Abstract
Although it has been proved that the epigenetic modification of DNA and histones is involved in the pathogenesis of systemic lupus erythematosus (SLE), there is no study to explore whether the modification of N6-methyladenosine (m6A) in RNA is involved. In this study, the mRNA levels of m6A “writers” (METTL3, MTEEL14, and WTAP), “erasers” (FTO and ALKBH5), and “readers” (YTHDF2) in peripheral blood were determined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The results demonstrated that the mRNA levels of METTL3, WTAP, FTO, ALKBH5, and YTHDF2 in peripheral blood from SLE patients were significantly decreased. The levels of ALKBH5 mRNA in SLE patients were associated with anti-dsDNA, antinucleosome, rash, and ulceration. Multivariate logistic regression analysis showed that the level of ALKBH5 mRNA in peripheral blood is a risk factor of SLE (P < 0.001). Moreover, our results suggested that there was a positive correlation between m6A“writers” (METTL3 and WTAP), “erasers” (FTO and ALKBH5), and “readers” (YTHDF2) in SLE patients. This study suggests that the mRNA level of ALKBH5 in peripheral blood may be involved in the pathogenesis of SLE.
1. Introduction
The systemic lupus erythematosus (SLE) is a chronic and incurable autoimmune disease characterized by intermittent episodes of increased disease activity that require treatment with immunosuppressive agents [1, 2]. Although there have been many studies trying to elucidate the pathogenesis of SLE, so far it has not been fully elucidated. Existing studies have demonstrated that the dysfunction of immune cells such as T cells, B cells, monocytes, neutrophils, and dendritic cells plays important roles in the pathogenesis of SLE [3–6]. Further elucidation of the aetiology of SLE is of great significance for the development of possible targeted and individualized therapy for SLE [7].
In recent years, epigenetic modifications have been demonstrated to play an important role in the genesis and development of SLE [8, 9]. N6-methyladenosine (m6A) modification is the most prevalent and evolutionarily conserved modification which occurs in nearly all types of RNAs and in most organisms [10]. This modification can be installed by adenosine methyltransferases, reversed by demethylases, and recognized by some RNA-binding proteins [11]. M6A methyltransferase complex, known as the m6A “writer” complex, contains methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumor 1-associating protein (WTAP), which functions by depositing the m6As in mammalian mRNA [12]. Fat mass and obesity-associated protein (FTO) and a-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) are selective demethylases capable of regulating gene expression and cell fate through oxidative removal of the methyl group in m6A-containing substrates, usually considered as m6A “erasers” [13]. Some RNA-binding proteins such as YT521-B homology domains 2 (YTHDF2) which can recognize m6A modification, decode the methylation code, and finally transform them into diverse functional signals are called m6A “readers” [14].
Recent studies have demonstrated that m6A modification is associated with various human diseases [15, 16]. However, there is no study to characterize m6A modification in patients with SLE. To investigate whether m6A modification plays a role in the genesis and development of SLE, the mRNA levels of METTL3, MTEEL14, WTAP, FTO, ALKBH5, and YTHDF2 in peripheral blood were detected in SLE patients and analyzed for their correlation with clinical variables.
2. Methods
2.1. Patient Variables and Controls
A total of 51 patients that fulfilled the revised American College of Rheumatology criteria for SLE [17] were recruited from the First Affiliated Hospital of Nanchang University from 2018.10 to 2019.3. Among them, 40 patients were new-onset SLE that first-time diagnosis of SLE and no history of immunosuppressive drug or corticosteroid use before recruitment. Among all new-onset SLE patients, 7 patients were reexamined after 15 days of regular treatment by using glucocorticoids and immunosuppressive agents. The other 11 patients were revisiting SLE patients receiving treatment. Disease activity was assessed by the SLE disease activity index (SLEDAI) [18]. 38 healthy controls (CON) without a clinical diagnosis of any inflammatory or autoimmune diseases and without relation to patients of autoimmune disease were enrolled from the First Affiliated Hospital of Nanchang University. In addition, 51 patients fulfilled the revised ACR 2010 criteria for rheumatoid arthritis (RA) [19], 30 patients were infected with hepatitis B virus (HBV) and 27 patients with tuberculosis (TB) were recruited from the First Affiliated Hospital of Nanchang University. The demographic characteristics of the study population are shown in Table 1. The study had approval from the Ethics Committee of the First Affiliated Hospital of Nanchang University (052) and complied with the Helsinki Declaration. All participants provided signed informed consent before they entered this study.
Table 1.
Clinical characteristics of SLE patients, RA patients, HBV-infected patients, TB patients, and CON.
| Categories | SLE patients | CON | RA patients | HBV | TB patients |
|---|---|---|---|---|---|
| n | 51 | 38 | 51 | 30 | 27 |
| Females (%) | 92.16 | 78.95 | 82.35 | 93.33 | 100 |
| Age (years) | 39.30 ± 14.37 | 39.71 ± 13.48 | 50.67 ± 12.67 | 35.93 ± 8.04 | 33.46 ± 12.05 |
| SLEDAI score | 13.09 ± 7.75 | ||||
| Anti-dsDNA (IU/mL) | 230.40 ± 342.73 | ||||
| Anti-ENA | |||||
| Anti-Sm (%) | 29.73 | ||||
| Anti-Ro52 (%) | 70.27 | ||||
| Anti-nRNP/Sm (%) | 59.46 | ||||
| Anti-RIB-P (%) | 24.32 | ||||
| Antinucleosome (%) | 32.43 | ||||
| Anti-SSA (%) | 64.86 | ||||
| Anti-SSB (%) | 18.92 | ||||
| C3 (g/L) | 0.59 ± 0.27 | ||||
| C4 (g/L) | 0.14 ± 0.11 | ||||
| IgG (g/L) | 18.66 ± 6.00 | ||||
| ESR (mm/h) | 62.07 ± 31.13 | ||||
| CRP (mg/L) | 18.63 ± 35.25 | ||||
| WBC (109/L) | 6.43 ± 3.90 | 5.84 ± 0.96 | |||
| RBC (1012/L) | 3.80 ± 0.67∗ | 4.59 ± 0.39 | |||
| HGB (g/L) | 124.41 ± 76.06∗ | 138.58 ± 11.50 | |||
| HCT (L/L) | 0.34 ± 0.06∗ | 0.41 ± 0.03 | |||
| PLT (109/L) | 195.83 ± 94.01∗ | 243.00 ± 46.32 | |||
| Lymphocytes (109/L) | 1.39 ± 0.62∗ | 1.86 ± 0.30 | |||
| Lymphocytes (%) | 25.08 ± 10.26∗ | 32.51 ± 6.30 | |||
| Monocytes (109/L) | 0.51 ± 0.34∗ | 0.36 ± 0.09 | |||
| Monocytes (%) | 8.09 ± 2.66∗ | 6.14 ± 1.43 | |||
| Neutrophils (109/L) | 4.45 ± 3.41 | 3.52 ± 0.82 | |||
| Neutrophils (%) | 65.49 ± 11.10∗ | 59.49 ± 6.00 | |||
| Clinical features | |||||
| Fever (%) | 44.44 | ||||
| Rash (%) | 36.11 | ||||
| Alopecia (%) | 17.14 | ||||
| Arthritis (%) | 47.22 | ||||
| NPLE (%) | 11.11 | ||||
| Ulceration (%) | 11.43 | ||||
| Pleuritis (%) | 45.71 | ||||
| Pericarditis (%) | 34.29 | ||||
| LN (%) | 54.90 |
∗ P < 0.05 SLE compared to CON. Anti-dsDNA: anti-double-stranded DNA; Anti-ENA: antiextractable nuclear antigen; Anti-nRNP/Sm: antinuclear ribonucleoprotein/Smith antibody; Anti-RIB-P: anti-ribosomal P-protein antibody; Anti-Sm: anti-Smith antibody; Anti-SSA: anti-Sjögren syndrome A antigen antibody; Anti-SS-B: anti-Sjögren syndrome B antigen antibody; HBV: hepatitis B virus (HBV); C3: complement 3; C4: complement 4; HC: healthy controls; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; HCT: hematocrit; HGB: hemoglobin; LN: lupus nephritis; IgG: immunoglobulin G; L: lymphocyte count; L%: lymphocyte percentage; M: monocyte count; M%: monocyte percentage; N: neutrophil count; N%: neutrophil percentage; NPLE: neuropathic lupus erythematosus; PLT: platelet count; RBC: red blood cell count; RA: rheumatoid arthritis; SLE: systemic lupus erythematosus; SLEDAI: SLE disease activity index; TB: tuberculosis; WBC: white blood cell count.
2.2. Blood Sample Collection and Total RNA Isolation
The peripheral blood collection and total RNA isolation were performed as described previously [20].
2.3. QRT-PCR Analysis
Complementary DNA (cDNA) was obtained from purified total RNA samples by reverse transcription using a PrimeScript™ RT reagent kit (Takara Bio Inc., Japan). METTL3, METTL14, WTAP, ALKBH5, FTO, and YTHDF2 transcripts were quantified with an ABI 7500 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) by using SYBR® Premix Ex Taq™ II (Takara Bio Inc., Japan). The sequences of amplification primers for METTL3, METTL14, WTAP, ALKBH5, FTO, YTHDF2, and GAPDH are listed in Table 2. The relative levels of transcripts were derived by the 2-ΔCt method [20, 21].
Table 2.
The amplification primers sequences.
| Gene name | Sequence (5′-3′) |
|---|---|
| METTL3 | F: AAGCTGCACTTCAGACGAAT |
| R: GGAATCACCTCCGACACTC | |
| METTL14 | F: AGAAACTTGCAGGGCTTCCT |
| R: TCTTCTTCATATGGCAAATTTTCTT | |
| WTAP | F: GGCGAAGTGTCGAATGCT |
| R: CCAACTGCTGGCGTGTCT | |
| ALKBH5 | F: CCCGAGGGCTTCGTCAACA |
| R: CGACACCCGAATAGGCTTGA | |
| FTO | F: TGGGTTCATCCTACAACGG |
| R: CCTCTTCAGGGCCTTCAC | |
| YTHDF2 | F: GGCAGCACTGAAGTTGGG |
| R: CTATTGGAAGCCACGATGTTA | |
| GAPDH | F: TGCACCACCAACTGCTTAGC |
| R: GGCATGGACTGTGGTCATGAG |
METTL3: methyltransferase-like 3; METTL14: methyltransferase-like 14; WTAP: Wilms tumor 1-associating protein; ALKBH5: a-ketoglutarate-dependent dioxygenase alkB homolog 5; FTO: fat mass and obesity-associated protein; YTHDF2: YT521-B homology domains 2.
2.4. Blood Routine, Urinary Routine, Serum Inflammatory Indicators, and Autoantibody Determination
The items in blood routine and urinary routine were detected using Sysmex-x2100 (SYSMEX, Kobe, Japan) and SysmexUF-1000i (SYSMEX, Japan). Erythrocyte sedimentation rate (ESR) was determined by the BlessLBY-XC40B automatic ESR analyzer (Bless, Beijing, China) according to the manufacturer's instructions. The levels of C-reactive protein (CRP), immunoglobulin G (IgG), complement 3 (C3), and complement 4 (C4) in serum were determined by nephelometry methods using IMMAGE800 system (Beckman, CA, USA). Anti-dsDNA of IgG in serum was measured by commercially available enzyme-linked immunosorbent assay (ELISA) kits (Kexin, Shanghai, China). Antiextractable nuclear antigen (Anti-ENA) was determined using line immunoassay kits (Euroimmun, Luebeck, Germany) according to the manufacturer's instructions.
2.5. Statistical Analysis
Student's t-test or the Mann-Whitney U-test was used to compare the data according to the normality. The Spearman method was used for correlation analysis. Logistic regression analysis was used for evaluating the risk factor. All the data were analyzed by GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA) and SPSS version 17.0 (SPSS Inc., Chicago, IL). P values < 0.05 were considered statistically significant.
3. Results
3.1. Differential Expression Screening of METTL3, METTL14, WTAP, ALKBH5, FTO, and YTHDF2 in Peripheral Blood of SLE Patients
To identify the expression levels of METTL3, METTL14, WTAP, ALKBH5, FTO, and YTHDF2 in patients with SLE, the mRNA levels of these genes in peripheral blood were determined by qRT-PCR in SLE patients and HCs. Data showed that the mRNA levels of METTL3, WTAP, ALKBH5, and FTO in peripheral blood of SLE patients significantly decreased compared to HCs (all P < 0.050) (Figure 1). The mRNA level of YTHDF2 trends to decrease in SLE patients, but a significant difference was not reached (P = 0.097) (Figure 1). The mRNA level of METTL14 was unchanged (P = 0.673) (Figure 1).
Figure 1.
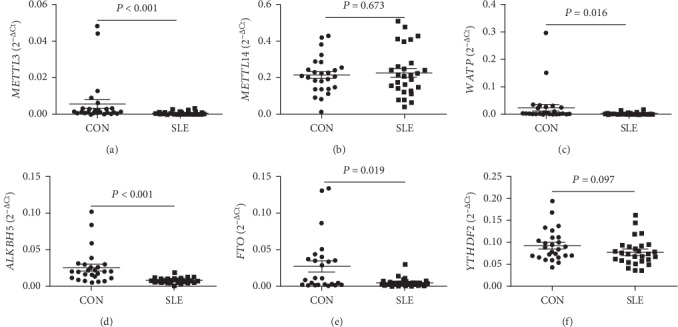
Differential expression screening of METTL3, METTL14, WTAP, ALKBH5, FTO, and YTHDF2 in peripheral blood of SLE patients. (a) The level of METTL3 in peripheral blood was significantly decreased in SLE patients (n = 28) compared to healthy controls (CON, n = 26). (b) The level of METTL14 showed no significant difference between SLE patients (n = 28) and CON (n = 26). (c) The level of WTAP in peripheral blood was significantly decreased in SLE patients (n = 28) compared to CON (n = 26). (d) The level of ALKBH5 in peripheral blood was significantly decreased in SLE patients (n = 28) compared to CON (n = 26). (e) The level of FTO in peripheral blood was significantly decreased in SLE patients (n = 28) compared to CON (n = 26). (f) The level of YTHDF2 trends to decline in SLE patients, but a significant difference was not reached.
3.2. Differential Expression Validation of METTL3, WTAP, ALKBH5, FTO, and YTHDF2
To further verify the differential expression of METTL3, WTAP, ALKBH5, FTO, and YTHDF2 in peripheral blood in the screening stage, an independent cohort including 23 SLE patients and 12 HCs were recruited and detected for the mRNA levels of these genes in peripheral blood. The data from all subjects, including all 51 SLE patients and 38 HCs in screening stage and validation stage, demonstrated that the mRNA levels of METTL3, WTAP, ALKBH5, and FTO in peripheral blood were significantly lower in SLE patients than those in HCs (all P < 0.050) (Figure 2). Inconsistent with the screening data, the level of YTHDF2 was also significantly decreased in SLE patients than that in HCs (P < 0.001) (Figure 2).
Figure 2.
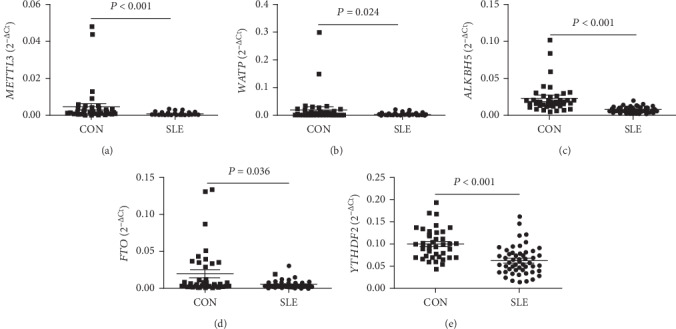
Differential expression validation of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2. (a) The level of METTL3 in peripheral blood was significantly decreased in SLE patients (n = 51) compared to healthy controls (CON, n = 38). (b) The level of WTAP in peripheral blood was significantly decreased in SLE patients (n = 51) compared to CON (n = 38). (c) The level of ALKBH5 in peripheral blood was significantly decreased in SLE patients (n = 51) compared to CON (n = 38). (d) The level of FTO in peripheral blood was significantly decreased in SLE patients (n = 51) compared to CON (n = 38). (e) The level of YTHDF2 in peripheral blood was significantly decreased in SLE patients (n = 51) compared to CON (n = 38).
3.3. Correlations between the mRNA Levels of Peripheral Blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 and Autoantibodies in SLE Patients
Production of multiple autoantibodies such as anti-double-stranded DNA (anti-dsDNA) and antiextractable nuclear antigens (Anti-ENAs) is one of the important characteristics of SLE. In 51 SLE patients, 44 SLE patients were tested for anti-dsDNA and 37 SLE patients were tested for anti-ENAs. The correlations between the mRNA levels of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 and autoantibodies were investigated in these SLE patients. As shown in Figure 3(a), the level of peripheral blood ALKBH5 was significantly decreased in patients with positive anti-dsDNA compared to patients with negative anti-dsDNA. Furthermore, the level of peripheral blood ALKBH5 in SLE patients negatively correlated with the level of anti-dsDNA (rs = −0.3062, P = 0.043) (Figure 3(b)). And, the level of peripheral blood ALKBH5 was significantly decreased in patients with positive antinucleosome compared to patients with negative antinucleosome (P = 0.003) (Figure 3(c)). The level of peripheral blood ALKBH5 did not correlate with other anti-ENAs (data not shown). No obvious correlation was observed between the levels of peripheral blood METTL3, WTAP, FTO, and YTHDF2 and autoantibodies in SLE patients (data not shown).
Figure 3.
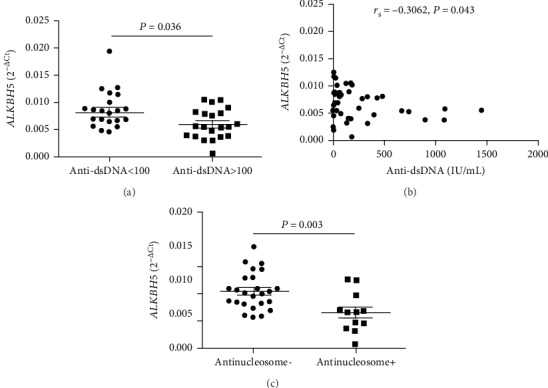
Correlations between the mRNA level of peripheral blood ALKBH5 and autoantibodies in SLE patients. (a) The mRNA level of peripheral blood ALKBH5 was significantly decreased in SLE patients with positive anti-double-stranded DNA (anti-dsDNA) compared to SLE patients with negative anti-dsDNA. (b) The mRNA level of peripheral blood ALKBH5 in SLE patients negatively correlated with the level of anti-dsDNA. (c) The level of peripheral blood ALKBH5 was significantly decreased in SLE patients with positive antinucleosome compared to SLE patients with negative antinucleosome.
3.4. Decreased mRNA Level of ALKBH5 in Peripheral Blood Was a Risk Factor for SLE
The aforementioned results demonstrated that the levels of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 in SLE patients were decreased and the level of ALKBH5 in peripheral blood was associated with autoantibody production. Considering that the production of autoantibodies plays an important role in the pathogenesis of SLE, the “enter method” of multivariate logistic regression was used to explore whether the expression of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 was risk factors for SLE. As shown in Table 3, the equations about the levels of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 were obtained, Y = −326.128 X1(METTL3) − 74.188 (WTAP) − 414.645 X3(ALKBH5) − 15.042 X4(FTO) − 0.106 X2(YTHDF2) + 6.197. The results demonstrated that the decreased expression of ALKBH5 in peripheral blood was a risk factor for SLE (P < 0.001), while other molecules are not (P > 0.050). Furthermore, the mRNA level of peripheral blood ALKBH5 in other inflammatory disorders including RA, TB, and HBV-infected patients was detected and compared with that in SLE patients. Results showed that the mRNA level of peripheral blood ALKBH5 in SLE patients was significantly lower compared to RA patients (P = 0.003) (Figure 4(a)), HBV-infected patients (P < 0.0001) (Figure 4(b)), and TB patients (P < 0.0001) (Figure 4(c)).
Table 3.
The expression of METTL3, WTAP, ALKBH5, FTO, and YTHDF2 in equation.
| B | S.E | Wald | df | P | Exp (B) | |
|---|---|---|---|---|---|---|
| METTL3 | -326.128 | 272.636 | 1.431 | 1 | 0.232 | 0.000 |
| WTAP | -74.188 | 66.854 | 1.231 | 1 | 0.267 | 0.000 |
| ALKBH5 | -414.645 | 108.988 | 14.474 | 1 | 0.000 | 0.000 |
| FTO | -15.042 | 58.027 | 0.067 | 1 | 0.795 | 0.000 |
| YTHDF2 | -0.106 | 14.726 | 0.000 | 1 | 0.994 | 0.899 |
| Constant | 6.197 | 1.408 | 19.373 | 1 | 0.000 | 491.119 |
METTL3: methyltransferase-like 3; WTAP: Wilms tumor 1-associating protein; ALKBH5: a-ketoglutarate-dependent dioxygenase alkB homolog 5; FTO: fat mass and obesity-associated protein; YTHDF2: YT521-B homology domains 2.
Figure 4.
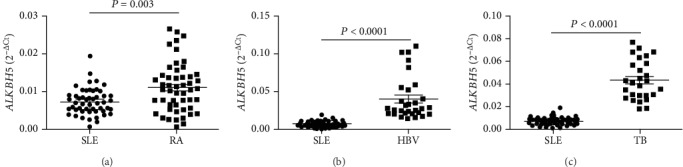
Validation of the abnormal expression of peripheral blood ALKBH5 in SLE patients, rheumatoid arthritis (RA) patients, hepatitis B virus- (HBV-) infected patients, and tuberculosis (TB) patients. (a) The peripheral blood level of ALKBH5 in SLE patients (n = 51) was significantly lower compared to RA patients (n = 51). (b) The peripheral blood level of ALKBH5 in SLE patients (n = 51) was significantly lower compared to HBV-infected patients (n = 30). (c) The peripheral blood level of ALKBH5 in SLE patients (n = 51) was significantly lower compared to TB patients (n = 27).
3.5. Correlations between the Levels of Peripheral Blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 and Clinical Variables in SLE Patients
The aforementioned results demonstrate that the level of peripheral blood ALKBH5 in SLE patients was negatively correlated with the level of anti-dsDNA. Anti-dsDNA was proved to be involved in the genesis of SLE and can be served as markers for evaluating ongoing disease activity in SLE and early relapse of SLE. And, the level of anti-dsDNA in SLE patients was found to negatively correlate with the level of C3 (r = −0.4244, P = 0.006) (Supplement Fig 1A) and C4 (r = −0.3905, P = 0.012) (Supplement Fig 1B) and positively correlate with SLEDAI (r = 0.3575, P = 0.024) (Supplement Fig 1C). Thus, the correlation test was performed to evaluate the correlations between the clinical variables of SLE and the levels of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2. Data showed that the mRNA levels of peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 in all SLE patients (including all new-onset and revisiting SLE patients) did not correlate with SLEDAI, CRP, ESR, IgG, C3, C4, WBC, RBC, HGB, HCT, PLT, L, M, or N (data not shown). However, further analysis in the new-onset SLE patients showed that the mRNA level of peripheral blood METTL3 positively correlated with PLT (rs = 0.3339, P = 0.035) (Figure 5(a)), the mRNA level of peripheral blood ALKBH5 positively correlated with WBC (rs = 0.3468, P = 0.028) (Figure 5(b)), and the mRNA level of peripheral blood WTAP positively correlated with M (rs = 0.3270, P = 0.039) (Figure 5(c)).
Figure 5.
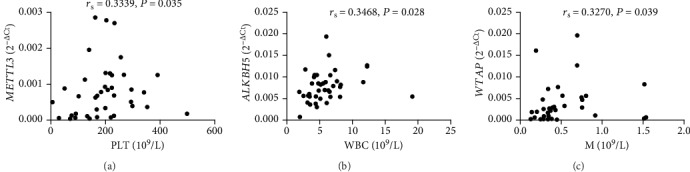
Correlations between peripheral blood METTL3, ALKBH5 and WTAP and clinical variables in new-onset SLE patients. (a) The level of peripheral blood METTL3 in new-onset SLE patients positively correlated with PLT (n = 40). (b) The level of peripheral blood ALKBH5 in new-onset SLE patients positively correlated with WBC (n = 40). (c) The level of peripheral blood WTAP in new-onset SLE patients positively correlated with M (n = 40).
Next, the relationship between the levels of these m6A modification-related molecules and drug therapy of SLE was analyzed. However, we found no significant difference between new-onset SLE patients and revisiting SLE patients (data not shown). Subsequently, a 15-day follow-up evaluation in 7 new-onset SLE patients who received regular treatment was performed, but there was still no difference (data not shown).
Moreover, the relationship between the clinical symptoms of SLE including LN, NPLE, arthritis, fever, rash, alopecia, ulceration, pleuritis, pericarditis, and the mRNA levels of these m6A modification-related molecules was analyzed. As shown in Figure 6, the mRNA levels of peripheral blood METTL3 and WTAP in SLE patients with alopecia was significantly decreased than that in SLE patients without alopecia (all P < 0.050); the mRNA level of peripheral blood ALKBH5 in SLE patients with rash and ulceration was significantly increased than that in SLE patients without rash and ulceration, respectively (all P < 0.050). No relationship was found between the levels of peripheral blood FTO, YTHDF2, and SLE symptoms (Figures 6(d) and 6(e)).
Figure 6.
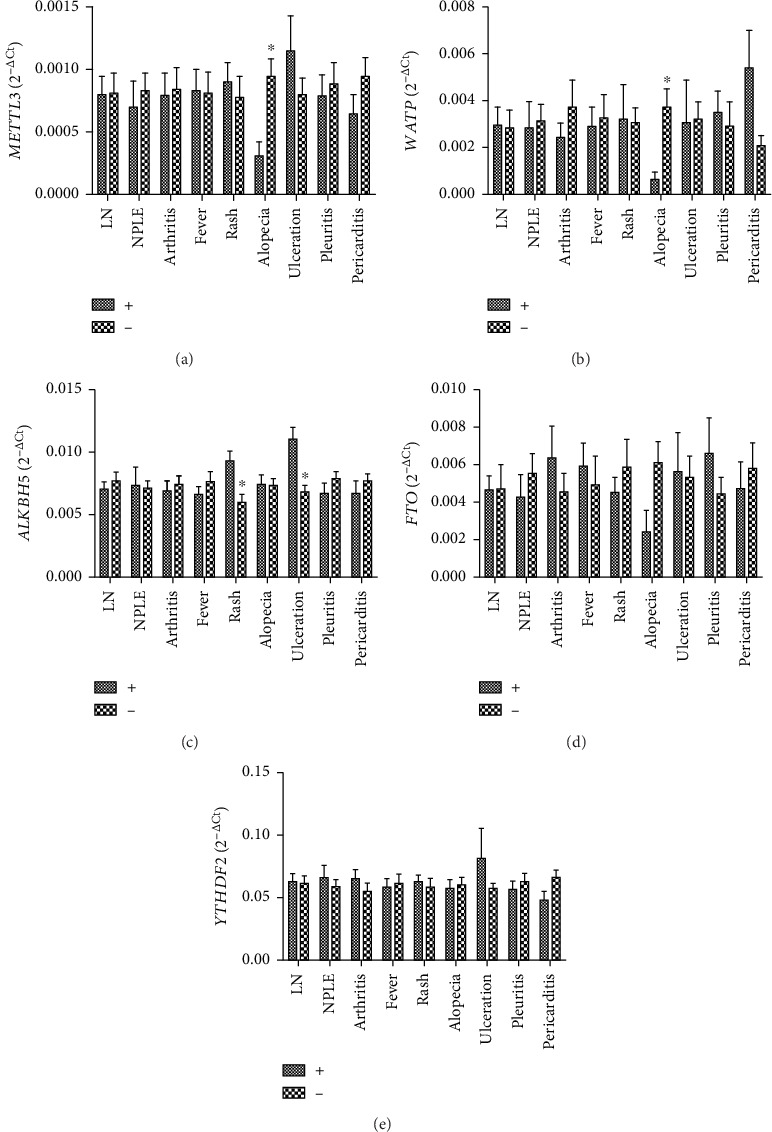
Correlations between peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 and clinical symptoms of SLE. (a) The level of peripheral blood METTL3 in SLE patients with alopecia was significantly decreased than that in SLE patients without alopecia. (b) The level of peripheral blood WTAP in SLE patients with alopecia was significantly decreased than that in SLE patients without alopecia. (c) The levels of peripheral blood ALKBH5 in SLE patients with rash and ulceration were significantly increased than those in SLE patients without rash and ulceration, respectively. (d) No correlation was found between peripheral blood FTO and clinical symptoms of SLE. (e) No correlation was found between peripheral blood YTHDF2 and clinical symptoms of SLE.
3.6. Correlations between the Levels of These m6A Modification-Related Molecules in SLE Patients
Considering that METTL3, WTAP, ALKBH5, FTO, and YTHDF2 are all key molecules in the process of m6A modification, we therefore want to know whether they are interrelated. As shown in Figure 5, the mRNA level of peripheral blood METTL3 positively correlated with the mRNA levels of WTAP (rs = 0.4595, P < 0.001) (Figure 7(a)), ALKBH5 (rs = 0.3393, P = 0.015) (Figure 7(b)), FTO (rs = 0.6025, P < 0.001) (Figure 7(c)), and YTHDF2 (rs = 0.3540, P = 0.011) (Figure 7(d)); the mRNA level of peripheral blood WTAP positively correlated with the mRNA level of FTO (rs = 0.3253, P = 0.020) (Figure 7(e)); and the mRNA level of peripheral blood YTHDF2 positively correlated with the mRNA levels of ALKBH5 (rs = 0.5770, P < 0.001) (Figure 7(f)) and FTO (rs = 0.3377, P = 0.015) (Figure 7(g)).
Figure 7.
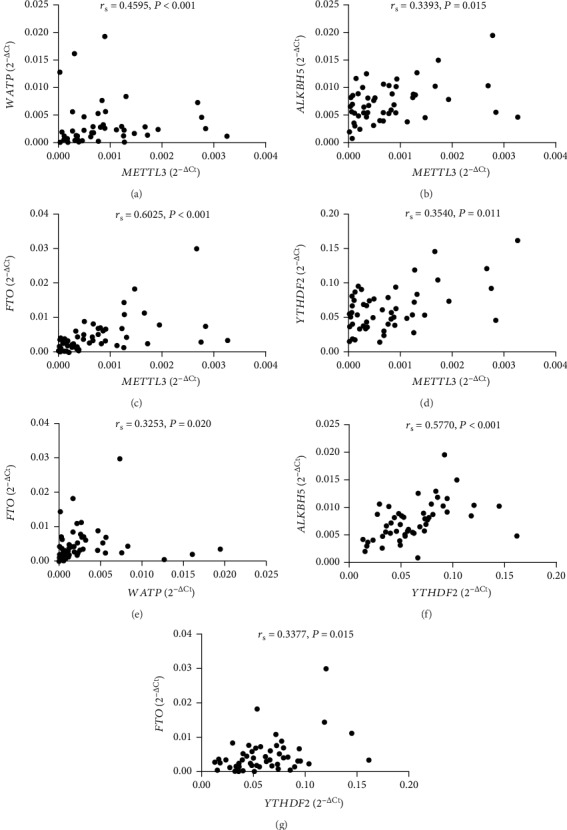
Interrelations between peripheral blood METTL3, WTAP, ALKBH5, FTO, and YTHDF2 in SLE patients. (a) The level of peripheral blood METTL3 positively correlated with WTAP. (b) The level of peripheral blood METTL3 positively correlated with ALKBH5. (c) The level of peripheral blood METTL3 positively correlated with FTO. (d) The level of peripheral blood METTL3 positively correlated with YTHDF2. (e) The level of peripheral blood WTAP positively correlated with FTO. (f) The level of peripheral blood YTHDF2 positively correlated with ALKBH5. (g) The level of peripheral blood YTHDF2 positively correlated with FTO.
4. Discussion
Epigenetic alterations, including DNA methylation and histone modifications, have been reported to contribute to SLE progression [8, 22, 23]. Zhao et al. showed that RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1, triggering autoimmune responses [24]. In recent years, the fields of m6A modification and epitranscriptomics have attracted much attention [25]. As an important form of posttranscriptional gene regulation, m6A modification was found to be involved in almost all aspects of RNA metabolism, which is widely involved in the T cell response to HIV infection [26], type I interferon production [27], and T cell differentiation and homeostasis [28]. Thus, it invites the speculation that m6A modification may be involved in the genesis and progression of SLE [13].
As the regulators of m6A methylation, METTL3, MTEEL14, WTAP, FTO, ALKBH5, and YTHDF2 play pivotal roles in the dynamic regulation of m6A modification. Nettersheim et al. showed that the mRNA levels of METTL3, MTEEL14, WTAP, and ALKBH5 were significantly decreased in testicular germ cell tumors and the m6A modification in RNA of testicular germ cell tumors was controlled by METTL3, ALKBH5, and YTHDF2 [29]. Tian et al. indicated that the mRNA expressions of METTL3 and FTO during the osteogenic induction of BMSCs were increased [30]. In addition, Yang et al. demonstrated that the mRNA levels of FTO, METTL3, METTL14, and WTAP were increased in patients with type-2 diabetes [16]. These studies indicated m6A methylation regulators including m6A methylation writers, erasers, and readers which were changed synchronously. In this study, we also found that the mRNA levels of m6A methylation writers (METTL3 and WTAP), erasers (ALKBH5 and FTO), and reader (YTHDF2) were all decreased in peripheral blood from SLE patients. In accordance with the results of Yang et al. [16], our results manifested that the mRNA levels of peripheral blood m6A methylation writers positively correlated with those of erasers and reader, and the mRNA levels of peripheral blood m6A methylation erasers positively correlated with that of m6A methylation reader in SLE patients. Considering the fact that the collaboration among writers, erasers, and readers sets up the m6A threshold and perturbs that m6A threshold leads to uncontrolled expression/activity of virulence gene and results in the occurrence and development of diseases [31], our study suggested that the decreased METTL3, WTAP, ALKBH5, FTO, and YTHDF2 may play an important role in the pathogenesis of SLE.
SLE is a systemic autoimmune disease characterized by elevated autoimmune antibodies, such as anti-dsDNA and antinucleosome. It is well known that anti-dsDNA and antinucleosome are pathogenic autoantibodies involved in organ damage of SLE [32–34]. In this study, we found that the level of peripheral blood ALKBH5 was significantly decreased in SLE patients compared to HCs, RA patients, TB patients, and HBV-infected patients. Furthermore, the level of peripheral blood ALKBH5 significantly decreased in SLE patients with positive anti-dsDNA or antinucleosome compared to SLE patients with negative anti-dsDNA or antinucleosome. And, the level of peripheral blood ALKBH5 in SLE patients was found to be negatively correlated with the level of anti-dsDNA. In addition, the level of peripheral blood ALKBH5 in SLE patients was associated with clinical symptoms of SLE such as rash, ulceration, and leukopenia. Logistic regression analysis revealed that decreased mRNA level of ALKBH5 in peripheral blood was a risk factor for SLE.
Considering that anti-dsDNA and antinucleosome are involved in the pathogenesis of SLE and are specific diagnostic biomarkers of SLE, our results suggested that the expression of peripheral blood ALKBH5 may be involved in the pathogenesis of SLE and may also serve as a potential biomarker for SLE diagnosis and severity evaluation.
In conclusion, it is the first study to our knowledge that explored the mRNA levels of peripheral blood METTL3, METTL14, WTAP, ALKBH5, FTO, and YTHDF2 in SLE patients. In this study, we found that the mRNA levels of METTL3, WTAP, ALKBH5, FTO, and YTHDF2 in peripheral blood from SLE patients were significantly decreased. Additionally, this study established the correlations between the mRNA level of ALKBH5 and the production of autoantibodies, clinical features, which might improve our understanding of the role of ALKBH5 in SLE.
Acknowledgments
This work was supported by the Key Research and Development Plan Project of Jiangxi Province (20181BBG70013), the Science and Technology Plan Project of the Education Department of Jiangxi Province (GJJ170008), the National Natural Science Foundation of China (81360459, 81560344, and 81660277), the Jiangxi Provincial Natural Science Foundation of China (20171BAB205113 and 20171ACB20032), and the Foundation for Distinguished Young Scientists of Jiangxi Province of China (20171BCB23087).
Contributor Information
Zikun Huang, Email: 491353062@qq.com.
Junming Li, Email: lisir361@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
QL and JML designed the study. BQF recruited the participants. JL and ZKH did the experimental work. LZ and YG collected the data and participated in the analysis of the data, supervised by QL which performed statistical analyses. QL, JML, JL, and ZKH drafted the manuscript, and all authors revised it critically for intellectual content. All authors have approved the final version. Qing Luo and Biqi Fu contributed equally to this study.
Supplementary Materials
Supplement Figure 1: correlations between anti-double-stranded DNA (anti-dsDNA) and complement 3 (C3), complement 4 (C4), and systemic lupus erythematosus disease activity index (SLEDAI) in systemic lupus erythematosus (SLE) patients. (a) The level of anti-dsDNA negatively correlated with that of C3. (b) The level of anti-dsDNA negatively correlated with that of C4. (c) The level of anti-dsDNA positively correlated with SLEDAI.
References
- 1.Frieri M. Mechanisms of disease for the clinician: systemic lupus erythematosus. Annals of Allergy, Asthma & Immunology. 2013;110(4):228–232. doi: 10.1016/j.anai.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 2.D'Cruz D. P., Khamashta M. A., Hughes G. R. Systemic lupus erythematosus. Lancet. 2007;369(9561):587–596. doi: 10.1016/S0140-6736(07)60279-7. [DOI] [PubMed] [Google Scholar]
- 3.Jenks S. A., Sanz I. Altered B cell receptor signaling in human systemic lupus erythematosus. Autoimmunity Reviews. 2009;8(3):209–213. doi: 10.1016/j.autrev.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crispin J. C., Kyttaris V. C., Terhorst C., Tsokos G. C. T cells as therapeutic targets in SLE. Nature Reviews Rheumatology. 2010;6(6):317–325. doi: 10.1038/nrrheum.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Q., Huang Z., Ye J., et al. PD-L1-expressing neutrophils as a novel indicator to assess disease activity and severity of systemic lupus erythematosus. Arthritis Research & Therapy. 2016;18(1):p. 47. doi: 10.1186/s13075-016-0942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsokos G. C. Systemic lupus erythematosus. The New England Journal of Medicine. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 7.La Paglia G. M. C., Leone M. C., Lepri G., et al. One year in review 2017: systemic lupus erythematosus. Clinical and Experimental Rheumatology. 2017;35(4):551–561. [PubMed] [Google Scholar]
- 8.Zhu H., Mi W., Luo H., et al. Whole-genome transcription and DNA methylation analysis of peripheral blood mononuclear cells identified aberrant gene regulation pathways in systemic lupus erythematosus. Arthritis Research & Therapy. 2016;18(1):p. 162. doi: 10.1186/s13075-016-1050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Zhang C., Wu Z., Chen Y., Shi W. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Research & Therapy. 2018;20(1):p. 118. doi: 10.1186/s13075-018-1618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Lu Z., Gomez A., et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinello N., Sun S., Wong J. J. Aberrant expression of enzymes regulating m6A mRNA methylation: implication in cancer. Cancer Biology & Medicine. 2018;15(4):323–334. doi: 10.20892/j.issn.2095-3941.2018.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuncel G., Kalkan R. Importance of m N6-methyladenosine (m6A) RNA modification in cancer. Medical Oncology. 2019;36(4):p. 36. doi: 10.1007/s12032-019-1260-6. [DOI] [PubMed] [Google Scholar]
- 13.Li L. J., Fan Y. G., Leng R. X., Pan H. F., Ye D. Q. Potential link between m6A modification and systemic lupus erythematosus. Molecular Immunology. 2018;93:55–63. doi: 10.1016/j.molimm.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Maity A., Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. The FEBS Journal. 2016;283(9):1607–1630. doi: 10.1111/febs.13614. [DOI] [PubMed] [Google Scholar]
- 15.Yang S., Wei J., Cui Y. H., et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nature Communications. 2019;10(1):p. 2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Shen F., Huang W., et al. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. The Journal of Clinical Endocrinology and Metabolism. 2019;104(3):665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 17.Tan E. M., Cohen A. S., Fries J. F., et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis and Rheumatism. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 18.Bombardier C., Gladman D. D., Urowitz M. B., et al. Derivation of the SLEDAI. A disease activity index for lupus patients. Arthritis and Rheumatism. 1992;35(6):630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 19.Aletaha D., Neogi T., Silman A. J., et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis and Rheumatism. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 20.Luo Q., Zhang L., Li X., et al. Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clinical and Experimental Immunology. 2018;194(1):118–124. doi: 10.1111/cei.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F., Wu L., Qian J., et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. Journal of Autoimmunity. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Wu H., Zhao M., Tan L., Lu Q. The key culprit in the pathogenesis of systemic lupus erythematosus: aberrant DNA methylation. Autoimmunity Reviews. 2016;15(7):684–689. doi: 10.1016/j.autrev.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Renaudineau Y., Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. The Keio Journal of Medicine. 2011;60(1):10–16. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]
- 24.Zhao M., Wu X., Zhang Q., et al. RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1. Arthritis Research & Therapy. 2010;12(6):p. R227. doi: 10.1186/ar3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Huang W., Huang J. T., et al. Increased N6-methyladenosine in Human Sperm RNA as a Risk Factor for Asthenozoospermia. Scientific Reports. 2016;6(1, article 24345) doi: 10.1038/srep24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichinchi G., Gao S., Saletore Y., et al. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nature Microbiology. 2016;1(4):p. 16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Q., Hou J., Zhou Y., Li Z., Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nature Immunology. 2017;18(10):1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- 28.Li H. B., Tong J., Zhu S., et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nettersheim D., Berger D., Jostes S., Kristiansen G., Lochnit G., Schorle H. N6-Methyladenosine detected inRNAof testicular germ cell tumors is controlled byMETTL3,ALKBH5,YTHDC1/F1/F2, andHNRNPCas writers, erasers, and readers. Andrology. 2019;7(4):498–506. doi: 10.1111/andr.12612. [DOI] [PubMed] [Google Scholar]
- 30.Tian C., Huang Y., Li Q., Feng Z., Xu Q. Mettl3 regulates osteogenic differentiation and alternative splicing of Vegfa in bone marrow mesenchymal stem cells. International Journal of Molecular Sciences. 2019;20(3):p. 551. doi: 10.3390/ijms20030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panneerdoss S., Eedunuri V. K., Yadav P., et al. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Science Advances. 2018;4(10, article eaar8263) doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yung S., Chan T. M. Mechanisms of kidney injury in lupus nephritis—the role of anti-dsDNA antibodies. Frontiers in Immunology. 2015;6:p. 475. doi: 10.3389/fimmu.2015.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gómez-Puerta J. A., Burlingame R. W., Cervera R. Anti-chromatin (anti-nucleosome) antibodies: diagnostic and clinical value. Autoimmunity Reviews. 2008;7(8):606–611. doi: 10.1016/j.autrev.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Manson J. J., Ma A., Rogers P., et al. Relationship between anti-dsDNA, anti-nucleosome and anti-alpha-actinin antibodies and markers of renal disease in patients with lupus nephritis: a prospective longitudinal study. Arthritis Research & Therapy. 2009;11(5):p. R154. doi: 10.1186/ar2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Figure 1: correlations between anti-double-stranded DNA (anti-dsDNA) and complement 3 (C3), complement 4 (C4), and systemic lupus erythematosus disease activity index (SLEDAI) in systemic lupus erythematosus (SLE) patients. (a) The level of anti-dsDNA negatively correlated with that of C3. (b) The level of anti-dsDNA negatively correlated with that of C4. (c) The level of anti-dsDNA positively correlated with SLEDAI.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


