Abstract
Objective
Coronavirus Disease 2019 (COVID-19) is a pandemic. This systematic review compares mortality risk factors including clinical, demographic and laboratory features of COVID-19, Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS). The aim is to provide new strategies for COVID-19 prevention and treatment.
Methods
We performed a systematic review with meta-analysis, using five databases to compare the predictors of death for COVID-19, SARS and MERS. A random-effects model meta-analysis calculated odds ratios (OR) and 95% confidence intervals (95% CI).
Results
845 articles up through 11/4/2020 were retrieved, but only 28 studies were included in this meta-analysis. The results showed that males had a higher likelihood of death than females (OR = 1.82, 95% CI 1.56–2.13). Age (OR = 7.86, 95% CI 5.46–11.29), diabetes comorbidity (OR = 3.73, 95% CI 2.35–5.90), chronic lung disease (OR = 3.43, 95% CI 1.80–6.52) and hypertension (OR = 3.38, 95% CI 2.45–4.67) were the mortality risk factors. The laboratory indicators lactic dehydrogenase (OR = 37.52, 95% CI 24.68–57.03), C-reactive protein (OR = 12.11, 95% CI 5.24–27.98), and neutrophils (OR = 17.56, 95% CI 10.67–28.90) had stronger correlations with COVID-19 mortality than with SARS or MERS mortality. Consolidation and ground-glass opacity imaging features were similar among COVID-19, SARS, and MERS patients.
Conclusions
COVID-19′s mortality factors are similar to those of SARS and MERS. Age and laboratory indicators could be effective predictors of COVID-19 mortality outcomes.
Keywords: COVID-19, SARS, MERS, Mortality, Risk factors, Meta-analysis
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a novel zoonotic pathogen. It is responsible for Coronavirus Disease 2019 (COVID-19).1 The COVID-19 pandemic has imposed a heavy burden on global health and medical systems.2 A whole genome scan has shown that SARS-CoV-2 has a 79% similarity to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), and a 50% similarity to Middle East Respiratory Syndrome Coronavirus (MERS-CoV).3 Currently, SARS-CoV-2 is classified as a SARS-associated coronavirus and taxonomically belongs to the subgenus of Sabek virus.3 SARS-CoV-2 is the 7th coronavirus outbreak, following HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV, and MERS-CoV.4 Of the 7 coronaviruses, the former four can cause common cold symptoms, but SARS-CoV-2, SARS-CoV, and MERS-CoV can lead to severe respiratory syndromes, with about 6.76%, 9.6%, and 35.5% mortality rates, respectively.2 , 4
Several similarities in the pathogenicity, clinical features and transmissibility of COVID-19, SARS, and MERS have been identified.5 The median age of COVID-19 cases ranges from 49 to 57 years, similar to those of SARS and MERS.5 The most common presenting symptom in COVID-19, SARS and MERS is fever, followed by cough, sore throat and dyspnea.5 Laboratory findings in patients diagnosed with COVID-19 differ little from those of patients diagnosed with SARS or MERS. Lymphopenia is the most common finding. Other findings included low platelet count, decreased albumin level and increased aminotransferases, lactic dehydrogenase (LDH), creatine kinase and C-reactive protein (CRP) levels5. Radiological presentation of COVID-19 differs little from the other two coronavirus-associated pneumonias.5 Furthermore, SARS-CoV-2 has the same human cell receptors as SARS-CoV, the angiotensin-converting enzyme 2 (ACE2).6 These features suggest that COVID-19, SARS, and MERS may have common mortality risk factors. There have only been a handful of case-control and cohort studies about COVID-19 mortality. This has resulted in poor COVID-19 mortality prognosis identification. Therefore, a systematic review of COVID-19, SARS, and MERS studies with mortality outcomes will provide perspective for understanding the clinical, laboratory and imaging features of COVID-19. In addition, a systematic review could compare and contrast the risk factors for in-hospital death among these three lethal coronaviruses. This will provide a new strategy for COVID-19 prevention and treatment.
Methods
Protocol and registration
This systematic review follows the methodological approaches outlined in the Cochrane Handbook for Systematic Reviews of Interventions version 6.0.7 It is described according to the PRISMA-P (Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocols) statement.8 This systematic review has been reported in the international Prospective Register of Systematic Reviews (PROSPERO) database (CRD42020180929).
Search strategy and selection criteria
We chose five online bibliographic databases (MEDLINE, EPISTEMONIKOS, COCHRANE, China National Knowledge Infrastructure and WANFANG STATA) as a basis for identifying the relevant literature up through April 11, 2020. The search string was: [“SARS” OR “Severe Acute Respiratory Syndrome virus” OR “Severe Acute Respiratory Syndrome related coronavirus” OR “MERS” OR “Middle Acute Respiratory Syndrome related coronavirus” OR “2019-nCoV” OR “novel coronavirus” OR “SARS-CoV-2″ OR “COVID-19″] AND [“prognosis” OR “prognostic factors” OR “factor, prognostic”]. In addition, we manually searched the included articles’ reference lists.
To determine eligibility, four independent researchers evaluated all studies retrieved by the search using their titles and abstracts. In cases of disagreement, a fifth investigator was consulted. A study's eligibility for inclusion in the meta-analysis was based on the following selection criteria: (I) Studies included patients infected by SARS-CoV-2, SARS-CoV, and MERS-CoV. (II) Patients in the studies had clinical outcomes including death. (III) Studies reported demographic characteristics, clinical manifestations, laboratory indicators and imaging. (IV) Articles were written in either Chinese or English. Conference papers, review, published letters, editorials, and studies including pregnant women or children were excluded. We also excluded studies if they were the only study reporting on a specific indicator. When there was overlap in studies’ patients (i.e. utilizing the same period, region, or hospital), we chose the study with the larger sample.
Data extraction and quality assessment
Four investigators independently extracted the following data from each included study and input them to a data extraction form: study name, author, research publication date, study design, study area and hospital or institute, number of patients, data source, demographic characteristics, clinical features, laboratory indicators and imaging, case definitions for COVID-19, SARS, and MERS. The fifth researcher checked the article list and data extraction to ensure there were no duplicate articles or duplicate patient data.
The quality of the studies included was independently appraised by four authors using the Newcastle-Ottawa Scale (NOS) for observational case-control and cohort studies.9 Observational studies were rated as high quality if they had a score of 6–9; moderate quality if they had a score of 4 or 5, and poor quality if their score was 3 or lower.
Statistical analysis
Results were merged across studies with STATA version 15.1 (Stata Corp MP., College Station, TX, USA).10 , 11 We applied a random-effects meta-analysis model, which accounted for the included studies’ populations being infected by different coronaviruses, as well as variation in the number of patients and their clinical stages. Estimating the common effect size was impossible in light of the studies’ heterogeneity. We calculated the standardized mean difference in laboratory indicators between the survivors and non-survivors of COVID-19, SARS and MERS, respectively. Then, to determine the strength of association for various indicators with clinical outcomes, estimates were pooled with generic inverse-variance, and expressed in odds ratios (OR) with 95% confidence intervals (CI) per factor. This follows the Cochrane handbook guidelines. Risk factors of COVID-19, SARS and MERS, including demographic characteristics, comorbidities, clinical manifestations and laboratory indicators, were ranked according to the absolute OR values shown in Table 1 . Subgroup analysis was done by COVID-19, SARS and MERS, respectively. Between-study heterogeneity was assessed using the I 2 statistic: I 2 values of 0%−39%, 40%−59%, 60%−90% were considered to indicate mild, moderate and severe heterogeneity, respectively. Forest plots are provided for indicators for which six or more studies were included in the meta-analysis. Publication bias was assessed with Egger's test.12 Sensitivity analysis was assessed by Duval and Tweedie's trim and fill.13 Exact p-value is given, unless it was less than 0.001. A p < 0.05 was considered statistically significant, except for Egger's regression test in which a p < 0.10 was considered statistically significant.
Table 1.
Risk factors for mortality of COVID-19, SARS, and MERS patients.
| Factors | COVID-19 |
SARS |
MERS |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of patients (No. of studies) | OR (95% CI)I2 | OR rank | No. of patients (No. of studies) | OR (95% CI)I2 | OR rank | No. of patients (No. of studies) | OR (95% CI)I2 | OR rank | |
| Demographical characteristics | |||||||||
| Age | 10,228 (9) | 6.45 (3.86, 10.77)c | – | 2439 (4) | 11.97 (8.82, 16.24)a | – | 360 (2) | 7.02 (4.59, 10.73)a | – |
| Gender | 927 (6) | 1.96 (1.43, 2.69)a | – | 2541 (6) | 1.81 (1.43, 2.30)a | – | 985 (2) | 1.74 (1.32, 2.30)a | – |
| Clinical manifestation | |||||||||
| Respiratory rate | 465 (2) | 4.90 (1.08, 22.24)c | – | 994 (2) | 8.88 (5.64, 13.97)a | – | – | – | – |
| Comorbidities | |||||||||
| Chronic lung disease | 2307 (5) | 4.75 (2.37, 9.52)a | 1 | 674 (2) | 3.09 (1.02, 9.35)a | 4 | 330 (1) | 1.38 (0.69, 2.74)NA | 4 |
| Comorbidity | 2517 (7) | 3.50 (2.35, 5.20)b | 2 | 3003 (3) | 6.47 (4.93, 8.50)a | 1 | 360 (2) | 3.08 (0.35, 27.01)c | 1 |
| Hypertension | 3342 (6) | 3.25 (2.15, 4.91)c | 3 | 1291 (1) | 4.70 (3.13, 7.07)NA | 2 | 128 (3) | 2.97 (1.32, 6.69)a | 2 |
| Diabetes | 2307 (5) | 2.63 (1.45, 4.76)c | 4 | 1986 (3) | 4.70 (3.13, 7.07)NA | 3 | 360 (2) | 2.57 (1.62, 4.10)a | 2 |
| Laboratory indicators | |||||||||
| LDH | 465 (2) | 37.52 (24.68, 57.03)a | 1 | 1962 (4) | 4.23 (2.22, 8.08)c | 4 | 330 (1) | 2.53 (1.67, 3.85)NA | 5 |
| Neutrophil | 274 (1) | 17.56 (10.67, 28.90)NA | 2 | 2032 (3) | 2.63 (2.06, 3.37)a | 6 | 48 (1) | 4.67 (1.60, 13.69)NA | 2 |
| CRP | 424 (2) | 12.11 (5.24, 27.98)c | 3 | 308 (1) | 8.47 (4.17, 17.21)NA | 1 | 30 (1) | 8.95 (1.60, 50.20)NA | 1 |
| WBC | 615 (3) | 9.13 (5.71, 14.59)b | 4 | 2039 (3) | 2.18 (1.46, 3.27)b | 8 | 360 (2) | 2.03 (1.36, 3.04)a | 7 |
| Albumin | 615 (3) | 0.11 (0.06, 0.19)c | 5 | 931 (1) | 0.13 (0.09, 0.20)NA | 2 | 30 (1) | 0.22 (0.04, 1.15)NA | 3 |
| BUN | 424 (2) | 8.49 (5.81, 12.40)a | 6 | 1089 (1) | 5.27 (3.59, 7.72)NA | 3 | 360 (2) | 3.89 (2.58, 5.88)a | 4 |
| Interleukin-6 | 615 (3) | 7.12 (3.65, 13.87)c | 7 | 50 (1) | 3.48 (1.11, 10.97)NA | 5 | – | – | – |
| Prothrombin time | 648 (3) | 7.08 (2.03, 24.75)c | 8 | 527 (1) | 2.61 (1.56, 4.38)NA | 7 | – | – | – |
| Lymphocyte | 615 (3) | 0.21 (0.12, 0.38)c | 9 | 2385 (4) | 0.55 (0.32, 0.95)c | 12 | – | – | – |
| Total bilirubin | 424 (2) | 4.07 (2.39, 6.94)b | 10 | 1451 (2) | 2.09 (0.88, 4.98)c | 9 | 330 (1) | 1.90 (1.26, 2.88)NA | 8 |
| Creatine | 615 (3) | 3.32 (2.24, 4.92)a | 11 | 520 (1) | 1.96 (1.37, 2.80)NA | 10 | 360 (2) | 2.09 (1.40, 3.13)a | 6 |
| AST | 424 (2) | 3.18 (0.68, 14.99)c | 12 | 520 (1) | 1.67 (1.17, 2.39)NA | 13 | 360 (2) | 1.32 (0.88, 1.97)a | 11 |
| Platelet | 615 (3) | 0.33 (0.24, 0.44)a | 13 | 1637 (2) | 0.81 (0.56, 1.18)b | 16 | 360 (2) | 0.60 (0.40, 0.89)a | 9 |
| ALT | 615 (2) | 2.26 (1.40, 3.63)b | 14 | 520 (1) | 1.45 (0.96, 2.18)NA | 14 | 360 (2) | 1.32 (0.88, 1.97)a | 10 |
| Creatine kinase | 615 (3) | 2.12 (1.07, 4.19)c | 15 | 674 (2) | 1.33 (0.65, 2.72)b | 15 | – | – | – |
| Hb | 615 (3) | 0.92 (0.68, 1.24)a | 16 | 1118 (1) | 0.52 (0.36, 0.76)NA | 11 | 360 (2) | 0.33 (0.22, 0.49)a | 5 |
| Serum ferritin | 615 (3) | 8.42 (4.21, 16.81)c | – | – | – | – | – | – | – |
| Procalcitonin | 465 (2) | 8.23 (5.20, 13.00)a | – | – | – | – | – | – | – |
| D-dimer | 648 (3) | 3.47 (1.75, 6.86)c | – | – | – | – | – | – | – |
| HSCT | 615 (3) | 2.43 (1.69, 3.51)a | – | – | – | – | – | – | – |
| APTT | 457 (2) | 1.61 (0.47, 5.54)c | – | – | – | – | – | – | – |
| CD3+ | – | – | – | 327 (2) | 0.20 (0.10, 0.38)a | – | – | – | – |
| CD4+ | – | – | – | 533 (3) | 0. 20 (0.12, 0.34)a | – | – | – | – |
| CD8+ | – | – | – | 533 (3) | 0.29 (0.15, 0.59)b | – | – | – | – |
OR = Odds Ratio. Pooled ORs are calculated on the bias of randomized effects. OR rank: Ranked according to the absolute OR values.
a = mild heterogeneity, b = moderate heterogeneity, c = severe heterogeneity, NA = Not Available.
LDH = Lactic Dehydrogenase, CRP = C-reactive Protein, WBC = White Blood Cell, BUN = Blood Urea Nitrogen, AST = Aspartate Transaminase, HSCT = High-sensitive Cardiac Troponin, ALT = Alanine Transaminase, Hb = Hemoglobin, APTT = Activated Partial Thromboplastin Time.
Results
Literature search, study characteristics and quality assessment
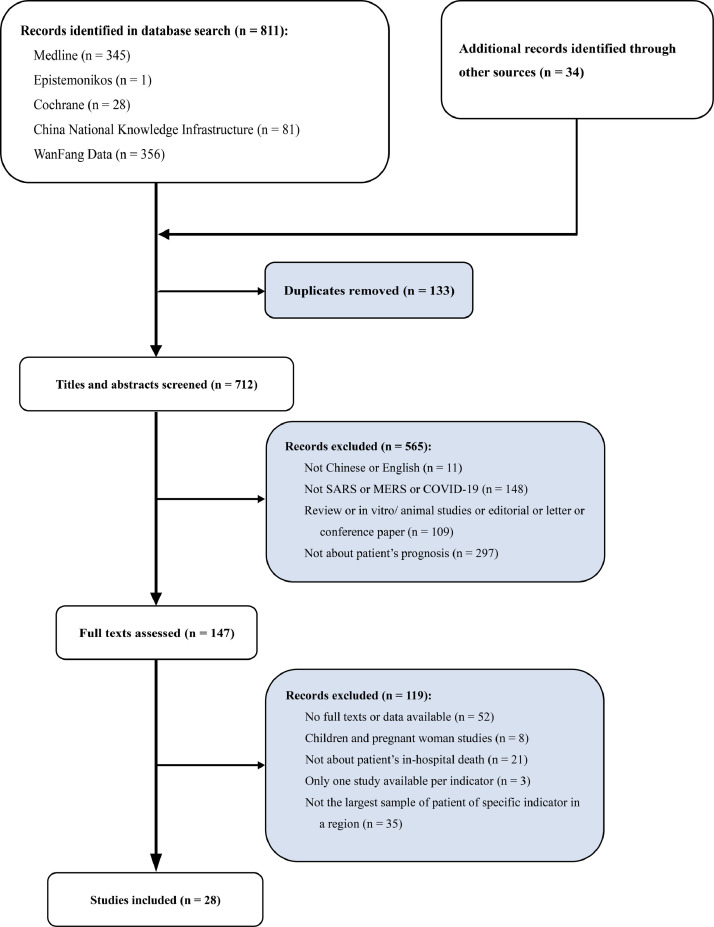
The database and manual searches identified 811 and 34 records, respectively. Subsequent screening of titles and abstracts led to 147 studies being deemed eligible for full texts assessment. Then, 119 publications were excluded because there was insufficient data (only median provided or p-value) and no full text available (n = 52), or the patients in the study had included children or pregnant women (n = 8), or they were unrelated to patient in-hospital death prognosis (n = 21), or the indicator was reported in one study (n = 3) (Fig. 1 ). Finally, there was considerable overlap in the studies in terms of regions and hospitals. In order to prevent research redundancy, we selected the 28 studies with the largest samples for our meta-analysis. This resulted in a total of 16,095 patients (COVID-19: 11,818 patients, 910 death; SARS: 3292 patients, 336 death; MERS: 985 patients, 465 death), including 1711 in-hospital death patients. The characteristics of the included 28 studies are summarized in supplementary Table 1.14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 These included studies were conducted in several coronavirus outbreak areas—Beijing, Guangdong, Shanxi, Hong Kong and Taiwan in China, and Toronto where SARS broke out (12 studies); Saudi Arabia and South Korea where MERS broke out (6 studies); China, Italy, South Korea and the United States where COVID-19 broke out (10 studies). Among these studies, 11 were case-control studies, 13 were cohort studies, and 4 were case series. The diagnostic criteria for COVID-19, SARS, and MERS were laboratory diagnostics, clinical characteristics and epidemiological investigation as issued by the World Health Organization (WHO). We analyzed 31 factors in the meta-analyses (Table 1). The NOS for observational case-control and cohort studies assessment were showed in supplementary Table 1. The 28 included studies scored between 5 and 9. None of the studies was considered to be poor quality.
Fig. 1.
Study selection flowchart, systematic review and meta-analysis of risk factors associated with COVID-19, SARS, and MERS patient death.
Factors associated with COVID-19, SARS, and MERS mortality demographical characteristics and comorbidities
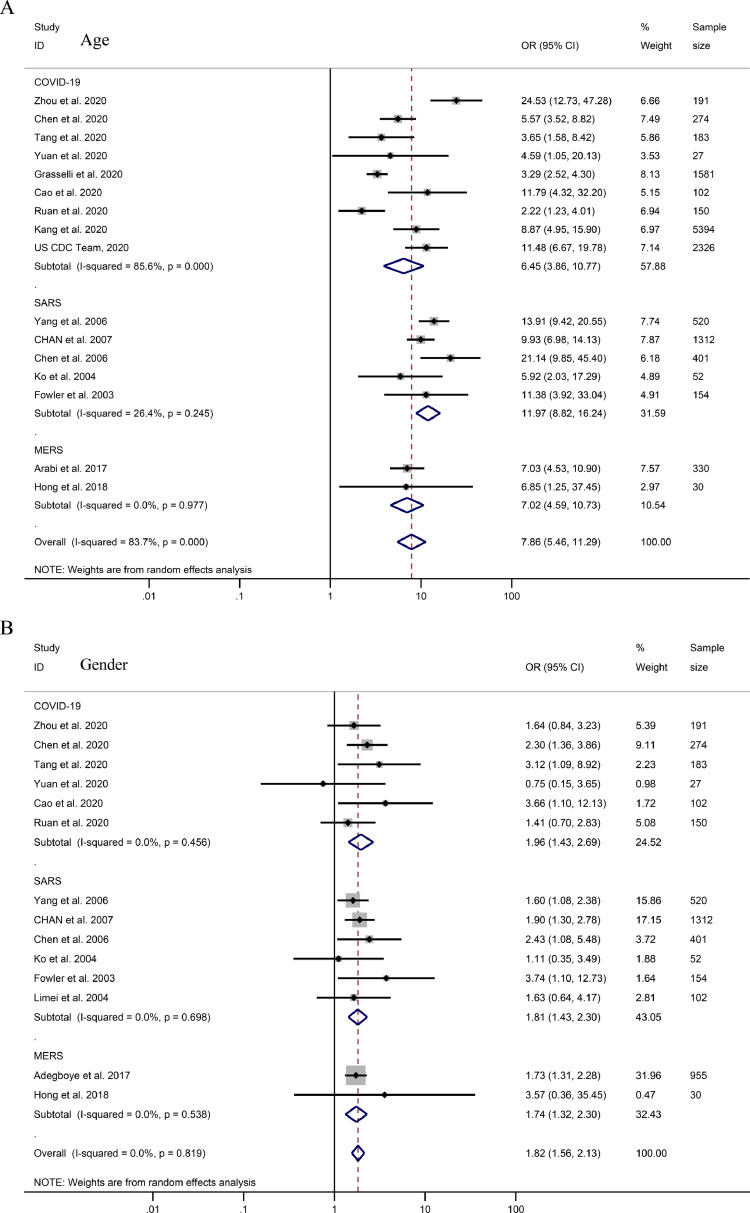
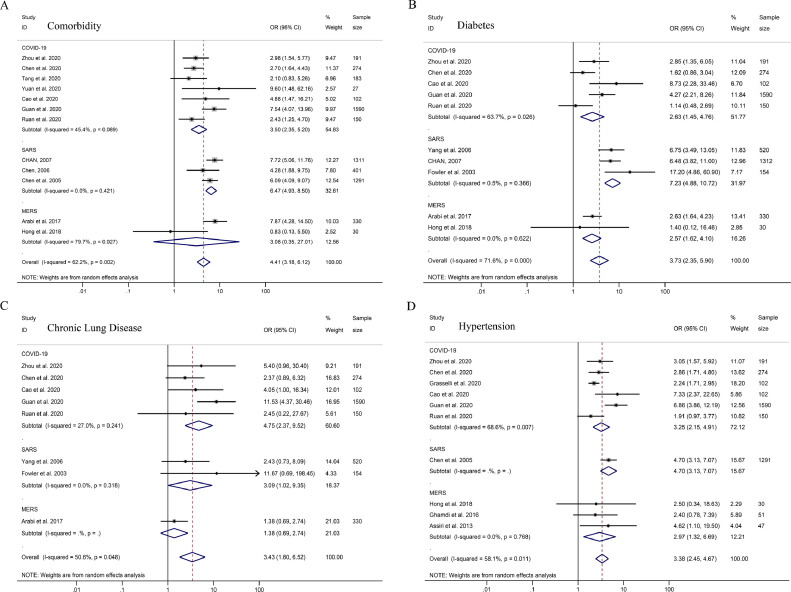
Elder patients were found to have higher mortality rates (Total OR = 7.86, 95% CI 5.46–11.29; COVID-19: OR = 6.45, 95% CI 3.86–10.77; SARS: OR = 11.97, 95% CI 8.82–16.24; MERS: OR = 7.02, 95% CI 4.59–10.73, Fig. 2 A and Table 1). Among patients died of COVID-19, SARS, or MERS, males had a higher mortality rate (total OR = 1.82, 95% CI 1.56–2.13; COVID-19: OR = 1.96, 95% CI 1.43–2.69; SARS: OR = 1.81, 95% CI 1.43–2.30; MERS: OR = 1.74, 95% CI 1.32–2.30, Fig. 2B). Patients with various comorbidities (Total OR = 4.41, 95% CI 3.18–6.12; COVID-19: OR = 3.50, 95% CI 2.35–5.20; SARS: OR = 6.47, 95% CI 4.93–8.50; MERS: OR = 3.08, 95% CI 0.35–27.01, Fig. 3 A) were found to have higher mortality rates for COVID-19, SARS, and MERS. The OR for patients’ mortality rates with comorbidities of diabetes, chronic lung disease and hypertension with COVID-19, SARS, and MERS are shown in Fig. 3. The comorbidities’ OR values (hypertension and diabetes) for COVID-19 mortality were lower than those for SARS (Fig. 3, Table 1).
Fig. 2.
ORs for demographic characteristics on COVID-19, SARS, and MERS mortality. Forest plots of pooled odds ratios for the strength of association between factors and mortality in COVID-19, SARS, and MERS patients. (A) Age, (B) Gender (male compare with female).
Fig. 3.
ORs for COVID-19, SARS, and MERS comorbidities. Forest plots of pooled odds ratios for the strength of association between factors and mortality in COVID-19, SARS, and MERS patients. (A) Comorbidity, (B) Diabetes, (C) Chronic Lung Disease, (D) Hypertension.
Clinical manifestation and laboratory indicators
Only clinical manifestations of respiratory rates were analyzed. The results showed that respiratory rate was a sensitive predictor of mortality for COVID-19 and SARS (COVID-19: OR = 4.90, 95% CI 1.08–22.24; SARS: OR = 8.88, 95% CI 5.64–13.97, Table 1). Laboratory indicators of mortality in COVID-19 patients are shown in Table 1. Mortality risk factors in COVID-19 patients included lower platelet (OR = 0.33, 95% CI 0.24–0.44) and lymphocyte counts (OR = 0.21, 95% CI 0.12–0.38), higher neutrophil (OR = 17.56, 95% CI 10.67–28.90) and white blood cell (WBC) counts (OR = 9.13, 95% CI 5.71–14.59), lower albumin levels (OR = 0.11, 95% CI 0.06–0.19) higher LDH (OR = 37.52, 95% CI 24.68–57.03), CRP (OR = 12.11, 95% CI 5.24–27.98), and blood urea nitrogen (BUN) (OR = 8.49, 95% CI 5.81–12.40) (Table 1).
We compared the six highest OR values for laboratory indicators of mortality from the non-survivors and survivors of COVID-19, SARS and MERS, respectively. This resulted in five similar laboratory indicators among these three coronavirus diseases, including LDH, neutrophils, CRP and BUN and albumin (Table 1). However, the WBC counts in the COVID-19 patients, the interleukin-6 (IL-6) levels in the SARS patients, and the creatine levels in the MERS patients varied between COVID-19, SARS and MERS. These results were statistically significant (Table 1). Moreover, the variation in laboratory indicators among the COVID-19 patients was higher than those of the SARS and MERS patients, with the exception of hemoglobin (Hb) (supplementary Figs. 1–2 and Table 1). Finally, lower lymphocyte subtype counts were mortality risk factors in SARS patients (CD3+: OR = 0.20, 95% CI 0.10–0.38; CD4+: OR = 0.20, 95% CI 0.12–0.34; CD8+: OR = 0.29, 95% CI 0.15–0.59).
Heterogeneity, publication bias and sensitivity analysis
The I2 statistics for age, respiratory rate, hypertension, diabetes and laboratory indicators of CRP, serum ferritin, IL-6, prothrombin time, d-dimer, aspartate transaminase (AST), creatine kinase, activated partial thromboplastin time (APTT), albumin and lymphocyte had severe heterogeneity between COVID-19 studies (Table 1). The laboratory indicators LDH, WBC and total bilirubin had severe heterogeneity among SARS studies (Table 1). Comorbidity had severe heterogeneity between MERS studies (Table 1). Publication bias was assessed with Egger's test. It suggested there was no evidence of publication bias (supplementary Table 2). For indicators ≥ 6 studies, sensitivity was analyzed with Duval and Tweedie's trim-and-fill test. The results showed that chronic lung disease and alanine aminotransferase (ALT) were not robust (supplementary Table 2).
Discussion
Millions of COVID-19 patients have been diagnosed worldwide, and it is a major burden on public health and medical systems around the world.2 Vaccines and specific drugs for COVID-19 are not yet available, and many countries have experienced collapses in their clinical medical systems. This has been a factor in COVID-19 becoming a pandemic.42 Therefore, to provide supportive treatment, COVID-19′s clinical manifestations, laboratory indicators and imaging features must be identified. This systematic review has included a large sample of case-control and cohort studies of COVID-19, SARS, and MERS. Its results are representative. We included 28 studies of COVID-19, SARS, and MERS with in-hospital death outcomes. This covered 16,095 patients and 1711 in-hospital deaths (COVID-19: 11,818 patients, 910 deaths; SARS: 3292 patients, 336 death; MERS: 985 patients, 465 deaths).
Advanced age has been reported as an independent predictor of death for SARS and MERS.25 , 41 This systematic review confirmed that advanced age is associated with COVID-19 mortality. Previous studies of macaques inoculated with SARS-CoV have found that older macaques have stronger innate host responses to virus infection than younger adults. These studies have also shown an increase in differential gene expression associated with inflammation, whereas type I interferon beta expression declined.43 The age-dependent defects in T-cell and B-cell function, and the excess production of type 2 cytokines could lead to a viral replication control deficiency and more prolonged pro-inflammatory responses, potentially causing poor outcomes.44 During systemic viral infection, aging alters the host-pathogen interaction to overproduce interleukin-17, which activates neutrophils. This contributes to liver injury and death.45 Moreover, advanced age was positively correlated with exaggerated pulmonary responses to injury. The inflammatory response is delayed, but aggravated, in the elderly.46 In addition, the average ages of both survivors and non-survivors of COVID-19 were higher than those of SARS and MERS (Supplementary Table 3). This might be due to SARS-CoV-2′s pathogenesis being milder than that of SARS-CoV.47
The results of this meta-analysis show that non-survivors of COVID-19, SARS, and MERS have a higher proportion of co-existing chronic diseases than survivors. COVID-19 patients with various comorbidities, including diabetes, hypertension and chronic lung disease were founded to have a higher likelihood of complications and death. Chronic inflammation, increased coagulation activity, immune response impairment, and potential direct pancreatic damage by SARS-CoV-2 might be among the underlying mechanisms of the association between diabetes and COVID-19.48 Pro-inflammatory cytokine storms have been shown to cause rapid deterioration in COVID-19 patients.49 Increases in systemic IL-2, IL-6, and IL-7, granulocyte colony-stimulating factor, C-X-C motif chemokine ligand 10, chemokine ligand 2, and tumor necrosis factor-α have been observed in COVID-19 patients. The same cytokines have been shown to be factors in the development of hypertension in experimental and clinical observations, as well as interventional studies.49 Research has found that chronic obstructive pulmonary disease (COPD) and current smokers have elevated expression of ACE-2—the entry receptor for the COVID-19 virus.50 It has been documented that SARS-CoV-2′s average age is higher than that of COVID-19 (see supplementary Table 3). This might contribute to the lower likelihood of death in COVID-19 patients with comorbidities of diabetes and hypertension, compared with SARS patients. Therefore, diabetes, hypertension and chronic lung disease are involved in the pathological mechanism of COVID-19. However, the progression of this disease needs further investigation.
The differences in laboratory indicators between COVID-19 survivors and non-survivors are substantial. Higher WBC counts and procalcitonin levels indicated that a large proportion of deceased COVID-19 patients might have had secondary bacterial infections. These secondary infections may be a factor in mortality. Deceased COVID-19 patients had more severe lymphopenia than patients who recovered, suggesting that a cellular immune deficiency state was a factor in poor prognosis. Additionally, other laboratory abnormalities in deceased patients included coagulation disorder (elevation of prothrombin time and d-dimer), impaired liver and kidney function (mild or moderate elevation of ALT, AST, total bilirubin, BUN, and creatinine), elevated inflammatory markers (high sensitivity CRP and ferritin), and cytokine storm. Most notably, deceased patients had higher LDH levels than patients who recovered. The D-dimer, high-sensitivity cardiac troponin I, serum ferritin, LDH, IL-6 levels and lymphocyte counts were all elevated in COVID-19 non-survivors, compared with survivors, throughout the clinical course and illness progression.14
Next, we identified increased LDH, CRP, and BUN levels and neutrophil count. Decreased albumin was correlated with mortality in COVID-19 patients. Neutrophils can form neutrophil extracellular traps which contribute to organ damage and mortality in COVID-19.51 The changes in CRP and albumin levels might be due to the coronavirus increasing inflammation in the body, leading to poor outcomes. The elevated LDH and BUN in deceased patients might be due to heart or kidney injury caused by the coronavirus disease. In addition, this systematic review has suggested that IL-6 level is an important mortality factor in COVID-19 and SARS patients. The role of IL-6 in predicting respiratory failure and identifying severe COVID-19 cases has been reported.52 , 53 The IL-6 level has also been positively associated with CRP, LDH, ferritin, and D-dimer levels in COVID-19 patients52 , 53. Furthermore, lower lymphocyte subtype counts are mortality risk factors in SARS patients. It has been shown that lymphocyte subsets can differentiate severe COVID-19 patients from non-severe COVID-19 patients54.
The degree of difference among COVID-19 laboratory indicators reviewed in this study was higher than that of SARS and MERS, with the exception of Hb. Accordingly, we speculated that the older patients with lower SARS-Cov-2 COVID-19 pathogenicity also contributed to the differences in laboratory indicators between COVID-19 and SARS, MERS patients. Stronger inflammatory effects, higher cytokine levels and cellular immune deficiency were found, and led to poor outcomes in older patients suffering from viral infections.43, 44, 45, 46 However, 81% of the confirmed Chinese COVID-19 cases were classified as mild (i.e., non-pneumonia or mild pneumonia), 14% as severe (i.e., dyspnea, respiratory frequency ≥ 30/min, blood oxygen saturation ≤ 93%, to inhaled oxygen ratio < 300, and/or lung infiltrates > 50% within 24 to 48 hrs.), and 5% as critically ill (i.e., respiratory failure, septic shock, and/or multiple organ dysfunction or failure).55 SARS patients’ respiratory distress and oxygen desaturation progressed rapidly with 20–36% requiring intensive care and 13–26% progressing to acute respiratory distress syndrome (ARDS) necessitating invasive ventilatory support.56 Among the 144 laboratory confirmed and 17 probable MERS patients from nine countries, 44.1% developed pneumonia; 51.6% required intensive care.57
In addition, the imaging features for consolidation and bilateral ground glass shadows in the lungs of COVID-19 patients were similar with those of SARS and MERS patients.17 The number of deceased COVID-19 patients who had suffered from pulmonary consolidation and had presented ground glass imaging characteristics exceeded that of those who had survived COVID-19 (consolidation: OR = 3.26, 95% CI 1.16–9.13; ground-glass opacity: OR = 1.45, 95% CI 0.47–4.49).14 , 17 The percentage of total lung opacity, no. of zones opacified and chest radiographic score (out of 24) in SARS non-survivors surpassed those of SARS survivors (Day 7 opacification ≥ 10%: OR = 26.31, 95% CI 10.73–64.52; Day 10 opacification ≥ 10%: OR = 17.50, 95% CI 6.09–50.33).58 , 59 Peak chest radiographic scores (out of 24) among MERS non-survivors were much higher than those of MERS survivors (OR = 15.23, 95% CI 4.90–47.31).60 Additionally, the imaging features were also correlated with initial presentation, start of ribavirin, first use of pulse corticosteroid, peak lung opacification, both before death and at discharge, for SARS patients. Imaging features were also correlated with the number of lymphocytes and neutrophils, oxygenation index and LDH levels59.
This systematic review and meta-analysis has several limitations. First, it analyzes a large sample of case-control and cohort studies of COVID-19 mortality risk factors. However, the patients’ COVID-19 laboratory mortality indicators included in this meta-analysis were limited to the Wuhan area. With the outbreak of the pandemic, studies from different regions are needed to validate the correlation between laboratory indicators and COVID-19 mortality. Secondly, one of the MERS mortality studies included in this systematic review covered critically ill patients. Thus, the OR values of the MERS mortality indicators may have been underestimated. Thirdly, transfer of patients between hospitals, variation in the types of patients admitted to the designated hospitals, and regional differences were all inevitable. Therefore, there was severe heterogeneity among the mortality risk factors in the COVID-19 studies included in this meta-analysis. In the future, more COVID-19 patients need to be investigated.
Conclusion
COVID-19 mortality factors are similar to those of SARS and MERS. Advanced age as well as comorbidities and laboratory indicators including LDH, CRP, neutrophil, BUN and albumin are correlated with COVID-19 mortality. Additionally, age and laboratory indicators would facilitate even better prediction of COVID-19 mortality outcomes.
Contributors
ZH and XZ conceived of the study and designed the protocol with YZ and BL. LL, WZ, ZB, ZL and KZ conducted study selection and data extraction. LL, MH, LL, JL, XL, YH, JJ, XY contributed to statistical analysis and data interpretation. LL, BL, YZ and ZH drafted the manuscript with all authors providing critical revision.
Supplementary materials
Supplementary materials to this article can be found online.
Declaration of Competing Interest
The authors declare having no conflict of interest related to this work.
Funding
This work was supported by National Key R&D Program of China (2018YFC1602103) Ministry of Science and Technology of China, National Natural Science Foundation of China (81872601), Guangdong Key R&D Program (2019B020210002) Ministry of Science and Technology of Guangdong Province, Natural Science Foundation of Guangdong Province (2018A030313068), Science and Technology Planning Project of Guangzhou City (201707010405), and the Guangdong Provincial Key Laboratory of Tropical Disease Research (2017B030314035).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.07.002.
Appendix. Supplementary materials
References
- 1.The Lancet Emerging understandings of 2019-nCoV. Lancet. 2020;395(10221):311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Coronavirus disease 2019 (COVID-19) situation report – 89. (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/) Accessed 18April 2020.
- 3.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., Li F., Shi Z. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrosillo N., Viceconte G., Ergonul O., et al. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.03.026. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan Y., Shang J., Graham R., et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins J.P.T., T.J. Chandler J, et al. Cochrane handbook for systematic reviews of interventions version 6.0 (updated July 2019) Cochrane. 2019 www.training.cochrane.org/handbook Available from. [Google Scholar]
- 8.Moher D., Shamseer L., Clarke M., et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1):1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyseshttp://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 19 Oct2018.
- 10.Harris R.J., D.J. Altman DG, Bradburn M.J., et al. Metan: fixed- and random-effects meta-analysis. Stata J. 2008;8(1):3–28. [Google Scholar]
- 11.Nyaga V.N., Arbyn M., Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health. 2014;72(1):39. doi: 10.1186/2049-3258-72-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egger M., Davey Smith G., Schneider M., et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duval S., Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T., Wu D., Chen H., et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang N., Li D., Wang X., et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan M., Yin W., Tao Z., et al. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan. China. PLoS One. 2020;15(3) doi: 10.1371/journal.pone.0230548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasselli G., Zangrillo A., Zanella A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA. 2020 doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao J., Tu W.J., Cheng W., et al. Clinical features and short-term outcomes of 102 patients with corona virus disease 2019 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan W.J., Liang W.H., Zhao Y., et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020 doi: 10.1093/cid/ciaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruan Q., Yang K., Wang W., et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang Y.J. Mortality rate of infection with COVID-19 in Korea from the perspective of underlying disease. Disaster Med Public Health Prep. 2020 doi: 10.1017/dmp.2020.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CDC COVID-19 Response Team Severe outcomes among patients with Coronavirus Disease 2019 (COVID-19) —United States, February 12–March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(12):343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J.K., Feng Y., Yuan M.Y., et al. Plasma glucose levels and diabetes are independent predictors for mortality and morbidity in patients with SARS. Diabet Med. 2006;23(6):623–628. doi: 10.1111/j.1464-5491.2006.01861.x. [DOI] [PubMed] [Google Scholar]
- 25.Chan J.C., Tsui E.L., Wong V.C. Prognostication in severe acute respiratory syndrome: a retrospective time-course analysis of 1312 laboratory-confirmed patients in Hong Kong. Respirology. 2007;12(4):531–542. doi: 10.1111/j.1440-1843.2007.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen R.C., Tang X.P., Tan S.Y., et al. Treatment of severe acute respiratory syndrome with glucosteroids: the Guangzhou experience. Chest. 2006;129(6):1441–1452. doi: 10.1378/chest.129.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko S.F., Lee T.Y., Huang C.C., et al. Severe acute respiratory syndrome: prognostic implications of chest radiographic findings in 52 Patients. Radiology. 2004;233(1):173–181. doi: 10.1148/radiol.2323031547. [DOI] [PubMed] [Google Scholar]
- 28.Fowler R.A., Lapinsky S.E., Hallett D., et al. Critically ill patients with severe acute respiratory syndrome. JAMA. 2003;290(3):367–373. doi: 10.1001/jama.290.3.367. [DOI] [PubMed] [Google Scholar]
- 29.Yam L.Y., Lau A.C., Lai F.Y., et al. Corticosteroid treatment of severe acute respiratory syndrome in Hong Kong. J Infect. 2007;54(1):28–39. doi: 10.1016/j.jinf.2006.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M., Li H., Li S., et al. Analysis of prognosis-related factors in critical SARS. Chin Shanxi Med J. 2004;33(01):6–8. [Google Scholar]
- 31.Wei J., Wang X., Zhang J., et al. Retrospective case study of death of 19 SARS patients. Chin J Capit Med Univ. 2003;24(04):374–379. [Google Scholar]
- 32.Yang M., Xu J., Li X., et al. Dynamic changes and significance of subsets of blood lymphocyte in 206 patients severe acute respiratory syndrome patients. Chin J Infect Dis. 2003;21(05):22–25. [Google Scholar]
- 33.Wu H., Zhang Y., Chen X., et al. Study of immunological function in severe acute respiratory syndrome. Chin J Infect Dis. 2003;21(05):29–32. [Google Scholar]
- 34.Chen X., Lin J., Su N., et al. Effect of co-existing illness on prognosis of severe acute respiratory syndrome. Chin J Emerg Med. 2005;14(01):53–56. [Google Scholar]
- 35.Zhang Y., Chen X., Yan H., et al. Discussion on clinical signification of cytokine in severe acute respiratory syndrome. Chin J Microbiol Immunol. 2004;03:18–20. [Google Scholar]
- 36.Arabi Y.M., Al-Omari A., Mandourah Y., et al. Critically ill patients with the Middle East respiratory syndrome: a multicenter retrospective cohort study. Crit Care Med. 2017;45(10):1683–1695. doi: 10.1097/CCM.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 37.Adegboye O.A., Gayawan E., Hanna F. Spatial modelling of contribution of individual level risk factors for mortality from Middle East respiratory syndrome coronavirus in the Arabian Peninsula. PLoS ONE. 2017;12(7) doi: 10.1371/journal.pone.0181215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al Ghamdi M., Alghamdi K.M., Ghandoora Y., et al. Treatment outcomes for patients with middle eastern respiratory syndrome coronavirus (MERS CoV) infection at a coronavirus referral center in the Kingdom of Saudi Arabia. BMC Infect Dis. 2016;16:174. doi: 10.1186/s12879-016-1492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garbati M.A., Fagbo S.F., Fang V.J., et al. A comparative study of clinical presentation and risk factors for adverse outcome in patients hospitalised with acute respiratory disease due to MERS coronavirus or other causes. PLoS ONE. 2016;11(11) doi: 10.1371/journal.pone.0165978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong K.H., Choi J.P., Hong S.H., et al. Predictors of mortality in Middle East Respiratory Syndrome (MERS) Thorax. 2018;73(3):286–289. doi: 10.1136/thoraxjnl-2016-209313. [DOI] [PubMed] [Google Scholar]
- 42.Cohen J. Vaccine designers take first shots at COVID-19. Science. 2020;368(6486):14–16. doi: 10.1126/science.368.6486.14. [DOI] [PubMed] [Google Scholar]
- 43.Smits S.L., de Lang A., van den Brand J.M., et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6(2) doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opal S.M., Girard T.D., Ely E.W. The immunopathogenesis of sepsis in elderly patients. Clin Infect Dis. 2005;41(suppl 7):S504–S512. doi: 10.1086/432007. [DOI] [PubMed] [Google Scholar]
- 45.Stout-Delgado H.W., Du W., Shirali A.C., et al. Aging promotes neutrophil-induced mortality by augmenting IL-17 production during viral infection. Cell Host Microbe. 2009;6(5):446–456. doi: 10.1016/j.chom.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schouten L.R., Schultz M.J., van Kaam A.H., et al. Association between maturation and aging and pulmonary responses in animal models of lung injury: a systematic review. Anesthesiology. 2015;123(2):389–408. doi: 10.1097/ALN.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 47.Benvenuto D., Giovanetti M., Ciccozzi A., et al. The 2019‐new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020;92(4):455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussain A., Bhowmik B., Moreira N.C.V., et al. COVID-19 and diabetes: knowledge in progress. Diabetes Res Clin Pract. 2020;162 doi: 10.1016/j.diabres.2020.108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreutz R., Algharably E.A.E., Azizi M., et al. Hypertension, the renin-angiotensin system, and the risk of lower respiratory tract infections and lung injury: implications for COVID-19. Cardiovasc Res. 2020 doi: 10.1093/cvr/cvaa097. cvaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung J.M., Yang C., Tam A., et al. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J. 2020 doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Betsy J.B., Jose M.A., Amelia B., et al. Targeting Potential Drivers of COVID-19: neutrophil Extracellular Traps. J Exp Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herold T., Jurinovic V., Arnreich C., et al. Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. medRxiv 2020.04.01.20047381; doi: 10.1101/2020.04.01.20047381. [DOI]
- 53.Liu T., Zhang J., Yang Y., et al. The potential role of IL-6 in monitoring severe case of coronavirus disease2019. medRxiv 2020.03.01.20029769; doi: 10.1101/2020.03.01.200297692020. [DOI]
- 54.Qin C., Zhou L., Hu Z., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 56.Hui D.S., Wong P., Wang C. SARS: clinical features and diagnosis. Respirology. 2003;8 doi: 10.1046/j.1440-1843.2003.00520.x. S20-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Who Mers-Cov Research Group State of knowledge and data gaps of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in humans. PLoS Curr. 2013;5 doi: 10.1371/currents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8. ecurrents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antonio G.E., Wong K.T., Tsui E.L., et al. Chest radiograph scores as potential prognostic indicators in Severe Acute Respiratory Syndrome (SARS) AJR Am J Roentgenol. 2005;184(3):734–741. doi: 10.2214/ajr.184.3.01840734. [DOI] [PubMed] [Google Scholar]
- 59.Antonio G.E., Ooi C.G., Wong K.T., et al. Radiographic-clinical correlation in severe acute respiratory syndrome: study of 1373 patients in Hong Kong. Radiology. 2005;237(3):1081–1090. doi: 10.1148/radiol.2373041919. [DOI] [PubMed] [Google Scholar]
- 60.Das K.M., Lee E.Y., Al Jawder S.E., et al. Acute middle east respiratory syndrome coronavirus: temporal lung changes observed on the chest radiographs of 55 patients. AJR Am J Roentgenol. 2015;205(3):267–274. doi: 10.2214/AJR.15.14445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.