Abstract
Symbiodiniaceae dinoflagellates are essential endosymbionts of reef building corals and some other invertebrates. Information of their genome structure and function is critical for understanding coral symbiosis and bleaching. With the rapid development of sequencing technology, genome draft assemblies of several Symbiodiniaceae species and diverse marine algal genomes have become publicly available but spread in multiple separate locations. Here, we present a Symbiodiniaceae and Algal Genomic Resource Database (SAGER), a user-friendly online repository for integrating existing genomic data of Symbiodiniaceae species and diverse marine algal gene sets from MMETSP and PhyloDB databases. Relevant algal data are included to facilitate comparative analyses. The database is freely accessible at http://sampgr.org.cn. It provides comprehensive tools for studying gene function, expression and comparative genomics, including search tools to identify gene information from Symbiodiniaceae species, and BLAST tool to find orthologs from marine algae and protists. Moreover, SAGER integrates transcriptome datasets derived from diverse culture conditions of corresponding Symbiodiniaceae species. SAGER was developed with the capacity to incorporate future Symbiodiniaceae and algal genome and transcriptome data, and will serve as an open-access and sustained platform providing genomic and molecular tools that can be conveniently used to study Symbiodiniaceae and other marine algae.
Database URL: http://sampgr.org.cn
Introduction
Symbiodiniaceae, symbiotic dinoflagellates, are well known as essential endosymbionts of reef building corals and some other invertebrates (1). They vary in the diversity and abundance with different hosts and environments (1–3). For example, the dominant Symbiodiniaceae species can shuffle during the process of coral bleaching (4, 5). Under environmental stress conditions, Symbiodiniaceae are expelled from their hosts resulting in coral bleaching (6). In face of stress resulting from climate change and anthropogenic disturbance, there has been increasingly widespread and severe coral degradation in recent decades, largely due to the disruption of the coral-dinoflagellate symbiosis (i.e. coral bleaching) (7, 8). To understand molecular mechanisms underpinning symbiosis and its disruption and develop strategies to conserve coral reefs, there have been dedicated efforts employing the high-throughput “omics” technologies to identify genomic and genetic elements associated with these processes (9–13). For example, studies have shown that Symbiodiniaceae evolutionarily expanded genes functioning in nutrient uptake (14), transmembrane transport, and combat of reactive oxygen species and UV radiations (9).
Next generation sequencing has benefited Symbiodiniaceae genomic and transcriptomic studies. So far, eight assemblies of six Symbiodiniaceae species have been reported, including the genome of Breviolum minutum (LaJeunesse, J.E.Parkinson & J.D.Reimer) J.E.Parkinson & LaJeunesse (formerly Clade B, Symbiodinium minutum) (15), Fugacium kawagutii (Trench & R.J.Blank ex LaJeunesse) LaJeunesse (formerly Clade F, S. kawagutii) (14, 16, 17), S. microadriaticum LaJeunesse (formerly Clade A) (18), Cladocopium goreaui (Trench & R.J.Blank ex LaJeunesse) LaJeunesse & H.J.Jeong (formerly Clade C, S. goreaui) (16), Symbiodinium sp. Trench & R.J.Blank ex LaJeunesse (formerly Clade A3) and Cladocopium sp. (Trench & R.J.Blank ex LaJeunesse) LaJeunesse & H.J.Jeong (formerly Clade C92) (19). However, these resources have not been integrated into a centralized database furnished with analysis tools that is publicly available for comparative genomics and other symbiosis-related studies.
In this paper, we present the Symbiodiniaceae and Algal Genomic Resource Database (SAGER), which integrates Symbiodiniaceae genome resources and provides homology search tools for marine algae and protists. SAGER is aimed to serve as a data resource and user-friendly platform for coral and algal research, and to facilitate comparative genomic analyses. Along with the database, tools such as keyword search, BLAST (20, 21), JBrowse (22) and download will allow Symbiodiniaceae and marine alga researchers to perform various tasks including manual check of gene model annotation and obtaining updates of marine algal genomic resources.
Database overview
The SAGER integrates Symbiodiniaceae genomic and transcriptomic datasets. This includes assembled genomes, CDS and amino acid sequences, re-annotated gene annotation and gene expression from different culture conditions of six Symbiodiaceae species (B. minutum, F. kawagutii, S. microadriaticum, C. goreaui, Symbiodinium sp. and Cladocopium sp). Keyword search using gene ID, located scaffold, and ID number or any keywords from functional annotation can facilitate users finding target genes in batch. These data can also be downloaded easily and explored online using JBrowse and BLAST tools. The interaction of the various tool pages facilitates researchers to study gene function, expression and comparative genomics. In addition, to meet the needs for comparative studies, the database also integrates two other existing marine plankton genomic resource database – Marine Microbial Eukaryotic Transcriptome Sequencing Project (MMETSP) database (23, 24) & PhyloDB (version 1.076), which can also be downloaded or explored via homology search with BLAST tool.
Database organization
The SAGER is an extensive open-access database that houses Symbiodiniaceae and marine algal genomic data. It was developed in a user-friendly and online mode, consisting of Search, BLAST, JBrowse and interlinked Gene Information components.
Gene information
The Gene Information section acts as a core linking component within SAGER, which can be accessed through Search, BLAST and JBrowse (Figure 1). This component displays information for each of the genes from all six Symbiodiniaceae species included in this work, including gene location, gene structure, gene description based on NR annotation, GO and Swiss-Prot annotation, gene expression under different culture conditions, sequences of gene, CDS and amino acid in FASTA format. The most recently available genome version 3 of F. kawagutii also includes annotations from databases of NT, KEGG pathway, eggnog (KOG) and PFAM. To better visualize gene structure, full-screen view of gene structure in Gene Information component is linked to JBrowse. Additionally, accession numbers included in the annotation information are linked to their corresponding source databases: NR, NT, Swiss-Prot, GO and KEGG.
Figure 1.
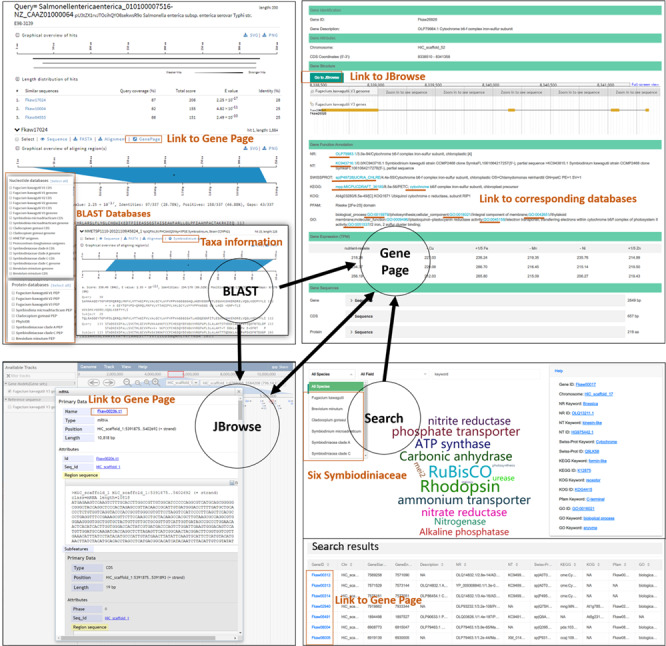
Database organization.
Search
The Search component allows users to retrieve gene information with keywords. The history of searched words is recorded, updated real time, and displayed in word cloud image in the middle of search page. The font size of the displayed word is proportional to the number of times the word has been searched for. Gene ID, scaffold ID, the functional description and accession number from NR, GO and Swiss-Psrot databases of genes from all Symbiodiniaceae species are provided in the Search component. For F. kawagutii genome information, users can search for genes using any keywords including accession number and functional annotations from other resources such as KEGG, NT, KOG and PFAM. The search results displayed include all returned genes with their information in list format which can be alternatively selected or filtered for download. The Detail in the column of the search results is linked to Gene Information.
BLAST server
To facilitate sequence similarity search, we integrated the SequenceServer (http://sequenceserver.com/) (25) into SAGER to enable BLAST analysis (20, 21) and visually display BLAST results. This tool displays alignment results in visual graphics which can be downloaded in SVG and PNG formats. It also provides direct download of sequence and alignment from the result page. Diverse datasets are provided in the BLAST server, including all Symbiodiniaceae genome assembly sequences and the two marine plankton genomic resources – augmented MMETSP (23, 24) and PhyloDB databases. To determine organism source of matched subject gene from MMETSP and PhyloDB, we developed a set of scripts using Ruby and Linux shell programming to list taxon information under the line of subject gene ID at the BLAST output page. This allows users to easily track the taxonomic affiliation of the source organism of the target gene. In addition, the subject ID from Symbiodiniaceae species on the result page can be linked to the corresponding Gene Information page.
Genome visualization
Symbiodiniaceae genomes are displayed using JBrowse, a tool designed for genome information visualization (22). We also integrated gene expression evidence of F. kawagutii version 3 from a transcriptome of mixed samples from different trace metal conditions. Users can conveniently view genomic scaffolds at any sequence location. This enables users to simultaneously view gene sets and gene expression evidence for manually checking gene structure.
Download page
In the Download page, Symbiodiniaceae genome datasets and metadata of the augmented MMETSP and PhyloDB are available there for users to download. The downloadable genome resources include genome assembly, GFF3 file, protein and CDS sequences, gene annotation files and gene expression from transcriptomes. The word ‘reference’ is linked to their corresponding publications.
Database sources
The SAGER database incorporates data of Symbiodiniaceae species and marine algae including: ① genomes (Table 1) and transcriptomes (Table S1) of six Symbiodiniaceae species including B. minutum (strain Mf1.05b, isolated from Florida Keys) (15, 26), F. kawagutii (strain CCMP2468, isolated from Hawaiian) (14, 16, 17, 27, 28), S. microadriaticum (strain CCMP2467, isolated from Gulf of Aqaba) (18, 29), C. goreaui (strain CCMP2466, isolated from Magnetic Island) (11, 16), Symbiodinium sp. (strain Y106, isolated from Okinawa) and Cladocopium sp. (strain Y103, isolated from Okinawa) (19); ② de novo transcriptome assembly of 302 marine microeukaryotic species from the MMETSP database (23, 24) and four dinoflagellates from our laboratory involving Prorocentrum shikokuense Hada (P. donghaiense Lu), Karenia mikimotoi (Miyake & Kominami ex Oda) Gert Hansen & Moestrup (30), Effrenium sp. (Trench & R.J.Blank ex LaJeunesse) LaJeunesse & H.J.Jeong (formerly Clade E) and Karlodinium veneficum (D.Ballantine) J.Larsen; and ③ Protein sequences of marine plankton from PhyloDB (version 1.076), which includes Eukaryota, Bacteria, Archaea and Viruses, and above-mentioned four dinoflagellates.
Table 1.
Publicly available genomes of Symbiodiniaceae
| Species | Strain | Isolation source | Clade | Assembly size (Mbp) | Gene No. | Gene symbol prefix | Reference |
|---|---|---|---|---|---|---|---|
| B. minutum | Mf1.05b | Florida Keys | B | 616 | 41 925 | symbB | (15) |
| F. kawagutii | CCMP2468, CS-156 | Hawaiian | F | 935 | 36 850 | Skaw | (14) |
| 1050 | 26 609 | SymbF | (16) | ||||
| 937 | 45 192 | Fkaw | (17) | ||||
| S. microadriaticum | CCMP2467 | Gulf of Aqaba | A | 808 | 49 109 | Smic | (18) |
| C. goreaui | CCMP2466, SCF055 | Magnetic Island | C1 | 1030 | 35 913 | SymbC1 | (16) |
| Symbiodinium sp. | Y106 | Okinawa | A3 | 767 | 71 632 | SymA3 | (19) |
| Cladocopium sp. | Y103 | Okinawa | C92 | 705 | 68 732 | SymC | (19) |
Symbiodiniaceae genome assemblies and transcriptomes
The first draft assembly of Symbiodiniaceae species was the genome of B. minutum (15), which was followed by that of F. kawagutii (14, 16, 17), S. microadriaticum (18), C. goreaui (16), Symbiodinium sp. and Cladocopium sp. (19) (Table 1). Among them, F. kawagutii has been updated after the initial release in 2015. Following its first revision (14), the second revision has just been published (16). In this version 3 (17), the genome assembly showed a N50 > 13 Mbp and the longest scaffold of 121 Mb, likely complete or nearly complete chromosome, and the number of predicted genes increased from 36 850 to 45 192. This is so far the best assembled genome of Symbiodiniaceae. Except F. kawagutii genes that already had the most recent annotation information, reannotation was carried out for all Symbiodiniaceae genes using Diamond blastx (E value =1E-5) to search against NCBI NR (updated on June 30, 2019) and Swiss-Prot databases (updated on March 20, 2020). And Blast2GO (31) was used to obtain the Gene Ontology (GO) annotation based on NR annotation. For those genes which have multiple transcript sequences, we selected the longest one to represent the gene in CDS and amino acid sequence files.
We also integrated published transcriptomic data of six Symbiodiniaceae species into SAGER (Table S1). Gene expression of each transcriptome was calculated based on raw sequencing data using same methods. Firstly, raw next-generation sequencing data were downloaded from the NCBI SRA database. Secondly, quality trimming was conducted on raw reads to remove poor quality data using Trimmomatic (32) with parameters setting as: LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 MINLEN:50. Finally, the trimmed clean reads were mapped to corresponding genome reference using Bowtie2 (33) and counted using RSEM software (34). Gene expression was normalized as Transcripts Per Million (TPM), and averaged across biological replicates. Totally, there are 66 transcriptomes of six Symbiodiaceae from different culture conditions (Table S1). B. minutum transcriptome was from cultures grown under normal conditions (L1 medium, 26°C) (26). Transcriptomes of F. kawagutii were from cultures grown under normal conditions (L1 medium, 25°C), heat stress, phosphate deprivation, organic phosphorus (OP) as P-source (Gro3P replacement) (27), and different trace metal conditions, including normal (L1 medium, 26°C) and reduced concentrations of Cu, Fe, Mn, Ni and Zn (28). S. microadriaticum transcriptomes represent samples grown under cold shock, cold stress, heat stress, heat shock, hyposalinity, hypersalinity, dark stress, dark cycle and control (12 h/12 h day/night cycle, 23°C) conditions (29). C. goreaui transcriptomes were from samples collected under control culture condition (27°C) on day 1, 32°C on day 9, 27°C on day 9, 32°C on day 13 and 27°C on day 13 (11). Symbiodinium sp. and Cladocopium sp. each has five transcriptomes under control culture condition (0 h and 48 h, 25°C), dark (48 h), heat stress (48 h) and heat stress in the dark (48 h) (19).
Marine phytoplankton transcriptome de novo assemblies
The MMETSP dataset contains transcriptomes from samples of marine eukaryotes representing more than 40 phyla (24). In SAGER, we integrated 678 transcriptomic de-novo re-assemblies from MMESTP representing 302 marine phytoplankton species (23). In addition, four dinoflagellate transcriptomes not covered in MMETSP that were sequenced in our laboratory were also included to expand species diversity, involving P. shikokuense (unpublished), K. mikimotoi (30), Effrenium sp. (unpublished) and K. veneficum (unpublished). P. shikokuense transcriptomes include non-redundant unigenes that merged with transcriptomic assemblies of culture samples collected at cell cycle phases S, M, G1 and G2, and grown under conditions of nitrogen (N)-replete, N-depleted, phosphorus (P)-depleted and P- replete using CD-HIT (35). K. mikimotoi unigenes were differentially expressed genes profiled from cultures grown in L1 medium (+P) and ATP-replacing-DIP medium (ATP) compared with DIP-depleted L1 medium (–P) using suppression subtractive hybridisation (SSH) followed by 454 pyrosequencing (30). Effrenium sp. transcriptomes combined unigenes data incorporating cultures grown at 20°C, 26°C and 33°C. Unigenes of K. veneficum were combination of splice leader-based transcriptomes of cultures grown in L1 medium and under mixotrophic condition, sampled during day (illuminated) and night (dark) time points. These resulted in 682 transcriptomic assembly of 306 phytoplankton species, containing 32 677 544 unigenes. These unigenes were annotated using Diamond blastx (E value =1E-5) against NCBI NR database (updated on June 30, 2019). Among them, 11 947 623 (36.56%) had functional annotation.
Marine plankton protein dataset
PhyloDB is a database suitable for comprehensive annotation of metagenomics and metatranscriptomics analyses (36), which is comprised of protein sequences from KEGG, GenBank, JGI, ENSEMBL, and initial assembled MMETSP databases (24). This dataset (version 1.076) was downloaded from https://scripps.ucsd.edu/labs/aallen/data/ (see “Databases and Collections”). Although the MMETSP assemblies in PhyloDB were older than the above re-assembled version, we still keep the old version in PhyloDB to maintain the species diversity in this dataset. However, we augmented the dataset by incorporating the protein sequences predicted from the above-mentioned four additional datasets of dinoflagellate unigenes using TransDecoder v5.5.0 (https://github.com/TransDecoder/TransDecoder/releases). As a result, the PhyloDB in SAGER consists of 29 690 621 protein sequences of 25 996 species, including 894 of Eukaryota, 4910 of Bacteria, 230 of Archaea and of 19 962 Viruses.
System implementation
SAGER was developed with combination of several tools and scripts (Figure 2). Genomic data were imported and managed with MySQL. The Nginx web server was used to construct the underlying web server. Tools of Joolma! Content Management System (CMS) and LayUI were used to build home and download pages, and new components of gene page and search tool. Third party tools of SequenceServer (25) and JBrowse (22) were used to construct BLAST and Genome Browser tools. PHP and JavaScript were applied to make the pages flexible and interactive.
Figure 2.
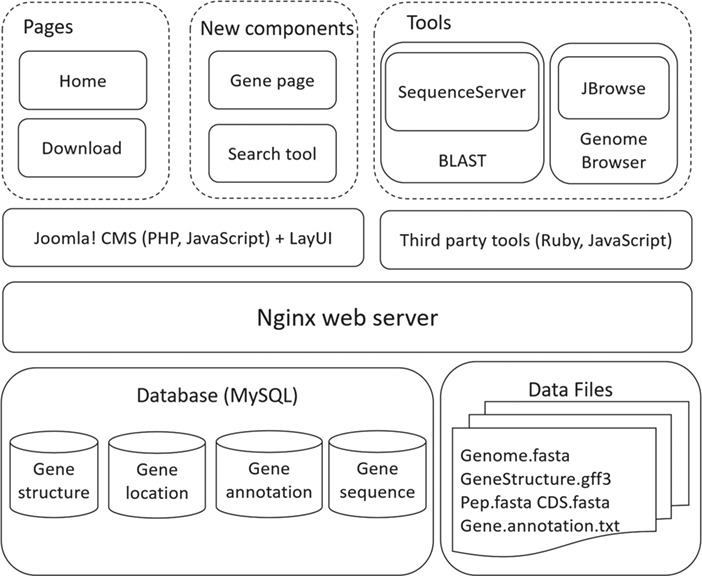
Architecture of SAGER.
Future work
SAGER is aimed to be a user-friendly database and tool resource, which integrates currently available Symbiodiniaceae genome data, marine phytoplankton genome resources, and analysis as well as visualization tools. It is worth noting that we used Symbiodiniaceae genomic resources that were generated by different research groups using slightly different approaches. Therefore, users of our integrated data should keep in mind that variations are likely to occur between datasets, which will affect comparative analyses between Symbiodiniaceae genomes, as recently demonstrated (37). Furthermore, the database was designed with room to accommodate and house newly generated data. We will continue to update and upgrade the data resources. Future updates will cover transcriptomes of Symbiodiniaceae and relevant cultured marine phytoplankton and field-collected samples such as coral holobiont (38–40) or harmful algal bloom metatranscriptomes (41, 42). To better support the capability of SAGER to serve the research community, new web tools will be developed to allow more efficient and effective use of this database.
Supplementary Material
Acknowledgements
We thank Likun Wang, Jiqiang Yan and Weiqi Tang for their assistance in database development. We also appreciate the availability of marine phytoplankton transcriptomes from the Marine Microbial Eukaryotic Transcriptome Sequencing Project (MMETSP) and these data’s reassembling by UC Davis as well as PhyloDB dataset from the laboratory of A.E. Allen (https://scripps.ucsd.edu/labs/aallen/data/).
Funding
This work was supported by State Key Laboratory of Marine Environmental Science, Xiamen University. This work was also supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) [No.2018SDKJ0406–3], and Natural Science Foundation of China [grant NSFC 31661143029 and 41776116].
Conflict of interest. None declared.
Reference
- 1. Baker A.C. (2003) Flexibility and specificity in coral–algal symbiosis, diversity, ecology and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst., 34, 661–689. [Google Scholar]
- 2. Quigley K.M., Davies S.W., Kenkel C.D. et al. (2014) Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS One, 9, e94297–e94297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LaJeunesse T.C., Bhagooli R., Hidaka M. et al. (2004) Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar. Ecol. Prog. Ser., 284, 147–161. [Google Scholar]
- 4. Baker A.C. (2001) Reef corals bleach to survive change. Nature, 411, 765–766. [DOI] [PubMed] [Google Scholar]
- 5. Grottoli A.G., Warner M.E., Levas S.J. et al. (2014) The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Glob. Chang. Biol., 20, 3823–3833. [DOI] [PubMed] [Google Scholar]
- 6. Hoegh-Guldberg O. (1999) Climate change, coral bleaching and the future of the world's coral reefs. Mar. Freshw. Res., 50, 839–866. [Google Scholar]
- 7. Hoegh-Guldberg O. (2011) The Impact of Climate Change on Coral Reef Ecosystems In: Dubinsky Z, Stambler N (eds). Coral Reefs: An Ecosystem in Transition. Springer Netherlands, Dordrecht, pp. 391–403. [Google Scholar]
- 8. Hoegh-Guldberg O. (2011) Coral reef ecosystems and anthropogenic climate change. Reg. Environ. Change, 11, 215–227. [Google Scholar]
- 9. Gonzalez-Pech R.A., Ragan M.A. and Chan C.X. (2017) Signatures of adaptation and symbiosis in genomes and transcriptomes of Symbiodinium. Sci. Rep., 7, 15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shinzato C., Mungpakdee S., Satoh N. et al. (2014) A genomic approach to coral-dinoflagellate symbiosis: studies of Acropora digitifera and Symbiodinium minutum. Front. Microbiol., 5, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levin R.A., Beltran V.H., Hill R. et al. (2016) Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol., 33, 2201–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer E. and Weis V.M. (2012) Study of cnidarian-algal symbiosis in the "omics" age. Biol. Bull., 223, 44–65. [DOI] [PubMed] [Google Scholar]
- 13. Xiang T., Nelson W., Rodriguez J. et al. (2015) Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J., 82, 67–80. [DOI] [PubMed] [Google Scholar]
- 14. Lin S., Cheng S., Song B. et al. (2015) The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science, 350, 691–694. [DOI] [PubMed] [Google Scholar]
- 15. Shoguchi E., Shinzato C., Kawashima T. et al. (2013) Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol., 23, 1399–1408. [DOI] [PubMed] [Google Scholar]
- 16. Liu H., Stephens T.G., González-Pech R.A. et al. (2018) Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol., 1, 95–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T., Yu L., Song B. et al. (2020) Genome Improvement and Core Gene Set Refinement of Fugacium kawagutii. Microorganisms, 8, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aranda M., Li Y., Liew Y.J. et al. (2016) Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep., 6, 39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shoguchi E., Beedessee G., Tada I. et al. (2018) Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics, 19, 458–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Camacho C., Coulouris G., Avagyan V. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S.F., Gish W., Miller W. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22. Buels R., Yao E., Diesh C.M. et al. (2016) JBrowse: a dynamic web platform for genome visualization and nalysis. Genome Biol., 17, 66–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson L.K., Alexander H. and Brown C.T. (2019) Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. GigaScience, 8, giy158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keeling P.J., Burki F. and lcox H.M., et al. (2014) The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol., 12, e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Priyam A., Woodcroft B.J., Rai V. et al. (2019) Sequenceserver: A modern graphical user interface for custom BLAST databases. Mol. Biol. Evol., 36, 2922–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parkinson J.E., Baumgarten S., Michell C.T. et al. (2016) Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome. Biol. Evol., 8, 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin S., Yu L. and Zhang H. (2019) Transcriptomic responses to thermal stress and varied phosphorus conditions in Fugacium kawagutii. Microorganisms, 7, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li T., Lin X., Yu L. et al. (2020) RNA-seq profiling of Fugacium kawagutii reveals strong responses in metabolic processes and symbiosis potential to deficiencies of iron and other trace metals. Sci. Total Environ., 705, 135767. [DOI] [PubMed] [Google Scholar]
- 29. Baumgarten S., Bayer T., Aranda M. et al. (2013) Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics, 14, 704–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luo H., Lin X., Li L. et al. (2017) Transcriptomic and physiological analyses of the dinoflagellate Karenia mikimotoi reveal non-alkaline phosphatase-based molecular machinery of ATP utilisation. Environ. Microbiol., 19, 4506–4518. [DOI] [PubMed] [Google Scholar]
- 31. Conesa A., Götz S., García-Gómez J.M. et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- 32. Bolger A.M., Lohse M. and Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B. and Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li B. and Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu L., Niu B., Zhu Z. et al. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kolody B.C., McCrow J.P., Allen L.Z. et al. (2019) Dieltranscriptional response of a California Current plankton microbiome to light, low iron, and enduring viral infection. ISME J., 13, 2817–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Y., González-Pech R.A., Stephens T.G. et al. (2020) Evidence that inconsistent gene prediction can mislead analysis of dinoflagellate genomes. J. Phycol., 56, 6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robbins S.J., Singleton C.M., Chan C.X. et al. (2019) A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nature Microbiol., 4, 2090–2100. [DOI] [PubMed] [Google Scholar]
- 39. Yuyama I., Ishikawa M., Nozawa M. et al. (2018) Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci. Rep., 8, 16802–16802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maor-Landaw K., Oppen M.J.H. and McFadden G.I. (2019) Symbiotic lifestyle triggers drastic changes in the gene expression of the algal endosymbiont Breviolum minutum (Symbiodiniaceae). Ecol. Evol., 10, 451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu L., Zhang Y., Li M. et al. (2020) Comparative metatranscriptomic profiling and microRNA sequencing to reveal active metabolic pathways associated with a dinoflagellate bloom. Sci. Total Environ., 699, 134323. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y., Lin X., Shi X. et al. (2019) Metatranscriptomic signatures associated with phytoplankton regime shift from diatom dominance to a dinoflagellate bloom. Front. Microbiol., 10, 590–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


