Significance
DNA replication in mammalian cells takes place at numerous replication origins clustered into thousands of replication domains (RDs) across the genome, which must be coordinated precisely to ensure that genomic information is duplicated once and only once during each cell cycle. Currently, how replication origins are chosen for firing within individual RDs remains poorly understood. We performed a quantitative characterization of the in situ epigenetic environment of replication foci across S-phase using superresolution microscopy. Our findings reveal a distinct pattern of replication propagation and point to a nonrandom mechanism for replication activation at early S-phase. These findings could have wide-ranging implications for our understanding of how local chromatin structures control dynamic processes within the cell nucleus.
Keywords: DNA replication, chromatin organization, superresolution microscopy, epigenetic environment, spatiotemporal dynamics
Abstract
Mammalian DNA replication is initiated at numerous replication origins, which are clustered into thousands of replication domains (RDs) across the genome. However, it remains unclear whether the replication origins within each RD are activated stochastically or preferentially near certain chromatin features. To understand how DNA replication in single human cells is regulated at the sub-RD level, we directly visualized and quantitatively characterized the spatiotemporal organization, morphology, and in situ epigenetic signatures of individual replication foci (RFi) across S-phase at superresolution using stochastic optical reconstruction microscopy. Importantly, we revealed a hierarchical radial pattern of RFi propagation dynamics that reverses directionality from early to late S-phase and is diminished upon caffeine treatment or CTCF knockdown. Together with simulation and bioinformatic analyses, our findings point to a “CTCF-organized REplication Propagation” (CoREP) model, which suggests a nonrandom selection mechanism for replication activation at the sub-RD level during early S-phase, mediated by CTCF-organized chromatin structures. Collectively, these findings offer critical insights into the key involvement of local epigenetic environment in coordinating DNA replication across the genome and have broad implications for our conceptualization of the role of multiscale chromatin architecture in regulating diverse cell nuclear dynamics in space and time.
DNA replication is a process of fundamental importance that maintains the integrity of genomic information in eukaryotic cells across the generations. After decades of biochemical and genetic studies, we now possess a comprehensive understanding of the mechanistic details of DNA replication and the various components of the cellular machinery that execute this process (1, 2). In addition, we have also gained substantial insights into the genomic organization of replication in sequence space, in which replication is initiated at multiple loci along the chromosomes termed replication origins, which demarcate the genome into numerous replicons (3, 4). Such organization is manifested in physical space as thousands of spatially clustered sites or puncta within the nucleoplasm, termed replication foci (RFi) or “factories,” as revealed by fluorescence imaging (4–6). Recent advances in high-throughput sequencing techniques have also enabled studies of DNA replication on a genome-wide scale, leading to the discovery of a replication timing program in which different regions of the genome tend to replicate in a temporally specific manner (7–9). Such temporally distinct regions, termed replication domains (RDs), could be considered as the counterpart of RFi in sequence space (6). In addition, cells have been found to respond and adapt to cellular environments by modulating the timing of RDs for specific genes, although the mechanisms of such modulation remain largely elusive (10, 11).
Several previous studies have suggested that the initiation of DNA replication is regulated at the RD level (9, 12, 13), while within an RD the activation of replication origins is rather random (14, 15). However, direct measurements on spread-out DNA fibers by DNA combing experiments have shown that an RD can harbor several replicons that fire synchronously (10, 16), thereby implicating a possible activation mechanism mediated by the spatial organization of the local chromatin environment. Such a proposal is also in line with the fact that the genomic boundaries of RDs align well with those of topologically associating domains (TADs), a prevalent structural feature of eukaryotic chromatin (17, 18).
In light of these findings, a fuller understanding of replication activation at the sub-RD level requires in situ visualization and characterization of individual DNA replication sites in the context of their surrounding chromatin environment, with single-cell sensitivity and ultrahigh spatiotemporal resolutions. Among the technical tools available to address this problem, sequencing-based methods are mostly limited to analyzing populations of cells, and thus are more suitable for studying replication at the RD level. On the other hand, the majority of previous imaging studies were limited in their spatial resolution as well as their characterization of related chromatin features and functions and were thus unable to provide details into the regulation of replication activation within RDs.
More recently, superresolution microscopy has emerged as a powerful tool for chromatin biology and has been used to study a variety of chromatin structures ranging from nucleosome heterogeneity to the morphology of chromatin DNA in different epigenetic states (19–22). In this study, we use stochastic optical reconstruction microscopy (STORM) to probe the spatiotemporal organization of DNA replication within individual RFi at subdiffraction-limit resolution in single human cells. Using metabolic labeling, we quantitatively characterized the morphology, distribution, sequence length, and replicon number of individual RFi across different stages of S-phase. We also characterized the in situ epigenetic signatures of individual RFi and demonstrated the correlation between replication timing and chromatin modifications and organization at the RD level. Importantly, we observed an intriguing spatiotemporal propagation pattern within individual RFi, with DNA that are replicated earlier spatially surrounded by DNA that are replicated later during early S-phase. The radial directionality of such a propagation pattern is reversed in late S-phase and diminished upon treatment with caffeine (an inhibitor of checkpoint kinase) or knockdown of CTCF (a key organizer of chromatin architecture). Together with simulations and bioinformatic analysis, our results point to a “CTCF-organized REplication Propagation” (CoREP) model, in which DNA replication activates nonrandomly within individual RFi, mediated by CTCF-organized chromatin structures at early S-phase. These findings offer critical insights into the regulation of origin activation at the sub-RD level and have broad implications for the spatiotemporal coordination of DNA replication across the genome, which may be applicable to a variety of mammalian systems.
Results
Superresolution Imaging and Quantitative Characterization of RFi across S-Phase.
To label newly replicated DNA in situ, we first synchronized HeLa-S3 cells to the G1/S boundary as previously described (23, 24). At specific stages of S-phase, a short 30-min pulse of dye-labeled dUTP (2′-deoxyuridine 5′-triphosphate) or EdU (5-ethynyl-2′-deoxyuridine) was supplied to the cells (see Materials and Methods for details), which incorporated these analog molecules into the DNA synthesized during the pulse (25). Imaging the labeled RFi with STORM (SI Appendix, Fig. S1A) revealed a punctate distribution as well as changing physical morphology across S-phase (Fig. 1A), in line with previous observations using immunogold electron microscopy (26), conventional optical microscopy (16, 27), and other superresolution microscopy methods (20, 28–31). However, the superior spatial resolution afforded by STORM (∼20 nm) enabled us to perform more accurate quantitative characterization of RFi, particularly for subsequent findings based on multicolor colocalization analysis within tens of nanometers.
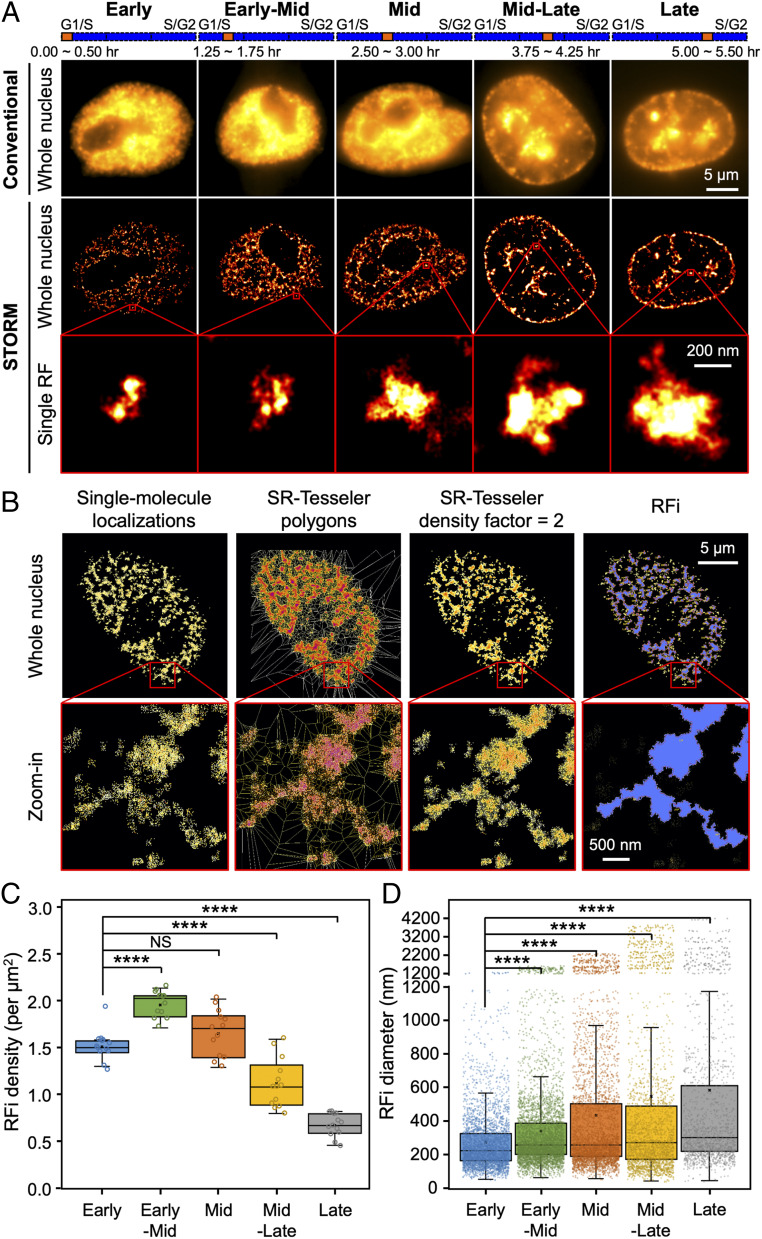
Fig. 1.
STORM imaging and quantitative characterization of RFi across S-phase. (A) Conventional and STORM images of RFi in single HeLa cell nuclei at five different stages (from left to right, each with 30 min of labeling) across S-phase. Insets (red boxes) show zoomed-in representative images of individual RFi at each stage. The orange bars at the top indicate the relative temporal position of the labeling periods during S-phase (blue bars, not drawn to scale). (B) Unbiased hierarchical identification of RFi with SR-Tesseler, starting from raw single-molecule localizations in STORM images, to polygons in SR-Tesseler, to polygons upon using density-based filtering, and finally to RFi. Insets (red boxes) show zoomed-in areas where individual foci can be discerned. (C and D) RFi density (C) and diameter (D) at each of the five stages imaged in A. Data are shown as box-and-whisker plots. Each dot denotes a single RF, and each circle denotes a single cell. P values are determined by two-tailed Student’s t test; ****P < 0.0001; NS: not significant. n = 10∼13 cells for C and 3,503, 4,556, 3,829, 2,566, and 1,509 RFi from 10∼13 cells at each stage for D. Source data are provided as Dataset S1.
In order to assign the single-molecule localizations detected to individual RFi in a robust and unbiased fashion, we adopted a recently developed segmentation strategy, SR-Tesseler (32) (Fig. 1B; see Materials and Methods for details). With the RFi obtained from the segmentation, we found that while RFi density peaks during early-mid S-phase and slowly declines toward late S-phase (Fig. 1C), the size of RFi increases steadily from a mean value of 275.7 nm in early S-phase to 582.7 nm in late S-phase (Fig. 1D and SI Appendix, Fig. S1B). As a proof of the validity of our approach, we also ascertained that our synchronization and labeling procedures minimally impacted the morphology of RFi as well as cell growth and division (SI Appendix, Fig. S1 C and D).
As a further validation of our superresolution imaging approach, we correlated the RFi observed in Cartesian space with the actual replicated DNA in sequence space using a DNA combing approach (33). By imaging single stretched DNA fibers in vitro, we found that each single-molecule localization in the STORM images corresponded to 51.6 ± 12.6 base pairs of replicated DNA (Fig. 2A and SI Appendix, Fig. S2A; see Materials and Methods for details). Since identical imaging conditions and analysis algorithm were applied consistently across all imaging experiments, both in situ and in vitro, any overcounting of localizations from the same molecule will be present to the same extent in the images of both RFi in situ and DNA fibers in vitro, thereby ensuring the validity and accuracy of our calibration between single-molecule localizations and RFi sequence length. Such calibration in turn allowed us to quantify the sequence length of RFi in situ (Fig. 2B). Importantly, we found that the sequence length of RFi produced during the 30-min pulse increases from a mean value of 177.8 kb in early S-phase to 950.0 kb in late S-phase, in good agreement with previous sequencing measurements of 400 to 800 kb per RD (17) and 880 kb per TAD (18).
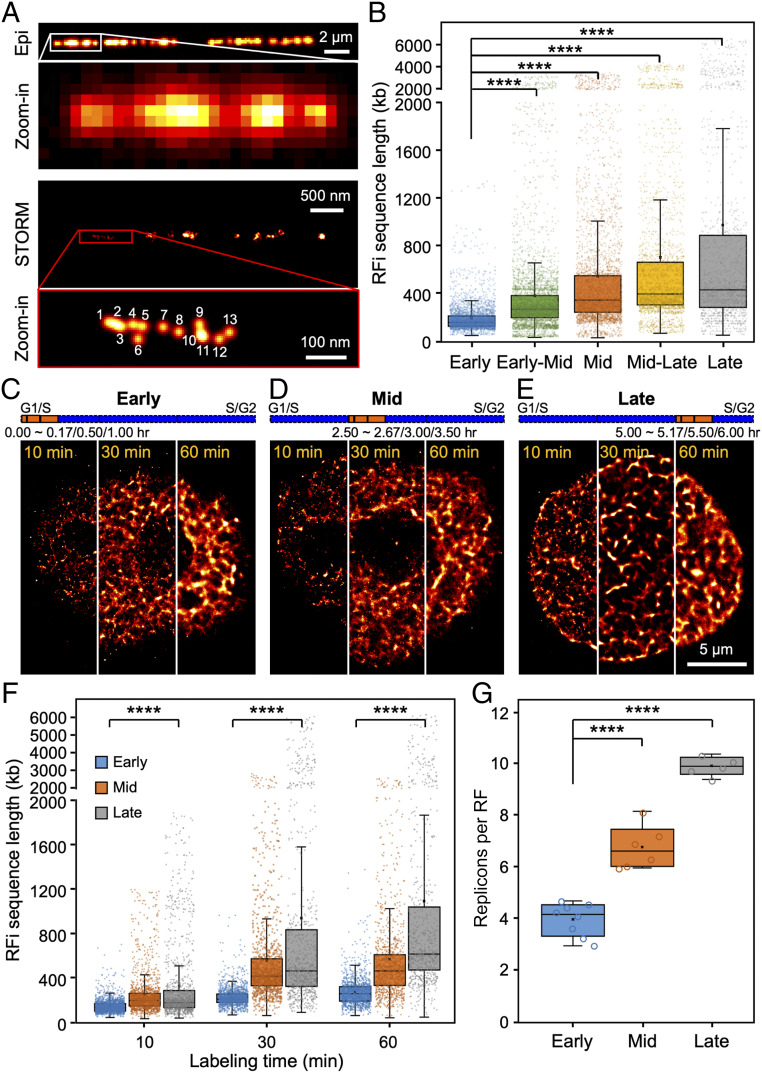
Fig. 2.
Quantification of sequence length and replicon number of individual RFi across S-phase. (A) Conventional and STORM images of Alexa 647-EdU–labeled replication sites on a single stretched DNA molecule; the physical length of the DNA molecule can be directly measured and converted into base pair length (29, 64). Inset in the STORM image (red box) shows a zoomed-in area, where single-molecule localizations can be directly counted. (B) Sequence length associated with individual RFi during each of the five stages imaged in Fig. 1A. (C–E) STORM images of cells with their newly replicated DNA labeled for 10 min, 30 min, or 60 min, respectively, at the beginning of early (C), mid (D), and late (E) S-phase. For more visual comparison, each image is stitched from three different cells, each labeled for 10 min, 30 min, and 60 min, respectively. The orange bars at the top indicate the relative temporal position of the labeling periods during S-phase (blue bars, not drawn to scale). (F) Sequence lengths of RFi replicated during different labeling durations at early, mid, and late S-phase. (G) Number of replicons per replication focus in early, mid, and late S-phase. Data are shown as box-and-whisker plots. Each dot denotes a single RF, and each circle denotes a single cell. P values are determined by two-tailed Student’s t test; ****P < 0.0001. n = 3,503, 4,556, 3,829, 2,566, and 1,509 RFi from 10∼13 cells at each stage for B; 1,962, 1,441, and 1,254 (10 min), 1,443, 1,214, and 1,125 (30 min), 1,146, 865, and 709 (60 min) RFi from 5∼8 cells at each stage for F; and 5∼8 cells for G. Source data are provided as Dataset S1.
Moreover, by varying the labeling pulse duration (from 10 min to 60 min), we monitored the rate of expansion of the size and sequence length of individual RFi across S-phase (Fig. 2 C–F and SI Appendix, Fig. S2B). We also estimated the in situ replication speed, defined as sequence length per replication focus per minute (dashed linear fits in SI Appendix, Fig. S2B). We found that while the size and sequence length of RFi are similar for short labeling duration (10 min), they increase much faster during late S-phase for longer labeling durations (30 and 60 min). In addition, using the replication fork speeds (defined as sequence length per fork per minute) obtained from previous DNA combing experiments (34), we further found that on average 3.8∼9.8 replicons exist within each replication focus at different stages of S-phase (Fig. 2G), and the number of replicons per focus increases from early to late S-phase, thereby lending support to the “increasing efficiency model” of DNA replication (35, 36).
RFi across S-Phase Are Associated with Distinct Stage-Specific Epigenetic Signatures In Situ.
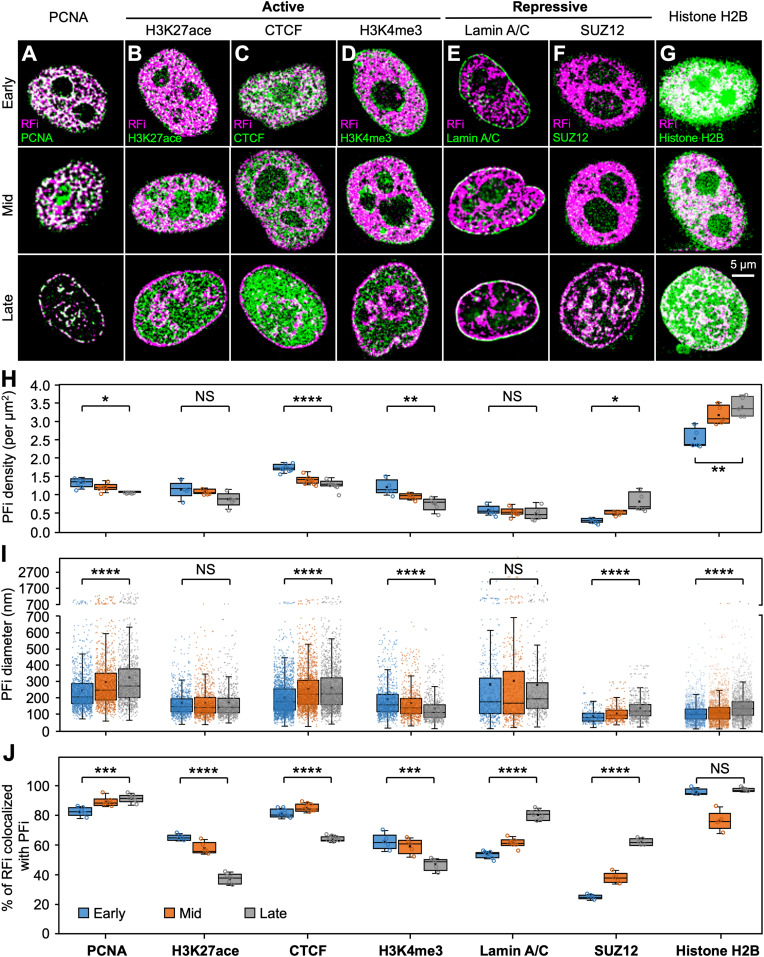
Even though the relationship between DNA replication timing and their chromatin environment has been studied using sequencing and biochemistry approaches, the measurements were often performed on cells in G1-phase (10, 21, 37, 38), and thus lacking in situ spatial and dynamic information for S-phase. To investigate the dynamic changes in the local chromatin environment of replicating DNA at subdiffraction-limit resolution, we colabeled RFi and seven key nuclear markers associated with a variety of epigenetic signatures in situ using two spectrally distinct dyes, Alexa 647 and Cy3B, and performed two-color STORM imaging to quantify their colocalization (Fig. 3 A–G and SI Appendix, Fig. S3A). Among these markers, CTCF is known to regulate three-dimensional (3D) chromatin architecture by forming chromatin loops and define the boundaries between euchromatic and heterochromatic DNA (39–41). Histone modifications H3K27ace and H3K4me3 are associated with transcriptionally active chromatin, with the former being a marker for active enhancers and the latter for active promoters (42, 43). In contrast, nuclear lamina-localized lamin A/C has been suggested to mostly associate with transcriptionally repressed DNA (44), while SUZ12 constitutes part of the polycomb repressive complex 2 (PRC2)/EED-EZH2 complex that methylates Lys-9 (H3K9me) and Lys-27 (H3K27me) of histone H3, leading to transcriptional repression of target genes (45, 46). Finally, proliferating cell nuclear antigen (PCNA) is an essential component of the mammalian replication fork and acts both as a processivity factor for DNA polymerase δ as well as a recruiter of other proteins involved in DNA replication (47). Thus, it serves as a positive control together with H2B, which provides an indication of the general histone background of the cell nucleus.
Fig. 3.
Dynamics of RFi-associated epigenetic signatures in situ across S-phase. (A–G) Colocalization analysis between RFi and seven key nuclear markers reveals distinct local chromatin states of RFi across S-phase. Cells with newly replicated DNA (pink) labeled at the beginning of early (Top), mid (Middle), and late (Bottom) S-phase were subsequently immunolabeled with antibody against each marker (green) and imaged with dual-color STORM. Contrast between the two colors is enhanced here for better visualization; white indicates colocalization between RFi and epigenetic marker. See SI Appendix, Fig. S3 for corresponding images with normal contrast as well as zoomed-in areas from each image. (H–J) Density (H) and diameter (I) of protein foci (PFi) associated with each of the seven markers imaged in A–G, as well as fraction of RFi that colocalize with the PFi of each marker (J) during early, mid, and late S-phase, respectively. Data are shown as box-and-whisker plots. Each dot denotes a single PF, and each circle denotes a single cell. P values are determined by two-tailed Student’s t test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS: not significant. n = 1,197, 1,088, and 969 (PCNA); 907, 859, and 699 (H3K27ace); 2,117, 1,712, and 1,570 (CTCF); 972, 765, and 608 (H3K4me3); 522, 473, and 431 (Lamin A/C); 253, 451, and 675 (SUZ12); and 1,804, 2,263, and 2,450 (Histone H2B) PFi from 5∼8 cells at each stage for I and 5∼8 cells for H and J. Source data are provided as Dataset S1.
Quantitative analysis of STORM images revealed that while the densities and sizes of the protein foci (PFi) do not vary significantly (Fig. 3 H and I), the extent to which RFi colocalize with each marker changes drastically across S-phase (Fig. 3J and SI Appendix, Fig. S3B). The relatively constant high colocalization ratio between RFi and PCNA throughout S-phase reflects the essential role of PCNA in DNA replication, and along with histone H2B lends support to the accuracy of our colocalization analysis. Specifically, the fractions of RFi that colocalize with H3K27ace, CTCF, or H3K4me3, all of which associate with the more open and transcriptionally active chromatin, show a marked decrease from early to late S-phase. In contrast, the fractions of RFi that colocalize with lamin A/C or SUZ12, which are involved in lamina-associating domains and transcriptional repression (48), display a pronounced increase from early to late S-phase. These results indicate that DNA near open and transcriptionally active chromatin tend to be replicated during early S-phase, while those near closed and transcriptionally inactive chromatin tend to be replicated during mid and late S-phase. Collectively, our imaging results constitute an in situ characterization of the dynamic epigenetic signatures of RFi, providing critical insights into the correlation between the timing of DNA replication and its local chromatin environment.
The Spatiotemporal Dynamics of RFi Propagation Are Directionally Specific.
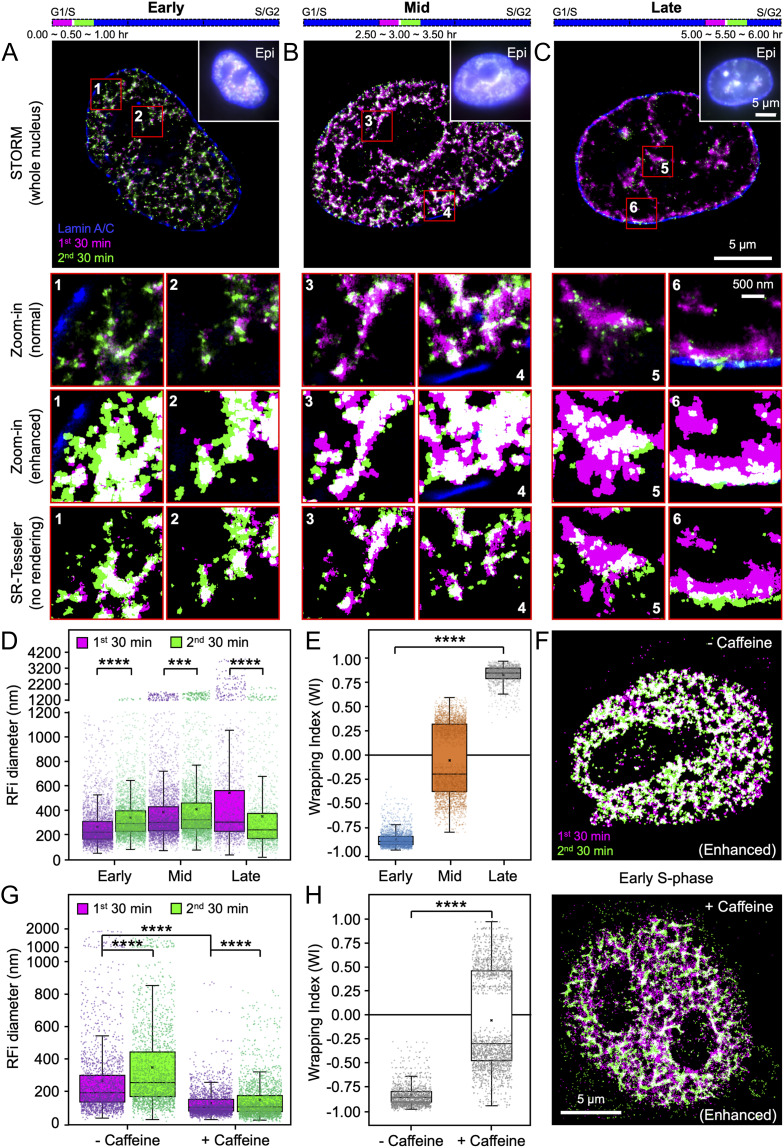
To probe the spatial organization and dynamics of individual RFi, we next set out to investigate the propagation pattern of DNA replication in space and time. To do so, newly replicated DNA during two consecutive 30-min time windows at three different stages of S-phase were labeled with two spectrally distinct dyes, Alexa 647 and Atto 550 (Materials and Methods and SI Appendix, Fig. S4A). In addition, lamin A/C in the nuclear lamina was immunofluorescently labeled with a third dye, Atto 488 (Fig. 4 A–C). Upon multicolor STORM imaging of the labeled cells, the relative spatial distributions of the foci during both time windows reveal a unique spatiotemporal pattern of RFi progression. In early S-phase, DNA synthesized during the second window occupies a larger area and envelopes DNA synthesized during the first window, a trend that could be better visualized by enhancing the contrast between the two colors or directly plotting the coordinates of RFi localizations output by SR-Tesseler without pixel rendering (Fig. 4 A, Insets). Such contrast enhancement serves solely as a visualization aid and does not introduce additional artifacts in STORM images (SI Appendix, Fig. S5); all subsequent quantitative analyses, however, were performed on coordinates of raw localizations output by SR-Tesseler. Three-dimensional STORM imaging excluded the possibility that the colocalization pattern observed in two-dimensional images results from structures that did not overlap in the z-direction (SI Appendix, Fig. S4B). Significantly, this spatial propagation pattern becomes less pronounced during mid S-phase (Fig. 4B) and reverses its radial directionality in late S-phase (Fig. 4C), during which the DNA synthesized in the first window envelopes the DNA synthesized in the second window and occupies a larger spatial spread.
Fig. 4.
RFi during early and late S-phase display opposing spatiotemporal patterns of propagation dynamics. (A–C) Newly replicated DNA during two consecutive 30-min windows (purple and green) at the beginning of early (A, 0∼1.0 h), mid (B, 2.5∼3.5 h), and late (C, 5.0∼6.0 h) S-phase were labeled (purple: first window; green: second window; white: colocalized RFi), together with lamin A/C (blue) to demarcate the nuclear boundary of each cell. Multicolor STORM images of single nuclei are shown in comparison with conventional images (Insets) of the same nuclei. The purple and green bars at the top indicate the relative temporal positions of the labeling periods during S-phase (blue bars, not drawn to scale). Insets (numbered 1 through 6) show zoomed-in areas (in red boxes), displaying both normal contrast (Top) and enhanced contrast (Middle) between the two colors for better visualization, as well as direct plotting of coordinates of RFi localizations output by SR-Tesseler without pixel rendering (Bottom) for comparison; white indicates colocalization. (D and E) Diameter (D) and WI (E) for RFi produced during the two labeling windows at early, mid, and late S-phase. (F) Images of RFi replicated in two consecutive 30-min windows (purple and green) at the beginning of early S-phase without (control, Top) and with (Bottom) 5 mM caffeine treatment. (G and H) Impact of caffeine treatment on RFi diameter (G) and WI (H). Data are shown as box-and-whisker plots. Each dot denotes a single RF. P values are determined by two-tailed Student’s t test; ***P < 0.001; ****P < 0.0001; n = 3,365 and 3,356 (early), 3,743 and 3,637 (mid), and 1,391 and 1,273 (late) RFi from 13 cells at each stage for D; 2,535, 3,241, and 1,180 RFi from 13 cells at each stage for E; 2,070 and 2,051 (− Caffeine) and 2,357 and 2,304 (+ Caffeine) RFi from 8 cells for G; and 1,446 and 1,954 RFi from 8 cells for H. Source data are provided as Dataset S1.
Such propagation dynamics can be more quantitatively illustrated by measuring the sizes of RFi in both time windows (Fig. 4D). To better quantify the radial directionality of RFi propagation, we further defined a wrapping index (WI) based on the extent of colocalization between the RFi from the two time windows as
where and denote the areas occupied by the RFi from the first and second time windows, respectively, and denotes their colocalized area. A WI of 1 indicates that the RFi in the first time window completely wraps around those in the second window, whereas a WI of −1 indicates the opposite (see SI Appendix, Fig. S4C for schematic illustration of this definition). From our STORM images, we found that the mean value of WI changed from −0.88 in early S-phase to +0.83 in late S-phase (Fig. 4E), clearly illustrating the reversal in the radial directionality of RFi propagation. A similar reversal was observed when the order of the two dye-labeled nucleotides used was swapped (SI Appendix, Fig. S4 D and E), thereby eliminating the possibility that such a propagation pattern could be the consequence of labeling and detection artifacts associated with specific dyes. Moreover, when cells were treated with 5 mM caffeine, a known inhibitor of cell cycle checkpoint that shifts origin firing from late to early S-phase (49), the RFi distribution pattern in early S-phase became similar to that in mid S-phase, especially around nuclear periphery and nucleoli (Fig. 4F). In addition, the size of RFi in both time windows decreased drastically (Fig. 4G), and the radial propagation pattern observed previously for early S-phase was abrogated, as indicated by a shift in the mean value of WI from −0.84 to −0.09 upon caffeine treatment (Fig. 4H).
CTCF Regulates RFi Morphology and Spatiotemporal Propagation Dynamics.
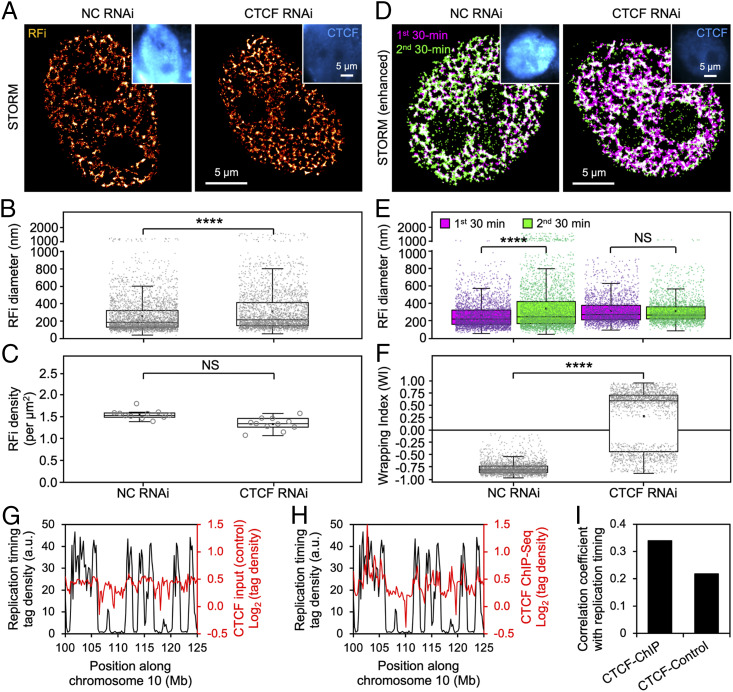
In order to understand the mechanistic basis underlying the unique pattern of RFi propagation, we remind ourselves of the prominent role that CTCF plays in regulating 3D genome architecture through forming chromatin loops and hypothesize that CTCF-organized loop structures could serve both as structural anchor for the replicating DNA as well as spatial constraints for their propagation. To that end, we knocked down CTCF expression in HeLa cells using RNA interference, which we confirmed at both messenger RNA and protein levels using qPCR, immunostaining, and Western blotting, respectively (Fig. 5A and SI Appendix, Fig. S6). Down-regulation of CTCF enlarged early S-phase RFi size without significantly reducing their density (Fig. 5 B and C). More importantly, the radially specific propagation pattern of RFi described in Fig. 4 was diminished upon CTCF knockdown (Fig. 5D), as evidenced by significant changes in the relative sizes of RFi (Fig. 5E) as well as WI (Fig. 5F), which exhibits a drastic reversal in mean value from −0.79 to +0.22. These results strongly suggest that CTCF and the chromatin loop structures it mediates act as a key regulator of RFi morphological and propagation dynamics.
Fig. 5.
CTCF regulates RFi morphology and spatiotemporal propagation dynamics. (A) STORM images of early S-phase RFi in the nuclei of cells transfected with either nonspecific control (NC) or CTCF siRNA. Insets show the successful down-regulation of CTCF (blue) upon RNA interference. (B and C) Diameter (B) and density (C) of RFi in cells treated with either NC or CTCF siRNA. (D) Dual-color STORM images of newly replicated DNA labeled in two consecutive 30-min windows (purple and green) at the beginning of early S-phase in the nuclei of cells treated with either NC or CTCF siRNA. (E and F) Diameter (E) and WI (F) for RFi produced during the two labeling windows in cells treated with either NC or CTCF siRNA. (G and H) Replication timing profile (black) along human chromosome 10 mapped onto ChIP-Seq data (red) of either control (G) or CTCF (H). (I) Correlation coefficient between replication timing profile and ChIP-Seq data for both control and CTCF at a binning size of 200 kb. Data are shown as box-and-whisker plots. Each dot denotes a single RF, and each circle denotes a single cell. P values are determined by two-tailed Student’s t test; ****P < 0.0001; NS: not significant. n = 3,089 and 2,661 RFi from 11 cells for B; 11 cells for C; 3,045, 3,059, 2,324, and 2,311 RFi from 11 cells for E; and 2,431 and 1,849 RFi from 11 cells for F. Source data are provided as Dataset S1.
To further correlate the RFi propagation dynamics mediated by CTCF-organized chromatin structures with replication timing, we mapped the replication timing profile along human chromosomes onto chromatin immunoprecipitation sequencing (ChIP-Seq) data for both control input and CTCF (Fig. 5 G and H; only a specific region along chromosome 10 is shown). The CTCF binding sequences exhibit a substantially higher level of correlation with the replication timing profile as compared to the control sequence (Fig. 5I). These bioinformatic analyses, along with our superresolution imaging results, point to the critical role played by CTCF-organized chromatin structures in facilitating the activation of DNA replication and its radial propagation within individual RFi.
CoREP Model for Nonrandom Replication Activation within Early S-Phase RD.
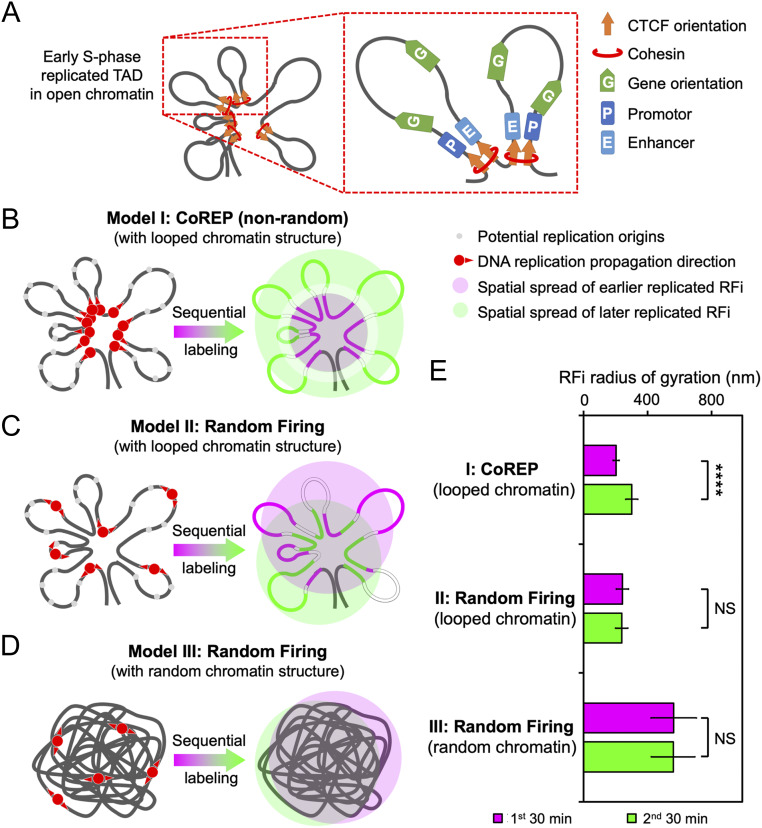
To account for the observed link between the propagation pattern of RFi and CTCF-mediated chromatin organization during early S-phase, we propose a spatiotemporal model, CoREP, for the nonrandom activation and propagation of DNA replication at the sub-RD level (Fig. 6). Our model is based on previous findings that an RD contains one or several TADs [each with a mean size of 880 kb (17)] and that each TAD contains 4∼6 CTCF-organized DNA loops [each with a mean size of 185 kb (40)]. These loops, occupied by promotors, enhancers, and active genes, are organized at their anchor sites by CTCF and cohesin (Fig. 6A). Our observed spatiotemporal pattern of RFi propagation suggests that replication activation events may take place preferentially at CTCF-mediated loop anchors within each RD, which then propagate to the periphery of the domain, thereby giving rise to the radially outward propagation pattern observed during early S-phase (Fig. 6B). Alternatively, it is also possible that DNA replication origins could be selectively activated at the periphery of each RD, and chromatin structural reorganization subsequently drives the DNAs that are replicated earlier toward the interior of the domain and those replicated later to the exterior (SI Appendix, Fig. S7). While our current data cannot differentiate between these two models, they unambiguously argue against the random activation of replication origins within RD, regardless of the structural architecture adopted by the chromatin (Fig. 6 C and D).
Fig. 6.
CoREP model for nonrandom replication activation and propagation. (A) Schematic diagram of an early S-phase TAD, organized into looped structures by CTCF and cohesin located at the base; each loop contains promotors, enhancers, and active genes. (B–D) Three potential models for the spatiotemporal propagation of DNA replication in early S-phase: CoREP model (B), in which replication is preferentially activated on CTCF-organized looped chromatin structures; random firing model with looped chromatin structures (C); and random firing model with random chromatin structures (D). (E) Comparison of simulated radii of gyration of RFi produced in two consecutive 30-min labeling windows, using the three models described in B–D. Error bars denote mean ± SD. P values are determined by two-tailed Student’s t test; ****P < 0.0001; NS: not significant. n = 500 chromatin configurations for E.
To further validate our model, we simulated the propagation of RFi for each of these models and calculated the radius of gyration of the simulated RFi (see Materials and Methods and SI Appendix, Figs. S8 and S9 for details). We found that the experimentally observed RFi size (Fig. 4D) agree much better with the simulated radius of gyration using the CoREP model as compared to those using random firing models (Fig. 6E). Collectively, these results strongly support a chromatin structure-mediated mechanism for the nonrandom activation and propagation of DNA replication.
Discussion
DNA replication in mammalian cells takes place at thousands of RDs across the genome and must be coordinated precisely to ensure the complete and faithful duplication of the genomic information in the cell. While previous studies have shed much light on the regulation of replication at the RD level and found the activation of replication to be correlated with a number of factors such as G-quadruplex motifs (14), chromatin modifications (10, 50, 51), transcriptional activity (9), and cis-regulatory elements (52), within an RD the choice of which replication origins to fire remains poorly understood. In this study, we fill a critical gap in our understanding of the spatiotemporal coordination of DNA replication by performing quantitative characterization of the in situ epigenetic environment of RFi in relation to replication activation and propagation across different stages of S-phase at the single-cell level. Our superresolution imaging data reveal a distinct radial pattern of replication propagation within individual RDs and strongly suggest a nonrandom selection mechanism for replication activation at early S-phase. The CoREP model we proposed further establishes the role that CTCF plays in this process and points to the key involvement of local chromatin structures in controlling replication activation at early S-phase.
While our findings are in line with previous reports of the role of CTCF and cohesin in organizing the genome into numerous “contact domains” (40, 41, 53, 54) as well as the enrichment of CTCF and cohesin at TAD boundaries (18) and replication sites (53), a recent study has found that CTCF depletion minimally impacts replication timing at the RD level (52). Such a seeming discrepancy could potentially be reconciled by the fact that the propagation pattern we have observed results from the cumulative DNA replicated in situ within individual RFi over a 30- to 60-min time window and thus may not fully correspond with the replication timing profile captured at rather different temporal and spatial (either Cartesian space or sequence space) scales using high-throughput sequencing. In fact, the distinct propagation pattern we revealed highlights the ability of in situ superresolution imaging in providing a powerful approach complementary to sequencing-based methods for probing DNA replication at unprecedented level of detail.
In summary, our study paves the way for further interrogations into the spatiotemporal organization and dynamics of DNA replication and other associated intranuclear processes (55). For instance, to pinpoint the precise relationship between the RFi observed in Cartesian space and the RDs in sequence space revealed by replication timing profiles, specific RD sequences could be colabeled using DNA fluorescence in situ hybridization and their colocalization with RFi quantitatively measured by STORM. The roles of CTCF, cohesin, as well as other factors implicated in replication timing during mid and late S-phase that have not been fully accounted for in our CoREP model, also warrant further systematic investigation, especially in light of the finding that cohesin and CTCF could differentially affect chromatin architecture (54). Moreover, as chromatin structures organized by CTCF/cohesin have been found to also spatially coordinate RNA polymerase II transcription (41), our findings also provide a framework for understanding the coupling between replication and transcription, since open chromatin around transcriptionally active promoters could facilitate the selection of replication origins in the vicinity, which might in turn explain the observation that transcriptionally active genes tend to replicate early while inactive ones tend to replicate late (41). In fact, replication initiation sites have often been found in close proximity to transcription initiation sites (14, 56). Combining the approaches developed here with those reported previously for probing RNA polymerase II-mediated transcription (57, 58) and transcription factor binding (59, 60) will allow us to elucidate the extent to which replication and transcription are correlated in space and time (61), thus shedding important light on the mechanistic cooperation between these two processes.
Materials and Methods
Cell Culture and Synchronization.
HeLa-S3 immortalized cell line (PubMed ID: 5733811) was obtained from Dr. Wei Guo, University of Pennsylvania, Philadelphia, PA, and cultured in high-glucose Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (Gibco), 100 U/mL penicillin (Thermo Fisher), and 100 mg/mL streptomycin (Thermo Fisher) at 37 °C with 5% CO2. The duration of the cell cycle was determined by counting the number of cells every 8 h for a continuous period of 72 h. Cells were synchronized to the G1/S boundary with sequential treatment of 2 mM thymidine (Sigma-Aldrich) for 15 h, normal culture medium for 10 h, and 2 μg/mL aphidicolin (Sigma-Aldrich) for 15 h. The synchronization efficiency was determined by measuring the cellular DNA content profile in a population of DAPI-stained cells, fixed at 30 min after release into S-phase, using fluorescence-activated cell sorter (MoFlo; Beckman Coulter) at excitation wavelength of 340 to 380 nm.
RNA Interference.
HeLa-S3 cells were transfected with the following small interfering RNA (siRNA) oligos (GenePharma) using Lipofectamine 2000 (Thermo Fisher) according to manufacturer’s instructions: Nonspecific control: CGUACGCGGAAUCUUCGATT (sense) and UCGAAGUAUUCCGCGUACGTT (anti-sense); CTCF: GGAGCCUGCCGUAGAAAUUTT (sense) and AAUUUCUACGGCAGGUCCTC (anti-sense).
Cells were harvested at 48 h after transfection, followed by Western blot and qPCR quantification of CTCF expression.
RFi Labeling.
At specific times after releasing the cells from G1/S boundary, RFi in the cell nucleus were labeled by introducing pulses of thymidine analogs of different durations (10, 30, or 60 min). Dye-labeled dUTPs (either with Alexa 647 [Invitrogen] or Atto 550 [MolBi Tech]) were delivered into the nucleus via transfection using FuGENE 6 reagent (Promega), while EdU (Life Technologies), being uncharged, was directly added into the culture medium and subsequently conjugated with a dye using the Click-iT EdU Imaging Kit (Life Technologies) according to the manufacturer’s instructions. When performing two-color labeling, the cells were washed with culture medium three times after incubating with the first dye before adding in the second dye. For caffeine treatment, a final concentration of 5 mM caffeine (Sigma-Aldrich) was added into the culture medium 30 min before the release and during the 30-min labeling time.
Upon labeling, the cells were incubated with extraction buffer (0.1 M Pipes at pH 7.0, 1 mM EGTA, 1 mM MgCl2, and 0.2% [vol/vol] Triton X-100 [all Sigma-Aldrich]) for 60∼70 s and fixed with 4% (wt/vol) paraformaldehyde (Electron Microscopy Science) and 0.1% (vol/vol) glutaraldehyde (Electron Microscopy Science) in phosphate-buffered saline (PBS) (Sigma-Aldrich) at room temperature for 15 min.
Immunofluorescence Labeling.
To label nuclear markers of interest, the cells were blocked and permeabilized with blocking buffer (5% [wt/vol] bovine serum albumin [Jackson Immuno Research] and 0.5% [vol/vol] Triton X-100 in PBS) for 30 min and then incubated with 1∼10 μg/mL (according to the manufacturer’s instructions) primary antibody against the protein of interest in blocking buffer for 60 min at room temperature (except for histone H2B antibody, for which 20 μg/mL with 10 h of incubation at 4 °C is needed). The following primary antibodies were used: PCNA (mouse monoclonal, sc-56; Santa Cruz), H3K27ace (rabbit polyclonal, 07-360; Millipore), H3K4me3 (rabbit polyclonal, 39159; Active Motif), CTCF (rabbit monoclonal, ab128873; Abcam), SUZ12 (rabbit polyclonal, a302-407a; Bethyl Laboratories), Lamin A/C (mouse monoclonal, ab40567; Abcam), and Histone H2B (goat polyclonal, sc8650; Santa Cruz). After washing with PBS three times, the cells were incubated with 2∼5 μg/mL fluorescently labeled secondary antibody against the primary antibody in blocking buffer for 40 min at room temperature. After washing with PBS three times, the cells were postfixed with 4% (wt/vol) paraformaldehyde in PBS for 10 min and stored in PBS for up to 1 wk before STORM imaging.
Secondary antibodies used were labeled with either Atto 488 (41698; Sigma-Aldrich), Cy3B (PA63101; GE Healthcare), or Alexa 647 (A20006; Thermo Fisher), all conjugated to a monofunctional N-hydroxysuccinimide ester, for 30 min at room temperature (62). Labeled antibody was removed from the free dye by gel filtration using a NAP5 column (17085302; GE Healthcare). The labeling ratio was controlled at 2∼5 and measured with NanoDrop (Thermo Fisher).
DNA Combing.
After incubation with EdU as described above, HeLa cells were harvested and resuspended in PBS to a concentration of 5 × 105 cells per mL and diluted fourfold with unlabeled cells at the same concentration. Cells were lysed with lysis buffer [200 mM Tris⋅HCl at pH 7.5, 50 mM ethylenediaminetetraacetic acid, and 0.5% (wt/vol) sodium dodecyl sulfate], and the lysate was added onto a coverslip and allowed to dry for 3∼5 min. The coverslip was then tilted at 15° to allow the DNA to flow down slowly and then air-dried and fixed with 3:1 methanol/acetic acid (vol/vol), followed by incubation with labeling reaction mixture from the Click-iT EdU Imaging Kit (Life Technologies) according to the manufacturer’s instructions. While potential difference in the efficiency of Click reaction between stretched DNA fibers and RFi in situ was not quantitatively assessed, such difference in exposedness between in vitro and the cellular environments should minimally impact Click reagents’ ability to access and label the RFi, and therefore the accuracy of our calibration. The physical lengths of the labeled DNA fibers were then measured with fluorescence microscopy and converted to base pairs using a stretching factor of 2 kb/μm determined by following the protocol detailed in ref. 33. The number of single-molecule localizations was obtained by direct counting of coordinates and used to derive the translation factor for the number of base pairs per localization. Identical microscope configurations (illumination mode, laser power, camera setting, etc.), imaging conditions, and analysis algorithm as those used for imaging RFi in situ were applied to ensure the accuracy of calibration between single-molecule localizations observed in vitro and RFi sequence length in situ.
STORM Imaging and Data Analysis.
STORM imaging and analysis were performed as previously described (47, 63). Superresolution images of Alexa 647-, Cy3B-, Atto 550-, and Atto 488-labeled RFi, DNA fibers, or nuclear markers of interest were continuously acquired for up to 100,000 frames under the excitation of 647-nm, 561-nm, or 488-nm lasers (MPB Communications) at a power density of 3∼5 kW/cm2 and photoactivated with a 405-nm laser (Coherent) at a power density of 0.5 kW/cm2. The excitation and photoactivation laser beams were reflected by a custom-designed polychromatic mirror (z405/488/561/640; Chroma). To correct for lateral drift between frames, bright-field images of 3-μm-sized glass beads (Weike Tech) placed in the sample dish prior to imaging were acquired for 100 ms every 1,000 ms (SI Appendix, Fig. S1A). An imaging buffer (100 mM Tris⋅HCl at pH 8.0, 20 mM NaCl, and 10% glucose, all from Sigma-Aldrich) with an oxygen scavenger system (60 mg/mL glucose oxidase and 6 mg/mL catalase, both from Sigma-Aldrich) was used for all STORM imaging.
STORM image analysis, drift correction, RFi quantification, and image rendering were performed using the Insight3 (63), custom-written codes in MATLAB (2011a; MathWorks), SR-Tesseler (Interdisciplinary Institute for Neuroscience) (32), and ImageJ (National Institutes of Health). For RFi identification, a density factor of 2 was applied to identify the RFi as the default parameter in SR-Tesseler with minimum bias (32). Coordinates of single-molecule localizations constituting RFi were output to MATLAB for further analysis and image reconstruction. The colocalization of two molecules in STORM images was defined as their centroid positions being not more than 100 nm apart from each other. Colocalization ratio in conventional images was analyzed with the Colocalization Analysis plugin in ImageJ. WI between two RFi was defined as
where and denote the areas occupied by the RFi from the first and second time windows, respectively, and denotes their colocalized area. It should be pointed out that when the value of Sc is very small, the computational algorithm will automatically reject such scenarios as being not colocalized, based on the definition of the colocalization threshold set; as such, a small fraction of WI values in the −0.2∼0.2 range will not be reflected in the WI histograms. This, however, does not significantly affect the mean values of WI calculated, since these unreflected WI values should be symmetrically distributed around 0.
Sequencing Data and Correlation Analyses.
Percentage-normalized replication timing profiles were downloaded from ENCODE at http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeUwRepliSeq. CTCF ChIP-Seq data were also downloaded from ENCODE, but remapped to hg19 using bowtie and reanalyzed using MACS. All chromosomes were divided into bins of different lengths (1 kb, 5 kb, 10 kb, 50 kb, 100 kb, 200 kb, 500 kb, 1 Mb, 2 Mb, and 5 Mb) at the same position. Median of each bin was calculated for both replication timing and CTCF ChIP-Seq profiles, and correlation coefficient was calculated for each length using the Pearson method in R (https://github.com/taoliu/MACS/wiki/Build-Signal-Track).
Modeling and Simulation.
The spatiotemporal spread of replicated chromatin was simulated in 3D using different models of replication activation. Briefly, coarse-grained models for nucleosome and linker DNA were first used to sample short chromatin chains, which were then used to construct long chromatin chains with internal loops to simulate chromatin structures organized by CTCF and cohesin. Such long chromatin chains were then split into two segments to represent DNA replicated during the two consecutive labeling time windows, and their radii of gyration were calculated for both the CoREP and random firing models. See SI Appendix for more details.
Statistical Analysis.
Statistical significance was assessed using two-tailed Student’s t test: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS: not significant. At least three experimental replicates were performed for all experiments.
Codes and Data Availability.
The localization coordinates used to generate the STORM images in Figs. 1–5 as well as all codes have been deposited in Zenodo (DOI: 10.5281/zenodo.3840656). Source data for statistics are provided as Dataset S1.
Supplementary Material
Acknowledgments
We thank Prof. Xiaowei Zhuang and Dr. Hazen Babcock (Harvard University) for help with optical setup of the STORM microscope, Prof. Bo Huang (University of California, San Francisco) for the gift of the Insight3 software, and the Core Facilities at School of Life Sciences, Peking University for imaging support. This work is supported by Australia National Health and Medical Council Grant APP1177374 and Australia National Heart Foundation Grant 102592 to Q.P.S.; National Medical Research Council of Singapore Open Fund-Young Individual Research Grant OFYIRG18nov-0019 and National University of Singapore Presidential Young Professorship start-up funds to Z.W.Z.; Beijing Advanced Innovation Center for Genomics at Peking University to X.S.X.; National Key R&D Program of China Grant 2017YFA0505300; and National Science Foundation of China Grant 21825401 to Y.S.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.L. is a guest editor invited by the Editorial Board.
Data deposition: The localization coordinates used to generate the STORM images in Figs. 1–5 as well as all codes have been deposited in Zenodo (DOI: 10.5281/zenodo.3840656).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001521117/-/DCSupplemental.
References
- 1.Perumal S. K., Yue H., Hu Z., Spiering M. M., Benkovic S. J., Single-molecule studies of DNA replisome function. Biochim. Biophys. Acta 1804, 1094–1112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Oijen A. M., Loparo J. J., Single-molecule studies of the replisome. Annu. Rev. Biophys. 39, 429–448 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Jacob F., The replicon: Thirty years later. Cold Spring Harb. Symp. Quant. Biol. 58, 383–387 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Leonard A. C., Méchali M., DNA replication origins. Cold Spring Harb. Perspect. Biol. 5, a010116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hozák P., Hassan A. B., Jackson D. A., Cook P. R., Visualization of replication factories attached to nucleoskeleton. Cell 73, 361–373 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Rhind N., Gilbert D. M., DNA replication timing. Cold Spring Harb. Perspect. Biol. 5, a010132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiratani I. et al., Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryba T. et al., Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 20, 761–770 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrova D. S., Gilbert D. M., The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4, 983–993 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Fragkos M., Ganier O., Coulombe P., Méchali M., DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 16, 360–374 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Besnard E. et al., Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 19, 837–844 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ferguson B. M., Fangman W. L., A position effect on the time of replication origin activation in yeast. Cell 68, 333–339 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Rivera-Mulia J. C., Gilbert D. M., Replicating large genomes: Divide and conquer. Mol. Cell 62, 756–765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cayrou C. et al., Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 21, 1438–1449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaykov A., Nurse P., The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 25, 391–401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson D. A., Pombo A., Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pope B. D. et al., Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon J. R. et al., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boettiger A. N. et al., Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang W. et al., Correlative live and super-resolution imaging reveals the dynamic structure of replication domains. J. Cell Biol. 217, 1973–1984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J. et al., Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells. Cell Rep. 24, 873–882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bintu B. et al., Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson D. A., S-phase progression in synchronized human cells. Exp. Cell Res. 220, 62–70 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Ma H. T., Poon R. Y. C., Synchronization of HeLa cells. Methods Mol. Biol. 761, 151–161 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Dimitrova D. S., Visualization of DNA replication sites in mammalian nuclei. Methods Mol. Biol. 521, 413–436 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Koberna K. et al., Electron microscopy of DNA replication in 3-D: Evidence for similar-sized replication foci throughout S-phase. J. Cell. Biochem. 94, 126–138 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Ma H. et al., Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 143, 1415–1425 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baddeley D. et al., Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res. 38, e8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chagin V. O. et al., 4D Visualization of replication foci in mammalian cells corresponding to individual replicons. Nat. Commun. 7, 11231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cseresnyes Z., Schwarz U., Green C. M., Analysis of replication factories in human cells by super-resolution light microscopy. BMC Cell Biol. 10, 88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triemer T. et al., Superresolution imaging of individual replication forks reveals unexpected prodrug resistance mechanism. Proc. Natl. Acad. Sci. U.S.A. 115, E1366–E1373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levet F. et al., SR-tesseler: A method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 12, 1065–1071 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Schwob E. et al., “Use of DNA combing for studying DNA replication in vivo in yeast and mammalian cells” in DNA Replication: Methods and Protocols, Vengrova S., Dalgaard Z. J., Eds. (Humana Press, Totowa, NJ, 2009), pp. 673–687. [DOI] [PubMed] [Google Scholar]
- 34.Malínsky J. et al., The supply of exogenous deoxyribonucleotides accelerates the speed of the replication fork in early S-phase. J. Cell Sci. 114, 747–750 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Rhind N., DNA replication timing: Random thoughts about origin firing. Nat. Cell Biol. 8, 1313–1316 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chagin V. O., Stear J. H., Cardoso M. C., Organization of DNA replication. Cold Spring Harb. Perspect. Biol. 2, a000737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S. et al., Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birney E. et al.; ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute , Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ong C. T., Corces V. G., CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao S. S. et al., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Z. et al., CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163, 1611–1627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barski A. et al., High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Creyghton M. P. et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peric-Hupkes D., van Steensel B., Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb. Symp. Quant. Biol. 75, 517–524 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Cao R., Zhang Y., SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15, 57–67 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Rinn J. L. et al., Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G. et al., PTEN regulates RPA1 and protects DNA replication forks. Cell Res. 25, 1189–1204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guelen L. et al., Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Marheineke K., Hyrien O., Control of replication origin density and firing time in Xenopus egg extracts: Role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 279, 28071–28081 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Berbenetz N. M., Nislow C., Brown G. W., Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet. 6, e1001092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eaton M. L., Galani K., Kang S., Bell S. P., MacAlpine D. M., Conserved nucleosome positioning defines replication origins. Genes Dev. 24, 748–753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sima J. et al., Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 176, 816–830.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillou E. et al., Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 24, 2812–2822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuin J. et al., Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 111, 996–1001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Z., “Probing the spatio-temporal organizations and dynamics of gene expression and DNA replication in the mammalian cell nucleus,” PhD dissertation, Harvard University, Cambridge, MA (2015).
- 56.Martin M. M. et al., Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 21, 1822–1832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Z. W. et al., Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy. Proc. Natl. Acad. Sci. U.S.A. 111, 681–686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X. et al., Study of RNA polymerase II clustering inside live-cell nuclei using Bayesian nanoscopy. ACS Nano 10, 2447–2454 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Gebhardt J. C. et al., Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods 10, 421–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Z. W. et al., Quantifying transcription factor-DNA binding in single cells in vivo with photoactivatable fluorescence correlation spectroscopy. Nat. Protoc. 12, 1458–1471 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Chakalova L., Debrand E., Mitchell J. A., Osborne C. S., Fraser P., Replication and transcription: Shaping the landscape of the genome. Nat. Rev. Genet. 6, 669–677 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Du W. et al., Kinesin 1 drives autolysosome tubulation. Dev. Cell 37, 326–336 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Huang B., Wang W., Bates M., Zhuang X., Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michalet X. et al., Dynamic molecular combing: Stretching the whole human genome for high-resolution studies. Science 277, 1518–1523 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.