Abstract
These data are related to the research article entitled “Induction of CCK and GLP-1 release in enteroendocrine cells by egg white peptides generated during gastrointestinal digestion”. In this article, the peptide and free amino acid profile of egg white gastrointestinal in vitro digests is shown. Egg white proteins were digested following the INFOGEST gastrointestinal digestion protocol. Different time points of gastric and intestinal digestion were characterized regarding protein, peptide and amino acid content. Protein degradation was followed by SDS-PAGE where some electrophoretic bands were identified by MALDI-TOF/TOF after tryptic digestion. Moreover, the molecular weight distribution of egg white peptides found at different times of gastrointestinal digestion was performed using MALDI-TOF. Peptides identified from the most abundant egg white proteins by tandem mass spectrometry were represented using a peptide profile tool and raw data are given in table format. These results reveal the protein regions resistant to digestion and illustrate the free amino acid profile of egg white protein at the end of the digestion process. These data can be used for nutritional purposes and to identify allergen epitopes or bioactive sequences.
Keywords: Egg white protein, Characterization, Mass spectrometry, Maldi-tof, Peptidomics
Specifications Table
| Subject area | Biochemistry |
|---|---|
| More specific subject area | Proteomics and biochemistry |
| Type of data | Figures, Tables |
| How data was acquired | High-pressure liquid chromatography coupled to electron spray ionization interface and ion trap. Matrix assisted Laser Desorption/Ionization coupled to a time of flight detector |
| Data format | Raw Analyzed |
| Parameters for data collection | Digested samples following INFOGEST protocol were freeze-dried and kept at −20 °C until analysis. Digests were analyzed by mass spectrometry after a reducing step with dithiothreitol. |
| Description of data collection | MS/MS raw files were processed by using Data Analysis (version 4.0 Bruker Daltonics) and Biotools version 3.2, and the identification search was achieved using Mascot v2.4 |
| Data source location | Data is collected and analysed at the Institute of Food Science Research, CIAL (CSIC-UAM). Nicolás Cabrera 9, 28049, Madrid, Spain |
| Data accessibility Related research article |
Data in this article M. Santos-Hernández, L. Amigo, Recio, I. “Induction of CCK and GLP-1 release in enteroendocrine cells by egg white peptides generated during gastrointestinal digestion”. |
Value of the data
-
•
The data provide the distribution of the nitrogen fraction into peptides and amino acids at the end of the gastrointestinal digestion of egg white. The profile of free amino acids that can be used for nutritional purposes is also given.
-
•
The here provided proteomic, peptidomic and amino acid profiles of egg white protein digests can be compared with in vivo data or with data obtained in dynamic systems.
-
•
Egg white protein domains resistant to gastrointestinal digestion are provided which could serve to detect allergen epitopes or peptides with biological activities.
-
•
This peptidomic characterization was useful to identify peptides as inducers of incretin hormones, being relevant to control food intake and diabetes.
1. Data description
Egg white protein was digested following a harmonized in vitro digestion protocol [1,2] where samples were taken at 30 and 120 min of gastric digestion and 30 and 120 min of gastrointestinal digestion. These digests were centrifuged at 5000 × g over 20 min to separate soluble and insoluble fraction, followed by snap freezing in liquid nitrogen and freeze-dried. All digests were characterized regarding their protein, peptide and amino acid composition.
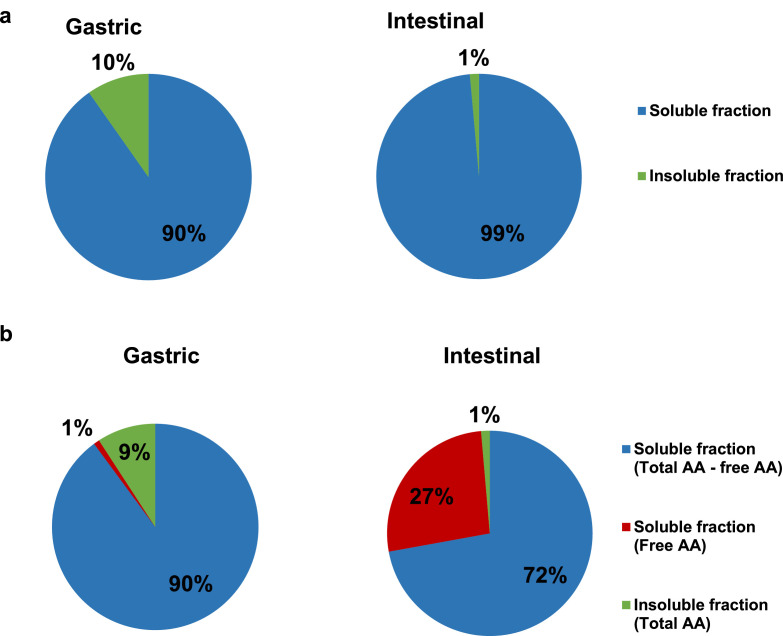
The distribution of the nitrogen fraction after egg white in vitro gastric and intestinal digestion was assessed by elemental analysis (Fig. 1a) and amino acid analysis (Fig. 1b). Around 90% of the total nitrogen content was collected in the soluble fraction after gastric digestion, determined by elemental analysis, and this percentage increased up to 99% at the end of the intestinal phase (Fig. 1a). Similar percentages were obtained by total amino acid analysis where the soluble fraction represented 91% and 99% at the end of the gastric and the intestinal phase, respectively (Fig. 1b). At the end of the gastric phase, just 1% of the total nitrogen fraction was in the form of free amino acids, increasing up to 27% at the end of the intestinal phase. The difference between total and free amino acids in the soluble part corresponded to the nitrogen fraction in the form of proteins and peptides which ranged from 90% at the end of the gastric phase to 72% at the end of the intestinal phase.
Fig. 1.
Distribution of the nitrogen content between soluble and insoluble fraction after gastric and intestinal digestion. Digests were centrifuged at 5000 × g for 20 min. Supernatant and pellet were freeze-dried and weighted. Nitrogen content in each fraction was determined by a) elemental analysis and b) amino acid analysis. Total and free amino acids were separately determined in the soluble fraction.
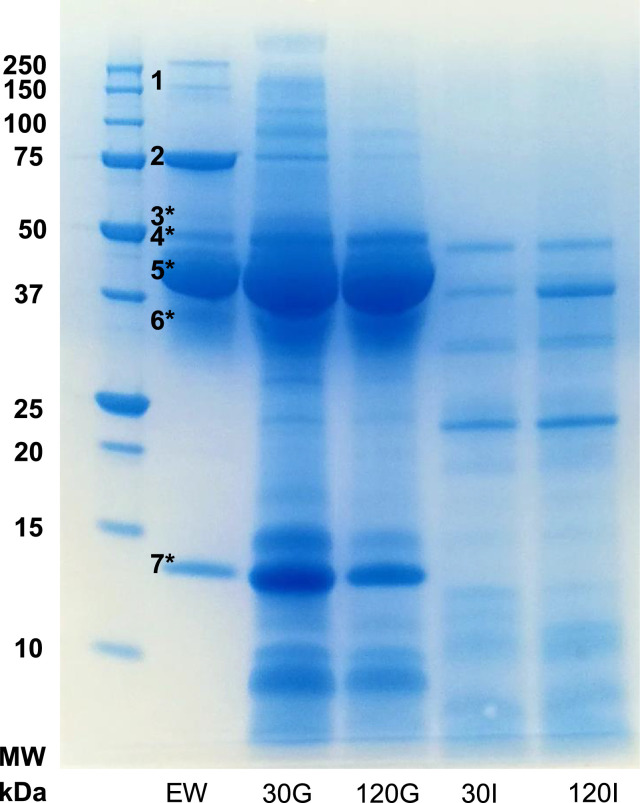
Protein degradation during egg white gastrointestinal digestion was followed by SDS-PAGE (Fig. 2). At the end of the gastric phase, main electrophoretic bands were identified as ovalbumin and ovalbumin-related protein Y and these were still detected at the end of the intestinal phase. Electrophoretic bands with * in Fig. 2 were identified by MALDI-TOF/TOF after in-gel digestion with trypsin.
Fig. 2.
SDS-PAGE protein profiles of egg white protein at different times of in vitro gastrointestinal digestion. MW, molecular weight marker; EW, egg white undigested protein; 30 G and 120 G, 30 and 120 min gastric digestion; 30I and 120I, 30 and 120 min intestinal digestion. The numbers correspond to an identified band. 1, Ovomucin; 2, Ovotransferrin; 3, Ovoinhibitor; 4, Ovalbumin-Y; 5, Ovalbumin; 6, Ovomucoid; 7, Lysozyme. The electrophoretic bands with * were identified by MALDI-TOF/TOF after hydrolysis with trypsin.
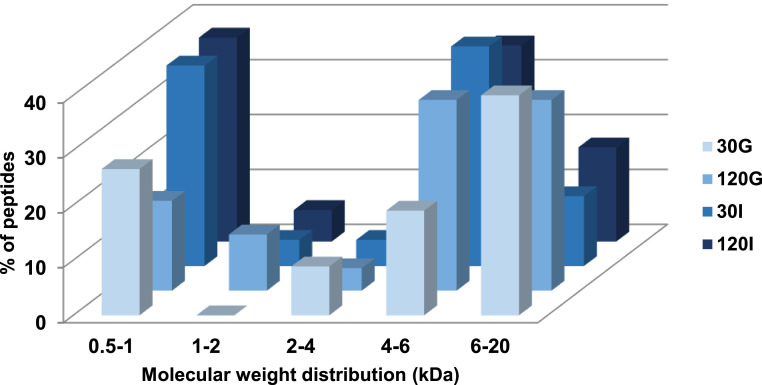
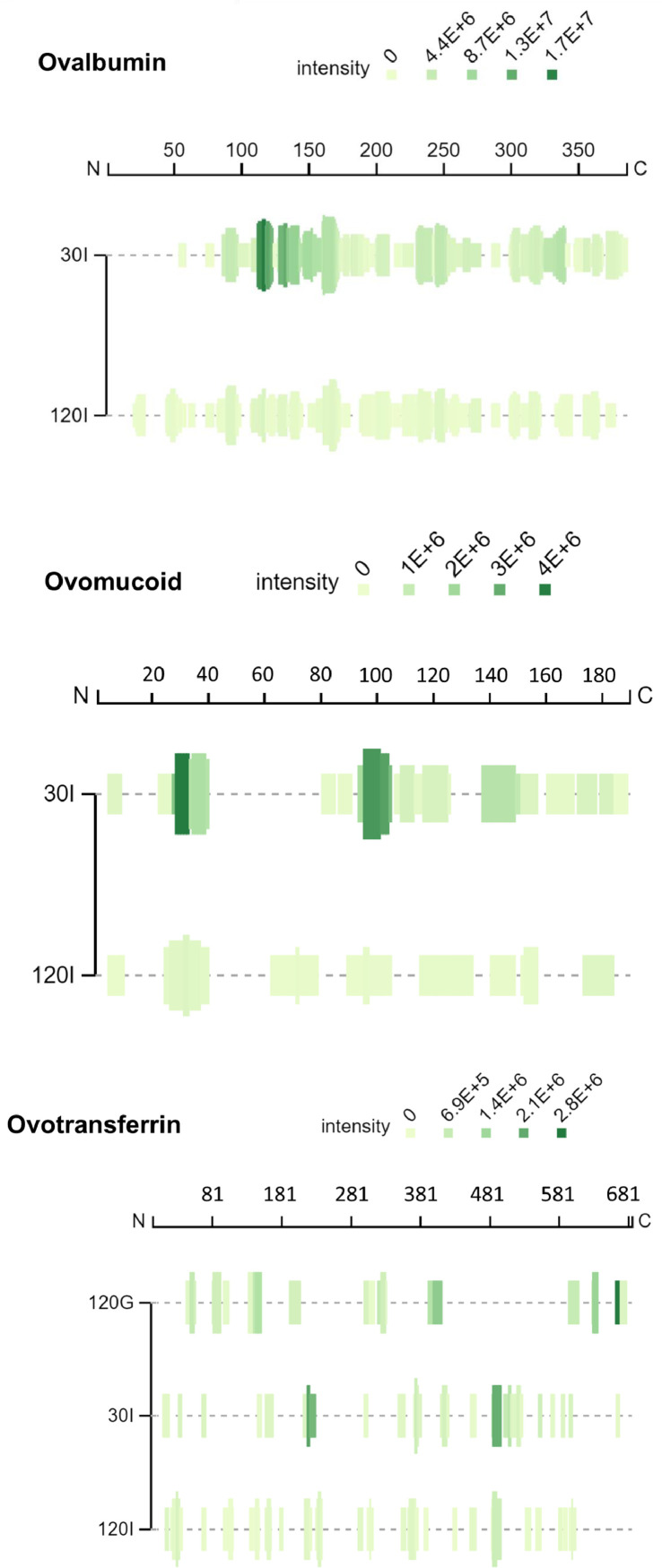
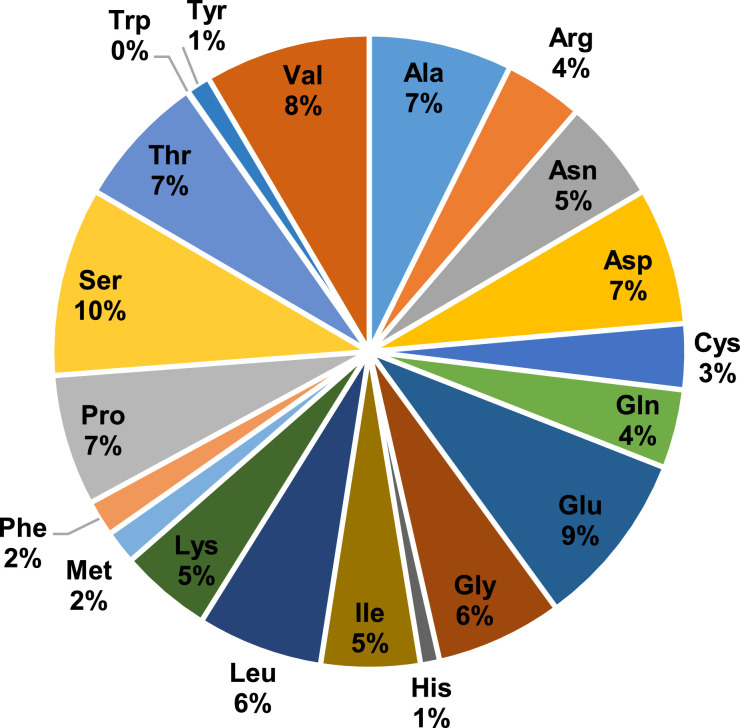
Egg white protein digests were also characterized by MALDI-TOF and the peptide profile is represented in Fig. 3, which describes the percentage of peptides within a given molecular weight range. As expected, peptides with a molecular weight lower than 2 kDa increased during gastrointestinal digestion and the amount of longer peptides decreased. However, it should be noted that peptides with a molecular weight higher than 6 kDa were still detected at the end of gastrointestinal digest. Peptidomic characterization of the digests was performed by HPLC tandem mass spectrometry (HPLC-MS/MS). Raw data of peptide sequences identified at the end of the gastrointestinal digestion by HPLC-MS/MS are given in Tables 1 to 5. The identified peptides from the major protein fractions, ovalbumin, ovomucoid and ovotransferrin, at different time points during gastrointestinal digestion, are also represented using the peptide profile tool from the Peptigram web-based application (Fig. 4). In these graphs, each vertical bar corresponds to an amino acid identified as part of a peptide sequence, the height of the bar is proportional to the number of peptides overlapping this position and the color intensity is proportional to the sum of the intensities of the peptides overlapping a given position. Under our mass spectrometry conditions, peptides with a molecular weight between 5 and 30 kDa are detected. The blank regions observed in the peptide profile during the gastric phase probably correspond to peptide fragments too long to be solubilized or ionized under our analysis conditions, while blank regions in the intestinal phase are more likely due to short peptides, free amino acids, or peptides with low ionization capacity. It has to be noted that peptide intensity depends on peptide ionization capacity and abundancy, and in consequence, peptide intensity cannot directly be transformed to peptide concentration. During the gastric phase, only peptides from ovotransferrin were identified, suggesting a higher susceptibility of this protein to the action of pepsin. Several ovalbumin and ovomucoid peptides were detected after 30 and 120 min intestinal digestion. In addition, the amino acid composition of the identified peptides was analysed by using the ExPASy-Protparam tool (Fig. 5). Resistant peptides identified at the end of intestinal digestion by mass spectrometry were rich in serine, valine and in negatively charges residues aspartic and glutamic acid.
Table 2.
Ovomucoid-derived peptides identified at the end of egg white gastrointestinal digestion.
| RANGEa | SEQUENCEb | |
|---|---|---|
| 1 | 6 | AEVDCS |
| 21 | 36 | VCNKDLRPICGTDGVT |
| 21 | 29 | VCNKDLRPI |
| 23 | 33 | NKDLRPICGTD |
| 28 | 36 | PICGTDGVT |
| 59 | 68 | DGECKETVPM |
| 68 | 75 | MNCSSYAN |
| 86 | 93 | LCNRAFNP |
| 92 | 101 | NPVCGTDGVT |
| 137 | 145 | DCSEYPKPD |
| 148 | 153 | AEDRPL |
| 149 | 153 | EDRPL |
| 170 | 180 | AVVESNGTLTL |
Range in the mature form of the protein. Uniprot accession number: P01005.
One letter amino acid code is used.
Table 3.
Ovotransferrin-derived peptides identified at the end of egg white gastrointestinal digestion.
| RANGEa | SEQUENCEb | RANGEa | SEQUENCEb | ||
|---|---|---|---|---|---|
| 13 | 17 | SSPEE | 304 | 308 | AIMLK |
| 21 | 30 | NNLRDLTQQE | 308 | 312 | KRVPS |
| 24 | 37 | RDLTQQERISLTCV | 353 | 359 | DEKSKCD |
| 29 | 33 | QERIS | 363 | 373 | VVSNGDVECTV |
| 66 | 71 | EAGLAP | 365 | 372 | SNGDVECT |
| 97 | 110 | VVKKGTEFTVNDLQ | 373 | 377 | VVDET |
| 105 | 109 | TVNDL | 386 | 391 | KGEADA |
| 135 | 139 | RGAIE | 428 | 432 | PASYF |
| 142 | 147 | GIESGS | 452 | 460 | KSCHTAVGR |
| 143 | 148 | IESGSV | 483 | 490 | YFSEGCAP |
| 157 | 165 | SASCVPGAT | 484 | 496 | FSEGCAPGSPPNS |
| 160 | 164 | CVPGA | 485 | 496 | SEGCAPGSPPNS |
| 178 | 182 | PKTKC | 532 | 539 | VEKGDVAF |
| 214 | 220 | NENAPDQ | 547 | 554 | ENTGGKNK |
| 214 | 223 | NENAPDQKDE | 580 | 584 | DYREC |
| 230 | 238 | DGSRQPVDN | 587 | 591 | AEVPT |
| 232 | 236 | SRQPV | 595 | 599 | VVRPE |
| 234 | 238 | QPVDN | 599 | 604 | EKANKI |
| 291 | 297 | KDPVLKD | |||
Range in the mature form of the protein. Uniprot accession number: P02789.
One letter amino acid code is used.
Table 4.
Lysozyme-derived peptides identified at the end of egg white gastrointestinal digestion.
| RANGEa | SEQUENCEb | |
|---|---|---|
| 18 | 25 | DNYRGYSL |
| 34 | 44 | FESNFNTQATN |
| 39 | 43 | NTQAT |
| 44 | 52 | NRNTDGSTD |
| 45 | 52 | RNTDGSTD |
| 47 | 52 | TDGSTD |
| 64 | 72 | CNDGRTPGS |
| 72 | 83 | SRNLCNIPCSAL |
| 85 | 90 | SSDITA |
| 85 | 89 | SSDIT |
| 97 | 105 | KIVSDGNGM |
| 99 | 105 | VSDGNGM |
| 117 | 122 | GTDVQA |
Range in the mature form of the protein. Uniprot accession number: P00698.
One letter amino acid code is used.
Fig. 3.
Molecular weight distribution of egg white peptides found at different times of gastrointestinal digestion. 30 G and 120 G, 30 and 120 min of gastric digestion, respectively; 30I and 120I, 30 and 120 min intestinal digestion, respectively.
Table 1.
Ovalbumin-derived peptides identified at the end of egg white gastrointestinal digestion.
| RANGEa | SEQUENCEb | RANGEa | SEQUENCEb | ||
|---|---|---|---|---|---|
| 20 | 28 | KVHHANENI | 188 | 199 | AFKDEDTQAMPF |
| 22 | 28 | HHANENI | 190 | 198 | KDEDTQAMP |
| 23 | 28 | HANENI | 200 | 209 | RVTEQESKPV |
| 44 | 50 | LGAKDST | 200 | 211 | RVTEQESKPVQM |
| 44 | 52 | LGAKDSTRT | 200 | 204 | RVTEQ |
| 45 | 50 | GAKDST | 200 | 210 | RVTEQESKPVQ |
| 48 | 52 | DSTRT | 205 | 209 | ESKPV |
| 48 | 55 | DSTRTQIN | 213 | 217 | YQIGL |
| 54 | 58 | INKVV | 219 | 228 | RVASMASEKM |
| 61 | 65 | DKLPG | 220 | 229 | VASMASEKMK |
| 74 | 79 | CGTSVN | 220 | 228 | VASMASEKM |
| 82 | 97 | SSLRDILNQITKPNDV | 230 | 234 | ILELP |
| 83 | 90 | SLRDILNQ | 230 | 240 | ILELPFASGTM |
| 89 | 97 | NQITKPNDV | 230 | 242 | ILELPFASGTMSM |
| 89 | 95 | NQITKPN | 230 | 241 | ILELPFASGTMS |
| 89 | 99 | NQITKPNDVYS | 232 | 240 | ELPFASGTM |
| 90 | 95 | QITKPN | 232 | 241 | ELPFASGTMS |
| 91 | 95 | ITKPN | 243 | 250 | LVLLPDEV |
| 107 | 117 | YAEERYPILPE | 244 | 252 | VLLPDEVSG |
| 108 | 117 | AEERYPILPE | 244 | 250 | VLLPDEV |
| 113 | 117 | PILPE | 244 | 253 | VLLPDEVSGL |
| 116 | 124 | PEYLQCVKE | 246 | 250 | LPDEV |
| 116 | 125 | PEYLQCVKEL | 254 | 260 | EQLESII |
| 121 | 125 | CVKEL | 255 | 260 | QLESII |
| 127 | 134 | RGGLEPIN | 257 | 261 | ESIIN |
| 127 | 133 | RGGLEPI | 261 | 267 | NFEKLTE |
| 128 | 133 | GGLEPI | 269 | 277 | TSSNVMEER |
| 136 | 142 | QTAADQA | 271 | 277 | SNVMEER |
| 136 | 143 | QTAADQAR | 286 | 291 | MKMEEK |
| 137 | 142 | TAADQA | 299 | 303 | MAMGI |
| 137 | 145 | TAADQAREL | 299 | 306 | MAMGITDV |
| 150 | 161 | VESQTNGIIRNV | 300 | 306 | AMGITDV |
| 156 | 160 | GIIRN | 302 | 306 | GITDV |
| 161 | 165 | VLQPS | 308 | 321 | SSSANLSGISSAES |
| 161 | 172 | VLQPSSVDSQTA | 314 | 319 | SGISSA |
| 161 | 170 | VLQPSSVDSQ | 314 | 322 | SGISSAESL |
| 161 | 169 | VLQPSSVDS | 314 | 321 | SGISSAES |
| 161 | 173 | VLQPSSVDSQTAM | 333 | 345 | AEINEAGREVVGS |
| 163 | 170 | QPSSVDSQ | 336 | 345 | NEAGREVVGS |
| 166 | 170 | SVDSQ | 337 | 345 | EAGREVVGS |
| 166 | 174 | SVDSQTAMV | 354 | 358 | SVSEE |
| 166 | 173 | SVDSQTAM | 354 | 365 | SVSEEFRADHPF |
| 166 | 172 | SVDSQTA | 354 | 364 | SVSEEFRADHP |
| 176 | 180 | VNAIV | 360 | 364 | RADHP |
| 188 | 198 | AFKDEDTQAMP | 360 | 365 | RADHPF |
Range in the protein. Uniprot accession number: P01012.
One letter amino acid code is used.
Table 5.
Mucin-5B-derived peptides identified at the end of egg white gastrointestinal digestion.
| RANGEa | SEQUENCEb | RANGEa | SEQUENCEb | ||
|---|---|---|---|---|---|
| 34 | 38 | GRSEC | 1064 | 1073 | EACHSKVNPI |
| 98 | 102 | VILEV | 1165 | 1169 | GCYPE |
| 125 | 129 | IEDTC | 1216 | 1223 | YPLNETIY |
| 125 | 131 | IEDTCAY | 1235 | 1241 | FCGPNGM |
| 134 | 141 | VTSKLGLT | 1326 | 1330 | EALET |
| 147 | 154 | ADTLLLDL | 1384 | 1388 | CLGEE |
| 200 | 204 | EKCPD | 1528 | 1532 | GIRIT |
| 272 | 277 | CICSTL | 1580 | 1584 | KSDDA |
| 293 | 298 | EWRTKE | 1581 | 1590 | SDDARKRNGE |
| 376 | 380 | STPCQ | 1596 | 1600 | KEMAL |
| 433 | 437 | TFVVI | 1648 | 1660 | PPQPYYEACVASR |
| 453 | 457 | KNVLV | 1690 | 1694 | RGQTN |
| 454 | 458 | NVLVT | 1721 | 1729 | REVIVDTLL |
| 545 | 552 | FRTATGAV | 1744 | 1751 | PDGNILLN |
| 546 | 551 | RTATGA | 1827 | 1834 | TETVCECD |
| 550 | 562 | GAVEDSAAAFGNS | 1827 | 1834 | TETVCECD |
| 657 | 662 | QGICDP | 1880 | 1884 | KPGAV |
| 753 | 761 | DCIGETVLV | 1880 | 1887 | KPGAVVPK |
| 901 | 908 | DAGTFRIV | 1886 | 1896 | PKSSCEDCVCT |
| 923 | 928 | LKITLI | 1910 | 1914 | CVPVK |
| 1012 | 1019 | GQSVEMSI | 1913 | 1917 | VKCQT |
| 1044 | 1051 | QPFKSALG | |||
Range in the mature form of the protein Uniprot accession number: Q98UI9.
One letter amino acid code is used.
Fig. 4.
Peptides from ovalbumin, ovomucoid and ovotransferrin identified in egg white gastrointestinal digests represented using the Peptigram Bioware tool. Each vertical bar corresponds to an amino acid identified as part of a peptide sequence. The height of the bar is proportional to the number of the peptides overlapping this position and the color intensity is proportional to the sum of the intensities of the peptides overlapping a given position. Each line corresponds to a different time point: 120 G, 120 min gastric digestion; 30I and 120I, 30 and 120 min intestinal digestion, respectively.
Fig. 5.
Amino acid composition of the identified peptide sequences at the end of the in vitro gastrointestinal digestion of egg white. Repeated sequences are avoided.
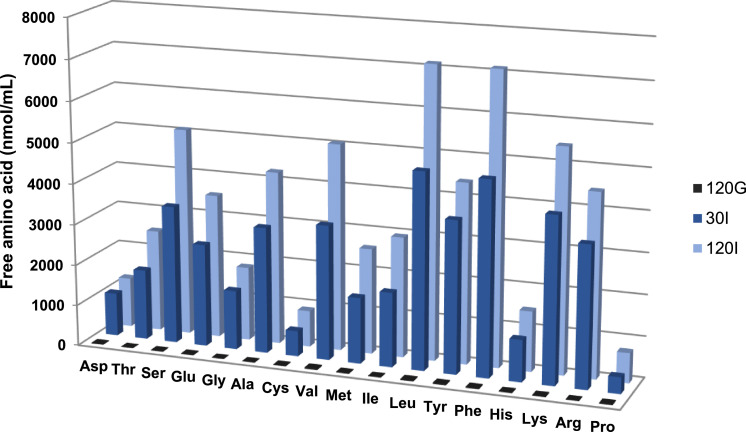
Free amino acids released after 120 min of gastric digestion and 30 and 120 min of intestinal digestion were followed by HPLC and post-column ninhydrin derivatization. As shown in Fig. 6, free amino acids were mainly released during intestinal digestion, with phenylalanine, leucine and lysine being the most abundant, followed by arginine, valine and serine.
Fig. 6.
Free amino acid profile in egg white protein digests at different time points. 120 G corresponds to 120 min of gastric digestion; 30I and 120I correspond to 30 and 120 min of intestinal digestion, respectively.
2. Experimental design, materials, and methods
2.1. Distribution of nitrogen content
Supernatant and pellet from gastric and intestinal digests were freeze-dried and weighted. Protein content in each fraction was measured by elemental analysis and by total amino acid analysis after acid hydrolysis with HCl 6 N at 110 °C for 24 h. In addition, free amino acids were also determined in the soluble part according to the method previously published [3]. The difference between total and free amino acids was ascribed to proteins and peptides.
2.2. Molecular weight distribution by MALDI-TOF
Samples were diluted with 33% acetonitrile and 0.1% of trifluoroacetic acid prior to spotted into a MALDI target plate with 2,5-dihydroxybenzoic acid matrix. Analyses were performed on an Autoflex SpeedTM (Bruker Daltonic, Bremen, Germany). Mass spectra were acquired in positive reflection mode and were collected from the sum of 1000 on average lasers shots. Monoisotopic peaks were generated by FlexAnalysis software. The monoisotopic peaks were organized and represented in a molecular weight distribution range. All other methods are described in [3].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relation-ships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work has received financial support from project AGL2015–66886-R from the Spanish Ministry of Economy and Competitiveness (MINECO). M. S-H is the recipient of an FPU grant (FPU15–03616) from the Ministry of Education, Culture and Sports.
References
- 1.Brodkorb A., Egger L., Alminger M., Alvito P., Assuncao R., Ballance S., Bohn T., Bourlieu-Lacanal C., Boutrou R., Carriere F., Clemente A., Corredig M., Dupont D., Dufour C., Edwards C., Golding M., Karakaya S., Kirkhus B., Le Feunteun S., Lesmes U., Macierzanka A., Mackie A.R., Martins C., Marze S., McClements D.J., Menard O., Minekus M., Portmann R., Santos C.N., Souchon I., Singh R.P., Vegarud G.E., Wickham M.S.J., Weitschies W., Recio I. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019;14:991–1014. doi: 10.1038/s41596-018-0119-1. [DOI] [PubMed] [Google Scholar]
- 2.Minekus M., Alminger M., Alvito P., Ballance S., Bohn T., Bourlieu C., Carriere F., Boutrou R., Corredig M., Dupont D., Dufour C., Egger L., Golding M., Karakaya S., Kirkhus B., Le Feunteun S., Lesmes U., Macierzanka A., Mackie A., Marce S., McClements D.J., Menard O., Recio I., Santos C.N., Singh R.P., Vegarud G.E., Wickham M.S.J., Weitschies W., Brodkorb A. A standardised static in vitro digestion method suitable for food - an international consensus. Food Funct. 2014;5:1113–1124. doi: 10.1039/c3fo60702j. [DOI] [PubMed] [Google Scholar]
- 3.Santos-Hernández M., Tomé D., Gaudichon C., Recio I. Stimulation of CCK and GLP-1 secretion and expression in STC-1 cells by human jejunal contents and in vitro gastrointestinal digests from casein and whey proteins. Food Funct. 2018;9:4702–4713. doi: 10.1039/c8fo01059e. [DOI] [PubMed] [Google Scholar]