Significance
Cells need to cope with oxidative stress caused by oxygen free radicals from mitochondrial oxidative phosphorylation. We identify a role for E2F1 to halt cell cycle progression for cell survival upon oxidative stress. Oxidative stress induces sumoylation of E2F1, primarily at lysine 266, which specifically modulates E2F1 transcriptional activity to enhance cell cycle arrest for cell survival. Furthermore, we identify SENP3 as an E2F1-interacting partner and a desumoylating enzyme for E2F1. Oxidative stress inhibits the interaction between E2F1 and SENP3, and leads to accumulation of sumoylated E2F1. These data demonstrate that sumoylation of E2F1 is important for cell cycle arrest to provide cells the opportunity to cope with oxidative stress and survive.
Keywords: E2F1, oxidative stress, SENP3, SUMO2, sumoylation
Abstract
Oxidative stress is a ubiquitous threat to all aerobic organisms and has been implicated in numerous pathological conditions such as cancer. Here we demonstrate a pivotal role for E2F1, a cell cycle regulatory transcription factor, in cell tolerance of oxidative stress. Cells lacking E2F1 are hypersensitive to oxidative stress due to the defects in cell cycle arrest. Oxidative stress inhibits E2F1 transcriptional activity, independent of changes in association with Rb and without decreasing its DNA-binding activity. Upon oxidative insult, SUMO2 is extensively conjugated to E2F1 mainly at lysine 266 residue, which specifically modulates E2F1 transcriptional activity to enhance cell cycle arrest for cell survival. We identify SENP3, a desumoylating enzyme, as an E2F1-interacting partner. Oxidative stress inhibits the interaction between E2F1 and SENP3, which leads to accumulation of sumoylated E2F1. SENP3-deficient cells exhibit hypersumoylation of E2F1 and are resistant to oxidative insult. High levels of SENP3 in breast cancer are associated with elevated levels of E2F targets, high tumor grade, and poor survival. Given the prevalence of elevated levels of SENP3 across numerous cancer types, the SENP3-E2F1 axis may serve as an avenue for therapeutic intervention in cancer.
Cell division is a tightly coordinated process between progrowth stimuli, responding signaling pathways, and downstream effectors that execute the proliferative program. One event that must occur for successful cell division is the precise replication of chromatin. Many components required for replication have their expression tightly linked to the cell cycle to coordinate replication only when cells have sufficient energy and macromolecule resources to complete mitosis. One part of the regulatory network involved is the E2F protein family, which consists of eight members that are key transcriptional regulators of cell cycle progression and cell proliferation (1). E2F1-3 have a canonical role in promoting G1- and S-phase progression, and are inhibited by the pocket family proteins (Rb, p130, and p107) in quiescent cells (2).
With integral functions in promoting cell proliferation, it is unsurprising that the E2F/Rb has long been implicated in cancer, even serving as the original impetus for the “two-hit hypothesis” (3). Alterations in the pathway take various forms, but it has been found to be deregulated across most, if not all, tumor types (4, 5). With a less restricted transcriptional growth program in place, E2F/Rb-altered cells are poised for a more drastic proliferative phenotype after subsequent ensuing “hits” such as protooncogene activation.
Cancer cells have long been observed to generate high levels of hydrogen peroxide and other reactive oxygen species (6). While the aberrant activation of a single oncogene in quiescent cells can itself enhance the formation of reactive oxygen species, there are many other contributing factors (7, 8). Cells produce ATP through oxidative phosphorylation, a pathway that can inherently generate reactive oxygen species due to oxygen’s role as the terminal electron acceptor in the electron transport chain. Oxidative stress can enhance genome instability and lead to a subsequent enhancement of mutation rate and tumor progression (9). In addition to playing an established role in promoting cell growth, E2F1 has been shown to play an important role in regulating the expression of oxidative phosphorylation genes (10). With E2F1 playing a role in oxidative phosphorylation, as well as the high incidence of alterations in the E2F/Rb pathway in cancer, we sought out to investigate what role, if any, E2F1 has in cells responding to oxidative stress.
Here we establish that E2F1 plays a critical role in facilitating cell survival after oxidative stress by enhancing cell cycle arrest at the G1/S-phase checkpoint. Cell cycle arrest coincides with repression of E2F1 transcriptional activity and the accumulation of highly modified species determined to be the extensive addition of SUMO2 to E2F1. Furthermore, E2F1’s function in transcriptional modulation and enhancing cell viability after oxidative insult requires the SUMO2 acceptor site on E2F1. Conjugation status of SUMO2 on E2F1 is regulated in part by the desumoylating enzyme SENP3, that when deleted, results in hypersumoylation on E2F1 as well as enhanced cell resistance to oxidative stress. These results suggest a mechanism by which cells can respond to oxidative stress by directly altering the proliferative transcriptional program to enhance cell cycle arrest and subsequently grant cells additional time to respond to oxidative insult before resuming growth.
Results
E2F1 Facilitates Cell Survival upon Oxidative Stress.
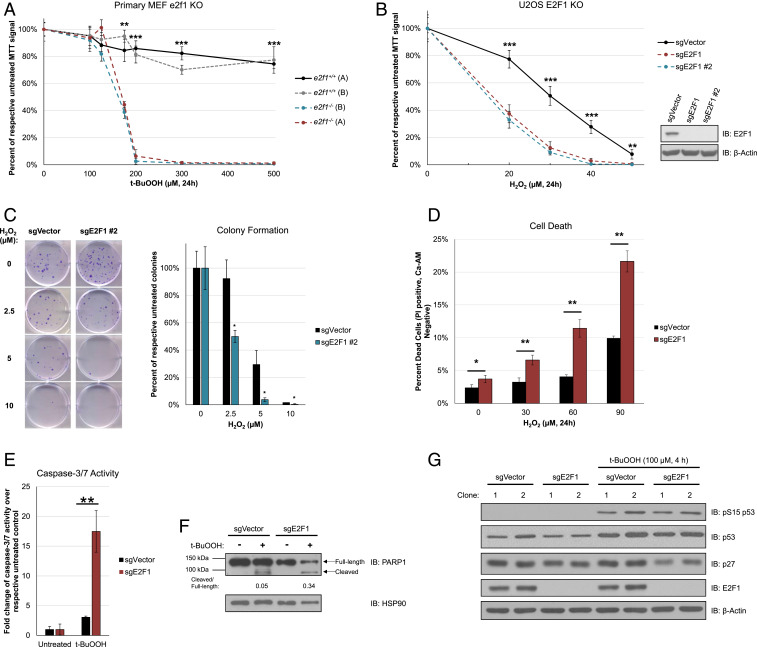
To first investigate how E2F1 functions in response to oxidative stress, we treated wild type or e2f1-null primary mouse embryonic fibroblasts (MEFs) (11, 12) with t-butyl hydroperoxide (t-BuOOH) and assessed cell viability via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Interestingly, e2f1-null primary MEFs were hypersensitive to t-BuOOH when compared to wild-type (WT) MEFs (Fig. 1A). E2F1 status in the primary MEFs was validated using qRT-PCR (SI Appendix, Fig. S1A). We sought to confirm these findings in CRISPR-Cas9-generated E2F1-knockout (KO) U2OS cells. Similar to the primary MEFs, E2F1-null U2OS cells (sgE2F1 and sgE2F1 #2, representing two different clones) exhibited diminished cell viability after oxidative stress insult when compared to the wild-type vector control for both hydrogen peroxide (Fig. 1B) and t-BuOOH (SI Appendix, Fig. S1B). We also wanted to assess if this phenomenon could impact clonogenic efficiency after insult with hydrogen peroxide. Indeed, E2F1-null cells formed fewer colonies compared to vector control, indicating hypersensitivity to oxidative stress (Fig. 1C). Finally, using calcein-AM and propidium iodide costaining, we determined that E2F1-null cells have a higher proportion of dead cells after treatment with hydrogen peroxide (Fig. 1D).
Fig. 1.
E2F1-deficient cells are hypersensitive to oxidative stress. (A) Cell viability of primary MEFs after 24 h t-BuOOH treatment at indicated concentrations as determined via MTT assay. Each cell line is normalized to respective untreated signal. Error bars represent mean ± SD (n ≥ 3). **P < 0.005, ***P < 5 × 10−6. (B) Cell viability of two E2F1-knockout U2OS cell lines, sgE2F1 and sgE2F1 #2, after 24 h H2O2 treatment at indicated concentrations as determined by MTT assay. Each cell line is normalized to respective untreated signal. A Western blot for E2F1 demonstrates knockout status. Error bars represent mean ± SD (n ≥ 3). **P < 0.05, ***P < 5 × 10−6. (C) Colony formation assay with representative images shown in sgE2F1 #2 cells after initial 24 h H2O2 treatment, followed by normal growth media. Error bars represent mean ± SD (n = 3). *P < 0.05. (D) Quantification of cell death in sgE2F1 cells after 24 h H2O2 treatment as determined by flow cytometry of calcein-AM and propidium iodide (PI) double-stained samples. Error bars represent mean ± SD (n = 3). *P < 0.05, **P < 0.005. (E) Caspase-3/7 activity in sgE2F1 cells after 8 h of 100 µM t-BuOOH treatment. (n = 3). **P < 0.005. (F) PARP1 expression in sgE2F1 cells after 8 h of 100 µM t-BuOOH treatment. Ratio is quantification of cleaved band divided by full-length band. (G) Expression of cell cycle inhibitors in sgE2F1 cells after 4 h of 100 µM t-BuOOH treatment. IB: immunoblotting for all relevant panels.
With an observed hypersensitivity to exogenous peroxides, we wanted to further explore what cellular mechanisms could be contributing to enhanced cell death in E2F1-deficient cells. Firstly, we wanted to determine if the observed cell death was through apoptosis. To first address this, we performed a Caspase-3/7 activity assay and determined that indeed there is enhanced Caspase-3/7 activity in E2F1-null cells when compared to the wild-type vector control (Fig. 1E). Additionally, we observed enhanced PARP1 cleavage in E2F1-null cells, a late stage marker for activated caspases during apoptosis (Fig. 1F). These data suggest that the observed oxidative stress-induced cell death in E2F1-null cells is through an induction of apoptosis.
With E2F1 having a role in the regulation of autophagy (13–15) and mitophagy (16, 17), we also tested whether the autophagy/mitophagy activation after oxidative stress is perturbed in E2F1-null cells. To examine this, we compared the expression of autophagy/mitophagy markers. While E2F1-null cells had a slightly higher level of basal autophagy/mitophagy, which is consistent with prior reports (15, 16), we observed no clear difference in the activation of autophagy/mitophagy after t-BuOOH treatment between two wild-type vector control clones, and two E2F1-null clones (SI Appendix, Fig. S2). We further examined the extent of p53 activation and observed no appreciable difference depending on E2F1 status. Interestingly, there was a marked reduction in p27Kip1 between the two E2F1-null clones when compared to wild-type vector controls (Fig. 1G). p27Kip1 is a key cell cycle inhibitor and also plays an active antiapoptotic role in some contexts (18–21). Taken together, these findings suggest a role for E2F1 in facilitating cell survival after oxidative stress insult.
E2F1 Potentiates Oxidative Stress-Induced G1/S-Phase Cell Cycle Arrest.
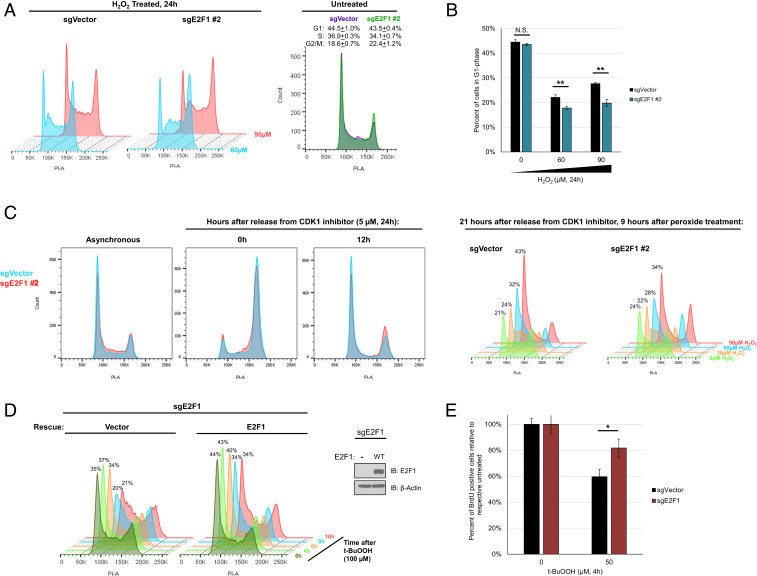
With E2F1 having a canonical role in regulating cell cycle progression, as well as the observed difference in p27Kip1 expression, we postulated that its function in regulating the cell cycle could be linked to the hypersensitivity of E2F1-null cells to oxidative stress. After treating cells for 24 h with hydrogen peroxide, E2F1-knockout cells had significantly less cells in the G1 fraction (Fig. 2 A and B). To further explore this finding we used CDK1 inhibitor RO-3306 to synchronize cells in G2/M phase, followed by release and subsequent treatment with hydrogen peroxide once cells were in G1 phase. Progression into S phase was then monitored 9 h after treatment across multiple concentrations of hydrogen peroxide. Interestingly, hydrogen peroxide-treated sgVector control cells retained a higher proportion of cells in G1 phase than E2F1-knockout cells, especially at higher doses of treatment (Fig. 2C). We believe this difference is due to a difference in the duration of G1/S-phase cell cycle arrest after cellular insult from oxidative stress. We further explored this idea with an additional cell cycle synchronization experimental design (SI Appendix, Fig. S3A), in which G1-phase cells were treated with t-BuOOH and then harvested at different time points after treatment. In this setting, sgVector cells had an obvious delay in G1/S-phase progression caused by t-BuOOH treatment as anticipated, but there was no appreciable delay in the G1/S progression of sgE2F1 cells after t-BuOOH treatment (SI Appendix, Fig. S3B).
Fig. 2.
E2F1-deficient cells show less efficient cell cycle arrest than WT controls in response to oxidative stress. (A) Overlay of representative cell cycle profiles after 24 h of H2O2 treatment (untreated, 60 µM, and 90 µM). The y axis scaling between treated sgVector and sgE2F1 #2 samples is identical. (B) Quantification of the percentage of cells in G1 phase of the cell cycle after 24 h of H2O2 treatment. Error bars represent mean ± SD (n = 3). **P < 0.005; N.S., not significant. (C) sgVector or sgE2F1 #2 cells were synchronized for 24 h with CDK1 inhibitor and subsequently released. The 0 h timepoint corresponds to the cell cycle status at time of release, while the 12 h timepoint corresponds to cell cycle status at time of t-BuOOH treatment. After t-BuOOH addition, cells were allowed to progress for 9 h prior to harvesting and assessing cell cycle progression. Percentage values shown are for amounts of cells in G1 phase. Scaling between 21 h sgVector and sgE2F1 #2 cells is identical. (D) sgE2F1 cells were stably rescued with an empty vector or WT E2F1, and subsequently treated with t-BuOOH. Cells were harvested at indicated timepoints and analyzed for cell cycle distribution. Percentage values shown are for the amounts of cells in G1 phase. A Western blot was performed to verify rescue of E2F1 expression, IB: immunoblotting. (E) BrdU incorporation assay in sgVector or sgE2F1 cells. Cells were treated with t-BuOOH for 2 h prior to the addition of BrdU for 2 additional hours. Samples were analyzed with flow cytometry, and values for each cell line were normalized to the percentage of BrdU-positive cells for the respective untreated samples. Error bars represent mean ± SD (n = 3). *P < 0.05.
To confirm that this phenomenon is indeed E2F1 dependent, we rescued E2F1 expression in the E2F1-null cells with a Cas9-resistant WT E2F1 construct. By treating asynchronous cells with t-BuOOH and assaying the cell cycle profiles at multiple time points, we were able to observe a synchronization phenomenon resulting from the transient cell cycle arrest and release in response to oxidative stress. The appearance of this synchronized population is evident at 8 h after treatment for the empty vector sgE2F1 cells, while those rescued with WT E2F1 exhibited an additional delay in progression (Fig. 2D). These data are consistent with the idea that E2F1 is responsible for prolonging oxidative stress-induced cell cycle arrest. Lastly, to reinforce this idea, we assayed E2F1-null cells for changes in BrdU incorporation after t-BuOOH treatment. Indeed, t-BuOOH treatment failed to inhibit bromodeoxyuridine (BrdU) incorporation in sgE2F1 cells to the same extent as in the E2F1-proficient sgVector cells (Fig. 2E). Taken together, these data indicate that E2F1 plays an important role in enhancing oxidative stress-mediated cell cycle arrest.
E2F1 Transcriptional Activity Is Repressed by Oxidative Stress while Its Interaction with Rb Is Not Altered and It Remains Bound to Chromatin.
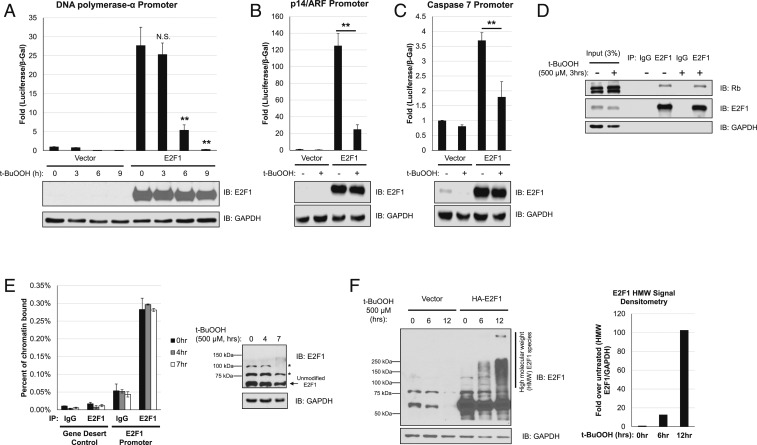
In light of E2F1 playing a key role in facilitating cell cycle arrest after oxidative stress insult, we sought to investigate if changes to E2F1 transcriptional activity could be contributing to this phenomenon. To first address this possibility, we utilized a DNA polymerase α promoter-luciferase reporter assay to evaluate E2F1 transcriptional activity (11) in U2OS cells. Strikingly, E2F1 transcriptional activity was inhibited following exposure to t-BuOOH, independent of changes in E2F1 protein level (Fig. 3A). Similar results were also observed with two other E2F1 activity reporter assays, p14/ARF promoter (11) and Caspase-7 promoter (22) luciferase reporter assays (Fig. 3 B and C, respectively). These data showed that E2F1 transcriptional activity can be modulated by oxidative stress.
Fig. 3.
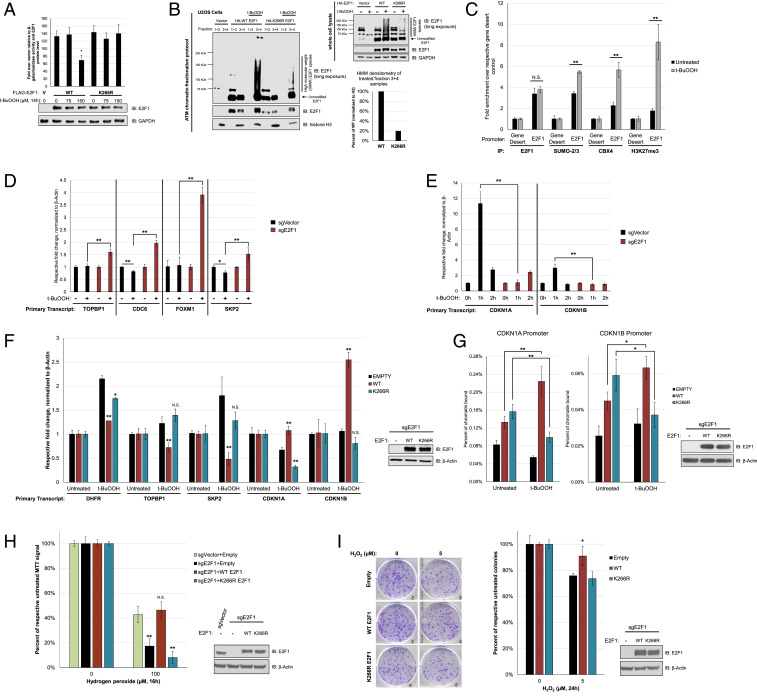
E2F1 transcriptional activity is altered by oxidative stress, which is independent of changes in association with Rb or chromatin. (A) U2OS cells were transiently transfected with a DNA polymerase-α promoter luciferase reporter and cytomegalovirus (CMV)-β-galactosidase, as well as E2F1 or an empty vector. Forty-eight hours after transfection, cells were subjected to 500 µM t-BuOOH, and harvested at indicated timepoints. Luciferase signal was normalized to internal β-galactosidase activity. Samples were additionally processed for Western blot analysis to validate E2F1 expression. Error bars represent mean ± SD (n = 3). Not significant, N.S., **P < 0.005. (B) E2F1 transcriptional activity assayed by a p14/ARF promoter luciferase reporter in U2OS cells overexpressing HA-E2F1, with or without 6 h of 500 µM t-BuOOH treatment. Luciferase signal was normalized to internal β-galactosidase activity. Error bars represent mean ± SD (n = 3). **P < 0.005. (C) E2F1 transcriptional activity assayed by a Caspase-7 promoter luciferase reporter in U2OS cells overexpressing HA-E2F1, with or without 6 h of 500 µM t-BuOOH treatment. Luciferase signal was normalized to internal β-galactosidase activity. Error bars represent mean ± SD (n = 3). **P < 0.005. (D) Coimmunoprecipitation of endogenous Rb with E2F1 in U2OS cells with or without t-BuOOH treatment. (E) Percentage of the E2F1 promoter bound by E2F1 as determined by chromatin-immunoprecipitation of E2F1 under varying lengths of t-BuOOH treatment in U2OS cells (Left) with a corresponding Western blot for E2F1 expression (Right). *: nonspecific bands. (F) Transient overexpression of HA-E2F1 in HEK 293T cells reveals the formation of extensively modified forms of E2F1 after t-BuOOH treatment. IB: immunoblotting for all panels.
With the observed repression of E2F1 transcriptional activity by oxidative stress, we postulated that changes in binding to Rb, a canonical transcriptional repressor of E2F1, occurs after exposure to t-BuOOH. Surprisingly, there was no appreciable change in the amount of Rb coimmunoprecipitated (CoIP) with E2F1 after t-BuOOH treatment (Fig. 3D). In light of this finding, we next wanted to determine if E2F1 actually bound to chromatin under t-BuOOH conditions. Utilizing chromatin immunoprecipitation (ChIP) of E2F1 we observed no appreciable change in the level of E2F1 at its own promoter after t-BuOOH treatment (Fig. 3E). Concurrent with E2F1 remaining bound to its promoter, we observed marked transcriptional down-regulation of E2F1 expression (SI Appendix, Fig. S4A). Moreover, we also determined that E2F1 still tightly associated with the p14/ARF, CASP7, and POLA2 promoters after oxidative insult, and its binding to CASP7 and POLA2 promoters actually increased at 7 h after t-BuOOH treatment (SI Appendix, Fig. S4B), suggesting a process of active repression. Interestingly, at 7 h after t-BuOOH treatment, we observed the formation of a high molecular weight species that was suspected to be E2F1 related (Fig. 3 E, Right). Upon examining exogenous E2F1 after t-BuOOH treatment, we also observed a strong induction of highly modified E2F1 species (Fig. 3F). With the observed t-BuOOH-mediated transcriptional repression of E2F1 while still binding chromatin, we decided to investigate the identity of this modification and any role it may play in regulating E2F1 transcriptional activity.
E2F1 Is Hypermodified by SUMO2 during Oxidative Stress, Resulting in Inhibition of Its Transcriptional Activity.
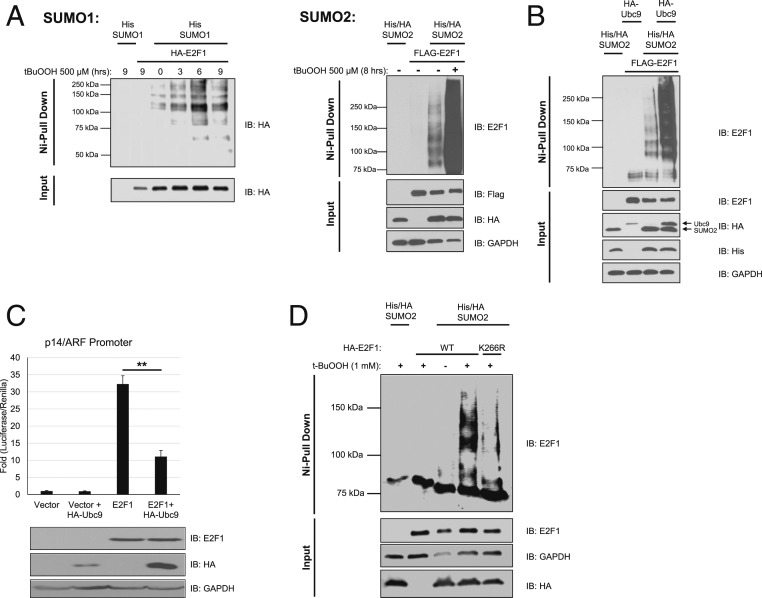
When considering what possible posttranslational modifications could be responsible for regulating E2F1 transcriptional activity as well as adding extensive molecular weight, sumoylation was of particular interest (23). As such, we decided to investigate if E2F1 sumoylation could be altered by oxidative stress. To first investigate this possibility, we utilized a sumoylation assay consisting of transient concurrent overexpression of E2F1 and His/HA-tagged SUMO1 or SUMO2 in HEK 293T cells that were subsequently treated with t-BuOOH. Sumoylated proteins were then isolated from the lysate using denaturing nickel-nitrilotriacetic acid (Ni-NTA) affinity purification, allowing the specific interrogation of E2F1 sumoylation status via a simple Western blot. Much to our surprise, SUMO2, but not SUMO1, conjugation of E2F1 was very strongly induced by t-BuOOH treatment (Fig. 4A). We also performed a sumoylation assay with various doses of hydrogen peroxide to confirm that the hypersumoylation was not specific to t-BuOOH (SI Appendix, Fig. S5). To verify that the observed signal was E2F1 highly modified by SUMO2, we repeated the sumoylation assay with concurrent overexpression of the sole known E2 conjugating enzyme for sumoylation, Ubc9. When overexpressing Ubc9 alone, we observed a similar pattern of highly modified E2F1 (Fig. 4B). Next, we wanted to investigate if changing the extent of E2F1 sumoylation via Ubc9 overexpression could impact E2F1 transcriptional activity. Indeed, overexpression of Ubc9 inhibited E2F1 transcriptional activity as determined by luciferase transcriptional reporter assay (Fig. 4C). To further investigate the role of sumoylation in regulating E2F1 activity, we set out to determine which lysine on E2F1 serves as the SUMO2 acceptor site. Previously, K266 has been identified as the SUMO1 acceptor site, and as such created a K266R mutant to determine what role this lysine plays in SUMO2 addition to E2F1 (23). A sumoylation assay with WT and K266R E2F1 revealed that mutating K266 alone blocks the majority of SUMO2 conjugation to E2F1 (Fig. 4D). Based on this observation, we believe that K266 serves as the primary acceptor lysine for SUMO2 addition to E2F1.
Fig. 4.
Oxidative stress induces hyperconjugation of SUMO2 to E2F1 at K266 residue. (A) Nickel pulldown of cell lysate from t-BuOOH-treated HEK 293T cells cotransfected with E2F1 and His-SUMO1 (Left) or His-SUMO2 (Right). Cells were treated with t-BuOOH (500 µM) for the indicated duration. (B) Nickel pulldown of HEK 293T cells co-overexpressing E2F1 and His-SUMO2, with or without HA-Ubc9. (C) p14/ARF-luciferase reporter assay in H1299 cells assessing transcriptional activity of overexpressed E2F1 in the presence of Ubc9 co-overexpression. Error bars represent mean ± SD (n = 3). **P < 0.005. (D) Nickel pulldown of WT and K266R E2F1 with t-BuOOH treatment in HEK 293T cells. IB: immunoblotting for all panels.
Lysine-266 Residue Is Required for E2F1 to Modulate Transcriptional Activity and Facilitate Cell Survival after Oxidative Stress Insult.
Since SUMO2 conjugation to E2F1 is induced by oxidative stress, and Ubc9 overexpression inhibits E2F1 transcriptional activity, we sought to further examine the role of sumoylation in enabling E2F1 to perform its cellular functions in response to oxidative stress. Interestingly, t-BuOOH treatment is able to inhibit the transcriptional activity of WT but not K266R E2F1 (Fig. 5A). With an apparent role in regulating E2F1 activity, we postulated that sumoylated E2F1 would be associated with chromatin. To test this hypothesis, we performed detergent-resistant fractionation with cells overexpressing WT or K266R E2F1. Upon t-BuOOH treatment, hypermodified species of E2F1 were formed more extensively for WT than K266R and associated with fractions 3 and 4, suggesting a tight association to chromatin (Fig. 5B). Based on the observed high-molecular-weight E2F1 species tightly associated with chromatin, we wanted to investigate how E2F1 could contribute to transcriptional changes after oxidative stress. Previous work has identified CBX4 (Pc2) as the E3 ligase for SUMO1 addition to E2F1, and given its known function in the epigenetic regulatory polycomb repressor complex, we postulated that epigenetic changes could be occurring under the context of oxidative stress response (23, 24). Chromatin immunoprecipitation was performed in U2OS cells with and without t-BuOOH treatment, and it was determined that indeed there was no change in E2F1 binding to the E2F1 promoter. However, SUMO2/3, CBX4, and H3K27me3, a marker of polycomb repressive complex 2 (PRC2) activity, were all enriched by t-BuOOH treatment (Fig. 5C) (25). These data suggest that there are indeed alterations in epigenetic regulation at an E2F1-responsive promoter after oxidative stress insult.
Fig. 5.
K266R fails to modulate E2F1 transcriptional activity or enhance cell viability in response to oxidative stress. (A) U2OS cells transfected with WT or K266R E2F1 were assayed for transcriptional activity after t-BuOOH treatment using a p14/ARF reporter luciferase construct. Error bars represent mean ± SD, normalized to internal β-galactosidase activity and E2F1 protein level (n = 3). *P < 0.05. (B) Detergent-resistant chromatin fractionation of U2OS cells overexpressing WT or K266R E2F1 (Left) and whole cell lysate and quantification of fractionation (Right). t-BuOOH treatment was given for 6 h at 500 µM prior to harvesting. (C) Chromatin-immunoprecipitation with indicated antibodies of lysate from U2OS cells with or without t-BuOOH treatment (500 µM, 3 h). Data shown are E2F1 promoter enrichment, normalized to respective gene desert signal. Error bars represent mean ± SD (n = 3). **P < 0.005. (D) Primary transcript analysis of proliferation-related target genes in sgVector or sgE2F1 cells harvested after 2 h of t-BuOOH treatment (100 µM). Error bars represent mean ± SD (n ≥ 2). **P < 0.005. (E) Primary transcript analysis of cell cycle inhibitors in sgVector or sgE2F1 cells were after 0 h, 1 h, and 2 h of t-BuOOH treatment (100 µM). Error bars represent mean ± SD (n ≥ 2). **P < 0.005. (F) Primary transcript analysis in untreated or t-BuOOH-treated (100 µM, 1 h) sgE2F1 cells stably rescued with an empty vector, WT, or K266R E2F1. Error bars represent mean ± SD (n ≥ 2). Not significant, N.S., *P < 0.05, **P < 0.005. (G) Chromatin-immunoprecipitation of E2F1 in sgE2F1 cells stably rescued with an empty, WT, or K266R E2F1 vector. Cells were untreated or exposed to 100 µM t-BuOOH for 1 h prior to harvesting. Error bar represent mean ± SD (n ≥ 3). *P < 0.05, **P < 0.005. (H) MTT viability assay after hydrogen peroxide treatment (16 h) in sgVector cells stably expressing an empty vector, and sgE2F1 cells stably rescued with an empty, WT, or K266R E2F1 vector. Error bars represent mean ± SD (n ≥ 3). Not significant, N.S., *P < 0.05. (I) Colony formation assay in sgE2F1 cells stably expressing an empty, WT, or K266R E2F1 vector. Error bars represent mean ± SD (n = 3). *P < 0.05. IB: immunobloting for all relevant panels.
Next, we sought to investigate what role E2F1 plays in transcriptional regulation of its target genes in response to oxidative stress. To assess dynamic changes in target gene expression in response to rapid changes in transcriptional activity, we specifically assayed primary transcript levels and validated primer pairs by requiring them to exhibit reduction in product when treated with Triptolide, a potent RNA polmerase inhibitor (SI Appendix, Fig. S6) (26). Via RT-qPCR of primary transcripts of E2F1 targets in sgVector or sgE2F1 cells, we found that after t-BuOOH treatment, sgVector cells exhibited no change for TopBP1 and FOXM1, and down-regulation for Cdc6 and Skp2, while sgE2F1 cells instead had an induction in primary transcripts for TopBP1, Cdc6, FOXM1, and Skp2 (Fig. 5D). With apparent differences in cell cycle arrest after oxidative stress insult, we also examined primary transcript levels for canonical cell cycle inhibitors CDKN1A and CDKN1B. After 1 h of t-BuOOH treatment, sgVector cells showed up-regulated expression of CDKN1A and CDKN1B, while sgE2F1 cells failed in this immediate response (Fig. 5E). In summary, these data suggest that there are dynamic, E2F1-dependent changes in expression of E2F1 target genes as a result of oxidative stress insult that could facilitate the observed potentiation of cell cycle arrest.
To further examine the function of K266 in E2F1-mediated cellular oxidative stress response, we established sgE2F1 cells stably expressing WT or K266R E2F1. When treated with t-BuOOH, WT but not K266R E2F1 was able to inhibit the expression of its proliferation-related target genes. Additionally, the expression of cell cycle inhibitors CDKN1A and CDKN1B were up-regulated only in the presence of WT E2F1 (Fig. 5F). These data suggest that K266 is required for E2F1 to modulate expression of its target genes after oxidative stress insult. To further investigate whether sumoylated E2F1 directly transactivates CDKN1A and CDKN1B upon oxidative stress, we performed ChIP of E2F1 in the sgE2F1 cells rescued with WT or K266R E2F1. Strikingly, both WT and K266R E2F1 were able to bind the CDKN1A and CDKN1B promoters under growing conditions; however, after t-BuOOH treatment, K266R E2F1 was deficient in remaining bound while WT E2F1 was significantly enriched (Fig. 5G). These data suggest that E2F1 is likely playing a direct role in transcriptionally regulating CDKN1A and CDKN1B in response to oxidative stress insult. More broadly, K266 is required for E2F1 to transcriptionally regulate both proliferative and cell cycle inhibitor target genes in response to oxidative stress.
With the observed deficiency of K266R E2F1 to rescue the transcriptional regulation after oxidative stress that WT E2F1 performs, we wanted to further examine what impact this would convey on cell survival. Before additional characterization of the rescue sgE2F1 cells, we verified that there was no significant difference in the basal cell cycle profile between sgE2F1 cells rescued with WT or K266R E2F1 (SI Appendix, Fig. S7). We further utilized this cell system to determine if K266 residue is also required for E2F1 to enhance cell survival after oxidative stress. After hydrogen peroxide treatment, cells rescued with WT but not K266R E2F1 survived to the same extent as sgVector control cells (Fig. 5H). In fact, K266R reconstitution rendered E2F1-KO cells more sensitive to oxidative stress, which is probably because its proapoptotic activity was not repressed. Furthermore, WT but not K266R E2F1 was able to enhance the clonogenic ability of sgE2F1 cells after exposure to hydrogen peroxide (Fig. 5I). Taken together, these data demonstrate that K266 is necessary for E2F1 to modulate expression of its target genes and promote cell survival under oxidative stress conditions.
SENP3 Regulates SUMO2 Conjugation to E2F1.
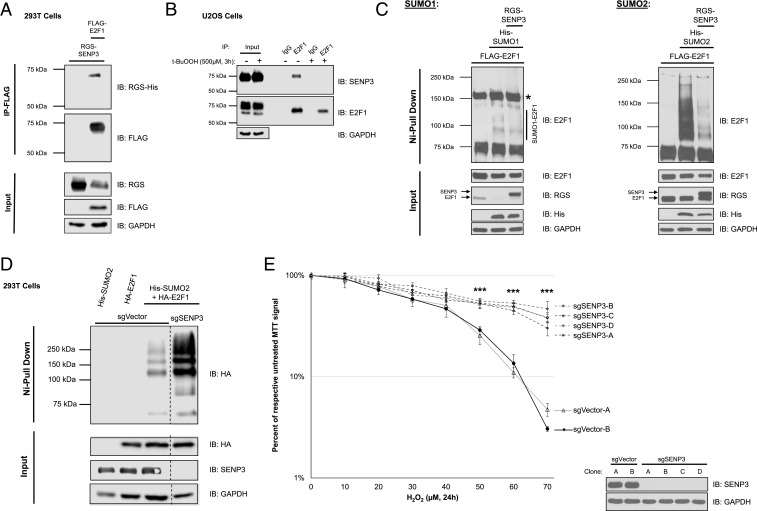
With the up-regulation of SUMO2 conjugation to E2F1 after oxidative stress insult, and the necessity of SUMO acceptor lysine-266 in preserving proper transcriptional regulation and cell survival functions, we wanted to investigate how SUMO2 conjugation of E2F1 is regulated. To this end, we performed CoIP-mass spectrometry of overexpressed E2F1 in HEK 293T cells to elucidate any potential SUMO-related E2F1 interacting partners. One potential binding partner of note was the desumoylating enzyme SENP3, which is known to be regulated by oxidative stress and removes SUMO2/3 conjugates from substrates (27, 28). To validate binding between SENP3 and E2F1, coimmunoprecipitation of both exogenous (Fig. 6A) and endogenous (Fig. 6B) proteins were performed. SENP3 interacted with E2F1 in both experimental designs, and endogenous protein binding was inhibited by t-BuOOH treatment.
Fig. 6.
SENP3 removes SUMO2 conjugates from E2F1. (A) Overexpressed SENP3 coimmunoprecipitates with FLAG-E2F1 in HEK 293T cells. (B) SENP3 coimmunoprecipitates with E2F1 in U2OS cells, and this interaction is greatly inhibited by t-BuOOH treatment. (C) Nickel pulldown of HEK 293T cells cotransfected with E2F1, WT, or C532A SENP3, and His/HA-SUMO1 or His/HA-SUMO2. *: nonspecific band. (D) Nickel pulldown of sgVector and sgSENP3 HEK 293T cells cotransfected with HA-E2F1 and His/HA-SUMO2. (E) MTT viability assay after 24 h H2O2 treatment in sgVector and sgSENP3 knockout clones. Error bars represent mean ± SD (n ≥ 4) ***P < 5 × 10−6. IB: immunoblotting for all panels.
Having confirmed SENP3-E2F1 binding, we sought to investigate if SENP3 can modulate the levels of SUMO conjugates on E2F1. A sumoylation assay was performed in cells co-overexpressing E2F1, SUMO1, or SUMO2, and either WT or catalytic mutant C532A SENP3. Strikingly, SENP3 only modulated SUMO2, but not SUMO1 conjugation of E2F1 (Fig. 6C). Moreover, C532A SENP3 failed to reduce SUMO2-E2F1 conjugates (SI Appendix, Fig. S8), suggesting that SENP3 has catalytic activity specifically toward SUMO2 linkages on E2F1. When performing the SUMO2 sumoylation assay in cells lacking SENP3 (sgSENP3), there was an accumulation in the levels of SUMO2-conjugated E2F1 (Fig. 6D), further supporting a role for SENP3 in regulating E2F1 sumoylation. With SENP3 deficiency enhancing the levels of E2F1-SUMO2 conjugates independent of oxidative stress, we wanted to determine if SENP3 deficiency changes cell sensitivity to hydrogen peroxide. All four sgSENP3 clones assayed for cell viability exhibited resistance to hydrogen peroxide when compared to sgVector controls (Fig. 6E). Collectively, SENP3 is an E2F1 interacting partner that functions to maintain lower levels of SUMO2-conjugated E2F1. Oxidative stress is able to inhibit this interaction, and cells lacking SENP3 exhibit hypersumoylation of E2F1 and resistance to oxidative stress.
Elevated SENP3 Levels Are Associated with Poorer Patient Outcomes.
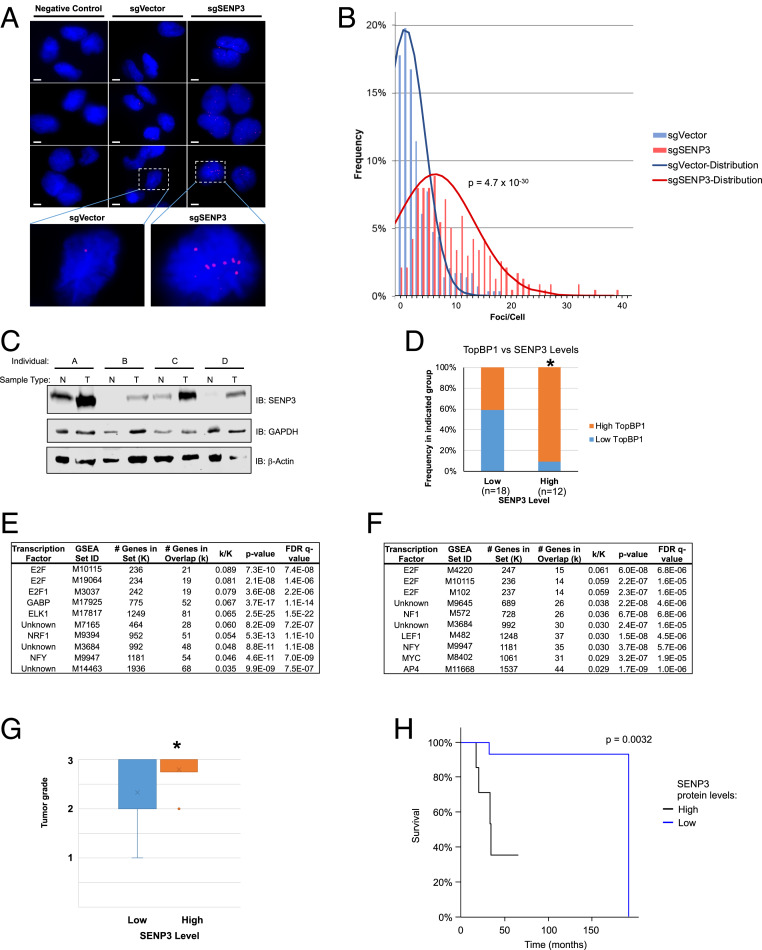
With SENP3 playing a clear role of maintaining E2F1 in a hyposumoylated state under normal growing conditions, we wanted to investigate if changes to endogenous E2F1 sumoylation could be observed in SENP3-knockout cells. To address this, we leveraged the sensitivity of proximity ligation assay (PLA) as a tool to detect and amplify otherwise low-abundance instances. Strikingly, far more PLA foci are observed in sgSENP3 cells when compared to sgVector control cells (Fig. 7A). When quantifying the foci per cell as means to quantify the extent to which E2F1 sumoylation is present, it is clear that sgSENP3 cells are markedly enriched in the number of foci compared to sgVector control (Fig. 7B). These data suggest that changes in SENP3 level are able to greatly impact the extent to which E2F1 is sumoylated in response to endogenous oxidative stress in the context of growing cells.
Fig. 7.
SENP3 regulates endogenous E2F1 sumoylation and its overexpression correlates with poor patient outcome. (A) E2F1-SUMO2/3 proximity ligation assay in sgVector control (clone sgVector-A) and sgSENP3 knockout (clone sgSENP3-A) cells. Positive PLA foci are red, while nuclear Hoechst staining is blue. (Scale bar, 5 µM.) (B) Quantification of PLA foci, where bars chart frequency of values and smoothed line represents theoretical normal distribution of data group. (n > 230) P = 4.7 × 10−30. (C) Western blot examining SENP3 expression of four patient-matched normal and breast tumor samples. IB: immunoblotting. (D) Correlation between SENP3 and TopBP1 protein expression in our cohort of breast tumor tissues. (n = 18 low, n = 12 high) *P < 0.05. (E) The dataset PXD000815 from deep proteomic profiling of luminal breast cancer progression (n = 88) (33) was analyzed for proteins coexpressed with SENP3 (Pearson correlation coefficient >0.4) and the protein list was analyzed with gene set enrichment analysis (GSEA) for overlap with transcription factor target genes. Only the top 10 transcription factors are shown. (F) Similar analysis was performed from the dataset PXD002619 which includes luminal, Her2 positive, and triple-negative breast cancer (n = 40) (34) and analyzed for proteins coexpressed with SENP3 (Pearson correlation coefficient >0.35). (G) Correlation between SENP3 expression and tumor grade in our breast cohort. *P < 0.05. (H) Patient overall survival as a function of low vs. high SENP3 expression in our cohort of breast cancer.
Having determined that SENP3 levels regulate the abundance of endogenous E2F1 sumoylation, we next wanted to investigate how SENP3 levels could differ between normal and cancerous breast tissue. When comparing four patient-matched, normal, and tumor samples it is clear that all four tumors assayed have elevated levels of SENP3 compared to the normal tissue (Fig. 7C). We then examined SENP3 protein levels by Western blot analysis in 30 primary breast tumors from a cohort of anonymized breast cancer patients for whom we have collected their clinical outcomes and the expression of TopBP1, an E2F1 target, in our prior studies (29, 30). Based on their relative SENP3 levels, these samples were assigned into either high or low SENP3 expression groups (SI Appendix, Table S1). Interestingly, high SENP3 levels correlate with a higher expression of TopBP1, a key proliferative transcriptional target of E2F1 (Fig. 7D) (31, 32). We further mined two breast cancer proteomic datasets (33, 34). Indeed, the proteins that coexpress with SENP3 protein are highly enriched for E2F target genes in both datasets examined (Fig. 7 E and F). Additionally, high SENP3 levels also correlate with a more advanced tumor grade (Fig. 7G) and a poorer survival outcome (Fig. 7H) in our cohort. Taken together, these data provide compelling evidence for the role that SENP3 expression, and potentially its regulation of E2F1 sumoylation, play in advancing tumor progression.
Discussion
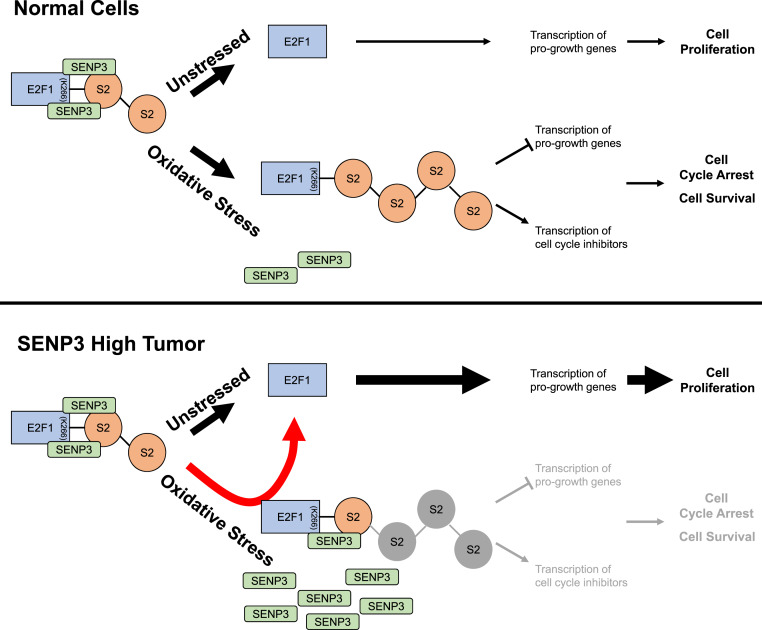
Oxidative stress is a reality that all eukaryotic organisms inherently face due to the integral role of oxygen in cellular respiration. We have demonstrated that E2F1 plays a key role in enhancing cell survival after oxidative stress insult by facilitating cell cycle arrest. Extensive SUMO2 conjugation on E2F1 allows for dynamic modulation of transcriptional activity in response to oxidative stress, as depicted in our proposed model (Fig. 8). During normal growing conditions, SENP3 maintains E2F1 in a hypomodified state to enable cell proliferation. Under enhanced levels of oxidative stress, SENP3 loses the ability to bind E2F1, and SUMO2 conjugates accumulate on E2F1 thus contributing to the observed changes in E2F1 transcriptional activity. Sumoylation of E2F1 upon oxidative stress can actively repress the expression of progrowth and proapoptotic genes and induce the expression of CDKN1A and CDKN1B, leading to G1/S cell cycle arrest, which is important for cells to repair the oxidative damage before S-phase entry and prevent cell death. Induction of CDKN1A and CDKN1B may also play an active role in cell survival (18–21, 35, 36). Thus, E2F1 is important for cell survival under oxidative stress.
Fig. 8.
A model for the proposed role of E2F1 sumoylation in cellular response to oxidative stress. In actively proliferating cells, E2F1 is constantly sumoylated and desumoylated. Under unstressed conditions, the poly-SUMO2 chains on E2F1 (mainly on K266 residue) are actively removed by SENP3 to promote cell proliferation. Upon oxidative stress, SENP3 can no longer bind and desumoylate E2F1, allowing the accumulation of sumoylated E2F1. SUMO2 modifications convert E2F1 from a transcriptional activator into a transcriptional repressor on the promoters of proliferative and apoptotic genes. Sumoylated E2F1 facilitates cell cycle arrest by actively repressing the expression of proliferative genes and also through activating CDKN1A and CDKN1B. The E2F1-mediated arrest allows cells to repair oxidative damage and up-regulates the antiapoptotic activities of CDKN1A and CDKN1B, therefore promoting cell survival. However, in SENP3 high tumors the sumoylated E2F1 is shunted back to unmodified E2F1 and therefore E2F1 sumoylation-mediated cell cycle arrest is negated by overexpressed SENP3.
Potential Cross-Talk with Additional Mechanisms to Modulate E2F1 Activity in Response to Oxidative Stress.
With how conserved the E2F pathway is in regulating cell proliferation in higher eukaryotes, it was initially surprising that loss of E2F1 alone contributes to such dramatic hypersensitivity to oxidative stress. It is possible that additional mechanisms unique to E2F1 could be at play in conjunction with sumoylation in responding to oxidative stress. E2F1 has been characterized as a dynamic transcription factor with a role in a multitude of cellular functions ranging from cell proliferation, DNA repair, and apoptosis, just to name a few (37, 38). With seemingly opposing cellular functions, how E2F1 is able to perform specific regulation on one pathway but not another has been a long-standing question in the field. A leading model to explain transcriptional specificity has been through a posttranslational modification “code,” much like one that exists for histones and chromatin regulation (39). Numerous posttranslational modifications have been found on E2F1 in both transcriptionally activating and repressing roles.
One striking observation from our study is the apparent role of K266 in E2F1-mediated transcriptional modulation, with simultaneous activation and repression functions depending on the target gene. One possible explanation for this phenomenon could be differentially modified pools of E2F1 resulting in different transcriptional outcomes. It should be noted that while K266 is the established acceptor lysine for SUMO1 conjugation to E2F1 and we cannot rule out contributions of SUMO1 addition to the observed transcriptional changes, the lack of extensive changes to SUMO1 conjugation of E2F1 with oxidative stress casts doubt on this possibility (23).
Moreover, it is possible that interplay exists between SUMO2 conjugation of E2F1 and other posttranslational modifications, and this could contribute to oxidative stress-induced transcriptional modulation. Two modifications of potential interest are neddylation and arginine methylation, both of which have been shown to inhibit E2F1 transcriptional activity (40, 41). Additional studies are needed to further explore this possible connection. Beyond changes in the posttranslational modification profile of E2F1 in response to oxidative stress, it is also likely that sumoylated E2F1 exhibits differential interaction patterns and affinities for binding partners when compared to unmodified E2F1. An interaction profile unique to sumoylated E2F1 would further enable specific regulation of an individual target gene(s) and should be investigated further.
Interplay between Oxidative Stress and SUMO Modifier Enzymes.
When considering that CBX4, the E3 ligase for SUMO1 conjugation to E2F1, is recruited to the E2F1 promoter after oxidative stress, as well as the same primary acceptor lysine (K266) being utilized for SUMO2 conjugation to E2F1 as previously described for SUMO1 addition, it seems likely that CBX4 is acting as the E3 ligase for SUMO2 conjugation under oxidative stress conditions (23). Additionally, CBX4 has been shown to have its activity altered by the redox-sensing kinase HIPK2 (42, 43). Further investigation of how SUMO2 conjugation to E2F1 is regulated by oxidative stress, and what redox response pathways are involved could provide clarity on the cellular processes upstream of the work described herein.
Oxidative stress response is also inherently linked to the removal of SUMO2 conjugates from E2F1 via the desumoylating enzyme SENP3. While under higher levels of oxidative stress, SENP3 loses the ability to bind E2F1. We believe this to be a key component to the robust accumulation of SUMO2 and not SUMO1 linkages on E2F1 after oxidative stress insult, which is independent of possible changes in E3 ligase activity or specificity. How the interaction of E2F1 with SENP3 is inhibited under oxidative stress conditions is presently unknown, and merits additional investigation. At low levels of oxidative stress, SENP3 has been shown to be stabilized and to promote transcriptional activation (27, 28). This presents the possibility that SENP3 could potentiate reactivation of E2F1 and subsequent resuming of cell cycle progression upon resolution of acute oxidative insult, in addition to its role in maintaining E2F1 in a hyposumoylated state to promote cell cycle progression. It is possible that even in the basal growing condition, the regulation by SENP3-E2F1 is required for optimal repair of endogenous oxidative stress; thus, we observed a slight increase of basal autophagy/mitophagy in E2F1-knockout cells.
SENP3 levels have been found to be elevated in multiple tumor types when compared to normal tissue, and this is associated with a poorer prognosis (44, 45). Our small breast cancer cohort provides another example of the important yet largely uncharacterized role SENP3 levels could play in predicting tumor aggressiveness and patient outcome. Based on our model, enrichment of SENP3 expression could contribute to a proliferative program leading to less regulated cell growth. Indeed, the observation that high SENP3 levels correlate with a high expression of TopBP1, a key E2F1 proliferative transcriptional target, and the correlation of SENP3 protein levels with E2F activities in two breast cancer proteomic datasets, further support this hypothesis. Interestingly, knockdown of SENP3 in ovarian cancer was shown to induce CDKN1A (44), which is also consistent with our data of the up-regulation of CDKN1A by sumoylated E2F1. Moreover, aberrant activation of a single protooncogene in normal fibroblasts can itself result in enhanced levels of reactive oxygen species (8). In light of this, our mechanism of E2F1 sumoylation to mediate cell cycle arrest could function as a portion of the first line of defense against aberrant proliferation after oncogene activation. Beyond the initial transforming event, by further understanding how cancer cells are deregulated to persistently proliferate despite their enhanced levels of oxidative stress, it becomes possible to therapeutically target these mechanisms to resensitize the cells to their inherent stress. Indeed, recent work has demonstrated that targeting antioxidant machinery has therapeutic potential in principle and provides further support for future investigations of this possibility (46).
Materials and Methods
Coimmunoprecipitation, Nickel Pulldowns, and Immunoblotting.
CoIP of proteins interacting with FLAG-E2F1 or endogenous E2F1 was performed as described previously (11) using anti-FLAG M2 monoclonal antibody-conjugated agarose beads or anti-E2F1 (KH95) mouse monoclonal antibody, respectively. Washes were performed as described, with the addition of a 0.5 M LiCl wash step to increase stringency. Samples were analyzed via sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) and indicated immunoblotting. Nickel pulldowns were performed as described previously (47).
BrdU Incorporation and Cell Cycle Analysis.
BrdU incorporation assay was performed as described previously (48), with a 2-h treatment of peroxide followed by 2 h of BrdU labeling prior to harvest. Cell cycle analysis was performed by staining ethanol-fixed cells with propidium iodide (5 µg/mL) followed by flow cytometry to assess DNA content.
Mass Spectrometry.
FLAG-E2F1 was transfected into HEK 293T cells and lysates were immunoprecipitated with anti-FLAG M2 monoclonal antibody-conjugated agarose beads. The immunoprecipitates were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) by the Baylor College of Medicine (BCM) Proteomics Core.
Frozen Breast Cancer Tissues.
Collection of frozen breast tumor tissues or healthy breast tissues with informed consent through the University of Alabama at Birmingham Tissue Procurement Core Facility has been described earlier (29, 30). Utilization of these samples and anonymized data are approved by an institutional review board protocol BCM H-26360. The frozen tissues were minced and lysed in 1% SDS (60 mM Tris HCl) and boiled for 5 min. The lysates were briefly sonicated and clarified by centrifugation to remove insoluble tissues, followed by Western blotting.
Cell culture, transfection, and treatments, plasmid construction, virus production and stable cell line generation, MTT assay, colony formation assay, Caspase activity assay, cell synchronization, Calcein-AM viability assay, flow cytometry, antibodies, chromatin binding assay, chromatin-immunoprecipitation, RNA extraction, and qRT-PCR, luciferase reporter assay, proximity ligation assay, and statistical analysis are described in SI Appendix, SI Materials and Methods.
Data Availability.
All pertinent data are included herein or in SI Appendix. The relevant clinical information concerning patient samples is given in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01CA100857 (to W.-C.L.) and R01CA203824 and Department of Defense Grants W81XWH-18-1-0329 and W81XWH-19-1-0369 (to W.-C.L. and F.-T.L.). J.D.G. was supported by T32 Fellowship T32DK060445. We also acknowledge support from the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921554117/-/DCSupplemental.
References
- 1.DeGregori J., Johnson D. G., Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6, 739–748 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Dyson N., The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Knudson A. G., Jr., Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U.S.A. 68, 820–823 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent L. N., Leone G., The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 19, 326–338 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Nevins J. R., The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10, 699–703 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Szatrowski T. P., Nathan C. F., Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 51, 794–798 (1991). [PubMed] [Google Scholar]
- 7.Maya-Mendoza A. et al., Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 9, 601–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vafa O. et al., c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol. Cell 9, 1031–1044 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Cooke M. S., Evans M. D., Dizdaroglu M., Lunec J., Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 17, 1195–1214 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Blanchet E. et al., E2F transcription factor-1 regulates oxidative metabolism. Nat. Cell Biol. 13, 1146–1152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K., Lin F. T., Ruppert J. M., Lin W. C., Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol. Cell. Biol. 23, 3287–3304 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu K., Paik J. C., Wang B., Lin F. T., Lin W. C., Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 25, 4795–4807 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polager S., Ofir M., Ginsberg D., E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 27, 4860–4864 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Wang B., Ling S., Lin W. C., 14-3-3Tau regulates Beclin 1 and is required for autophagy. PLoS One 5, e10409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H. et al., The RB-E2F1 pathway regulates autophagy. Cancer Res. 70, 7882–7893 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bucha S., Mukhopadhyay D., Bhattacharyya N. P., E2F1 activates MFN2 expression by binding to the promoter and decreases mitochondrial fission and mitophagy in HeLa cells. FEBS J. 286, 4525–4541 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Yurkova N. et al., The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ. Res. 102, 472–479 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Baldassarre G. et al., Key role of the cyclin-dependent kinase inhibitor p27kip1 for embryonal carcinoma cell survival and differentiation. Oncogene 18, 6241–6251 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Eymin B. et al., p27Kip1 induces drug resistance by preventing apoptosis upstream of cytochrome c release and procaspase-3 activation in leukemic cells. Oncogene 18, 1411–1418 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Hiromura K., Pippin J. W., Fero M. L., Roberts J. M., Shankland S. J., Modulation of apoptosis by the cyclin-dependent kinase inhibitor p27(Kip1). J. Clin. Invest. 103, 597–604 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii T. et al., Effects of p27Kip1 on cell cycle status and viability in A549 lung adenocarcinoma cells. Eur. Respir. J. 23, 665–670 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Paik J. C., Wang B., Liu K., Lue J. K., Lin W. C., Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J. Biol. Chem. 285, 6348–6363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L. et al., ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margueron R. et al., Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon J. A., Kingston R. E., Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Vispé S. et al., Triptolide is an inhibitor of RNA polymerase I and II-dependent transcription leading predominantly to down-regulation of short-lived mRNA. Mol. Cancer Ther. 8, 2780–2790 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Huang C. et al., SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 28, 2748–2762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan S. et al., Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90. EMBO J. 29, 3773–3786 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K. et al., Regulation of p53 by TopBP1: A potential mechanism for p53 inactivation in cancer. Mol. Cell. Biol. 29, 2673–2693 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B. et al., 14-3-3Tau regulates ubiquitin-independent proteasomal degradation of p21, a novel mechanism of p21 downregulation in breast cancer. Mol. Cell. Biol. 30, 1508–1527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K., Luo Y., Lin F. T., Lin W. C., TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 18, 673–686 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida K., Inoue I., Expression of MCM10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene 23, 6250–6260 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Pozniak Y. et al., System-wide clinical proteomics of breast cancer reveals global remodeling of tissue homeostasis. Cell Syst. 2, 172–184 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Tyanova S. et al., Proteomic maps of breast cancer subtypes. Nat. Commun. 7, 10259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Javelaud D., Besancon F., Inactivation of p21WAF1 sensitizes cells to apoptosis via an increase of both p14ARF and p53 levels and an alteration of the Bax/Bcl-2 ratio. J. Biol. Chem. 277, 37949–37954 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Poluha W. et al., The cyclin-dependent kinase inhibitor p21 (WAF1) is required for survival of differentiating neuroblastoma cells. Mol. Cell. Biol. 16, 1335–1341 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswas A. K., Mitchell D. L., Johnson D. G., E2F1 responds to ultraviolet radiation by directly stimulating DNA repair and suppressing carcinogenesis. Cancer Res. 74, 3369–3377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iaquinta P. J., Lees J. A., Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19, 649–657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munro S., Carr S. M., La Thangue N. B., Diversity within the pRb pathway: Is there a code of conduct? Oncogene 31, 4343–4352 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Cho E. C. et al., Arginine methylation controls growth regulation by E2F-1. EMBO J. 31, 1785–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loftus S. J., Liu G., Carr S. M., Munro S., La Thangue N. B., NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 13, 811–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de la Vega L. et al., A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell 46, 472–483 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Roscic A. et al., Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell 24, 77–89 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Cheng J. et al., Upregulation of SENP3/SMT3IP1 promotes epithelial ovarian cancer progression and forecasts poor prognosis. Tumour Biol. 39, 1010428317694543 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Zhou Z. et al., SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene 35, 5826–5838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujitani N. et al., Silencing of glutathione S-transferase Pi inhibits cancer cell growth via oxidative stress induced by mitochondria dysfunction. Sci. Rep. 9, 14764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Mahanic C. S., Budhavarapu V., Graves J. D., Li G., Lin W. C., Regulation of E2 promoter binding factor 1 (E2F1) transcriptional activity through a deubiquitinating enzyme, UCH37. J. Biol. Chem. 290, 26508–26522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu K., Lin F. T., Graves J. D., Lee Y. J., Lin W. C., Mutant p53 perturbs DNA replication checkpoint control through TopBP1 and Treslin. Proc. Natl. Acad. Sci. U.S.A. 114, E3766–E3775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All pertinent data are included herein or in SI Appendix. The relevant clinical information concerning patient samples is given in SI Appendix, Table S1.