Abstract
Contralaterally positioned maxillary (upper jaw) venom glands in snakes are mechanically independent, being able to discharge venom from either gland separately. This has led some studies to test venom function and composition of each contralaterally positioned venom gland to investigate any differences. However, the data on the subject to-date derives from limited sample sizes, appearing somewhat contradictory, and thus still remains inconclusive. Here, we tested samples obtained from the left and right venom glands of four N. siamensis specimens for their relative binding to the orthosteric site of amphibian, lizard, snake, bird, and rodent alpha-1 nicotinic acetylcholine receptors. We also show the relative proteomic patterns displayed by reversed phase liquid chromatography – mass spectrometry. Our results indicate that three of the venom gland sets showed no difference in both functional binding and composition, whilst one venom gland set showed a slight difference in functional binding (but not in specificity patterns between prey types) or venom composition. We hypothesise that these differences in functional binding may be due to one gland having previously ejected venom at some time prior to venom extraction, whilst its contralateral counterpart did not. This might cause the differential rate of toxin replenishment to be unequal between glands, thus instigating the difference in potency, likely due to uneven toxin proportions between glands at the time of venom extraction. These results demonstrate that the separate venom producing glands in snakes remain under the same genetic control elements and produce identical venom components.
Keywords: Naja siamensis, Alpha-neurotoxins, Neurotoxicity, Venom, Venom glands, Nicotinic acetylcholine receptor, Liquid chromatography – mass spectrometry
Highlights
-
•
No differences in venom profile by contralateral venom glands of Naja siamensis.
-
•
No differences in alpha-1 binding potency by contralateral N. siamensis venom glands.
-
•
A single outlier in the data showed a small difference in relative potency.
-
•
Potency is likely affected by differential replenishment rates.
1. Introduction
Many advanced snakes (Caenophidia) utilise venom delivery systems that vary across the group in both structure, topography and complexity (Fry et al., 2008, Fry et al., 2013, Jackson et al., 2017), with venom glands either being solely contralateral maxillary glands or containing both maxillary and mandibular venom glands (upper and lower jaw, respectively) (Fry et al., 2013, Fry et al., 2015a). The venom systems of Elapidae and Viperidae are the most widely studied, with sophisticated delivery apparatus consisting of hollow fangs connected to pressurised post-orbital maxillary venom glands (Kardong, 1982, Kochva, 1987). Due to the contralateral positioning of the glands, they are mechanically independent of each other, being able to separately control the ejection of venom from either venom gland (Hayes et al., 2008, Young and Zahn, 2001).
Despite the physiological independence of contralateral glands, they are expected to be under the same genetic control, producing equal ratios of specific toxins and thus having the same relative venom potency and specificity of toxins in each gland. Yet, surprisingly, few studies provide direct evidence in this regard, with any study that has compared contralateral venom glands in snakes has not focused on this aspect as the primary aim of the study.
In regards to contralateral maxillary glands, one study found no discernible compositional difference between protein/toxin content of left and right gland extractions of Daboia palastinae using gel electrophoresis (Oron et al., 1978). Further, the expression levels of left and right venom glands revealed no significant difference in Bothrops atrox, when comparing their cDNA libraries (Amazonas et al., 2018). Another study assessing the diversification of venom systems in reptiles showed that there were identical toxin transcripts between the maxillary and mandibular venom glands of snakes that contain both these glandular systems (Fry et al., 2013). This is consistent with the Toxicofera hypothesis which proposes that the mandibular venom glands in Anguimorpha lizards and maxillary glands in advanced snakes are derivations of a plesiomorphic system present in the last common ancestor which had toxin secreting glands on both the upper and lower jaw (Fry et al., 2006). Although the maxillary and mandibular venom glands are not anatomical counterparts, they still produce identical toxin transcripts (Fry et al., 2013), which further suggests that genetic control of separate venom glands appear consistent between their contralateral equals.
A further study discovered no significant difference in LD50 values for left and right venom gland extractions of seven snake species found across Iran (Latifi, 1984). The study tested 10 individuals of each species; Echis carinatus, Gloydius halys (Agkistrodon halys in the mentioned study), Macrovipera lebetina (Vipera lebetia in the mentioned study), Montivipera latifii (Vipera latifii in the mentioned study), Montivipera xanthina (Vipera xanthina in the mentioned study), Naja oxiana (Naja naja oxiana in the mentioned study), and Pseudocerastes persicus. These data indicate no overall difference in functional lethality between the glands. However, the study stated that there was a single individual outlier that had a significant difference in LD50 between left and right gland venom, yet the authors failed to identify which species this individual belonged to and the data as presented in the tables did not reveal such a difference in samples. Thus, the reasoning behind this statement is enigmatic based upon what data is actually presented in the manuscript.
Another study showed the converse in that there were distinct differences between venom composition and functional potency of left and right venom gland extractions of Crotalus viridis helleri (Johnson et al., 1987). This study also noted that the left gland venom extraction was yellow-coloured, and the right gland venom extraction was white-coloured, suggesting a difference in venom composition between both venom glands. Indeed, the left gland venom was high in Phospholipase A2 (PLA2s) and L-amino acid oxidase (LAAO), while the right gland venom contained relatively lower molecular weight components than the left and also lacked LAAO and PLA2 activity. Despite the compositional differences in left/right fang extractions, there were also many functional differences. The left gland venom was lethal to mice, caused intradermal haemorrhage and myonecrosis. The right gland venom also caused intradermal haemorrhaging and myonecrosis but was not lethal to mice, with all test subjects surviving intravenous injections.
Two of the studies (Johnson et al., 1987, Oron et al., 1978) only used a single individual (N = 1 per gland), whilst the another study (Latifi, 1984) only found a single outlier from 70 individuals tested. The small number of individuals showing venom gland differences may call into question the robustness of the data. Additionally, some of these studies were conducted 30+ years ago with what may now be seen as basic, outdated biochemical and functional assays. Thus, it is possible that composition and functional differences observed between glands were due to failings in the techniques. Based on these methodological short-comings, and the notion that there seems to be some contradiction in the literature as to whether contralateral maxillary glands can differ at both the molecular and functional levels, further investigation is needed.
We set out to understand if individual venom glands can produce venom toxins asymmetrically (i.e., left vs right) and how this might affect relative potency, if at all. To do this, we tested the relative neurotoxic potency and venom composition of left and right venom extractions from four individuals of Naja siamensis (Indochinese spitting cobra). To assess orthosteric (active site) binding of α-neurotoxins (three finger toxins; 3FTxs in the case for N. siamensis) to α-1 nicotinic acetylcholine receptor (nAChR) mimotopes (Harris et al., 2020a, Harris et al., 2020b, Zdenek et al., 2019), we used a validated biolayer interferometry (BLI) assay (Zdenek et al., 2019) to test for differential binding on amphibian, lizard, snake, bird, and rodent targets. The composition of both the left and right venom gland extractions was determined using reverse phase liquid-chromatography tandem mass spec (RP-LC/MS).
2. Materials and methods
2.1. Venom collection and preparation
Venoms were obtained from left and right fang extractions of Naja siamensis from captive specimens at Venom Supplies Pty Ltd. All venom samples were lyophilised and reconstituted in double deionised water (ddH2O), and centrifuged (4 °C, 10 min at 14,000 relative centrifugal force (RCF)). The supernatant was made into a working stock (1 mg/mL) in 50% glycerol to prevent freezing at −20 °C. The concentrations of working stocks were determined in triplicate using a NanoDrop 2000 UV–Vis Spectrophotometer (Thermo Fisher, Sydney, Australia) at an absorbance wavelength of 280 nm.
2.2. Mimotope production and preparation
Following methods from a previously developed assay (Harris et al., 2020b, Zdenek et al., 2019), a 13–14 amino acid mimotope of the vertebrate α-1 nAChR orthosteric site was developed by GenicBio Ltd. (Shanghai, China) designed upon specification. The design was based from publicly available sequences of the cholinergic receptors (Chrna1) from GenBank and UniProt databases.
Accession codes for the amino acid sequences of the α-1 nAChR orthosteric site for each taxa were as follows: amphibian α-1 (uniprot F6RLA9), lizard α-1 (genbank XM_015426640), avian α-1 (uniprot E1BT92), rodent α-1 (uniprot P25108). The only exception was the α-1 sequence for the snake α-1 (Coelognathus radiatus), which was Sanger sequenced in a previous study (Zdenek et al., 2019).
Mimotope dried stocks were solubilised in 100% dimethyl sulfoxide (DMSO) and diluted in ddH2O at 1:10 dilution to obtain a stock concentration of 50 μg/mL. Stocks were stored at −80 °C until required.
2.3. Biolayer interferometry (BLI)
Full details of the developed assay, including all methodology and data analysis, can be found in the validated protocol (Zdenek et al., 2019) and further data using this protocol (Harris et al., 2020a, Harris et al., 2020b). In brief, the BLI assay was performed on the Octet RED 96 system (ForteBio, Fremont, CA, USA). Venom (analyte) samples were diluted 1:20, making a final experimental concentration of 50 μg/mL per well. Mimotope aliquots were diluted 1:50, with a final concentration of 1 μg/mL per well. The assay running buffer was 1X DPBS with 0.1% BSA and 0.05% Tween-20. Preceding experimentation, Streptavidin biosensor were hydrated in the running buffer for 30–60 min, whilst being agitated on a shaker at 2.0 revolutions per minute (RPM). The dissociation of analytes occurred using a standard acidic solution glycine buffer (10 mM glycine (pH 1.5–1.7) in ddH2O). Raw data are provided in Supplementary File 2.
2.4. Sample preparation for RP-LC/MS
Powdered crude venom samples were brought up in 0.1% TFA. The samples were filtered via Millipore ZipTips® using the following protocol: tips were wetted with 100% ACN (3 × 10 μL) then equilibrated with 10 μL 5% ACN/0.1% TFA (3 × 10 μL). Samples were then loaded and pipetted up and down twice the volume of sample. Tips were then washed with 5% ACN/0.1% TFA (3 × 10 μL). Samples were eluted with 80% ACN/0.1% TFA (10 μL) in a new tube. Samples were then lypholised and later resuspended in 20 μL of 0.1% TFA.
LC-MS analysis was conducted using reversed-phase chromatography on a Dionex Ultimate 3000 RSLC nano system. Using a flow rate of 30 μL/min, protein was first desalted on a Thermo PepMap 100 C18 trap (0.3 × 5 mm, 5 μm) for 5 min using 1% ACN, 0.1% FA, followed by separation on a Vydac Everest C18 column (5 μm, 0.75 × 150 mm) at a flow rate of 300 nl/min. A gradient of 1–60% buffer B over 60 min, followed by 60–98% B over 3 min, held at 98% B was used where buffer A = 1% ACN, 0.1% FA and buffer B = 80% ACN, 0.1% FA. Eluted proteins were directly analysed on an Orbitrap Elite mass spectrometer (Thermo) using a NSI electrospray interface. Source parameters included a capillary temperature of 275 °C; S-Lens RF level at 70%; source voltage of 2.4 kV and maximum injection times of 200 ms for MS and 150 ms for MS2. Proteins were analysed with an FTMS scan across m/z range 600–3000 at 60,000 resolution.
2.5. Data processing and analyses
All data were obtained from an Octet RED 96 system (ForteBio, Fremont, CA, USA) and processed in exact accordance to the validation of this assay (Zdenek et al., 2019). The association step data (in triplicate) were obtained in a.csv file extracted from raw outputs of the Octet Red 96 system, tidied in Excel, and then imported into Prism 7.0 software (GraphPad Software Inc., La Jolla, CA, USA) where area under the curve (AUC) calculations were made and graphs produced.
Intact masses were determined using the Intact Protein Analysis tool in ThermoFisher Scientific BioPharma Finder™ software version 3.2.46.13. Reconstruct settings included mass range of 1000–15,000 Da, using MS data between 600 and 2000 m/z, with resolution set to 15,000, and protein charged by H+. The masses returned were the uncharged monoisotopic values. Chromatograms were then saved from ThermoFisher Scientific Xcaliber FreeStyle™ software version 1.6. All chromatograms were then annotated in Adobe Photoshop with the deconvoluted monoisotopic masses to their corresponding peaks previously attained from ThermoFisher Scientific BioPharma Finder™.
3. Results and discussion
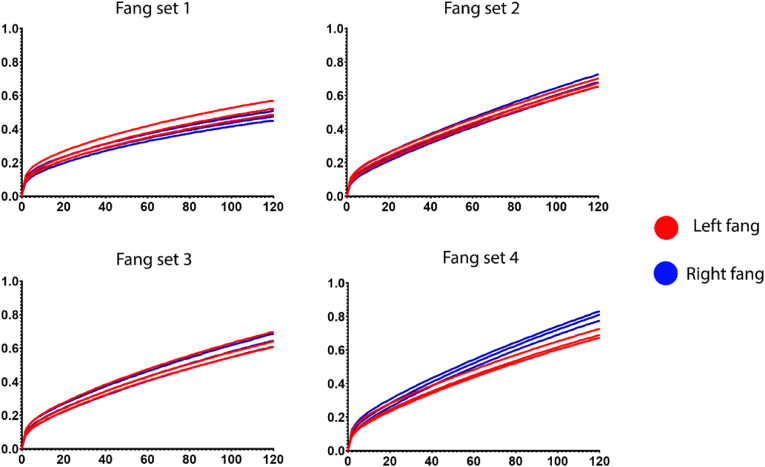
The overall results from the neurotoxic functional testing indicate that there is no significant difference between the α-1 nAChR binding of venom from the left or right gland of most fang sets (1–3) between all prey types tested (Fig. 1 showing the most effectively targeted taxa [amphibian], consistent with previous studies (Harris et al., 2020b); with supplementary file 1 showing the results for the other prey types). Only fang set 4 (Fig. 1.; supplementary file 1) had a slight binding difference between each fang, with the right fang showing a marginally higher overall binding than the left. However, a difference of ~0.1 nm shift is not likely to cause any biochemically significant difference during an envenomation scenario.
Fig. 1.
A binding comparison of Naja siamensis venom from the left and right venom gland to the amphibian α-1 nAChR mimotope. The Y-axis shows wavelength (nm) detection of the association step (Ka binding step); the X-axis displays seconds (for the 120 s assay run period). All venoms were tested against amphibian, lizard, bird, rodent and snake mimotopes in triplicate. Only the amphibian data have been represented here since this was the highest binding by all fang sets compared to all other peptides, and there was no difference between the left and right fang binding of the other mimotopes tested (see supplementary file 1), producing the same overall outcome. All triplicates (N = 3) are shown.
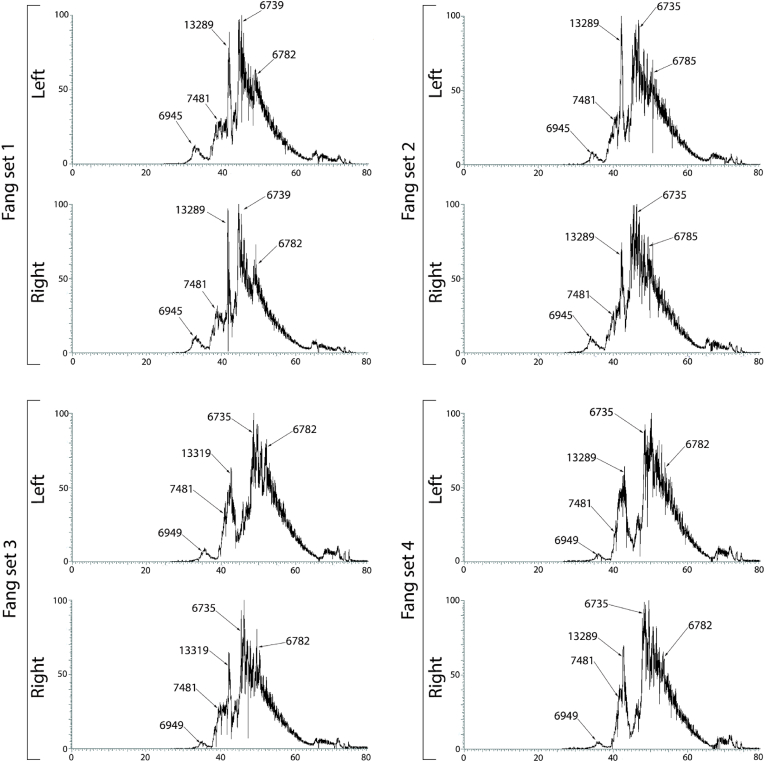
Examination of the venom sets by RP-LC/MS to ascertain the elution patterns and dominant masses within each peak revealed the left and right fangs to be identical in composition (Fig. 2). The results for RP-LC/MS revealed there is no difference in the major toxin components of all fang sets (Fig. 2.). The major masses that were detected are likely 3FTxs, being the peaks identified as 6735, 6782, 6785, 6949 and 7481. These molecular masses are in the range of 3FTxs, a ubiquitous toxin class within Elapidae venoms (Barber et al., 2013, Nirthanan and Gwee, 2004), with similar 3FTx molecular masses detected in N. siamensis (Karlsson et al., 1971, Ohkura et al., 1988). The masses of 13,289 and 13,319 are likely to be Phospholipase A2 (PLA2) toxins (Sunagar et al., 2015), particularly since PLA2s have been shown to be major components in Naja venoms (Bittenbinder et al., 2018). Any marginal differences of masses between fang sets (e.g. 13,319 in fang set 3 and 13,289 from all other fang sets) is likely due to both isoforms being present within the venom, however due to ion suppression (a reduced detector response), different isoform masses are being deconvoluted within different fang sets (Fry et al., 2015b). We acknowledge that the resolution of the RP-LC/MS toxin resolution is not the best quality, however we ascertain that since the purposes of this study was to simply determine if there were any differences between the contralateral venoms glands and not to deconvolute every toxin mass within the venom, then this is adequate. Thus, it is clear from the elution patterns that the venoms are virtually identical.
Fig. 2.
RP-LC/MS profiles of four different fang sets (left and right fangs) of Naja siamensis. Deconvoluted monoisotopic masses are annotated to their corresponding peak by an arrow. X-axis is retention time in minutes (0–80) and the Y-axis is the relative abundance (0–100%).
Since the RP-LC/MS venom profiles for fang set 4 had the same major mass peaks (Fig. 2.) and thus the same toxic components, this suggests that the difference in functional neurotoxic nAChR binding (Fig. 1.) is likely due to an unequal proportion of the specific toxins between each gland. A logical explanation for this functional difference between venom extractions from both fangs may be due to an unequal discharge of venom from each fang occurring prior to the extractions for this study, resulting in a concomitant unequal replenishment rate of venom in each gland. For example, either during feeding or a defensive spray prior to venom extraction, one gland but not the other might have been stimulated (by the vaginal sheath around the fang) to eject venom, causing an unequal replenishment state between the venom glands, with the one gland replenishing venom components not the other. Partial discharge of one gland and complete discharge of another occurring at the same time is also an alternate explanation for an unequal state of replenishment. Such a dichotomous extraction of venom between the two venom glands in the one snake may explain the yellow and white venom extractions from the fangs of Crotalus viridis helleri in reference (Johnson et al., 1987) and the data-free outlier species in reference (Latifi, 1984) having different left and right fang LD50 values.
The replenishment rate of different venom components has been shown to occur asynchronously within snake venom glands (Brown et al., 1975, Oron and Bdolah, 1973, Oron et al., 1978), meaning that different toxin components are replenished at different rates over time within the venom gland. Thus, resulting in an unequal proportion of different venom components over time in one gland compared to the other. This might also explain the differences in molecular components in the case of C. viridis helleri (Johnson et al., 1987) and also the differences seen with venom function and lethality (Johnson et al., 1987, Latifi, 1984). Yet some evidence has refuted the asynchronous replenishment of toxins, finding that re-synthesis of all toxin family transcripts occur in parallel (Currier et al., 2012). It may be that the venom gland synthesis of non-toxic, structural/stabilising proteins are produced in the gland before the synthesis of the toxic components. However, regardless of the debate on toxin replenishment, it does not directly apply if one fang ejects all the venom and the other ejects none. This is simply because if one gland does not need to replenish its venom components whilst the other requires a full suite of toxin replenishment, with the empty gland having a different concentration of toxic components over time until the full replenishment is achieved. Thus, it is possible that there may be a lower relative concentration of venom proteins and a higher concentration of non-toxin elements in the venom (e.g. stabilising salts). Consequently, if the venom extractions for N. siamensis fang set 4 (Fig. 1.; supplementary file 1) occurred when the left fang was not fully replenished, then the replenishment of the 3FTxs in this instance may not be equally replenished to the same proportions as the right gland.
Although this disparity between glands seems like a rare occurrence, it should be considered when testing for individual functional differences in venom, and that difference in relative potency may occur as a consequence. This supports pooling venom extraction samples from multiple individuals as being the more effective method when investigating snake venom, unless specifically testing for individual venom variation.
4. Conclusions
This research has shown that, although there may be no discernible differences in venom components between contralateral fangs, there may be some functional difference due to differential rates of replenishment. We have provided evidence that supports the notion that there is no difference between contralateral maxillary venom glands, however, in line with other studies we did have an outlier within our data. Thus, we have provided some logical hypotheses for the results, including the outlier in our data and other outliers within the literature. These differences in contralateral glands are possibly due to a rare occurrence in which one fang may eject more venom than its contralateral counterpart, and due to replenishment occurring in one fang and not the other, then certain toxin types may be in much lower proportions in subsequent venom extractions occurring before both venom glands reach the state of full replenishment, possibly causing this functional difference. However, it is unlikely that this would affect the fitness of the snake or alter any functional affects during envenomation scenarios. Future transcriptomic research should attempt to investigate if our hypotheses on how these venom gland differences occur is valid. Comparing venom from one fully replenished gland with a series of time period venom extractions will be able to shed light on the hypothesis proposed here.
Funding
R.J.H and C.N.Z was supported by the University of Queensland International PhD scholarship fund. B.G.F was funded by Australian Research Council Discovery Project DP190100304.
Ethical statement
Ethical statement: no live animals were used in this study and thus animal ethics approval was not needed. Venom samples were obtained from a licenced commercial supplier.
Author contributions
Conceptualisation, B.G.F.; data curation, R.J.H; formal analysis, R.J.H.; funding acquisition, B.G.F.; investigation, R.J.H. and C.N.Z.; methodology, R.J.H., C.N.Z., A.N. and B.G.F.; project administration, B.G.F.; resources, C.S. and N.D.; supervision, B.G.F.; writing—original draft, R.J.H.; writing—review and editing, R.J.H, C.N.Z. and B.G.F.
Declaration of competing interest
The authors declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.toxcx.2020.100050.
Contributor Information
Richard J. Harris, Email: rharris2727@gmail.com.
Christina N. Zdenek, Email: christinazdenek@gmail.com.
Amanda Nouwens, Email: a.nouwens@uq.edu.au.
Charlotte Sweeney, Email: csweeney@vaxxas.com.
Nathan Dunstan, Email: venoms@venomsupplies.com.
Bryan G. Fry, Email: bgfry@uq.edu.au.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Amazonas D.R., Portes-Junior J.A., Nishiyama M.Y., Jr., Nicolau C.A., Chalkidis H.M., Mourão R.H., Grazziotin F.G., Rokyta D.R., Gibbs H.L., Valente R.H. Molecular mechanisms underlying intraspecific variation in snake venom. J. Proteom. 2018;181:60–72. doi: 10.1016/j.jprot.2018.03.032. [DOI] [PubMed] [Google Scholar]
- Barber C.M., Isbister G.K., Hodgson W.C. Alpha neurotoxins. Toxicon. 2013;66:47–58. doi: 10.1016/j.toxicon.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Bittenbinder M.A., Zdenek C.N., Youngman N.J., Dobson J.S., Naude A., Vonk F.J., Fry B.G. Coagulotoxic cobras: clinical implications of strong anticoagulant actions of African spitting Naja Venoms that are not neutralised by antivenom but are by LY315920 (Varespladib) Toxins. 2018;10:516. doi: 10.3390/toxins10120516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.S., Brown M.B., Bdolah A., Kochva E. Accumulation of some secretory enzymes in venom glands of Vipera palaestinae. Am. J. Physiol. Legacy Content. 1975;229:1675–1679. doi: 10.1152/ajplegacy.1975.229.6.1675. [DOI] [PubMed] [Google Scholar]
- Currier R.B., Calvete J.J., Sanz L., Harrison R.A., Rowley P.D., Wagstaff S.C. Unusual stability of messenger RNA in snake venom reveals gene expression dynamics of venom replenishment. PloS One. 2012;7 doi: 10.1371/journal.pone.0041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry B., Sunagar K., Casewell N., Kochva E., Roelants K., Scheib H., Wüster W., Vidal N., Young B., Burbrink F. The origin and evolution of the Toxicofera reptile venom system. In: Fry B., editor. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. 2015. pp. 1–31. [Google Scholar]
- Fry B., Undheim E., Jackson T., Roelants K., Georgieva D., Vetter I., Calvete J., Scheib H., Cribb B., Yang D. 2015. Research Methods. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery; pp. 153–214. Fry, BG, Ed. [Google Scholar]
- Fry B.G., Scheib H., van der Weerd L., Young B., McNaughtan J., Ramjan S.R., Vidal N., Poelmann R.E., Norman J.A. Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia) Mol. Cell. Proteomics. 2008;7:215–246. doi: 10.1074/mcp.M700094-MCP200. [DOI] [PubMed] [Google Scholar]
- Fry B.G., Undheim E.A., Ali S.A., Jackson T.N., Debono J., Scheib H., Ruder T., Morgenstern D., Cadwallader L., Whitehead D. Squeezers and leaf-cutters: differential diversification and degeneration of the venom system in toxicoferan reptiles. Mol. Cell. Proteomics. 2013;12:1881–1899. doi: 10.1074/mcp.M112.023143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry B.G., Vidal N., Norman J.A., Vonk F.J., Scheib H., Ramjan S.R., Kuruppu S., Fung K., Hedges S.B., Richardson M.K. Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- Harris R.J., Zdenek C.N., Debono J., Harrich D., Fry B.G. Evolutionary interpretations of nicotinic acetylcholine receptor targeting venom effects by a clade of asian Viperidae snakes. Neurotox. Res. 2020 doi: 10.1007/s12640-020-00211-2. [DOI] [PubMed] [Google Scholar]
- Harris R.J., Zdenek C.N., Harrich D., Frank N., Fry B.G. An appetite for destruction: detecting prey-selective binding of α-neurotoxins in the venom of Afro-Asian elapids. Toxins. 2020;12:205. doi: 10.3390/toxins12030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes W.K., Herbert S.S., Harrison J.R., Wiley K.L. Spitting versus biting: differential venom gland contraction regulates venom expenditure in the black-necked spitting cobra, Naja nigricollis nigricollis. J. Herpetol. 2008;42:453–460. [Google Scholar]
- Jackson T.N., Young B., Underwood G., McCarthy C.J., Kochva E., Vidal N., van der Weerd L., Nabuurs R., Dobson J., Whitehead D. Endless forms most beautiful: the evolution of ophidian oral glands, including the venom system, and the use of appropriate terminology for homologous structures. Zoomorphology. 2017;136:107–130. [Google Scholar]
- Johnson E.K., Kardong K.V., Ownby C.L. Observations on white and yellow venoms from an individual southern Pacific rattlesnake (Crotalus viridis helleri) Toxicon. 1987;25:1169–1180. doi: 10.1016/0041-0101(87)90135-8. [DOI] [PubMed] [Google Scholar]
- Kardong K.V. The evolution of the venom apparatus in snakes from colubrids to viperids and elapids. Mem. Inst. Butantan (Sao Paulo) 1982;46:105–118. [Google Scholar]
- Karlsson E., Arnberg H., Eaker D. Isolation of the principal neurotoxins of two Naja naja subspecies. Eur. J. Biochem. 1971;21:1–16. doi: 10.1111/j.1432-1033.1971.tb01433.x. [DOI] [PubMed] [Google Scholar]
- Kochva E. The origin of snakes and evolution of the venom apparatus. Toxicon. 1987;25:65–106. doi: 10.1016/0041-0101(87)90150-4. [DOI] [PubMed] [Google Scholar]
- Latifi M. Variation in yield and lethality of venoms from Iranian snakes. Toxicon. 1984;22:373–380. doi: 10.1016/0041-0101(84)90081-3. [DOI] [PubMed] [Google Scholar]
- Nirthanan S., Gwee M.C. Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty years on. J. Pharmacol. Sci. 2004;94:1–17. doi: 10.1254/jphs.94.1. [DOI] [PubMed] [Google Scholar]
- Ohkura K., Inoue S., Ikeda K., Hayashi K. Amino-acid sequences of four cytotoxins (cytotoxins I, II, III and IV) purified from the venom of the Thailand cobra, Naja naja siamensis. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1988;954:148–153. doi: 10.1016/0167-4838(88)90065-9. [DOI] [PubMed] [Google Scholar]
- Oron U., Bdolah A. Regulation of protein synthesis in the venom gland of viperid snakes. J. Cell Biol. 1973;56:177–190. doi: 10.1083/jcb.56.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron U., Kinamon S., Bdolah A. Asynchrony in the synthesis of secretory proteins in the venom gland of the snake Vipera palaestinae. Biochem. J. 1978;174:733–739. doi: 10.1042/bj1740733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Jackson T., Reeks T., Fry B. Oxford University Press; New York, NY, USA: 2015. Group I Phospholipase A2 Enzymes, Venomous Reptiles and Their Toxins; pp. 327–334. [Google Scholar]
- Young B.A., Zahn K. Venom flow in rattlesnakes: mechanics and metering. J. Exp. Biol. 2001;204:4345–4351. doi: 10.1242/jeb.204.24.4345. [DOI] [PubMed] [Google Scholar]
- Zdenek C.N., Harris R.J., Kuruppu S., Youngman N.J., Dobson J.S., Debono J., Khan M., Smith I., Yarski M., Harrich D. A taxon-specific and high-throughput method for measuring ligand binding to nicotinic acetylcholine receptors. Toxins. 2019;11:600. doi: 10.3390/toxins11100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.