Abstract
Background
A significant gap in pancreatic ductal adenocarcinoma (PDAC) patient's care is the lack of molecular parameters characterizing tumours and allowing a personalized treatment.
Methods
Patient-derived xenografts (PDX) were obtained from 76 consecutive PDAC and classified according to their histology into five groups. A PDAC molecular gradient (PAMG) was constructed from PDX transcriptomes recapitulating the five histological groups along a continuous gradient. The prognostic and predictive value for PMAG was evaluated in: i/ two independent series (n = 598) of resected tumours; ii/ 60 advanced tumours obtained by diagnostic EUS-guided biopsy needle flushing and iii/ on 28 biopsies from mFOLFIRINOX treated metastatic tumours.
Findings
A unique transcriptomic signature (PAGM) was generated with significant and independent prognostic value. PAMG significantly improves the characterization of PDAC heterogeneity compared to non-overlapping classifications as validated in 4 independent series of tumours (e.g. 308 consecutive resected PDAC, uHR=0.321 95% CI [0.207–0.5] and 60 locally-advanced or metastatic PDAC, uHR=0.308 95% CI [0.113–0.836]). The PAMG signature is also associated with progression under mFOLFIRINOX treatment (Pearson correlation to tumour response: -0.67, p-value < 0.001).
Interpretation
PAMG unify all PDAC pre-existing classifications inducing a shift in the actual paradigm of binary classifications towards a better characterization in a gradient.
Funding
Project funding was provided by INCa (Grants number 2018–078 and 2018–079, BACAP BCB INCa_6294), Canceropole PACA, DGOS (labellisation SIRIC), Amidex Foundation, Fondation de France, INSERM and Ligue Contre le Cancer.
Keywords: Pancreatic cancer, Transcriptomic signature, Chemosensitivity prediction, Prognostic, Translational medicine, Precision medicine
Research in context.
Evidence before this study
Despite being an overall dismal cancer, the clinical outcome of pancreatic ductal adenocarcinoma is difficult to anticipate, with newly diagnosed patients having a potential life expectancy ranging from 3 months to more than 5 years. Previous studies of the inter-patient heterogeneity of pancreatic ductal adenocarcinoma based on large scale molecular profiles have proposed molecular classifications. These findings can be summarized into a molecular dichotomic perspective with the classical subtype and the more aggressive basal-like. However, an increasing body of evidence is highlighting the intra-tumour heterogeneity of pancreatic ductal adenocarcinoma, including mix tumours with both basal-like and classical cancer cells. These results suggest that a two-class stratification of patients with a pancreatic ductal adenocarcinoma is ill-suited.
Added value of this study
The RNA-based signature identified in this work is different to all molecular classification schemas already proposed and validated by other investigators. This signature does not classify tumours in a non-overlapping subtyping but grades them along a continuum. This fact significantly improves the accuracy of individual patient's tumours characterization with a higher prognostic value that is also highly reproducible. The signature represents also a major technical improvement since it is independent of the sample type or RNA measurement platform.
Implications of all the available evidence
This method will unify pre-existing classifications inducing a shift in the actual paradigm of binary classifications towards the characterization of patients’ tumours into a gradient that better considers the real complexity of tumour phenotype.
Alt-text: Unlabelled box
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive gastrointestinal tumours. While activating mutations in KRAS are the most common genetic alterations 1, mutations in other driver genes such as CDKN2A, TP53 or SMAD4 are randomly associated to KRAS mutations, generating a heterogeneous genetic landscape between patients. However, these mutations do not predict patient outcome or tumour drug sensitivity and PDAC patients with similar clinical presentation show high variability in overall survival (OS), ranging from 3 months to >5–6 years after diagnosis. While histopathological analyses of tumours revealed OS is shorter in patients presenting with aggressive poorly-differentiated tumours relative to patients with well-differentiated ones 2, this analysis required large amounts of undamaged tumour tissue. Such samples are only available from resected tumours, representing as few as 15% of PDAC cases. For resectable PDAC, the current recommendation is upfront surgical resection followed by systemic chemotherapy with or without radiation 3. However, this strategy can fail in patients with biologically aggressive disease that do not benefit from resection. Therefore, an accurate molecular characterization of tumour phenotype will help in predicting prognosis and chemotherapy sensitivity, as well as inform decisions regarding upfront resection and the most appropriate drug choice for chemotherapy. Deep tumour molecular profiling constitutes an important source of information regarding tumour phenotype and biology, with impact on the choice of available therapeutic strategies. This information will increase the likelihood of success and also spare patients from unnecessarily aggressive therapeutic interventions.
Recent reports indicate PDAC can be classified into distinct, biologically relevant categories based on histological and molecular analysis 4,5. However, relatively few patients (15%) undergo resection that allows this analysis, and high intra-tumour heterogeneity and the limited amount of material obtained from EUS-FNA diagnostic biopsies prevent a precise classification of all PDAC tumours. One solution to circumvent these problems is transplantation of PDAC tumours into immunodeficient mice to produce patient-derived xenografts (PDX). This process makes it possible to obtain PDXs from EUS-FNA diagnostic biopsies providing adequate material to determine PDAC histological classes for locally advanced or metastatic tumours. We observed that PDXs are less complex and heterogeneous tumours, but faithfully recapitulate the molecular profiles and histology of the original patient tumours6. Another important point that conducts us to choose PDX as model is that it offers the possibility to distinguish between the tumour and stromal cells. In fact, sequencing profiles of a mix of human grafted cancerous and infiltrating mouse stromal cells can be analysed separately in silico by unambiguously assigning each sequence to the human or mouse genome 7. Therefore, we generated PDX samples for a cohort of patients (PaCaOmics) to define histological and molecular grades for each sample. In this study we take advantage of these PDX characteristics and used this model to identify a molecular signature based on the transcriptomic profiles of PDAC patients that would allow for prediction of tumour progression and response to therapy.
To obtain an unbiased predictor of tumour aggressiveness, we established a series of patient-derived xenografts (PDX) from a multi-centric clinical trial that included resectable, locally advanced and metastatic PDAC patients. From these PDX samples, a transcriptomic signature (indicated as pancreatic adenocarcinoma molecular gradient; PAMG) was developed that accurately predicted tumour aggressiveness and resistance to mFOLFIRINOX, and could be applied to small amount of fine needle biopsies from EUS and formalin-fixed paraffin-embedded obtained tissue.
Materials and methods
PaCaOmics patient derived tumour xenograft and RNA-sequencing
All animal experiments were conducted in accordance with institutional guidelines and were approved by the “Plateforme de Stabulation et d'Expérimentation Animale” (PSEA, Scientific Park of Luminy, Marseille). Resected PDAC tissue was fragmented, mixed with 100 μL of Matrigel and implanted with a 10-gauge trocar (Innovative Research of America, Sarasota, FL) in the subcutaneous right upper flank of an anesthetized male NMRI-nude mouse (Swiss Nude Mouse Crl: NU(lco)-Foxn1nu; Charles River Laboratories, Wilmington, MA). Alternatively, samples obtained from direct tumour endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) were mixed with 100 μL of Matrigel (BD Biosciences, Franklin Lakes, NJ) and injected as above. Once xenografts reached 1 cm3, they were removed and passed to NMRI-nude mice. After 3 passages, tumors were isolated and RNA extracted using the miRneasy mini kit (Qiagen). RNA-seq was performed as previously described 7,8 using Illumina's TrueSeq Stranded RNA LT protocol to obtain 100b paired-end reads. RNA-seq reads were mapped using STAR and SMAP on the human hg19 and mouse mmu38 genomes. Gene expression profiles were obtained using FeatureCount and normalized using the upper-quartile approach 9. tumour differentiation was defined based on the following established criteria, briefly: tumors were considered poorly differentiated when tissue architecture is solid, forming massive structures or with isolated cells without visible glandular structures in more than 50% of the tissue. This group included two classes (I and II) based on the degree of cyto-nuclear atypia and degree of mitosis. Class I tumors showed high nucleo-cytopasmic ratios (>0.5), and nuclei with irregular contours, dense chromatin, and/or prominent nucleolus. A high proportion of mitoses (>5 per 10 high-power field [HPF]) was also visible in this subgroup. Class II includes tumors with fewer atypia with a nucleo-cytoplasmic ratio < 0.5, regular-contoured nuclei, fine chromatin and a fine nucleolus. Mitoses were less frequent than in class I (< 5 mitosis/10 HPF). Class III includes tumors that were moderately differentiated with both types of architectures, glands made up 50–95% of the tumour, massive structures and nucleo-cytoplasmic atypia were less frequent (approximately 50% of nuclei) than in class I and II. Class IV and V were included in well differentiated PDX. They present a glandular architecture without solid component in more than 95%. In this group, class IV presents glands with cubic or short cylindrical cells with low or absent mucus secretion. The nuclei remain predominantly polarized and the atypia are more marked than in class V (looser chromatin, increase in the size of the nuclei when compared with class V). Mitoses were more frequent than in class V (2–5 mitosis / 10 HPF). Class V corresponds to the most differentiated tumors, the glands secrete mucin and cells present a cylindrical form, the nucleus was localized at the basal pole of the cell (polarized). Nuclei were small, with regular contours and mature chromatin without visible nucleolus. Mitoses were less frequent (0–1 mitosis / 10 HPF) that in class IV.
PaCaOmics patient's cohort
Seventy-six patients with a confirmed PDAC diagnosis were included in this study. Clinical data was collected until July 2017 (supplementary Tables I and II). Tumour samples were obtained from pancreatectomy in 40 patients (52.6%), EUS-FNA in 25 patients (32.9%) and carcinomatosis or liver metastasis during explorative laparotomy in 11 patients (14.5%). The study was approved by the local ethics committee following patient informed consent. The PaCaOmics study is registered at www.clinicaltrials.gov with registration number NCT01692873. All samples were xenografted in immunocompromised mice producing PDX samples. Animal experiments were approved by the local ethics committee and performed following the guidelines of our centre (CRCM).
BACAP patient's cohort
The BACAP (Base Clinico-Biologique de l'Adénocarcinome Pancréatique) cohort is a prospective multicenter pancreatic cancer cohort (ClinicalTrials.gov Identifier: NCT02818829. Registration date: June 30, 2016) with a biological clinical database. Treatment naive tumour biological samples from endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) were available for 60 patients. Survival analysis was performed on the 47 patients with locally-advanced or metastatic diseases that subsequently received chemotherapy. BACAP project was approved by the “Sud-Ouest et Outre-Mer I” March 2014 ethics committee. All the patients were informed of the study and voluntarily agreed to participate. All the patients who agreed to participate provided written consent.
Transcriptomic profiling and analysis to derive the PAMG
RNA was obtained from all PDX and BACAP cohort samples, for more details see supplementary material and methods. Next Generation Sequencing (RNA-seq) was performed on these samples. The Pancreatic Adenocarcinoma Molecular Gradient (PAMG) was derived from the PDX RNA-seq profiles using an Independent Component Analysis (ICA) on 50% most variant genes (n = 20,434), after gene-wise zero-centring (no unit scaling). The independent component that best correlated with PDX histology was identified as the PAMG. Further details are available in the supplementary information. RNA sequencing of pancreatic adenocarcinoma (PDAC) xenograft samples accession number E-MTAB-5039. The PAMG is available as an online application (http://cit-apps.ligue-cancer.net//pancreatic_cancer/pdac.molgrade) and as an R package (https://github.com/RemyNicolle/pdacmolgrad).
Statistical analysis
Categorical variables were associated to continuous variables using an ANOVA. Survival analysis were performed using a Cox proportional hazards regression model. Wald's p-value are reported unless otherwise specified. All analyses were performed in R.
Results
Using pdx to define the molecular diversity of PDAC
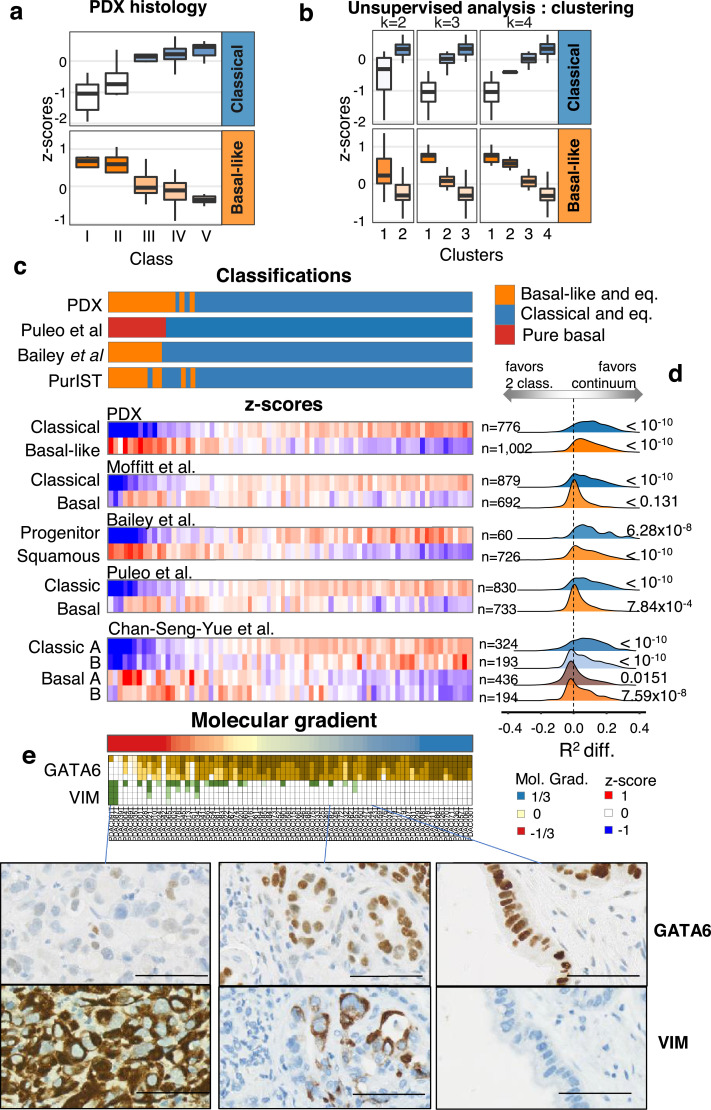
First, we assessed the histology of PDX using the entire cohort of 76 patients. PDX were ranked into five different histological classes by two blinded expert pathologists ranging from the less differentiated PDX (class I), which is associated with the most aggressive phenotype, to the most differentiated PDX (class V; Figure S1). The here described five histological classes of PDX strongly correlates with the expression of genes defining the already described molecular subtypes 7,10, 11, 12 as higher expression of genes linked to the classical PDAC subtype is correlated with increased differentiation of PDX samples, combined with lower expression of genes linked to basal-like subtype (Fig. 1a and Figure S1b). Interestingly, the variation in the expression of the classical genes towards the basal-like genes vary gradually from the more differentiate to the less differentiate histological classes respectively. Therefore, the precise histological analysis of PDX suggests that molecular classification of PDAC is more complex than a two-class dichotomy (i.e. basal-like and classical). We next employed a consensus clustering approach on whole-transcriptome with increased subtypes splitting. Fig. 1b shows the clustering results in 2 to 4 subtypes which, similarly to histological classification, demonstrate a gradual increase and decrease in genes of the classical and basal-like subtypes respectively.
Fig. 1.
PDAC gene signatures and classification in PDX. a. Normalized and averaged expression of genes specific to the classical and basal-like subtypes in PDX (n = 76) grouped by a five-subtype histological classification. b. Unsupervised classifications in k classes by consensus clustering (with k from 2 to 4) and association of each cluster to basal-like and classical gene expression. On a. and b. boxplots are coloured by the median z-score of each group. c. Heatmap representation of the transcriptomic characterization of the PDX (n = 76) with each PDX as a column. Previously published classifications were applied to the human transcriptome profiles of the PDX. Non-tumour driven classifications were applied (ADEX, Immunogenic, desmoplastic, activated stroma, Immune classical), however, no PDX were assigned to any of them. The z-score of each of the published classification gene sets is represented. The number of genes of each signature is annotated on the right of the heatmap. PDX were ordered by their value on the molecular gradient. d. Distribution of the differences in the coefficient of determination (R2) between two generalized linear models associating the expression of each gene in each signature with either the two-class classification from PurIST or the Molecular Gradient. The distribution of R2 differences was compared to that of other genes (not found in any other subtype signatures) using Welch's t-test. e. GATA6 and Vimentin (VIM) immunohistochemical quantification. Four levels of staining were used to quantify the proportion of cells at each four levels of GATA6 or VIM protein expression.
Histological and molecular classifications of PDX suggest PDAC diversity may be better represented by a continuum of differentiation that is as also followed at the molecular level. To establish a robust continuous molecular description of PDAC, we applied an unsupervised approach termed independent component analysis (ICA) previously shown to derive highly reproducible signatures from transcriptome profiles by extracting biologically relevant components 13,14. Figure S2 illustrates the procedure used to uncover an RNA signature which, in essence, builds on the blind deconvolution of the PDX transcriptomic profiles to generate component spaces. The component (and its associated space) that best correlated to the PDX histological classification was selected and, in analogy to histological grading, was termed the pancreatic adenocarcinoma molecular gradient (PAMG). The PAMG is computed from a weighted combination of the expression values of all variable genes (n = 20,434) providing for each sample a standardized score around zero with non-outlier values between −1 and +1. The PAMG summarizes all previous epithelial molecular classification of PDAC whether at the level of sample characterization or by the sets of genes defining each classification (Fig. 1c and Figure S2). To evaluate whether a continuous or dichotomous description of PDAC epithelial diversity is more relevant, gene expression in each of these signatures was fitted with the proposed PAMG and with the latest basal-like/classical classifier PurIST. The difference in the coefficient of determination (R2) of the two models was compared to the background (genes not in any of the assessed signatures, n = 7393) showing overall that a continuum is likely to be a more reliable description of PDAC molecular diversity. We observed that PAMG produces a better description on 11 out of 12 signatures tested by a Welch's t-test (Fig. 1d).
A continuum of phenotypes would predict that extreme cases would be more homogeneous, composed of a high proportion of highly specified epithelial cells from the corresponding end of the spectra (i.e. basal-like or classical). PDAC cases in the middle of the spectrum could either be the result of a homogeneous intermediate epithelial phenotype or a mixture of extreme phenotypes of which bulk tumour analysis would result in an intermediary phenotype. To evaluate these non-mutually exclusive hypotheses, we performed immunostaining for GATA6, which we previously showed to be a major driver of the classical phenotype 8, and Vimentin (VIM) in a tissue microarray containing all 76 xenograft tumours. VIM is a marker of mesenchymal differentiation and carcinomas with more aggressive behaviour and poor histological differentiation 15. Fig. 1e shows quantitative results and representative examples of expression. While some GATA6+/VIM+ stained tumours exist, we generally observed a continuum of differentiation defined by increases in the level and proportion of expression of GATA6 along the PAMG that correlated with increased differentiation. Conversely, we observed VIM expression increasing gradually towards low differentiated phenotypes.
Reproducibility of the pamg in resectable human primary PDAC
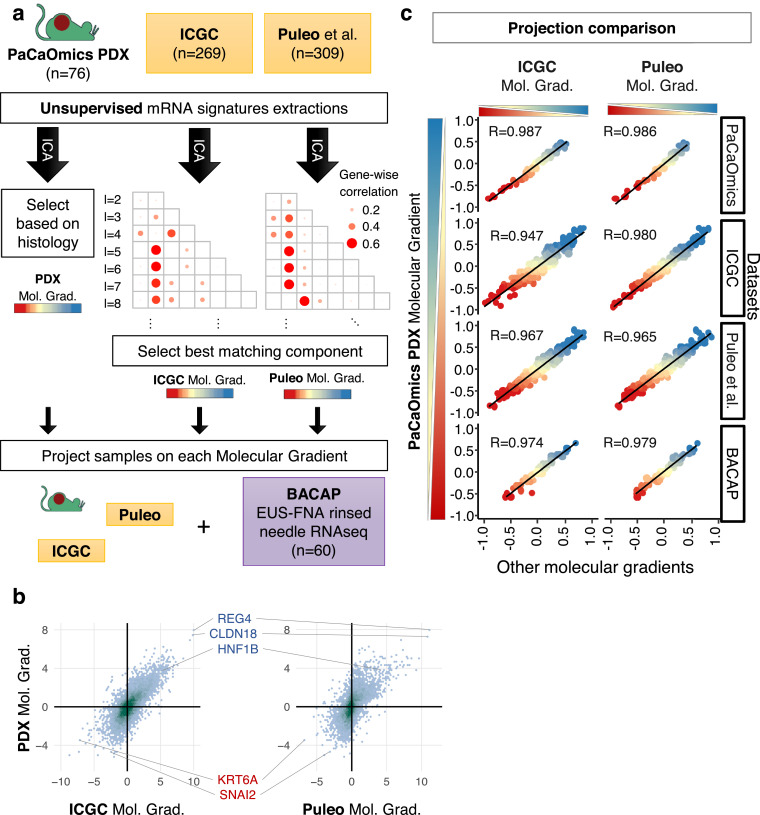
To evaluate the robustness of the PAMG, we tested whether an equivalent RNA signature could be blindly reproduced in independent PDAC series with transcriptomic data. Two large series of PDAC were used for this purpose 269 resected tumours from the Australian ICGC 16 profiled on Illumina microarrays from frozen samples, and the multi-centric cohort of 309 consecutive patients from Puleo et al. 10, profiled on Affymetrix arrays from paraffin-embedded samples. To assess the reproducibility of the PAMG in these series of samples, a blind deconvolution of the transcriptomes was performed using ICA with increasing number of components resulting in ICA spaces of up to 25 unsupervised independent components (Fig. 2a). Once components were extracted, a component matching the PAMG from the PDX was sought by correlating gene weights of both the reference PDX ICA space and the new ICA spaces to be evaluated. This analysis aimed at evaluating whether a component biologically similar to the PAMG could be extracted from the human tumour datasets. A molecular component equivalent to the PDX-derived PAMG was found in virtually all ICA component spaces in both datasets despite the difference in measurement technologies and in tissue preservation (Figure S3). The component with the highest gene weight correlation to the PDX-based PAMG was selected from each dataset. Fig. 2b illustrates the overall consistency in the gene weights defining each of the components of three PAMGs. Overall, three components were selected from an unsupervised gene-expression deconvolution analysis applied to three independent datasets representing diverse technological (microarrays and RNAseq) and tissue (FFPE, Frozen, PDX) options to profile PDAC resulting in three biologically equivalent implementations of the PAMG.
Fig. 2.
Reproducibility of the PAMG in PDAC. a. Schematic illustration of the identification of the PAMG in public datasets. ICA (independent component analysis) blind deconvolution was used on three different datasets of whole transcriptome profiling, generating spaces of independent components of increasing sizes (2 ≤ l ≤ 25). The PAMG was first obtained from PDX by selecting the component most associated with PDX histology. The gene weights of this initial PDX-based independent component was then correlated to the gene weights of all extracted independent components in the other datasets, with the spearman correlation represented in a grid. The highest correlating component of each dataset was selected as the PAMG. b. Density plot of the PAMG gene weights of common genes found in each pair of datasets. Marker genes are highlighted. c. Scatter plots comparing the three versions of the Molecular Gradient (PDX, ICGC and Puleo) on four datasets. Each point is a sample, coloured by its PAMG score as defined by the PDX version. Pearson correlation is shown.
While the three independently identified PAMGs share a similar gene expression basis, we next sought to evaluate the extent they define the same PDAC heterogeneity. The samples from the three different datasets were each projected on all three PAMGs. Fig. 2c shows a high correlation between the three PAMGs in all three datasets, demonstrating that the signatures measure a common biological diversity independent of the types of samples profiled and the technologies used. To validate this high reproducibility, the same analyses were applied to a PDAC cohort consisting of 60 RNAseq profiles from RNA obtained by rinsing EUS-FNA diagnostic biopsies. The three versions of the PAMG gave highly similar results on FNA-derived samples (R>0.97). Overall, these results show that the three versions of the PAMG are essentially identical biological signatures despite the broad differences between the samples in each dataset (e.g. primary tumour with typical stromal content versus PDX).
The pamg is associated to tumour aggressiveness
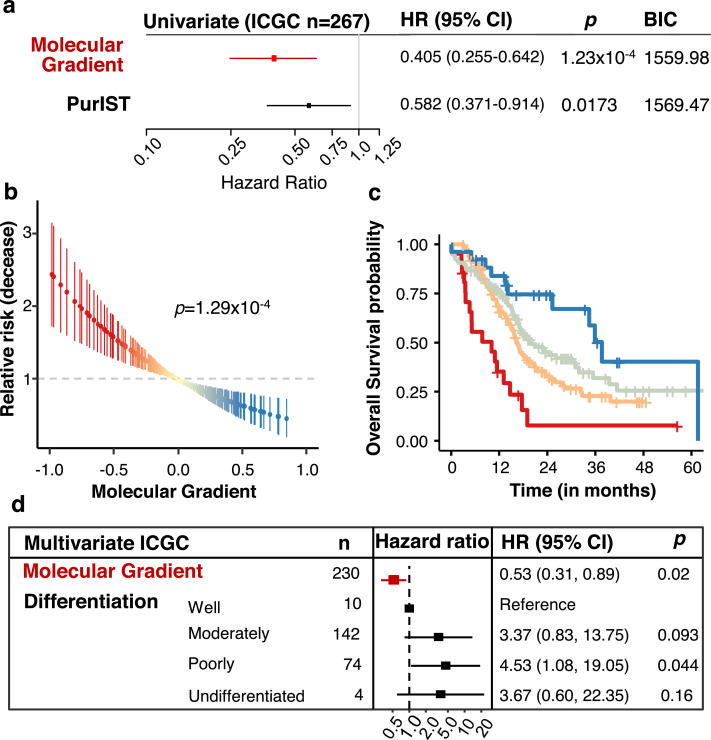
Several studies using only resectable tumours show molecular diversity of the epithelial compartment of PDAC is associated with tumour aggressiveness and patient prognosis 10,11,16. Our next goal was to determine if the PAMG could be predictive in primary PDAC tumours by analysing large series of patients with thorough clinical data and follow-up. To assess the prognostic value of the PAMG, association with overall survival was first evaluated on the ICGC series 16 which consisted of 267 resected patients with follow-up, and 230 samples with histological characterization. The continuous value of the PAMG (as extracted from the ICGC transcriptome dataset) was strongly associated to patient's overall survival (univariate Hazard Ratio: uHR=0.405, 95% CI [0.255–0.642]; p = 1.23 × 10−4 and compared favourably to the basal-like/classical dichotomous classification (Fig. 3a and Figure S4). A virtually identical result was obtained with the other PAMGs derived from the PDX and Puleo et al. cohorts (Figure S4). The continuous characterization of patients in the ICGC series by the PAMG showed a positive correlation with significant increase in OS (Fig. 3b) also illustrated in a Kaplan-Meier analysis (Fig. 3c) after splitting the PAMG using three arbitrary thresholds (−0.5, 0 and 0.4; selected on the basis of the separation of histological classes of PDX). A significant yet weak association was found between the PAMG and the histological differentiation of these tumours (Figure S4a), suggesting a partial relationship between molecular classification of PDAC and traditional histological classes 10. In a multivariate analysis including the PAMG and the histology of these tumours, the PAMG was an independent predictor of OS (Fig. 3d).
Fig. 3.
Prognostic value of the PAMG in the ICGC series. a. Univariate survival analysis using the overall survival (OS) of 260 patients associated with either the PAMG or the PurIST two-subtype classification. b. Univariate relative risk for OS associated with the PAMG. Each point is a patient's relative risk of disease with error bars corresponding to a 95% confidence interval. c. Kaplan-Meier plot of survival using arbitrary cuts of the Molecular Gradient. d. Multivariate survival analysis forest plot. Univariate: n = 267. Multivariate: n = 230. Wald's test p-values are shown.
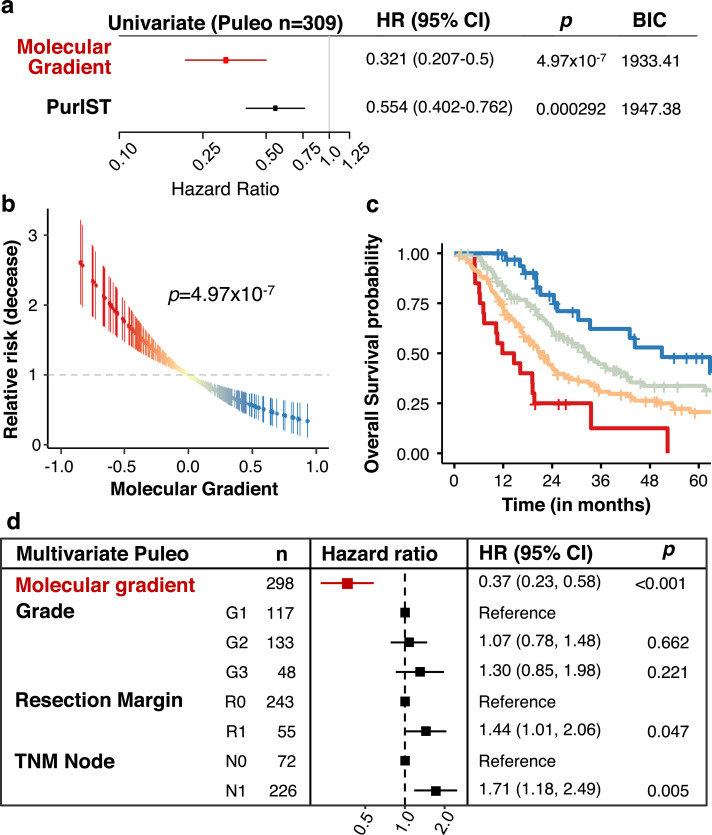
To further assess the value of the PAMG in a more reliable cohort of patients, the multicentric cohort of 309 consecutive patients from Puleo et al. 10 was used. This very complete cohort contains whole follow up for 308/309 patients (median follow-up 51.4 months) and with a majority (298/309) also having data on extended clinical and pathological characterization. The PAMG was associated with patients OS (uHR = 0.321, 95% CI [0.207–0.5];p = 4.97 × 10−7) and compares favourably to the basal-like/classical classification (Fig. 4a and Figure S5). The PAMG was correlated to a positive outcome in Puleo cohort, with a progressive improvement of OS coinciding with higher PAMG levels (Figs. 4b and 4c). A multivariate analysis including resection margins, histological grading and TNM Node status demonstrated the PAMG is an independent prognostic factor in resected PDAC (Fig. 4d).
Fig. 4.
Prognostic value of the PAMG in the Puleo cohort. a. Univariate survival analysis using the OS of 308 patients associated with either the PAMG or the PurIST two-subtype classification. b. Univariate relative risk for OS associated with the PAMG. Each point is a patient's relative risk of decease with error bars corresponding to a 95% confidence interval. c. Kaplan-Meier plot of survival using arbitrary cuts of the PAMG. d. Multivariate survival analysis forest plot. Univariate: n = 308. Multivariate: n = 298. Wald's test p-values are shown.
PAMG predicts the clinical outcome of advanced pdac patients
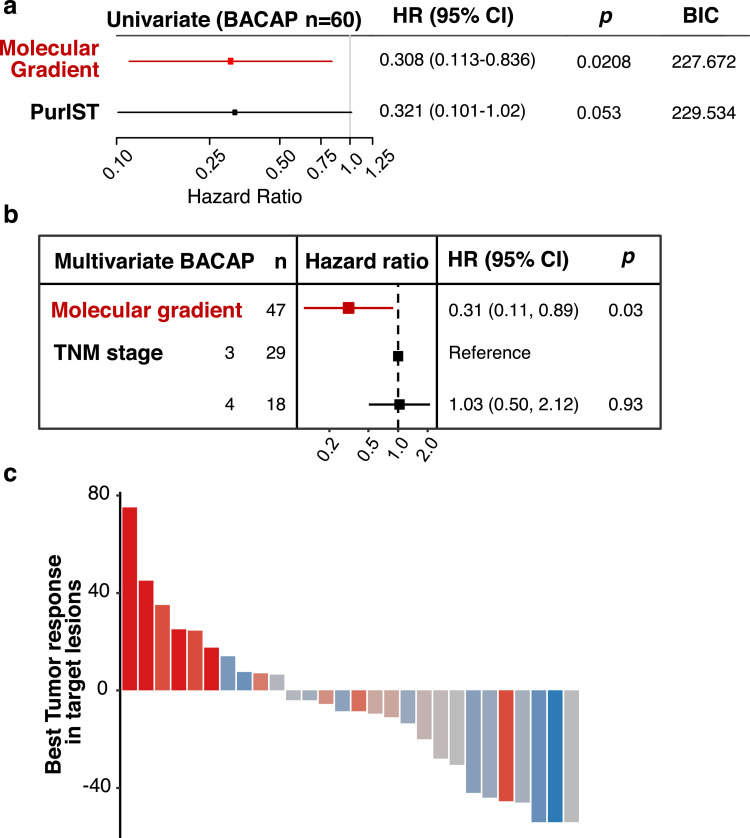
The clinical relevance of the PAMG is dependant on its applicability to work on biopsy samples obtained prior to treatment. In the BACAP cohort, RNA was extracted from 60 samples obtained by rinsing the echo endoscopy-guided fine needles. The original aspirate was used for diagnosis. Fig. 2c shows all three versions of the PAGM gave the same result on these small sample biopsies. The PAGM was also associated with the OS of the 47 patients with advanced diseases (uHR=0.308, 95% CI [0.113–0.836]; p = 0.0208, Fig. 5a) and, similar to resectable tumours, compared favourably to the PurIST two-subtype classification. The PAGM was also associated to survival in a multivariate model including the tumour stage (Fig. 5b).
Fig. 5.
Evaluation of the PAMG in advanced disease. a. Univariate survival analysis using the OS of 47 patients in the BACAP cohort associated with either the PAMG or the PurIST two-subtype classification. b. Multivariate survival analysis forest plot for the BACAP cohort. c. Waterfall plot illustrating the change in tumour size induced by mFOLFIRINOX treatment evaluated by RECIST 1.1 in the COMPASS cohort (n = 28). Annotated Pearson's correlation between RECIST 1.1 and PAMG is shown.
PAMG predicts the response to mFOLFIRINOX of advanced pdac patients
It was previously suggested that molecular subtypes of PDAC were associated with responses to chemotherapy, in particular FOLFIRINOX 11,17,18. Therefore, to evaluate the predictive value of the PAGM to chemotherapy response, it was applied to metastatic patients in the COMPASS trials for which transcriptomic profiles and tumour responses to mFOLFIRINOX were available 18. The objective response was significantly associated with the PAMG (Fig. 5c, R= −0.67; p < 0.001), with more aggressive tumours (i.e. low on the PAMG) showing little to no response to mFOLFIRINOX.
Discussion
An important factor in determining treatment options for PDAC involves the ability to accurately classify the tumour and predict the aggressiveness of the disease. However, resolving the diversity of molecular tumour phenotypes in PDAC is a complex issue involving the necessary distinction of transformed and non-transformed cells as well as a multiscale integration in which microscopic cellular phenotypes are considered with macroscopic phenotypes of the whole-tumour tissue. Previous work has mainly focused on resected primary PDAC tumours, often resulting in classifications that considers all of the cell types within the tumour (e.g. the infiltrated Immunogenic subtype), and delineated a consensual basal-like versus classical dichotomy. However, this two-subtype classification system of PDAC has recently been challenged by several studies showing the coexistence of basal-like and classical cells in the same tumours as well as to the likely existence of intermediate cellular phenotypes 17,19. In this study, we have used a gradient system that takes this into consideration to molecularly grade PDAC. The resulting PAMG signature is more informative and clinically relevant than a binary non-overlapping method.
Single cell RNA sequencing 17, 19 and immunohistochemistry 20 of PDAC revealed intra-tumour heterogeneity where both types of cells (basal-like and classical) frequently co-exist. RNA profiling of multiple regions or multiple lesions from the same patient also demonstrated intra-tumour heterogeneity of the transformed epithelial compartment 4. Using single cell RNA-seq, Chan-Seng-Yue et al. in 2020 17 confirmed the presence of several subpopulations with differential proliferative and migratory potentials in PDAC. In particular, they observed two ductal subtypes with abnormal and malignant gene expression17. In another single cell approach Juiz et al. 19 identified four common cell clusters in patients with a classical PDAC. These four clusters were present in different proportions in all tumours examined, with one of these clusters corresponding to a basal-like phenotype, even though the tumours were classified as classical by global RNAseq analysis. We have made similar observations in this study. VIM, which is mainly expressed in basal-like subtype, was detected by immunohistochemistry in almost all classical tumours, with variable levels of expression 19. We detected few VIM+ cells in tumours presenting an intermediate PAMG. In other words, very classical or very basal-like subtypes are mainly composed by pure cells, but the intermediate subtype is the consequence of a mix of classical and basal-like subtypes and/or an intermediate phenotype. These observations question the relevance of a dichotomous model of PDAC diversity and make the molecular description a different and complex scenario for every tumour. Since PDAC tumours are heterogeneous, this must be taken into consideration for classification and treatment purposes making protocols characterizing the proportion of intermediate cell types or tumour heterogeneity are necessary.
In this work, we developed a molecular gradient that defines a continuum of PDAC phenotypes. We developed 76 PDX, obtained from resectable and unresectable PDAC, since they offer a platform with an incomparable discrimination of transformed and non-transformed cells RNA. First, we applied a deconvolution algorithm (ICA) to the transformed epithelial RNA profiles to identify in an unsupervised manner the RNA signatures that best defined the heterogeneity of PDX and, in particular, its aggressiveness. This approach extracted a specific RNA signature robustly identified in PDX and human primary tumours with a minor effect of tissue preservation (FFPE vs. frozen), RNA profiling platform (microarrays or RNAseq) or of the algorithm's parameter (the total number of extracted components). The use of a deconvolution approach to define a gene signature is especially appropriate for highly heterogeneous and stromal tumours as it is able to extract signatures independently of other biological signals (e.g. stroma, normal tissue). This RNA signature, termed PAMG, provides a score measuring the molecular level of differentiation of a given sample derived from a whole-transcriptome profile. We show the robustness of our approach to derive an RNA-phenotyping signature by consistently identifying the same signature from multiple independent dataset, all providing highly similar results (average correlation of >97%). Previous studies focusing on PDAC phenotype dichotomy11,21 actually provide a continuous scoring scheme which underlies their classification method. While these may hold valuable clinical value, potentially more than the initially proposed dichotomy, the PAMG remains less affected by technological aspects such as RNA-measuring platform (Figure S6). One possible limitations of molecular analysis in PDAC is the low level of cellularity usually found in these tumours. While this is not fully addressed in this work and warrants further investigation, the PAMG is shown to be effective in unselected rinsed EUS-FNA biopsies, contaminated not only by stromal and normal pancreatic tissue but also by blood. Studies including different proportions of cellularity in the same tumour needs to be analysed to answer this question.
The PAMG introduces a simple framework, based on a simple RNA signature compatible with all previously proposed PDAC classifications. The genes previously described as defining PDAC subtypes were in fact better explained by the PAMG than by the two-class classifications themselves. Molecular classifications of PDAC and, in particular, the basal-like/classical dichotomy, are a major prognostic factor in most datasets and are typically shown to correlate with response to FOLFIRINOX. Our results showed the PAMG holds superior clinical value that could be ascertained prior to entering any curative protocols, using any current diagnostic material including EUS-guided biopsy needle flushing. The PAMG can be applied to a single PDAC sample, resulting in a score which is an estimation of the overall survival, with a low PAMG corresponding to poor outcome (high relative risk) and a high PAMG corresponding to an improved outcome (low relative risk). Despite the high prognostic value of the PAMG, the clinico-pathological characteristics typically used to inform on the patients’ outcome, such as the resection margins or the spread to lymph node, remain informative in multivariate models (e.g. Fig. 4d). For instance, in the TCGA series with a less complete clinical follow up, the PAMG was significantly associated with overall survival in a model including T, N status and resection margin (Figure S7).
This model could have a major impact on patients who are cleared for resection by identifying patients that will have an unfavourable disease evolution and may benefit from initial neoadjuvant therapy prior to upfront surgery. Another group of patient the PAMG could impact is the 20 to 30% percent of patients diagnosed with a locally-advanced disease. If pancreatectomy and simultaneous arterial resection has traditionally been considered as a general contraindication to resection 22, some of these patients with good prognosis might indeed benefit from aggressive surgical approaches 23.
In conclusion we propose a transcriptomic signature that unifies all previous molecular classifications of PDAC under a continuous gradient of tumour aggressiveness that can be performed on FFPE samples and EUS-guided biopsies. In addition to its strong prognostic value, it may predict mFOLFIRINOX responsiveness.
This work is part of the national program Cartes d'Identité des Tumeurs (CIT) funded and developed by the Ligue Nationale Contre le Cancer. This work was supported by INCa (Grants number 2018–078 and 2018–079, BACAP BCB INCa_6294), Canceropole PACA, DGOS (labellisation SIRIC), Amidex Foundation, Fondation de France and INSERM. The funders do not participate in study design, data collection, data analysis, interpretation or writing of the report. Authors wish to thanks Christopher Pin for critically reading the manuscript and IPC/CRCM Experimental Pathology platform for TMA and immunohistochemistry performing. This research was supported by the CRCM Integrative Platform (Cibi) and the CRCM's DataCentre for IT and Scientific Computing (DISC).
Conflicts of interest
The authors declare no competing financial interests
Acknowledgments
The BACAP consortium
1 Barbara Bournet, Cindy Canivet, Louis Buscail, Nicolas Carrère, Fabrice Muscari, Bertrand Suc, Rosine Guimbaud, Corinne Couteau, Marion Deslandres, Pascale Rivera, Anne-Pascale Laurenty, Nadim Fares, Karl Barange, Janick Selves, Anne Gomez-Brouchet. 2 Bertrand Napoléon, Bertrand Pujol, Fabien Fumex, Jérôme Desrame, Christine Lefort, Vincent Lepilliez, Rodica Gincul, Pascal Artru, Léa Clavel, Anne-Isabelle Lemaistre. 3 Laurent Palazzo; 4 Jérôme Cros; 5 Sarah Tubiana; 6 Nicolas Flori, Pierre Senesse, Pierre-Emmanuel Colombo, Emmanuelle Samail-Scalzi, Fabienne Portales, Sophie Gourgou, Claire Honfo Ga, Carine Plassot, Julien Fraisse, Frédéric Bibeau, Marc Ychou; 7 Pierre Guibert, Christelle de la Fouchardière, Matthieu Sarabi, Patrice Peyrat, Séverine Tabone-Eglinger, Caroline Renard; 8 Guillaume Piessen, Stéphanie Truant, Alain Saudemont, Guillaume Millet, Florence Renaud, Emmanuelle Leteurtre, Patrick Gele; 9 Eric Assenat, Jean-Michel Fabre, François-Régis Souche, Marie Dupuy, Anne-Marie Gorce-Dupuy, Jeanne Ramos; 10 Jean-François Seitz, Jean Hardwigsen, Emmanuelle Norguet-Monnereau, Philippe Grandval, Muriel Duluc, Dominique Figarella-Branger; 11 Véronique Vendrely, Clément Subtil, Eric Terrebonne, Jean-Frédéric Blanc, Etienne Buscail, Jean-Philippe Merlio; 12 Dominique Farges-Bancel, Jean-Marc Gornet, Daniela Geromin; 13 Geoffroy Vanbiervliet, Anne-Claire Frin, Delphine Ouvrier, Marie-Christine Saint-Paul; 14 Philippe Berthelémy, Chelbabi Fouad; 15 Stéphane Garcia, Nathalie Lesavre, Mohamed Gasmi, Marc Barthet; 16 Vanessa Cottet; 17 Cyrille Delpierre.
1 The CHU and the University of Toulouse, Toulouse, France; 2 Jean Mermoz Hospital, Lyon, France; 3 Trocadéro Clinic, Paris, France; 4 The Department of Pathology, Beaujon Hospital and Paris 7 University, Clichy, France.5 The Biobank, Bichat Hospital and Paris 7 University, Paris, France; 6 The Cancer Institute and the University of Montpellier, Montpellier, France; 7 The Léon Bérard Cancer centre, Lyon, France;8 The Department of Digestive Surgery, the CHU and the University of Lille, Lille, France;9 The CHU and the University of Montpellier, Montpellier, France.10 La Timone Hospital and the University of Marseille, Marseille, France;11 The CHU and the University of Bordeaux, Bordeaux, France;12 Saint Louis Hospital and Paris 7 Diderot University, Paris, France;13 The CHU and the University of Nice, Nice, France;14 Pau Hospital, Pau, France;15 The CHU Nord Hospital and the University of Marseille, Marseille, France;16 INSERM UMR866 and the University of Dijon, Dijon, France;17 INSERM UMR1027 and the University of Toulouse, Toulouse France.
Author contributions
Conception and design: Nelson Dusetti, Rémy Nicolle and Juan Iovanna
Provision of study materials or patients: Marine Gilabert, Vincent Moutardier, Stephane Garcia, Pauline Duconseil, Charles Vanbrugghe, Philippe Grandval, Marc Giovannini, Cindy Canivet, Barbara Bournet and Louis Buscail.
Histological classification and analysis: Nicolas Brandone, Flora Poizat, Marion Rubis; Jérôme Cros.
Collection and assembly of data: Martin Bigonnet, Odile Gayet, Julie Roques, Pauline Duconseil, Nabila Elarouci, Lucile Armenoult, Mira Ayadi and Aurélien de Reyniès.
Data analysis and interpretation: Nelson Dusetti, Rémy Nicolle, Yuna Blum and Juan Iovanna.
Manuscript writing: Nelson Dusetti, Rémy Nicolle, Yuna Blum and Juan Iovanna
Final approval of manuscript: All Authors
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102858.
Appendix. Supplementary materials
Bibliography
- 1.Deramaudt T., Rustgi A.K. Mutant KRAS in the initiation of pancreatic cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2005;1756(2):97–101. doi: 10.1016/j.bbcan.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Lenz J., Karasek P., Jarkovsky J. Clinicopathological correlations of nestin expression in surgically resectable pancreatic cancer including an analysis of perineural invasion. Journal of gastrointestinal and liver diseases: JGLD. 2011;20(4):389–396. [PubMed] [Google Scholar]
- 3.Li D., O'Reilly E.M. Adjuvant and neoadjuvant systemic therapy for pancreas adenocarcinoma. Semin. Oncol. 2015;42(1):134–143. doi: 10.1053/j.seminoncol.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi A., Fan J., Chen R. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nature Cancer. 2020;1(1):59–74. doi: 10.1038/s43018-019-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N Kalimuthu S., Wilson G.W., Grant R.C. Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut. 2019 doi: 10.1136/gutjnl-2019-318217. gutjnl-2019-318217. [DOI] [PubMed] [Google Scholar]
- 6.Duconseil P., Gilabert M., Gayet O. Transcriptomic analysis predicts survival and sensitivity to anticancer drugs of patients with a pancreatic adenocarcinoma. Am J Pathol. 2015;185(4):1022–1032. doi: 10.1016/j.ajpath.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Nicolle R., Blum Y., Marisa L. Pancreatic Adenocarcinoma Therapeutic Targets Revealed by Tumor-Stroma Cross-Talk Analyses in Patient-Derived Xenografts. Cell Rep. 2017;21(9):2458–2470. doi: 10.1016/j.celrep.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomberk G., Blum Y., Nicolle R. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullard J.H., Purdom E., Hansen K.D., Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11(1) doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puleo F., Nicolle R., Blum Y. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology. 2018;155(6):1999–2013. doi: 10.1053/j.gastro.2018.08.033. .e3. [DOI] [PubMed] [Google Scholar]
- 11.Rashid N.U., Peng X.L., Jin C. Purity Independent Subtyping of Tumors (PurIST), A Clinically Robust, Single-sample Classifier for Tumor Subtyping in Pancreatic Cancer. Clinical Cancer Research. 2019 doi: 10.1158/1078-0432.CCR-19-1467. 1078-0432.CCR-19-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffitt R.A., Marayati R., Flate E.L. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biton A., Bernard-Pierrot I., Lou Y. Independent Component Analysis Uncovers the Landscape of the Bladder Tumor Transcriptome and Reveals Insights into Luminal and Basal Subtypes. Cell Rep. 2014;9(4):1235–1245. doi: 10.1016/j.celrep.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Sompairac N., Nazarov P.V., Czerwinska U. Independent Component Analysis for Unraveling the Complexity of Cancer Omics Datasets. Int J Mol Sci. 2019;20(18):4414. doi: 10.3390/ijms20184414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satelli A., Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Australian Pancreatic Cancer Genome I., Bailey P., Chang D.K. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 17.Chan-Seng-Yue M., Kim J.C., Wilson G.W. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020;52(2):231–240. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 18.Aung K.L., Fischer S.E., Denroche R.E. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: early Results from the COMPASS Trial. Clinical Cancer Research. 2018;24(6):1344–1354. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juiz N., Elkaoutari A., Bigonnet M. Basal-like and Classical cells coexistence in pancreatic cancer revealed by single cell analysis. bioRxiv. 2020 doi: 10.1096/fj.202000363RR. [DOI] [PubMed] [Google Scholar]
- 20.Porter R.L., Magnus N.K.C., Thapar V. Epithelial to mesenchymal plasticity and differential response to therapies in pancreatic ductal adenocarcinoma. Proceedings of the National Academy of Sciences. 2019;116(52):26835–26845. doi: 10.1073/pnas.1914915116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng X.L., Moffitt R.A., Torphy R.J., Volmar K.E., Yeh J.J. De novo compartment deconvolution and weight estimation of tumor samples using DECODER. Nat Commun. 2019;10(1):4729. doi: 10.1038/s41467-019-12517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollberg N., Rahbari N.N., Koch M. Arterial Resection During Pancreatectomy for Pancreatic Cancer: a Systematic Review and Meta-Analysis. Ann. Surg. 2011;254(6):882–893. doi: 10.1097/SLA.0b013e31823ac299. [DOI] [PubMed] [Google Scholar]
- 23.Bachellier P., Addeo P., Faitot F., Nappo G., Dufour P. Pancreatectomy With Arterial Resection for Pancreatic Adenocarcinoma: how Can It Be Done Safely and With Which Outcomes? Ann. Surg. 2018:1. doi: 10.1097/SLA.0000000000003010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.