Abstract
Improving catalytic ability of enzymes is critical to the success of many metabolic engineering projects, but the search space of possible protein mutants is too large to explore exhaustively through experiments. To some extent, highly soluble enzymes tend to exhibit high activity due to their better folding quality. Here, we demonstrate that an optimization algorithm based on a regression model can effectively design short peptide tags to improve solubility of a few model enzymes. Based on the protein sequence information, a support vector regression model we recently developed was used to evaluate protein solubility after small peptide tags were introduced to a target protein. The optimization algorithm guided the sequences of the tags to evolve towards variants that had higher solubility. The optimization results were validated successfully by measuring solubility and activity of the model enzyme with and without the identified tags. The solubility of one protein (tyrosine ammonia lyase) was more than doubled and its activity was improved by 250%. This strategy successfully increased solubility of another two enzymes (aldehyde dehydrogenase and 1-deoxy-D-xylulose-5-phosphate synthase) we tested. The presented optimization methodology thus provides a valuable tool for improving enzyme performance for metabolic engineering and other biotechnology projects.
Keywords: Protein solubility, Protein activity, Machine learning, Optimization, Peptide tags
Highlights
-
•
Short tags were added to proteins to improve their protein solubility.
-
•
Machine learning model was used to optimize amino acid composition of the tags.
-
•
Multiple proteins’ solubility was substantially improved.
-
•
Catalytic activity of one enzyme was increased when its solubility was enhanced.
1. Introduction
The exploration of expressing recombinant proteins started in 1976, when human peptide hormone Somatostatin was produced in Escherichia coli (Itakura et al., 1977). As the most commonly used expression host, E. coli was investigated intensively to improve the expression and activity of recombinant proteins (W.-C. Chan et al., 2010; Fang et al., 2018; Lempp et al., 2019). Various experimental strategies, such as using protein fusion partners, co-expressing chaperones, choosing suitable promoters, optimizing codon usage, changing culture conditions, and using directed evolution (Esposito and Chatterjee, 2006; Ganesan et al., 2016; Idicula-Thomas and Balaji, 2005; Magnan et al., 2009; Reyes et al., 2017; Trésaugues et al., 2004), were adopted to improve protein expression. For example, the expression of human recombinant enzyme N-acetylgalactosamine-6-sulfatase (rhGALNS) in E. coli was unsatisfactory due to protein aggregation. Several methods including the use of physiologically-regulated promoters, overexpression of native chaperones and applying osmotic shock were investigated to improve the production and activity of rhGALNS (Reyes et al., 2017). Protein activity, a phenotype representing the catalytic ability of a protein if it is an enzyme, is partly determined by its genotype (sequence of its coding gene). Directed evolution can effectively improve protein activity through changing the associated genotype, but this approach is resource-intensive. In the process of improving protein activity via directed evolution, mutagenesis is performed to change gene sequence and the mutated genes are inserted into plasmid used for transformation of a microbe, such as E. coli. Additional techniques are employed to screen a large number of transformed cells for those that have higher protein activity. Since most of the protein directed evolution studies were only interested in the mutants with the highest activity, they did not reveal the genotype of most proteins that had lower activity. This fact has caused the challenge that very few databases of protein activity were available for training computational models that can predict protein activity from protein sequence. Such models would greatly assist protein engineering by evaluating protein sequences in silico. A suitable dataset for training the model should contain both protein activity data and the associated sequence data, and should be large enough.
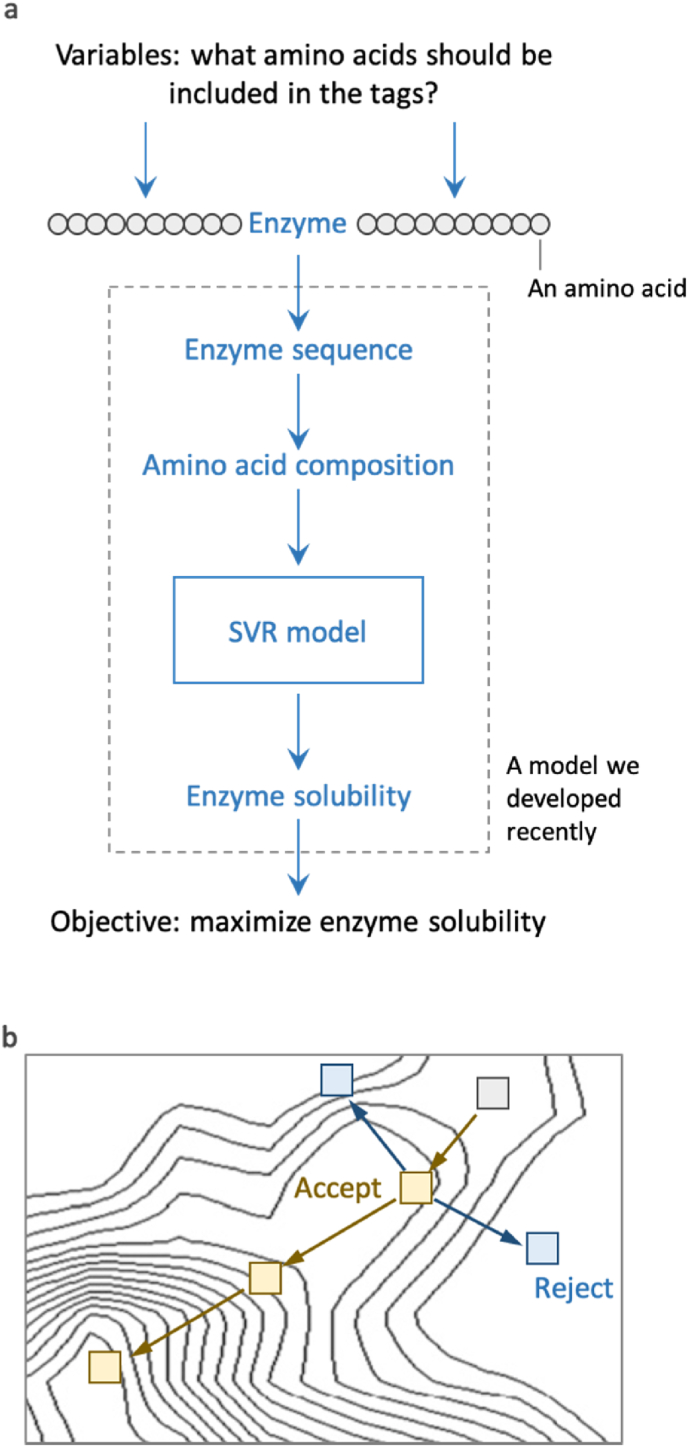
Protein activity data cannot be easily pooled together for model training if they are related to enzymes that catalyze different chemistries, which is another reason why it is difficult to generate the aforementioned datasets for model training. The data of protein solubility from most types of proteins, however, can be compiled into one dataset, because protein solubility is a basic protein property. In this study and the relevant literature, protein solubility is defined as the percentage of a protein’s soluble fraction (Niwa et al., 2009). It is a metric that is often used to assess the folding quality of a protein, under the assumption that incorrectly folded proteins form aggregates and are insoluble in aqueous solution. Protein activity is thus correlated with protein solubility to some extent, because protein solubility may indicate the quality of protein folding which influences protein tertiary structure and activity, i.e. proteins with higher solubility likely exhibit higher activity (Zhou et al., 2012). Thus, protein solubility may be used as a proxy for protein activity to develop predictive models that use protein sequence as input. With such a model, it would be possible to optimize the sequence of a protein in silico for improving its solubility and activity. For example, a Monte Carlo optimization method can be used as the procedures as demonstrated in Fig. 1: (1) a random change is introduced to the protein sequence, (2) the new protein sequence is evaluated by the model, and (3) if the predicted solubility is lower than that of the parent sequence, the change would be rejected, otherwise it would be accepted and used to initiate the subsequent iteration. This in silico optimization process may identify promising protein sequences to improve the success rate of the time-consuming and labor-intensive experiments. If the protein activity heavily depends on its solubility, the experiment would identify new protein mutant that has higher solubility and activity.
Fig. 1.
Machine learning model assisted optimization of protein solubility. (a) Illustration of the decision variables, optimization objective and the objective function. SVR: support vector regression. A SVR model we recently developed was used in this study (Han et al., 2019). (b) Illustration of the optimization algorithm. Genetic algorithm was used in this study.
Recently, Machine Learning has gained increasing attention in various fields, such as internet commerce, autonomous vehicles, and image recognition (Bojarski et al., 2016; Ferrucci et al., 2013; Godec et al., 2019; LeCun et al., 2015; Li et al., 2019; Silver et al., 2016; Weber et al., 2019; Y. Wu et al., 2016; Zador, 2019). Until now, a large number of machine learning methods have been explored to predict protein solubility from amino acid sequence (Agostini et al., 2012; Diaz et al., 2010; Idicula-Thomas and Balaji, 2005; Niwa et al., 2009; Xiaohui et al., 2014). Among the previous studies, we developed regression models that can predict protein solubility in continuous values (Han et al., 2019). Classification models which only label a protein as soluble or insoluble were developed in other studies but cannot be used in further in silico optimization, because it would mistakenly reject most changes that can result in a small but important increase in the protein solubility. So far, very few studies performed experimental validation of their solubility-prediction models and no study used such models to improve protein properties through the in silico optimization of protein sequence.
To improve protein solubility, some trial-and-error procedures were developed by introducing small polyionic tags (Bianchi et al., 1994; P. Chan, Curtis and Warwicker, 2013; Nguyen et al., 2019), because they were short and less likely to interfere with protein structure (Bianchi et al., 1994). One study found that non-polar surface and positively-charged patches were important factors in determining protein solubility (P. Chan et al., 2013). In another study, a negatively charged fusion tag, NT11, was developed to enhance protein expression in E. coli (Nguyen et al., 2019). These previous studies explored negatively charged tags by trial and error and cannot provide a generally useful quantitative model which can forecast performance of tags with proteins which have not been tested.
In our present study, based on a regression model that can predict protein solubility from protein sequence (Han et al., 2019), we developed optimization algorithms to increase predicted solubility under constraints that have been set after considering experimental feasibility and impact on protein function. We found that adding short peptide rich in negatively charged amino acids was effective in improving solubility of a few proteins. More importantly, we also verified that activity of some proteins was indeed substantially improved when their solubility was increased. The short peptide tags characterized in this study and the workflow used to design them should be useful to metabolic engineers who need to improve activity of rate-limiting enzymes.
2. Materials and Methods
2.1. Protein database
All the information of protein solubility used in our study is from the eSol database (Niwa et al., 2009) which is a unique database containing continuous values of protein solubility. After removing items without sequence information according to the previous study (Han et al., 2019), 3148 proteins in the eSol dataset were used for this study. In the study which generated the dataset, the values of protein solubility were measured by synthesizing the recombinant proteins via the cell-free protein expression technology. The expressed proteins were then separated into soluble and insoluble fractions with centrifugation (Niwa et al., 2009). Solubility was defined as the ratio of supernatant protein to total protein which was quantified by SDS-PAGE.
2.2. Training flowsheet
The whole process of rationally engineering proteins with higher solubility includes data pre-processing, training the Support Vector Regression (SVR) prediction model, constructing an optimization methodology, and validating the methodology. As the first step, amino acid composition was extracted from protein sequences by using Amino Acid Composition Descriptor in protr package (Xiao et al., 2014) within R software, which converted characters of amino acids into numeric values indicating amino acid composition. For the second part, the SVR model was built in MATLAB and trained following the same procedure described in the previous study (Han et al., 2019). Then SVR was trained with the whole dataset to predict continuous values of protein solubility from amino acid composition. For the third step, we selected 58 proteins with solubility of 0.1 in the original dataset. Proteins with long sequences are more challenging to synthesize in experiments, therefore the protein sequences were further filtered to have less than 333.3 amino acids (1 kb, gene length), which selected 27 proteins. Among the 27 proteins, the one with the minimum difference between the predicted value and the real value of protein solubility, named GLCE, was selected as the sample protein to build a methodology for further optimizing protein solubility. Genetic algorithm (GA), an optimization method, was explored to search for maximum predicted solubility with constraints for the sample protein. The difference between protein solubility before and after mutagenesis was used to evaluate the optimization performance. Subsequently, besides the sample protein, ten proteins with solubility of 0.1 which have the least differences between predicted and original solubility among the 27 proteins mentioned above were selected for testing the optimization methodology. Six proteins commonly used in our laboratory were also investigated for improving protein solubility. Finally, among the 16 proteins selected for optimization, four proteins that bear low solubility before adding the tags were chosen for experimental validation. Among the four proteins, TAL, DXS and VALC were from the six proteins commonly used in the lab and AGAW was from the eSol database. The original and mutated proteins were expressed in E. coli to validate the predicted change of protein solubility.
2.3. Optimization algorithms
Genetic algorithm (GA), one of the metaheuristic optimization algorithms, is inspired by the process of natural selection observed in nature (Mitchell, 1996). It is frequently utilized as a randomized algorithm for solving optimization problems with constrained conditions. GA essentially simulates the way in which life evolves to find solutions to real world problems. In GA, the solutions to a problem are represented as a population of chromosomes evolving through successive generations. The offspring chromosomes are generated by merging two parent chromosomes by crossover or modifying a chromosome by mutation. The offspring chromosomes are evaluated according to the fitness as defined by the objective function in each generation. Chromosomes with higher fitness values have higher possibility to survive and the process will stop when the offspring chromosomes are almost identical or the termination conditions set are reached. Strong individuals will dominate the generation through iterations in the process of mutation, crossover and selection. The final chromosome represents an optimal or near-optimal solution for the optimization problem. In our problem, the chromosomes are the amino acid compositions of peptide tags and the fitness function is the predicted solubility for proteins after adding tags. Several hyperparameters can be tuned for the optimization algorithm, such as the population size, the number of iterations for evolution and the number of individuals mutating in each generation. We used a MATLAB Toolbox to implement the optimization (iteration number = 1,000, other parameters are provided in Supplementary Table S1). The generic structure of GA in our study can be described as follows:
- begin:
- initiate a tag representing by a 20-dimensional vector with constrained conditions (sum of the vector is 20 and the value of each dimension is within the range of 0-20);
- evaluate the protein sequence after adding the tag;
- while (if termination conditions are not met):
- do crossover and mutate parent tag sequences to yield offspring sequences;
- evaluate the protein solubility for the proteins with offspring sequences;
- select and generate offspring sequence with higher solubility;
- end while
end
2.4. Data visualization
The heat map in Fig. 6 was plotted by using the “cmap” function of the matplotlib package in Python. The violin plot of the amino acid compositions was made by using the “violinplot” function of the seaborn package in Python. Violin plot featured a kernel density estimation of the underlying distribution. Spearman’s rank correlation between amino acid composition and solubility was computed using the “spearmanr” function of the scipy.stats package in Python. The equation used was
| (1) |
where the subscript i denoted the ranks, and x and y represented amino acid composition and solubility respectively.
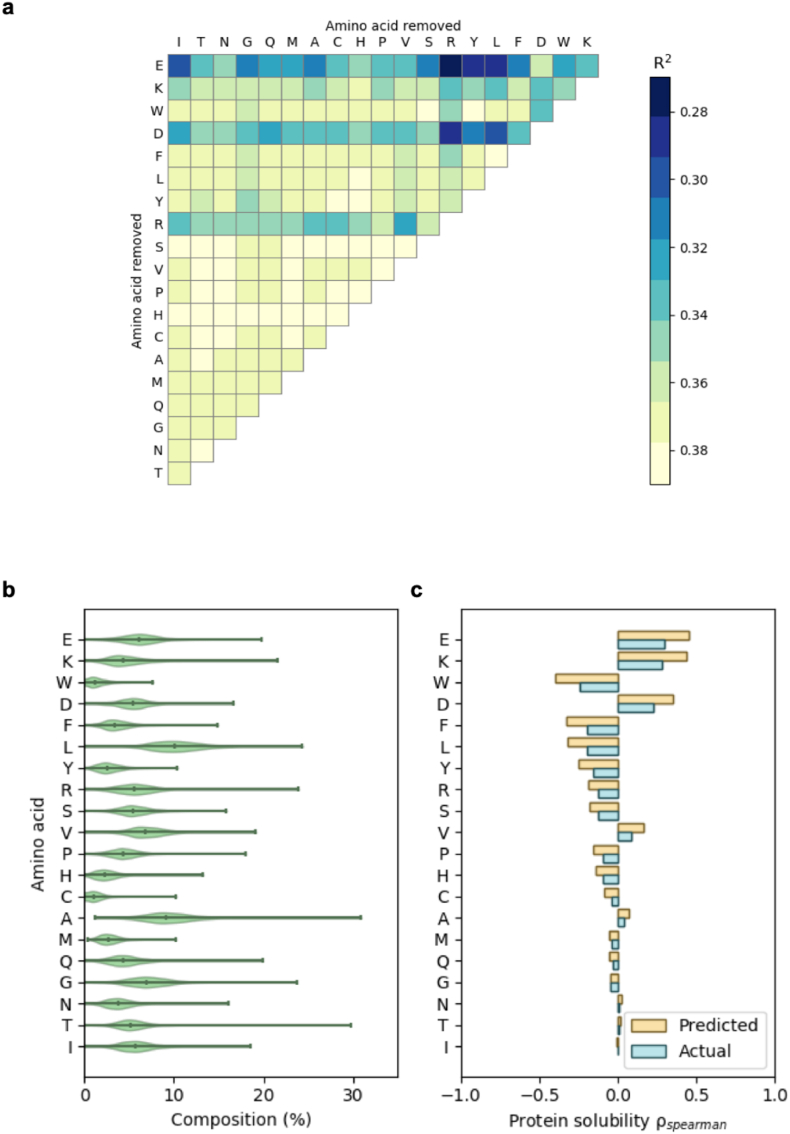
Fig. 6.
(a) Importance of various amino acids in determining the accuracy of the SVR model. The R2 of the SVR model was shown by using a heat map after removing the information of two types of amino acids. Model training is described in Materials and Methods. Single letter amino acid abbreviations are used in this figure. All the combinations of removing two types of amino acids are tested and the performance of the resulting models is presented in the upper triangular matrix. Performance of the models was gauged by using R2, which is presented here by using color (a color bar is provided). The darker the color is, the more important the related amino acids are to the model performance. (b) The distribution of amino acid composition (the input variables of the SVR model we used) among all the proteins in the eSol database (the date source we used to train the SVR model). The violin plot showed the mean value and the range of the amino acid composition used to train the SVR model. (c) The Spearman’s rank correlation between actual/predicted protein solubility and various amino acids. Spearman’s correlation, , is a measure of monotonicity and represents the general sensitivity of solubility to amino acid composition. A comparison between the Spearman’s rank correlation tornado plot for actual solubility and predicted solubility depicted how the model captured and magnified general trends between amino acid composition and solubility. For example, for both the actual and predicted solubility of proteins in the eSol dataset, the composition of D, E, or K was positively correlated with solubility.
2.5. Chemicals in experimental validation
All the chemicals used in this study were purchased from Sigma-Aldrich unless otherwise stated. All reagents used were of analytical grade. The DNA oligonucleotides used in this study were synthesized by Integrated DNA Technologies.
2.6. Plasmid construction
All the plasmids used in this work were constructed according to GT standard (Ma et al., 2019) (Supplementary Tables S7–S10).
2.7. Cell culture and SDS-PAGE analysis of protein solubility
Each plasmid was introduced into E. coli BL21 (DE3) (C2530H, New England Biolabs) for SDS-PAGE analysis by using standard heat shock protocol. In order to test the resulting strains, single colony was inoculated into 1 mL of LB with 100 μg/mL of ampicillin, and was cultured overnight at 37 °C/250 rpm. Fifty microliters of the overnight grown cell suspension were inoculated into 5 mL of K3 medium (Ma et al., 2019) with 100 μg/mL of ampicillin. When cell was grown to 0.4–0.6 (optical density at 600 nm), isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM. After incubated overnight at 30 °C/250 rpm, the cell culture was diluted to OD600 = 2.0, and centrifuged at 5000 g for 10 min. The obtained cell pellets were resuspended in 100 μL of B-PER II reagent (78248, Thermo Fisher Scientific). The mixtures were incubated for 15 min at room temperature with gentle shaking, and centrifuged at 16,000 g for 20 min. The obtained supernatant contained soluble cell lysates. The insoluble cell pellets were resuspended in 100 μL of 2% w/v SDS. Both soluble and insoluble cell pellets were analyzed by using SDS-PAGE (Mini-PROTEAN® TGX™ Precast Protein Gels, 4561083, Bio-Rad). The image of the gel was processed and quantified by Gel Doc EZ Gel Documentation System (Bio-Rad).
2.8. TAL activity assay
One milliliter of obtained supernatant containing soluble cell lysates was added to 4 mL of PBS buffer (pH = 9.0) with 1 g/L tyrosine (final concentration) in 50 mL falcon tube and incubated at 30 °C/250 rpm. Three hundred microliters of samples were taken at 0 h, 1 h, 3 h and 12 h after incubation, and mixed with 700 μL of acetonitrile to dissolve the produced p-courmaric acid (PCA). The mixture was incubated at 30 °C/250 rpm for 1 h, and then centrifuged at 13,500 g for 5 min. Two microliters of the obtained supernatant was analyzed by using HPLC (Agilent 1260 Infinity HPLC) based on a previously reported method (Ma et al., 2019).
3. Results
3.1. Design of the optimization methodology
In order to improve protein solubility by in silico mutagenesis, we needed to address several questions regarding how to change the protein sequence. One can change a protein sequence by adding amino acids to the sequence (addition), replacing amino acids in the sequence (mutation) and/or removing amino acids from the sequence (deletion). The protein functions may be frequently abolished by mutation and deletion as the core protein structure and active sites may be changed. To avoid such detrimental change to the original function of the protein, addition was used in our study to change protein sequence for improving protein solubility. The subsequent decision to make was how many amino acids should be added. Adding too many amino acids would make experimental validation more expensive and may also negatively affect the protein function. Adding too few amino acids may not be able to improve protein solubility substantially. We decided to evaluate adding 20 or 30 amino acids because adding more than 30 amino acids to a protein by using synthetic oligonucleotides was much more expensive.
To optimize the sequence of the amino acids to be added, we designed an algorithm based on the support vector regression (SVR) prediction model we previously developed (Han et al., 2019). The independent variable in the optimization function was the amino acid composition of the short peptides to be added, expressed as number of each amino acid in a vector (Fig. 1). The SVR model we developed only accepted amino acid composition of a protein as input, so we did not consider the full sequence information during the optimization. Then the amino acid composition of a model protein with the added amino acids was calculated and used as input for the SVR model. We used the genetic algorithm (GA) with the objective function defined as the predicted protein solubility from the SVR model in the format of continuous values between 0 and 1. The sum of the number of amino acids added was set as 20 or 30 and the searching range for the number of each amino acid added was from 0 to 20 or 30.
3.2. Optimizing protein sequence in silico for improving protein solubility
After designing this optimization algorithm, ten proteins with low solubility (0.1) in the eSol database (we had used this database to train our machine learning model (Han et al., 2019)) were selected as model proteins to test the algorithm (information of these proteins is provided in Supplementary Table S2). The predicted solubility of all the ten proteins was improved after adding 30 amino acids as peptide tags (Supplementary Fig. S2). One protein’s solubility (name: AGAW, N-acetylgalactosamine-specific enzyme IIC component of PTS) was improved to 0.9951 from 0.1 after adding the designed short peptide tags. When we allowed adding only 20 instead of 30 amino acids, the improvement of predicted solubility slightly decreased (Supplementary Fig. S2). Since it is easier and cheaper to add 20 amino acids in experiments than 30, we adopted adding 20 amino acids as the constraint in the rest of this study.
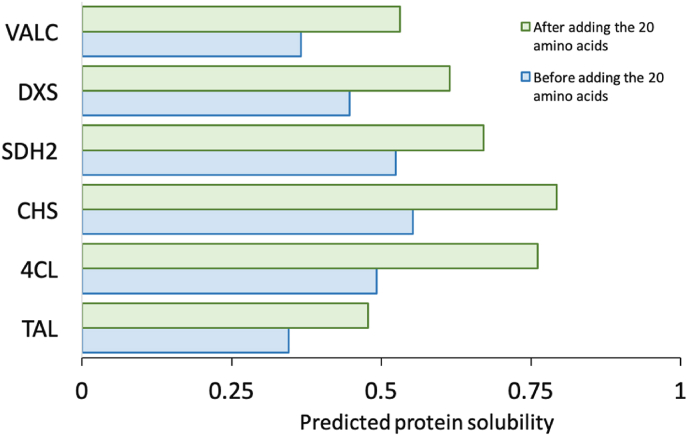
To make this study more relevant to useful applications of recombinant enzymes, we selected six proteins which were important in engineering metabolic pathway of E. coli to produce valuable metabolites (information of these proteins is provided in the caption of Fig. 2). These proteins’ predicted solubility was all lower than 0.6. Adding 20 amino acids also substantially improved the predicted solubility of all the six proteins (Fig. 2). Three proteins (TAL, DXS and VALC) were chosen to experimentally validate the optimization results since their original predicted solubility was low.
Fig. 2.
The predicted solubility before and after adding 20 amino acids for six proteins commonly used in metabolic engineering projects. The six proteins were VALC (valencene synthase), DXS (1-deoxy-D-xylulose-5-phosphate synthase), ADH2 (alcohol dehydrogenase), CHS (chalcone synthase), 4CL (4-coumarate-CoA ligase) and TAL (tyrosine ammonia-lyase). The sequences of oligos used to amplify these proteins are listed in Supplementary Tables S7–S10. Before adding the tags, the protein solubility of each of them was predicted by using SVR and recorded. Then Genetic Algorithm was used to improve their solubility by adding 20 amino acids. The protein solubility after adding the tags was also recorded for comparison.
We also included AGAW in the test because of the large improvement we observed in the in silico optimization. The number of the amino acids to be added was allowed to be decimal during the optimization and was rounded for experimental validation. The predicted solubility after rounding the number of the amino acids added was very similar to that before rounding for all the tested proteins (Supplementary Table S6). To generate sequence of the two tags to be added to a protein from the number of amino acids we minimized the occurrence of amino acid repeats, which reduced the difficulty in synthesizing the related DNA oligonucleotides. The sequence of the tags for those four proteins is listed in the Supplementary Tables S7–S10.
3.3. Experimental validation of the optimized protein sequence
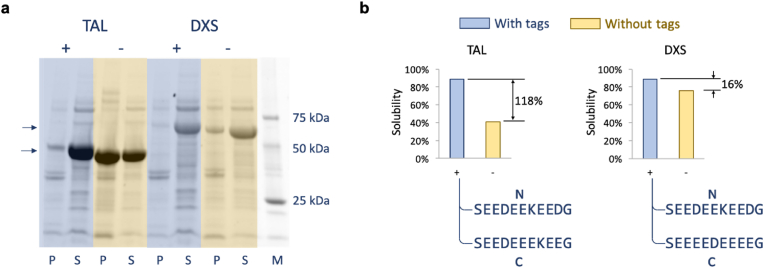
We constructed expression vectors to express the four proteins with and without the optimized tags. Among them, protein AGAW cannot be expressed (as determined by using SDS-PAGE) with and without the tags, which may be caused by the unstable protein structure or unsuitable experimental conditions. Protein VALC can be expressed only without the peptide tags which may have impaired the protein stability. Protein TAL and DXS were expressed with and without the tags (Fig. 3). Protein solubility of TAL and DXS was increased by 118% and 16% respectively by adding the tags.
Fig. 3.
(a) The SDS-PAGE analysis of protein TAL and DXS expressed in E. coli with and without tags designed by our optimization algorithm. “+” and “-” indicate having or not having the peptide tags respectively. “P” and “S” indicate the pellet fraction (insoluble) and supernatant fraction (soluble) respectively. Molecular weight of TAL and DXS were 53.85 kDa and 67.49 kDa respectively (two arrows were used to indicate them in the figure). Protein TAL and DXS were expressed in K3 medium with 20 g/L glucose at 30 °C. This experiment was repeated four times and the other SDS-PAGE images were shown in the Supplementary Figs. S3a–c. (b) Quantitative presentation of the SDS-PAGE images in a. Solubility of a protein is defined as the fraction of the soluble protein molecules among all the protein molecules. The protein amount was estimated by using band intensity on SDS-PAGE images. The sequences of the designed tags for N-terminal and C-terminal were shown. The amino acid S and G on the two ends of the tags were the linkers for GT DNA assembly standard, which was used to guide plasmid construction in this study (Ma et al., 2019).
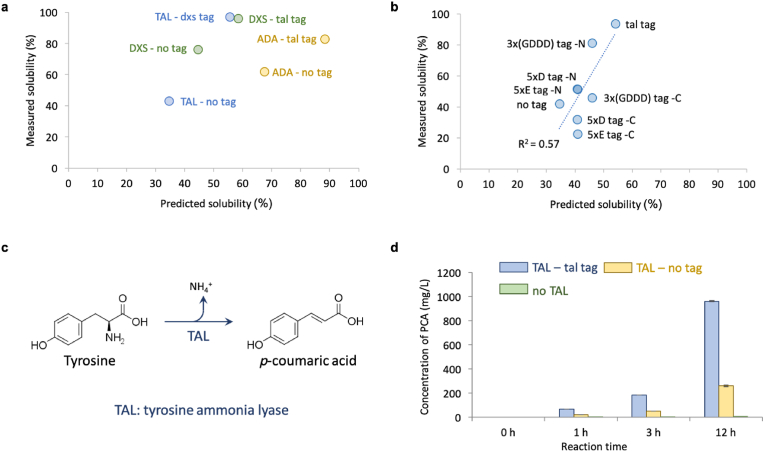
By observing the amino acids added to DXS and TAL (Fig. 3b and Supplementary Table 5), it can be found that their peptide tags were dominated by aspartic acid (D) and glutamic acid (E). Aspartic acid and glutamic acid are the two negatively charged amino acids among the 20 amino acids. Adding them may introduce repulsive electrostatic interactions between protein molecules to prevent aggregation and to provide sufficient time for correct folding of proteins (Paraskevopoulou and Falcone, 2018). The similarity of the peptide tags inspired us to test whether one tag designed for one protein can be used to improve solubility of another protein. We found that the tags optimized for improving solubility of TAL could also increase both predicted and measured solubility of DXS, and vice versa (Fig. 4a). Another protein (name: ADA, aldehyde dehydrogenase) used in a project of our laboratory was also tested with the tag designed for TAL and its predicted and measured solubility were also enhanced (Fig. 4a). The results of switching tags suggested that the tags we designed may be generally effective in improving protein solubility.
Fig. 4.
(a) The predicted and measured solubility of TAL, DXS and ADA after adding tags designed for other proteins. The purpose of switching tags was to test if the solubility-enhancing tags were generally effective in improving protein solubility. The same protein was labelled by using the same color to highlight the data before and after adding tags. In the data labels, the text before “-” indicates protein name and the text after “-” indicates the tags used if any. In the process of measuring the solubility, the protein expression condition was K3 medium with 20 g/L glucose at 30 °C. The SDS-PAGE images were shown in the Supplementary Fig. S3d. (b) The comparison of the tags designed in this study with tags used in previous studies. Protein TAL was the only model protein used in this plot. No tag: solubility of TAL without any tag. Tal tag: solubility of TAL when we added the tags that were designed by our optimization algorithm for TAL. 5xE tag –N/C: solubility of TAL when 5xE tag (EEEEE) was added to its N- or C-terminus. 5xD tag –N/C: solubility of TAL when 5xD tag (DDDDD) was added to its N- or C-terminus. 3x(GDDD) –N/C: solubility of TAL when 3x(GDDD) tag (GDDDGDDDGDDD) was added to its N- or C-terminus. 5xD, 5xE and 3x(GDDD) were three tags used in a previous study and used here for comparison (Paraskevopoulou and Falcone, 2018). Since in previous studies, only one tag was added to one protein, either at N- or C-terminus, we tested both cases for each tag. The two tags we designed for TAL were added to both ends of TAL (Fig. 1, Fig. 3b). The sequences of all the tags are provided in Supplementary Tables S7–S10. The SDS-PAGE images were shown in the Supplementary Fig. S3f. (c) The reaction catalyzed by protein TAL. (d) The enzymatic activity of protein TAL before and after introducing the tal tag. A control was included to show that there was no reaction if TAL protein was absent. The product of the reaction catalyzed by protein TAL was p-coumaric acid (PCA) and its concentration was used to indicate the activity of protein TAL. Cell lysate containing TAL was used in the reaction. TAL – tal tag: the strain containing TAL with the tags designed in this study. Tal – no tag: the strain containing TAL without any tag. No TAL: the strain that did not express TAL. Each bar indicates the mean value of six replicates. The error bars indicate standard error (n = 6).
The dxs tag was slightly better than the tal tag in terms of improving the solubility of TAL (Figs. 3b and 4a), although the tal tag was designed specifically for TAL. One reason is that the optimization algorithm we used may have stopped at local optima rather than global optima, although it was designed to search for the global optima. Therefore, it was possible that some better solutions could exist. GA does not guarantee finding the global optima for all the types of functions, and the function in our problem was a black-box model which might not be continuous. The other reason is that measured solubility from experiments might be affected by multiple factors, such as the sequence of amino acids in the tags, which we did not consider during the in silico optimization.
The ultimate goal of this project was to improve activity of enzymes. Following the success of improving protein solubility, we measured activity of TAL with and without the tags. Protein TAL is tyrosine ammonia lyase which can deaminate tyrosine to produce coumaric acid (Fig. 4c), which is an important precursor of flavonoids (Jendresen et al., 2015; Rodriguez et al., 2015; Santos et al., 2011). TAL activity was increased by 269% by adding the tags we designed for it (Fig. 4d, based on 12 h reaction). The extent of the increase in activity was even larger than that in solubility, suggesting that adding the tags may also increase the expression level and/or specific activity of soluble TAL. Estimation of the protein expression level based on band intensity of the SDS-PAGE gels revealed that the total expression level was reduced by adding the tags (Fig. S5), suggesting that the additional increase should be attributed to the changes of specific activity. This result proved that our optimization scheme for protein solubility was also effective for improving protein activity - at least for this protein - and using protein solubility as a proxy to increase protein activity was reasonable.
3.4. Design of tags under more constrained conditions
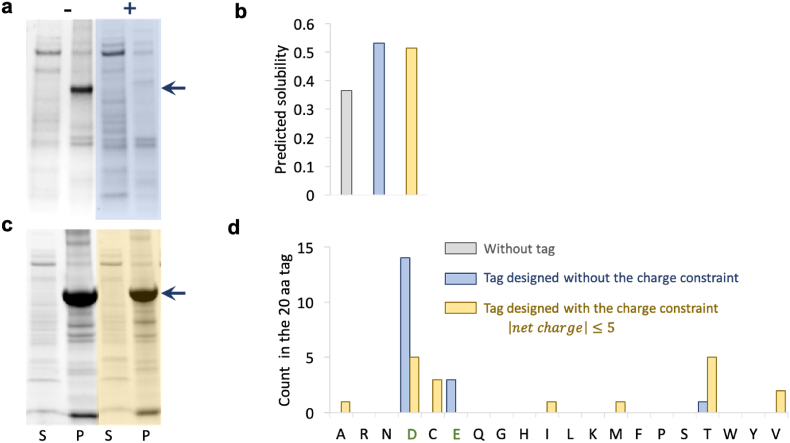
Among the four proteins selected for experimental validation, protein VALC (valencene synthase) cannot be synthesized only after the tags were added. This may be caused by the fact that the stability of VALC was damaged after adding the highly negatively charged tags. Our prediction model and optimization algorithm only took the protein solubility into account. However, other properties of the protein may be changed during the addition of highly charged tags, such as the protein stability. Therefore, we explored whether the peptide tags that contained mainly aspartic acid and glutamic acid can be replaced by tags that contain less charged amino acids to improve protein solubility.
The constrained condition that the number of aspartic acid and glutamic acid cannot be more than a threshold was therefore set in the optimization algorithm. The threshold was from 0 to 10 with step size of 1 for aspartic acid and glutamic acid respectively (Supplementary Table S11). When the maximal number of aspartic acid and glutamic acid allowed to be added was reduced gradually from 10, the predicted solubility was decreasing but the change was small. With the decrease in the number of aspartic acid and glutamic acid, the number of lysine (K) increased substantially. Other amino acids only had a relatively small increase in the optimization solutions. When the constrained condition was very strict, for example, no aspartic acid and glutamic acid were allowed, the amino acids introduced were mostly alanine (A).
Another constrained condition was explored which limited the net charge of the peptide tags. In this case, the upper bound for the absolute value of the net charge of the tag was set as 5, 4, 3, 2, 1, and 0, respectively (Supplementary Table S12) and it could be observed that the number of alanine increased substantially with the decrease of net charge, which was consistent with the results obtained under the other constraint and supported that introducing alanine may be beneficial for the dissolution of protein or the optimization failed to find a feasible solution under such stringent constraints.
The tags with net charge 1, 3, and 5 (Supplementary Fig. S3e, Supplementary Table S12) were used with VALC. These new tags did not abolish protein expression, confirming the hypothesis that excessive amount of aspartic acid and/or glutamic acid destabilized the protein (Fig. 5 and Supplementary Fig. S3). However, the solubility of protein VALC was not improved by the tags. VALC may have strong affinity to cellular membranes and thus cannot be solubilized by the designed tags.
Fig. 5.
(a) The SDS-PAGE analysis of protein VALC expressed in E. coli without tag (”-“) and with the tag designed without the charge constraint (”+“). “P” and “S” represented the pellet (insoluble) fraction and soluble fraction respectively. (b) The predicted solubility of protein VALC without tag (grey), with the tag designed without the charge constraint (blue) and with the tag designed with the charge constraint (yellow). (c) The SDS-PAGE analysis of protein VALC expressed in E. coli without tag (”-“) and with the tag designed with the charge constraint (”+“). (d) The number of amino acid contained in the 20-amino-acid tag designed for protein VALC.
3.5. Comparison with previous studies
To find out if the tags we obtained from our optimization were more effective than published ones (Paraskevopoulou and Falcone, 2018), we compared them by using our predictive model and by conducting experiments. We used TAL as the model protein here, because its solubility was experimentally confirmed to be low and its measured solubility can be substantially improved by adding tags. All the three previously known polyanionic tags increased solubility of TAL when added to its N-terminus (Fig. 4b). But none of them outperformed the tags identified in our optimization, supporting the usefulness of the tags and the optimization procedure we reported here. In addition, there was a desirable correlation between the predicted protein solubility and measured protein solubility based on all the solubility data of TAL we obtained (Fig. 4b). The linear correlation between predicted solubility and measured solubility was quantified by R2 with a value of 0.57. Although the previous study explored tags including aspartic acid and glutamic acid by trial and error, our study provided better optimization performance and a generally effective quantitative model.
4. Discussions
4.1. Using machine learning for optimizing protein properties
Using machine learning to assist the selection of proteins with specific properties has been explored recently (Heckmann et al., 2018; Z. Wu, Kan, Lewis, Wittmann and Arnold, 2019; Yang et al., 2019). Heckmann et al. utilized machine learning to predict the turnover number of enzymes in E. coli, which were further used to parameterize two mechanistic genome-scale models. The machine learning model was trained by using the information of protein structure, biochemistry properties and assay conditions in the study (Heckmann et al., 2018), whereas protein sequences were used to train the predictive model in our study. Therefore, it would be more difficult to use their model to optimize protein sequence for improving protein activity. Wu et al. incorporated machine learning into the directed evolution workflow to help them identify proteins with high fitness value (Z. Wu et al., 2019). Then it was applied to engineering an enzyme for stereodivergent carbon–silicon bond formation, a new-to-nature chemical transformation. However, their training data for machine learning only included variants mutated at four amino acid residues. A protein might include multiple positions for mutagenesis and information of four positions is not representative enough to train a machine learning model to handle other positions. The selection of mutagenesis positions needs to be customized based on prior knowledge of the protein structure. Compared with the studies mentioned above, we do not need to train our prediction model again when we handle a new protein. In our study, we utilized the machine learning model to identify proteins with a generic protein property, protein solubility. Our training dataset was obtained by using various proteins of E. coli and the optimization methodology did not need any customization and knowledge in biochemistry for new target proteins. With only the sequence information, our optimization model can provide effective guide for improving protein solubility and activity. We foresee that our generic model can be combined with the protein-specific models to achieve additive or even synergistic effect in metabolic engineering projects.
4.2. The contribution of aspartic acid and glutamic acid
In this study, we applied a predictive model of protein solubility to improve protein solubility by adding short peptide tags. Aspartic acid and glutamic acid dominated the tags that were obtained by using our optimization strategy. This finding was consistent with the conclusion of an experiment we did to determine which amino acids were the most important in determining accuracy of our solubility prediction model. In the experiment, we removed the percentage information of two amino acids and evaluated the negative impact on the performance of the predictive model. The model’s input was a protein’s amino acid composition, among which the percentages of 19 amino acids were independent. As a result, removing information of only one amino acid would have no impact on model performance and we had to remove the percentages of two amino acids. We evaluated all the combinations of two amino acids. After removing aspartic acid or glutamic acid, the decrease in the prediction performance was the most substantial (Fig. 6a), indicating that they were the most important ones to the accuracy of the model. The causal relationship between the observations from this experiment (Fig. 6a) and the finding of the optimization experiment could be that aspartic acid and glutamic acid had large positive influence on protein solubility (as seen in the optimization experiment), so they were important to the accuracy of the model (as observed in the importance analysis experiment). Arginine, which also showed some influence on the prediction performance when it was removed (Fig. 6a), was not selected during the optimization process. This might be caused by that arginine negatively affected the protein solubility and this hypothesis was tested (Supplementary Fig. S4). After adding 20 arginine residues to six proteins, the predicted solubility of all of them was decreased. The aforementioned effects of glutamic acid, aspartic acid and arginine were also supported by their Spearman correlation coefficients (Fig. 6c), which were obtained by analyzing the dataset we used to train our model. There were some amino acids that were identified to be important by Spearman coefficient (Fig. 6c) but were not found to be critical to model performance (Fig. 6a), such as tryptophan and phenylalanine. It may be due to that Spearman coefficient alone is not sufficient to quantitatively describe the effects of amino acid on protein solubility because of its qualitative nature and because it did not consider abundance of other amino acids (Fig. 6b). In this study, we have shown that our machine learning model is able to quantitatively describe the relationship and guide optimization of protein sequence.
When we trained the solubility prediction model through machine learning, we did not use any biochemistry knowledge. The optimization of protein tag to maximize protein solubility was also purely mathematical without any dependence on prior knowledge. Yet, the identified most beneficial amino acids and their influence on protein solubility can be explained by using known biochemistry knowledge (electrostatic repulsion). As to why the best tags were dominated by negatively charged amino acids rather than positively charged ones, the reason might be that positively charged amino acids may also improve protein solubility but their influence is less than those of negatively charged amino acids. When the number of the negatively charged amino acids was constrained, the optimization algorithm used positively charged amino acid (lysine) to improve protein solubility, which led to less improvement in solubility than using the negatively charged ones (Supplementary Tables S11 and S12).
5. Conclusions
In this study, we adopted the strategy of adding small peptides tags (each tag contained 20 amino acids) to less soluble proteins to improve protein solubility, which was less likely to disturb the active sites of target proteins. In an in silico optimization experiment based on GA and a machine learning model in silico, the amino acid composition of the short tags were varied to maximize the protein solubility. With the tags designed by this procedure, the solubility of three enzymes, TAL, DXS and ADA, was improved substantially in subsequent experimental validation. Protein TAL, with 118% increase in protein solubility, was selected as the model protein to test if its catalytic activity was also improved. An 250% improvement was observed. In addition, we have also experimentally demonstrated that our peptide tags outperformed the commonly used polyanionic tags. The tags identified in this study and the related algorithms should be useful to metabolic engineers who need to debottleneck the rate-limiting reaction in a metabolic pathway, and/or to balance activity of multiple enzymes for production of value-added chemicals.
Codes availability
We present the optimization workflow as a series of notebooks hosted on GitHub (https://github.com/KangZhouGroupNUS/optimization_protein-solubility). The workflow can be used as a template for analysis of other expression and solubility datasets.
Funding
Xi Han was supported by a Singapore Ministry of Education (MOE) PhD Scholarship. This project was financially supported by research grants from the following agencies: Singapore MOE (R-279-000-452-133), Singapore National Research Foundation (R-279-000-512-281), and Disruptive & Sustainable Technologies for Agricultural Precision.
CRediT authorship contribution statement
Xi Han: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing - original draft, Writing - review & editing. Wenbo Ning: Investigation, Validation, Formal analysis, Writing - original draft. Xiaoqiang Ma: Investigation, Validation, Formal analysis. Xiaonan Wang: Conceptualization, Supervision, Writing - review & editing. Kang Zhou: Conceptualization, Supervision, Writing - review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Cortes-Pena Yoel Rene for providing data visualization for data distribution and Spearman’s rank correlation tornado plot.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00138.
Contributor Information
Xiaonan Wang, Email: chewxia@nus.edu.sg.
Kang Zhou, Email: kang.zhou@nus.edu.sg.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agostini F., Vendruscolo M., Tartaglia G.G. Sequence-based prediction of protein solubility. J. Mol. Biol. 2012;421(2–3):237–241. doi: 10.1016/j.jmb.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Bianchi E., Venturini S., Pessi A., Tramontano A., Sollazzo M. High level expression and rational mutagenesis of a designed protein, the minibody: from an insoluble to a soluble molecule. J. Mol. Biol. 1994;236(2):649–659. doi: 10.1006/jmbi.1994.1174. [DOI] [PubMed] [Google Scholar]
- Bojarski M., Del Testa D., Dworakowski D., Firner B., Flepp B., Goyal P.…Zhang J. 2016. End to End Learning for Self-Driving Cars. arXiv preprint arXiv:1604.07316. [Google Scholar]
- Chan P., Curtis R.A., Warwicker J. Soluble expression of proteins correlates with a lack of positively-charged surface. Sci. Rep. 2013;3:3333. doi: 10.1038/srep03333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.-C., Liang P.-H., Shih Y.-P., Yang U.-C., Lin W.-c., Hsu C.-N. Learning to predict expression efficacy of vectors in recombinant protein production. BMC Bioinf. 2010;11(1):S21. doi: 10.1186/1471-2105-11-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A.A., Tomba E., Lennarson R., Richard R., Bagajewicz M.J., Harrison R.G. Prediction of protein solubility in Escherichia coli using logistic regression. Biotechnol. Bioeng. 2010;105(2):374–383. doi: 10.1002/bit.22537. [DOI] [PubMed] [Google Scholar]
- Esposito D., Chatterjee D.K. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006;17(4):353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Fang H., Li D., Kang J., Jiang P., Sun J., Zhang D. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B 12. Nat. Commun. 2018;9(1):4917. doi: 10.1038/s41467-018-07412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci D., Levas A., Bagchi S., Gondek D., Mueller E. Watson: beyond jeopardy! Artif. Intell. 2013;199:93–105. [Google Scholar]
- Ganesan A., Siekierska A., Beerten J., Brams M., Van Durme J., De Baets G. Structural hot spots for the solubility of globular proteins. Nat. Commun. 2016;7:10816. doi: 10.1038/ncomms10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godec P., Pančur M., Ilenič N., Čopar A., Stražar M., Erjavec A. Democratized image analytics by visual programming through integration of deep models and small-scale machine learning. Nat. Commun. 2019;10(1):1–7. doi: 10.1038/s41467-019-12397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang X., Zhou K. Develop machine learning based regression predictive models for engineering protein solubility. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz294. [DOI] [PubMed] [Google Scholar]
- Heckmann D., Lloyd C.J., Mih N., Ha Y., Zielinski D.C., Haiman Z.B. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models. Nat. Commun. 2018;9(1):5252. doi: 10.1038/s41467-018-07652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idicula-Thomas S., Balaji P.V. Understanding the relationship between the primary structure of proteins and its propensity to be soluble on overexpression in Escherichia coli. Protein Sci. 2005;14(3):582–592. doi: 10.1110/ps.041009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura K., Hirose T., Crea R., Riggs A.D., Heyneker H.L., Bolivar F., Boyer H.W. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977;198(4321):1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- Jendresen C.B., Stahlhut S.G., Li M., Gaspar P., Siedler S., Förster J. Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015;81(13):4458–4476. doi: 10.1128/AEM.00405-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lempp M., Farke N., Kuntz M., Freibert S.A., Lill R., Link H. Systematic identification of metabolites controlling gene expression in E. coli. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-12474-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ruan H., Liu C., Li Y., Shuang Y., Alù A. Machine-learning reprogrammable metasurface imager. Nat. Commun. 2019;10(1):1082. doi: 10.1038/s41467-019-09103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Liang H., Cui X., Liu Y., Lu H., Ning W. A standard for near-scarless plasmid construction using reusable DNA parts. Nat. Commun. 2019;10(1):3294. doi: 10.1038/s41467-019-11263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan C.N., Randall A., Baldi P. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics. 2009;25(17):2200–2207. doi: 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- Mitchell M. 1996. An introduction to genetic algorithms mit press. Cambridge, Massachusetts. London, England. [Google Scholar]
- Nguyen T.K.M., Ki M.R., Son R.G., Pack S.P. The NT11, a novel fusion tag for enhancing protein expression in Escherichia coli. Appl. Microbiol. Biotechnol. 2019:1–12. doi: 10.1007/s00253-018-09595-w. [DOI] [PubMed] [Google Scholar]
- Niwa T., Ying B.-W., Saito K., Jin W., Takada S., Ueda T., Taguchi H. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106(11):4201–4206. doi: 10.1073/pnas.0811922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou V., Falcone F. Polyionic tags as enhancers of protein solubility in recombinant protein expression. Microorganisms. 2018;6(2):47. doi: 10.3390/microorganisms6020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes L.H., Cardona C., Pimentel L., Rodríguez-López A., Alméciga-Díaz C.J. Improvement in the production of the human recombinant enzyme N-acetylgalactosamine-6-sulfatase (rhGALNS) in Escherichia coli using synthetic biology approaches. Sci. Rep. 2017;7(1):5844. doi: 10.1038/s41598-017-06367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Kildegaard K.R., Li M., Borodina I., Nielsen J. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 2015;31:181–188. doi: 10.1016/j.ymben.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Santos C.N.S., Koffas M., Stephanopoulos G.J. M.e. Optimization of a heterologous pathway for the production of flavonoids from glucose. 2011;13(4):392–400. doi: 10.1016/j.ymben.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Silver D., Huang A., Maddison C.J., Guez A., Sifre L., Van Den Driessche G.…Lanctot M. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529(7587):484. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- Trésaugues L., Collinet B., Minard P., Henckes G., Aufrère R., Blondeau K., van Tilbeurgh H. Refolding strategies from inclusion bodies in a structural genomics project. J. Struct. Funct. Genom. 2004;5(3):195–204. doi: 10.1023/B:JSFG.0000029017.46332.e3. [DOI] [PubMed] [Google Scholar]
- Weber T., Wiseman N.A., Kock A. Global ocean methane emissions dominated by shallow coastal waters. Nat. Commun. 2019;10(1):1–10. doi: 10.1038/s41467-019-12541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Schuster M., Chen Z., Le Q.V., Norouzi M., Macherey W. 2016. Google’s Neural Machine Translation System: Bridging the Gap between Human and Machine Translation. arXiv preprint arXiv:1609.08144. [Google Scholar]
- Wu Z., Kan S.J., Lewis R.D., Wittmann B.J., Arnold F.H. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116(18):8852–8858. doi: 10.1073/pnas.1901979116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N., Xu Q., Cao D. 2014. Protr: Protein Sequence Descriptor Calculation and Similarity Computation with R. R Package Version 0. 2-1. [Google Scholar]
- Xiaohui N., Feng S., Xuehai H., Jingbo X., Nana L. Predicting the protein solubility by integrating chaos games representation and entropy in information theory. Expert Syst. Appl. 2014;41(4):1672–1679. [Google Scholar]
- Yang K.K., Wu Z., Arnold F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods. 2019;1 doi: 10.1038/s41592-019-0496-6. [DOI] [PubMed] [Google Scholar]
- Zador A.M. A critique of pure learning and what artificial neural networks can learn from animal brains. Nat. Commun. 2019;10(1):1–7. doi: 10.1038/s41467-019-11786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Zou R., Stephanopoulos G., Too H.-P. Enhancing solubility of deoxyxylulose phosphate pathway enzymes for microbial isoprenoid production. Microb. Cell Factories. 2012;11(1):148. doi: 10.1186/1475-2859-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.