Summary
Background
The NEJ026 Phase 3 study demonstrated that erlotinib and bevacizumab (BE)-treated NSCLC patients with EGFR mutations had significantly better progression-free survival (PFS) than those treated with erlotinib alone (E). This study included a prospective analysis of the relationship between the mutational status of EGFR in plasma circulating tumor DNA (ctDNA) and the efficacy of TKI monotherapy or combination therapy. We describe these results herein.
Methods
Plasma samples were collected from patients enrolled in NEJ026 at the start of treatment (P0), 6 weeks after the start of treatment (P1), and upon confirmation of progressive disease (P2). Plasma ctDNA was analyzed using a modified PNA-LNA PCR clamp method. PFS and OS according to EGFR status at the time of plasma collection were evaluated.
Findings
Plasma activating EGFR mutation (aEGFR) at P0 was detected in 68% of cases; patients without plasma aEGFR had longer PFS. The frequency of T790M mutation at P2 was similar in both arms: 8 (19.0%) in BE and 11 (20.8%) in E. Based on the aEGFR profiles, PFS was evaluated among three groups: type A [P0(-), P1(-)], type B [P0(+), P1(-)], and type C [P0(+), P1(+)]. This revealed that BE was more efficacious than E, and that BE was associated with improved PFS in all types.
Interpretation
Pre-treatment plasma aEGFR status have a potential of early predictor of response of TKI efficacy. Monitoring plasma aEGFR mutation will contribute to selection and continuation of treatment with BE or E.
Funding
Chugai Pharmaceutical.
Research in Context section.
Evidence before this study from systematic review
We performed a PubMed search for articles published in English with restrictions for clinical trials up to March 14, 2020 using the search terms “NSCLC” and “EGFR” and (“liquid biopsy” or “ctDNA” or “cfDNA”). We identified many clinical trials demonstrated the usefulness of detection of plasma EGFR activating mutation at baseline and/or EGFR T790M resistant mutation at progressive disease. There was no Phase 3 trial designed to evaluate the utility of monitoring plasma EGFR mutational status in order to predict the efficacy of each treatment arm.
Added value of this study
As far as we know, our study is the first to suggest a treatment strategy using plasma EGFR mutational analysis in a Phase 3 trial. In addition to the analyses at pretreatment and at disease progression, the analyses at 6 weeks after the start of treatment were able to provide more information to select the best treatment.
Implication of all the available evidence
Plasma EGFR analyses of cell-free DNA at pretreatment and at disease progression can be used to predict the prognosis of each patient. Additionally, the monitoring of the plasma EGFR mutational status during treatment may provide important information to assess the efficacy of the current treatment.
Alt-text: Unlabelled box
1. Background
EGFR (epidermal growth factor receptor) mutation is a major oncogenic driver in NSCLC (non-small cell lung carcinoma), and inhibition of EGFR signaling by EGFR-TKIs (tyrosine kinase inhibitors) is the optimal clinical treatment strategy for NSCLC patients with EGFR mutations [1], [2], [3], [4]. Currently, the clinically available EGFR-TKIs include gefitinib, erlotinib, afatinib, dacomitinib, and osimertinib. In addition, the combination of gefitinib plus platinum chemotherapy is also associated with favorable progression free survival (PFS) and overall survival (OS) [5]. In the NEJ026 study, we demonstrated that PFS following erlotinib plus bevacizumab (BE) treatment was significantly superior to erlotinib alone (E) in NSCLC patients harboring EGFR mutation [6]. While there are many treatment choices for NSCLC with EGFR mutation, there are no predictive markers available for guiding the selection of treatment regimen. If a such indicators were available, patients with poor prognosis could be treated more effectively, and a higher incidence of adverse events might be acceptable. Conversely, if a good prognosis was indicated for other patients, they could be spared treatment with aggressive agents that are associated with high toxicity. Tumors can acquire resistance to EGFR-TKIs by mutation of codon 790 (T790M), and in these cases second-line treatment with the next generation EGFR-TKI, osimertinib, is recommended [7]. However, the frequency at which T790M during a BE regimen remains unclear.
This is important, since although BE treatment showed excellent efficacy and safety, resistance does develop after about 16 months treatment. Unfortunately, it is not possible to obtain tumor tissue re-biopsies from all the patients who become refractory to treatment. This is because the success of tissue re-biopsy is dependent on tumor size, location, and patient general conditions in each case.
In recent years, the utility of EGFR mutation analysis of plasma circulating tumor DNA (ctDNA) has been demonstrated in many reports [8], [9], [10], [11], [12], [13]. The NEJ026 biomarker study contains a preplanned analysis of plasma EGFR mutations at pretreatment, at 6 weeks after starting first-line and second-line treatment, and at PD of first-line and second-line. Here we report on the results of this study, and describe the relationship between activating EGFR mutations (aEGFRs) and clinical response, as well as the frequency of T790M mutations.
2. Methods
2.1. Sample collection
The NEJ026 biomarker study was preplanned in the study protocol in order to evaluate one of the exploratory endpoints (Supplementary Figure 1). Tissue samples were collected at three time points as follows. First, between enrollment and the start of study treatment (T0). Second, at the end of study treatment from PD 1 until the date of the start of second-line treatment (T1). Third, at the end of second-line treatment from PD 2 until the date of the start of third-line treatment (T2). Plasma samples were collected at five time points. First, between enrollment and the start of study treatment (P0). Second, during study treatment from the date of the start of study treatment until 6 weeks ± 21 days after (P1). Third, at the end of study treatment from PD 1 until the date of the start of second-line treatment (P2). Fourth, during second-line treatment from the date of the start of second-line treatment until 6 weeks ± 21 days after (P3). Fifth, at the end of second-line treatment from PD 2 until the date of the start of third-line treatment (P4). The data cutoff was 31st March 2019.
2.2. EGFR mutation analysis
Plasma ctDNA and tissue DNA analysis for detection of the aEGFR and T790M mutation were performed using an improved peptide nucleic acid–locked nucleic acid (PNA-LNA) PCR clamp method as previously described (LSI Medience Corporation, Tokyo, Japan) [11]. In brief, whole blood samples (14 mL) were collected in ethylenediaminetetraacetic acid (EDTA) tubes. DNA was extracted from plasma samples with the QIAamp Circulating Nucleic Acid kit or from cytohistological samples with the QIAamp DNA FFPE Tissue kit (QIAGEN, Hilden, Germany). CtDNA was extracted from 5 ml of plasma and finally dissolved in 50 μl of solvent. The EGFR mutants were then detected using the PNA-LNA PCR clamp method. By using smaller PCR products and by increasing the number of cycles from 45 to 50, this PCR system was able to detect mutations present at a frequency of less than 0.1%. In this study, the analyzed EGFR subtypes were activating mutations (aEGFR): Exon 19 E746-A750(2235–2249)del, E746-A750(2236–2250)del, other Exon 19 del variants and Exon 21 L858R, the resistant mutation, Exon 20 T790M, and other mutations including Exon 18 G719S, G719A, G719C, and Exon 21 L861Q. Other Exon 19 del variants were detected by fragment analysis. In actual measurement, 5 μl (corresponding to 0.5 ml of plasma) was used for each reaction. Investigators in the central laboratory performed a blinded analysis of the mutational data obtained from cytohistological samples in order to avoid biases.
2.3. Statistical analysis
The efficacy data of each case were transferred from the NEJ026 main study [6]. Each case was divided into subgroups according to the results of EGFR mutation analysis at P0 / P1 / P2 /T0 /T1. The data were compared between BE and E group or compared the data of each subgroup. Kaplan-Meier survival curves were drawn for PFS and compared using a stratified log-rank test. Hazard ratio and its confidence interval (CI) were calculated using Cox proportional hazard analysis. The 95% CI for median PFS was calculated using the method of Brookmeyer and Crowley [14]. A multivariate Cox regression model was used for the adjusted comparison of PFS between treatment groups. Tumor response (the proportion of patients with an objective response and disease control, and duration of response) were compared between BE and E using a chi-squared test with one degree of freedom. Statistical analyses were performed with SAS for Windows release 9.4 (SAS Institute Inc. Cary, NC, USA).
3. Results
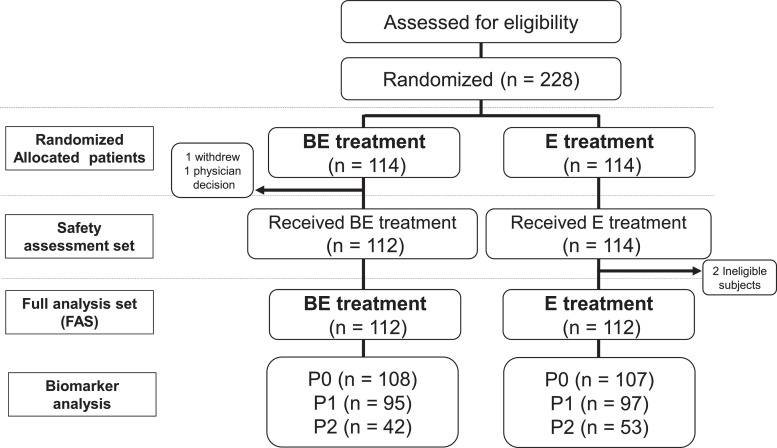
The total number of collected plasma samples in BE and E were 108 (96.4%) and 107 (95.5%) at P0, 95 (84.8%) and 97 (86.6%) at P1, 42 (37.5%) and 53 (47.3%) at P2, 25 (22.3%) and 36 (32.1%) at P3, and 19 (17.0) and 15 (13.4%) at P4, respectively (Fig. 1 and Table 1). In contrast to the plasma samples, we could not collect enough tissue samples to perform a statistically meaningful analysis.
Fig. 1.
Study profile depicting patient randomization and selection. BE, bevacizumab and erlotinib combination therapy arm; E, erlotinib monotherapy arm. P0, Plasma samples taken before the start of study treatment; P1, Plasma samples taken 6 weeks from the start of study treatment; P2, Plasma samples taken at the time of disease progression while on treatment.
Table 1.
Collection rates of plasma and tissue samples.
| N(%) | BE | E | Total |
|---|---|---|---|
| Full Analysis Set | 112 | 112 | 224 |
| P0 | 108 (96.4) | 107 (95.5) | 215 (96.0) |
| P1 | 95 (84.8) | 97(86.6) | 192 (85.7) |
| P2 | 42 (37.5) | 53 (47.3) | 95 (42.4) |
| P3 | 25 (22.3) | 36 (32.1) | 61 (27.2) |
| P4 | 19 (17.0) | 15 (13.4) | 34 (15.2) |
| T0 | 2 (1.8) | 3 (2.7) | 5 (2.2) |
| T1 | 7 (6.3) | 9 (8.0) | 16 (7.1) |
| T2 | 3 (2.7) | 3 (2.7) | 6 (2.7) |
Plasma samples, P0: Before the start of study treatment, P1: 6 weeks from the start of study treatment, P2: At the time of definite disease progression during initial treatment, P3: 6 weeks from the start of second-line treatment, and P4: At the time of definite disease progression during second-line treatment. Tissue samples were not mandatory, T0: Before the start of study treatment, T1: At the time of definite disease progression during initial treatment and T2: At the time of definite disease progression during second-line treatment.
In eligible patients with aEGFR mutations identified by cytohistology, the frequency of plasma aEGFR mutations before study treatment (P0) was 68.4% (Table 2). Age, sex, smoking status, histology, and EGFR subtype were not related to the rate at which plasma aEGFR was detected. Lower detection rates of EGFR mutation were seen in PS 0, M1a, and in tumors that recurred after surgery, suggesting that the mutation rate in the plasma was lower in patient with non-advanced compared to advanced disease. In terms of metastatic site, the mutation rate was lower in pulmonary, pleural, or brain metastases, which is consistent with previous reports [11].
Table 2.
Frequency of detectable plasma EGFR mutations at P0 in each patient characteristics.
| P0 activating mutation | + | – | Total | Frequency(%) |
|---|---|---|---|---|
| Total | 147 | 68 | 215 | 68.4 |
| Age | ||||
| 75< | 123 | 52 | 175 | 70.3 |
| >=75 | 24 | 16 | 40 | 60.0 |
| Sex | ||||
| Male | 57 | 21 | 78 | 73.1 |
| Female | 90 | 47 | 137 | 65.7 |
| Smoking status | ||||
| Non smoker | 84 | 39 | 123 | 68.3 |
| Former light smoker | 7 | 6 | 13 | 53.8 |
| Other smoker | 56 | 23 | 79 | 70.9 |
| PS | ||||
| 0 | 75 | 52 | 127 | 59.1 |
| 1 | 70 | 16 | 86 | 81.4 |
| 2 | 2 | 0 | 2 | 100.0 |
| Histology | ||||
| adeno | 145 | 68 | 213 | 68.1 |
| large | 1 | 0 | 1 | 100.0 |
| other | 1 | 0 | 1 | 100.0 |
| EGFR subtype | ||||
| Exon 19 del | 76 | 30 | 106 | 71.7 |
| Exon 21 L858R | 71 | 38 | 109 | 65.1 |
| Stage | ||||
| IIIB | 12 | 2 | 14 | 85.7 |
| IV (M1a) | 26 | 23 | 49 | 53.1 |
| IV (M1b) | 93 | 18 | 111 | 83.8 |
| Recurrence | 16 | 25 | 41 | 39.0 |
| Metastatic sites (Stage IV) | ||||
| PUL only | 6 | 8 | 14 | 42.9 |
| PLE only | 17 | 15 | 32 | 53.1 |
| BRA only | 9 | 4 | 13 | 69.2 |
| Others | 87 | 14 | 101 | 86.1 |
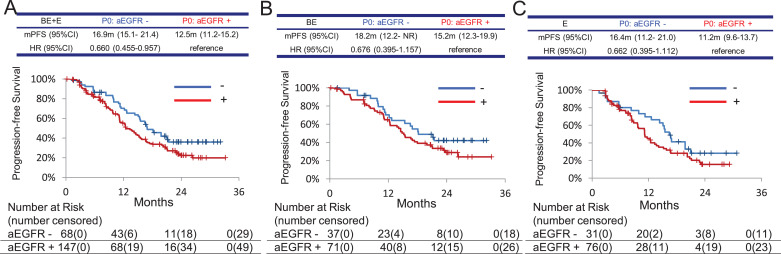
When comparing detectable and undetectable aEGFR mutations at P0, the undetectable group was associated with a longer PFS (16.9 months [95%CI: 15.1–21.4] vs. 12.5 months [11.2–15.2], hazard ratio [HR] 0.660 [95% CI 0.455–0.957]) in the BE plus E groups (Fig. 2A). The same tendencies of longer PFS in patients with undetectable EGFR mutation at P0 were observed in the BE plus E group (Fig. 2B and C). On the other hand, when comparing BE treatment to E treatment in each plasma EGFR positive group and negative group at P0, BE treatment showed longer PFS than E treatment in both groups and clear separation of K-M curve was observed in the EGFR positive group. These results might be due to frequency of mutations being associated with the extent of disease progression. (Supplement Figure 2)
Fig. 2.
Kaplan–Meier curves for PFS among patients classified with detectable (+) or undetectable (-) plasma activating EGFR mutation (aEGFR) at pretreatment. HR, hazard ratio. BE, bevacizumab and erlotinib combination therapy arm; E, erlotinib monotherapy arm. A, both BE and E; B, BE; C, E.
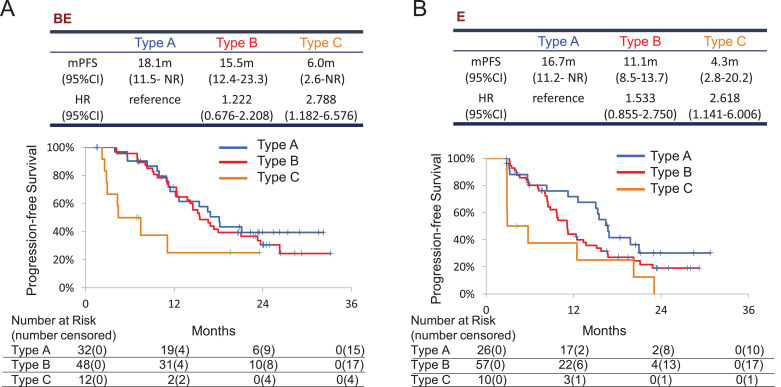
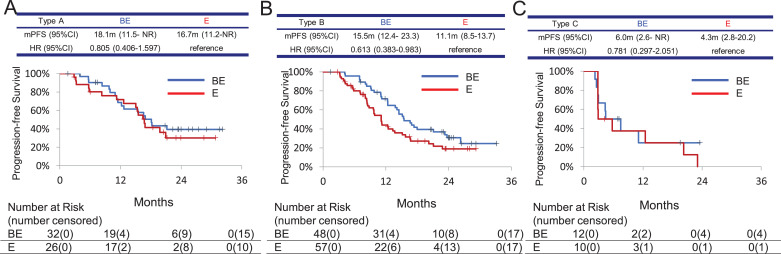
Based on the detection of plasma EGFR mutations before treatment (P0) and 6 weeks after initiation of treatment (P1), plasma positivity types were classified into the following 4 groups: type A, P0 (-) and P1 (-); type B, P0 (+) and P1 (-); type C, P0 (+) and P1 (+); and type D, P0 (-) and P1 (+). The number of patients in each of these groups is shown in Table 3. Only one patient was classified as Type D. In the BE arm, the PFS of each group was as follows. Type A: 18.1 months [95%CI: 11.5–upper limit not reached (NR)], Type B: 15.5 months [12.4–23.3], HR 1.222 [0.676–2.208], and Type C: 6.0 months [2.6-NR], HR 2.788 [1.182–6.576] (Fig. 3A). In the E arm, the PFS of each group was as follows. Type A: 16.7 months [11.2–NR], Type B: 11.1 months [8.5–13.7], HR 1.533 [0.855–2.750], and Type C: 4.3 months [2.8–20.2], HR 2.618 [1.141–6.006] (Fig. 3B). Type A patients had the longest PFS of all types in both treatment arms, while the difference in median PFS between the two arms was not significant (HR 0.805 [0.406–1.597]) (Fig. 4A). Type C patients had the shortest PFS and the difference between the two treatment arms in this group was not significant (HR 0.781 [0.297–2.051]) (Fig. 4C). On the other hand, the BE arm showed significantly longer PFS in Type B compared to the E arm (HR 0.613 [95% CI 0.383–0.983]) (Fig. 4B). BE treatment achieved response rates of 65.5% in Type A, 89.6% in Type B, and 41.7% in Type C. E treatment elicited response rates of 57.7% in Type A, 75.4% in Type B, and 30.0% in Type C (Table 4).
Table 3.
The classification of plasma activating EGFR mutations from P0 to P1 within each group.
| N(%) | BE | E | Total | |
|---|---|---|---|---|
| Type A | P0- ➡ P1- | 32 (34.4) | 26 (28.0) | 58 (31.2) |
| Type B | P0+ ➡ P1- | 48 (51.6) | 57 (61.3) | 105 (56.5) |
| Type C | P0+ ➡ P1+ | 12 (12.9) | 10 (10.8) | 22 (11.8) |
| Type D | P0- ➡ P1+ | 1 (1.1) | 0 (0.0) | 1 (0.5) |
| Total | 93 | 93 | 186 | |
+:Detectable, -:Undetectable.
Fig. 3.
Kaplan–Meier curves for PFS among patients classified by patterns of detection of plasma EGFR both at pretreatment (P0) and at 6 weeks from the start of treatment (P1). HR, hazard ratio. Type A, neither detectable at P0 nor at P1; Type B, detectable at P0 and disappeared at P1; Type C, detectable both at P0 and P1. A. bevacizumab and erlotinib combination therapy arm; B. erlotinib monotherapy arm.
Fig. 4.
Kaplan–Meier curves for PFS of patients classified by the presence or absence of plasma EGFR mutations both at pretreatment (P0) and at 6 weeks from the start of study treatment (P1). HR, hazard ratio. BE, bevacizumab and erlotinib combination therapy arm; E, erlotinib monotherapy arm. A. type A, neither detectable at P0 nor at P1; B. type B, detectable at P0 and disappeared at P1; C. type C, detectable both at P0 and P1.
Table 4.
Response rate of each group.
| Response rate (%) | BE | E | |
|---|---|---|---|
| Type A | P0- ➡ P1- | 62.5 | 57.7 |
| Type B | P0+ ➡ P1- | 89.6 | 75.4 |
| Type C | P0+ ➡ P1+ | 41.7 | 30.0 |
+:Detectable, -:Undetectable.
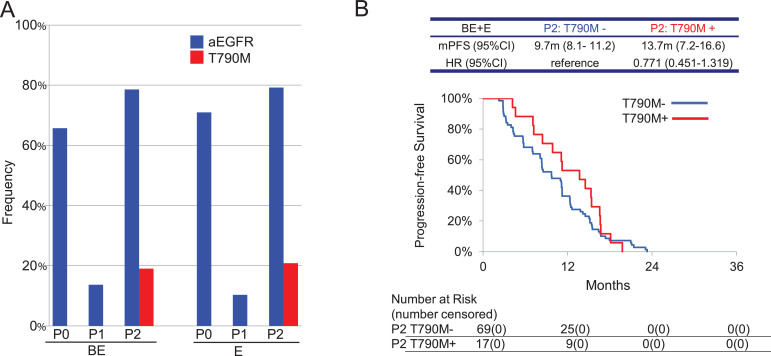
The frequency of plasma activating EGFR mutations at progression of disease (P2) increased compared to P1 and were almost similar in both arms: 33 (78.6%) in BE and 42 (79.2%) in E (Table 5 and Fig. 5A). The frequency at which the T790M resistance mutation emerged at P2 was similar in both arms: 8 (19.0%) in BE and 11 (20.8%) in E. Patients who had the T790M resistance mutation at P2 in both arms tended to have longer PFS compared to patients without T790M [13.7 months (95% CI: 7.2–16.6) vs 9.7 months (95% CI: 8.1–11.2), respectively; the HR was 0.771 (0.451–1.319)] (Fig. 5B).
Table 5.
Detection of the T790M resistance mutation in each sample.
| Plasma | ||||||
|---|---|---|---|---|---|---|
| N (%*) | BE |
E |
||||
| T790M | EGFR⁎⁎ | Total | T790M | EGFR | Total | |
| P0 | 0 (0.0) | 71 (65.7) | 108 | 0 (0.0) | 76 (71.0) | 107 |
| P1 | 0 (0.0) | 13 (13.7) | 95 | 0 (0.0) | 10 (10.3) | 97 |
| P2 | 8 (19.0) | 33 (78.6) | 42 | 11 (20.8) | 42 (79.2) | 53 |
| P3 | 1 (4.0) | 15 (60.0) | 25 | 1 (2.8) | 17 (47.2) | 36 |
| P4 | 1 (5.3) | 14 (73.7) | 19 | 1 (6.7) | 14 (93.3) | 15 |
| Tissue | ||||||
|---|---|---|---|---|---|---|
| N (%*) | BE |
E |
||||
| T790M | EGFR⁎⁎ | Total | T790M | EGFR | Total | |
| T0 | 0 (0.0) | 2 (100.0) | 2 | 0 (0.0) | 3 (100.0) | 3 |
| T1 | 4 (57.1) | 7 (100.0) | 7 | 4 (44.4) | 9 (100.0) | 9 |
| T2 | 1 (33.3) | 3 (100.0) | 3 | 0 (0.0) | 3 (100.0) | 3 |
percentage of the total collected samples with each mutation.
EGFR activating mutations (L858R or exon 19 deletion).
Fig. 5.
A. The frequency of activating EGFR mutation (aEGFR) and EGFR T790M resistance mutations at the following times. P0, Plasma samples taken before the start of study treatment; P1, Plasma samples taken 6 weeks after the start of study treatment; P2, Plasma samples taken at the time of disease progression. BE, bevacizumab and erlotinib combination therapy arm; E, erlotinib monotherapy arm. B. Kaplan–Meier curves for PFS of all patients classified by the presence or absence of plasma T790M mutations at P2. HR, hazard ratio.
4. Discussion
In this study, we found that plasma EGFR activating mutations were present at a frequency of 68.4% in NSCLC patients before study treatment. Cases with intrathoracic or brain metastases had lower frequencies of detectable plasma EGFR activating mutations compared to the cases with other metastases. The patients with longer PFS did not have detectable plasma EGFR mutations at the pretreatment stage. Cases with in which EGFR activating mutations were cleared by 6 weeks of treatment also had a good prognosis, especially in the BE treatment arm. On the other hand, patients with sustained detectable plasma EGFR activating mutations had a poor prognosis. There was no difference between BE and E cohorts with regard to the frequency of plasma T790M mutations upon progression of disease.
The baseline plasma EGFR (P0) prevalence was 68%. In the FLAURA study, plasma EGFR positivity was 359 (74.3%) out of 483 cases using the Cobas plasma test 15. In the BELIEF study, plasma EGFR positivity was 55 in 91 cases (60.4%) using the PNA-Q-PCR method [16]. In this study, improved PNA-LNA clamp PCR method could be detected activated EGFR mutation equally. The reason why the plasma EGFR frequency in this article is slightly lower than that in the FLAURA, is supposed to the difference of the proportion of stage IV at which plasma EGFR tends to be detectable. In the FLAURA trial, Stage IV cases were 95%, compared to 74% in the NEJ026 trial.
Undetectable cases of plasma EGFR activating mutations before treatment were related to metastatic sites. For example, plasma EGFR activating mutations were not detected before treatment in patients with the M classifications M1a when compared with M1b/c. The same was true for patients with metastatic sites restricted to the lungs, pleura and brain compared with other organs, and for patients with ECOG PS 0 versus PS 1. These results are compatible with several previous reports [11,12,17,18]. Interestingly, Aldea et al. also reported ctDNA from isolated CNS metastases group was lower mutation positivity than other metastasis, and negative ctDNA at the time of isolated CNS metastases shifted to positive when the patient had a systemic progression [19]. In our study, we speculate that the absence of detectable plasma EGFR mutations is indicative of relatively low distal metastatic activities and low systemic tumor burden. It also appears that patients with undetectable plasma EGFR mutations before treatment is initiated can expect an improved prognosis following either BE or E treatment. Our results are consistent with those of previous reports [15,17,20,21].
By adding plasma EGFR data 6 weeks after the start of the study treatment to the pre-treatment data, we could classify the detectable or undetectable group at P0 as type A (undetectable for 6 weeks), type B (mutations detectable before treatment disappeared within 6 weeks) and type C (mutations were still detectable at 6 weeks). Type A and B patients had a superior response compared to type C patients in both the BE and E arms, consistent with previous reports [8,17,[20, [21, [22]. In the present study, BE induced a better response in type B patients than in those of the A group, whereas there was no difference between the BE group and E group in type A and C. Although various resistance mechanisms of erlotinib have been reported, it is suggested that addition of bevacizumab could not overcome the initial resistance mechanism corresponding to Type C. However, it should be noted that efficacies of BE treatment might be underestimated in Type A and C, which have smaller numbers of cases compared to Type B. From our study results, we propose a treatment model that can be based on the ctDNA assay at baseline and 6 weeks post-treatment. Specifically, if EGFR TKI-naïve patients have detectable plasma EGFR mutation at the pretreatment stage, we recommend more erlotinib plus bevacizumab treatment than erlotinib treatment due to more solid efficacy of erlotinib plus bevacizumab treatment for patients with plasma mutation positive compared to with mutation negative. The subsequent efficacy of this combination should be estimated by presence or absence of plasma EGFR mutation at 6 weeks post-treatment.
Compared to E treatment, BE treatment induced a similar frequency of acquired T790M mutations that were detectable in the plasma. We therefore infer that, following a BE regimen, a similar number of patients will therefore receive osimertinib, which is currently the best second-line treatment for T790M mutant tumors. Similarly, in the Phase 3 RELAY trial (which compared of erlotinib plus ramucirumab, VEGFR2 inhibitor, and erlotinib plus placebo), Ramucirumab + Erlotinib combination therapy and Erlotinib monotherapy showed similar frequency of T790M detection in plasma [23]. In the RELAY study, T790M was observed in 43% of the Ramucirumab + Erlotinib group and 47% of the Erlotinib group in cases with activating EGFR-positive at progression. In current study, Detection rates of T790M / activating mutation were 24.2% in BE arm and 26.1% in E arm. The frequencies of T790M detection in both treatments in RELAY study were higher than in this study. This may be derived from difference in sensitivity of the test methods between next generation sequence in RELAY and PNA-LNA PCR clamp in current study. Anyway, the main point is that treatment of erlotinib plus antiangiogenic agent induces T790M resistance as frequently as 1st-generation EGFR-TKI.
A limitation of this study was the small number of tissue samples collected. This was especially an issue with samples needed at the time of disease progression in order to confirm ctDNA mutation. Furthermore, at the time of writing, OS data were not available to us for analysis.
The improved PNA-LNA PCR clamp is an allele-specific PCR method that is limited to the evaluation of known EGFR mutations and could not be able to quantitative analysis. If some quantitative analysis was available, it may help to understand the relation plasma EGFR status and efficacy of EB or E treatment. Actually, the newer technologies of digital droplet PCR and next generation sequencing can be used to evaluate alterations in many genes simultaneously However, the advantages of PNA-LNA PCR clamp method include lower practical costs and shorter turn-around time over next-generation sequencing.
Conclusion
Cases with restricted intrathoracic or brain metastases tend to have a lower frequency of detectable plasma EGFR activating mutations when compared to cases with other metastases. Patients with undetectable plasma EGFR activating mutations at the pretreatment stage had a longer PFS following first-line treatment. Clearance of EGFR activating mutations after 6 weeks’ treatment was also related to good prognosis, especially in the BE cohort. The frequency of plasma T790M mutations at the time of disease progression were similar in the BE and E cohorts. Analysis of ctDNA obtained at the pretreatment stage and at 6 weeks after treatment provided a robust early predictor of response model and is therefore a candidate approach for prediction of prognosis.
Data sharing
The data collected for this study can be made available to others in de-identified form after all primary and secondary endpoints have been published and in the presence of a data transfer agreement.
Role of the funding source
The funding source gave its approval at the conception of this study and was involved in the provision of information; however, it was not involved in the study design, the data collection, the analysis, the interpretation of the data, or the writing of the report. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Declaration of Competing Interest
TF reports personal fees from Chugai Pharma., during the conduct of the study; personal fees from AstraZeneca, personal fees from Boehringer Ingelheim, outside the submitted work. HS reports grants from Chugai Pharmaceutical, grants from AstraZeneca, personal fees from Ono Pharmaceutical, personal fees from Nippon Boehringer Ingelheim, grants from MSD, personal fees from Novartis Pharma, outside the submitted work. . NF reports personal fees from Eli Lilly Japan, personal fees from Chugai, personal fees from AstraZeneca, personal fees from Bristol Myers Squibb, personal fees from Boehringer Ingelheim Japan, personal fees from Taiho, personal fees from Ono, personal fees from Pfizer Japan, outside the submitted work. KW has nothing to disclose. SS reports personal fees from Chugai Pharma, personal fees from AstraZeneca, personal fees from Nippon Boehringer Ingelheim, personal fees from MSD, personal fees from Bristol-Myers Squibb, personal fees from Ono Pharmaceutical, personal fees from Pfizer, personal fees from Taiho Pharmaceutical, personal fees from Eli Lilly and Company, personal fees from Novartis, personal fees from Kyowa Hakko Kirin, outside the submitted work. SI reports personal fees from Chugai Pharmaceutical Co., Ltd., during the conduct of the study; personal fees from AstraZeneca K.K., grants from ONO PHARMACEUTICAL CO.,LTD., outside the submitted work. NT has nothing to disclose. OY reports other from Ono Pharmaceutical Co., Ltd, other from Bristol-Myers Squibb, other from Taiho Pharmaceutical, other from MSD, other from Chugai Pharmaceutical Co., Ltd., other from AstraZeneca, during the conduct of the study. MO has nothing to disclose. KY has nothing to disclose. IN has nothing to disclose. AG reports personal fees from Chugai, outside the submitted work.
KA has received honoraria from Ono Pharmaceutical, Bristol-Myers Squibb, AstraZeneca, MSD and Chugai Pharmaceutical. FK has nothing to disclose.
Yukari T has nothing to disclose. YF reports personal fees from Lilly, personal fees from Chugai Pharma, personal fees from AstraZeneca, personal fees from Bristol-Myers Squibb Company, outside the submitted work. HN has nothing to disclose. GA has nothing to disclose. SW reports personal fees from AstraZeneca, personal fees from Chugai Pharma, personal fees from Ono Pharmaceutical, personal fees from Bristol-Myers, personal fees from Boehringer Ingelheim, personal fees from Eli Lilly, personal fees from MSD, personal fees from Taiho Pharmaceutical, personal fees from Pfizer, personal fees from Novartis, personal fees from Daiichi Sankyo, outside the submitted work. MasakiM has nothing to disclose. KH reports personal fees from Astra Zeneca, personal fees from Chugai Pharmaceutical, grants and personal fees from Taiho Pharmaceutical, grants and personal fees from Ono Pharma, personal fees from Boehringer-Ingelheim, personal fees from Novartis, personal fees from Kyorin pharmaceutical, personal fees from Bristol Myers Squibb, personal fees from MSD, during the conduct of the study; In addition, KH has a patent EGFR mutation test licensed to LSI medience. TN has nothing to disclose. SM reports personal fees from AstraZeneca K.K, personal fees from Bristol-Myers Squibb Company, personal fees from Chugai Pharmaceutical Co. Ltd, personal fees from Eli Lilly Japan K.K, personal fees from MSD K.K, personal fees from Pfizer Japan Inc, personal fees from Taiho Pharmaceutical Co. Ltd, grants and personal fees from Nippon Boehringer Ingelheim Co. Ltd., personal fees from Ono Pharmaceutical Co. Ltd., outside the submitted work. KK reports personal fees from AstraZeneca, personal fees from Bristol-Myers Squibb Japan, personal fees from Ono Pharmaceutical, personal fees from Boehringer Ingelheim, personal fees from Taiho Pharmaceutical Company, outside the submitted work. Maemondo reports grants from Chugai pharma during the conduct of the study; personal fees from Chugai pharma, personal fees from AstraZeneca, grants and personal fees from Boehringer Ingelheim, personal fees from Ono pharma, personal fees from Bristol Myers Squibb, personal fees from MSD, personal fees from Pfizer, personal fees from Lilly, outside the submitted work.
Acknowledgments
We thank all patients, their families, and the site investigators who participated in the study. Authors also thank Ms. Hiromi Odagiri and Mr. Yukio Kakehashi in NEJSG office for their technical support and Dr Koji Ohba, Dr Yasuo Saijo, and Dr Fumikazu Sakai for assistance as the independent Data Monitoring Committee.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102861.
Appendix. Supplementary materials
References
- 1.Maemondo M., Inoue A., Kobayashi K. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C., Wu Y.L., Chen G. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R., Carcereny E., Gervais R. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T., Morita S., Yatabe Y. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 5.Hosomi Y., Morita S., Sugawara S. Gefitinib Alone Versus Gefitinib Plus Chemotherapy for Non-Small-Cell Lung Cancer with Mutated Epidermal Growth Factor Receptor: NEJ009 Study. J Clin Oncol. 2020;38:115–123. doi: 10.1200/JCO.19.01488. [DOI] [PubMed] [Google Scholar]
- 6.Saito H., Fukuhara T., Furuya N. Erlotinib plus bevacizumab versus erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomised, multicentre, phase 3 trial. Lancet Oncol. 2019;20:625–635. doi: 10.1016/S1470-2045(19)30035-X. [DOI] [PubMed] [Google Scholar]
- 7.Mok T.S., Wu Y.L., Ahn M.J. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oxnard G.R., Paweletz C.P., Kuang Y. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reck M., Hagiwara K., Han B. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: the ASSESS study. J Thorac Oncol. 2016;11:1682–1689. doi: 10.1016/j.jtho.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 10.Sueoka-Aragane N., Katakami N., Satouchi M. Monitoring EGFR T790M with plasma DNA from lung cancer patients in a prospective observational study. Cancer Sci. 2016;107:162–167. doi: 10.1111/cas.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe K., Fukuhara T., Tsukita Y. EGFR Mutation Analysis of Circulating Tumor DNA Using an Improved PNA-LNA PCR Clamp Method. Can Respir J. 2016;2016 doi: 10.1155/2016/5297329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwama E., Sakai K., Azuma K. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann Oncol. 2017;28:136–141. doi: 10.1093/annonc/mdw531. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y.L., Lee V., Liam C.K. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer. 2018;126:1–8. doi: 10.1016/j.lungcan.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Brookmeyer R., Crowley J. A confidence interval for the median survival time. Biometrics. 1982;38:29–41. [Google Scholar]
- Gray J.E., Okamoto I., Sriuranpong V. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin Cancer Res. 2019;25:6644–6652. doi: 10.1158/1078-0432.CCR-19-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Vila M.A., Stahel R.A., Dafni U. Evolution and clinical impact of EGFR mutations in circulating free DNA in the BELIEF trial. J Thorac Oncol. 2020;15:416–425. doi: 10.1016/j.jtho.2019.11.023. [DOI] [PubMed] [Google Scholar]
- Tseng J.S., Yang T.Y., Tsai C.R. Dynamic plasma EGFR mutation status as a predictor of EGFR-TKI efficacy in patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol. 2015;10:603–610. doi: 10.1097/JTO.0000000000000443. [DOI] [PubMed] [Google Scholar]
- Wu Y.L., Sequist L.V., Hu C.P. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer. 2017;116:175–185. doi: 10.1038/bjc.2016.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldea M., Hendriks L., Mezquita L. Circulating tumor DNA analysis for patients with oncogene-addicted NSCLC with isolated central nervous system progression. J Thorac Oncol. 2020;15:383–391. doi: 10.1016/j.jtho.2019.11.024. [DOI] [PubMed] [Google Scholar]
- Iwama E., Sakai K., Hidaka N. Longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer. 2020;126:219–227. doi: 10.1002/cncr.32481. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Qing X., Xiumin W. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02) Oncotarget. 2016;7:6984–6993. doi: 10.18632/oncotarget.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cheng Y., An T. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. 2018;6:681–690. doi: 10.1016/S2213-2600(18)30264-9. [DOI] [PubMed] [Google Scholar]
- Nakagawa K., Garon E.B., Seto T. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:1655–1669. doi: 10.1016/S1470-2045(19)30634-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.