Abstract
The global pandemic of coronavirus disease 2019 (COVID-19), caused by novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in over 7,273,958 cases with almost over 413,372 deaths worldwide as per the WHO situational report 143 on COVID-19. There are no known treatment regimens with proven efficacy and vaccines thus far, posing an unprecedented challenge to identify effective drugs and vaccines for prevention and treatment. The urgency for its prevention and cure has resulted in an increased number of proposed treatment options. The high rate and volume of emerging clinical trials on therapies for COVID-19 need to be compared and evaluated to provide scientific evidence for effective medical options. Other emerging non-conventional drug discovery techniques such as bioinformatics and cheminformatics, structure-based drug design, network-based methods for prediction of drug-target interactions, artificial intelligence (AI) and machine learning (ML) and phage technique could provide alternative routes to discovering potent Anti-SARS-CoV2 drugs. While drugs are being repurposed and discovered for COVID-19, novel drug delivery systems will be paramount for efficient delivery and avoidance of possible drug resistance. This review describes the proposed drug targets for therapy, and outcomes of clinical trials that have been reported. It also identifies the adopted treatment modalities that are showing promise, and those that have failed as drug candidates. It further highlights various emerging therapies and future strategies for the treatment of COVID-19 and delivery of Anti-SARS-CoV2 drugs.
Keywords: COVID-19, SARS-CoV2, Re-purposing, Drug targets, Clinical trials, Vaccines
Graphical abstract
Highlights
-
•
COVID-19 is a global pandemic with no approved cure and vaccine.
-
•
For expedited approval, repurposing of available drugs has been an option explored.
-
•
High volume of emerging therapies for COVID-19 reported need to be compared and evaluated.
-
•
Emerging non-conventional drug discovery strategies shows potential for developing drugs for COVID-19.
-
•
Novel drug delivery systems can improve drug targeting and prevent resistance.
List of abbreviations/acronyms
- 2019-nCoV
2019 Novel Coronavirus
- AAK-1
Adaptor-associated protein kinase 1
- ACE2
Angiotensin Converting Enzyme 2
- ACEI
Angiotensin converting enzyme inhibitor
- ARB
Angiotensin receptor blocker
- ARDS
Acute Respiratory Distress Syndrome
- ARI
Acute Respiratory Infection
- CDC
Centers for Disease Control and Prevention
- CoV
Coronavirus
- COVID-19
Coronavirus Disease 2019
- CQ
Chloroquine phosphate
- CRS
Cytokine release syndrome
- CYP 3A4
Cytochrome 3A4
- DNA
Deoxyribonucleic acid
- ECMO
Extracorporeal Membrane Oxygenation
- Epi curve
Epidemic curve
- FDA
Food and Drug Administration
- FFP
Filtering facepiece
- HCQ
Hydroxycholoroquine
- HEPA
High-efficiency particulate air
- HR2
Heptad repeat 2
- ICU
Intensive care unit
- IFN
Interferon
- IL
Interleukin
- LMWH
Low molecular weight heparin
- LPV/r; Kaletra
Lopinavir/ritonavir
- mAbs or moAbs
Monoclonal antibodies
- MERS-CoV
Middle East Respiratory Syndrome Coronavirus
- mRNA
Messenger ribonucleic acid
- NSAIDs
Non-steroidal anti-inflammatory drugs
- NSPS
Non-structural proteins
- ORF
Open reading frames
- PP
Polyprotein
- PPE
Personal Protective Equipment
- R0
Reproductive number
- RBD
Receptor binding domain
- RBV
Ribavarin
- RCT
Replication-transcription complex
- RDV
Remdesivir
- RNA
Ribonucleic acid
- RT-PCR
Reverse Transcription Polymerase Chain Reaction
- S protein
Spike protein
- SARS-CoV
Severe Acute Respiratory Syndrome Coronavirus
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus II
- TCM
Traditional Chinese Medicine
- TMPRSS2
Transmembrane Serine Protease 2
- WHO
World Health Organization
1. Introduction
In December 2019, patients in Wuhan, China started experiencing a disease with acute respiratory attacks. Investigations revealed that the disease was due to a novel virus that was later designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The disease was assigned as the novel coronavirus disease 2019 (COVID-19) and its global spread has led to the World Health Organization (WHO) declaring it as a global pandemic (Yuen et al., 2020). SARS-CoV-2 is a member of the coronavirus family which has threatened the lives and health of millions of people around the world. As per the WHO situational report 143 on COVID-19, the number of infection cases, globally, were over 7,273,958. The number of deaths is increasing exponentially, with over 413,372 worldwide thus far (WHO, 2020ba, WHO, 2020bb, d).
Coronaviruses are single stranded positive sense RNA viruses with genome sizes of between 28 and 32 kb. These viruses have the ability to infect both animals and humans. They cause an array of diseases, including and not limited to; respiratory, neurologic, hepatic and gastrointestinal disease (Su et al., 2016). Based on its phylogenetic relationships and genomic structure, the novel virus belongs to the genera, Betacoronavirus, which has a close similarity to that of severe acute respiratory syndrome-related coronaviruses (SARSr-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). Importantly, the genomic sequence of SARS-CoV-2 and its replication cycle has been elucidated, and together with our evolving understanding of the host immune response, these factors have been the basis for investigating diagnostic and prognostic testing, epidemiologic tracking and the initiation of therapeutic strategies (Wang et al., 2020a; Wu et al., 2020a).
The threat of the virus has reached unimaginable proportions and has exposed the unpreparedness of national healthcare systems and has triggered international solidarity. Despite COVID-19 being declared a global pandemic by WHO, no drug regimen has been shown to be effective in the treatment of the virus. Moreover, numerous completed randomized clinical trials have not produced clear guidelines on treatment options and prophylactic therapy yet. Currently several active clinical trials for treatment are ongoing. Other emerging, non-conventional drug discovery techniques provides alternative faster and less costly routes to discovering potent Anti-SARS-CoV2 drugs. Furthermore, while drugs are being repurposed and discovered for COVID-19, novel drug delivery systems will play a paramount role in designing efficient delivery systems that have virus targeting ability, improved pharmacokinetic profiles and avoidance of possible drug resistance leading to superior therapies. This review summarizes proposed drug targets for therapy and reviews the various clinical trials that have been reported, as well as vaccines that are under development. It also highlights emerging therapies and future perspectives for the treatment and prevention of COVID-19.
2. Pathophysiology and transmission
2.1. Virology and pathophysiology
The novel coronavirus belongs to the Coronaviridae family of the order Nidovirales which are divided into four genera viz. α, β, γ, and δ. The Coronaviridae family (CoVs) have an outer envelope and the genetic material consists of positive sense RNA (Gorbalenya et al., 2020). They have been reported to be the largest known viruses with a size of 28–32 kb (Bosch et al., 2003). The International Committee on Taxomony of Viruses (ICTV) classified the virus as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) (Gorbalenya et al., 2020). The SARS-CoV-2 has been identified as belonging to the β genus CoVs family that contains at least four structural proteins (spike, envelope, membrane and nucleocapsid) (Bosch et al., 2003).WHO named the disease that is caused by SARS-CoV-2 virus as COVID-19. The spikes on the viral surface are composed of homotrimers of the S protein that acts as a link to host receptors. Furthermore, the spike glycoproteins have two subunits, S1 and S2, mediating attachment and membrane fusion, respectively. The S2 subunit contains a fusion peptide, a cytoplasmic domain and a transmembrane domain. The S protein-receptor interaction is the main determinant for the infection of a host species (Letko et al., 2020). The viral entry is a complex sequence of events that includes attachment to the cell surface, receptor engagement, protease processing and membrane fusion. The spike (S) protein is responsible for cellular entry as it binds to the receptors of the target cell, and via this interaction virus-cell fusion occurs. Virus-cell fusion occurs via angiotensin converting enzyme 2 (ACE2) of the susceptible cell. ACE2 is part of the renin–angiotensin–aldosterone system (RAAS) pathway responsible for cleaving vasoconstrictor octapeptide Ang II to the vasodilatory Ang 1–7 (Vaduganathan et al., 2020). ACE 2 are abundantly found in the lungs, moreover, they are also widely distributed in the digestive system, kidneys, the heart, the liver, endothelial cells and smooth muscle cells of various organs (Bavishi et al., 2020). S protein priming by the serine protease transmembrane protease serine 2 (TMPRSS2) is essential for SARS-CoV-2 infection of target cells and spreading throughout the host (Zhang et al., 2020b).
The endothelium is arguably the largest ‘organ’ in the body and this perhaps explains why the viral effects spread to extra-pulmonary organs once it enters the blood circulation. Given that there is such a wide number of targets in the human body, from a pathophysiological approach, it explains why COVID 19 patients present with cardiovascular and other diverse complications (Hamming et al., 2004; Zhang et al., 2020b). Exceptionally high proportion of aberrant coagulation was seen in severe and critical patients with COVID-19, revealing a hypercoagulable state, elevated levels of D-dimer and fibrinogen, near normal activated partial thromboplastin time, with some patients progressing to overt disseminated intravascular coagulation (DIC) (Cao and Li, 2020).
Recent studies investigating the expression of viral entry-associated genes, using single-cell RNA-sequencing data from multiple tissues from healthy human donors, have identified transcripts in more tissues and cells, not previously analysed, including its co-expression with TMPRSS2 (Sungnak et al., 2020). Importantly, Sungnak, et al., 2020 co-detected these transcripts in specific cells in the respiratory, corneal and intestinal epithelium. Furthermore, they found that these genes are co-expressed, in nasal epithelial cells, with genes involved in innate immunity, which impacts upon initial viral infection, spread and clearance. Their work is in accordance with previous studies (Qi et al., 2020; Zou et al., 2020). They found that ACE2 was expressed in cells from airways, cornea, oesophagus, ileum, colon, liver, gallbladder, heart, kidney and testis while TMPRSS2 was highly expressed with a broader distribution, suggesting that ACE2 may be a limiting factor for viral entry at the initial infection stage, rather than TMPRSS2 (Sungnak et al., 2020). Cells from the respiratory tree, cornea, oesophagus, ileum, colon, gallbladder and common bile duct expressed both genes (ACE2 and TMPRSS2) in the same cells. In particular, the upper and lower respiratory tract was assessed for ACE2, indicating a low level of expression overall, in multiple epithelial cell types across the airways (Sungnak et al., 2020). Of importance, regarding nasal epithelial cells, two previously described clusters of goblet cells and one cluster of ciliated cells, showed the highest expression among all investigated cells in the respiratory tree (Sungnak et al., 2020).
After binding, the virus enters the cell and once infected viral replication starts inside the host cell.Inside the host cell, the synthetic process of polyprotein 1a/1 ab (pp1a/b) is realized and once the virus uncoats it releases the viral RNA material. Transcription of the virus occurs via the replication-transcription complex (RCT), which is organized within the double membrane vesicles through the synthesis of subgenomic RNAs(Lei et al., 2018). Transcription terminates at transcription regulatory sequences, which are found in between the open reading frames (ORF), which are templates for subgenomic mRNAs. ORF guides the production of pp1a and pp1b polypeptides; which are then processed by chymotrypsin like protease and papain proteases to produce 16 nonstructural proteins (nsps) (Wrapp et al., 2020). It has been demonstrated that the nsps inhibit the hosts innate immune response (Li et al., 2020b) through the effects of the envelope. After transcription, viral assembly occurs; thereafter it leads to release and the cycle repeats with attachment to the next ciliated epithelial cell (Zhang et al., 2020b).
2.1.1. Host response
An understanding of the host response is crucial to implement valid therapeutic strategies as stated previously, and it is now apparent that severity of the disease is dependent on both the virus itself and host response. Host response to SARS-CoV-2 has been described in the literature, however key aspects are still being elucidated. Notably symptoms of patients range from minimal to severe respiratory failure with multiple organ failure, complicating linking pathophysiology with symptoms in a complete picture. Hence, we can report on key findings in each aspect to date (Berlin et al., 2020; Magro et al., 2020; Tay et al., 2020). Importantly, SARS-CoV-2 is transmitted mainly via respiratory droplets, with a possible, but unproven, faecal–oral transmission route (Tay et al., 2020). The median incubation period is approximately 4–5 days before symptom onset, with 97.5% of symptomatic patients developing symptoms within 11.5 days (Lauer et al., 2020). The most common symptoms of COVID -19 are reported to be fever, cough, fatigue, anorexia, myalgia and diarrhea (CDC guidelines) (Backer et al., 2020; CDC, 2020). Dyspnoea (often accompanied by hypoxemia) is a common symptom of severe illness (Wang et al., 2020d; Zhou et al., 2020a) which may progress to ARDS. Other manifestations of severe COVID-19 are lymphopenia, disorders of the central or peripheral nervous system, acute cardiac, kidney, and liver injury, in addition to cardiac arrhythmias, rhabdomyolysis, coagulopathy, and shock (Berlin et al., 2020).
Notably SARS-CoV-2 viral load reaches its peak within 5–6 days of symptom onset, and severe COVID-19 cases progress to acute respiratory distress syndrome (ARDS), on average around 8–9 days after symptom onset (Tay et al., 2020). A timeline for symptoms of the severe disease is described by Berlin et al., (2020).
The virus enters the cells via ACE 2, is internalized as described above and triggers a series of events intracellularly, leading to disease manifestation, based on the targeting of key regulatory pathways, cells and organs. However, the main target is the respiratory tree and lung tissue and therefore this will be described in more detail. Notably, the characteristic pulmonary ground glass opacification is seen even in asymptomatic patients (Mason, 2020). Based on the cells that are likely infected, COVID-19 can be divided into three phases that correspond to different clinical stages of the disease (Mason, 2020). COVID-19 confined to the conducting airways should be mild and treated symptomatically at home, when it has progressed to the gas exchange units of the lung it must be monitored carefully with the best support (Mason, 2020).
Tay et al. (2020) describes the chronology of events upon cell entry, describing interaction of SARS-CoV-2 with the immune system leading to either a normal ‘healthy response’ or a dysfunctional immune response (IR). In brief the virus enters the cell (which has both ACE2 and TMPRRSS2), and replicates leading to more viral release. Thereafter, the cell undergoes pyroptosis, and releases damage-associated molecular patterns (DAMPS). This triggers the release of pro-inflammatory cytokines and chemokines (including IL-6, IP-10, macrophage inflammatory protein 1α (MIP1α), MIP1β and MCP1). Other immune cells are attracted to the site (viz. monocytes, macrophages and T cells) which produce a proinflammatory loop. This can lead to viral clearance and minimal lung damage (healthy immune response) or a series of events which can culminate in multi-organ damage (dysfunctional immune response) (Tay et al., 2020).
An overproduction of pro-inflammatory cytokines, resulting in cytokine storm damages the lung structure and leads to multi-organ damage as it spreads to other parts of the body. The cytokine release syndrome (CRS) or storm is characterized by multiorgan pathology and fever (Lin et al., 2020). Importantly, non-neutralizing antibodies may further exacerbate organ damage. However, in a healthy immune response, the initial inflammation attracts the following; T cells to the site of infection, antibodies (neutralizing) which can block viral infection, and alveolar macrophages which phagocytise neutralized viruses and apoptotic cells. This contributes to viral clearance, minimal lung damage and recovery (Tay et al., 2020).
The symptoms and pathogenesis are similar to those of SARS-CoV and MERS-CoV infections (Rockx et al., 2020). This has helped in understanding of the disease despite a lack of knowledge on SARS-CoV-2. Data attained based upon the first cases in Wuhan, China indicate that the incubation time is generally between 2 and 7 days, however the longest incubation time was seen to be 12.5 days (Backer et al., 2020; Yuen et al., 2020). However, data from the United States of America has shown some patients have up to 17 days of incubation (Lauer et al., 2020).
Based on the life cycle, transduction and signaling process of the virus as well as patient responses, various targets can be exploited to combat the virus. The following section describes the specific targets and highlights the drugs being investigated to exploit them for overcoming viral infection. It also describes the possible targets which can be explored to create vaccines against the virus.
3. Drug targets
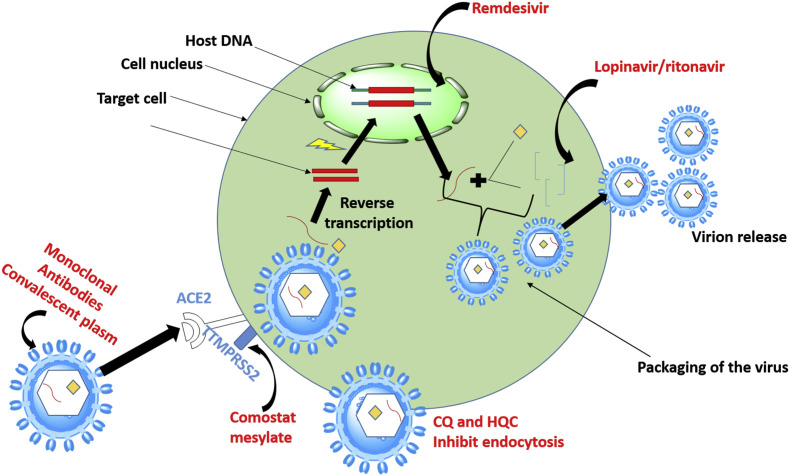
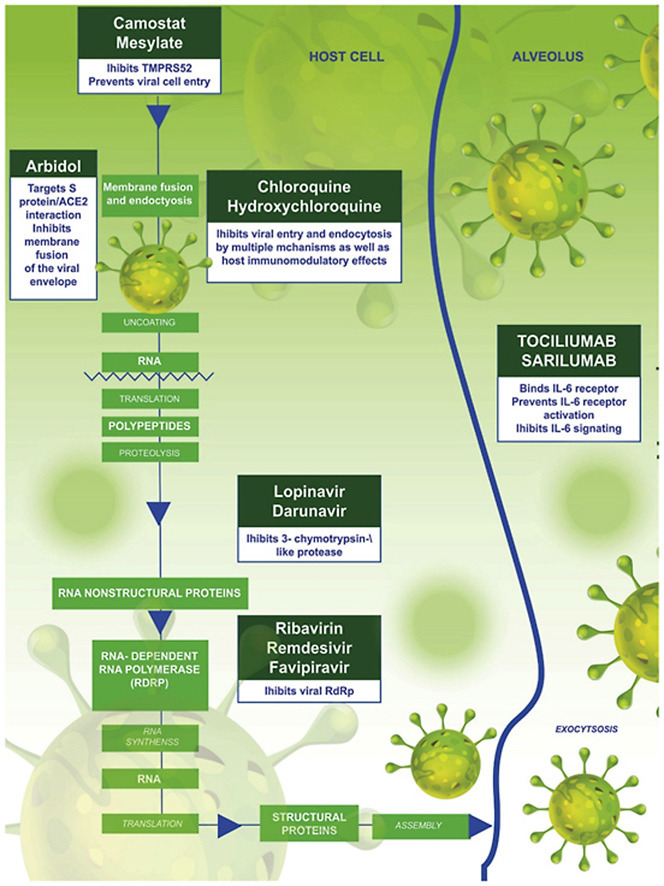
As of April 2020, there was no recommended antiviral treatment and no vaccine for SARS-CoV-2 (Mahase, 2020). Despite the fact that some studies are showing some promise (Colson et al., 2020; Gao et al., 2020; Keyaerts et al., 2004; Vincent et al., 2005), successful drug therapy to control coronavirus infection has yet to materialize. Fig. 1 depicts the major pathways that can be targeted by therapeutic agents for treatment of SARS-CoV-2 as discussed above. The current management of infected persons is primarily symptomatic treatment, while oxygen therapy is employed for intervention in patients with severe infection. These factors have triggered a global pursuit towards the urgent discovery and repurposing of drugs in order to end the pandemic (Dong et al., 2020). With the increasing rise in the number of infected persons and death rate, more drugs/therapeutic agents and treatment for SARS-CoV-2 virus are required.
Fig. 1.
SARS-CoV-2 replication circle involves cellular entry which is via fusion and endocytosis, after entry translation occurs as the virus uses host cells enzyme it creates its DNA. This is followed proteolysis then translation to back to RNA after that packaging and viral release occurs. The circles.
Anti-coronavirus therapies so far have been classified into two categories depending on the target i.e those that target the immune system and infected human cells, and those that target the virus itself. The severity of COVID-19 is dependent on the ability of the human innate immune system to control the replication and infection. Blocking the signaling pathways in cells that are required for viral replication is a strategy in mediating antiviral effects. The infection pathway of the virus often involves the interactions of the virus and the receptor proteins of the target cells. This has been found to be the same for SARS-CoV-2 which binds to the ACE2 receptors. The second group of anti-corona therapies acts on the coronavirus itself, by inhibiting critical enzymes for viral genome replication, the virus's self-assembly process and preventing viral entry by blocking the binding of the virus to human cell receptors ( Fig. 1 ).
3.1. Inhibition of the SARS-CoV-2 fusion/entry
Inhibition of the SARS-CoV-2 fusion/entry is an important strategy in drug targeting and approximately over 23 drugs that cause inhibition of viral entry are being evaluated (Gysi et al., 2020). The interaction linking the virus and host cell receptor can be inhibited in variety of ways (Ou et al., 2020), (Zhang et al., 2020a). One approach that has been proposed involves targeting the ACE2 and S protein association. In particular, the receptor binding domain (RBD), which is within the S protein and is an essential target to develop antibodies (Tai et al., 2020).
According to Zheng et al. more than 85% of the RBD antibody epitopes in SARS-CoV-2 exhibited significantly higher binding affinity to ACE2 receptor than the SARS-CoV. This provides a platform to develop anti-SARS-CoV-2 drugs that target the RBD. Hence, there is a need to develop novel fusion inhibitors targeting SARS-CoV-2 (Shanmugaraj et al., 2020). Moreover, patients suffering from severe SARS-CoV-2 have shown that cell entry depends on serine protease TMPRSS2, apart from S protein. The drug targets being used inhibit entry and fusion of SARS-CoV-2 into the cells are described below.
3.1.1. Inhibitors of transmembrane protease serine 2 (TMPRSS2)
By understanding cellular factors that aid the entry and fusion of the SARS-CoV-2 virus it has provided therapeutic targets for drug development against the virus. The virus has been reported to use the serine protease TMPRSS2 for S protein priming and binding to ACE2 for cellular entry. A TMPRSS2 inhibitor can block viral entry, constituting a treatment option (Hoffmann et al., 2020b). Computational methods on repurposing existing candidates, have revealed candidate inhibitors for TMPRSS2 (Rensi et al., 2020). Amongst these, camostat mesylate, a clinically proven inhibitor of TMPRSS2, was used in Japan, to treat unrelated diseases (Zhang et al., 2020b). It has been associated with reduced lung cell line infection with SARS-CoV-19 and could therefore be used as a therapeutic option in the management of COVID-19 (Hoffmann et al., 2020a).
3.1.2. HR2 derived peptides
SARS-CoV-2 has utilized protein-mediated cell-viral fusion as an entry mechanism into the cell, providing another potential drug target for management of the SARS-CoV-2 (Zhu et al., 2020). Hence another option may be to target AP-2 associated protein kinase 1 (AAK1). It is a host kinase that is involved in the regulation of clathrin-mediated endocytosis (Yang and Shen, 2020). Based upon artificial intelligence, subsets of approved drugs were evaluated. The janus kinase (JAK) inhibitor, baricitinib, is a suitable candidate to inhibit AAKI (Favalli et al., 2020; Marotto and Sarzi-Puttini, 2020). It demonstrates antiviral as well as anti-inflammatory activity (Richardson et al., 2020a). In addition, peptides such as the HR2 derived peptides can be potential drug targets for COVID-19 as they could inhibit viral entry.
3.1.3. Angiotensin receptor blockers (ARBs) and angiotensin converting enzyme inhibitors (ACEIs)
Angiotensin-converting enzyme 2 (ACE2) and SARS-CoV-2 interactions suggest that antihypertensive drugs ACEI and ARBs may reduce the risk and severity of COVID-19. As mentioned previously, SARS-CoV-2 exploits ACE2 as a co-receptor to gain entry into susceptible cells via the transmembrane spike (S) glycoprotein and cleavage activates the viral protein for membrane fusion via extensive irreversible conformational changes (Walls et al., 2020). The involvement of RAAS in attachment makes the system a drug target for management of COVID-19, and has raised debates on the benefits and downsides associated with ACEI/ARBs (Vaduganathan et al., 2020). Also since ACE2 is used for viral entry, it has raised concerns on its impact on other components of RAAS.
The debates concern firstly, a purported upregulation in ACE2 due to ACEI use since their mechanism of action ultimately leads to decreased Ang II, the main effector octapeptide of RAAS. The same would apply to ARBs. This would then predispose the patient to infection due to increased ACE2. However a review by Vaduganathan et al., (2020), reports that animal studies using ARBs and ACEI are found to be inconclusive, and not enough data is available to support translation to humans (Vaduganathan et al., 2020).
Secondly, it is contended that since the function of ACE2 is to convert Ang II to Ang 1–7, if it is unavailable/downregulated (due to viral binding), this would then lead to increased Ang II which could then impact on lung pathology. This would also lead to lower Ang 1–7 levels. Notably, ACEI/ARBs would then be expected to ameliorate the resultant pathology since it reduces Ang II levels or action. The role of Ang II was postulated because of studies, for example, in patients with COVID-19 who appeared to have elevated levels of plasma angiotensin II, which correlated with total viral load and degree of lung injury (Li et al., 2020a) The connection between AT1/Ang II and lung pathology is reinforced by others (Kai and Kai, 2020). However, others found that restoration of ACE2 through the administration of recombinant ACE2 appeared to reverse lung-injury in preclinical models of other viral infections (Gu et al., 2016; Zou et al., 2014). Multicenter, double-blinded placebo-controlled, randomized trials (RCTs) are currently being conducted to investigate the effects of losartan on mortality and hospital admission in COVID-19 patients (Kai and Kai, 2020).
Vaduganathan et al. (2020) do not recommend withdrawal of the drugs by patients of heart failure and myocardial infarction. Importantly, they recommend that, until further data are available, RAAS modulators should be continued in patients in an otherwise stable condition who are at risk for, being evaluated for, or with COVID-19 (Vaduganathan et al., 2020). It is clear that more research needs to be conducted into the impact of ACE 2 on infectivity within this context, the role at Ang II on lung pathology and role of ACEI/ARBs in COVID given the complexity of RAAS from a systemic and tissue specific perspective.
In addition, ACEI is known to potentiate the vasodilator bradykinin (BK). Roche et al. (2020) used established and emerging evidence to propose a testable hypothesis that, a vicious positive feedback loop involving dysregulated BK signaling is involved in inflammation, which likely responsible for the severe respiratory complications in COVID-19. Hence, they propose the FDA-approved molecule, icatibant, that might be able to interrupt this feedback loop and, thereby, improve the clinical outcomes (Roche and Roche, 2020).
3.1.4. Endosome membrane fusion inhibition
It is widely reported that the host cellular entry involves the endocytic pathway and non-endosomal pathway. Inhibition of the pathway could be a strategy to inhibit the viral entry and infection. Mechanism of the drug targets could include drugs that have the ability to penetrate the lysosomes, accumulating inside and neutralizing the acidic pH of endosomes and lysosome. Such targets could inhibit the viral entry. The second possible mechanism involves protease inhibitors targeting clathrin-mediated endocytosis (Zhu et al., 2020; Zumla et al., 2016). These targets could play an important role in developing novel therapeutic strategies for treatment of COVID-19.
3.2. Disruption of the SARS-CoV-2 replication
Multiple antiviral agents are developed in order to disrupt viral replication such as DNA methyltransferase (MTases), polymerases and other entry proteins. Currently, the antiviral with the most potential is remdesivir (Mahase, 2020). It is a synthetic molecule which targets RNA synthesis, since it acts as an analogue of a DNA nucleoside, adenosine. Remdesivir is a competitive inhibitor of adenosine-triphosphate for the incorporation into viral RNA strands. Once it gets incorporated into the viral RNA at position I, it terminates RNA synthesis. The anti-viral effects have also been investigated in the rhesus macaque which was infected with MERS-CoV (Smith and Prosser, 2020a).
3.3. Suppression of inflammatory response
Beta coronavirus have the ability to infect immune cells such as monocytes, macrophages, and dendritic cells, resulting in the activation and secretion of IL-6 and other cytokines that have proinflammatory properties. Severe form of COVID-19 disease is characterized by fever and pneumonia that further develops to acute respiratory distress syndrome (ARDS). Elevation of serum concentrations interleukin-6 (IL-6) and other inflammatory cytokines results in CRS that induces ARDS, a hallmark of SARS-CoV2 infections. Developing effective IL-6 antagonists inhibits the cytokine-driven hyperinflammatory syndrome for the management of COVID-19 (Moore and June 2020).
4. Clinical application of drug targets and status of the outcomes
The various targets highlighted above have led to the exploration of various drugs either as mono or combination therapy. The outcomes and clinical significance for further application as treatment regimen of these drugs are discussed hereunder. Table 1 summarizes the clinical trials of the repurposed FDA approved drugs for treating COVID-19.
Table 1.
Drug molecules that are being studied for potential antiviral efficacy against SARS-CoV-2 virus.
| Therapy | Target | Therapeutic status | Reference |
|---|---|---|---|
| Remdesivir | RNA-dependent RNA polymerases | Under clinical trial for repurposing | (ClinicalTrials.gov, 2020e, ClinicalTrials.gov, 2020k; Guo, 2020; Sheahan et al., 2020; Wang et al., 2020; WHO, 2020b) |
| Chloroquine | Endosome/ACE2/pH-dependent enzymes such as proteases or glycosyltransferases | Under clinical trial for repurposing | (ANZCTR, 2020; ClinicalTrials.gov, 2020a; Guo, 2020; Wang et al., 2020) |
| Hydroxychloroquine | virus/cell fusion and cell try (Mild/Moderate Cases, Severe Cases) | Under clinical trial for repurposing | (ClinicalTrials.gov, 2020f; Gao et al., 2020), (clinicaltrials. gov, 2020c, clinicaltrials. gov, 2020g, clinicaltrials. gov, 2020h, clinicaltrials. gov, 2020i) |
| Hydroxychloroquine and Azithromycin | CYP2D6 (virus/cell fusion) | Under clinical trial for repurposing | (clinicaltrials gov, 2020b, clinicaltrials. gov, 2020l; clinicaltrialsregister. eu, 2020a, clinicaltrialsregister. eu, 2020b; Gautret et al., 2020a, Gautret et al., 2020b) |
| Camostat mesylate | TMPRSS2 (transmembrane protease, serine 2) | Under clinical trial for repurposing | (clinicaltrials.gov, 2020d; Hoffmann et al., 2020) |
| Arbidol | ACE2 and S protein | Under clinical trial for repurposing | Zhu et al. (2020) |
| Pirfenidone | IL-1β and IL-4 | Under clinical trial for repurposing | Driggin et al. (2020) |
| Baricitinib | JAK kinase | Under clinical trial for repurposing | Richardson et al. (2020) |
| Lopinavir/ritonavir | CYP3A4 | Under clinical trial for repurposing | (Driggin et al., 2020; Maxmen, 2020; Scott, 2020; Sheahan et al., 2020) |
| Favipiravir | RdRp (RNA-dependent RNA polymerase) | Under clinical trial for repurposing | (Guo, 2020; Mifsud et al., 2019) |
| Galidesivir | RNA-dependent RNA polymerases | Under clinical trial for repurposing | Warren et al. (2014) |
| BCX-4430 (salt form of galidesivir) | RNA-dependent RNA polymerases | Under clinical trial for repurposing | Warren et al. (2014) |
| Fingolimod | Lymphocytes | Under clinical trial for repurposing | Driggin et al. (2020) |
| Tocilizumab | Interleukin-6 | Under clinical trial for repurposing | Scott (2020) |
| Ribavirin | RdRp (RNA-dependent RNA polymerase) | Under clinical trial for repurposing | (Arabi et al., 2019; Guo, 2020; Wu et al., 2020) |
| Nitazoxanide | Unknown | Under clinical trial for repurposing | Guo (2020) |
| Ritonavir | viral proteases: 3CLpro or PLpro | Under clinical trial for repurposing | (Sheahan et al., 2020; Zeldin and Petruschke, 2004) |
| Darunavir | viral proteases: 3CLpro or PLpro | Under clinical trial for repurposing | Liu et al. (2020) |
| Methylprednisolone IV | Immunoglobulin M antibodies | Decreased death risk. Prospective clinical trial is planned | Driggin et al. (2020) |
| Convalescent plasma | spike protein | Recommended for clinical trials | Shen et al. (2020) |
| L-163491 | angiotensin AT2 receptor | Under clinical trial for repurposing | De Witt et al. (2000) |
| Bevacizumab | VEGF | Under clinical trial | Driggin et al. (2020) |
4.1. Repurposed FDA registered drugs
New drugs are needed urgently for management of the disease. However, due to the length of time it takes for final approval of a drug, as a matter of urgency, drug development has also focused on repurposing approved drugs already in the market. Sections below and Table 1 discuss those drugs and the phases of clinical trials.
4.1.1. Anti-helminthic drugs
Various parasitic drugs are being investigated at different stages of study for combating SARS-CoV-2. In a recent study, ivermectin, which is known to inhibit the in vitro replication of several positive stranded RNA viruses such as dengue, zika, chikungunya and yellow fever, could be a potential treatment options for COVID-19 (Leon et al., 2020). In vitro data shows 50% inhibition of SARS-CoV-2 replication after 48 h incubation with Vero cells at 2.8 μM/mL (equivalent to a plasma concentration of 2100 mg/mL), a concentration of 50-fold less than the concentration of ivermectin proven to reduce viral replication by half. This antiviral activity may lead to carefully conducted, controlled trials.
However, ivermectin can cross into the brain and react with GABA-gated chlorine channels causing neurotoxicity. This is usually prevented by the blood brain barrier (BBB), particularly by the P-glycoprotein (P-gp), hence, ivermectin may not be considered a safe drug if the BBB is compromised in the inflammatory state (Høy et al., 1990). Due to the leaky BBB caused by SARS-CoV-2 (Wu et al., 2020b), the use of ivermectin in severely ill patients should be administered with caution. Interestingly, if ivermectin is used in combination with other drugs, such as ritonavir, which is a very good CYP3A4 inhibitor with some effect on the P-gp as well (Li et al., 2019), this can boost ivermectin concentration, increasing Cmax and causing further toxicity by crossing the BBB leading to accumulation in the brain (Edwards, 2003; Kennedy et al., 2014).
4.1.2. Antimicrobial drugs
Chloroquine (CQ) and hydroxychloroquine (HCQ) are antiplasmodium drugs approved for malaria treatment. They are also a part of the model list of essential medicines declared by WHO (WHO, 2017). CQ and HCQ antiviral activity is attributed to their ability to elevate the pH of acidic intracellular organelles, such as endosomes and lysosomes that are known to be essential for viral-host cell membrane fusion. The second mode of action of CQ has been reported to be via the change of glycosylation of ACE2 receptor and spike protein (Liu et al., 2020b).
Chloroquine, is currently under intense investigation, albeit controversial, as a potential COVID-19 treatment (Wang et al., 2020b). Several in vitro, in vivo and clinical tests are being carried out for COVID-19. In vitro studies using infected Vero E6 cells treated with chloroquine, revealed that it CQ targets the virus at the entry and post-entry stages during infection. Furthermore, an EC90 value of 6.90 μM against the 2019-nCoV in Vero E6 cells after 48 h, in addition, an EC50 of 1.13 μM, CC50 > 100 μM, and SI > 88.50, was reported for CQ. This was in keeping with clinical studies where the plasma of rheumatoid arthritis patients were administered a daily dose of 250 mg of CQ (Bjelle et al., 1983; Rynes, 1997). Some cytotoxicity studies confirmed the biosafety of chloroquine (Cortegiani et al., 2020). Chloroquine may be potentially clinically applicable against the SARS-CoV-2, because of the cost and a known safety profile due to long usage for malaria treatment but requires intense scientific scrutiny for COVID-19, before use.
The immune-modulating activity of CQ was hypothesized to enhance its antiviral effect in vivo. However, its toxicity and side effects such as prolonging QT interval can trigger tachycardias such as Torsades de Pointes. This was proven in Brazil where 81 patients of COVID-19 were evaluated viz. a high-dose group receiving 12 g of chloroquine over 10 days, and a low-dose group of 2.7 g over 5 days (Borba et al., 2020). Twenty five % of high dose CQ group displayed 500 ms of QT elongation, and in 17% of them the QT interval elongation was fatal, in comparison to the lower dosage group. The toxic effects resulted in termination of the trial, and indicate that higher dosage and long duration of the treatment courses (10-day regimen) should not be recommended for management COVID-19.
Hydroxychloroquine (HCQ), a derivative of chloroquine, is a less toxic option that has been widely touted as a treatment option for SARS-CoV-2, based on various recent trials, studies and anecdotal reports. In one such study, the efficacy of HCQ was evaluated in a randomized clinical trial of 62 SARS-CoV-2 patients (Chen et al., 2020c), where the experimental group received 400 mg/d HCQ treatment for 5 days vs. control. Notably, more patients treated with HQC had improved pneumonia (25 of 31) vs. untreated groups, which showed 54.8% improvement (17 of 31). Notably 2 patients reacted mildly to the adverse effect of HCQ in the treatment group, while 4 patients progressed to severe illness in the control. Some showed significant relief through reduced cough and fever, supporting HCQ as an option in SARS-CoV-2 infections. Also, a key observation in this study is that by administering HCQ, the time to clinical recovery (TTCR) was significantly shortened and promoted the resolution of pneumonia in the group administered with HCQ. However, the methodology was not described, the study design was questionable, and outcomes were subjective, based on an individual clinical opinion.
Despite HCQ efficacy against SARS-CoV-2, the low cost and ease to access, its potential adverse side effects in viral diseases must also be carefully considered. Major side effects of HCQ includes elongation of the QT interval, thus being contraindicated in patients with underling cardiovascular disease. Other side effects include liver dysfunction, CNS effects like headache, dizziness, insomnia, peripheral neuropathy, retinopathy associated with long-term therapy, gastrointestinal reactions and itching (Bordy et al., 2018; Leon et al., 2020).
The combination of azithromycin and HCQ has been proposed to have better treatment outcomes, creating a buzz in mainstream media after reports that it may be a treatment option for SARS-CoV-2 (Gautret et al., 2020a, Gautret et al., 2020b). However, shortcomings included lack of randomization of the groups and a very small patient cohort, and the efficacy of the combination is therefore inconclusive. In contrast, in another study with similar shortcomings that assessed only 11 patients, HCQ and azithromycin did not show rapid viral clearance, with little clinical benefit (10 out of 11 patients had fever and received nasal oxygen therapy, 1 patient died, 2 were transferred to the ICU). In one of the patients, HCQ and azithromycin had to be discontinued after 4 days because of a prolongation of the QT interval. Nasopharyngeal swabs in 10 patients, 5–6 days after treatment showed that they were still positive of SARS-CoV-2 (Molina et al., 2020).
The side effects associated with HCQ treatment are still a major cause of concern. On 9th April, the Le Monde newspaper, France reported since March 27, of the 54 reported cases of cardiac disorders, there were 7 sudden cardiac arrests, 37 cases of prolonged QT and 10 arrhythmias accompanied with syncope, including 4 fatalities, in patients taking HCQ, and in some cases in combination with azithromycin (Cabut, 2019). Based on anecdotal reports, HCQ is promising for COVID-19, however, to date, there are no robust studies of HCQ which adequately demonstrates efficacy, absence of toxicity or proper clinical guidelines for usage. Currently studies examining hydroxychloroquine and chloroquine are underway in order to determine the efficacy and safety, and it does not seem promising as some institutions like National Institute for Health are advising against its use in their guidelines for treatment (NIH, 2020). Moreover, patients currently on HCQ and azithromycin treatment need therapeutic drug monitoring to avoid toxic effects associated with the drug.
4.1.3. Antiviral drugs
Based on the pathophysiology of SARS-CoV-2, and the viral life cycle, several antiviral drugs have been screened and repurposed as targets against COVID-19.
Such drugs include remdesivir, an analogue of adenosine, which interacts with the viral RNA chains and leads to premature termination (Warren et al., 2016). The drug has completed phase III clinical trials for the treatment of Ebola. Thus, the data on its safety for use in humans is available, and it can be accelerated to clinical trials, if preclinical efficacy against SARS-CoV-2 is proven (Lu et al., 2020). Studies have shown remdesivir to have potent antiviral activity against a clinical isolate of SARS-CoV-2. Preliminary trials have indicated that it was effective against SARS-CoV-19 in vitro ( Lai et al., 2020 ) and in Vero E6 cells results indicated that it acted at the post virus entry stage (Wang et al., 2020b). An EC50 of 0.77 μM, CC50 > 100 μM, and SI > 129.87 was reported in Vero E6 cells that were exposed to remdesivir treatment. The EC90 value of remdesivir against SARS-CoV-2 in Vero E6 cells was reported to be 1.76 μM. This implies that at this concentration, remdesivir may work in clinical models, and that at 10 μM, it completely (100%) prevented Ebola virus infection (Warren et al., 2016). The drug was then accelerated to clinical trials by U.S. Food and Drug Administration's (FDA) and two Phase 3 clinical studies began to evaluate the safety and efficacy. A clinical trial showed that three quarters of patients that were on the drug had better outcomes than those who were not (Gilead, 2020). However, setbacks such as cohort size, the duration of the follow up, lack of information about the patients initially treated, as well as lack of a control group hindered this study. In a recent well designed trial that was randomized, double-blind and with a placebo control, no statistically significant clinical benefits of remdesivir administration was observed when adult patients were admitted to hospital with severe COVID-19 (Wang et al., 2020e). However, there was reduction in time to clinical improvement in patients given early treatment. Although the study showed optimism, there is a need for studies with a larger group of patients to confirm the results.
Another antiviral drug being considered as anti-SARS-CoV-2 is camostat mesylate (CM), which is a clinically proven serine protease inhibitor. In vitro studies investigated the antiviral efficacy of CM in Caco-2, 293T, Vero and Vero-TMPRSS2 cells lines, respectively (Hoffmann et al., 2020b). Its ability to inhibit the entry of SARS-CoV-2 was shown. This inhibitory property of the drug was enhanced when it was combined with a CatB/L inhibitor (E−64d), indicating that SARS-2 can utilize CatB/L and TMPRSS2 for priming in these cell lines. The study also showed that CM treatment significantly reduced Calu-3 infection with SARS-CoV-2. This suggests that SARS-CoV-2 can use TMPRSS2 for S protein priming and that CM is a good inhibitor of TMPRSS2, as it blocked the lung infection. These findings are in keeping with studies where SARS-2-CoV-2 exploits ACE2 receptor for cellular entry (Zhou et al., 2020b). These results provide an understanding of the transmissibility and pathogenesis of SARS-CoV as well as major targets for therapeutic interventions. Nafamostat mesylate, a drug used to treat acute pancreatitis, has shown some potential to inhibit fusion of the envelope of the virus to the host cell surface membranes. It has been utilized in Japan for many years and there is adequate data to ensure safety (Hoffmann et al., 2020b).
Another drug that is FDA approved and is currently being considered as a treatment as an anti-SARS-CoV-2 is lopinavir/ritonavir. Ritonavir and lopinavir, a combination drug, which is sold under the brand name Kaletra®, showed promise as a target of the coronavirus protease enzyme. The protease enzyme is responsible for the cleavage of the gag-pol polyproteins which plays an important role in the virus replication cycle (Cao et al., 2020) The main role of ritonavir in the combination is to enhance protease inhibitor (PI) regimens in order to improve the pharmacokinetics of the lopinavir. The bioavailability of the second lopinavir is greatly improved by the interaction with CYP3A4 and twice daily dosing is made possible. This boosted PI regimen is evident by high levels of viral suppression among antiretroviral naïve and prior PI treated patients (Zeldin and Petruschke, 2004). The clinical benefits of administering lopinavir/ritonavir to patients with COVID-19, was evaluated via a clinical trial involving 199 hospitalized adult patients. The outcomes of the study showed there was no difference in the mortality after 28 days of treatment in the group receiving lopinavir/ritonavir and the standard-care groups. Moreover, gastrointestinal adverse events were more common in the lopinavir–ritonavir group. However, serious effects were experienced in the standard-care group. Lopinavir–ritonavir treatment was stopped early in 13.8% of the patients because of adverse events, leading to the conclusion that no benefit was observed with lopinavir–ritonavir treatment beyond standard-care (Cao et al., 2020). Similar results have been reported in other clinical trials (Colson et al., 2020). However, more studies involving high sample sizes of patients at different stages of the disease need to be performed to conclusively determine the efficacy of the drug.
By tampering with vRNA synthesis, it is possible to eliminate viruses. Hence, therapeutic drugs such as favipiravir, which acts by preventing RNA strand elongation and viral proliferation via purine nucleoside inhibition (Furuta et al., 2005), is being considered for SARS-CoV-2. Broad-spectrum activity of favipiravir against RNA viruses such as West Nile virus, poliovirus, Ebola virus and norovirus (Rocha-Pereira et al., 2012) makes it a suitable candidate for SARS-CoV-2. Phase I clinical trials to evaluate its efficacy against SARs-CoV-2 have begun and doses ranging from 30 to 1600 mg/kg have shown no serious adverse effects (Kobayashi et al., 2008). Unfortunately, a more recent study (Furuta et al., 2017), has shown that favipiravir has teratogenic and/or embryotoxic effects. Favipiravir use has been linked with other mild adverse effects such as diarrhea, nausea, vomiting and elevation of blood acid levels (Health, 2018). Based on these studies, favipiravir may be recommended for patients whose condition is not complicated. A phase II Chinese clinical trial on the pharmacokinetics and efficacy of favipiravir was conducted last year, and it is envisaged that the results of this study will shed more light on favipiravir for SARS-CoV-2.
Combinations of different classes of antiviral drugs are being proposed to manage COVID-19. Such initiatives include clinical trials to evaluate the efficacy of lopinavir plus ritonavir and arbidol combination against the novel coronavirus infection. The outcome of a randomized clinical trial involving 125 participants is expected in May 2020 (ClinicalTrials.gov, 2020a). It is proposed and hypothesized that the combination of these drugs will provide better efficacy against SARS-CoV-2 as a synergistic effect is expected.
At the moment, most of these therapeutic drugs are still under clinical trials and their efficacy against SARS-CoV-2 have not been determined holistically. Other drugs that are currently being investigated at various stages of clinical trials for potential application in SARS-CoV-2 treatment include pirfenidone (targets IL-1β and IL-4), rintatolimod (targets TLR-3), plitidepsin (targets EF1A), anti-SARS-CoV-2 polyclonal hyperimmune globulin (targets immunoglobulin), kevzara (targets interleukin-6), brilacidin (targets immunoglobulin), TAK-888 (Takeda) and Gimsilumab (Roivant), amongst many others (Driggin et al., 2020). Most of the proposed treatment approaches have involved targeting the replication cycle of the virus.
4.2. Convalescent plasma therapy
Convalescent plasma therapy involves antibodies (Ab) transfusion a person who has recovered into an infected person, to initiate an immune system response against the disease. The possible sources of SARS-CoV-2 Abs are human convalescent sera, obtained from individuals who have fully recovered from SARS-CoV-2 through humoral immunity, monoclonal antibodies, or they can be acquired from animal hosts, such as genetically engineered cows who produce human antibodies (Beigel et al., 2018). The convalescent blood products are able to cause neutralization of the virus, and ultimately lead to eradication from the blood circulatory system through induction of artificially acquired immunity (Marano et al., 2016). There are ongoing trials of convalescent plasma in patients with SARS-CoV-2 (Smith and Prosser, 2020a, 2020b). Notably, on the March 24, 2020, the FDA granted clinicians the permission to make use of investigational convalescent plasma for single patient use under emergency Investigational New Drug Applications (FDA, 2020). Eligible patients have to be positive for SARS-CoV-2, the disease must be severe and life threatening, and they must be able to provide informed consent.
A more recent study on convalescent plasma treatment for SARS-CoV-2 (Shen et al., 2020) was conducted in 5 patients with SARS-CoV-2, who were transfused with convalescent plasma containing SARS-CoV-2– specific antibody (IgG) binding with titer greater than 1:1000. This was obtained from 5 patients who just recovered from SARS-CoV-2, and the newly infected patients were administered the convalescent plasma 10 and 22 days after admission. Observations after treatment included normalization temperature within 3 days in 4 out of 5 patients. Increased PAO2/FIO2, and decreased viral load which subsequently became negative within 12 days, was observed. In addition, after 12 days, ARDS was resolved in 4 patients and mechanical ventilation was stopped for 3 patients within 14 days. Three patients were discharged after 55 days. In another study in China, 10 patients who had undergone convalescent therapy showed a shortening of the duration of symptoms, improved oxygen levels and a drop in the viral load (Duan et al., 2020). These results show that the administration of convalescent plasma containing neutralizing antibody lead to improved clinical status. Due to the limited sample size, it is suggested that these observations be evaluated in randomized clinical trials with a larger number of participants.
4.3. Adjuvant therapies
Adjuvant therapy refers to treatment that is given, in addition to the primary (initial) treatment, to lower the risk of the disease during management. COVID-19 patients presenting with bacterial co-infection are associated with serious outcomes. In a retrospective study on 191 patients from Jinyintan Hospital and Wuhan Pulmonary Hospital, half of the deaths experienced a secondary infection (Zhou et al., 2020a). In Wuhan China again, another cohort of severely ill patients displayed bacterial and fungal coinfections, and common secondary infections included A baumannii, K pneumoniae, A flavus, C glabrata, and C albicans (Chen et al., 2020b). The co-infections have necessitated the inclusion of antibiotics as adjuvant therapy for COVID-19 patients, which can be utilized based upon institutional antibiograms, and as well as after culture sensitivity testing. Standard therapy has included, ceftriaxone 1–2 g IV daily in combination with azithromycin 500 mg IV for 3 days (Ruan et al., 2020).
Non-steroidal anti-inflammatory drugs (NSAIDs) have also been found to be relevant in the symptomatic treatment of SARS-CoV-2. Paracetamol is indicated to manage the high fever associated with SARS-CoV-2. NSAIDs such as Ibuprofen have been hypothesized to exacerbate the viral activity as it ‘dampens’ the immune system, increases risk of stroke, myocardial infarction and nephrotoxicity. Moreover, acute respiratory tract infections are usually associated with increased risk from those effects. Therefore, even short-term use of NSAIDs by patients with COVID-19 was thought to worsen the disease (Day, 2020). However, European Medicines Agency (EMA) has recently stated that currently there is no scientific evidence for the link between the worsening of COVID-19 and Ibuprofen (EMA, 2020). WHO supported this after a systemic review o NSAIDS usage which indicated no evidence of harm in patients with COVID-19 (WHO, 2020c).
A coordinated cytokine response forms part an effective host immune response. However, a dysregulated pathological outcome in some patients manifests as a hyperinflammatory response (Fig. 3), and some patients infected with SARS-CoV-2 experience this phenomenon known as a ‘cytokine storm’ (Li et al., 2020b). The use of corticosteroids has been pharmacologically effective in reducing the inflammation. As of March 2020, there have been no clinical trials on the use of corticosteroids for the treatment of SARS-CoV-2. However, corticosteroids are administered in high doses for a longer treatment duration, resulting in side effects (Yasir and Sonthalia, 2019). Nonetheless in April, Zhan et al. reported the first case of a COVID-19 patient with multiple myeloma being successfully treated with tocilizumab (Zhang et al., 2020d). The known effects of cytokine storm by SARS-CoV-2 infection have been attributed to IL-6, therefore the management by tocilizumab is considered as a better option than corticosteroids (Kotch et al., 2019; Luo et al.). Reports for successful use of tocilizumab in the management of the ‘cytokine storm’ call for randomized clinical trials with high number of participants so that clinical guidelines can be drawn (Mehta et al., 2020).
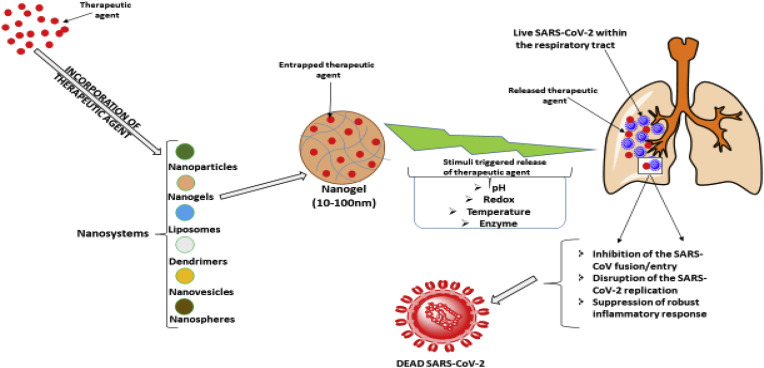
Fig. 3.
Mechanisms of nano-encapsulated therapeutic molecules against SARS-CoV-2 virus. Different drugs can be encapsulated in nanodrug delivery system with ability to specifically target the virus or its infection sites. Enzymes specific to the lungs, conditions peculiar to COVID-19 such as pH and receptors can be employed to specifically increase the drug concentration at the SARS-CoV-2 infection sites.
Other immune modulators and viral entry inhibitors include monoclonal and polyclonal antibodies. In addition, sirolimus and everolimus are macrolides that have been used to prevent organ rejection. Their target protein is the mTOR which belongs to the family of protein kinases (Kindrachuk et al., 2014). They may be used to prevent a cytokine storm experienced by SARS-CoV-19 patients. Kiniksa Pharmaceuticals has announced early confirmation of treatment response with mavrilimumab. It is a fully human monoclonal antibody that is still under investigatory stages. It has been shown to target granulocyte macrophage colony stimulating factor alpha (GM-CSFα) (Trus et al., 2020). The basis of this is the involvement of GM-CSFα in the mechanism of over activation cellular immune response in the lungs. It is thought to be the major contributor of the mortality associated with the disease. Recent reports suggest that patients diagnosed with SARS-CoV-2 have higher serum levels of proinflammatory cytokines such as GM-CSFα and interferon gamma (Tsantikos et al., 2018). Kiniksa is on track to begin phase 2 and 3 clinical development of mavrilimumab for SARS-CoV-2 pneumonia (Crotti et al., 2019). It is intended to be used in patients suffering from severe SARS-CoV-2 pneumonia and hyperinflammation. Thus far, 6 patients have been treated with mavrilimumab, and all the patients exhibited improved oxygenation, as well as early resolution of fever. In addition, none of the patients required mechanical ventilation. Furthermore, mavrilimumab has shown a high therapeutic index as it was well tolerated (Bloomberg, 2020).
Severe cases of SARS-CoV-2 have been associated with a cytokine storm that leads to activation of the coagulation cascade, leading to thrombotic phenomena (Henry et al., 2020; Iba et al., 2020). Furthermore, there are reports of an association between abnormal coagulation parameters that causes coagulopathy and DIC which is linked to mortality in COVID-19 patients. In a clinical trial of 449 patients with severe COVID-19, 99 of them received low molecular weight heparin, (LMWH) for 7 days or longer. The results indicated a better prognosis in severe COVID-19 patients with markedly elevated D-dimer on LMWH than non-coagulant users (Tang et al., 2020a). The use of anticoagulant therapy with heparin has been recommended as it has shown to decrease mortality in COVID-19 patients (Coppola et al., 2020).
5. Emerging strategies
While the discovery and identification of drug targets are still underway, there is an urgent need for a cure for SARS-CoV-2. There are several clinical trials still ongoing and more information from those studies will provide treatment guidelines and safe and effective therapeutic options. Several drugs such as remdesivir, favipiravir and ribavirin have been selected for the SOLIDARITY trial that is currently taking place globally (WHO, 2020aa, WHO, 2020ab). However, most of the drugs on trial and those from anecdotal reports have been sourced largely from repurposing, and they were not specifically designed to target SARS-CoV-2 virus.
This SARS-CoV-2 pandemic has resulted in massive investment into biomedical and pharmaceutical research accelerating the development of drugs for COVID 19. However, approval rate for new drugs by the U.S. FDA has remained relatively low and limited in recent times.
Single stranded positive sense RNA viruses like the SARS-CoV-2 are prone to mutation, hence as expected, there are current reports of various strains of the virus worldwide (Tang et al., 2020b). This could imply resistance, also to the repurposed drugs. Thus, even though repurposing of drugs gathers pace there is a need for drugs specifically designed to combat the virus and with the ability to avoid resistance. These compounding factors in the search for new drugs, calls for a novel, ‘outside of the box’ approach. Some of the non-traditional methods for drug delivery and design are discussed below.
5.1. Bioinformatics and cheminformatics
Bioinformatics and cheminformatics are newer strategies to screen and design various drug candidates for SARS-CoV-2. Computation chemistry analytical tools have found relevance in predicting the interactions between therapeutic agents and their binding affinity/sites within the human body and the disease-causing organisms. This strategy could lead to the generation of more accurate approaches and techniques for SARS-CoV-2 management. For instance, a simulation study by Kadam and Wilson revealed that arbidol, (umifenovir) could be a broad-spectrum antiviral agent against both enveloped and non-enveloped viruses (Kadam and Wilson, 2017). Aribidol is an anti-influenza drug and it's anti-SARS-CoV-2 effects were determined in predictive in silico studies (Vankadari, 2020) and its efficacy was reinforced via in vitro studies (Wang et al., 2020c). Such predictive studies make it possible for faster and cost-effective development of drugs. After efficacy was proved, the drug has been registered for clinical trials (Kadam and Wilson, 2017; Mina et al., 2020). Moreover, it has now been included in guidelines for the Prevention, Diagnosis, and Treatment of Novel Coronavirus by the National Health Commission (NHC) of the People's Republic of China (Wang et al., 2020c).
Baricitinib, which is a janus inhibitor was also subjected to molecular modelling predictions for its potential in the treatment of SARS-CoV-2b (Richardson et al., 2020b). Amongst the six drugs with high binding affinity to AAK-1, baricitinib which also binds the cyclin G-associated kinase (endocytosis regulator), was shown to be a promising drug target for SARS-CoV. Baricitinib is now in clinical trials for SARS-CoV in a patient population as it has proven to reduce inflammation and inhibit viral entry (Richardson et al., 2020b).
5.2. Structure-based drug design
Studies to date indicate that drugs that have a similar grouping to chloroquine and hydroxychloroquine, may have some activity against SARS-CoV-2 (Hu and Bajorath, 2012). Chlorpromazine as well as dasatinib have a similar structure to chloroquine. Both drugs are also chlorobenzenes, and thus can be exploited for potential activity against SARS-CoV-2. Combinations of two or more drugs have a higher probability of achieving the desired targets of viral replication, entry and modulation of the host immune response.
5.3. Network-based methods for prediction of drug-target interactions
Another non-conventional method used to search for drug targets of COVID-19 is the use of Network-based drug methodology for predicting drug targets (Wu et al., 2018). Through development of systems biology and network pharmacology, there is a paradigm shift in drug discovery from a linear mode of one drug for one target and one disease, to mapping multiple drug targets for multiple diseases via the network based method (Anighoro et al., 2014). Network based methodologies apply the power of networks to establish correlations between various data sets of interest for drug discovery. For SARS-CoV-2, the methodology can combine systems pharmacology-based network platforms, to identify the various interactions between the virus, the host cells and proteins that could be possible drug targets.
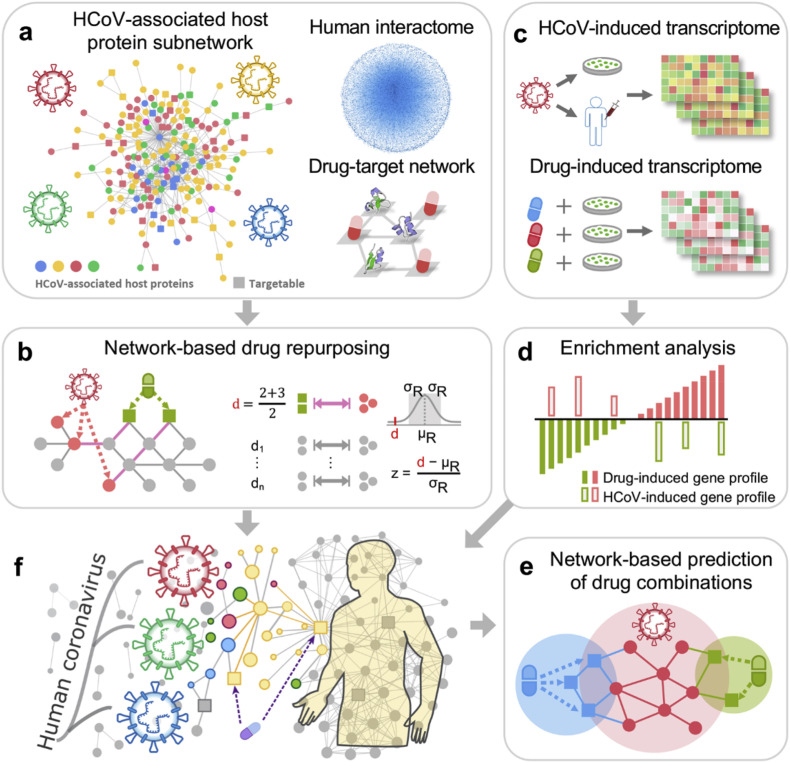
Zhou et al. built the host-virus interactome network of various viruses such as HCoV-229E, HCoV-NL63, MERS-CoV and SARS-CoV, mouse hepatitis virus (MHV), avian infectious bronchitis with virus nucleocapsid protein (N protein) and 119 host proteins associated with corona virus infection signaling pathways obtained from various experimental studies ( Fig. 2 .) (Zhou et al., 2020c). The study found that 47 human proteins could be targeted by at least one FDA approved and experimental medicines under clinical trials. To further refine the results, drug–target networks were built to generate information related to more than 2000 anti CoV drugs that are both approved by FDA and those in experimental phase. From the network, 135 drugs were identified as possible candidates to manage the virus. From the dataset, 200 were further optimized, and the top 16 most repurposable drugs for COVID-19, with least expected side-effects and targeting specificity, were identified. The drugs included irbesartan, toremefene, camphor, equilin, mesalazine, mecaptopurine, paroxetine, sirolimus, carvedilol, colcochine, dactinomycin, melatonin, quinacrine, eplerenone, emodin and oxymetholone. Most of the drugs that have been shown clinically to have antiviral activity, act via different mechanisms. Moreover, integration of drug–target networks of the top 16 drug candidates produced of potent drug combinations using a complementary exposure technique (Chen et al., 2020b). Other studies have also shown the potential of using a network based methodology to screen possible drug targets for SARS-CoV-2 (Hofmarcher et al., 2020; Kumar et al., 2020; Li et al., 2020c).The method is a quick way of identifying drug targets. However, it is not designed to produce drugs, but is a useful tool for prediction of new drugs, and combinations, and reduces the period of drug screening and development. Once prediction is done, extensive in vitro, in vivo and human trial studies are still required.
Fig. 2.
Network-based methodology combining pharmacology-based network medicine platform to quantify the interplay between the virus–host interactome and drug targets in the human protein-protein interactions network. a Human coronavirus (HCoV) associated host protein sourced from literature and pooled to generate a protein subnetwork. b Network proximity between drug targets and HCoV-associated proteins that were calculated to screen for candidate repurposability. c, d Gene set utilized to validate the network-based prediction. e Prioritization of top candidates for drug combinations using network based. Reproduced with permission from (Zhu et al., 2020).
5.4. Artificial intelligence and machine learning
Artificial intelligence (AI) and machine learning (ML) can contribute to drug development for COVID-19, by improving the speed and efficiency of repurposing and proposing new potent molecules to inhibit SARS-CoV-2. Both AL and ML can also be employed to predict the spread of the disease. Application of AI and ML for the development of diagnostic and monitoring techniques for COVID-19 has proven to be successful, through increasing accuracy, speed and protecting healthcare workers from exposure (Gozes et al., 2020; Wang et al., 2020d). With no effective treatment regimen for COVID-19, it is paramount to design therapeutic approaches that repurpose clinically approved drugs by designing new therapies via AI and ML. Zhavoronkov et al. recently reported new druglike molecules against SARS-CoV-2 targets using machine deep learning (Zhavoronkov et al., 2020). AI and ML were used to screen the possibility of repurposing poly-ADP-ribose polymerase 1 (PARP1) inhibitor, CVL218, which is currently in Phase I clinical trial to inhibit SARS-CoV-2 (Ge et al., 2020). Using AI, it was discovered that Baricitinib could be used in the treatment of COVID-19, and have now started a randomized-controlled trial with the US National Institute of Allergy and Infectious Diseases (NIAID, to evaluate its efficacy to inhibit COVID-19 infection and reduce inflammatory damage due to the disease (BenevolentAI, 2020; Russell et al., 2020). Google also recently developed a deep learning system, AlphaFold, which was released to predict protein structures associated with COVID-19. This usually takes several months via conventional experimental methods to determine and has proven to be a valuable resource for developing COVID-19 drug targets and vaccines (Senior et al., 2020). With increased amount of research towards COVID-19 and the voluminous data being published; this information can be crucial in feeding into AI and ML system for developing potent anti- SARS-CoV-2 drugs.
5.5. Phage technique
Presently, antivirals are used to attack viruses after the lungs have been affected. It would be advantageous to prevent infection rather than to treat it. A novel study has taken on an approach that includes a phage capsid that enveloped flu viruses perfectly, so that they cannot infect cells (Lauster et al., 2020). The SARS-CoV-2 needs a protease called Mpro or 3CLpro to form its viral replication complex. This enzyme can be blocked by a drug which provides a complimentary 3D shape. Scientists at University of Lübeck used high intensity x-rays to elucidate the proteases 3D structure. The X-ray structures were reported for SARS-CoV-2 Mpro with α-ketoamide inhibitor and resulted in a lead compound that was further developed to produce a potent inhibitor of SARS-CoV-2. The inhibitor, 13b, showed that it could be well tolerated in mice via the inhalation route (Zhang et al., 2020c). Direct administration of the molecule to the lungs has some possibility. However, a pharmaceutical input is needed and it may be several years before it can be used as a coronavirus treatment (ter Meulen et al., 2004; Walter et al., 2020).
5.6. Novel drug delivery systems approach
In addition to the development of new drugs, the design of novel drug delivery systems such as nano-sized systems, need to be considered, as they have been shown to be superior in the treatment of disease conditions and overcoming drug resistance as compared to conventional dosage forms. The identification of suitable drugs will therefore require innovative and superior delivery approaches to maximise their therapeutic efficacy. Nanosystems provide increased drug accumulation at the target site of the disease, increase intracellular penetration, protect drugs from degradation, increase stability and can prevent mechanisms of drug resistance (Fig. 3 ). In addition, nanosystems have the ability to incorporate different therapeutic agents which can be released in a controlled manner to the specific target site. Nanosystems being used for various antiviral drugs can be similarly explored for SARS-CoV-2. S protein of CoV forms the characteristic corona of the virus, with distinctive spikes on the viral envelope (Holmes, 2003) as well as complex surface projections with sizes of about 20–40 nm (Ksiazek et al., 2003). Therefore, targeting of the S protein of CoV using nanosystems with sizes of about 10–100 nm may be a promising approach for early diagnosis as well as treatment. Hence, it is highly recommended that alternate drug delivery systems, such as the use of nanomaterials, be developed in order to more effectively deliver current and new drugs showing promise in eradicating SARS-CoV-2. Fig. 3 below shows a possible mechanism by which nanosystems can be used to target specific sites associated with SARS-CoV-2 infections.
5.7. Plant-based remedies
According to available literature, plant based remedies have been used for centuries to tackle respiratory tract infections and may therefore be helpful in the treatment of SARS-CoV-2 (Ren et al., 2020). Chinese plant based therapy consisting of a mixture of Chinese herbs such as San Wu Huangqin Decoction, Lianhuaqingwen Capsules, and Yinhuapinggan granules have shown to display antiviral properties (Li et al., 2020d). The mechanisms of action of such plants are associated with inhibition of replication of viral particles as well as blocking proliferation (Ren et al., 2020). Diammonium glycyrrhizinate, which is an extract of Glycyrrhiza glabra (Liquorice) has been recommended to treat COVID-19 (Chen et al., 2020a; Murck, 2020); however, no official guidelines have been issued yet. The other commonly used herbs in COVID-19 management include, Glycyrrhizae Radix, Et Rhizoma (Gancao), Atractylodis Macrocephalae Rhizoma (Baizhu), Saposhnikoviae Radix (Fangfeng), Lonicerae Japonicae Flos (Jinyinhua), and Forsythiae Fructus (Lianqiao) Astragali Radix. Astragali Radix (Huangqi), Saposhnikoviae Radix (Fangfeng), and Atractylodis Macrocephalae Rhizoma (Baizhu) which are all components of a traditional herbal formula called Yupingfeng Powder (Xu and Zhang, 2020). Certain studies have demonstrated that Yupifeng powder has anti-inflammatory, antiviral and immunoregulatory effects (Li et al., 2020a). A review conducted by Cochrane, found that the combination of Traditional Chinese Medicine with Western Medicine demonstrated a great improvement in symptoms of SARS-CoV-2 (Liu et al., 2004, 2020c). Stilbene-based natural compounds were reported to be effective in inhibiting spike protein and human ACE2 receptor complex SAR-COV2 in silico (Wahedi et al., 2020). Unlike conventional medicines, herbal and plant products are not well understood and must be used with extreme caution, to prevent herb-drug interactions. As of now, there are no rigorous peer reviewed clinical trials of plant-based therapies. Most approvals and use are based upon anecdotal clinical reports. Based upon historical records and ethnomedical sources, plant-based therapies could be an alternative source of an anti-SARS-CoV2 potent drug.
5.8. Vaccines
For an innate immune response by the body to viruses to be initiated, the cells must have the ability to identify the pathogen trying to invade it. Currently, there are several clinical trials being directed to develop a vaccine, based on several different strategies against SARS-CoV-2 as illustrated in Table 2 . Such approaches include those that target the spike glycoprotein (S protein) as the major inducer of the immune response. Numerous strategies have been trialled, which include and are not limited to the use of the full length S protein, S1 receptor binding domain (RBD), DNA or viral vectors and finally expression of virus like proteins (VLP) (Dhama et al., 2020). Phase 1 human clinical trials are anticipated to begin in April 2020, and many have commenced. The Bill and Melinda Gates Foundation have provided a grant to Inovio for a large-scale intradermal vaccine entitled INO-4800. On 16th March 2020, Moderna Inc started phase 1 trials in 45 healthy volunteers at the Kaiser Permanente Washington Health Research Center. As of April 2020, there are more than 115 possible vaccine candidates of SARS-CoV-2 that are being developed globally (Smith and Prosser, 2020a), (Le et al., 2020).
Table 2.
Status of vaccines in development against SARS-CoV-2.
| Developer | Molecular target | Phase of clinical trial | References |
|---|---|---|---|
| Biontech | mRNA Vaccine | Phase I | Biontech (2020) |
| Hoth Therapeutics | Self-assembling Vaccine | Pre-Clinical | Hoth (2020) |
| Moderna | mRNA Vaccine | Phase I | clinicaltrials.gov (2020j) |
| Johnson&Johnson | COVID-19 Vaccine | Pre-Clinical | Chen et al. (2020) |
| CanSinoBIO | COVID-19 Vaccine | Phase I | CanSinoBIO (2020) |
| Inovio | COVID-19 Vaccine | Phase I | (Amanat and Krammer, 2020; WHO, 2020a) |
| Janssen Pharmaceutical | Non-replicating viral vector | Pre-clinical | WHO (2020a) |
| Clover Biopharmaceuticals Inc./GSK | Protein subunit; S trimer | Pre-clinical | GSK (2020) |
| University of Oxford | Non-replicating viral vector; ChAdOx1 | Phase I | Lane (2020) |
| Sinovac | Formaldehyde in activated + alum | Phase I | sinovac (2020) |
| Codagenix/Serum Institute of India | Deoptimized live attanuated vaccines | Pre-clinical | WHO (2020a) |
| Novavax | Full length S trimers/nanoparticle + Matrix M | Pre-clinical | Novavax (2020) |
| Vaxart | Non-replicating viral vector; | Pre-clinical | Vaxart (2020) |
Some of the approaches that have been involved in creating a vaccine have also included the development of attenuated vaccines. By introducing a mutation into ORF1a/b polyprotein, the lack of virulence of the mouse coronavirus was demonstrated (MHV-A59) due to attenuation of the virus (Liu et al., 2020a). DNA based vaccines include patent WO2005081716, which induces and enhances immune responses, in particular against antigens of the SARS coronaviruses. The immune response is induced by chimeric nucleic acids which encodes for an endoplasmic reticulum polypeptide such as calreticulin. Employing gene gun delivery to deliver DNA-coated gold particles, it was shown that vaccinated animals displayed both humoral and T cell mediated immune response (Liu et al., 2020a).
Another approach involves protein-based vaccines, capable of invoking an immune response against SARS. The one patented by GlaxoSmithKline contains an S protein immunogen together with oil in water emulsion which is also an adjuvant. The vaccine showed induction of high levels of anti-SARS-CoV IgG2a and IgG2b antibodies and neutralizing antibodies in animal testing. Virus-like particle vaccines are being tested by Moderna. They report that mRNA-1273 is an mRNA vaccine which targets a prefusion stabilized form of an S protein which is associated with SARS-CoV-2 (http://clinicaltrials.gov/ clinicaltrials. gov, 2020b).
Although, the use of vaccines is a generally a better preventive approach, unfortunately, they are currently undergoing development. The few that have been developed are still under different stages of pre-clinical and clinical trials. This implies that there is no certainty in respect to their efficacy against SARS-CoV-2, hence, the disease is still a threat globally.
6. Concluding remarks
COVID-19 is a pandemic which is currently threatening the health of the entire human race, in addition to the global economy. Therefore, providing a solution and cure has become a matter of urgency which may not allow for the development and evaluation of new biomolecules, the approval of which is a lengthy process. Hence, a better alternative is to repurpose therapeutic drugs or approaches that can target pathophysiological pathways that are associated with the disease. Several potential therapeutic agents and approaches against SARS-CoV-2 have been highlighted and discussed in detail. Various successful and unsuccessful in vitro and in vivo studies have been reported for drugs that are currently repurposed to treat SARS-CoV-2. Some of these repurposed drugs have shown some encouraging clinical outcomes and have been taken further to different stages of clinical trials. However, due to the dire need for therapeutic options, some of the reported studies are preliminary pilot studies with lower sample sizes, have a poor study design such as lack of randomization and do not include controls and placebos in the evaluations. There is a need for further investigations that include larger number of participants, evaluate long-term implications on human health and identify the toxicity profile of dosages proposed to be effective against the virus. Future newly designed therapeutic agents or/and approaches must be target specific, effective and safe to treat the infected patient. The development of novel drug delivery systems that target the virus could go a long way in preventing possible viral resistance. The development of various vaccines against this deadly virus as a preventative therapeutic is also ongoing, as the world is desperately in need of a solution to this virus.
Despite significant progress that has been made by scientists based on in vitro, animal, cohort studies and clinical trials, there is currently no definitive cure and vaccine. However, some studies appear to be very promising. A major limitation observed in all of these studies is that no long-term experimental protocol has been undertaken in order to evaluate the long-term safety and efficacy of the proposed anti SARS-CoV-2 drugs. It is therefore, strongly advised that long term studies be conducted for each of the therapeutic approaches and vaccines.
Authors contribution
Calvin A. Omolo: Conceptualization, methodology, writing of the original draft, data curation and revision of the manuscript. Nikki Soni: Writing of the original draft, data curation. Victoria Fasiku: Writing, reviewing and editing, data curation. Irene Mackraj: Writing and reviewing of the manuscript. Thirumala Govender: Conceptualization, supervision, reviewing and editing. All authors read and approved the final manuscript.
Declaration of competing interest
Authors declare no conflict of interest.
Acknowledgments
The authors acknowledge the University of KwaZulu-Natal (UKZN) South Africa and United States International University Africa Kenya for financial support.
References
- Amanat F., Krammer F. SARS-CoV-2 vaccines: Status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anighoro A., Bajorath J., Rastelli G. Polypharmacology: challenges and opportunities in drug discovery: miniperspective. J. Med. Chem. 2014;57:7874–7887. doi: 10.1021/jm5006463. [DOI] [PubMed] [Google Scholar]
- Anzctr . anzctr.org; 2020. Chloroquine Chemorophylaxis Countermeasure against COVID-19. [Google Scholar]
- Arabi Y.M., Shalhoub S., Mandourah Y., Al-Hameed F., Al-Omari A., Al Qasim E., Jose J., Alraddadi B., Almotairi A., Al Khatib K. Ribavirin and interferon therapy for critically ill patients with Middle East respiratory syndrome: a multicenter observational study. Clin. Infect. Dis. 2019 doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020;25:2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavishi C., Maddox T.M., Messerli F.H. Coronavirus disease 2019 (COVID-19) infection and renin angiotensin system blockers. JAMA Cardiology. 2020 doi: 10.1001/jamacardio.2020.1282. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Voell J., Kumar P., Raviprakash K., Wu H., Jiao J.-A., Sullivan E., Luke T., Davey R.T., Jr. Safety and tolerability of a novel, polyclonal human anti-MERS coronavirus antibody produced from transchromosomic cattle: a phase 1 randomised, double-blind, single-dose-escalation study. Lancet Infect. Dis. 2018;18:410–418. doi: 10.1016/S1473-3099(18)30002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BenevolentAI Potential treatment for COVID-19 identified by BenevolentAI using artificial intelligence enters randomised clinical trial. BenevolentAI. 2020 [Google Scholar]
- Berlin D.A., Gulick R.M., Martinez F. Severe covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- Biontech BioNTech and Pfizer announce regulatory approval from German authority Paul-Ehrlich-Institut to commence first clinical trial of COVID-19 vaccine candidates. BioNTech. 2020 [Google Scholar]
- Bjelle A., Björnham A., Larsen A., Mjörndal T. Chloroquine in long-term treatment of rheumatoid arthritis. Clin. Rheumatol. 1983;2:393–399. doi: 10.1007/BF02041561. [DOI] [PubMed] [Google Scholar]
- Bloomberg Kiniksa announces early evidence of treatment response with mavrilimumab in 6 patients with severe COVID-19 pneumonia. Bloomberg. 2020 [Google Scholar]
- Borba M.G.S., Val F.d.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., Mouratildeo M.P.G., Brito Sousa J., Eacute D., Baia-da-Silva D.C., Guerra M.V.F., Hajjar L.A., Pinto R.C., Balieiro A.A.S., Naveca F.G., Xavier M.S., Salomão A., Siqueira A.M., Schwarzbolt A., Croda J.H.R., Nogueira M.L., Romero G.A.S., Bassat Q., Fontes C.J., Albuquerque B.C., Daniel-Ribeiro C.T., Monteiro W.M., Lacerda M.V.G. Chloroquine diphosphate in two different dosages as adjunctive therapy of hospitalized patients with severe respiratory syndrome in the context of coronavirus (SARS-CoV-2) infection: preliminary safety results of a randomized, double-blinded, phase IIb clinical trial (CloroCovid-19 Study) MedRxiv. 2020 2020.2004.2007.20056424. [Google Scholar]
- Bordy R., Totoson P., Prati C., Marie C., Wendling D., Demougeot C. Microvascular endothelial dysfunction in rheumatoid arthritis. Nat. Rev. Rheumatol. 2018;14:404–420. doi: 10.1038/s41584-018-0022-8. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabut S. 2019. Coronavirus: Serious Side Effects Accumulate on Hydroxychloroquine. (Le Monde, France) [Google Scholar]
- CanSinoBIO . 2020. CanSinoBIO's Investigational Vaccine against COVID-19 Approved for Phase 1 Clinical Trial in China. [Google Scholar]
- Cao W., Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020;30:367–369. doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Centers for Disease Control and Prevention; 2020. Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID-19) [Google Scholar]
- Chen W.-H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Current tropical medicine reports. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hu C., Hood M., Zhang X., Zhang L., Kan J., Du J. A novel combination of vitamin C, curcumin and glycyrrhizic acid potentially regulates immune and inflammatory response associated with coronavirus infections: a perspective from system biology analysis. Nutrients. 2020;12:1193. doi: 10.3390/nu12041193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J.a., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hu J., Zhang Z., Jiang S.S., Han S., Yan D., Zhuang R., Hu B., Zhang Z. Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial. MedRxiv. 2020 [Google Scholar]
- clinicaltrials gov . U.S National Library of Medicine; 2020. Safety and Immunogenicity Study of 2019-nCoV Vaccine (mRNA-1273) for Prophylaxis SARS CoV-2 Infection (COVID-19) [Google Scholar]
- clinicaltrials gov . national Library of Medicine; 2020. Efficacy and Safety of Hydroxychloroquine and Azithromycin for the Treatment of Ambulatory Patients with Mild COVID-19. [Google Scholar]
- clinicaltrials gov . National Library of medicine; 2020. Hydroxychloroquine in Outpatient Adults with COVID-19. [Google Scholar]
- clinicaltrials gov . National Library of medicine; 2020. The Impact of Camostat Mesilate on COVID-19 Infection (CamoCO-19) [Google Scholar]
- clinicaltrials gov . Nataional library of science; 2020. Outcomes Related to COVID-19 Treated with Hydroxychloroquine Among In-Patients with Symptomatic Disease (ORCHID) [Google Scholar]
- clinicaltrials gov . national Library of science; 2020. PATCH 2&3:Prevention & Treatment of COVID-19 (Severe Acute Respiratory Syndrome Coronavirus 2) with Hydroxychloroquine. [Google Scholar]
- clinicaltrials gov . 2020. Post-exposure Prophylaxis/Preemptive Therapy for SARS-Coronavirus-2 (COVID-19 PEP) [Google Scholar]
- clinicaltrials gov . U.S National Library of Medicine; 2020. Safety and Immunogenicity Study of 2019-nCoV Vaccine (mRNA-1273) for Prophylaxis SARS CoV-2 Infection (COVID-19) [Google Scholar]
- clinicaltrials gov . National Library of Medicine; 2020. Treatment in Patients with Suspected or Confirmed COVID-19 with Early Moderate or Severe Disease (RCT) [Google Scholar]
- ClinicalTrialsgov . U.S National Library of Medicine; 2020. The Efficacy of Lopinavir Plus Ritonavir and Arbidol against Novel Coronavirus Infection (ELACOI) [Google Scholar]
- ClinicalTrialsgov . National Library of Medicine; 2020. Mild/Moderate 2019-nCoV Remdesivir RCT Clinical Trials. [Google Scholar]
- ClinicalTrialsgov . National Library of Medicine; 2020. Norwegian Coronavirus Disease 2019 Study (NO COVID-19) [Google Scholar]
- ClinicalTrialsgov . Natiotional Library of Medicine; 2020. Severe 2019-nCoV Remdesivir RCT Trials. [Google Scholar]
- clinicaltrialsregister eu . EU Clinical Trials Register; 2020. COVID-19 - Epidemiology of SARS-CoV-2 and Mortality to Covid19 Disease upon Hydroxychloroquine and Azithromycin Therapy in French Cancer Patients. [Google Scholar]
- clinicaltrialsregister eu . EU Clinical Trials Register; 2020. Randomized Clinical Trial to Evaluate the Efficacy of Hydroxychloroquine Associated or Not with Azithromycin as a Treatment for COVID-19 Infection. [Google Scholar]
- Colson P., Rolain J.-M., Lagier J.-C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;105932 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola A., Lombardi M., Tassoni M.I., Carolla G., Tala M., Morandini R., Paoletti O., Testa S.J.B.T. COVID-19, thromboembolic risk and thromboprophylaxis: learning lessons from the bedside. awaiting evidence. 2020;18:226. doi: 10.2450/2020.0113-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020 doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti C., Agape E., Becciolini A., Biggioggero M., Favalli E.G. Targeting granulocyte-monocyte colony-stimulating factor signaling in rheumatoid arthritis: future prospects. Drugs. 2019:1–15. doi: 10.1007/s40265-019-01192-z. [DOI] [PubMed] [Google Scholar]
- Day M. Covid-19: European drugs agency to review safety of ibuprofen. BMJ. 2020;368:m1168. doi: 10.1136/bmj.m1168. [DOI] [PubMed] [Google Scholar]
- De Witt B.J., Garrison E.A., Champion H.C., Kadowitz P.J. L-163,491 is a partial angiotensin AT1 receptor agonist in the hindquarters vascular bed of the cat. Eur. J. Pharmacol. 2000;404:213–219. doi: 10.1016/s0014-2999(00)00612-9. [DOI] [PubMed] [Google Scholar]
- Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. Human Vaccines & Immunotherapeutics; 2020. COVID-19, an Emerging Coronavirus Infection: Advances and Prospects in Designing and Developing Vaccines, Immunotherapeutics, and Therapeutics; pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discoveries & Therapeutics. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Driggin E., Madhavan M.V., Bikdeli B., Chuich T., Laracy J., Bondi-Zoccai G., Brown T.S., Der Nigoghossian C., Zidar D.A., Haythe J. Cardiovascular considerations for patients, health care workers, and health systems during the coronavirus disease 2019 (COVID-19) pandemic. J. Am. Coll. Cardiol. 2020 doi: 10.1016/j.jacc.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Yu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X.-x., Yang X. The feasibility of convalescent plasma therapy in severe COVID-19 patients: a pilot study. medRxiv. 2020 doi: 10.1073/pnas.2004168117. 2020.2003.2016.20036145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G. Ivermectin: does P-glycoprotein play a role in neurotoxicity? Filaria J. 2003;2(Suppl. 1) doi: 10.1186/1475-2883-2-S1-S8. S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA EMA advice on the use of NSAIDs for Covid-19. Drug Therapeut. Bull. 2020 doi: 10.1136/dtb.2020.000021. dtb-2020-000021. [DOI] [PubMed] [Google Scholar]
- Favalli E.G., Biggioggero M., Maioli G., Caporali R. 2020. Baricitinib for COVID-19: a Suitable Treatment? the Lancet Infectious Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . US Food and Drug Adimistration; 2020. Recommendations for Investigational COVID-19 Convalescent Plasma. [Google Scholar]
- Furuta Y., Takahashi K., Kuno-Maekawa M., Sangawa H., Uehara S., Kozaki K., Nomura N., Egawa H., Shiraki K. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proceedings of the Japan Academy, Series B. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends. 2020 doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ge Y., Tian T., Huang S., Wan F., Li J., Li S., Yang H., Hong L., Wu N., Yuan E. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv. 2020 doi: 10.1038/s41392-021-00568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilead . 2020. Remdesivir Clinical Trials. [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of, V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid Ai Development Cycle for the Coronavirus (Covid-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning Ct Image Analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- GSK . GSK; 2020. Clover and GSK Announce Research Collaboration to Evaluate Coronavirus (COVID-19) Vaccine Candidate with Pandemic Adjuvant System. [Google Scholar]
- Gu H., Xie Z., Li T., Zhang S., Lai C., Zhu P., Wang K., Han L., Duan Y., Zhao Z. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 2016;6:19840. doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D. Old weapon for new enemy: drug repurposing for treatment of newly emerging viral diseases. Virol. Sin. 2020:1–3. doi: 10.1007/s12250-020-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysi D.M., Valle Í.D., Zitnik M., Ameli A., Gan X., Varol O., Sanchez H., Baron R.M., Ghiassian D., Loscalzo J. 2020. Network Medicine Framework for Identifying Drug Repurposing Opportunities for COVID-19. arXiv preprint arXiv:2004.07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol.: A Journal of the Pathological Society of Great Britain and Ireland. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health M.o. 2018. Report on the Deliberation Results. [Google Scholar]
- Henry B.M., Vikse J., Benoit S., Favaloro E.J., Lippi G. Hyperinflammation and derangement of renin-angiotensin-aldosterone system in COVID-19: a novel hypothesis for clinically suspected hypercoagulopathy and microvascular immunothrombosis. Clin. Chim. Acta. 2020 doi: 10.1016/j.cca.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 2020.2001.2031.929042. [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmarcher M., Mayr A., Rumetshofer E., Ruch P., Renz P., Schimunek J., Seidl P., Vall A., Widrich M., Hochreiter S. 2020. Large-scale Ligand-Based Virtual Screening for SARS-CoV-2 Inhibitors Using Deep Neural Networks. Available at SSRN 3561442. [Google Scholar]
- Holmes K.V. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Hoth . 2020. Hoth Therapeutics to Host Shareholder Update Conference Call with Dr. Mark Poznansky to Discuss Covid-19 Vaccine Candidate. (vaxcelerate) [Google Scholar]
- Høy T., Horsberg T.E., Nafstad I. The disposition of ivermectin in Atlantic salmon (Salmo salar) Pharmacol. Toxicol. 1990;67:307–312. doi: 10.1111/j.1600-0773.1990.tb00835.x. [DOI] [PubMed] [Google Scholar]
- Hu Y., Bajorath J. Many structurally related drugs bind different targets whereas distinct drugs display significant target overlap. RSC Adv. 2012;2:3481–3489. [Google Scholar]
- Iba T., Levy J.H., Levi M., Connors J.M., Thachil J. 2020. Coagulopathy of Coronavirus Disease 2019. Critical Care Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020:1–7. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C.J., Tierney K.B., Mittelstadt M. Inhibition of P-glycoprotein in the blood–brain barrier alters avermectin neurotoxicity and swimming performance in rainbow trout. Aquat. Toxicol. 2014;146:176–185. doi: 10.1016/j.aquatox.2013.10.035. [DOI] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk J., Ork B., Hart B.J., Mazur S., Holbrook M.R., Frieman M.B., Traynor D., Johnson R.F., Dyall J., Kuhn J.H. The antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for MERS-CoV infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 2014 doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi O., Noto M., Sakurai T., Iwamoto A. T-705, A novel anti-influenza virus compound-the safety, tolerability and pharmacokinetics in human. Int. J. Infect. Dis. 2008;12:e299. [Google Scholar]
- Kotch C., Barrett D., Teachey D.T. Tocilizumab for the treatment of chimeric antigen receptor T cell-induced cytokine release syndrome. Expet Rev. Clin. Immunol. 2019;15:813–822. doi: 10.1080/1744666X.2019.1629904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kumar N., Mishra B., Mehmood A., Athar M., Mukhtar M.S. Integrative network biology framework elucidates molecular mechanisms of SARS-CoV-2 pathogenesis. bioRxiv. 2020 doi: 10.1016/j.isci.2020.101526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;105924 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane R. Sarah Gilbert: carving a path towards a COVID-19 vaccine. Lancet. 2020;395:1247. doi: 10.1016/S0140-6736(20)30796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020 doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster D., Klenk S., Ludwig K., Nojoumi S., Behren S., Adam L., Stadtmüller M., Saenger S., Zimmler S., Hönzke K. Phage capsid nanoparticles with defined ligand arrangement block influenza virus entry. Nat. Nanotechnol. 2020:1–7. doi: 10.1038/s41565-020-0660-2. [DOI] [PubMed] [Google Scholar]
- Le T.T., Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020 doi: 10.1038/d41573-020-00073-5. 10. [DOI] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon C., Julian D., Mike C., Jans A.D., Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sparidans R.W., Wang Y., Lebre M.C., Beijnen J.H., Schinkel A.H. Oral coadministration of elacridar and ritonavir enhances brain accumulation and oral availability of the novel ALK/ROS1 inhibitor lorlatinib. Eur. J. Pharm. Biopharm. 2019;136:120–130. doi: 10.1016/j.ejpb.2019.01.016. [DOI] [PubMed] [Google Scholar]
- Li L., Li R., Wu Z., Yang X., Zhao M., Liu J., Chen D. Therapeutic strategies for critically ill patients with COVID-19. Ann. Intensive Care. 2020;10:1–9. doi: 10.1186/s13613-020-00661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yu J., Zhang Z., Ren J., Peluffo A.E., Zhang W., Zhao Y., Yan K., Cohen D., Wang W. 2020. Network Bioinformatics Analysis Provides Insight into Drug Repurposing for COVID-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu X., Guo L., Li J., Zhong D., Zhang Y., Clarke M., Jin R. Traditional Chinese herbal medicine for treating novel coronavirus (COVID-19) pneumonia: protocol for a systematic review and meta-analysis. Syst. Rev. 2020;9:75. doi: 10.1186/s13643-020-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection——a review of immune changes in patients with viral pneumonia. Emerg. Microb. Infect. 2020:1–14. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang M., He L., Li Y. Chinese herbs combined with western medicine for severe acute respiratory syndrome (SARS) Cochrane Database Syst. Rev. 2004 doi: 10.1002/14651858.CD004882.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S. ACS Publications; 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S. ACS Publications; 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery. 2020;6:1–4. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Gao Y., Yuan Y., Yang K., Shi S., Zhang J., Tian J. Efficacy and safety of integrated traditional Chinese and western medicine for corona virus disease 2019 (COVID-19): a systematic review and meta-analysis. Pharmacol. Res. 2020;158:104896. doi: 10.1016/j.phrs.2020.104896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, P., Liu, Y., Qiu, L., Liu, X., Liu, D., Li, J., Tocilizumab treatment in COVID-19: a single center experience. Journal of Medical Virology n/a. [DOI] [PMC free article] [PubMed]
- Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020 doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: what treatments are being investigated? British Medical Journal Publishing Group. 2020 doi: 10.1136/bmj.m1252. [DOI] [PubMed] [Google Scholar]
- Marano G., Vaglio S., Pupella S., Facco G., Catalano L., Liumbruno G.M., Grazzini G. Convalescent plasma: new evidence for an old therapeutic tool? Blood Transfus. 2016;14:152–157. doi: 10.2450/2015.0131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotto D., Sarzi-Puttini P. What is the role of rheumatologists in the era of COVID-19? Autoimmun. Rev. 2020 doi: 10.1016/j.autrev.2020.102539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020:2000607. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxmen A. More than 80 clinical trials launch to test coronavirus treatments. Nature. 2020;578:347. doi: 10.1038/d41586-020-00444-3. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud E.J., Hayden F.G., Hurt A.C. Antiviral research; 2019. Antivirals targeting the polymerase complex of influenza viruses; p. 104545. [DOI] [PubMed] [Google Scholar]
- Mina F., Rahman M., Das S., Karmakar S., Billah M. 2020. Potential Drug Candidates Underway Several Registered Clinical Trials for Battling COVID-19. [Google Scholar]
- Molina J.M., Delaugerre C., Goff J.L., Mela-Lima B., Ponscarme D., Goldwirt L., de Castro N. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Maladies Infect. 2020 doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B.J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020 doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Murck H. Symptomatic protective action of glycyrrhizin (licorice) in COVID-19 infection? Front. Immunol. 2020;11:1239. doi: 10.3389/fimmu.2020.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH . National Institute for Health; 2020. COVID-19 Treatment Guidelines. [Google Scholar]
- Novavax . 2020. Novavax Identifies Coronavirus Vaccine Candidate; Accelerates Initiation of First-In-Human Trial to Mid-may. [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.-l., Zhang A.-H., Wang X.-J. Traditional Chinese medicine for COVID-19 treatment. Pharmacol. Res. 2020;155:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensi S., B Altman R., Liu T., Lo Y.-C., McInnes G., Derry A., Keys A. Homology modeling of TMPRSS2 yields candidate drugs that may inhibit entry of SARS-CoV-2 into human cells. ChemRxiv. 2020 [Google Scholar]
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395:e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Pereira J., Jochmans D., Dallmeier K., Leyssen P., Nascimento M., Neyts J. Favipiravir (T-705) inhibits in vitro norovirus replication. Biochem. Biophys. Res. Commun. 2012;424:777–780. doi: 10.1016/j.bbrc.2012.07.034. [DOI] [PubMed] [Google Scholar]
- Roche J.A., Roche R. A hypothesized role for dysregulated bradykinin signaling in COVID‐19 respiratory complications. Faseb. J. 2020 doi: 10.1096/fj.202000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Munnink B.B.O., de Meulder D., van Amerongen G., van den Brand J., Okba N.M. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020 doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020:1–3. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B., Moss C., George G., Santaolalla A., Cope A., Papa S., Van Hemelrijck M. Associations between immune-suppressive and stimulating drugs and novel COVID-19—a systematic review of current evidence. eEcancer Med. Sci.cancermedicalscience. 2020;14 doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynes R. Antimalarial drugs in the treatment of rheumatological diseases. Rheumatology. 1997;36:799–805. doi: 10.1093/rheumatology/36.7.799. [DOI] [PubMed] [Google Scholar]
- Scott B. Treatment of coronavirus disease 2019 (COVID-19): investigational drugs and other therapies. Medscape. 2020 [Google Scholar]
- Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W., Bridgland A. Improved protein structure prediction using potentials from deep learning. Nature. 2020:1–5. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- sinovac . 2020. Sinovac Announces Approval of Human Clinical Trial for a Vaccine Candidate against COVID-19. [Google Scholar]
- Smith T., Prosser T. 2020. COVID-19 Drug Therapy–Potential Options. Clinical Drug Information/Clinical Solutions. [Google Scholar]
- Smith T., Prosser T. Elsevier; 2020. COVID-19 Drug Therapy–Potential Options. Clinical Drug Information| Clinical Solutions. [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemostasis. 2020;18:1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. National Science Review. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., Bakker A.B.H., van den Brink E.N., Weverling G.J., Martina B.E.E., Haagmans B.L., Kuiken T., de Kruif J., Preiser W., Spaan W., Gelderblom H.R., Goudsmit J., Osterhaus A.D.M.E. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus E., Basta S., Gee K. Who's in charge here? Macrophage colony stimulating factor and granulocyte macrophage colony stimulating factor: competing factors in macrophage polarization. Cytokine. 2020;127:154939. doi: 10.1016/j.cyto.2019.154939. [DOI] [PubMed] [Google Scholar]
- Tsantikos E., Lau M., Castelino C.M., Maxwell M.J., Passey S.L., Hansen M.J., McGregor N.E., Sims N.A., Steinfort D.P., Irving L.B., Anderson G.P., Hibbs M.L. Granulocyte-CSF links destructive inflammation and comorbidities in obstructive lung disease. J. Clin. Invest. 2018;128:2406–2418. doi: 10.1172/JCI98224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduganathan M., Vardeny O., Michel T., McMurray J.J., Pfeffer M.A., Solomon S.D. Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking the trimerization of viral spike glycoprotein? Int. J. Antimicrob. Agents. 2020:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaxart 2020. https://investors.vaxart.com/press-releases
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H.M., Ahmad S., Abbasi S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1762743. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J.D., Hutter C.A., Zimmermann I., Earp J., Egloff P., Sorgenfrei M., Hürlimann L.M., Gonda I., Meier G., Remm S. Synthetic nanobodies targeting the SARS-CoV-2 receptor-binding domain. BioRxiv. 2020 [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS‐CoV‐2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cao R., Zhang H., Liu J., Xu M., Hu H., Li Y., Zhao L., Li W., Sun X., Yang X., Shi Z., Deng F., Hu Z., Zhong W., Wang M. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hu M., Li Q., Zhang X.-P., Zhai G., Yao N. 2020. Abnormal Respiratory Patterns Classifier May Contribute to Large-Scale Screening of People Infected with COVID-19 in an Accurate and Unobtrusive Manner. arXiv preprint arXiv:2002. [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2017. WHO Model List of Essential Medicines. [Google Scholar]
- WHO . World Health Organization; 2020. Coronavirus Disease 2019 (COVID-19): Situation Report; p. 72. [Google Scholar]
- WHO . 2020. DRAFT Landscape of COVID-19 Candidate Vaccines World Health Organisation. [Google Scholar]
- WHO . 2020. Coronavirus Disease (COVID-19) Situation Report – 143 World Health Organisation. [Google Scholar]
- WHO . World Health Organization; 2020. Public Health Emergency SOLIDARITY Trial of Treatments for COVID-19 Infection in Hospitalized Patients. [Google Scholar]
- WHO . World Health Organisation; 2020. The Use of Non-steroidal Anti-inflammatory Drugs (NSAIDs) in Patients with COVID-19. [Google Scholar]
- WHO . 2020. WHO Coronavirus Disease (COVID-19) Dashboard. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Li W., Liu G., Tang Y. Network-based methods for prediction of drug-target interactions. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu X., Chen Z., Duan J., Hashimoto K., Yang L., Liu C., Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;S0889–1591(820) doi: 10.1016/j.bbi.2020.03.031. 30357-30353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Xu X., Chen Z., Duan J., Hashimoto K., Yang L., Liu C., Yang C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020;S0889–1591(820) doi: 10.1016/j.bbi.2020.03.031. 30357-30353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang Y. Traditional Chinese medicine treatment of COVID-19. Complementary Therapies in Clinical Practice. 2020:101165. doi: 10.1016/j.ctcp.2020.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Shen H.-M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasir M., Sonthalia S. 2019. Corticosteroid Adverse Effects. [PubMed] [Google Scholar]
- Yuen K.-S., Ye Z.-W., Fung S.-Y., Chan C.-P., Jin D.-Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:1–5. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldin R.K., Petruschke R.A. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J. Antimicrob. Chemother. 2004;53:4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- Zhang D., Guo R., Lei L., Liu H., Wang Y., Wang Y., Dai T., Zhang T., Lai Y., Wang J. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. MedRxiv. 2020 [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020:1–5. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Song K., Tong F., Fei M., Guo H., Lu Z., Wang J., Zheng C. First case of COVID-19 in a patient with multiple myeloma successfully treated with tocilizumab. Blood Adv. 2020;4:1307–1310. doi: 10.1182/bloodadvances.2020001907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A., Aladinskiy V., Zhebrak A., Zagribelnyy B., Terentiev V., Bezrukov D.S., Polykovskiy D., Shayakhmetov R., Filimonov A., Orekhov P. Potential COVID-2019 3C-like protease inhibitors designed using generative deep learning approaches. Insilico Medicine Hong Kong Ltd A. 2020;307:E1. [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 2020 [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. BioRxiv. 2020;2020 doi: 10.1128/JVI.00635-20. 2003.2026.009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., Ju X., Liang Z., Liu Q., Zhao Y., Guo F., Bai T., Han Z., Zhu J., Zhou H., Huang F., Li C., Lu H., Li N., Li D., Jin N., Penninger J.M., Jiang C. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]