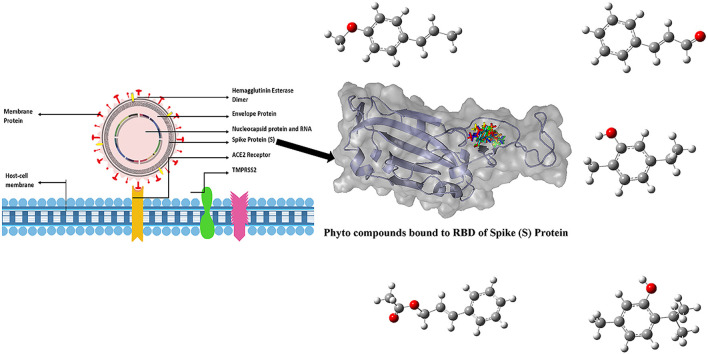
Abstract
COVID-19, caused by SARS-CoV-2 has recently emerged as a global pandemic. Intense efforts are ongoing to find a vaccine or a drug to control the disease across the globe. Meanwhile, alternative therapies are also being explored to manage the disease. Phytochemicals present in essential oils are promising candidates which have been known to possess wide range of therapeutic activities. In this study, major components of several essential oils which are known for their antimicrobial properties have been docked against the S1 receptor binding domain of the spike (S) glycoprotein, which is the key target for novel antiviral drugs, to ascertain their inhibitory effects based on their binding affinities. It has been found that some monoterpenes, terpenoid phenols and phenyl propanoids such as anethole, cinnamaldehyde, carvacrol, geraniol, cinnamyl acetate, L-4-terpineol, thymol and pulegone from essential oils extracted from plants belonging to families such as Lamiaceae, Lauraceae, Myrtaceae, Apiaceae, Geraniaceae and Fabaceae are effective antiviral agents that have potential to inhibit the viral spikeprotein.
Keywords: COVID-19, SARS-CoV-2, Phytochemicals, Essential oils, Antiviral
Graphical abstract
1. Introduction
A novel coronavirus (nCoV19) was first reported to have infected humans in Wuhan city of Hubei province, China in the late 2019, which was later termed as severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) [1,2]. Coronavirus disease 19 (COVID-19) caused by this virus has been declared a global pandemic and Public Health Emergency of International Concern (PHEIC) by WHO since it is affecting millions of people across the world in alarming proportions [3]. The symptoms and manifestations of this disease is similar to respiratory distress indicated previously by beta coronaviruses which were responsible for causing Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) [4,5]. Currently, America and Europe are worst affected with about 1.3 and 1.4 million cases respectively. Cases in Eastern Mediterranean, West Pacific, South-East Asia and Africa are also increasing steadily [6].
Presently, efforts are being intensified by scientists across the world for development of vaccine and drugs to treat COVID-19. FDA approved drugs are being repurposed to test their efficacy against SARS-CoV-2 to address the urgency of the situation [7,8]. Simultaneously alternative plant-based therapies are also being explored to either find a cure or manage the disease. Although, drugs used in western medicine system are predominantly synthetic compounds, over the past few decades natural phyto compounds are being researched owing to their enormous structural and chemical diversity [9,10]. Bioactive phyto compounds which were used to treat ailments traditionally, have now become indispensable source for many drugs that prevent, manage or treat a plethora of diseases [[11], [12], [13]] Plant essential oils are being studied extensively since they are known to possess numerous versatile properties such as antimicrobial, antibacterial, antiviral, antiparasitic and insecticidal. They are a diverse mixture of organic compounds predominantly composed of terpenes, terpenoids, phenylpropanoids and aldehydes [[14], [15], [16]]. The chemical composition and structural components of essential oils majorly contribute to their activity [17]. Essential oils fall under ‘GRAS’ (generally regarded as safe) category [18], therefore, they have an edge over synthetic drugs with an added advantage of being safe and non-toxic. The role of essential oils in treating infectious, acute and chronic diseases has been well documented [[19], [20], [21]]. Though synthetic drugs will be tested for toxicity, minimal side effects can definitely be expected. Hence, essential oil based therapeutic approach can be prioritised in case the efficacy is comparable with synthetic drugs. Previous studies indicate that many essential oils have been effective in inhibiting wide array of microorganisms. In the context of this study related to antiviral properties, a few essentials oils have been effective against RNA and DNA viruses such as avian influenza A virus [22], herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2), dengue virus type 2, Junin virus, influenza virus adenovirus type 3, poliovirus, and coxsackievirus B1 [23]. Since there are very few drug compounds available for treating viral diseases, more insight into drug development based on phyto compounds could be considered. Synergism between these molecules may also be explored with an ultimate goal to find novel antiviral lead molecules.
In silico studies play a significant role in accelerating the drug discovery process. To assess the therapeutic potential of certain compounds, it is essential to know the interaction between the ligands and the protein at molecular level. An initial in silico study involving many techniques considerably reduces the time needed for lead molecule identification [[24], [25], [26]]. For any computational study, the structure of the protein under consideration is of prime importance to understand the interactions and the effects that follow. The protein structure can be modelled through other ways too, in case of unavailability of a 3D protein crystal structure [[27], [28], [29]]. In this case, the crystal structure of the SARS-CoV-2 spike glycoprotein (S) and main protease (Mpro) has been recently deduced [30,31]. The S protein is further comprised of two units namely S1, which is also called as receptor binding domain (RBD) as it is involved in binding to the host Angiotensin converting enzyme 2 (ACE2) and S2, which is responsible for fusion of viral and cellular membrane [32]. The S1 subunit of S protein is of clinical importance because inhibiting the RBD can lead to conformational changes in it, as a result, the first step of viral infection is obstructed.
In the present study, molecular docking and the conceptual density functional theory (DFT) approach has been utilized to assess the antiviral properties of major components of certain essential oils derived from plants that are notable for antimicrobial activity. The interaction between RBD and essential oil components were assessed based on their binding modes and affinities. Hydrogen bonding and hydrophobic interactions also suggest stable interaction between the protein and ligand. The major objectives of this study were to dock essential oil components against the RBD of the S protein and to identify potential antiviral compounds that specifically inhibit viral attachment and replication in the host. The conceptual DFT study has also been performed to understand and validate the chemical nature of the phytochemicals based on the various molecular descriptors which are derived from the electron density of the molecules.
2. Materials and methods
2.1. Preparation of protein structure
The protein target is the receptor binding domain (RBD) of S1 subunit of SARS-CoV-2 ‘S’ glycoprotein (PDB ID: 6M0J). The crystallographic 3D structure of the protein was particularly chosen since RBD is the key site for targeting inhibitory ligands. It was retrieved from protein data bank (http://www.rcsb.org/). The protein was visualised using PyMol (http://www.pymol.org) and the water molecules, co-crystal ligands like NAG, Cl, Zn and ACE2 structure that were bound to the RBD were removed. Further, the protein was prepared using Autodock vina in PyRx open source software by adding charges and minimizing the energy of the protein and subsequently converting it to pdbqt format [33,34].
2.2. Selection and preparation of ligands
The components of essential oils are well known for a variety of activities including antimicrobial, antiviral, antifungal and antibacterial. Major components from certain essential oils such as thyme, cinnamon, clove, star anise, basil, holy basil, eucalyptus, geranium, oregano and ajwain, have been selected as ligands since the efficacy of these components are well established against various organisms [17,35].
The 3D structure of the ligands were downloaded from PUBCHEM database (https://pubchem.ncbi.nlm.nih.gov/). It is a chemical database which contains exhaustive information about chemical compounds such as their 2D and 3D structures, properties, bioactivity and safety. The ligands were loaded into the PyRx virtual screening software using OpenBabel control. The OpenBabel software converts the SDF files of the ligands to PDB format [36]. Further, the ligands are prepared by detecting the torsion root, correcting the torsion angles, assigning charges, optimizing using UFF (Universal force field) and finally converting them to pdbqt format to generate 3D atomic coordinates of the molecules. The 2D images of these ligands are presented in Table 1 .
Table 1.
Components of essential oils with their 2D structures.
| Compound name | Structure | Compound name | Structure |
|---|---|---|---|
| Thymol | 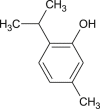 |
l-α-terpineol | 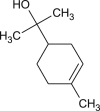 |
| Carvacrol | 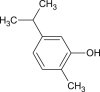 |
Cinnamyl acetate | 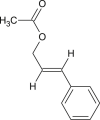 |
| Eugenol | 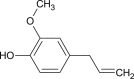 |
Limonene | 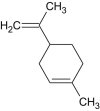 |
| Eucalyptol | 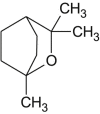 |
γ-terpinene | 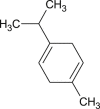 |
| Cinnamaldehyde | 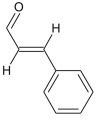 |
α-terpinene | 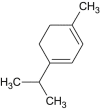 |
| Anethole | 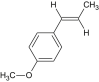 |
Camphene | 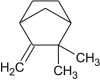 |
| Camphor | 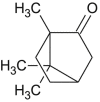 |
Menthol | 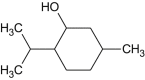 |
| Sabinene | 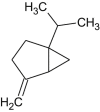 |
Menthone | 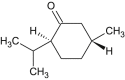 |
| Pulegone | 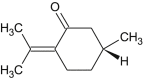 |
p-menthan-1-ol | 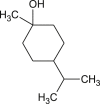 |
| Ocimene | 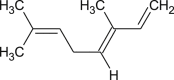 |
p-cymene | 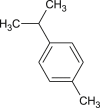 |
| Thujene | 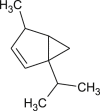 |
o-cymene | 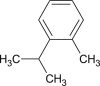 |
| Geraniol | 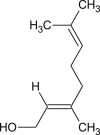 |
linalool | 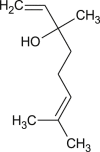 |
| L-4-terpineol | 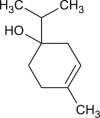 |
2.3. Active site prediction
There could be many sites suitable for ligand binding, but the most appropriate site would be the radial area around the co-crystal ligand site with about 25.0 Å from the ligand coordinates [37]. ACE2 was complexed with RBD in the 3D crystal structure of protein 6M0J. Therefore the site at and around which ACE2 was bound would be the active site of interest. Recently, the crystal structure of SARS-CoV-2 RBD bound to ACE2 has been characterized. The study suggests that the overall binding mode of SARS-CoV-2 RBD- ACE2 is almost similar to SARS-CoV RBD- ACE2. Small structural changes in SARS-CoV-2 such as insertion of β5, β6 strands, α4, α5 helices and loops in between β4 and β7 contain most of the contacting residues such as Tyr449, Tyr453, Asn487, Phe486, Tyr489, Gln493, Gly496, Gln498 Thr500, Gly502, Tyr505 [30]. To further validate the active sites, the protein structure was submitted to active site prediction server maintained by SCFBio, IIT-D, to predict all possible sites (http://www.scfbio-iitd.res.in/dock/ActiveSite.jsp). About nine cavities were suggested by the server.
2.4. Molecular docking
Molecular docking is an in silico technique that predicts the interactions between a protein and small molecules based on the geometry and the scores [38]. In this study, docking was performed using Autodock vina in PyRx virtual screening open source software [39]. Autodock vina has better performance in comparison to Autodock 4.0 in terms of average accuracy of binding mode prediction, speed and also automatic pre-calculation of grid maps which is done internally [40,41]. The protein and ligand molecules to be docked are selected under the vina wizard control. A grid appears over the protein structure. The grid size can be adjusted according to the active site residues that are selected and autodock vina is run. The docking results can be viewed under ‘analyse results’ tab.
2.5. Conceptual DFT
Density functional theory (DFT) is helpful in analyzing the molecular and/or atomic structure using the energies of their molecular orbitals [42] and can provide crucial insights on structure-activity relationship of molecules. The theory is based on the Hohenberg-Kohn theorem [43] A subfield of DFT, known as Conceptual DFT, has been used in this study to observe the chemical behavior of a molecule using electron density relevant concepts [44]. Ten different molecular descriptors have been calculated that are derived from electron density of molecules. The descriptors include total energy, lowest unoccupied molecular orbital (LUMO), highest occupied molecular orbital (HOMO), energy gap, global softness, absolute hardness, molecular dipole moment, electronegativity, electrophilicity index and chemical potential.
3. Results
3.1. Binding site characterization
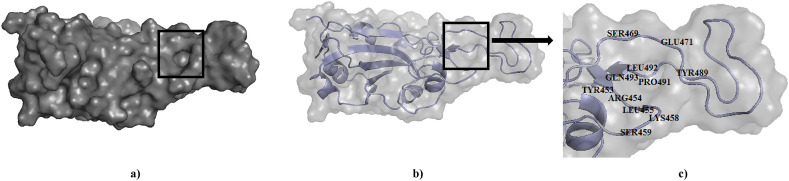
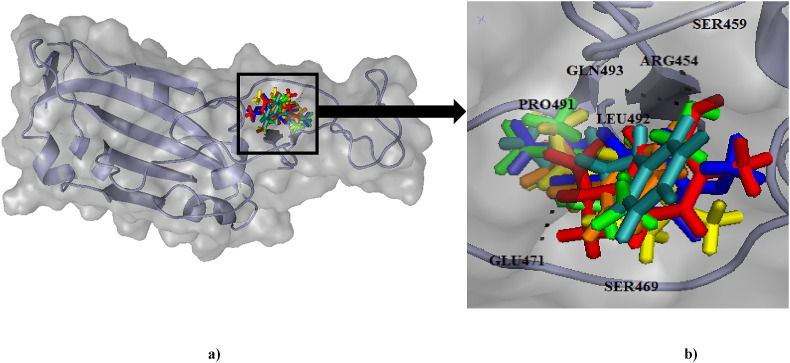
The ACE2 binding site in the complex protein and residues suggested by literature and the sites predicted by active site prediction server by SCFBio were scrutinized and compared. Out of the nine cavities suggested by SCFBio server, four of them were located in the distal end of the RBD and the residues in that area almost matched the ones suggested by literature. The residues proposed to be a part of binding site are Tyr453, Arg454, Leu455, Lys458, Ser459, Ser469, Glu471, Pro491, Leu492, Gln493 and Tyr489. Fig. 1 represents the binding site determined by this study.
Fig. 1.
Binding site for ligands in RBD of spike (S) protein. The active site has been highlighted (a) surface and (b) cartoon representation in a black box and the (c) cavity has been magnified to display the residues.
3.2. Molecular docking
Docking technique essentially aids in identifying putative inhibitors to a particular protein. In this case, the ligands are docked against RBD of S protein to identify potential antiviral compounds. All the ligands were docked in the binding pocket that was proposed. Carvacrol, cinnamaldehyde, cinnamyl acetate, geraniol, L-4-terpineol and anethole displayed better binding affinity with high docking scores compared to the rest of the ligands. Out of all the conformations generated during docking, one conformation each for every ligand with least RMSD were selected.These ligands displayed one or more hydrogen bonding interaction with residues Arg454, Lys458, Ser459, Ser469, Glu471, Leu492 and Tyr505. The hydrogen bonding and hydrophobic interaction between the ligands and protein along with their binding affinities has been tabulated in Table 2 . Though, thymol, camphene, pulegone, ocimene and menthol also displayed good binding affinity, they lacked hydrogen bonding interactions with protein. Since there are many hydrophobic residues around the ligands in the binding pocket, hydrophobic interactions between them may contribute to the stability of the complex. The docked poses of each of the top ligands in RBD domain are depicted in Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7 . They have been represented in distinct colours for better visualisation. All the selected ligands docked together in the binding site characterized has been represented in Fig. 8 .
Table 2.
Binding affinities of the ligands with the RBD of S protein along with the H-bond and hydrophobic interactions with the amino acid residues in the binding site.
| Compound name | Binding affinity (kcal/mol) | H-bond interactions | Hydrophobic interactions |
|---|---|---|---|
| Thymol | −5.4 | – | |
| Carvacrol | −5.2 | SER459 | |
| Eugenol | −4.9 | – | |
| Eucalyptol | −4.3 | – | |
| Cinnamaldehyde | −5.0 | GLU471, ARG454 | |
| Anethole | −5.2 | SER459 | |
| l-α-terpineol | −4.9 | – | |
| Cinnamyl acetate | −5.2 | GLU471, ARG454, SER469 | |
| Limonene | −5.1 | – | TYR449, TYR451, TYR453, LEU455, PHE456, LEU461, ILE468, THR470, ILE472, PRO491, LEU492, TYR505 |
| γ-terpinene | −4.8 | – | |
| α-terpinene | −5.0 | – | |
| Camphene | −5.2 | – | |
| Camphor | −4.8 | – | |
| Sabinene | −4.9 | – | |
| Pulegone | −5.4 | – | |
| Ocimene | −5.2 | – | |
| Thujene | −4.9 | – | |
| Geraniol | −5.0 | LYS458, SER459 | |
| L-4-terpineol | −5.1 | LEU492, TYR505 | |
| Menthol | −5.2 | – | |
| Menthone | −5.0 | – | |
| p-menthan-1-ol | −5.1 | – | |
| p-cymene | −4.9 | – | |
| o-cymene | −4.9 | – | |
| linalool | −4.8 | – |
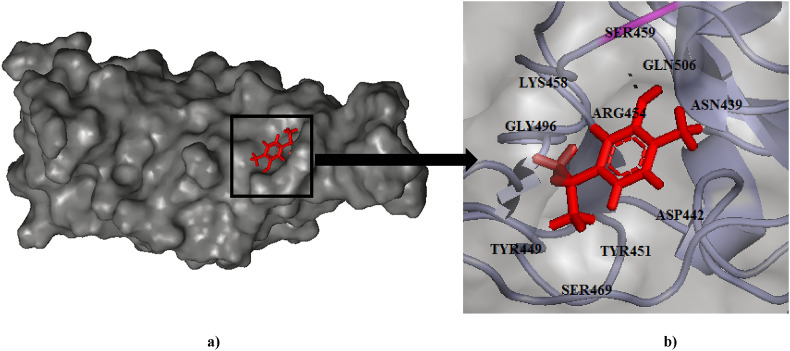
Fig. 2.
(a) Binding pose of Carvacrol (red) in RBD of spike (S) protein. (b) The hydrogen bonding with residue Ser459 has been depicted in pink
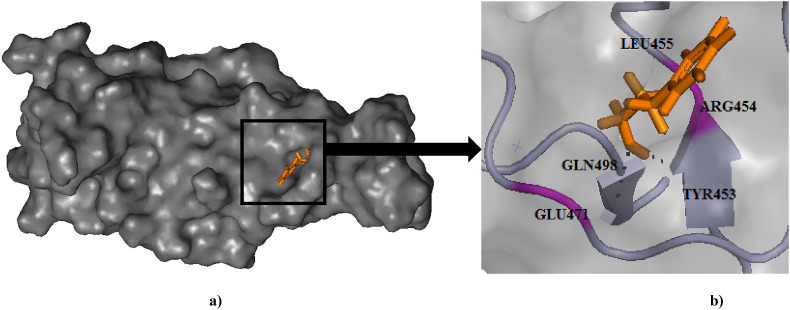
Fig. 3.
(a) Binding pose of Cinnamaldehyde (orange) in RBD of spike (S) protein. (b) The hydrogen bonding with active site residues Arg454 and Glu471 has been depicted in pink
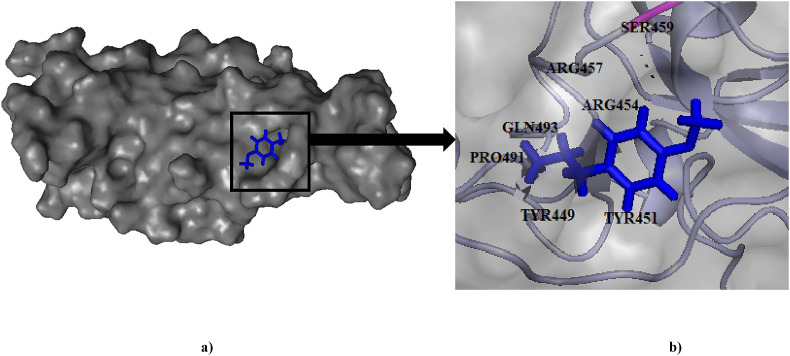
Fig. 4.
(a) Binding pose of Anethole (blue) in RBD of spike (S) protein. (b) The hydrogen bonding with active site residue Ser459 has been depicted in pink.
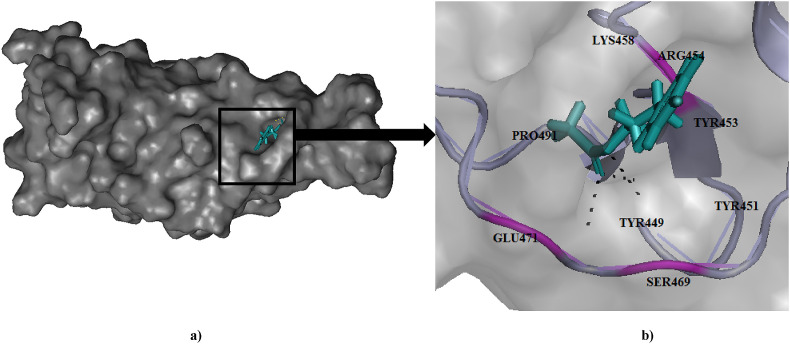
Fig. 5.
(a) Binding pose of Cinnamyl acetate (teal) in RBD of spike (S) protein. (b) The hydrogen bonding with active site residues Ser469, Arg454 and Glu471 has been depicted in pink.
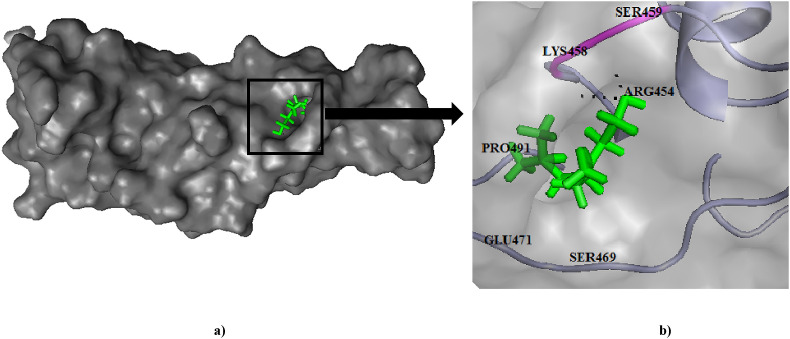
Fig. 6.
(a) Binding pose of Geraniol (green) in RBD of spike (S) protein. (b) The hydrogen bonding with active site residues Lys458, Ser459 has been depicted in pink.
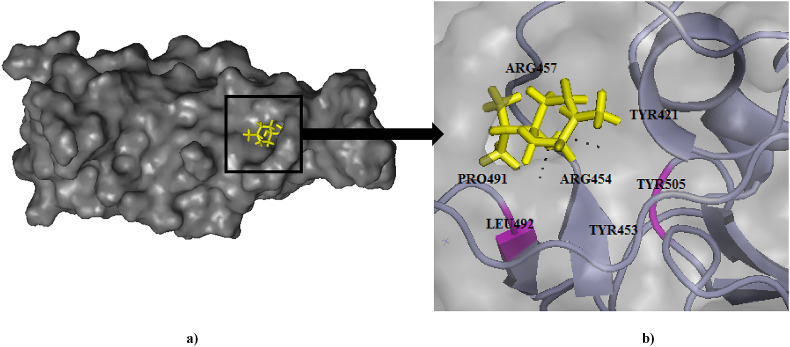
Fig. 7.
(a) Binding pose of L-4-terpineol (yellow) in RBD of spike (S) protein. (b) The hydrogen bonding with active site residues Leu492, Tyr505 has been depicted in pink.
Fig. 8.
(a) Binding poses of the selected ligands (represented in distinct colours) in the binding pocket of RBD of spike (S) protein has been depicted (b) The binding pocket area has been zoomed in, to visualise the residues in the vicinity.
3.3. Conceptual DFT
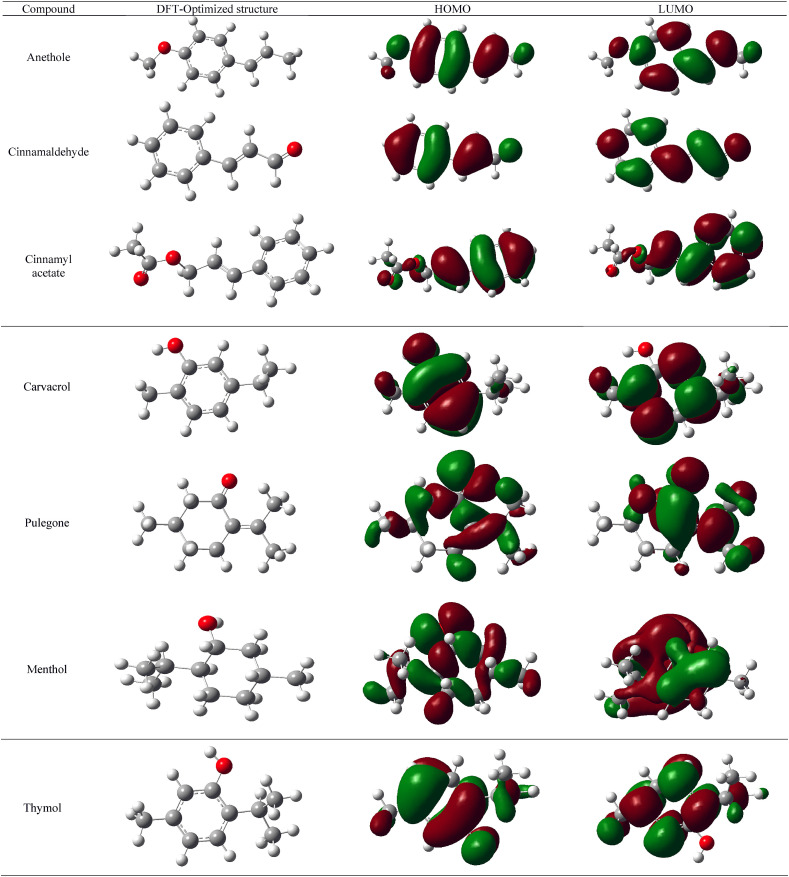
Initially, the phytochemicals were optimized using B3LYP function [45,46] with a 6-31G (d) basis in Gaussian 16 [47] to estimate their molecular properties using Fukui’s molecular orbital theory [48] Molecular orbital energies like HOMO energy (E HOMO) and LUMO energy (E LUMO) were calculated. E HOMO and E LUMO are important descriptors which stands for ability of the molecules to donate and accept electrons, respectively. Maps representing the density of electrons in different regions of the molecules at HOMO and LUMO were generated and analyzed. Table 3 represents the E HOMO and E LUMO values of the phytochemicals. Fig. 9 represents the electron density maps of molecular orbitals of selected phytochemicals. Density maps of the molecular orbitals were constructed and analyzed. Energy gap (ΔE) between the molecular orbitals of the molecules were estimated with the formula: ΔE = E LUMO - E HOMO. It is important to mention that energy gap is directly proportional to the reactivity of the molecules which can be correlated to the transition of molecules from HOMO to LUMO [49]. Hence, we have calculated the band energy gap of phytochemicals and plotted their distribution in Fig. 9 α-terpinene showed the lowest energy gap between HOMO and LUMO with a ΔE value of 2.58 eV. o-Cymene has the maximum energy difference between orbitals for all the compounds with a value of 12.57 eV. Compounds like cinnamaldehyde, anethole, cinnamyl actetate, pulegone, ocimene, thymol, carvacrol and menthol scored ΔE lesser than 6.0 eV.
Table 3.
Statistics of DFT based molecular descriptors of phytochemicals.
| Compound | Total Energy(ET) (in eV) | Molecular dipole moment(Debye) | EHOMO (eV) | ELUMO (eV) | HOMO/LUMO Gap | Absolute hardness(η) | Global softness(σ) | Electro negativity (χ) | Chemical potential (μ) | Electrophilicity index(ω) |
|---|---|---|---|---|---|---|---|---|---|---|
| Thymol | −12646.53 | 1.47 | −5.73 | 0.15 | 5.88 | 2.94 | 0.17 | −2.79 | 2.79 | 1.32 |
| Carvacrol | −12646.54 | 1.44 | −5.75 | 0.18 | 5.93 | 2.97 | 0.17 | −2.79 | 2.79 | 1.31 |
| Eugenol | −14659.05 | 2.21 | −5.73 | 0.60 | 6.33 | 3.16 | 0.16 | −2.56 | 2.56 | 1.04 |
| Eucalyptol | −12712.11 | 1.29 | −6.24 | 1.86 | 8.10 | 4.05 | 0.12 | −2.19 | 2.19 | 0.59 |
| Cinnamaldehyde | −11510.10 | 4.53 | −6.59 | −2.10 | 4.49 | 2.24 | 0.22 | −4.34 | 4.34 | 4.21 |
| Anethole | −12612.72 | 1.18 | −5.33 | −0.41 | 4.92 | 2.46 | 0.20 | −2.87 | 2.87 | 1.67 |
| l-α-terpineol | −12711.76 | 1.53 | −5.98 | 1.11 | 7.08 | 3.54 | 0.14 | −2.44 | 2.44 | 0.84 |
| Cinnamyl-acetate | −15697.18 | 2.21 | −6.14 | −1.06 | 5.08 | 2.54 | 0.20 | −3.60 | 3.60 | 2.55 |
| Limonene | −10631.48 | 0.46 | −6.17 | 0.81 | 6.98 | 3.49 | 0.14 | −2.68 | 2.68 | 1.03 |
| γ-terpinene | −10631.64 | 0.03 | −5.92 | 0.84 | 6.76 | 3.38 | 0.15 | −2.54 | 2.54 | 0.95 |
| α-terpinene | −10631.73 | 0.52 | −5.24 | −2.66 | 2.58 | 1.29 | 0.39 | −3.95 | 3.95 | 6.03 |
| Camphene | −10631.47 | 0.56 | −6.36 | 0.74 | 7.10 | 3.55 | 0.14 | −2.81 | 2.81 | 1.11 |
| Camphor | −12679.05 | 2.96 | −6.23 | −0.23 | 6.01 | 3.00 | 0.17 | −3.23 | 3.23 | 1.74 |
| Sabinene | −10631.02 | 0.82 | −6.03 | 0.76 | 6.78 | 3.39 | 0.15 | −2.64 | 2.64 | 1.02 |
| Pulegone | −12679.00 | 2.81 | −6.19 | −1.00 | 5.19 | 2.60 | 0.19 | −3.59 | 3.59 | 2.48 |
| Ocimene | −10630.83 | 0.84 | −5.82 | −0.37 | 5.46 | 2.73 | 0.18 | −3.10 | 3.10 | 1.75 |
| Thujene | −10630.98 | 0.18 | −6.07 | 0.62 | 6.69 | 3.35 | 0.15 | −2.72 | 2.72 | 1.11 |
| Geraniol | −12710.80 | 1.78 | −6.12 | 0.42 | 6.54 | 3.27 | 0.15 | −2.85 | 2.85 | 1.24 |
| L-4-terpineol | −12711.78 | 1.61 | −6.39 | 0.68 | 7.07 | 3.53 | 0.14 | −2.86 | 2.86 | 1.15 |
| Menthol | −12745.17 | 1.49 | −6.92 | 2.04 | 8.96 | 4.48 | 0.11 | −2.44 | 2.44 | 0.66 |
| Menthone | −12712.26 | 2.98 | −6.24 | −0.27 | 5.96 | 2.98 | 0.17 | −3.26 | 3.26 | 1.78 |
| p-Menthan-1-ol | −12745.21 | 1.72 | −6.88 | 1.73 | 8.61 | 4.30 | 0.12 | −2.58 | 2.58 | 0.77 |
| p-Cymene | −10599.69 | 0.09 | −6.16 | 0.16 | 6.32 | 3.16 | 0.16 | −3.00 | 3.00 | 1.42 |
| o-Cymene | −10527.11 | 0.46 | −8.50 | 4.07 | 12.57 | 6.29 | 0.08 | −2.22 | 2.22 | 0.39 |
| Linalool | −12710.80 | 1.68 | −6.19 | 0.38 | 6.58 | 3.29 | 0.15 | −2.90 | 2.90 | 1.28 |
Fig. 9.
Electron density maps of HOMO and LUMO of phytochemicals which showed better activity.
Molecular dipole moment of a molecule can be correlated with its chemical reactivity, as the two are directly proportional to each other [50]. Compounds like cinnamaldehyde, menthone, camphor, pulegone, cinnamylacetate and eugenol scored molecular dipole moments greater than 2.0 debye, with cinnamaldehyde being the highest with 4.53 debye. From the values of E HOMO and E LUMO, we derived the values of other descriptors like Electronegativity (χ), Electrophilicity index (ω), Chemical potential (μ), Global softness (σ) and Absolute hardness of the phytochemicals. Electronegativity of a compound is a key descriptor as it influences the ability of a molecule to accept electrons. Lower the electronegativity of a molecule, higher will be its efficiency of inhibition [51]. Cinnamaldehyde has the lowest electronegativity value among the phytochemicals with a score of −4.34. Other compounds like α-terpinene, methone, camphor, pulegone, cinnamylacetate, ocimene and p-cymene scored values lesser than −3.0. The values of descriptors of the phytochemicals calculated based on conceptual DFT are correlated with the docking analysis. We have identified that cinnamaldehyde, one of the phytochemicals with a high docking score, has been the best scoring molecule, considering all the descriptors. The high electronegativity of cinnamaldehyde compared to other phytochemicals correlates well with the docking analysis. Other compounds with good docking scores like carvacrol, cinnamylactetate, anethole, thymol, pulegone and menthol also scored better values.
4. Discussion
In silico techniques have a primary role in early drug discovery process. A computational analysis of drug-protein interactions gives a fair picture of probable lead molecules which can further be tested in vitro. In case of COVID-19, the scientific fraternity is under immense pressure to develop a drug or vaccine. Since identifying a novel antiviral drug candidate particularly for SARS-CoV-2 and conducting clinical trials for the same will be time consuming, drugs are being repurposed to address the situation. Hydroxychloroquine sulphate which was used to treat malaria has been proposed to treat COVID-19 since it has an immunomodulatory effect that stops the cytokine storm generation in the host system [52,53]. Similarly, Remdesvir was originally meant to treat ebola virus disease but was found to be ineffective. It was repurposed to treat SARS and MERS and was found to be effective, but in case of COVID-19, it failed in the phase 3 clinical trials [54,55]. Ribavirin, which was an antiviral drug to treat RSV (Respiratory Syncytial Virus) is also being considered to treat this infection [56]. Synergestic effect of lopinavir, oseltamivir and ritonavir have been found to be highly effective against SARS-CoV-2 protease [57].
Exploring plant-based drugs may also be considered because phyto drugs have been known to treat many diseases. They represent about 60% of all the drugs in western medicine [58]. Medicinal herbs contain numerous active ingredients that may be used for the development of therapeutic agents. Essential oils are plant-based volatile liquids. Some oils and their components are known for their antiviral properties. Clove and oregano oils could inhibit polio virus, coxsackie virus B1 and adeno virus type 3 [14]. Similarly, essential oils such as thyme, eucalyptus, tea tree could inhibit herpes simplex virus (HSV) by 96% and some monoterpene compounds could inhibit the same by 80% [59,60]. Eugenol, which is a terpene present in clove essential oil also exhibited virucidal effect against human herpes virus [61]. Likewise, many essential oils have displayed antiviral properties. Therefore in our study, major components of essential oils known for antiviral or rather antimicrobial activity in general have been selected as a ligand set.
The Docking analysis and DFT calculation of these ligands against RBD of S protein has resulted in some components showing better inhibitory effects against the target, though almost all were competent to be potent antiviral agents. Anethole which is a phenol methyl ether is abundantly present in star anise essential oil (∼80%), fennel oil (∼60%) and some other plants belonging to Apiaceae, Myrtaceae and Fabaceae families [62]. Oseltamivir contains shikimic acid, which is also derived from star anise. Thymol, carvacrol, which are monoterpenoids and geraniol, which is an acyclic monoterpene alcohol possess a broad range of activities. They are majorly present in essential oil extracted from plants belonging to Lamiaceae and Geraniaceae family respectively. The phenol ring structure with an addition of hydroxyl group renders them highly efficient antivirals [63]. Cinnamaldehyde and cinnamyl acetate present in cinnamon essential oil are phenylpropanoids with versatile properties known to possess high antioxidant, anti inflammatory, antifungal, antiviral, anticancer and antibacterial properties [64]. L-4-terpineol is present in ample amounts in tea tree and lavender essential oils. Tea tree oil is a notable antiviral essential oil, and also, it has the ability to suppress the production of inflammatory mediators, thereby reducing post-infection inflammation in lungs [65]. Other terpenes such as pulegone, camphene, menthol and ocimene are also efficient antiviral components since their binding affinities are high and hydrophobic interactions are strong enough for stable complex formation, even though they lacked hydrogen bond formation with the target protein. The molecular descriptors calculated by conceptual DFT technique has further complemented the docking results to prove that the above mentioned compounds are highly potent antiviral agents.
The RBD comprises of about 188 amino acid residues.The residues in the binding site characterized in this study are located in the cavity which is at the distal end of the RBD. The important residues for interaction range from Tyr449 to Tyr505. The residues in the RBD that interact with ACE2 also fall in the above mentioned range. About 14 hydrogen bonds are involved in RBD-ACE2 longitudinal interaction as pointed out by Lan et al. since ACE 2 is a large protein, whereas ligands are small molecules that have stabilized with a couple of hydrogen bond interactions [30]. The stability of the ligands may also be attributed to strong hydrophobic interactions in the cavity owing to amino acids in the vicinity as depicted in Table 2. The binding site characterized in this study could be considered an important site for targeting drugs in vitro with antiviral potential since it has been validated thoroughly through in silico methods.
5. Conclusion
COVID-19 is a highly contagious viral respiratory disease which has to be controlled to prevent further spread and fatalities. The potent antiviral terpenes and phenylpropanoids identified through this study could play a vital role in inhibiting the viral replication in host system and thereby halting further damage. The binding site presented in this study is of utmost importance to elicit the determined action and this site could play vital role in targeting other drugs too. The conceptual DFT study has rendered more insight into the chemical nature of the phytochemicals based on the molecular descriptors which are derived from the electron density of the molecules. In silico methods may also be used to discover other phyto-compounds, suitable for treatment, through virtual screening.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
This study was supported by research facility lab funded by SERB Young scientist grant (SB/YS/LS-128/2013).
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., F Gao G., Tan W. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan W., Zhao S., Yu B., Chen Y., Wang W., Song Z., Hu Y., Tao Z., Tian J., Pei Y., Yuan M., Zhang Y., Dai F., Liu Y., Wang Q., Zheng J., Xu L., Holmes E.C., Zhang Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 4.Drosten C., Guenther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., ouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. https://doi:10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. https://doi: 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 6.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200502-covid-19-sitrep-103.pdf?sfvrsn=d95e76d8_4
- 7.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., Tummino T.A. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci. 2013;34:267–272. doi: 10.1016/j.tips.2013.03.004. https://doi: 10.1016/j.tips.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 9.Egbuna C., Kumar S., Ifemeje J.C., Ezzat S.M., Kaliyaperumal S. first ed. Elsevier; 2019. Phytochemicals as Lead Compounds for New Drug Discovery. [Google Scholar]
- 10.Bai W., Xiao J. Third Int. Symp. Phytochem. Med. Food. 2019;59:S1–S3. doi: 10.1080/10408398.2019.1601857. [DOI] [Google Scholar]
- 11.Roy M., Datta A. Cancer Genetics and Therapeutics. Springer; Singapore: 2019. Drugs and phytochemicals; pp. 83–109. [DOI] [Google Scholar]
- 12.Xiao J., Bai W. Bioactive phytochemicals. Crit. Rev. Food Sci. Nutr. 2019;59(6):827–829. doi: 10.1080/10408398.2019.1601848. [DOI] [PubMed] [Google Scholar]
- 13.Baptista-Silva S., Borges S., Ramos O.L., Pintado M., Sarmento B. The progress of essential oils as potential therapeutic agents: a review. J. Essent. Oil Res. 2020:1–17. doi: 10.1080/10412905.2020.1746698. [DOI] [Google Scholar]
- 14.Swamy M.K., Akhtar M.S., Sinniah U.R. Antimicrobial properties of plant essential oils against human pathogens and their mode of action: an updated review. Evid. Based Complem. Altern. Med. 2016 doi: 10.1155/2016/3012462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdivieso-Ugarte M., Gomez-Llorente C., Plaza-Díaz J., Gil A. Antimicrobial, antioxidant and immunomodulatory properties of essential oils: a systematic review. Nutrients. 2019;11(11):2786. doi: 10.3390/nu11112786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey A.K., Kumar P., Singh P., Tripathi N.N., Bajpai V.K. Essential oils: sources of antimicrobials and food preservatives. Front. Microbiol. 2017;7:2161. doi: 10.3389/fmicb.2016.02161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koroch A.R., Juliani H.R., Zygadlo J.A. Flavours and Fragrances. Springer; Berlin, Heidelberg: 2007. Bioactivity of essential oils and their components; pp. 87–115. [DOI] [Google Scholar]
- 18.El Omari K., Hamze M., Alwan S., Osman M., Jama C., Chihib N.E. In-vitro evaluation of the antibacterial activity of the essential oils of Micromeria barbata, Eucalyptus globulus and Juniperus excelsa against strains of Mycobacterium tuberculosis (including MDR), Mycobacterium kansasii and Mycobacterium gordonae. J. Infect. Pub. Health. 2019;12(5):615–618. doi: 10.1016/j.jiph.2019.01.058. [DOI] [PubMed] [Google Scholar]
- 19.Elshafie H.S., Camele I. An overview of the biological effects of some mediterranean essential oils on human health. BioMed Res. Int. 2017 doi: 10.1155/2017/9268468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stea S., Beraudi A., De Pasquale D. Essential oils for complementary treatment of surgical patients: state of the art. Evid. Based Complem. Altern. Med. 2014 doi: 10.1155/2014/726341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mossa A.T.H., Refaie A.A., Ramadan A., Bouajila J. Antimutagenic effect of Origanum majorana L. essential oil against prallethrin-induced genotoxic damage in rat bone marrow cells. J. Med. Food. 2013;16(12):1101–1107. doi: 10.1089/jmf.2013.0006. [DOI] [PubMed] [Google Scholar]
- 22.Pourghanbari G., Nili H., Moattari A., Mohammadi A., Iraji A. Antiviral activity of the oseltamivir and Melissa officinalis L essential oil against avian influenza A virus (H9N2) Virusdisease. 2016;27(2):170–178. doi: 10.1007/s13337-016-0321-0. https://DOI: 10.1007/s13337-016-0321-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tariq S., Wani S., Rasool W., Shafi K., Bhat M.A., Prabhakar A., Rather M.A. A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug-resistant microbial pathogens. Microb. Pathog. 2019;134:103580. doi: 10.1016/j.micpath.2019.103580. https://DOI:10.1016/j.micpath.2019.103580 [DOI] [PubMed] [Google Scholar]
- 24.Terstappen G.C., Reggiani A. In silico research in drug discovery. Trends Pharmacol. Sci. 2001;22(1):23–26. doi: 10.1016/s0165-6147(00)01584-4. https://DOI: 10.1016/s0165-6147(00)01584-4 [DOI] [PubMed] [Google Scholar]
- 25.Ekins S., Mestres J., Testa B. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br. J. Pharmacol. 2007;152(1):9–20. doi: 10.1038/sj.bjp.0707305. https://DOI: 10.1038/sj.bjp.0707305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava P., Tiwari A. Critical role of computer simulations in drug discovery and development. Curr. Top. Med. Chem. 2017;17(21):2422–2432. doi: 10.2174/1568026617666170403113541. https://DOI: 10.2174/1568026617666170403113541 [DOI] [PubMed] [Google Scholar]
- 27.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Šali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. 2000;29(1):291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 28.Hardin C., Pogorelov T.V., Luthey-Schulten Z. Ab initio protein structure prediction. Curr. Opin. Struct. Biol. 2002;12(2):176–181. doi: 10.1016/S0959-440X(02)00306-8. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee S., Zhang Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;9(7):955–966. doi: 10.1016/j.str.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. https://DOI: 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trott A.J., Olson O. Auto Dock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dallakyan S., Olson A.J. Chemical Biology. Humana Press; New York, NY: 2015. Small-molecule library screening by docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 35.Dhifi W., Bellili S., Jazi S., Bahloul N., Mnif W. Essential oils’ chemical characterization and investigation of some biological activities: a critical review. Medicines. 2016;3(4):25. doi: 10.3390/medicines3040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashraf Z., Saeed A., Nadeem H. Design synthesis and docking studies of some novel isocoumarin analogues as antimicrobial agents. RSC Adv. 2014;4(96):53842–53853. doi: 10.1039/C4RA07223E. [DOI] [Google Scholar]
- 38.Leach A.R., Shoichet B.K., Peishoff C.E. Prediction of protein− ligand interactions. Docking and scoring: successes and gaps. J. Med. Chem. 2006;49(20):5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 39.Kitchen D.B., Decornez H., Furr J.R., Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 40.Vieira T.F., Sousa S.F. Comparing AutoDock and vina in ligand/decoy discrimination for virtual screening. Appl. Sci. 2019;9(21):4538. doi: 10.3390/app9214538. [DOI] [Google Scholar]
- 41.Chang M.W., Ayeni C., Breuer S., Torbett B.E. Virtual screening for HIV protease inhibitors: a comparison of AutoDock 4 and Vina. PloS One. 2010;5(8) doi: 10.1371/journal.pone.0011955. https://doi:10.1371/journal.pone.0011955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geerlings P., Chamorro E., Chattaraj P.K., De Proft F., Gázquez J.L., Liu S., Ayers P. Conceptual density functional theory: status, prospects, issues. Theor. Chem. Acc. 2020;139(2):36. doi: 10.1007/s00214-020-2546-7. [DOI] [Google Scholar]
- 43.Hohenberg P., Kohn W. Density functional theory (DFT) Phys. Rev. 1964;136:B864. [Google Scholar]
- 44.Domingo L., Ríos-Gutiérrez M., Pérez P. Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules. 2016;21(6):748. doi: 10.3390/molecules21060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becke A. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 1988;38(6):3098–3100. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- 46.Lee C., Yang W., Parr R. Development of the Colle-Salvetti -energy formula into a functional of the electron density. Phys. Rev. B. 1988;37(2):785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 47.Frisch M., Trucks G., Schlegel H., Scuseria G., Robb M., Cheeseman J., Fox D. Gaussian, Inc.; Wallingford CT: 2016. Gaussian 16 (Version Revision B.01) [Linux] [Google Scholar]
- 48.Fukui K. The role of frontier orbitals in chemical reactions (nobel lecture) Angew Chem. Int. Ed. Engl. 1982;21(11):801–809. doi: 10.1002/anie.198208013. [DOI] [Google Scholar]
- 49.Bostan R., Varvara S., Găină L., Mureşan L. Evaluation of some phenothiazine derivatives as corrosion inhibitors for bronze in weakly acidic solution. Corrosion Sci. 2012;63:275–286. doi: 10.1016/j.corsci.2012.06.010. [DOI] [Google Scholar]
- 50.Mert B., Erman Mert M., Kardaş G., Yazıcı B. Experimental and theoretical investigation of 3-amino-1,2,4-triazole-5-thiol as a corrosion inhibitor for carbon steel in HCl medium. Corrosion Sci. 2011;53(12):4265–4272. doi: 10.1016/j.corsci.2011.08.038. [DOI] [Google Scholar]
- 51.Zhan C., Nichols J., Dixon D. Ionization potential, electron affinity, electronegativity, hardness, and electron excitation energy: molecular properties from density functional theory orbital energies. J. Phys. Chem. 2003;107(20):4184–4195. doi: 10.1021/jp0225774. [DOI] [Google Scholar]
- 52.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Zhan S. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah S., Das S., Jain A., Misra D.P., Negi V.S. A systematic review of the prophylactic role of chloroquine and hydroxychloroquine in Coronavirus Disease-19 (COVID-19) Int. J. Rheum. Disease. 2020 doi: 10.1111/1756-185X.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng C.Y., Lee Y.L., Chen C.P., Lin Y.C., Liu C.E., Liao C.H., Cheng S.H. Lopinavir/ritonavir did not shorten the duration of SARS CoV-2 shedding in patients with mild pneumonia in Taiwan. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. https://doi: 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Diaz G. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. J. Biomol. Struct. Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 58.Pan S.Y., Zhou S.F., Gao S.H., Yu Z.L., Zhang S.F., Tang M.K., Ko K.M. New perspectives on how to discover drugs from herbal medicines: CAM’s outstanding contribution to modern therapeutics. Evid. Based Complem. Altern. Med. 2013 doi: 10.1155/2013/627375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Astani A., Reichling J., Schnitzler P. Comparative study on the antiviral activity of selected monoterpenes derived from essential oils. Phytother Res. 2010;24(5):673–679. doi: 10.1002/ptr.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koch C., Reichling J., Schneele J., Schnitzler P. Inhibitory effect of essential oils against herpes simplex virus type 2. Phytomedicine. 2008;15(1–2):71–78. doi: 10.1016/j.phymed.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 61.Benencia F., Courreges M.C. In vitro and in vivo activity of eugenol on human herpesvirus. Phytother Res. 2000;14(7):495–500. doi: 10.1002/1099-1573(200011)14:7%3C495::AID-PTR650%3E3.0.CO. [DOI] [PubMed] [Google Scholar]
- 62.Vázquez-Fresno R., Rosana A.R.R., Sajed T., Onookome-Okome T., Wishart N.A., Wishart D.S. Herbs and spices-biomarkers of intake based on human intervention studies–A systematic review. Genes nutr. 2019;14(1):18. doi: 10.1186/s12263-019-0636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saad N.Y., Muller C.D., Lobstein A. Major bioactivities and mechanism of action of essential oils and their components. Flavour Fragrance J. 2013;28(5):269–279. doi: 10.1002/ffj.3165. [DOI] [Google Scholar]
- 64.Rao P.V., Gan S.H. Cinnamon: a multifaceted medicinal plant. Evid. Based Complem. Altern. Med. 2014 doi: 10.1155/2014/642942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carson C.F., Hammer K.A., Riley T.V. Melaleuca alternifolia (tea tree) oil: a review of antimicrobial and other medicinal properties. Clin. Microbiol. Rev. 2006;19(1):50–62. doi: 10.1128/CMR.19.1.50-62.2006. https://doi:10.1128/CMR.19.1.50-62.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]