Abstract
SAD kinases regulate presynaptic vesicle clustering and neuronal polarization. A previous report demonstrated that Sada−/− and Sadb−/− double-mutant mice showed perinatal lethality with a severe defect in axon/dendrite differentiation, but their single mutants did not. These results indicated that they were functionally redundant. Surprisingly, we show that on a C57BL/6N background, SAD-A is essential for cortical development whereas SAD-B is dispensable. Sada−/− mice died within a few days after birth. Their cortical lamination pattern was disorganized and radial migration of cortical neurons was perturbed. Birth date analyses with BrdU and in utero electroporation using pCAG-EGFP vector showed a delayed migration of cortical neurons to the pial surface in Sada−/− mice. Time-lapse imaging of these mice confirmed slow migration velocity in the cortical plate. While the neurites of hippocampal neurons in Sada−/− mice could ultimately differentiate in culture to form axons and dendrites, the average length of their axons was shorter than that of the wild type. Thus, analysis on a different genetic background than that used initially revealed a nonredundant role for SAD-A in neuronal migration and differentiation.
Keywords: axon/dendrite differentiation, neuronal migration, SAD kinase
Introduction
In the developing mammalian cerebral cortex, most neuronal precursor cells proliferate in the ventricular zone (VZ) or subventricular zone (SVZ) and migrate radially toward the pial surface (Rakic 1972). After their exit from the cell cycle in the VZ, postmitotic neurons move radially to the SVZ or intermediate zone (IZ) where they become multipolar (MP) (multipolar migration) (Tabata and Nakajima 2003; Noctor et al. 2004). Eventually, these MP cells extend an axon tangentially, transform into bipolar (BP) cells, and migrate radially in the cortical plate (CP) using a radial glial fiber as a scaffold (locomotion mode) (Rakic 1972; Nadarajah et al. 2001; Tabata and Nakajima 2003; Hatanaka and Yamauchi 2013). The migrating neocortical neurons travel past their predecessors forming layers in an “inside-out” manner, resulting in the positioning of the earliest-born neurons in the deepest layers with later-born neurons ultimately residing in the more superficial layers (Angevine and Sidman 1961; McConnell 1988). This neuronal migration is crucial for brain organization and defects during embryogenesis give rise to brain malformations such as lissencephaly, heterotopia, and polymicrogyria (Verrotti et al. 2010), resulting in severe epilepsy and mental retardation. In addition, aberrant neuronal migration is also implicated in the pathogenesis of not only brain malformations but also psychiatric disorders such as schizophrenia and autism (Kähler et al. 2008; Li et al. 2011).
A multitude of molecular mechanisms underlying neuronal migration have been verified (for review, Bielas et al. 2004; Cooper 2014). Reelin, a glycoprotein that Cajal–Retzius cells secrete, and Disabled-1 (Dab1), an adaptor protein that is an obligate effector of the Reelin-signaling pathway, have been shown to be critical for neuronal migration (for review, Honda et al. 2011). The small GTPases, which regulate the actin cytoskeleton and cell polarity, including Rap1, Rac1, and Cdc42, also play a crucial role in the MP–BP transition (Kawauchi et al. 2003; Jossin and Cooper 2011), indicating that the regulation of cytoskeletal dynamics is important for morphological transition. Several protein kinases including cyclin-dependent kinase 5 (Cdk5), glycogen synthase kinase (GSK3β), and c-Jun N-terminal kinases (JNKs) are also involved in neuronal migration (Kawauchi et al. 2003; Ohshima et al. 2007; Morgan-Smith et al. 2014; for review, Ohshima 2014). Although increasing evidence suggest the orchestration of various molecules in neuronal migration, the precise mechanism by which a highly organized brain structure is developed in the mammalian cerebral cortex is not fully understood.
SAD kinase, a member of the cAMP-activated protein kinase (AMPK) family, was first identified as SAD-1 in C. elegans, in which mutant neurons failed to engage in presynaptic vesicle clustering (Crump et al. 2001). Its mouse orthologs, mSAD-A and mSAD-B, are required for neuronal polarization; double-mutant neurons showed polarization and axon outgrowth defects both in vivo and in vitro (Kishi et al. 2005). This role appears to be redundant, in that deletion of either Sada or Sadb alone had no detectable effect, even though lack of one kinase had no apparent compensatory effect on the expression of the other. Consistent with these results, LKB1, a master upstream kinase of the AMPK subfamily, phosphorylates and activates SAD-A (BRSK2) and SAD-B (BRSK1) kinases (Lizcano et al. 2004), leading to axon/dendrite specification of polarization through phosphorylating tau in cortical neurons (Barnes et al. 2007). SAD kinases are also required for structural and functional maturation of synapses (Lilley et al. 2014), and SAD-B has been implicated in regulation of neurotransmitter release at mature synapses (Inoue et al. 2006). We have previously found that human SAD1 kinase is involved in UV-induced DNA damage checkpoint function (Lu et al. 2004).
In the present study, we generated Sada-deficient mice in a mixed background, similar to that reported previously (Kishi et al. 2005) but then backcrossed them to C57BL/6N mice. Very surprisingly, we found that the majority of Sada−/− mice with a C57BL/6N genetic background died within a few days after birth, whereas Sadb−/− mice with the same background survived into adulthood. Neuronal migration is impaired in Sada−/− mice but is apparently normal in Sadb−/− mice. Our study thus indicates that SAD-A has a distinct, nonredundant role in neuronal migration during the development of cerebral cortex.
Materials and Methods
Animals
Experimental animal protocols in the present study were approved by the Review Board of the Institute for Developmental Research, Aichi Human Service Center and the Nagoya City University Medical School, or by the Institutional Animal Use and Care Committee at Harvard University. They were carried out according to the guidelines for animal experimentation of the Japan Neuroscience Society and the NIH to minimize the number of animals used as well as their suffering. C57BL/6N mice were provided by Japan SLC (Shizuoka, Japan). The original Sada−/− mice (Kishi et al. 2005) are denoted here as Sada∆ex1/∆ex1, due to the deletion of its exon 1. Sadb-deficient mice and Sada/Sadb double mutants were also described previously (Kishi et al. 2005). Both knockout strains were backcrossed with C57BL/6N mice for at least 6 generations before analyses for the present report.
Generation of Sada Exon 2-Deficient Mice
Isolation of Sada Genomic Clones and Construction of the Targeting Vector
A probe spanning nucleotides 1–493 of human testis SAD1 cDNA (Lu et al. 2004) was used to screen a 129 Sv mouse genomic library in Lambda FIX II vector according to the manufacturer’s instructions (Stratagene). Among the 3 clones isolated, clone #18 contained exons 2–12, encompassing 31–408 amino acids. The second exon encoded a part of the ATP-binding site. The clone #18 was used to prepare the targeting vector. A 0.8-kb SacI-BamHI fragment from #18 was inserted into the XhoI site and a 5.5-kb NcoI-BamHI fragment was subcloned into the SalI site of pLNTK vector. The targeting vector was used to replace exon 2 of the Sada mouse genome with a neomycin-resistant gene (Fig. 1A).
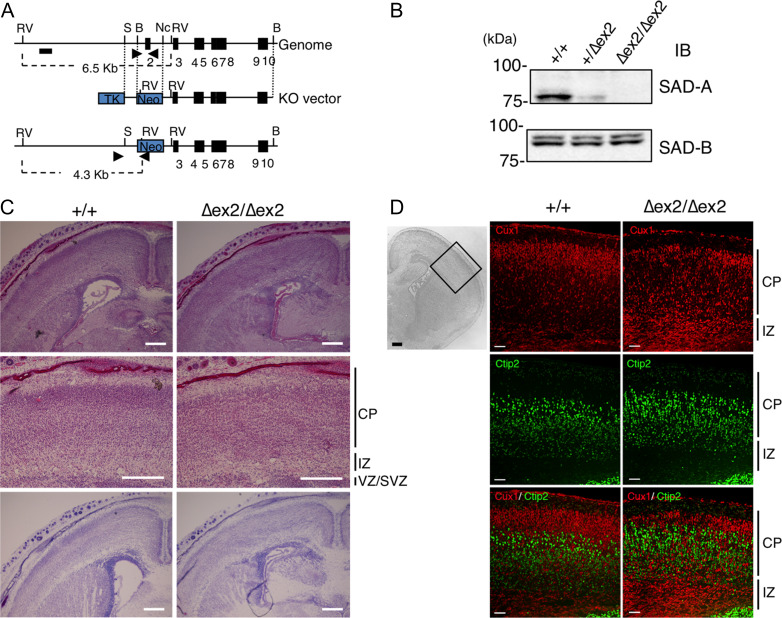
Figure 1.
Cortical lamination of SAD-A-deficient mice is disorganized. (A) Schematic representation of the methods used for gene targeting of the Sada genome. Arrowheads indicate the primers used in the PCR for determining genotypes. (B) Lysates of the cerebral cortex of Sada wild type (+/+), heterozygous (+/∆ex2), and homozygous (∆ex2/∆ex2) mutant mice at postnatal day 0 (P0) were analyzed by immunoblotting using the indicated antibodies. (C) HE staining (upper and middle panels) and Nissl staining (lower panels) of the brains of wild-type (+/+) and Sada homozygous (∆ex2/∆ex2) mutant mice at P0. Middle panels show higher magnification views. Bar, 300 μm. (–) Immunostaining of wild-type (+/+, middle) and homozygous (∆ex2/∆ex2, right) cerebral cortex at P0 with layer-specific markers. Left panel indicates the cortical area analyzed (DAPI staining). Cux1 (II–IV layer marker, red, upper panels) staining, Ctip2 (Vb-VI layer marker, green, middle panels) staining and their overlay (lower panels). Bar: 100 μm (left panel), 50 μm (middle and right panels), CP, cortical plate, IZ, intermediate zone, VZ/SVZ, ventricular zone/subventricular zone.
Isolation of the Targeted Embryonic Stem Clones and Generation of the Sada+/∆ex2 Mice
To generate heterozygous Sada-targeted embryonic stem (ES) cell clones, the targeting vector was linearized with SalI and transfected into ES cells (E14 cells supplied by Dr KI Nakayama). Drug selection was carried out with 0.3 mg/mL G418 (Gibco BRL) and 1.5 μM gancyclovir (Sigma-Aldrich). Drug-resistant clones were screened for homologous recombination of the targeting vector by PCR analysis with primers G7 (5′-GGGTGGGGTGGGATTAGATAAATG-3′) and SC03 (5′- TCCTGAGGGGACGGACACATTG-3′) yielding a 1.3-kb product (Supplementary Fig. 1A). For Southern blotting, a 1-kb XbaI fragment derived from the clone #18 was hybridized to genomic DNA digested with HindIII. Two independent ES cell clones containing a correctly targeted Sada mutation were selected and used to generate Sada knockout mice. Establishing the founder mice of Sada exon 2 deletion was based on the standard procedures by the use of chimeric mice derived from the ES cells (Nagy et al. 2003). The resulting germ line chimeric mice were backcrossed with C57BL/6N mice for at least 6 generations before analysis.
The Sada exon 2 mutation was confirmed by PCR using primers specific for the Sada wild-type allele (G01-F, 5′-GGCTGAATCTGGCCTCTCTCCT-3′; G02-R, 5′-CTATCCAGCCCTCTTCATGGCA-3′), resulting in an 0.48-kb PCR product, and with primers specific for the Sada exon 2-null allele (G7 and SC03), resulting in a 1.3-kb product (Supplementary Fig. 1A). These Sada exon 2-deleted heterozygous and homozygous mice are referred to as Sada+/∆ex2 and Sada∆ex2/∆ex2, respectively, hereafter.
Immunoblotting and Immunoprecipitation
SDS-PAGE and immunoblotting were performed as described by Nakanishi et al. (2006). Briefly, cerebral cortices of each pup (P0) were lysed in IP kinase buffer (50 mM HEPES, pH 8.0, 150 mM NaCl, 2.5 mM EGTA, 1 mM EDTA, 1 mM DTT, 0.1% Tween 20, 10% Glycerol) containing 5 μg/mL leupeptin, 2 μg/mL aprotinin, 2 mg/mL PMSF, 20 μg/mL trypsin inhibitors, 50 mM NaF, 0.1 mM Na3VO4, 80 mM β-glycerophosphate, and 15 mM p-nitrophenylphosphate. After sonication, lysates were centrifuged at 15 000× g for 10 min, and the supernatants (20–50 μg each) were used for western blot analyses.
To confirm the knockdown of SAD-A or SAD-B protein, cells were directly lysed with Laemmeli buffer (2% SDS, 10% glycerol, 5% mercaptoethanol, 0.002% bromophenol blue, and 62.5 mM Tris HCl at pH 6.8) and subjected to immunoblotting (Johmura et al. 2016).
For immunoprecipitation, cerebral cortices were lysed in TBSN buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% NP-40) containing 10 μg/mL leupeptin, 2 μg/mL aprotinin, 2 mg/mL PMSF, 20 μg/mL trypsin inhibitors, 100 mM NaF, 1 mM Na3VO4, 40 mM β-glycerophosphate, 10 mM sodium pyrophosphate, and 15 mM p-nitrophenylphosphate. After sonication, the resulting lysates were clarified by centrifugation at 15 000× g for 15 min at 4 °C before immunoprecipitation with the specified antibody (Johmura et al. 2016).
Tissue Preparation
Timed embryos were deeply anesthetized by placement on ice or using isoflurane inhalation and were intracardially perfusion-fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB, pH 7.4) at designated embryonic or postnatal stages as described previously (Tabata and Nakajima 2001). Each brain was dissected out and postfixed in the same fixative for 2 h. After washing in 0.1 M PB, the samples were equilibrated in 30% sucrose in 0.1PB, embedded in OCT compound (Sakura), cryopreserved at −80 °C, and cut into 15-μm-thick coronal sections.
Immunohistochemistry
Histological and immunohistochemical procedures were performed as described previously (Nakanishi et al. 2017). After the sections were subjected to hematoxylin–eosin (HE) or Nissl staining (0.5% cresyl violet) to compare the histology of the cortex between homozygotes and wild type/heterozygotes, some were processed for immunostaining.
After blocking nonspecific binding using goat serum, the sections were incubated overnight at 4°C with primary antibodies and subsequently incubated with the appropriate secondary antibodies. Fluorescent images were obtained using an FV-1000 confocal laser scanning microscope (Olympus) or a fluorescence microscope (Eclipse E800, Nikon Instruments). For double staining with Cux1 and Ctip2, antigen retrieval was performed by heating the sections at more than 95 °C for 20 min in 10 mM citrate buffer (pH 6.0). For BrdU staining after antigen retrieval, the sections were incubated with 2N HCl at 37 °C for 15 min and neutralized by immersing in borate buffer (0.1 M, pH 8.5) for 20 min. After blocking endogenous peroxidase with 3% H2O2 in methanol for 30 min, sections were incubated in biotinylated anti-BrdU antibody for 1 h, followed by treatment with a streptavidin–HRP (BD Pharmingen BrdU in situ detection kit, BD Biosciences).
Antibodies
Rabbit polyclonal antibodies against SAD-A peptide (aa 441–455) were prepared and purified, and detected only SAD-A protein but not SAD-B protein. (Supplementary Fig. 1B). Rabbit polyclonal anti-GFP (Code No. 598, MBL), anti-Cux1 (CDP, sc-13 024, Santa Cruz BioTechnology, Inc.), anti-SAD-B (Brsk1, #5935, Cell Signaling Technology), anti-pJNK (#9251, Thr183/Tyr185, Cell Signaling), rat monoclonal anti-Ctip2 (ab18465, Abcam), chicken polyclonal anti-MAP2 (ab5392, Abcam), anti-Brn1 (Goat IgG, Santa Cruz), anti-Tbr1 (ab31940, abcam), and anti-Calbindin D-28K (#AB1778, Millipore) were used. For immunoblotting, anti-Dab1 (AB5840, EMD Millipore Corp.), anti-phosphospecific monoclonal (4G10, #05–1050, Millipore), anti-Tau (ab32057, abcam), anti-Tau[pS262] phosphospecific (44–750G, Invitrogen Corporation), mouse anti-Tau-1 monoclonal antibody (MAB3420, Chemicon), anti-MAP1b (ab11266, abcam), MAP1b-pT1265 (ABH58, Millipore), MAP2 (ab32454, abcam), MAP2-pS136 (ab96378, abcam), LKB1(D60C5, #3047, Cell Signaling), and phospo-LKB1(Ser428, C67A3, #3482, Cell Signaling) were used. Anti-SAD-A antibody (anti-Brsk2, PAB6896, Abnova) was also used (Supplementary Fig. 6B,C). For secondary antibody, Alexa-488-conjugated anti-rabbit IgG (711-546-152, Jackson ImmunoResearch Laboratories, Inc. or A11034, Molecular Probes), Alexa488 or Cy3-conjugated anti-rat IgG (A21208, Molecular Probes or 712-165-153, Jackson ImmunoResearch) and Alexa-568-conjugated anti-chicken IgY H&L (ab175711, abcam) were used. For immunoblotting, HRP-conjugated anti-rabbit IgG, anti-mouse IgG (GE healthcare), or HRP–Protein A (Invitrogen) was used.
BrdU Injection
Female Sada+/∆ex2 mice crossed with the Sada+/∆ex2 male were peritoneally injected with bromodeoxyuridine (BrdU, 100 μg/g body weight, Sigma) on the 14th or 15th day of pregnancy and the brains of the pups were analyzed at P0.
In Utero Electroporation
Female Sada+/∆ex2 mice crossed with the Sada+/∆ex2 male or female Sadb+/− mice crossed with an Sadb+/− male were subjected to in utero electroporation as described previously (Tabata and Nakajima 2001). At E14.5, pregnant mice were deeply anesthetized with 3 types of mixed anesthetic agents (medetomidine chloride 0.3 mg/kg, midazolam 4 mg/kg, and butorphanol tartrate 5 mg/kg), and their abdomen was opened. Approximately 1 μL of plasmid solution (pCAG-EGFP, 0.5 μg/μl) was introduced into the lateral ventricles of the embryos, followed by electroporation using a CUY21 electroporator (NEPA gene). Most experiments using in uteroelectroporation were performed with Sada∆ex2/∆ex2 mice, but similar results were obtained with Sada∆ex1/∆ex1 mice.
For RNA interference, the pSUPER-gfp/neo vector (OligoEngine) was designed to target coding sequences in mSada (pSUPER-mSada, 5′-GCTAGAGCACATTCAGAAA-3′, 1118–1136 in NM_029 426.2) and mSadb (pSUPER-mSadb, 5′-GAGCTGTGGAGTCATCCTA-3′, 797–817 in NM_001 003 920.3) and prepared according to the manufacturer’s instructions (Yamada et al. 2010). The plasmid solution (pSUPER-mSada, pSUPER-mSadb, or empty pSUPER, 1 μg/μL; and pCAG-EGFP, 0.5 μg/μL) was cointroduced at E14.5 and the embryos were sacrificed at E19.5/P0. To visualize the centrosome in migrating neurons, pCAG-PACT-mKO1 vector (0.5 μg/μL, kindly provided by Dr F Matsuzaki, Riken CDB, Konno et al. 2007) was introduced with pCAG-EGFP 0.5 μg/μl at E14.5 and the embryos were analyzed at E17.5.
Time-Lapse Imaging
Time-lapse imaging of brain slices was conducted as described previously (Tabata and Nakajima 2003). Briefly, E14.5 embryos from the Sada+/∆ex2 female crossed with the Sada+/∆ex2 male were electroporated with pCAG-EGFP and the brains were excised at E16.5, embedded in 3% low melting agarose, and sectioned with a vibratome (Microtome HyRax V50). Coronal brain slices (250 μm) from the anterior one-third of the cerebral cortex on a permeable filter (Millicell-CM, 0.4 μm pore size, Millipore) were placed on a glass-bottom dish in neurobasal medium containing 10% FCS and 2% B27 (Invitrogen) and incubated at 37°C in 5% CO2 on a laser scanning microscope (FV1000, Olympus). Images in 19 areas of 6 embryos were captured every 30 min and imaging continued for 40 h.
Quantification of Neuronal Migration
After the brain slices were stained with anti-BrdU or anti-GFP antibodies, the distribution of BrdU- or GFP-positive neurons was quantified as described (Ohshima et al. 2007). Briefly, coronal sections of cerebral cortices were divided into 8 or 4 equally spaced bins from the ventricles to the pial surface for BrdU- or GFP-positive neurons, respectively. The percentage of labeled cells in each bin was calculated for each individual. For BrdU-positive cells, usually 100–200 cells per individual (both sides of the somatosensory cortex) were counted. For GFP-positive cells, usually 50–200 cells per individual (somatosensory cortex) were analyzed.
Quantification of Centrosome Position
After the coronal sections of cerebral cortices were stained with anti-GFP antibodies and DAPI, fluorescent images of migrating neurons introduced with both pCAG-mPACT-mKO and pCAG-EGFP were captured using a fluorescence microscope (Eclipse E800, Nikon). Cells were visually divided into 4 quadrants as shown in Fig. 4B. The centrosomal (Kusabira orange fluorescence) position of GFP-positive migrating neurons in the IZ close to the CP was plotted within the quadrants and the percentage in each quadrant was calculated. Usually 30–80 cells per individual were analyzed.
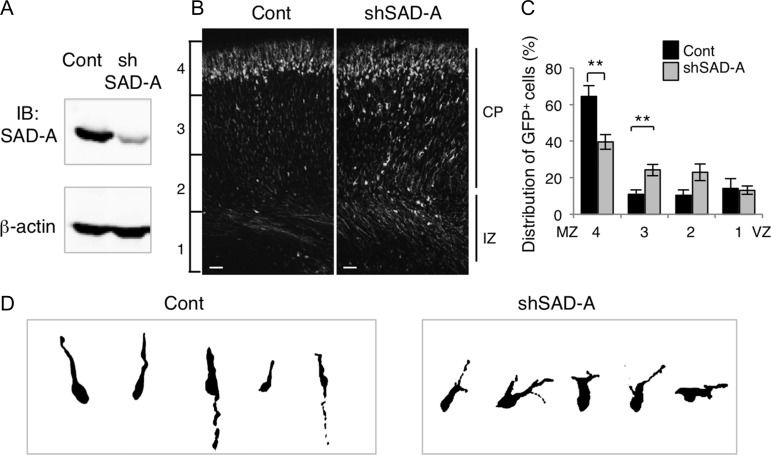
Figure 4.
SAD-A affects radial migration in a cell-autonomous manner. (A) Characterization of mSAD-A knockdown (pSUPER-mSAD-A) vectors. pCAG-GS-mSada vector was cotransfected into HEK293T cells with control pSUPER vector (Cont) or pSUPER-mSAD-A (shSAD-A). After 48 h, cells were harvested and subjected to western blotting with anti-SAD-A antibody. Anti-β-actin was used as a loading control. (B) Migratory defect in SAD-A knockdown neurons. pCAG-EGFP was coelectroporated with control pSUPER (Cont) or pSUPER-mSAD-A (shSAD-A) into cerebral cortices at E14.5, followed by fixation at P0. CP, cortical plate, IZ, intermediate zone, Bar, 50 μm. (C) Quantification of the distribution of GFP-positive neurons in distinct regions of the cerebral cortex (bins 1–4). Closed bars, control (Cont), open bars, shSAD-A, **P < 0.01. (Cont, n = 5; shSAD-A, n = 8; bin 4, P = 0.0026, bin 3, P = 0.0081, t-test). (D) Morphology of neurons transfected with control (Cont) or pSUPER-mSAD-A (shSAD-A) vectors in the CP.
Cell Culture and Transfection
To confirm the knockdown of SAD-A, HEK293T cells were cultured in a 12-well plate and transfected with pSUPER-mSada or pSUPER (0.9 μg) together with pCAG-GS-mSada vector (0.1 μg) using Lipofectamine 2000 (Invitrogen). For the knockdown of SAD-B, pSUPER-mSadb or pSUPER (0.9 μg) together with pCAG-GS-mSadb vector (0.1 μg) were used. Two days later, cells were subjected to immunoblotting.
Hippocampal neurons were cultured as described previously (Nakanishi et al. 2006). Briefly, hippocampi of E17.5 embryos were digested with 1.4 U/mL papain (Worthington) and 0.01% DNase I (Roche Life Science) at 37°C for 60 min. Cells were plated on poly-l-lysine-coated coverslips in 24-well plates at a density of 1.0 × 104 cells/cm2 and maintained in neurobasal medium (Invitrogen) containing 2% B27, 2 mM glutamine, 50 units/mL penicillin, 25 μg/mL streptomycin, 25 μM glutamate, 25 μM 2-mercaptoethanol, and 1% FCS in a 95% air, 5% CO2, H2O-saturated atmosphere at 37°C for 7 days. Cells were fixed with 3% paraformaldehyde in PBS containing 1% sucrose, 1 mM MgCl2, and 0.1 mM CaCl2 for 20 min and then subjected to immunostaining.
Immunocytochemistry and Quantification of Neurite Length
Immunocytochemical procedures were performed as described previously (Nakanishi et al. 2006). Briefly, after blocking nonspecific binding with goat serum, cells were incubated overnight at 4°C with primary antibodies and subsequently incubated with the appropriate secondary antibodies. Fluorescent images were captured using a fluorescence microscope (Eclipse E800, Nikon). Fluorescent images of 10 random fields on each coverslip were taken of triplicate coverslips on which cells had been exposed to each condition. The neurites of more than 30 neurons subjected to each condition were traced and their lengths were analyzed using the Public Domain Image J Program (Rasband 1997; Schneider et al. 2012). Three separate culture preparations were performed and similar results were obtained.
Statistical Analysis
For the migration assay, at least 3 litters and 3 brains (either sex) per experiment were used. When wild-type mice could not be obtained in the litter (Fig. 2A left and Fig. 6), heterozygous mice were included as a control because we had not observed any difference in phenotype between wild-type and heterozygous mice. Numerical data are presented as means ± standard error (SEM). Statistical differences between 2 groups were compared for each condition using the unpaired t-test or Mann–Whitney Utest. A P-value of <0.05 was considered statistically significant. All the analyses were performed using Instat 3 statistics software (GraphPad Software Inc.).
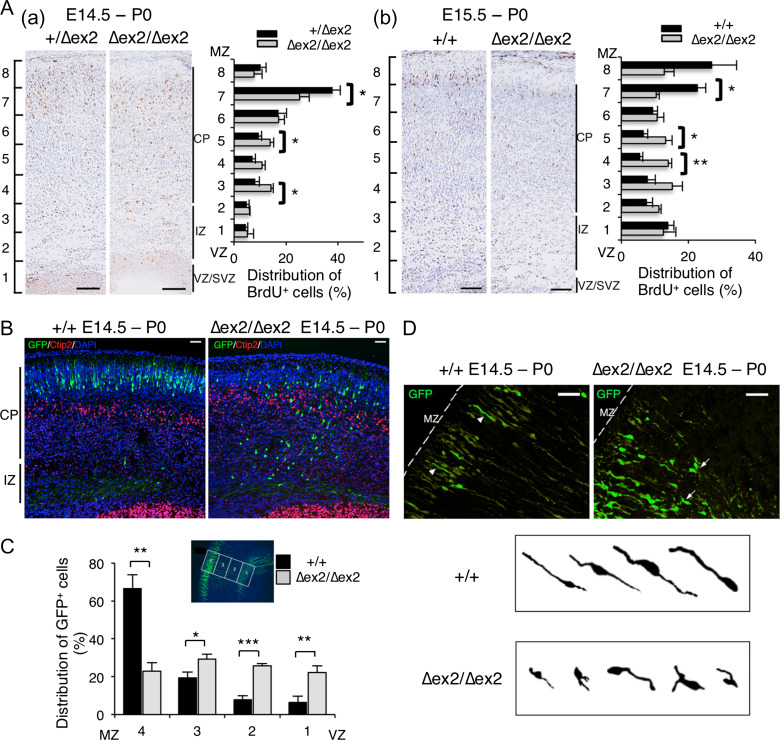
Figure 2.
Radial migration of Sada∆ex2/∆ex2 neocortical neurons is impaired. (A) Birth date analysis. BrdU was injected into pregnant mice at E14.5 (a, left panels) or E15.5 (b, right panels), followed by fixation at P0. The brain sections were immunostained with anti-BrdU antibody. The distribution of BrdU-positive cells was quantified by dividing the cerebral cortex into 8 parts (bins 1–8). The number of BrdU-positive cells in each bin was counted and represented as a percentage. Data are expressed as means ± SEM. *P < 0.05; **P < 0.01. (E14.5, Sada+/∆ex2, n = 5; Sada∆ex2/∆ex2, n = 3; bin 7, P = 0.038, bin 5, P = 0.046, bin 3, P = 0.031; E15.5, Sada+/+, n = 4; Sada∆ex2/∆ex2, n = 3; bin 7, P = 0.012, bin 5, P = 0.035, bin 4, P = 0.0024, t-test). Bar, 100 μm. (B) In utero electroporation. pCAG-EGFP vector was electroporated into the cerebral cortices at E14.5, followed by fixation at P0. The brain sections were immunostained with anti-GFP (green), anti-Ctip2 (red) antibody, and DAPI (blue). Photomicrographs show an overlay of the staining of wild-type (+/+, left) and homozygous (∆ex2/∆ex2, right) mutant brains. CP, cortical plate, IZ, intermediate zone, Bar, 50 μm. (C) Quantification of the distribution of GFP-positive cells in 4 distinct parts of the cerebral cortex (bins 1–4 as indicated in the inset) of Sada+/+ (closed bars) or Sada∆ex2/∆ex2 (open bars) mice. Data are expressed as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (Sada+/+, n = 6; Sada∆ex2/∆ex2, n = 4; bin 4, P = 0.0012, bin 3, P = 0.0331, bin 2, P = 0.001, bin 1, P = 0.0098, t-test). (D) Photomicrographs of GFP staining. Arrowheads in left panel(+/+) and arrows in right panel (∆ex2/∆ex2) indicate the cells enlarged. Morphology of migrating neurons in Sada+/+ (upper panel) or Sada∆ex2/∆ex2 (lower panel) mice. MZ, marginal zone, Bar, 50 μm.
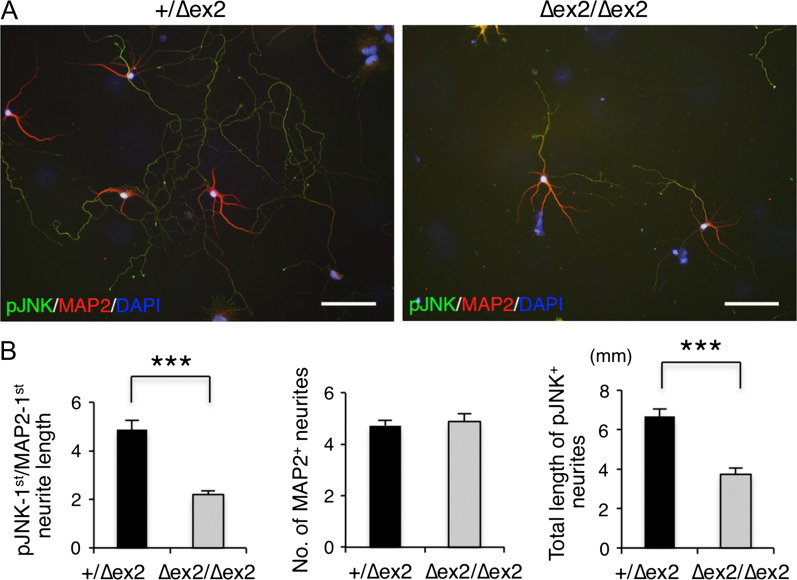
Figure 6.
Axonal development of SAD-A-deficient hippocampal neurons. (A) Representative images of hippocampal neurons cultured from Sada heterozygous (+/∆ex2, left) and homozygous (∆ex2/∆ex2, right) mouse brains. Hippocampal neurons were cultured for 7 days and stained with anti-pJNK (axonal marker, green), anti-MAP2 (dendrite marker, red), and DAPI (blue). Bar, 100 μm. (B) Quantification of neurite length. The ratio of the longest pJNK-positive/MAP2-positive neurite length (left), the total number of MAP2-positive neurites (middle), and the total length of pJNK-positive neurites (right). Closed bar, +/∆ex2, n = 58 neurons; open bar, ∆ex2/∆ex2, n = 35 neurons. ***P < 0.001.
Results
SAD-A Deficiency Causes Neonatal Lethality Under a C57BL/6 Background
To investigate the role of SAD-A kinase during the development of the brain, we generated Sada-deficient ES cells from a 129SvEv strain using the targeting vector illustrated in Figure 1A. The targeting vector was designed to delete exon 2 of mSAD-A, which contains a conserved kinase catalytic domain, generating Sada-null mutant mice (Sada∆ex2/∆ex2). Heterozygote matings of the first- or second-generation yielded Sada+/+, Sada+/∆ex2, and Sada∆ex2/∆ex2 offspring at roughly the expected Mendelian ratio, indicating no significant embryonic lethality. Sada∆ex2/∆ex2 grew to adulthood without any apparent abnormalities or any remarkable neuronal phenotypes as described by Kishi et al. (2005) for Sada∆ex1/∆ex1mice. Western blot analyses showed that SAD-A protein was not present in the homozygous (Sada∆ex2/∆ex2) brain, while the level of SAD-B protein was comparable to that of the wild type (Fig. 1B). Given that many neuronal phenotypes are affected by the genetic background (Kunst et al. 2000), we backcrossed Sada+/∆ex2 mice to C57BL/6N mice to obtain mice with a uniform genetic background. Very surprisingly, after the sixth generation or greater, the majority of Sada∆ex2/∆ex2 mice died within a few days of birth (Table 1). To ask whether this lethality was specific to the new allele, we also backcrossed the Sada∆ex1/∆ex1 mice (Kishi et al. 2005) to C57BL/6N mice for 6 generations. Again, we observed neonatal lethality. Thus, the genetic background has a profound effect on the Sada phenotype. In contrast, Sadb−/− mice (Kishi et al. 2005) survived into adulthood on both a C57BL/6N background and a mixed background (Table 1). These results indicate that an SAD-A deficiency specifically causes neonatal lethality in mice with a C57BL/6N genetic background, indicating that SAD-A has an isozyme-specific function.
Table 1.
The proportion of Sada or Sadb +/+, +/−, −/− offspring by crossing with heterozygous mice
| +/+ | +/− | −/− | |
|---|---|---|---|
| Expected % | 25% | 50% | 25% |
| Sada ∆exon 2 (F5-F8) | +/+ | +/∆ex2 | ∆ex2/∆ex2 |
| E16 | 28 (26.2%) | 55 (51.4%) | 24 (22.4%) |
| E18-P0 | 33 (26.6%) | 63 (50.8%) | 28 (22.6%) |
| P21 | 8 (34.8%) | 15 (65.2%) | 0 (0%) |
| Sada ∆exon 1 (F6) | +/+ | +/∆ex1 | ∆ex1/∆ex1 |
| E18-P0 | 5 (15.2%) | 21 (63.6%) | 7 (21.1%) |
| P21 | 9 (33.3%) | 18 (66.7%) | 0 (0%) |
| Sadb (F6) | +/+ | +/− | −/− |
| E18-P0 | 13 (37.1%) | 15 (42.9%) | 7 (20%) |
| P21 | 3 (10%) | 18 (60%) | 9 (30%) |
Cortical Lamination in Sada∆ex2/∆ex2 Mice is Disorganized
SAD-A and SAD-B proteins were detected in the developing brain at E13.5 and their levels gradually increased with age (Supplementary Fig. 1C). In situ hybridization confirmed the existence of Sada and Sadb in the developing cerebral cortex (Supplementary Fig. 1D). At postnatal day 0 (P0), HE and Nissl staining revealed that the CP in Sada∆ex2/∆ex2 mice lacked a discrete layered structure compared with the wild type (Fig. 1C). Immunostaining with antibodies against layer-specific markers showed that some Cux1 (a specific marker of layer II-IV)-positive cells were present in the deeper side of the Ctip2 (a specific marker of layer Vb-VI)-positive cell layer in the Sada∆ex2/∆ex2 brain (Fig. 1D). Similar results were obtained in Sada∆ex1/∆ex1 (data not shown). Thus, radial migration of neocortical neurons is impaired in SAD-A-deficient mice.
An initial study reported that Sada−/−:Sadb−/− double-mutant cortex (on the mixed genetic background) was abnormally thin with disordered segregation of Tbr1-positive neurons, which are normally expressed in the MZ, layer VI, and subplate (Kishi et al. 2005). We reexamined cortical development in the double mutants in the mixed background (Supplementary Fig. 2). By the time of birth (E18.5), the CP appeared homogeneous (Supplementary Fig. 2A). Markers for specific layers II-IV (Cux1 and Brn1, Feng and Walsh 2004; Molyneaux et al. 2007) showed abnormal distributions (Supplementary Fig. 2B,C), consistent with the results on Tbr1 (Kishi et al. 2005). Together, these results show that defects in Sada−/−:Sadb−/− double mutants in the mixed background are similar to those in Sada−/− single mutants in the C57BL/6N background, which suggests a possible predominant role of SAD-A in cortical development as compared with SAD-B.
Aberrant Radial Migration in Sada∆ex2/∆ex2 Neocortical Neurons
To further investigate radial migration in Sada∆ex2/∆ex2 mice, we first performed birth date analyses of neocortical neurons. We injected BrdU into pregrant mice at E14.5 or E15.5. The pups were fixed at P0, and the brain slices were immunostained with anti-BrdU antibody. We divided the cortex into 8 bins from VZ to the marginal zone (MZ) and counted the BrdU-positive cells in each bin. The percentage of BrdU-positive cells in the superficial layer (bin 7) of Sada∆ex2/∆ex2 mice labeled at E14.5 was significantly decreased but that in the middle layer (bins 3 and 5) was increased when compared with rates in Sada+/∆ex2 mice (Fig. 2A, left panel). In the case of BrdU labeling at E15.5, similar results were obtained (Fig. 2A, right panel). We then performed double staining with Cux1 plus EdU labeling at either E12.5 (the time layer V–VI neurons were born) or E14.5 (the time layer II–IV neurons were born). Around one-third (36.2 ± 4.6%) of EdU-positive cells labeled at E14.5 were Cux1-positive in the lower CP of Sada∆ex2/∆ex2 mice (Supplementary Fig. 3A right upper panel and Supplementary Fig. 3C), whereas few labeled at E12.5 were Cux1-positive (0.42 ± 0.37%, Supplementary Fig. 3A right lower panel and Supplementary Fig. 3C). This result indicates that some Cux1-positive cells in the lower CP of Sada∆ex2/∆ex2 mice were layer II–IV neurons born at E14.5 with a migratory defect, but not layer V–VI neurons born at E12.5 with improper expression of Cux1.
To confirm the aberrant radial migration in the mutant brains, we next examined radial migration using an in utero electroporation technique (Tabata and Nakajima 2001). pCAG-EGFP vector was electroporated into progenitor cells in VZ at E14.5, and the localization of transfected cells was analyzed at E19.5/P0. The representative photomicrographs of GFP-labeled neurons in Sada+/+ and Sada∆ex2/∆ex2 mice are shown (Fig. 2B). GFP-positive neurons dispersed within the CP in the Sada∆ex2/∆ex2 mouse, while those in wild type were located in the upper part of the CP (Fig. 2B). We then performed the same analysis, this time by dividing the cortex into 4 bins from VZ to MZ. Again, the percentage of GFP-positive cells in the superficial layer (bin 4) and middle layers of Sada∆ex2/∆ex2 brains was smaller and larger compared with rates in Sada+/+ brains, respectively (Fig. 2C). The migrating neurons in Sada+/+ mice had elongated leading processes oriented toward the pial surface, whereas those of Sada∆ex2/∆ex2 were shorter and sometimes branched or bifurcated (Fig. 2D). These results further confirm the aberrant radial migration of Sada∆ex2/∆ex2 neurons. On the contrary, the number of neurons positive for Calbindin D, a specific marker for interneurons, was similar in Sada∆ex2/∆ex2 mice and wild type (Supplementary Fig. 4), suggesting that tangential migration of interneurons was not impaired in Sada∆ex2/∆ex2 mice.
Migration of Sada∆ex2/∆ex2 Neurons is Slow
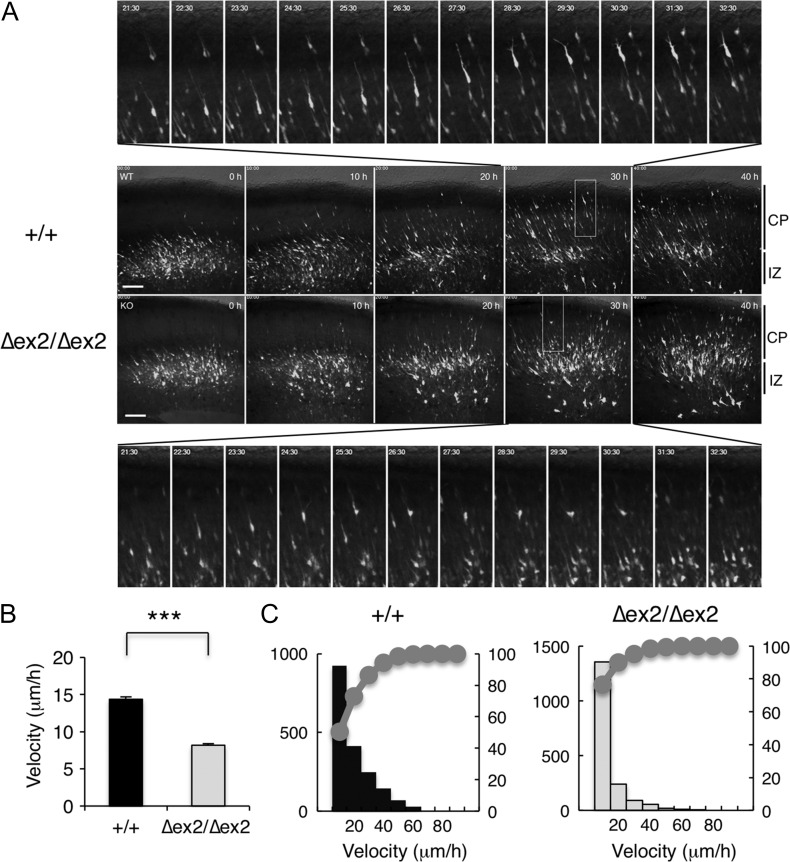
To elucidate the mechanism underlying the aberrant radial migration of SAD-A-deficient neurons, we performed time-lapse imaging of EGFP-positive cells (Tabata and Nakajima 2003). During the culture, most wild-type neurons labeled at E14.5 quickly migrated radially toward the pial surface in an inchworm-like fashion in the CP one after another (Supplementary Movie 1 and Fig. 3A upper panels). In contrast, the movement of most Sada∆ex2/∆ex2 neurons was slow and their leading processes were thin with only a small portion of neurons migrating quickly (Supplementary Movie 2 and Fig. 3A lower panels). Some bipolar neurons in the Sada∆ex2/∆ex2 brain sometimes retracted their leading processes and reversed their direction toward the ventricle even in the CP (Supplementary Movie 2 and Fig. 3A lower panels, lower insets). The average velocity of migration of Sada∆ex2/∆ex2 bipolar neurons in the CP was significantly slower than that of Sada+/+ neurons (+/+, 14.4 ± 0.30 μm/h, n = 1834 cells, vs. ∆ex2/∆ex2, 8.2 ± 0.22 μm/h, n = 1769 cells, P < 0.0001, t-test, Fig. 3B). A cumulative curve showed that slowly migrating neurons were dominant among Sada∆ex2/∆ex2 neurons compared with the wild type (Fig. 3C).
Figure 3.
Migration velocity determined by time-lapse imaging. (A) Images from Supplementary Movies 1 and 2. pCAG-EGFP vector was electroporated at E14.5. The brain slices were excised at E16.5, and cultured for 40 h. Middle panels represent the photomicrographs of wild-type (+/+, upper panels) and SAD-A-deficient (Δex2/Δex2, lower panels) slices in the indicated time during time-lapse imaging. Most upper panels and the lowest panels represent the higher magnification views from 21.5 to 32.5 h of the rectangle in the panel of wild-type (+/+) and SAD-A-deficient (Δex2/Δex2) slices at 30 h, respectively. Bar, 100 μm. (B, C) Average (B), histogram, and cumulative distribution curves (C) of migration velocity of wild-type (+/+, closed bar, left) and SAD-A-deficient (∆ex2/∆ex2, open bar, right) bipolar neurons in the CP. Data are expressed as means ± SEM. ***P < 0.001. (Sada+/+, n = 1834 cells; Sada∆ex2/∆ex2, n = 1769 cells).
Sada Regulates Neuronal Migration in a Cell-Autonomous Manner
Co-staining with Tbr1 (a marker of MZ, layer VI, and subplate) and Sada mRNA revealed that Sada was expressed by many Tbr1-positive neurons (Supplementary Fig. 5), indicating that Sada is not restricted to migrating neurons but persists following migration. To verify the involvement of SAD-A in neuronal migration, we examined the effect of SAD-A knockdown. Depletion of mSAD-A in HEK293 cells was confirmed (Fig. 4A). In control experiments, most neurons migrated radially and reached the superficial layer of the CP at P0 (Fig. 4B, left panel). In contrast, a considerable number of mSAD-A-depleted neurons remained in the lower zone of CP and/or IZ (Fig. 4B, right panel). Quantification of the distribution of migrating neurons revealed that the percentage of SAD-A-knocked down cells in the superficial layer (bin 4) was smaller compared with that of the control (Fig. 4C). The leading processes of some mSAD-A-depleted neurons in the lower CP were shorter and occasionally branched (Fig. 4D) as seen in Sada∆ex2/∆ex2 neurons. The consistent results observed in Sada∆ex2/∆ex2 mutants and following mSAD-A knockdown further confirm that SAD-A regulates neuronal migration in a cell-autonomous fashion.
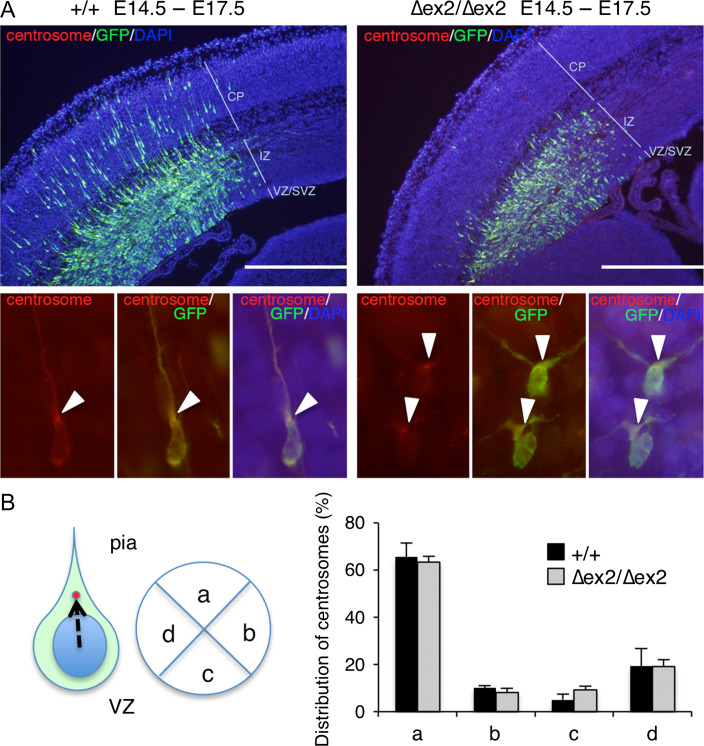
Centrosome of Sada∆ex2/∆ex2-Migrating Neurons in the IZ Orients Toward the Pia
In migrating neurons, the centrosome is positioned ahead of the nucleus with the perinuclear microtubule cage converging into the centrosome and projecting into the leading process from the centrosome. This nuclear–centrosome coupling is critical for neuronal migration (Tanaka et al. 2004). In order to examine whether SAD-A is involved in this nuclear–centrosome coupling, we identified centrosome localization of Sada∆ex2/∆ex2-migrating cells in the outer zone of IZ. Although many wild-type neurons labeled at E14.5 migrated into the CP at E17.5 (Fig. 5A, left panel), most Sada∆ex2/∆ex2 neurons labeled at E14.5 failed to enter the CP and remained in the IZ (Fig. 5A right panel). The centrosomes of wild-type neurons localized at the base of the leading process in the IZ near the CP, which was oriented toward the pial surface (arrowheads in Fig. 5A left lower panels). In Sada∆ex2/∆ex2 neurons, the leading processes of migrating neurons in the IZ near the CP were often bifurcated, but their centrosomes were directed toward the pial surface (arrowheads in Fig. 5A right lower panels). To determine the centrosome position, cells were visually divided into 4 quadrants (Fig. 5B, left panel). Centrosomes were localized in the upper (pial) side of the quadrant in more than 60% of migrating neurons of both wild-type and Sada∆ex2/∆ex2 brains. There was no significant difference in each quadrant between these 2 groups (for example, quadrant a, +/+, 65.6 ± 5.9%, n = 4, vs. ∆ex2/∆ex2, 63.4 ± 2.4%, n = 4, P = 0.743, t-test, Fig. 5B, right panel). These results suggest that Sada∆ex2/∆ex2-migrating neurons in the IZ reorient their centrosome toward the pia as wild-type neurons do.
Figure 5.
Centrosomal localization of Sada∆ex2/∆ex2-migrating neurons in the IZ. (A) Representative photographs of wild-type (+/+, left panels) or SAD-A-deficient (∆ex2/∆ex2, right panels) cerebral cortex cotransfected with pCAG-PACT-mKO (a marker for centrosomes) and pCAG-EGFP at E14.5. GFP (green), centrosome (red), and DNA (DAPI, blue) were stained in coronal sections at E17.5. CP, cortical plate, IZ, intermediate zone, VZ/SVZ, ventricular zone/ subventricular zone, Bar, 500 μm. Lower panels show high magnification views. Left, centrosome (red), middle, GFP (green), and centrosome (red), right, overlay. Arrowheads indicate the position of the centrosome. (B) Quantification of the distribution of centrosomes in the cell. Cells were visually divided into 4 quadrants. The centrosome position against the nucleus of each migrating neuron in the IZ was plotted and the number was represented as percentage. More than 30 neurons per individual were counted. Quadrant (a) represents the upper quadrant toward the pial surface. Closed bars, wild type (+/+), n = 4; open bars, SAD-A-deficient (∆ex2/∆ex2) mice, n = 4.
Neurites of Hippocampal Neurons in Sada∆ex2/∆ex2 Differentiate into Axons and Dendrites
We then examined whether SAD-A regulates axon-dendrite polarity. Neurites of hippocampal neurons of Sada∆ex2/∆ex2 in culture differentiated into pJNK-positive axons and MAP2-positive dendrites, although the length of some were shorter and less complex (Fig. 6A, right panel). The ratio of the longest pJNK+/MAP2+ neurite length was significantly smaller in Sada∆ex2/∆ex2 neurons than in wild-type neurons (+/+, 4.88 ± 0.39, n = 58, vs. ∆ex2/∆ex2, 2.20 ± 0.16, n = 35, P < 0.0001, t-test, Fig. 6B left panel). The total length of pJNK+ neurites of Sada∆ex2 /∆ex2 neurons was significantly shorter compared with the wild type (+/+, 6.67 ± 0.37 mm, n = 58, vs. ∆ex2/∆ex2, 3.73 ± 0.33 mm, n = 35, P < 0.0001, t-test, Fig. 6B right panel). These results suggest that although axon-dendrite polarization is established in Sada∆ex2/∆ex2 neurons, their axon growth is impaired.
To elucidate the mechanism underlying SAD-A-mediated radial migration, phosphorylation of various candidate proteins was analyzed. Tyrosine phosphorylation of Dab1, a key molecule in the reelin-signaling pathway, did not vary in the cerebral cortices of Sada+/+, Sada+/∆ex2, and Sada∆ex2/∆ex2 mice at P0 (Supplementary Fig. 6A). The level of phosphorylation of MAP1b (pT1265) and MAP2 (pS136) was also comparable among the 3 groups (Supplementary Fig. 6B,C). In addition, we could not detect any significant differences in the level of dephosphorylated tau (Tau-1) and phosphorylated tau (pS262) among the 3 groups (Supplementary Fig. 6D).
LKB1, a master kinase of AMPK family, is viewed as an essential activator of SAD-A/BRSK2 and is known to phosphorylate both SAD-A and SAD-B (Lizcano et al. 2004; Barnes et al. 2007), but the possibility that SAD-A can also phosphorylate LKB1 has not been tested. However, the phosphorylation level of LKB1 in Sada∆ex2/∆ex2 cortex was similar to that of wild type (Supplementary Fig. 6E). We also assayed stable interaction between LKB1 and SAD-A and were unable to detect endogenous binding between SAD-A and LKB1 protein by immunoprecipitation (Supplementary Fig. 6F).
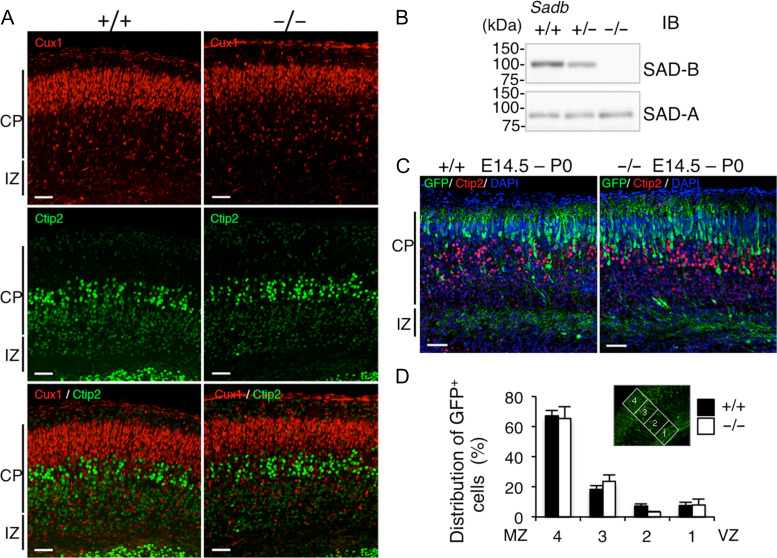
Radial Migration of Sadb−/− Neurons Appears to be Normal
Western blot analyses confirmed that SAD-B protein was not present in the Sadb−/− brain, while the amount of SAD-A protein was comparable with that of the wild type (Fig. 7B). Most Cux1-positive cells were found outside of the Ctip2-positive cell layer in both wild-type and Sadb−/− brains (Fig. 7A). In addition, the majority of cortical neurons labeled at E14.5 migrated and reached the superficial layer in the Sadb−/− brain (Fig. 7C). There was no significant difference in the percentage of GFP-positive cells in each bin between the control and Sadb−/− groups (Fig. 7D). In addition, knockdown of Sadb did not affect radial migration (Supplementary Fig. 7). In a hippocampal neuronal culture, Sadb−/− neurons exhibited axon-dendrite differentiation and elongated pJNK+ axons with a similar complexity to that of wild-type neurons (Supplementary Fig. 8). These results suggest that SAD-B is not involved in the regulation of radial neuronal migration during the development of cerebral cortex.
Figure 7.
Radial migration of Sadb-deficient neurons is not impaired. (A) Immunostaining of Sadb wild-type (+/+, left) and homozygous (−/−, right) cerebral cortex at P0 with layer-specific markers. Cux1 (II–IV layer marker, red, upper panels), Ctip2 (Vb–VI layer marker, green, middle panels) staining, and their overlay (lower panels). Bar: 50 μm. (B) Confirmation of Sadb−/− brains. Lysates of the cerebral cortex of Sadb wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mutant mice at P0 were analyzed by immunoblotting using the indicated antibodies. (C) In utero electroporation. pCAGGS-EGFP vector was electroporated into the cerebral cortices at E14.5, followed by fixation at P0. The brain sections were immunostained with anti-GFP (green), anti-Ctip2 (red) antibody, and DAPI (blue). Representative images of the staining of wild-type (+/+, left) and homozygous (−/−, right) brains. CP, cortical plate, IZ, intermediate zone. Bar, 50 μm. (D) Quantification of the distribution of GFP-positive cells in 4 distinct parts of the cerebral cortex (bins 1–4 as indicated in the inset) of Sadb+/+ (closed bars, n = 5) or Sadb−/− (open bars, n = 5) mice. Data are expressed as means ± SEM. There were no significant differences between the 2 groups (bin 4, P = 0.850; bin 3, P = 0.333; bin 2, P = 0.054; bin 1, P = 0.927).
Discussion
mSAD-A and mSAD-B have been proposed to function redundantly based on the observation that defective neuronal polarization was detected in double-mutant (Sada−/−;Sadb−/−) but not single-mutant (Sada−/− or Sadb−/−) mice (Kishi et al. 2005). In analyzing the roles of these genes in cortical development, we crossed them to C57BL/6N mice, which had not been used in the original study. Serendipitously, we found neonatal lethality and profound cortical defects in the single Sada−/− mutants on the C57BL/6N background. Further analysis on this background provided clear evidence for the essential role of SAD-A but not SAD-B in neuronal radial migration during the development of the cerebral cortex.
Neonatal lethality might be due to a disconnection between the cortex and subcortical areas since few axonal tracts have descended from the cortex to the thalamus in Sada−/−:Sadb−/− double mutants in the mixed background (Kishi et al. 2005). The same disconnection might explain the lethality of Sada∆ex2/∆ex2 and Sada∆ex1/∆ex1 mice in a C57BL/6N genetic background based on the similarities of the cortical defects. It is possible, however, that subcortical defects contribute to lethality in both cases. Further analysis will be required to answer the relationship between lethality and cortical defects.
Nissl and layer-specific marker staining showed that cortical defects in Sada−/−:Sadb−/− double mutants in the mixed background appear to be more severe than those in Sada−/− single mutants in the C57BL/6N background. Although it is not conclusive because the genetic background and the assay are different between the 2 strains, both SAD kinases might be required for cortical development, with the role of SAD-A predominating. This might be an example of what has been called unequal redundancy (Briggs et al. 2006).
Strain-dependent differences in phenotype have been observed in several cases. They include a mouse model of Alport syndrome, a human hereditary glomerulonephritis with a collagen mutation (Andrews et al. 2002), and of amyotrophic lateral sclerosis (ALS) in which the Cu/Zn superoxide dismutase (SOD1) gene was mutated (Kunst et al. 2000). In these cases, the existence of modifier genes was postulated (Kunst et al. 2000; Andrews et al. 2002). It is possible that some strain-specific modifier also affect the function of Sada. Identification of such modifiers could provide novel insights into the regulation of cortical development.
Migration of neocortical projection neurons has been considered to be divided into 4 phases based on results of detailed histological and live imaging studies (Shoukimas and Hinds 1978; O’Rourke et al. 1992; Nadarajah et al. 2001; Hatanaka et al. 2004; Tabata and Nakajima 2003; Noctor et al. 2004; de Anda et al. 2010; for review, Cooper 2014). In phase 1, neuronal progenitors in the VZ are divided asymmetrically into new postmitotic neurons and intermediate progenitors (IPs). After these cells reach the SVZ/IZ and become MP (phase 2, stage 2A), a ventricle- or horizontally oriented process near the centrosome begins to extend and ultimately becomes the axon (stage 2B). The MP cell reorients the Golgi and centrosome toward the pia, establishes a dominant pia-directed leading process, assumes a bipolar morphology, and commences locomotion along the glia, trailing the axon behind (stage 2C, Hatanaka et al. 2004; de Anda et al. 2010). This is known as the multipolar to bipolar (MP–BP) transition. Neurons then rapidly exit from the IZ by locomotion along the radial glia (phase 3) and finally reach the top of the CP (terminal translocation, phase 4). We found that Sada∆ex2/∆ex2 neurons with bipolar shape migrated slowly in the CP (Fig. 3 and Supplementary Movie 2) and their centrosome was localized toward the pia (Fig. 5). On the other hand, Sada∆ex2/∆ex2 hippocampal neurons underwent axon-dendrite differentiation and elongated their axons (Fig. 6). Considering these findings, many Sada∆ex2/∆ex2 neurons appear to be able to enter phase 3 and their migration defects appear to occur after the switch from MP to BP. That is, SAD-A deficiency may give rise to an impairment of BP migration in the CP. Given that the leading processes of Sada∆ex2/∆ex2-migrating neurons were thin and easy to be branched in our time-lapse imaging (Supplementary Movie 2), SAD-A might be involved in the stabilization of the leading process.
SAD kinase is a member of the AMPK family. LKB1, a master kinase of the AMPK family, has been reported to phosphorylate and activate SAD-A/BRSK2 and SAD-B/BRSK1 (Lizcano et al. 2004), regulating neuronal polarity (Barnes et al. 2007). Conditional LKB1-deficient neurons were found to lack axons, but they migrated radially and reached the CP as well as wild-type neurons did (Barnes et al. 2007), which was in contrast to our current observations. However, their results were obtained by performing organotypic slice culture for 3 days after EGFP plasmid electroporation at E14.5 (Barnes et al. 2007). Their experimental conditions were obviously different from those of in vivo migration. On the other hand, the leading processes of conditional LKB1-deficient neurons were branched or bifurcated (Barnes et al. 2007), which was similar to the appearance of Sada∆ex2/∆ex2 neurons in the present study (Fig. 2D). Acute knockdown of LKB1 in migrating immature neurons resulted in aberrant neuronal migration (Asada et al. 2007). Taken together, it is possible that the LKB1-SAD-A pathway might regulate not only axon specification but also stabilization of the leading processes. Consequently, LKB1 activity might be altered in Sada mutant mice. However, we could detect neither increased phosphorylation of LKB1 in Sada∆ex2/∆ex2 brain nor endogenous binding between SAD-A and LKB1 (Supplementary Fig. 6E,F). Alternatively, SAD-A kinase might be regulated by another upstream pathway involved in neuronal migration.
SAD kinases are required for the formation of central axonal arbors built up by subsets of sensory neurons (Lilley et al. 2013), which is consistent with our results showing that Sada∆ex2/∆ex2 hippocampal axons were shorter and less complex than normal (Fig. 6). SAD-A might regulate not only stabilization of the leading processes but also microtubule organization in multiple processes including axons. Tau, MAP1b, and MAP2 are the main neuronal microtubule-associated proteins (MAPs) and are implicated in putative functional redundancy (Takei et al. 2000), given that tau/map1b and map1b/map2 double-deletion delayed neuronal migration (Takei et al. 2000; Teng et al. 2001). In the CP of double-mutant (Sada−/−;Sadb−/−) mice, the level of dephosphorylated tau increased and that of tau-p(S262) decreased, suggesting that SAD kinases might affect neuronal polarity by regulating phosphorylation of MAP family proteins (Kishi et al. 2005). However, in the present study, we could not detect any significant reduction in the levels of dephosphorylated tau (Tau1) and tau-p(S262), or MAP1b(pT1265) and MAP2(pS136) in the cerebral cortices of Sada∆ex2/∆ex2 mice (Supplementary Fig. 6B–D), suggesting that SAD-A-mediated radial migration might be independent of tau (S262) phosphorylation. Reelin induces Dab1 tyrosine phosphorylation, and hyperphosphorylation of tau and disorganized neuronal migration was observed in the brain of reeler and vldlr/apoER2 (the receptors of reelin) double-deficient mice (Hiesberger et al. 1999). However, our present results show that the level of Dab1 tyrosine phosphorylation in the Sada∆ex2/∆ex2 brain was comparable with that of the wild type (Supplementary Fig. 6A) and that hyperphosphorylation of tau was not detected in the Sada∆ex2/∆ex2 brain (Supplementary Fig. 6D).
Neuronal migration is critical for the formation of the laminated structure of the cerebral cortex, and defects in migration during embryogenesis give rise to brain malformations, resulting in severe epilepsy and mental retardation. We clearly show here that SAD-A is involved in neuronal migration and that SAD-A has a nonredundant role in the developing cerebral cortex. Elucidating the mechanism of mediation of neuronal migration and identifying downstream targets of SAD-A could contribute to the future understanding of migratory disorders.
Supplementary Material
Notes
We are grateful to Drs Fumio Matsuzaki, Masato Asai, and the members of the Dept. of Perinatology for their helpful discussions and to Ms Naomi Nakayama and Ms Yoshie Chiba for their skillful technical assistance. Conflict of Interest: None declared.
Funding
Japan Society for the Promotion of Science (JSPS), Grants-in-Aid for Scientific Research (KAKENHI, grant numbers JP25461659, JP16K10119).
References
- Andrews KL, Jacqueline LM, Cong L, Miner JH. 2002. Quantitative trait loci influence renal disease progression in a mouse model of Alport syndrome. Am J Pathol. 160:721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angevine JB, Sidman RL. 1961. Autoradiographic study of cell migration during histogenesis of cerebral cortex in mouse. Nature. 192:766–768. [DOI] [PubMed] [Google Scholar]
- Asada N, Sanada K, Fukada Y. 2007. LKB1 regulates neuronal migration and neuronal differentiation in the developing neocortex through centrosomal positioning. J Neurosci. 27:11769–11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AP, Lilley BN, Pan A, Plummer LJ, Powell AW, Raines AN, Sanes JR, Polleux F. 2007. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 129:549–563. [DOI] [PubMed] [Google Scholar]
- Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG. 2004. Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Biol. 20:593–618. [DOI] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. 2006. Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends Plant Sci. 11:492–498. [DOI] [PubMed] [Google Scholar]
- Cooper JA. 2014. Molecules and mechanisms that regulate multipolar migration in the intermediate zone. Front Cell Neurosci. 8:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JG, Zhen M, Jin Y, Bargmann CI. 2001. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron. 29:115–129. [DOI] [PubMed] [Google Scholar]
- de Anda FC, Meletis K, Ge X, Rei D, Tsai LH. 2010. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci. 30:10391–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. 2004. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 14:279–293. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Hisanaga S, Heizmann CW, Murakami F. 2004. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J Comp Neurol. 479:1–14. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Yamauchi K. 2013. Excitatory cortical neurons with multipolar shape establish neuronal polarity by forming a tangentially oriented axon in the intermediate zone. Cereb Cortex. 23:105–113. [DOI] [PubMed] [Google Scholar]
- Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumby MC, Cooper JA, Herz J. 1999. Direct binding of reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 24:481–489. [DOI] [PubMed] [Google Scholar]
- Honda T, Kobayashi K, Mikoshiba K, Nakajima K. 2011. Regulation of cortical neuron migration by the Reelin signaling pathway. Neurochem Res. 36:1270–1279. [DOI] [PubMed] [Google Scholar]
- Inoue E, Mochida S, Takagi H, Higa S, Deguchi-Tawarada M, Takao-Rikitsu E, Inoue M, Yao I, Takeuchi K, Kitajima I, et al. 2006. SAD: a presynaptic kinase associated with synaptic vesicles and the active zone cytomatrix that regulates neurotransmitter release. Neuron. 50:261–275. [DOI] [PubMed] [Google Scholar]
- Johmura Y, Sun J, Kitagawa K, Nakanishi K, Kuno T, Naiki-Ito A, Sawada Y, Miyamoto T, Okabe A, Aburatani H, et al. 2016. SCFFBXO22-KDM4A targets methylated p53 for degradation and regulates senescence. Nature Commun. 7:10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Cooper JA. 2011. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci. 14:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. 2003. The in vivo roles of STEF/Tiam1, Rac1, and JNK in cortical neuronal migration. EMBO J. 22:4190–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi M, Pan A, Crump JG, Sanes JR. 2005. Mammalian SAD kinases are required for neuronal polarization. Science. 307:929–932. [DOI] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. 2007. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 10:93–101. [DOI] [PubMed] [Google Scholar]
- Kunst CB, Messer L, Gordon J, Haines J, Patterson D. 2000. Genetic mapping of a mouse modifier gene that can prevent ALS onset. Genomics. 70:181–189. [DOI] [PubMed] [Google Scholar]
- Kähler AK, Djurovic S, Kulle B, Jönsson EG, Agartz I, Hall H, Opjordsmoen S, Jakobsen KD, Hansen T, Melle I, et al. 2008. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 147B:1089–1100. [DOI] [PubMed] [Google Scholar]
- Li W, Song X, Zhang H, Yang Y, Jiang C, Xiao B, Li W, Yang G, Zhao J, Guo W, et al. 2011. Association study of RELN polymorphisms with schizophrenia in Han Chinese population. Prog Neuropsychopharmacol Biol Psychiatry. 35:1505–1511. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Krishnaswamy A, Wang Z, Kishi M, Frank E, Sanes JR. 2014. SAD kinases control the maturation of nerve terminals in the mammalian peripheral and central nervous systems. Proc Natl Acad Sci USA. 111:1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Pan A, Sanes JR. 2013. SAD kinases sculpt axonal arbors of sensory neurons through long- and short-term responses to neurotrophin signals. Neuron. 79:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Göransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Mäkelä TP, Hardie DG, et al. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Niida H, Nakanishi M. 2004. Human SAD1 kinase is involved in UV-induced DNA damage checkpoint function. J Biol Chem. 279:31164–31170. [DOI] [PubMed] [Google Scholar]
- McConnell SK. 1988. Development and decision-making in the mammalian cerebral cortex. Brain Res Rev. 13:1–23. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. 2007. Neuronal subtype specification in the cerebral cotex. Nature Rev. 8:427–437. [DOI] [PubMed] [Google Scholar]
- Morgan-Smith M, Wu Y, Zhu X, Pringle J, Snider WD. 2014. GSK-3 signaling in developing cortical neurons is essential for radial migration and dendritic orientation. Elife. 3:e02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. 2001. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 4:143–150. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. 2003. Manipulating the mouse embryo. A laboratory manual. Third ed Cold spring harbor (New York): Cold Spring Harbor Laboratory Press. [Google Scholar]
- Nakanishi K, Aono S, Hirano K, Kuroda Y, Ida M, Tokita Y, Matsui F, Oohira A. 2006. Identification of neurite outgrowth-promoting domains of neuroglycan C, a brain-specific chondroitin sulfate proteoglycan, and involvement of phosphatidylinositol 3-kinase and protein kinase C signaling pathways in neuritogenesis. J Biol Chem. 281:24970–24978. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Sato Y, Mizutani Y, Ito M, Hirakawa A, Higashi Y. 2017. Rat umbilical cord blood cells attenuate hypoxic-ischemic brain injury in neonatal rats. Sci Rep. 7:44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 7:136–144. [DOI] [PubMed] [Google Scholar]
- Ohshima T. 2014. Neuronal migration and protein kinases. Front Neurosci. 8:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Hirasawa M, Tabata H, Mutoh T, Adachi T, Suzuki H, Saruta K, Iwasato T, Itohara S, Hashimoto M, et al. 2007. Cdk5 is required for multipolar- to-bipolar transition during radial neuronal migration and proper dendritic development of pyramidal neurons in the cerebral cortex. Development. 134:2273–2282. [DOI] [PubMed] [Google Scholar]
- O’Rourke NA, Dailey ME, Smith SJ, McConnell SK. 1992. Diverse migratory pathways in the developing cerebral cortex. Science. 258:299–302. [DOI] [PubMed] [Google Scholar]
- Rakic P. 1972. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 145:61–83. [DOI] [PubMed] [Google Scholar]
- Rasband WS. 1997. –2012. Image J, U.S. National Institute of Health, Bethesda, Maryland, USA. Available at: http://imagej.nih.gov/ij/
- Schneider CA, Rasband WS, Eliceiri K. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukimas GM, Hinds JW. 1978. The development of the cerebral cortex in the embryonic mouse: an electron microscopic serial section analysis. J Comp Neurol. 179:795–830. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. 2001. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 103:865–872. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. 2003. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci. 23:9996–10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N. 2000. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J Cell Biol. 150:989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. 2004. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 165:709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J, Takei Y, Harada A, Nakata T, Chen J, Hirokawa N. 2001. Synergistic effects of MAP2 and MAP1b knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J Cell Biol. 155:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrotti A, Spalice A, Ursitti F, Papetti L, Mariani R, Castronovo A, Mastrangelo M, Iannetti P. 2010. New trends in neuronal migration disorders. Eur J Paediatr Neurol. 14:1–12. [DOI] [PubMed] [Google Scholar]
- Yamada K, Fukushi D, Ono T, Kondo Y, Kimura R, Nomura N, Kosaki KJ, Yamada Y, Mizuno S, Wakamatsu N. 2010. Characterization of a de novo balanced t(4;20)(q33;q12) translocation in a patient with mental retardation. Am J Med Genet A. 152A:3057–3067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.