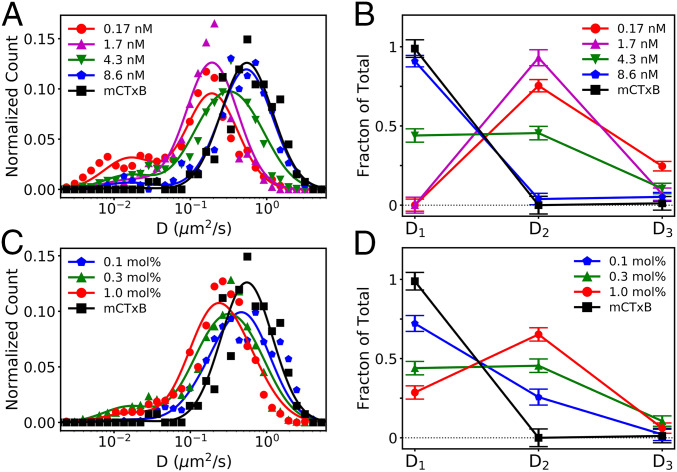
Fig. 3.
Single-particle tracking of CTxB reveals a distribution of diffusion rates suggestive of the presence of three distinct subpopulations of CTxB bound to differing numbers of GM1s. Single-particle tracking analysis was conducted on individual, membrane-bound CTxB molecules in bilayers containing (A and B) constant 0.3 mol % GM1 and varying CTxB concentrations or (C and D) constant 4.3 nM CTxB and varying GM1 concentrations, as also imaged in Fig. 1. For comparison, single-particle tracking was also performed for 1.7 nM mCTxB. (A and C) Histograms report the distribution of D values obtained under each condition. (B and D) The histograms were fit using Eq. 1 to quantify the diffusion coefficient and frequency of the three observed CTxB subpopulations. Fits of individual subpopulations are shown in SI Appendix, Fig. S5. The slower diffusing subpopulations were more abundant at lower CTxB concentrations or higher GM1 concentrations, which correspond to higher GM1:CTxB ratios and to greater membrane bending.
To discern the underlying subpopulations, the experimentally acquired distributions of D were fit to a probability distribution (P) composed from a sum of subpopulations of diffusers (Eq. 1), with each subpopulation contributing a log-normal distribution of D (Eq. 2).
| [1] |
| [2] |
The width of the log-normal distribution (σ) was assumed to be as theoretically expected from the distribution of trajectory lengths analyzed and constant for all subpopulations (31); σ = 0.78. The mean diffusion coefficient for each subpopulation (Di) is equal to exp(μi+σ2/2).
Three subpopulations were sufficient to fit all of our data of CTxB diffusing on model membranes. Di values of D1 = 0.75, D2 = 0.26, and D3 = 0.022 μm2/s were identified by fitting the eight experimentally acquired datasets simultaneously (SI Appendix, Fig. S5). We interpret Di as the mean diffusion coefficient for CTxB with i GM1 per CTxB, that is, D1, D2, and D3, corresponding to the diffusion rates of CTxB bound to one, two, and three GM1, respectively. The subpopulations of four or five GM1-to-CTxB were not apparent in our superresolution observation (i.e., A4 = A5 = 0). The sum of the three fit Ai values were normalized to represent the relative fraction of CTxB with one, two, or three GM1 per CTxB in each experiment (Fig. 3 B and D). As expected, the experiments with mCTxB, concentrated CTxB, or dilute GM1 each yielded A1 close to unity, confirming one GM1 per CTxB. In contrast, with decreasing CTxB concentration or increasing GM1 concentration, the subpopulations that represented more GM1-per-CTxB became prevalent.
In addition to confirming the presence of multiple stoichiometries of CTxB−GM1 complexes, the results of these experiments show the diffusion coefficients do not scale linearly with the number of bound GM1 molecules. The dramatic slowing of CTxB diffusion with an increasing GM1:CTxB ratio suggests that CTxB either progressively penetrates the membrane or induces local membrane deformations that slow their diffusion. Taking these results together with the PLM results, we conclude that the binding of CTxB to just two GM1 was sufficient to induce membrane curvature. Further increasing the binding stoichiometry resulted in increasing the radii of the buds.
Varying Ceramide Structure or Cholesterol Content Has Minimal Effects.
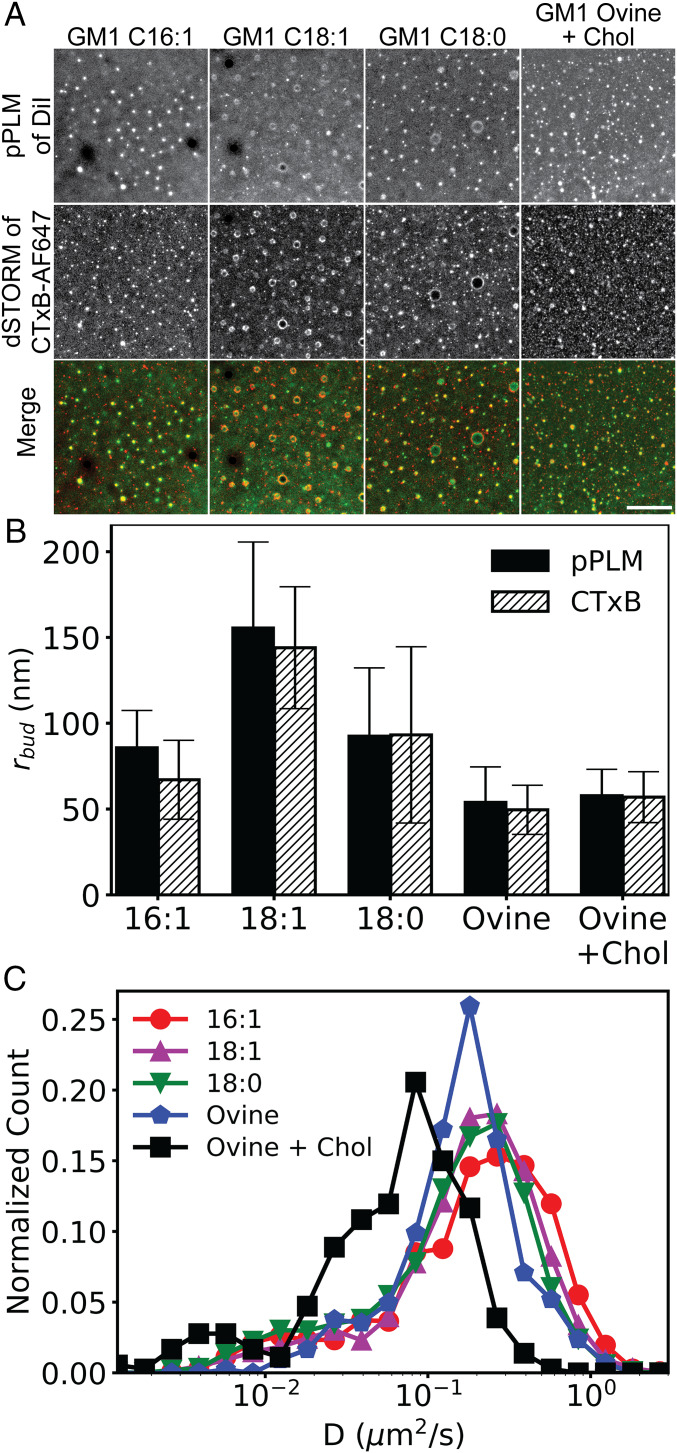
The ceramide structure of GM1 functions as a key regulator of intracellular trafficking of both GM1 itself and CTxB−GM1 complexes (32), suggesting it plays an essential function in sorting as the toxin moves from one intracellular compartment to another. Furthermore, GM1 and Gb3 structures have been directly linked to curvature induction by SV40 and STx, respectively (3–5). We thus tested the hypothesis that the fatty acyl chains of the ceramide domain of GM1 are important for toxin-induced membrane curvature. Three custom gangliosides were used to address this question, recognizing that longer and saturated acyl tails are more correlated with ordered lipid phases (SI Appendix, Fig. S6). In our model systems, membrane bending was detected at similar levels for GM116:1, GM118:1, and GM118:0 upon the addition of CTxB, and there was no systematic change in the radii of the buds correlated with the expected lipid phase preferences of the tails (Fig. 4).
Fig. 4.
Curvature induction in response to CTxB binding is not dependent on the GM1 acyl chain structure or affected by the presence of cholesterol; 4.3 nM CTxB was bound to planar lipid bilayers containing 0.3 mol % of synthetic GM1 with acyl chains of 16:1, 18:1, or 18:0 or commercially available ovine GM1 plus 30 mol % cholesterol. (A) Samples were subsequently imaged using pPLM to detect membrane curvature or dSTORM to image CTxB clustering. Merged images of the DiI (green) and CTxB (red) show strong colocalization of clustered CTxB at sites of membrane bud formation regardless of the GM1 acyl tails or cholesterol content. Additional CTxB concentrations are shown, for cholesterol-containing membranes, in SI Appendix, Fig. S7. (Scale bar, 2 μm.) (B) Curvature was quantified by measuring the mean and SD of membrane bud sizes from three repeats. The bud sizes for GM118:1 were significantly larger than that of GM1Ovine in the presence or absence of cholesterol (P < 0.05), but no other statistical differences were present. (C) Histograms of the distribution of D from single-molecule trajectories of CTxB bound to planar membranes containing the indicated GM1 species or a combination of ovine GM1 and 30 mol % cholesterol. The CTxB diffusion slowed with increasing length and saturation of the GM1 acyl tails or in the presence of cholesterol.
Cholesterol levels strongly regulate endocytosis of CTxB (33). We thus wondered whether cholesterol levels might control the ability of CTxB to induce membrane curvature. To test this, we added 30 mol % cholesterol to the supported bilayers. The bud radii obtained in samples with and without cholesterol were 50 ± 14 nm and 57 ± 15 nm via dSTORM and 54 ± 14 nm and 58 ± 15 nm via pPLM, respectively, when 1.7 nM CTxB and 0.3 mol % GM1Ovine were used (Fig. 4). Just as in the absence of cholesterol, in the presence of cholesterol, higher CTxB concentrations resulted in less membrane bending (SI Appendix, Fig. S7). Thus, neither the ceramide structure nor cholesterol inclusion impacted the capacity of CTxB to induce curvature in artificial membranes under the conditions of our experiments.
Diffusion coefficient of CTxB when bound to GM116:1, GM118:1, GM118:0, or GM1Ovine or in the presence of cholesterol was 0.29 ± 0.27, 0.25 ± 0.23, 0.22 ± 0.21, 0.21 ± 0.17, and 0.081 ± 0.08 μm2/s, respectively. The trend observed in the distribution of diffusion coefficients is consistent with slower diffusion for longer or more saturated acyl tails or by increasing acyl tail ordering through the addition of cholesterol (Fig. 4C).
GM1 Binding Stoichiometry Affects Membrane Bending in Cells.
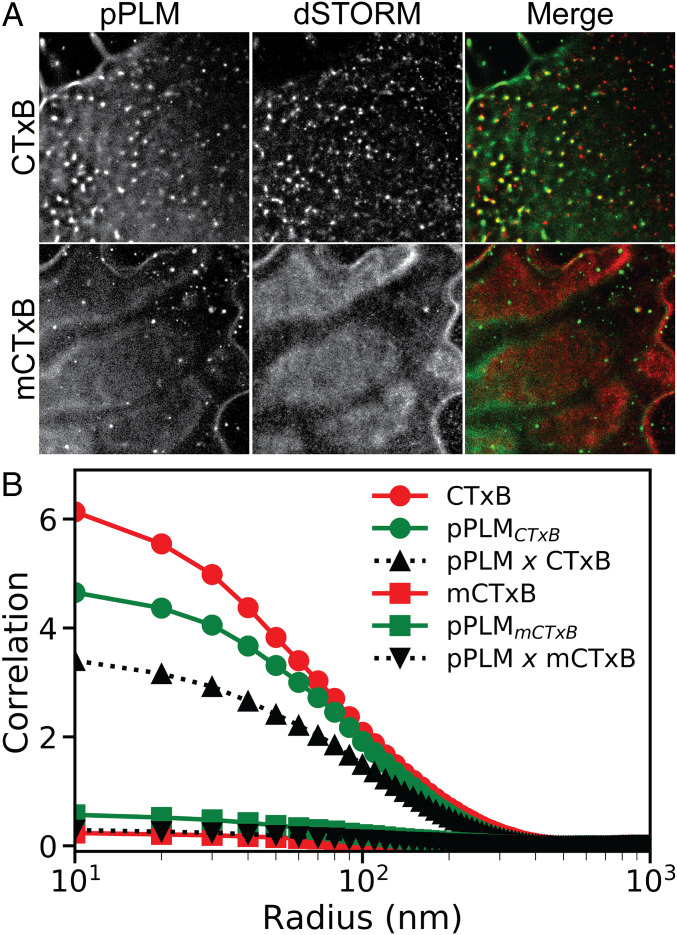
Finally, we tested whether the requirement for multivalent binding by CTxB to induce curvature is also conserved in cellular membranes. We assayed for curvature induction upon CTxB binding in COS-7 cells using PLM (28); 1.7 nM CTxB or mCTxB was applied to live COS-7 cells stained with DiI (Fig. 5). Cells labeled with CTxB displayed punctate accumulations of DiI and CTxB, with a mean bud radius of 220 ± 160 nm and 260 ± 150 nm as detected by pPLM and dSTORM, respectively. There were no apparent puncta in cells labeled with both DiD and mCTxB, indicating that multivalent GM1 binding was also required to generate membrane curvature in cells.
Fig. 5.
CTxB clusters and induces curvature in the plasma membranes of living cells, whereas mCTxB undergoes minimal clustering and fails to generate curvature in cell membranes. COS-7 cells were labeled with DiI or DiD and 1.7 nM CTxB or mCTxB. (A) They were subsequently imaged using pPLM and dSTORM. Merged images show areas where clustered CTxB or mCTxB (red) colocalize with sites of induced curvature (green). (Scale bar, 2 μm.) (B) Spatial autocorrelations were performed to quantify the magnitude of the clustering of CTxB (red circles) or mCTxB (red squares) and the magnitude of nanoscale membrane budding induced by either CTxB (green circles) or mCTxB (green squares). Cross-correlation analysis was performed to quantify the magnitude of the sorting of CTxB (black triangles) or mCTxB (black inverted triangles) to sites of membrane bending. Cells labeled with CTxB show an increase in nanoscale membrane bending that is spatially correlated to the CTxB clustering, whereas those labeled with mCTxB show minimal membrane curvature induction and negligible sorting of the mCTxB to curvature sites.
To quantify this difference, we performed spatial cross-correlation analysis and assessed the characteristic abundance, size, and colocalization of the punctate structures (Fig. 5B). Significant cross-correlation between pPLM and CTxB was observed, demonstrating that the protein localization was spatially coincident with curved membranes with a characteristic radius of 85 nm to 105 nm. In contrast, pPLM and mCTxB showed negligible cross-correlation. The amplitude of spatial autocorrelations of CTxB and pPLM after CTxB addition were 14-fold higher than those for mCTxB and pPLM after mCTxB addition. We conclude there is a substantial increase in nanoscopic membrane bending events when CTxB, but not mCTxB, is added to cells.
To confirm the differential membrane binding induced by CTxB and mCTxB binding to cells, single-molecule diffusion coefficients were again examined (SI Appendix, Fig. S8). On live cells, CTxB and mCTxB displayed D of 0.33 ± 0.40 and 0.17 ± 0.20 μm2/s, respectively.
Discussion
The clustering of lipids by AB5 toxins and SV40 virus is thought to induce compression of the lipids as a means of bending membranes (3, 4, 34) via a process linked to the geometry of receptor binding sites (16, 35). However, much of this process warrants further characterization, including the role of the clustering and leaflet asymmetry in the generation of curvature (36). In this study, we investigated the factors that drive membrane bending by CTxB, a prototypical cargo for clathrin-independent endocytosis, using superresolution PLM.
There are two important considerations that must be emphasized to understand our results. First, this curvature is induced on supported lipid bilayers (SLBs), which have adhesion to the underlying support that reduces the diffusion of lipid domains, imposes membrane tension, and reduces shape fluctuations (37). Our method of creating SLB patches via GUV fusion facilitates membrane bending by apparently trapping the center of the patch with a lower tension and less adhesion than the perimeter of the patch (i.e., Fig. 1B) (17). Secondly, CTxB is exposed to the distal side of the SLB such that membrane curvature can only form by bending the membrane away from the substrate and toward the CTxB. This direction of bending is the opposite of that observed on cells and GUVs (i.e., SI Appendix, Fig. S1) but potentially consistent with the preference for CTxB to bind to negative principal curvatures (SI Appendix, Fig. S9) (17). The SLBs used in this study provide control of the lipid and protein constituents such that the mechanisms and stoichiometric dependence CTxB membrane bending could be ascertained.
Our data support the hypothesis that toxin-induced glycolipid cross-linking induces membrane curvature in both model membranes and cellular environments. We also show that the multivalent binding of GM1 by CTxB is necessary for the membrane curvature and experimentally demonstrate the importance of GM1-to-CTxB stoichiometry in the membrane bending capability of CTxB. Importantly, lipid scaffolding on its own is not sufficient to induce membrane bending, and more dramatic membrane shape changes occurred at lower CTxB concentrations. Thus, the mechanism by which CTxB induces curvature likely does not arise from protein crowding effects (38) but does arise as a consequence of the spatial organization of glycolipids induced by multivalent binding to pentavalent CTxB.
We also determined the likely stoichiometry of GM1 binding to CTxB by fitting the histogram of the diffusion rate to a sum of log-normal Gaussians (Eq. 1). This single-particle diffusion analysis directly reports on the number of GM1s bound to CTxB and enables correlation of membrane bending activity to structured lipid cross-linking. While mCTxB engineered with a single binding site had only a single population of diffusers, wtCTxB had multiple species at the concentrations tested. Increasing the wtCTxB concentration resulted in faster wtCTxB diffusion, allowing us to exclude CTxB aggregation as a likely mechanism underlying our observations.
The distribution of D of wtCTxB was well fit with three populations representing CTxB bound to one, two, or three GM1 per CTxB (Fig. 3 and SI Appendix, Fig. S5). This interpretation is consistent with computational simulations (39), plasmonic nanocube membrane binding assays (40), flow cytometry (41), and mass spectrometry (42) that show concentration-dependent binding stoichiometries and that saturating the CTxB binding sites is unexpected for the concentrations used here. The CTxB concentration-dependent dissociation rate constants measured by flow cytometry show that 20 nM CTxB results in one GM1 per CTxB with an off rate of 5.2 min, and lower concentrations of CTxB displayed slower off rates (41). A single CTxB maintains a binding conformation for a longer duration than our single-molecule trajectories. Maximum membrane coverage occurred when ≤1 mol % GM1 was exposed to 20 nM CTxB (40, 41) and each GM1 was bound to a separate CTxB. Prior studies are consistent with our observed variations in GM1:CTxB ratios.
The diffusion rates observed here are consistent with the variety of previously published average diffusion rates for CTxB (17, 20, 43–49). The diffusion of mCTxB and the fastest CTxB diffusion are consistent with the diffusion of unbound GM1 (50), implying that monovalent CTxB binding provides no significant membrane disruption (i.e., curvature) or resistance to GM1 diffusion. A nonlinear slowing of the diffusion coefficient was observed upon increasing the stoichiometry of GM1 per CTxB. CTxB bound to one GM1 diffuses 34× faster than CTxB bound to three GM1. This slowdown with lipid binding is more than is expected by the free draining limit (51, 52) and reveals that increased GM1 per CTxB is correlated with membrane perturbations in addition to GM1 cross-linking (53). Slowed CTxB diffusion reduces the probability of saturating the GM1 binding sites, because protein and glycolipid diffusion affects their association rate, which is key in determining the observed binding stoichiometries for SV40 (9) and CTxB (54).
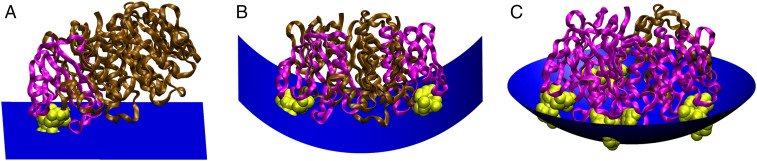
Based on these findings, we propose a model for how the stoichiometry of GM1 binding to CTxB can be mechanistically linked to curvature generation (Fig. 6 and SI Appendix, Fig. S9). With zero GM1 bound per CTxB, the CTxB is not membrane bound, and there is no curvature generation. With one GM1 bound, the CTxB is attached to the membrane, but there is no curvature generation (Fig. 6A). This single-bound GM1 conformation likely includes the CTxB sitting tilted on the membrane to juxtapose the single GM1 binding pocket close to the lipids with minimal membrane shape dependence, as observed previously in computational models (16, 39, 55, 56). With two GM1s bound, however, CTxB has a strong preference for negative curvature along one dimension and a weak preference for negative membrane curvature along the perpendicular dimension (Fig. 6B). Further increasing the GM1:CTxB ratio increasingly perturbs the membrane, giving rise to an enhanced preference toward negative curvature, which is more available on larger membrane buds (SI Appendix, Fig. S9).
Fig. 6.
Model for how the stoichiometry of binding of CTxB to GM1 generates membrane curvature. The ratio of CTxB subunits bound (pink) and unbound (brown) to GM1 (yellow) affects the shape of the membrane surface (blue). (A) Binding CTxB to a single GM1 has no effect on membrane curvature. (B) Binding CTxB to two GM1s generates negative curvature in one dimension, such as occurs at the neck of a membrane bud (SI Appendix, Fig. S9). (C) Binding of CTxB to three or more GM1s induces negative membrane curvature in two dimensions. This results in a wrapping of the membrane around the CTxB, such as within an endocytic pit. The schematics were created in Visual Molecular Dynamics (75) by building upon a crystal structure of CTxB (76).
Our results provide insights into the steps in the endocytosis and trafficking of CTx that rely on the ability of the toxin to induce curvature. CTxB can be internalized through different endocytic pathways (21, 25, 57), including via tubular clathrin-independent carriers generated in response to toxin binding (3, 4, 58–60). Our results support the notion that wt toxin is inherently capable of initiating membrane bending events and that curvature induction requires the toxin to bind multiple GM1. However, monovalent variants of CTxB are efficiently sorted into surface-attached endocytic tubules (58). Furthermore, “chimeric” toxins with a differential number of GM1 binding sites can undergo endocytosis, albeit less efficiently compared to wtCTx (61), and binding of CTxB to a single GM1 is sufficient to complete the intoxification pathway (8). Given that the ability of CTxB to induce curvature is dependent on multivalent binding to GM1, these findings suggest the CTxB-induced curvature is not required for sorting of toxin into nascent endocytic structures, entry of the toxin into cells by endocytosis, or directing its intracellular transport into pathways that leads to toxicity. However, the conditions under which CTxB induces curvature also enhance the efficacy of the toxin, perhaps due to the recruitment of ancillary host proteins that facilitate endocytosis (59, 60) or the increased affinity of the CTB−GM1 complex for curved membrane structures (i.e., sorting tubules or endocytic pits).
It is well known that the ceramide structure is important for intracellular lipid sorting pathways and influences the ability of AB5 toxins to bend membranes. Unsaturated GM1 is a better receptor for retrograde trafficking of CTx to the ER than saturated GM1 (32). In the case of another AB5 toxin, STx, the side chains of its glycolipid receptor are required to be unsaturated for tubulation in artificial vesicles (4). Furthermore, long-chain but not short-chain GM1 is essential for SV40 to generate membrane invaginations (3). Motivated by these findings, we tested the hypothesis that the ceramide structure of GM1 would be relevant to the initial membrane bending process using both synthesized and naturally occurring GM1 variants. Our experiments showed there was minimal change in membrane topography in model membranes containing GM1 variants. This result suggests that CTxB-induced membrane bending does not rely on the acyl chain identity of GM1, at least within the range of the ceramide structures tested in this study. Our experiments were performed only in artificial membranes, and we do not rule out the possibility that ceramide structure may impact the ability of CTxB to generate membrane curvature in the cell surface or intracellular membranes of intact cells. In phase-separated vesicles, for instance, ceramide tail structure is critical for raft partitioning of both un−cross-linked and CTxB−cross-linked GM1 (32). Thus, ceramide structure could impact the raft association of cross-linked GM1, which is credited with the membrane bending and internalization of CTxB in cells (62–65). Variation in the ceramide tail structure may alter the composition and acyl-tail order of GM1-rich nanoscopic domains (43, 66, 67). Accordingly, the spontaneous recruitment of liquid-order preferring lipids and the line tension surrounding the GM1-rich domain are likely to depend on the ceramide structure. Given the importance of ceramide structure in various physiological contexts, we hypothesize that the role of the GM1 ceramide domain is to recruit the downstream endocytic machinery, such as clathrin, caveolin, flotillin, or dynamin.
These results also have implications for the use of CTx as a marker for the liquid-ordered (raft) lipid phase. CTxB is not a nonperturbing marker for ordered domains; it can induce phase separation in membranes close to a demixing point (7, 63), stabilize ordered phases (7, 68), sort to already curved membranes (13, 17, 18), and induce membrane curvature (3, 17). Our current findings reveal that any measurements of the impact of CTxB on a membrane require precise control of the CTxB and GM1 concentrations, because the diffusive, sorting, and curvature-inductive properties of CTxB depend on its binding stoichiometry.
Materials and Methods
Custom GM1 Synthesis.
GM1 variants in the ceramide were synthesized in dimethyl sulfoxide (DMSO) solvent containing 1 μmol of lyso-GM1, 10 μmol of fatty acid, 10 μmol of dicyclohexyl-carbodiimid, and 10 μmol of sulfo-N-hydroxysuccinimide for 3 h at room temperature, as described previously (32). The DMSO was evaporated using a Speed Vac (Thermo Fisher Scientific) and purified following established methods via a silica column, preparative thin-layer chromatography plates, and reversed-phase Bond-Elut cartridges (69). The products were analyzed by electrospray ionization mass spectrometry with a quadrupole orthogonal time-of-flight instrument (Q-Star Pulsar I, Applied Biosystems).
SLB Preparation and Formation.
SLBs were made from bursting GUVs that included POPC (Avanti Polar Lipids), DiI (Life Technologies), and GM1. Commercially available Ovine GM1 (Avanti Polar Lipids) or GM1 with custom acyl tails were used. All SLBs were primarily POPC, with cholesterol (Sigma-Aldrich) only included when indicated. All SLBs included 0.3 mol % of DiI and DiD for CTxB-AF647 and mCTxB-AF488 experiments, respectively. For streptavidin-biotin binding experiments, GM1 was replaced with DPPE-Biotin (Avanti Polar Lipids).
GUVs were prepared by electroformation, as described previously (70). Briefly, a hydration solution of 200 mM sucrose was added to the dried lipids on an ITO slide. A chamber was created with a silicone frame and a second ITO plate enclosing the lipids and sucrose solution. The two ITO plates were connected to a sine wave function generator at 2 Vpeak-to-peak and 10 Hz for 3 h at 50 °C.
wtCTxB and mCTxB.
Alexa Fluor 647-wtCTxB (CTxB) was obtained from Invitrogen. Alexa Fluor 488-mCTxB was made as described previously (8, 62). Briefly, an E. coli expression strain containing the plasmids encoding native CTxB-G33D (nonbinding mutant) and C-terminal GS-H6−tagged wtCTxB was induced. Each subunit was secreted to the periplasm where mixed pentamers assembled. The mixture of assembled pentamers was purified from a cell extract by Talon affinity chromatography and separated into individual species by ion-exchange chromatography. Further purification was performed by cationic resin, HQ20 anion exchange column, and anion exchange prior to buffer exchange into phosphate-buffered saline by ultrafiltration using an Amicon Ultra-4 (Millipore) 10-K cutoff centrifugal filter. Labeling was performed by reacting Alexa Fluor 488-succinimidyl ester (Invitrogen) to 300 µg of mCTxB in 100 mM sodium bicarbonate buffer pH 8.3 for 1 h under stirring at room temperature and purified using provided size exclusion chromatography resin. DiI replaced DiD in the experiments on model membranes with the mCTxB-AF488 addition to decrease channel cross-talk. Model membranes were labeled by CTxB and mCTxB with 0.5 min of incubation, after which the unbound CTxB or mCTxB was rinsed away. The resulting membrane budding showed no change 20 min after CTxB was added; imaging was performed ∼60 min after CTxB addition.
Cell Labeling.
DiI or DiD (Sigma-Aldrich) was dissolved in ethanol at 1 mM. Cells were rinsed twice with Hepes-based cell imaging buffer (Sigma-Aldrich), and the 1-mM DiI-C18 stock solution was added to COS-7 cells in Hepes at 1:100 dilution for 10 s prior to washing with excess Hepes buffer. Cells were exposed to 1.7 nM CTxB-AF647 or mCTxB-AF488 for 5 min at room temperature prior to rinsing twice with Hepes imaging buffer prior. Seven individual cells were analyzed per condition.
Anti-CTxB Antibody Addition.
The mCTxB cross-linking was induced by the addition of anti-CTxB antibodies (Calbiochem, 227040 goat pAb). Antibodies were added to the membrane at the 90-min time point following mCTxB addition at final dilutions of 1:10,000, 1:1,000, 1:500, and 1:100 and allowed to incubate for 30 min prior to imaging.
Imaging Optics.
Dual-color imaging was performed with a custom IX83 (Olympus). The excitation light polarization was controlled by a liquid crystal wave plate (LCC1111-A; Thorlabs). The excitation lasers were steered through a periscope and a four-band notch filter (ZET405/488/561/640m; Chroma) onto the back of a 100×, 1.49 numerical aperture objective (Olympus). The emitted light passes through emission filters (BrightLine single-band band-pass filters; Semrock) within an OptoSplit with a 2.5× magnification lens (Cairn Research) before being collected by an iXon-897 Ultra EMCCD camera (Andor Technology).
Samples were exposed to high laser power (>80 mW) with λex = 488 nm (mCTxB-AF488), λex = 561 nm (DiI), or λex = 647 nm (CTxB-AF647 or DiD) to provide a steady rate of photoswitching by the fluorescent probes. Sample imaging was performed 90 min after CTxB addition; 40,000 frames of blinking fluorophores were acquired for each two-color channel with an 18-ms exposure time at a 50-Hz frame rate.
Imaging Buffer.
PLM and dSTORM were performed on SLBs surrounded by an oxygen-scavenging buffer (150 mM NaCl, 50 mM Tris, 0.5 mg/mL glucose oxidase, 20 mg/mL glucose, 40 μg/mL catalase, and 1% β-mercaptoethanol at pH 8). This buffer encourages the reversible blinking of fluorophores in a low oxygen concentration. Buffer proteins were purchased from Sigma-Aldrich, and salts were purchased from Thermo Fisher Scientific.
Single-Molecule Localization Microscopy.
Single-fluorophore localizations were identified by the ImageJ plugin ThunderSTORM using bilinear interpolation to achieve subpixel precision (71). Single-fluorophore localizations with intensities of <100 photons or location uncertainty of >45 nm were excluded from the analysis. The separate channel images were overlaid via a custom-made MATLAB routine (The MathWorks) by aligning the TetraSpeck nanoparticles (100-nm diameter; Life Technologies) that were visible in all channels and maximizing the image cross-correlation. The bright TetraSpecks localizations reduced the probability of finding a single fluorophore in their immediate proximity. The bright TetraSpeck localizations were hidden from the reconstructed images for clarity and removed prior to analysis of the single-molecule data.
Bud Identification and Size Evaluation.
The detection of membrane buds in each color channel was performed by a custom-made MATLAB program that masks regions with a low local density of localizations to identify regions of curvature as described previously (28). Two-dimensional (2D) Gaussian fitting for small buds (<150 nm) and 2D toroidal fitting for buds (>150 nm) that had a ring shape were performed for center estimation. The size of the bud was calculated by evaluating the mean distance of all localizations at the bud location while considering the additional localizations from the surrounding flat membrane, as done previously (28). The number of localizations expected within a radius R if no bud was present (NSLB) is NSLB = ρπR2 = Nall − Nbud, where ρ is the uniform density of localizations on a flat membrane, and Nbud is the number of extra localizations due to the presence of the bud. The mean distance of the localizations from the center of the bud expected for the flat SLB within R is 2R/3. By analyzing all collected localizations within R and subtracting the expected localizations from the flat SLB, the size of a single budding event (rbud) was calculated according to
| [3] |
Single-Particle Tracking.
Single fluorophores were analyzed to reveal the diffusion rates of the lipid and toxin. Blinking fluorophores were linked based on their proximal distance in consecutive frames via u-track (72), and single trajectories that lasted ≥15 frames were further analyzed. The slopes of the single-molecule MSD vs. Δt for Δt from two to four frames were fit to obtain the single-molecule diffusion rates according to
| [4] |
The localization imprecision (s) and the motion blur coefficient (B) were equal to 15 nm and 1/6, respectively, for the imaging parameters used here (73).
Image Correlation Analysis.
Spatial cross-correlations of two images, I1 and I2, were computationally calculated according to Eq. 5 and azimuthally averaged, as done previously (74).
| [5] |
FFT represents the fast Fourier transform, conj represents the complex conjugate, and Re represents taking only the real component. The correlations are normalized by the average localization density (ρ) and the azimuthally averaged autocorrelation of the region of interest (W). The autocorrelation of an image, I1, was calculated from Eq. 5 by setting I2 = I1.
Data Availability.
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Eric Stimpson, KeVaughna Patrick, Sonali Gandhi, Meshari Al Rafidi, Xin Xin Woodward, and Stephanie Schmieder for helpful discussions and comments. A.M.K. was funded by a Thomas C. Rumble Fellowship Award. Financial support was provided by Wayne State University laboratory start-up funds and the Richard J. Barber Interdisciplinary Research Program. K.R. was supported, in part, by Children’s Hospital of Pittsburgh of the University of Pittsburgh Medical Center Health System. This material is based upon work supported by the NSF under Grant DMR-1652316 and the National Institute of General Medical Sciences of the NIH under Award R01GM106720. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.J. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001119117/-/DCSupplemental.
References
- 1.Ali M., Nelson A. R., Lopez A. L., Sack D. A., Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9, e0003832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beddoe T., Paton A. W., Le Nours J., Rossjohn J., Paton J. C., Structure, biological functions and applications of the AB5 toxins. Trends Biochem. Sci. 35, 411–418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewers H. et al., GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12, 11–18 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Römer W. et al., Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Pezeshkian W. et al., Membrane invagination induced by Shiga toxin B-subunit: From molecular structure to tube formation. Soft Matter 12, 5164–5171 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Pezeshkian W. et al., Mechanism of Shiga toxin clustering on membranes. ACS Nano 11, 314–324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghunathan K. et al., Glycolipid crosslinking is required for cholera toxin to partition into and stabilize ordered domains. Biophys. J. 111, 2547–2550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jobling M. G., Yang Z., Kam W. R., Lencer W. I., Holmes R. K., A single native ganglioside GM1-binding site is sufficient for cholera toxin to bind to cells and complete the intoxication pathway. MBio 3, e00401-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk O. M. et al., Receptor concentration and diffusivity control multivalent binding of Sv40 to membrane bilayers. PLoS Comput. Biol. 9, e1003310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wernick N. L. B., Chinnapen D. J.-F., Cho J. A., Lencer W. I., Cholera toxin: An intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins 2, 310–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinnapen D. J.-F., Chinnapen H., Saslowsky D., Lencer W. I., Rafting with cholera toxin: Endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol. Lett. 266, 129–137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day C. A., Kenworthy A. K., Tracking microdomain dynamics in cell membranes. Biochim. Biophys. Acta 1788, 245–253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian A., Baumgart T., Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 96, 2676–2688 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh W.-T. et al., Curvature sorting of peripheral proteins on solid-supported wavy membranes. Langmuir 28, 12838–12843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaizuka Y., Groves J. T., Bending-mediated superstructural organizations in phase-separated lipid membranes. New J. Phys. 12, 95001 (2010). [Google Scholar]
- 16.Pezeshkian W., Nåbo L. J., Ipsen J. H., Cholera toxin B subunit induces local curvature on lipid bilayers. FEBS Open Bio 7, 1638–1645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabbani A. M., Kelly C. V., Nanoscale membrane budding induced by CTxB and detected via polarized localization microscopy. Biophys. J. 113, 1795–1806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu Y.-S. et al., Reconstituting ring-rafts in bud-mimicking topography of model membranes. Nat. Commun. 5, 4507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu Y.-S. et al., Curvature elasticity-driven leaflet asymmetry and interleaflet raft coupling in supported membranes. Adv. Mater. Interfaces 5, 1801290 (2018). [Google Scholar]
- 20.Day C. A., Kenworthy A. K., Mechanisms underlying the confined diffusion of cholera toxin B-subunit in intact cell membranes. PLoS One 7, e34923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torgersen M. L., Skretting G., van Deurs B., Sandvig K., Internalization of cholera toxin by different endocytic mechanisms. J. Cell Sci. 114, 3737–3747 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Singh R. D. et al., Selective caveolin-1-dependent endocytosis of glycosphingolipids. Mol. Biol. Cell 14, 3254–3265 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen G. H. et al., Cholera toxin entry into pig enterocytes occurs via a lipid raft- and clathrin-dependent mechanism. Biochemistry 44, 873–882 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Broeck D. V., Lagrou A. R., De Wolf M. J., Distinct role of clathrin-mediated endocytosis in the functional uptake of cholera toxin. Acta Biochim. Pol. 54, 757–767 (2007). [PubMed] [Google Scholar]
- 25.Pang H., Le P. U., Nabi I. R., Ganglioside GM1 levels are a determinant of the extent of caveolae/raft-dependent endocytosis of cholera toxin to the Golgi apparatus. J. Cell Sci. 117, 1421–1430 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Ewers H., Helenius A., Lipid-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 3, a004721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannes L., Parton R. G., Bassereau P., Mayor S., Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 16, 311–321 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Kabbani A. M., Kelly C. V., The detection of nanoscale membrane bending with polarized localization microscopy. Biophys. J. 113, 1782–1794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Linde S. et al., Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Manley S., Horton M. R., Lecszynski S., Gast A. P., Sorting of streptavidin protein coats on phase-separating model membranes. Biophys. J. 95, 2301–2307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxton M. J., Single-particle tracking: The distribution of diffusion coefficients. Biophys. J. 72, 1744–1753 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chinnapen D. J.-F. et al., Lipid sorting by ceramide structure from plasma membrane to ER for the cholera toxin receptor ganglioside GM1. Dev. Cell 23, 573–586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orlandi P. A., Fishman P. H., Filipin-dependent inhibition of cholera toxin: Evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 141, 905–915 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watkins E. B. et al., Shiga toxin induces lipid compression: A mechanism for generating membrane curvature. Nano Lett. 19, 7365–7369 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Johannes L., Shiga toxin-A model for glycolipid-dependent and lectin-driven endocytosis. Toxins 9, 340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dasgupta R., Miettinen M. S., Fricke N., Lipowsky R., Dimova R., The glycolipid GM1 reshapes asymmetric biomembranes and giant vesicles by curvature generation. Proc. Natl. Acad. Sci. U.S.A. 115, 5756–5761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert U., Lipowsky R., Adhesion of vesicles. Phys. Rev. A 42, 4768–4771 (1990). [DOI] [PubMed] [Google Scholar]
- 38.Stachowiak J. C. et al., Membrane bending by protein-protein crowding. Nat. Cell Biol. 14, 944–949 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Basu I., Mukhopadhyay C., Insights into binding of cholera toxin to GM1 containing membrane. Langmuir 30, 15244–15252 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Worstell N. C., Krishnan P., Weatherston J. D., Wu H.-J., Binding cooperativity matters: A GM1-like ganglioside-cholera toxin B subunit binding study using a nanocube-based lipid bilayer array. PLoS One 11, e0153265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauer S., Goldstein B., Nolan R. L., Nolan J. P., Analysis of cholera toxin-ganglioside interactions by flow cytometry. Biochemistry 41, 1742–1751 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Lin H., Kitova E. N., Klassen J. S., Measuring positive cooperativity using the direct ESI-MS assay. Cholera toxin B subunit homopentamer binding to GM1 pentasaccharide. J. Am. Soc. Mass Spectrom. 25, 104–110 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Štefl M. et al., Dynamics and size of cross-linking-induced lipid nanodomains in model membranes. Biophys. J. 102, 2104–2113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang M., Day C. A., Kenworthy A. K., DiBenedetto E., Simplified equation to extract diffusion coefficients from confocal FRAP data. Traffic 13, 1589–1600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenworthy A. K. et al., Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165, 735–746 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bacia K., Scherfeld D., Kahya N., Schwille P., Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys. J. 87, 1034–1043 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moens P. D. J., Digman M. A., Gratton E., Modes of diffusion of cholera toxin bound to GM1 on live cell membrane by image mean square displacement analysis. Biophys. J. 108, 1448–1458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spillane K. M. et al., High-speed single-particle tracking of GM1 in model membranes reveals anomalous diffusion due to interleaflet coupling and molecular pinning. Nano Lett. 14, 5390–5397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh C.-L., Spindler S., Ehrig J., Sandoghdar V., Tracking single particles on supported lipid membranes: Multimobility diffusion and nanoscopic confinement. J. Phys. Chem. B 118, 1545–1554 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Goins B., Masserini M., Barisas B. G., Freire E., Lateral diffusion of ganglioside GM1 in phospholipid bilayer membranes. Biophys. J. 49, 849–856 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gambin Y. et al., Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. U.S.A. 103, 2098–2102 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knight J. D., Lerner M. G., Marcano-Velázquez J. G., Pastor R. W., Falke J. J., Single molecule diffusion of membrane-bound proteins: Window into lipid contacts and bilayer dynamics. Biophys. J. 99, 2879–2887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziemba B. P., Falke J. J., Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids 172–173, 67–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee D., Mohr A., Kwon J. S.-I., Wu H.-J., Kinetic Monte Carlo modeling of multivalent binding of CTB proteins with GM1 receptors. Comput. Chem. Eng. 118, 283–295 (2018). [Google Scholar]
- 55.Sridhar A., Kumar A., Dasmahapatra A. K., Multi-scale molecular dynamics study of cholera pentamer binding to a GM1-phospholipid membrane. J. Mol. Graph. Model. 68, 236–251 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Rissanen S. et al., Phase partitioning of GM1 and its bodipy-labeled analog determine their different binding to cholera toxin. Front. Physiol. 8, 252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massol R. H., Larsen J. E., Fujinaga Y., Lencer W. I., Kirchhausen T., Cholera toxin toxicity does not require functional Arf6- and dynamin-dependent endocytic pathways. Mol. Biol. Cell 15, 3631–3641 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Day C. A. et al., Microtubule motors power plasma membrane tubulation in clathrin-independent endocytosis. Traffic 16, 572–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renard H.-F. et al., Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 517, 493–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boucrot E. et al., Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Wolf A. A. et al., Attenuated endocytosis and toxicity of a mutant cholera toxin with decreased ability to cluster ganglioside GM1 molecules. Infect. Immun. 76, 1476–1484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Day C. A., Kenworthy A. K., Functions of cholera toxin B-subunit as a raft cross-linker. Essays Biochem. 57, 135–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammond A. T. et al., Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 102, 6320–6325 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lingwood D., Ries J., Schwille P., Simons K., Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. U.S.A. 105, 10005–10010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johannes L., Mayor S., Induced domain formation in endocytic invagination, lipid sorting, and scission. Cell 142, 507–510 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Yuan C., Furlong J., Burgos P., Johnston L. J., The size of lipid rafts: An atomic force microscopy study of ganglioside GM1 domains in sphingomyelin/DOPC/cholesterol membranes. Biophys. J. 82, 2526–2535 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun H., Chen L., Gao L., Fang W., Nanodomain formation of ganglioside GM1 in lipid membrane: Effects of cholera toxin-mediated cross-linking. Langmuir 31, 9105–9114 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Johnson S. A. et al., Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim. Biophys. Acta 1798, 1427–1435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Panasiewicz M., Domek H., Hoser G., Kawalec M., Pacuszka T., Structure of the ceramide moiety of GM1 ganglioside determines its occurrence in different detergent-resistant membrane domains in HL-60 cells. Biochemistry 42, 6608–6619 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Veatch S., “Electro-formation and fluorescence microscopy of giant vesicles with coexisting liquid phases” in Lipid Rafts, McIntosh T., Ed. (Methods in Molecular Biology, Humana, 2007), Vol. 398, pp. 59–72. [DOI] [PubMed] [Google Scholar]
- 71.Ovesný M., Křížek P., Borkovec J., Svindrych Z., Hagen G. M., ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jaqaman K. et al., Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berglund A. J., Statistics of camera-based single-particle tracking. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82, 11917 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Veatch S. L. et al., Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7, e31457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 76.Merritt E. A. et al., Structural studies of receptor binding by cholera toxin mutants. Protein Sci. 6, 1516–1528 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.